Abstract
Sufficient data with complete annotation is essential for training deep models to perform automatic and accurate segmentation of CT male pelvic organs, especially when such data is with great challenges such as low contrast and large shape variation. However, manual annotation is expensive in terms of both finance and human effort, which usually results in insufficient completely annotated data in real applications. To this end, we propose a novel deep framework to segment male pelvic organs in CT images with incomplete annotation delineated in a very user-friendly manner. Specifically, we design a hybrid loss network derived from both voxel classification and boundary regression, to jointly improve the organ segmentation performance in an iterative way. Moreover, we introduce a label completion strategy to complete the labels of the rich unannotated voxels and then embed them into the training data to enhance the model capability. To reduce the computation complexity and improve segmentation performance, we locate the pelvic region based on salient bone structures to focus on the candidate segmentation organs. Experimental results on a large planning CT pelvic organ dataset show that our proposed method with incomplete annotation achieves comparable segmentation performance to the state-of-the-art methods with complete annotation. Moreover, our proposed method requires much less effort of manual contouring from medical professionals such that an institutional specific model can be more easily established.
Keywords: Image Segmentation, Male Pelvic Organ, Deep Learning, Incomplete Annotation, CT
I. INTRODUCTION
PROSTATE cancer is a main male cancer in the world [1]. According to the World Cancer Research Fund International (WCRF), there are approximately 1,300,000 new cases and 360,000 deaths of prostate cancer around the world in 2018 [1]. Nowadays, external beam radiotherapy is a major treatment way for prostate cancer. For the sake of radiotherapy, prostate and nearby organs (e.g., bladder and rectum) need to be accurately segmented to optimize the radiation onto the prostate while sparing the organs at risk. Currently, these pelvic organs are manually segmented by experienced oncologists, which is time-consuming and may suffer from large intra- and inter-observer variability. Hence an automatic segmentation method is highly desired in clinical applications.
However, accurate and automatic segmentation of the male pelvic organs (i.e., prostate, bladder, and rectum particularly) in CT images is very challenging, as illustrated in Fig. 1. First, the male pelvic organs are rendered with blurred boundaries due to the limited contrast of CT images. Second, the organs bear a large shape variation across different patients, which is often difficult to encode. Finally, the appearance of the rectum may drastically change due to bowel gas and filling, incurring additional difficulty in image segmentation. Moreover, the fiducial markers that are often used to guide dose delivery in radiotherapy may compromise the image quality as well.
Fig. 1.

Typical CT images and their corresponding male pelvic organ labels. The first and second rows show the same sagittal slices without and with manual annotation. The last row visualizes the segmentation results in 3D. Red denotes the prostate, green denotes the bladder, and blue denotes the rectum. The yellow rectangle and circles in the first row indicate bowel gas and fiducial markers, respectively.
Recently, supervised deep learning methods have achieved great successes in many semantic segmentation tasks [2]–[6]. However, it is generally expensive to acquire sufficient organ annotations for medical images, which are critical to supervise the deep learning methods for CT pelvic organ segmentation. First, annotating CT pelvic images requires the professional knowledge of clinicians and is much more challenging than natural images. Second, the 3D organs are annotated slice by slice, which is laborious and inefficient to human experts. Finally, complete organ annotation is often difficult for clinicians owing to their unavailability. Then the insufficient organ annotations usually become a bottleneck to supervised deep learning-based segmentation methods.
In this paper, we propose a novel deep framework to utilize incomplete annotation for segmenting male pelvic organs in CT images. Different from complete image annotation, the proposed incomplete annotation means that the clinicians only need to manually delineate a few sampled 2D slices from a 3D organ for saving time and efforts (e.g., Fig. 2). That is, the experts can choose to annotate the organ on the slices only when they are confident and comfortable. Since clinicians are not asked to perform the whole-organ delineation, it is flexible for them to annotate the images according to their availability. Our new method is to address real medical image segmentation applications in a real clinical setting.
Fig. 2.

Different forms of annotation used in our proposed incomplete annotation. In particular, for a given image, oncologists can delineate (a) the whole image (W-Image), (b) a set of consecutive slices (C-Slice), or (c) a set of typical separate slices (S-Slice). The form of annotation depends on the confidence and availability of oncologists. The annotation is performed along with the axial view while displayed in the sagittal view for checking of clarity, as shown in the second row of this figure. Red indicates the prostate, green indicates the bladder, blue indicates the rectum, white indicates the background, and gray indicates the unannotated voxels.
Relying on the deep learning backbone of the fully convolutional network (FCN) [2], our method particularly adopts U-net [3] due to its excellent performance in medical image segmentation. Considering the fact that the FCN model trained by incomplete annotation can hardly compete with the model trained by complete annotation, we propose a hybrid loss network for the U-net backbone [3] to jointly perform voxel classification and organ boundary regression. Moreover, we propose a novel label completion strategy to infer the labels of unannotated data with high conhdence before the hrst iteration and also after each iteration. The inferred voxels can then augment the training data and contribute to the iterative optimization of the segmentation network.
In summary, our main contributions are listed below.
We propose a novel deep learning-based segmentation method for male pelvic organs in CT images using incompletely annotated data, which helps reduce the labeling cost and facilitates the data collection in real applications.
We design a pelvis localization method based on salient bone structures to focus on the regions of interested pelvic organs, which signihcantly reduces the computational complexity and improves the segmentation performance.
To enhance the model capability, we propose the hybrid loss network to jointly perform voxel segmentation and organ boundary regression. Also, for enriching the training set, we propose a label completion strategy to make full use of unannotated data.
Using incomplete annotation only, our method achieves comparable segmentation performance with respect to the model that is trained by completely annotated data.
II. Related Work
A. CT Male Pelvic Organ Segmentation
Previous automatic segmentation methods for CT male pelvic organ typically fall into three categories: multiatlas-based methods, deformable-model-based methods, and learning-based methods [7]–[11].
Multi-atlas-based methods hrst register atlases with manual labeling onto a target image. Then label fusion is performed to achieve the segmentation of the target image [12]. For example, Davis et al. [13] established the correspondence from the planning and previous treatment images to the current treatment image using deformable image registration after reducing the effect of bowel gas. Liao et al. [14] proposed a hierarchical sparse label propagation framework based on patch-based representation for prostate localization. Oscar et al. [15] presented a detailed explanation about atlas-based segmentation and evaluated different strategies in segmenting prostate, bladder, and rectum in CT images. These approaches have obtained attractive segmentation performance, but the cost of registration is often high.
Deformable-model-based methods solve image segmentation under the guidance of the statistical models of organ shape and appearance [16], [17]. Costa et al. [18] adopted coupled 3D deformable models for the localization and segmentation of lower abdomen structures. Feng et al. [19] leveraged both population and patient-specific information to capture the variation of the prostate. Chen et al. [20] segmented the prostate and rectum based on a cost function involving intensity, shape and anatomy information. Gao et al. [21] learned a displacement regressor to guide the deformable model for organ segmentation. Since deformable-model-based methods are sensitive to the initialization, they often suffer from the large appearance and shape variation of organs.
Learning-based methods adopt machine learning, such as K-Nearest Neighbor (KNN), Random Forest (RF), and Support Vector Machine (SVM), to segment target organs. Lu et al. [22] used marginal space learning to estimate the organ location and then utilized Jensen-Shannon Divergence (JSD) to refine the organ boundary. Shi et al. [23] proposed a prostate-likelihood predictor to propagate the physician’s simple manual specification to unlabeled voxels. Ma et al. [24] incorporated a population model and a patient-specific model in the training of pelvic organ segmentation model.
Recently deep learning methods are popular in pelvic organ segmentation due to their excellent performance in feature representation and mapping [5], [6]. For example, Cha et al. [25] utilized FCNs to generate the initial contour for the bladder and then refined the segmentation using level-sets. Shi et al. [26] proposed a cascaded deep domain adaptation model to segment the prostate from easy to hard. He et al. [27] introduced the distinctive curve information into the learning of deep features and then applied them to guide the segmentation of the prostate, bladder, and rectum. Despite the great successes, deep learning methods heavily depend on the quantity and quality of the training data. However, it is often difficult to acquire sufficient medical images with high-quality annotation.
B. Incomplete Annotation for Medical Image Segmentation
To deal with data insufficiency of deep learning, some state-of-the-art methods attempt to segment target organs upon incompletely annotated data [28]–[31]. Different from the complete annotation, only part of the organ needs to be manually annotated. Therefore, the cost of image annotation can be greatly reduced. For example, Çiçek et al. [32] proposed a 3D network to generate dense volumetric segmentation from sparse annotation, in which uniformly sampled slices were selected for manual annotation. Goetz et al. [33] selectively annotated unambiguous regions and employed domain adaptation techniques to correct the differences between the training and test data distributions caused by sampling selection errors. Bai et al. [34] first propagated the label map of a specific time frame to the entire longitudinal series based on the motion estimation, and then combined FCN with a Recurrent Neural Network (RNN) for longitudinal segmentation. Lejeune et al. [35] introduced a semi-supervised framework for video and volume segmentation, which iteratively refined the pixel-wise segmentation within an object of interest.
Even though these works can effectively reduce the time cost for manual annotation, there are still two main limitations with these methods. 1) They impose some restrictions on the sampling manner of acquiring incomplete annotation, such as annotating uniformly-distributed slices [32] or some preset slices [33], [34]. These restrictions are inconvenient to oncologists as they need to deliberately select the slices for annotation. 2) These methods do not fully utilize and exploit a large amount of unannotated data. While the labels of unannotated data are unavailable in the beginning, the labels can still be inferred and updated gradually to help training. Therefore, we propose a method to adopt user-friendly incomplete annotation and also simplify the labeling process to well fit the real clinical applications. In our method, both the annotated and unannotated data can be jointly utilized to achieve the final segmentation.
III. Method
A. Overview
In this paper, we propose a novel deep learning-based method for the automatic segmentation of male pelvic organs in CT images. our method is featured by training the deep network with incomplete image annotation, which can be acquired in a low-cost and user-friendly manner. The pipeline of the proposed method is shown in Fig. 3. Prior to the deep segmentation network, we design a pelvis localization module based on salient bone structures, which helps the following segmentation network to focus on candidate segmentation regions and thus reduces computational complexity. With limited and incomplete annotation for training, we propose a hybrid loss to optimize network parameters, which accomplishes voxel classification and boundary regression jointly. The two tasks contribute discriminative features, thus facilitating both tasks to be better solved. Moreover, a label completion strategy, including a pre-processing before the first iteration and a post-processing after each iteration, is performed to infer the labels of the voxels without manual annotation. And, if the inferred labels are highly confident as revealed in label completion, the voxels and their inferred labels are then fed back to augment the training data. In this way, the network can be iteratively optimized to improve segmentation performance.
Fig. 3.
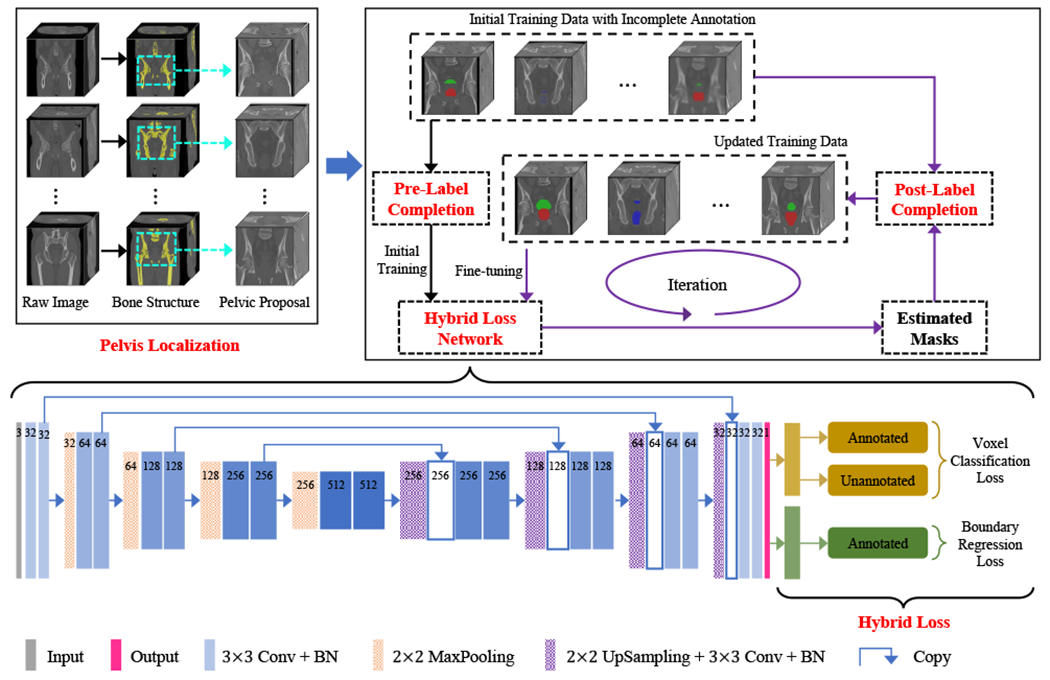
Overview of our proposed framework. The backbone structure can be any fully convolution-style network.
B. User-Friendly Incomplete Annotation
In this paper, we design a more user-friendly manner to collect incomplete image annotation for the training of the deep segmentation network. Specially, an oncologist can delineate the organ of interest in the image under consideration in three different manners as indicated in Fig. 2: the whole 3D organ (W-Image), a set of consecutive slices (C-Slice), or a set of typical separate slices (S-Slice). The manner adopted by an oncologist in real applications depends on his/her confidence in labeling a specific image as well as his/her time available for annotation. This flexible annotation method reduces restrictions and requirements for data collection and thus saves the cost in terms of human effort and finance. In our implementation, we adopt a mixed manner to generate the incompletely annotated data, i.e., randomly selecting images from the whole training dataset for annotation by W-Image, C-Slice, or S-Slice. And the image annotation is performed on the axial view in experiments, as well as the sagittal and coronal views.
Therefore, in our incompletely annotated data, there are two types of slices: selected and unselected slices for manual annotation. For selected slices, their voxels are manually annotated with semantic labels, i.e., belonging to a certain organ or the background. For unselected slices, their voxels are not manually annotated, so their semantic labels are unknown. Especially, for the image annotated by W-Image, because the whole organ has been delineated, all non-labeled voxels belong to the background. But, for images annotated by C-Slice or S-Slice, only the voxels in the selected slices have their annotated semantic labels, while the borders of the organ are unavailable. Here, we denote I as a 3D CT image and G as its corresponding manually delineated label mask. For the i-th voxel, gi = 1 means the voxel belonging to a certain organ under consideration, and gi = 0 means the voxel corresponding to the background, while gi = −1 means the voxel not manually annotated. Compared with the complete annotation, our proposed incomplete annotation is user-friendly and flexible.
C. Pre-localized Pelvic Proposals
The CT pelvic image usually covers much larger fleld-of-view than target organs. Thus, it is necessary to localize the organs under consideration and alleviate the challenges caused by nearby tissues in the image. In general, bone has rather large CT values compared with other tissues and the pelvis is associated with a distinct bone structure. That is, the bones superior to the pelvis mainly belong to the lumbar vertebra while they are located in the center of the body, and the bones inferior to the pelvis mainly belong to the legs while they are located on two sides of the body. Thus, we locate the apex and base of the pelvis based on the proportion of bone voxels near the center to all bone voxels of the corresponding whole-body region. The details are listed as follows:
3D Image Binarization. The raw 3D CT image is binarized using a simple threshold to extract the bones, as shown in Fig. 4(b).
3D-to-2D Mapping. The 3D binary image is converted to a 2D binary image by summing up along the anterior-posterior direction followed by a binarization operation. Moreover, only the maximally connected region is preserved by removing other small yet isolated regions, as shown in Fig. 4(c).
Apex and Base Localization. Based on the distinct bone structure, two sliding windows along the superior-inferior midline of the 2D binary image is used to compute the proportion of bone voxels near the center to all bone voxels of the corresponding whole-body region. The apex of the pelvis is found to have the minimum ordinate value with the proportion greater than a large threshold and the base is the maximum ordinate value with the proportion less than a small threshold, as shown in Fig. 4(d).
Pelvic Proposal Extraction. Based on the located apex and base points, the pelvic proposal is cropped from the raw image and used in the subsequent organ segmentation.
Fig. 4.
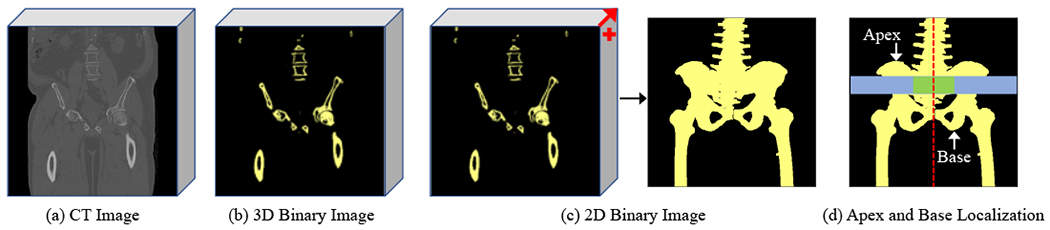
An example to extract the pelvic proposal. A CT image (a) is first converted to a 3D binary image (b) to reveal salient bones. Then, the 3D binary image is mapped to 2D (c) by summing up along the anterior-posterior direction followed by a binarization operation. Finally, the apex and base points are determined based on the proportion of the bone voxels near the center (green window) to all bone voxels of the corresponding whole-body region (blue window) (d).
D. Label Completion
The performance of deep learning in medical image segmentation is closely related to the amount and quality of the data annotation, especially for the cases of solving the challenging CT male pelvic organ segmentation problem. Unfortunately, in our study, manual annotation is insufficient and also incomplete. To tackle these challenges, we attempt to infer the semantic labels of unannotated voxels before the first iteration, and also facilitate the training of our proposed hybrid loss network after every iteration.
1). Label Propagation Using Spatial Cues:
This pre-processing step is performed before the first iteration, based on the spatial relationship between slices. For a given image, we assume that an organ is continuous in 3D space, which allows us to complete the labels of the unannotated slices from adjacent yet annotated slices of the same image. Let Ic denote an unannotated slice under consideration, denote its corresponding inferred label mask, and Gp (Gs) denote the manually annotated label mask of its adjacent (previous/subsequent) slice if existing. Here, considering shape variation and slice thickness, we perform a morphological operation to suppress their effects to ensure that correct labels are propagated as much as possible. Especially, the contour line Lp (Ls) is first extracted from Gp (Gs) and then performed with the dilation operation using morphological kernel size K (K = 5 here) to generate the contour region Cp (Cs). Let C∪ denote the union of Cp and Cs if one or all of them exist(s). The labels of Gp (Gs) in C∪ will not be used for spatial propagation, while the variation between adjacent slices in this region is uncertain. Let B∩ (F∩) denote the intersection of background (foreground) of Gp and Gs if one or all of them exist(s). We complete the labels of the unannotated slice under consideration from its adjacent annotated slices as:
| (1) |
From Eq. 1, we can see that only the labels in the confidence regions of the adjacent annotated slices are propagated to complete the labels of the unannotated slice under consideration. For voxels in the unreliable region (C∪) caused by shape variation, they will not be given semantic labels to avoid introducing label errors to mislead the training.
2). Label Update Using Iterative Learning:
Although the unannotated voxels are not manually labeled, their labels can be updated by the tentatively trained model. Suppose that we have a trained model (with sigmoid activation function for binary segmentation) and an unannotated voxel v, if the output probability for v based on is near 1, v can be labeled as belonging to the organ under consideration with high confidence. On the contrary, if the output probability is near 0, v can be labeled as the background with high confidence. Even though the labels of the unannotated voxels with the probability around 0.5 can also be estimated by a hard threshold, they are not reliable enough to guide the training of the next iteration. Therefore, after each iteration, we update the labels of unannotated data based on their probabilities under the current model, as shown in Eq. 2:
| (2) |
where T1 and T2 are the confidence thresholds that correspond to the background and the organ under consideration, respectively. If the output probability of one unannotated voxel falls in (T1, T2), it will not be assigned a semantic label. Here we use T1 = 0.25 and T2 = 0.75.
In label completion, label propagation is performed as a pre-processing step for a single time before the first iteration, while label updating is performed as a post-processing step after each iteration to infer the labels of the unannotated voxels.
E. Hybrid Loss Network for Incompletion Annotation
The CT male pelvic organ segmentation is challenging due to the blurred organ boundaries and large shape variation. In this work, it is even more difficult as our method is based on incompletely annotated data. Here, we design a hybrid loss to supervise the network for the segmentation of the pelvic organs. Specifically, we adopt U-net [3] liked architecture as the backbone of the network. The hybrid loss is derived from two joint tasks of voxel classification and boundary regression.
1). Voxel Classification Loss:
Because there are two different types of data in the training phase, i.e., annotated and unannotated, we separate the voxel classification loss into two components for the annotated and unannotated data, respectively.
For Annotated Data.
The voxels with ground-truth labels are insufficient to train a satisfactory network. In training, more attention should be devoted to the falsely segmented voxels. Thus, inspired by the focal loss [36], we impose a confidence weight map when computing the cross-entropy loss:
| (3) |
where pi ∈ [0, 1] is the probability of the i-th image voxel belonging to the organ under consideration, gi ∈ {0, 1} is the ground-truth label of the i-th voxel, and λl denotes the tuning parameter. From Eq. 3 we can see that the more difficult the voxels are to be segmented, the greater the weights become.
For Unannotated Data.
Compared with the annotated voxels, the amount of unannotated data is rich. Even though the unannotated voxels are with unknown semantic labels in the beginning, we can use the label completion to update the labels for them. But the confidence of the tentatively estimated labels may vary, and should be treated differently from the annotated voxels. We hence define the confidence weight map for the unannotated voxels as:
| (4) |
where pi ∈ [0, 1] is the predicted probability of the i-th voxel belonging to the organ under consideration, T1 and T2 are the thresholds that preserve the unannotated voxels with high confidence only. Specially, before the first iteration, the weights of the inferred labels of unannotated voxels by label propagation are set as 1 while no probability output can be referenced.
Incorporating the confidence weight map of the annotated and unannotated data into the cross-entropy loss function, our voxel classification loss can be formulated as:
| (5) |
where Nl is the number of the annotated image voxels, Nu is the number of the unannotated voxels whose labels are updated using label completion, are their tentatively estimated labels, and α is the tuning parameter that balances the contributions of the annotated and the unannotated voxels. We set λl = λu = 1 and α = 0.5 in our implementation. Note that can be updated offline in the interval of two iterations in optimizing the network, while can be updated not only offline but also online in each forward-propagation within a certain training iteration.
2). Boundary Regression Loss:
For CT pelvic organ segmentation, the voxels near the organ boundaries are most difficult to classify due to the low contrast in CT images and large shape variation of the organs. To this end, inspired by [37] and [27], a regression branch is introduced to the segmentation network, which predicts the boundary heatmap of the organ under consideration. Here, the heatmap aims to encode the distance from each voxel to its nearest voxel on the contour line:
| (6) |
where ri is the coordinate of the i-th voxel, C is the set of the voxels on the contour line of the organ under consideration, δh is a controlling parameter. Based on Eq. 6, the loss function for the boundary regression task is
| (7) |
where is the estimate of the network for the i-th voxel.
3). Hybrid Loss:
We combine the voxel classification loss and the boundary regression loss to derive the loss that supervises the training of our segmentation network:
| (8) |
where the hyper-parameter β (= 0.5) balances the voxel classification and boundary regression tasks. For these two tasks, they share the low-level and mid-level features and are separated after the last deconvolutional layer.
IV. Experiments
A. Dataset, Implementation, and Metrics
Dataset
Our study comprises of 313 planning CT images acquired from different prostate cancer patients of the North Carolina Cancer Hospital. Two radiation oncologists delineate the prostate, bladder, and rectum with voxel-wise segmentation labels. And the results after their careful cooperative correction are used as the ground-truth. These images are acquired from different CT scanners with different protocols, which leads to different in-plane resolutions (0.932mm ~ 1.365 mm) and inter-slice thicknesses (1mm ~ 3 mm). Therefore, we resample all images and their corresponding ground-truth into the same resolution of 1 mm × 1 mm × 1 mm.
Implementation
In our implementation, we adopt U-net [3] liked architecture as the backbone but with the following modification: 1) the number of feature maps starts from 32, instead of 64, for the first convolutional block; 2) each convolutional layer, except the last one for the prediction, is followed by a batch normalization layer; 3) the number of feature maps in the last convolutional layer is 1 with the sigmoid activation function, instead of 2 with the soft-max activation function. Please note any FCN model can replace U-net [3] liked architecture as the backbone. Here, we denote our trained model directly using initially incompletely annotated data as Naive-FCN (N-FCN for short) and the final fine-tuned model as Enhanced-FCN (E-FCN for short).
All networks are implemented with Keras [38] using TensorFlow [39] as the backend in Python. All experiments are conducted on an Nvidia GeForce GTX 1080 Ti with 11G Memory. The batch size is set to 24, and the initial learning rate to 10−3. The pelvis localization is implemented in volumes for all images. For training, patches of size 128 × 128 × 3, randomly sampled from the resulting pelvic proposals, are input to predict the segmentation map of the corresponding middle slices. For testing, the averaged predictions of the overlapped patches in pelvic proposals with a constant stride are used to achieve the final segmentation. To perform a robust non-biased evaluation of the proposed method, five-fold crossvalidation is conducted. In each round, four folds are used for training and validation (with a ratio of 2:1) and the remaining fold is used for testing. Five repeated rounds are performed until each of the five folds is traversed as the testing data.
Metrics
The segmentation performance is measured with the Dice Similarity Coefficient (DSC), defined as:
| (9) |
and Average Symmetric Surface Distance (ASD), defined as:
| (10) |
where Vgt and Vseg are the voxel sets of ground-truth volume and automatically predicted volume, Sgt and Sseg are their corresponding surface voxel sets, and d(p, Sgt) is the minimum Euclidean distance from the voxel p ∈ Sseg to Sgt.
B. Incomplete Annotation for Training
To be in line with real applications, we adopt a mixed manner to generate the required incompletely annotated dataset. There are three manners to randomly select images/slices for annotation, i.e., W-Image, C-Slice, and S-Slice. Obviously, the amount of annotated data is critical to the segmentation performance. Therefore, in order to verify the effectiveness of our proposed method, three sets of data with different levels of incompleteness are generated, i.e., 10% annotated and 90% unannotated data (10% in short), 20% annotated and 80% unannotated data (20% in short), and 30% annotated and 70% unannotated data (30% in short). Here, d% annotated data means that d% slices of the entire training set are manually labeled (with semantic labels, 0 or 1), while 1-d% unannotated data means that the remaining 1-d% slices are not manually labeled (marked with the unknown label, −1). Specially, if there are N images available, to generate an incompletely annotated dataset with d% annotated slices and 1-d% unannotated slices, d% × N/3 images are randomly selected for annotation by W-Image, N/3 images are selected for annotation by C-Slice, and N/3 images are for S-Slice. Moreover, for each image annotated by C-Slice and S-Slice, the number of annotated slices accounts for d% of the total slices of the image. Therefore, the amount of the annotated data accounts for d% of the training data in slices, if all training images are of the same size.
C. Compared with Complete Annotation Based Approach
In this section, four popular FCN architectures, i.e., FCN-8s [2], SegNet [40], Dilation8 [41], and U-net [3] trained using complete annotation are compared with our models trained using 10%, 20%, and 30% annotated data, respectively.
Table I shows the mean DSC and ASD values with standard deviations of all comparison methods for segmenting the prostate, bladder, and rectum. It can be seen that with the growth of annotated data size, the performance of our method is gradually improving. Especially when the annotated data accounts for 30% of the whole training data, our method can already achieve comparable results with the FCN approaches that are trained using completely annotated data. In the remainder, if not stated otherwise, the results mentioned are corresponding to using 30% annotated data.
TABLE I.
Quantitative comparison of popular FCN models and other existing works trained using completely annotated data and our proposed model trained using incompletely annotated data in segmenting three male pelvic organs. (* indicates that the method is based on FCN and bold indicates the best result achieved among FCN-based methods)
| Metric | DSC | ASD (mm) | ||||
|---|---|---|---|---|---|---|
| Prostate | Bladder | Rectum | Prostate | Bladder | Rectum | |
| *FCN-8s [2] | 0.848±0.076 | 0.931±0.048 | 0.858±0.053 | 1.790±0.801 | 1.209 ±0.705 | 1.769±0.882 |
| *SegNet [40] | 0.853±0.067 | 0.939±0.053 | 0.870±0.045 | 1.828±1.054 | 1.170±0.734 | 1.563±0.594 |
| *Dilation8 [41] | 0.808±0.039 | 0.912±0.045 | 0.826±0.052 | 2.095±0.420 | 1.456±0.411 | 1.934±0.608 |
| *U-net [3] | 0.859±0.058 | 0.935±0.041 | 0.863±0.043 | 1.740±0.812 | 1.174±0.611 | 1.639±0.523 |
| *30% U-net [3] | 0.745±0.054 | 0.856±0.081 | 0.763±0.068 | 3.727±1.257 | 2.681±0.959 | 3.059±1.462 |
| Shao [17] | 0.880±0.020 | 0.860±0.080 | 0.840±0.050 | 1.860±0.210 | 2.220±1.010 | 2.210±0.500 |
| Gao [42] | 0.860±0.050 | 0.910±0.100 | 0.790±0.200 | 1.850±0.740 | 1.710±3.740 | 2.130±2.970 |
| Gao [21] | 0.864±0.041 | 0.921±0.047 | 0.884±0.048 | 1.770±0.660 | 1.370±0.820 | 1.380±0.750 |
| *10% Ours | 0.811±0.105 | 0.880±0.102 | 0.821±0.073 | 2.386±1.772 | 2.373±1.394 | 2.491±1.534 |
| *20% Ours | 0.833±0.095 | 0.909±0.084 | 0.832±0.062 | 2.153±1.441 | 2.202±0.800 | 2.328±1.397 |
| *30% Ours | 0.842±0.062 | 0.924±0.049 | 0.846±0.055 | 1.931±0.868 | 1.242±0.595 | 1.771±0.807 |
For prostate segmentation, U-net [3] achieves the best DSC and ASD. Even though our method obtains the worst results except Dilation8 [41], the performance gap between ours and the best one is very slight (i.e., less than 0.017 for DSC and 0.191 mm for ASD). For bladder segmentation, the results of all methods are excellent which is mainly due to the clear bladder wall. And the performance of U-net [3] and ours is very close (0.935 vs 0.924 for DSC and 1.174 mm vs 1.242 mm for ASD). For rectum segmentation, SegNet [40] obtains the best results followed by U-net [3]. Because rectum segmentation is confronted with more severe challenges, the DSC of ours is 0.024 lower than the best one. Yet it is still comparable with others. Moreover, we compare our method using 30% annotated data with the U-net [3] trained directly using 30% annotated data. We can see that our method can make significant improvements in terms of segmentation results of three organs, compared with the U-net [3] directly using the incompletely annotated data. While the performance is comparable with approaches trained using completely annotated data, our method can facilitate oncologists to perform data collection and reduce the cost in terms of both finance and human effort.
In Fig. 5, we show several representative 3D segmentation results of the prostate, bladder, and rectum obtained by the FCN approaches and our proposed method under comparison. From Fig. 5, we can see that our proposed method yields satisfactory results for both the relatively easy bladder segmentation and relatively difficult prostate and rectum segmentation. Moreover, to achieve these similar visualization results, we just use incompletely annotated data while others use completely annotated data with much more effort to collect than ours.
Fig. 5.
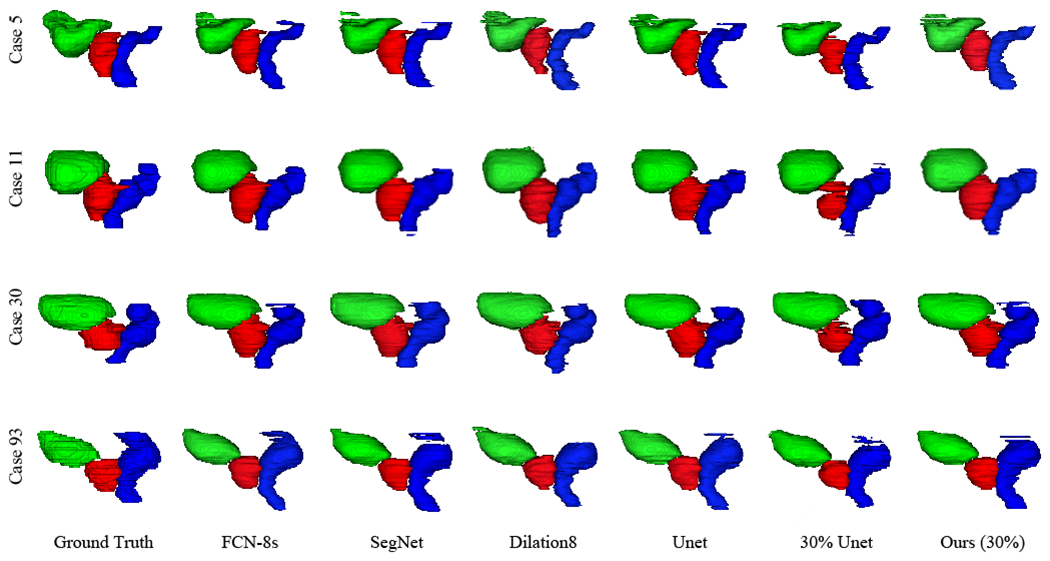
3D visualization of the segmentation results of the prostate, bladder, and rectum obtained by the compared FCN approaches trained using the completely annotated data and the incompletely annotated data with 30% ground-truth labels, respectively. Four rows indicate the results of four patients. Red denotes the prostate, green denotes the bladder, and blue denotes the rectum.
Fig. 6 shows some representative comparisons between the ground-truth and automatic segmentation of our method for three male pelvic organs separately. As can be observed, our method can delineate organ boundaries with high accuracy compared with the ground-truth, despite using such incompletely annotated data to deal with the unclear boundaries and large shape variation.
Fig. 6.

Comparisons between the ground-truth (red) and automatic segmentations of our method (yellow) trained using incompletely annotated with 30% annotated data for the prostate, bladder, and rectum separately.
Moreover, we also compare our method with non-deep learning-based methods trained using completely annotated data based on their reported performance as shown in Table I. The results of Shao [17], Gao [42], and Gao [21] are obtained in the same dataset as ours but may use different parameter settings. It can be observed that our method can also achieve comparable results to other state-of-the-art approaches in terms of both DSC and ASD for all three organs.
D. Comparison between Different Incomplete Annotation Manners
To evaluate the efficacy of the proposed method, we report the experimental results of using only one of the three annotation manners. Specially, we use W-Image, C-Slice, and S-Slice to generate the incompletely annotated training datasets and train the corresponding models, respectively. To acquire a dataset with d% annotated and 1-d% unannotated, we have randomly selected d% of images for annotation using W-Image, or d% of slices for annotation using C-Slice or S-Slice. Fig. 7 shows the DSC values of the segmentation models when trained with the above different annotation manners and the varying d%. we can see only a slight difference among the performance of using different annotation manners for given d%. These results verify the efficacy of our method in flexibly handling different annotation manners and their combinations. It is true that the optimal annotation manner may be related to many factors including the particular organ under consideration, the randomization to determine images/slices to annotate, and the percentage of images/slices being annotated, but our method is overall stable and effective.
Fig. 7.
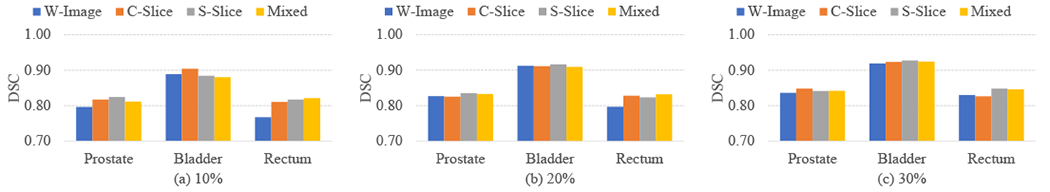
DSC values of using different incomplete annotation manners with different levels of incompleteness.
V. Ablation Studies
A. Effectiveness of Pelvis Localization
We introduce two other metrics to verify the effectiveness of our pelvis localization technique, i.e., target-to-target ratio (TTR) which measures the ratio of the number of organ voxels before and after pelvis localization and target-to-background ratio (TBR) which measures the ratio of the number of organ voxels to the number of background voxels, defined as [43], [44]:
| (11) |
where gi is the ground-truth label of the i-th voxel, Pro denotes the pelvic proposal extracted from the raw image Img based on the proposed pelvis localization technique, I denotes one CT volume (Pro or Img), and ⟦·⟧ is the indicator function. TTR measures whether the pelvis localization can preserve all voxels belonging to male pelvic organs, while TBR measures how much distractive background can be dropped.
Two U-net [3] based models are trained using raw images and extracted pelvic proposals separately, to demonstrate the effectiveness of our pelvis localization technique. Table II reports DSC values of two U-net models, TTR value computed between raw images and pelvic proposals, and their respective TBR values. We can observe that the segmentation performance can be significantly improved after focusing on the candidate segmentation regions for all three organs in terms of DSC. Moreover, the extracted pelvic proposals can absolutely cover the entire organ volumes in terms of TTR while much distractive background can be effectively dropped in terms of TBR. These pieces of evidence demonstrate that the proposed pelvis localization can effectively improve model accuracy and reduce computational complexity.
TABLE II.
DSC values of U-net trained on raw images and pelvic proposals separately, TTR value computed between raw images and pelvic proposals, and their respective TBR values.
| DSC | TTR | TBR | |||
|---|---|---|---|---|---|
| Prostate | Bladder | Rectum | |||
| Raw | 0.73±0.10 | 0.84±0.08 | 0.72±0.13 | 1.0 | 0.002 |
| Pro | 0.86±0.06 | 0.94±0.04 | 0.86±0.04 | 0.031 | |
B. Effectiveness of Label Completion
To make full use of unannotated data, we design a label completion strategy, including the pre-label completion before the first iteration and the post-label completion after each iteration, to augment the training data. To verify the effectiveness of label completion, we report the DSC values of the models trained with only incomplete annotation (N-FCN), with pre-label completion (N-FCN+Pre), and with single-iteration post-label completion (N-FCN+Pre+Post), as shown in Fig. 8. The comparison of results by N-FCN and N-FCN+Pre verifies the effectiveness of pre-label completion. Whereas the comparison of results by N-FCN+Pre and N-FCN+Pre+Post verifies the effectiveness of post-label completion after the first iteration. Moreover, to intuitively demonstrate the effectiveness of label completion in augmenting the training data, we report the DSC values obtained by comparing the initial incomplete annotation (30% Annotated), the initial incomplete annotation with pre-label completion (+Pre), the prediction of N-FCN+Pre, and the prediction of N-FCN+Pre+Post with the complete annotation on the training dataset, respectively, in Fig. 9. Specifically, the DSC values of 30% Annotated and +Pre are computed only based on the data with semantic labels (0/+1) while ignoring the data with unknown labels (−1). We can see that both the pre- and post-label completions can effectively augment the training data in terms of the comparison between 30% Annotated and +Pre and also between N-FCN+Pre and N-FCN+Pre+Post, respectively.
Fig. 8.

Comparisons between the models trained without and with label completion. N-FCN stands for the model trained using only incomplete annotation; N-FCN+Pre includes pre-label completion; N-FCN+Pre+Post further integrates single-iteration post-label completion. The number at the bottom of each bar indicates the corresponding mean DSC value.
Fig. 9.

Comparison of DSC values obtained by comparing the initial incomplete annotation (30% Annotated), the initial incomplete annotation with pre-label completion (+Pre), the prediction of N-FCN+Pre, and the prediction of N-FCN+Pre+Post with the complete annotation on the training dataset, respectively.
Fig. 10 shows the 3D segmentation results of the prostate, bladder, and rectum obtained by N-FCN, N-FCN+Pre, and N-FCN+Pre+Post, respectively. It is evident that the segmentation results are well refined after introducing confidence unannotated data.
Fig. 10.

3D visualization of the segmentation results of the prostate, bladder and rectum obtained by N-FCN, N-FCN+Pre, and N-FCN+Pre+Post, respectively.
C. Effectiveness of Hybrid Loss
While male pelvic organs are with unclear boundaries and large shape variation, the incompletely annotated data in clinical setting makes it more challenging to develop an accurate segmentation algorithm. To overcome this, we introduce a hybrid loss function to enhance the performance of the trained model. To verify its effectiveness, we replace our proposed hybrid loss with conventional cross-entropy loss to achieve the segmentation. We denote the final trained model corresponding to E-FCN but using conventional cross-entropy loss as E-FCNCE. Table III reports the mean DSC and ASD values of E-FCNCE and E-FCN. Instead of cross-entropy loss, our proposed hybrid loss can significantly improve the capability of the trained model, while E-FCN achieves better results than E-FCNCE with a large margin.
TABLE III.
Mean DSC and ASD (mm) values of E-FCNCE and E-FCN. Asterisks indicate E-FCN is better than E-FCNCE with statistical significance (p < 0.05).
| Metric | E-FCNCE | E-FCN | |
|---|---|---|---|
| Prostate | DSC | 0.788 | 0.842* |
| ASD | 2.481 | 1.931* | |
| Bladder | DSC | 0.901 | 0.924* |
| ASD | 2.938 | 1.242* | |
| Rectum | DSC | 0.808 | 0.846* |
| ASD | 2.966 | 1.771* | |
Fig. 11 shows the 3D segmentation results of the prostate, bladder, and rectum obtained by E-FCNCE and E-FCN. For better observation, the regions with significant improvement have been zoomed in.
Fig. 11.

3D visualization of the segmentation results of the prostate, bladder and rectum obtained by E-FCNCE and E-FCN. The regions with significant improvement are zoomed in for better observation.
D. Effectiveness of Iterative Learning
Iterative training is an effective mechanism to refine segmentation gradually. To verify its efficacy in our case, we report the DSC values of our method over the training iterations, as shown by segmentations results of three organs in Fig. 12. With more iterations, the segmentation performance for all three organs can be improved gradually. After the third iteration, the perfonnance tends to become stable or even slightly worse, since no more unannotated data can be correctly inferred and used for training.
Fig. 12.

DSC values of the proposed method for the prostate, bladder, and rectum segmentation with respect to the number of training iterations used.
VI. Conclusion and Discussion
In this paper, we present a new deep learning-based framework for the segmentation of CT male pelvic organs, in which only incompletely annotated data collected in a user-friendly manner is available for model training. To deal with great challenges of low contrast, large shape variation, and also incomplete annotation, we propose a hybrid loss network to enhance the model capability in an iterative way. And also, we design a label completion strategy to make full use of rich unannotated data. Given that too much background can interfere with the segmentation of target organs, we exploit the salient bone structures to locate the pelvic regions instead of raw images for segmentation. Experiments on the incompletely annotated data demonstrate that our proposed method achieves comparable segmentation performance on male pelvic organs, compared with the state-of-the-art methods trained on completely annotated data. Moreover, our method is user-friendly for data collection in real applications.
Deep learning has achieved excellent performance in medical image segmentation. However, its performance depends on the quality and scale of the dataset, which often brings a bottleneck to the training of the deep learning model. Compared with the complete annotation, our proposed incomplete annotation can reduce the cost, in terms of human effort and finance, but it cannot achieve satisfactory results if using conventional deep learning models based on the annotated data only (since CT pelvic images have indistinguishable shape and border).
In this study, we design a label propagation module as a pre-processing step to infer the incomplete annotation before the first iteration. Its effectiveness can be diminished by large shape variation and high decaying rate. So we adopt a large morphological kernel size and normalize all images into the same resolution for the used dataset in advance. In our future work, we will make this module more flexible and adaptable. For example, we can take one annotated adjacent slice around the unannotated slice under consideration as the reference to train a deep network for the prediction of the shape variation, and then, based on the prediction result, an energy minimization based method (i.e., GrabCut [45]) can be used to refine the results.
In this work, we use a hybrid loss network to iteratively update the labels of unannotated data and further introduce the inferred labels with high confidence into the training set for the next iteration of training. But the unavoidable cumulative errors could mislead the iterative model training. In the future, a self-correction strategy needs to be developed for improving the performance of this iterative learning method.
Acknowledgments
This work was supported in part by a NIH grant (5R01CA206100).
Contributor Information
Shuai Wang, Department of Radiology and Biomedical Research Imaging Center, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599 USA.
Dong Nie, Department of Radiology and Biomedical Research Imaging Center, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599 USA.
Liangqiong Qu, Department of Radiology and Biomedical Research Imaging Center, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599 USA.
Yeqin Shao, School of Transportation and Civil Engineering, Nantong University, Jiangsu 226019, China.
Jun Lian, Department of Radiation Oncology, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599 USA.
Qian Wang, Institute for Medical Imaging Technology, School of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai 200030, China.
Dinggang Shen, Department of Radiology and Biomedical Research Imaging Center, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599 USA; Department of Brain and Cognitive Engineering, Korea University, Seoul 02841, South Korea.
References
- [1].Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, and Jemal A, “Global cancer statistics 2018: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries,” CA: a cancer journal for clinicians, 2018. [DOI] [PubMed] [Google Scholar]
- [2].Long J, Shelhamer E, and Darrell T, “Fully convolutional networks for semantic segmentation,” in Proceedings of the IEEE conference on computer vision and pattern recognition, 2015, pp. 3431–3440. [DOI] [PubMed] [Google Scholar]
- [3].Ronneberger O, Fischer P, and Brox T, “U-net: Convolutional networks for biomedical image segmentation,” in International Conference on Medical image computing and computer-assisted intervention. Springer, 2015, pp. 234–241. [Google Scholar]
- [4].Men K, Dai J, and Li Y, “Automatic segmentation of the clinical target volume and organs at risk in the planning ct for rectal cancer using deep dilated convolutional neural networks,” Medical physics, vol. 44, no. 12, pp. 6377–6389, 2017. [DOI] [PubMed] [Google Scholar]
- [5].Shen D, Wu G, and Suk H-I, “Deep learning in medical image analysis,” Annual review of biomedical engineering, vol. 19, pp. 221–248, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M et al. ,, “A survey on deep learning in medical image analysis,” Medical image analysis, vol. 42, pp. 60–88, 2017. [DOI] [PubMed] [Google Scholar]
- [7].Ghose S, Oliver A, Martí R, Llado X, Vilanova JC, Freixenet J et al. , “A survey of prostate segmentation methodologies in ultrasound, magnetic resonance and computed tomography images,” Computer methods and programs in biomedicine, vol. 108, no. 1, pp. 262–287, 2012. [DOI] [PubMed] [Google Scholar]
- [8].Shi Y, Liao S, Gao Y, Zhang D, Gao Y, and Shen D, “Prostate segmentation in ct images via spatial-constrained transductive lasso,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2013, pp. 2227–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lay N, Birkbeck N, Zhang J, and Zhou SK, “Rapid multi-organ segmentation using context integration and discriminative models,” in International Conference on Information Processing in Medical Imaging. Springer, 2013, pp. 450–462. [DOI] [PubMed] [Google Scholar]
- [10].Martínez F, Romero E, Dréan G, Simon A, Haigron P, De Crevoisier R et al. , “Segmentation of pelvic structures for planning ct using a geometrical shape model tuned by a multi-scale edge detector,” Physics in Medicine & Biology, vol. 59, no. 6, p. 1471, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Balagopal A, Kazemifar S, Nguyen D, Lin M-H, Hannan R, Owrangi A et al. , “Fully automated organ segmentation in male pelvic ct images,” Physics in Medicine & Biology, vol. 63, no. 24, p. 245015, 2018. [DOI] [PubMed] [Google Scholar]
- [12].Park SH, Gao Y, and Shen D, “Multiatlas-based segmentation editing with interaction-guided patch selection and label fusion,” IEEE Transactions on Biomedical Engineering, vol. 63, no. 6, pp. 1208–1219, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Davis BC, Foskey M, Rosenman J, Goyal L, Chang S, and Joshi S, “Automatic segmentation of intra-treatment ct images for adaptive radiation therapy of the prostate,” in International Conference on Medical Image Computing and Computer-Assisted Intetvention. Springer, 2005, pp. 442–450. [DOI] [PubMed] [Google Scholar]
- [14].Liao S, Gao Y, Lian J, and Shen D, “Sparse patch-based label propagation for accurate prostate localization in ct images,” IEEE transactions on medical imaging, vol. 32, no. 2, pp. 419–434. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Acosta O, Dowling J, Drean G, Simon A, De Crevoisier R, and Haigron P, “Multi-atlas-based segmentation of pelvic structures from ct scans for planning in prostate cancer radiotherapy,” in Abdomen and Thoracic Imaging. Springer, 2014, pp. 623–656. [Google Scholar]
- [16].Zhan Y and Shen D, “Automated segmentation of 3d us prostate images using statistical texture-based matching method,” in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2003, pp. 688–696. [Google Scholar]
- [17].Shao Y, Gao Y, Wang Q, Yang X, and Shen D, “Locally-constrained boundary regression for segmentation of prostate and rectum in the planning ct images,” Medical image analysis, vol. 26, no. 1, pp. 345–356, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Costa MJ, Delingette H, Novellas S, and Ayache N, “Automatic segmentation of bladder and prostate using coupled 3d deformable models,” in International conference on medical image computing and computer-assisted intervention. Springer, 2007, pp. 252–260. [DOI] [PubMed] [Google Scholar]
- [19].Feng Q, Foskey M, Chen W, and Shen D, “Segmenting ct prostate images using population and patient-specific statistics for radiotherapy,” Medical Physics, vol. 37, no. 8, pp. 4121–4132, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chen S, Lovelock DM, and Radke RJ, “Segmenting the prostate and rectum in ct imagery using anatomical constraints,” Medical image analysis, vol. 15, no. 1, pp. 1–11, 2011. [DOI] [PubMed] [Google Scholar]
- [21].Gao Y, Shao Y, Lian J, Wang AZ, Chen RC, and Shen D, “Accurate segmentation of ct male pelvic organs via regression-based deformable models and multi-task random forests,” IEEE transactions on medical imaging, vol. 35, no. 6, pp. 1532–1543, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lu C, Zheng Y, Birkbeck N, Zhang J, Kohlberger T, Tietjen C et al. , “Precise segmentation of multiple organs in ct volumes using learning-based approach and information theory,” in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2012, pp. 462–469. [DOI] [PubMed] [Google Scholar]
- [23].Shi Y, Gao Y, Liao S, Zhang D, Gao Y, and Shen D, “A learning-based ct prostate segmentation method via joint transductive feature selection and regression,” Neurocomputing, vol. 173, pp. 317–331, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ma L, Guo R, Zhang G, Schuster DM, and Fei B, “A combined learning algorithm for prostate segmentation on 3d ct images,” Medical physics, vol. 44, no. 11, pp. 5768–5781, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Cha KH, Hadjiiski L, Samala RK, Chan H-P, Caoili EM, and Cohan RH, “Urinary bladder segmentation in ct urography using deeplearning convolutional neural network and level sets,” Medical physics, vol. 43, no. 4, pp. 1882–1896, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Shi Y, Yang W, Gao Y, and Shen D, “Does manual delineation only provide the side information in ct prostate segmentation?” in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2017, pp. 692–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].He K, Cao X, Shi Y, Nie D, Gao Y, and Shen D, “Pelvic organ segmentation using distinctive curve guided fully convolutional networks,” IEEE transactions on medical imaging, 2018. [DOI] [PubMed] [Google Scholar]
- [28].Jin D, Xu Z, Harrison AP, George K, and Mollura DJ, “3d convolutional neural networks with graph refinement for airway segmentation using incomplete data labels,” in International Workshop on Machine Learning in Medical Imaging. Springer, 2017, pp. 141–149. [Google Scholar]
- [29].Wang S, Cong Y, Fan H, Yang Y, Tang Y, and Zhao H, “Computer aided endoscope diagnosis via weakly labeled data mining,” in 2015 IEEE International Conference on Image Processing (ICIP). IEEE, 2015, pp. 3072–3076. [Google Scholar]
- [30].Taha A, Lo P, Li J, and Zhao T, “Kid-net: convolution networks for kidney vessels segmentation from ct-volumes,” in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2018, pp. 463–471. [Google Scholar]
- [31].Rackerseder J, Gobl R, Navab N, and Hennersperger C, “Fully automatic segmentation of 3d brain ultrasound: Learning from coarse annotations,” arXiv preprint arXiv:1904.08655, 2019. [Google Scholar]
- [32].Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, and Ronneberger O, “3d u-net: learning dense volumetric segmentation from sparse annotation,” in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2016, pp. 424–432. [Google Scholar]
- [33].Goetz M, Weber C, Binczyk F, Polanska J, Tarnawski R, Bobek-Billewicz B et al. , “Dalsa: domain adaptation for supervised learning from sparsely annotated mr images,” IEEE transactions on medical imaging, vol. 35, no. 1, pp. 184–196, 2016. [DOI] [PubMed] [Google Scholar]
- [34].Bai W, Suzuki H, Qin C, Tarroni G, Oktay O, Matthews PM et al. , “Recurrent neural networks for aortic image sequence segmentation with sparse annotations,” in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2018, pp. 586–594. [Google Scholar]
- [35].Lejeune L, Grossrieder J, and Sznitman R, “Iterative multi-path tracking for video and volume segmentation with sparse point supervision,” Medical image analysis, vol. 50, pp. 65–81, 2018. [DOI] [PubMed] [Google Scholar]
- [36].Lin T-Y, Goyal P, Girshick R, He K, and Dollár P, “Focal loss for dense object detection,” arXiv preprint arXiv:1708.02002, 2017. [DOI] [PubMed] [Google Scholar]
- [37].Pfister T, Charles J, and Zisserman A, “Flowing convnets for human pose estimation in videos,” in Proceedings of the IEEE International Conference on Computer Vision, 2015, pp. 1913–1921. [Google Scholar]
- [38].Chollet F et al. , “Keras,” 2015. [Online]. Available: https://github.com/fchollet/keras [Google Scholar]
- [39].Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J et al. , “Tensorflow: a system for large-scale machine learning.” in OSDI, vol. 16, 2016, pp. 265–283. [Google Scholar]
- [40].Badrinarayanan V, Kendall A, and Cipolla R, “Segnet: A deep convolutional encoder-decoder architecture for image segmentation,” IEEE transactions on pattern analysis and machine intelligence, vol. 39, no. 12, pp. 2481–2495, 2017. [DOI] [PubMed] [Google Scholar]
- [41].Yu F and Koltun V, “Multi-scale context aggregation by dilated convolutions,” in ICLR, 2016. [Google Scholar]
- [42].Gao Y, Lian J, and Shen D, “Joint learning of image regressor and classifier for deformable segmentation of ct pelvic organs,” in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2015, pp. 114–122. [Google Scholar]
- [43].Ju W, Xiang D, Zhang B, Wang L, Kopriva I, and Chen X, “Random walk and graph cut for co-segmentation of lung tumor on pet-ct images,” IEEE Transactions on Image Processing, vol. 24, no. 12, pp. 5854–5867, 2015. [DOI] [PubMed] [Google Scholar]
- [44].Wang S, He K, Nie D, Zhou S, Gao Y, and Shen D, “Ct male pelvic organ segmentation using fully convolutional networks with boundary sensitive representation,” Medical image analysis, vol. 54, pp. 168–178, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Rother C, Kolmogorov V, and Blake A, “Grabcut: Interactive foreground extraction using iterated graph cuts,” in ACM transactions on graphics (TOG), vol. 23, no. 3. ACM, 2004, pp. 309–314. [Google Scholar]


