Graphical abstract
Keywords: SARS-CoV-2 spike protein, Cholesterol, Lipids, Neutron reflection, Infrared spectroscopy
Abstract
Cholesterol has been shown to affect the extent of coronavirus binding and fusion to cellular membranes. The severity of Covid-19 infection is also known to be correlated with lipid disorders. Furthermore, the levels of both serum cholesterol and high-density lipoprotein (HDL) decrease with Covid-19 severity, with normal levels resuming once the infection has passed. Here we demonstrate that the SARS-CoV-2 spike (S) protein interferes with the function of lipoproteins, and that this is dependent on cholesterol. In particular, the ability of HDL to exchange lipids from model cellular membranes is altered when co-incubated with the spike protein. Additionally, the S protein removes lipids and cholesterol from model membranes. We propose that the S protein affects HDL function by removing lipids from it and remodelling its composition/structure.
1. Introduction2
In 2020, over just a few months, the SARS-CoV-2 pandemic reshaped the world as we knew it. Great effort has been invested in understanding the nature of the virus, its host interactions, the transmission routes, and the reasons why different parts of the population are afflicted so differently. Early on, inverse correlation was established between the degree of severity of Covid-19 and patient serum levels of both high-density lipoprotein (HDL) and cholesterol in patients [1], [2], [3], [4], with normal cholesterol profiles re-established during recovery [1], [3]. The SARS-CoV-2 virus particles are contained by a lipid bilayer from which the spike (S) protein protrudes [5]. The S protein was found to contain a pocket into which fatty acids bind, locking it in a more invasive conformation [6]. A cholesterol 25-hydroxylase was found to inhibit SARS-CoV-2 by depleting cholesterol from plasma membranes; this occurs as a result of over-activation of the ER-localized acyl-CoA:cholesterol acyltransferase (ACAT) [7]. ACAT mediates intracellular cholesterol esterification, which controls the cholesterol levels in the cell membrane [8].
At the same time, cholesterol was found to mediate Dengue virus binding to host membranes: cholesterol was shown to be a key component of the virion envelope however, it was not required as a part of the host membrane itself [9]. In a more recent study, cholesterol in the host membrane was shown to indirectly facilitate the binding of the envelope protein of Dengue virus: this fusion trimeric protein was shown to penetrate into the headgroup region of the model membrane adopting a tilted configuration along the membrane [10].
Lipoproteins are known to be involved in infection, by binding and neutralizing liposaccharides and lipoteichoic acid on the surface of bacteria [11]. Viral infection also leads to alteration of lipoprotein levels [12], with dramatic effects observed for Covid-19 patients [1]. Since cholesterol and HDL were found to bind S protein, it is possible that the function of HDL (related, among others, to inverse cholesterol transport [13] for cholesterol elimination in the liver) is altered. This may explain why patients with low HDL plasma levels are at higher risk of developing severe Covid-19 symptoms [2].
Recently two independent research groups showed that HDL binds and deposits its lipid cargo at model membranes in the absence of receptors [14], [15], [16]. In particular, HDL was found to deposit and remove lipids to an extent that depended on the type of lipids present in the model membranes [16], [17]: cholesterol dramatically decreases both processes when mixed with saturated fats. In contrast, cholesterol decreased only lipid removal when mixed with unsaturated fats [16].
Here the effects of co-incubation of the S protein and HDL on HDL’s ability to exchange lipids are described. This study was instigated in order to investigate the possibility that the interaction between the S protein and HDL could lead to an imbalance between lipid metabolism and the regulation of serum lipid and lipoprotein concentrations. Model membranes were used in the form of supported lipid bilayers composed of deuterated 1,2-dimyristoyl-D54-3-sn-glycerophosphatidylcholine (dDMPC) and perdeuterated cholesterol (dcholesterol [18]) at a molar ratio of 80:20 mol%, and were exposed to the S protein, HDL or a mixture of the S protein and HDL in physiologically relevant conditions (saline buffer, pH 7.4 and 37 °C). Under these conditions, the model membrane is fluid [16]. Neutron Reflection (NR) and Attenuated Total Reflection – Fourier Transform Infrared Spectroscopy (ATR-FTIR) were used to follow both lipid removal and lipid deposition by HDL and the S protein. Cholesterol was included in the model membranes since it has been shown to be essential for the fusion of the SARS-CoV virus to cellular membranes and for entry into the cell [19]. The urgent need to study the role of plasma membrane cholesterol for SARS-CoV-2 infection has recently been discussed [20].
2. Materials and methods
2.1. Materials
dDMPC (>99%) and dcholesterol/hcholesterol (>99%) were purchased from Avanti or produced according to previous procedures [18]. D2O (99,9%, VWR Chemicals) and TBS tablets pH 7.4 were from Sigma Aldrich. Centrifugal devices, Microsep™ Advance for sample buffer exchange were acquired from VWR.
2.2. Spike protein expression and purification
SARS-CoV-2 spike proteins (2P constructs [21]) were expressed in EXPI293 cells and purified, as described previously [22]. Final gel filtration buffers were 20 mM Tris-HCl pH 7.5, 150 mM NaCl for spike 2P and 25 mM Tris-HCl pH 8, 150 mM NaCl buffer for spike 6P. Both constructs do not include the transmembrane domain of the S protein.
2.3. HDL purification
HDL preparation was carried out using human plasma from three healthy males, donated from the blood bank of Malmö University Hospital. No personal information was used in this study. The plasma was purified using sequential ultracentrifugation resulting in isolated HDL at a density of 1.065 g/mL, these purified samples were then pooled and stored at –80 °C in 50% sucrose, 150 mM NaCl, 24 mM EDTA, pH7.4. Before use, HDL was further purified via size exclusion chromatography (Superose 6 Increase 10/300 GL column, GE Healthcare). The peak corresponding to pure HDL was then collected and stored in 50 mM Tris-HCl 150 mM NaCl, pH 7.4 at 4 °C away from light in an inert atmosphere and used within a week. This method was demonstrated to give pure HDL fractions earlier [14], [23]. The HDL was used at a concentration of 0.132 mg/mL as determined by Bradford assay.
2.4. Model Membrane Formation
Model membranes were prepared using published protocols [16], [24]. Briefly, lipid films for NR and ATR-FTIR experiments were prepared in the same way, from chloroform stocks of dDMPC and dcholesterol. The films were dried under a stream of nitrogen and put under vacuum overnight to ensure the full removal of chloroform and stored at –20 °C until use. To prepare the vesicles for injection, the lipid films were hydrated in MilliQ water (for NR) or in D2O (for ATR-FTIR) and kept in a bath sonicator for 1 h at 40–50 °C. Suspensions were tip sonicated immediately before use for 5 min until a clear dispersion was obtained (under low power). Equal volumes of 0.2 mg/mL vesicles and 4 mM CaCl2 (prepared in H2O for NR and D2O for ATR-FTIR) were mixed immediately prior to injection, the vesicle solutions were introduced to the cells via syringe pump at 1 mL/min at a concentration of 0.1 mg/mL to aid in vesicle fusion. In the NR experiments the vesicles were incubated for 20 min before rinsing with MilliQ water followed by h-TBS buffer at 1 mL/min. For the ATR-FTIR the vesicles were incubated for 1 h to ensure no further changes were seen in the IR signal, then rinsed with D2O followed by d-TBS buffer. For ATR-FTIR solutions were injected slowly by hand.
2.5. Neutron Reflection Experiment
NR data were collected on the D17 instrument [25] at the Institut Laue Langevin (Grenoble, France) and fitted using the MOTOFIT package [26]. The Monte Carlo error analysis using genetic optimisation within the MOTOFIT software was used to determine the error of the fits. Significance of pairwise parameter differences were calculated using an F-test assuming a Gaussian distribution of the errors. The experiment was performed using a typical setup for solid/liquid interfaces. A silicon substrate was sealed inside a cell, with the polished surface in contact with a liquid reservoir of 1 mL. The cell was installed on a sample stage and was connected to a high pressure liquid chromatography (HPLC) pump to produce contrast variation by solvent exchange with D2O/H2O mixtures of different D/H ratios. Two configurations were used to cover a momentum transfer range 0.006 ≤ q ≤ 0.3 Å−1, with an incident neutron beam of wavelength 3 ≤ λ ≤ 30 Å, where the angle of the incoming beam was set to 0.8° and 3.0° and surface footprint of 35 × 65 mm. The wavelength-dependent spatial resolution (Δq/q) was varying between 3 and 9%.
The silicon (1 1 1) blocks were cleaned with chloroform, ethanol, acetone and finally MilliQ water, sonicating in each solvent for 20 min. After the final sonication in MilliQ water, the blocks were thoroughly rinsed, dried under nitrogen and exposed to ozone for 30 min. The blocks were rinsed again and assembled in wet conditions. The polyether ether ketone (PEEK) and O-ring components were extensively cleaned in Hellmanex 2% (v/v) solution and MilliQ water twice with bath sonication, and rinsed with MilliQ between each sonication.
Three contrasts were used throughout the experiment, and changed in situ via HPLC pump. The contrasts used were h-TBS (made in H2O), d-TBS (made in D2O) and cmSi-TBS (made in 38:62 D2O:H2O (v/v)), to contrast-match the Silicon block. The bespoke solid/liquid cells were pre-equilibrated to 37 °C and characterized to measure the thickness and roughness of the silicon oxide layer in H2O and D2O before the introduction of samples, and to aid in the fitting of the supported lipid bilayers. The clean bilayers were characterized in three contrasts and fitted simultaneously using a 5-layer model corresponding to the oxide layer, a small solvent layer and three layers analogous to the heads-tails-heads of the lipid membrane. The symmetrical bilayer allowed for further constraints to be put on the fit whereby the thickness, coverage and scattering length density (SLD) of the heads were the same for both, and the roughness was constrained to be the same across the whole bilayer.
The introduction of Spike protein and/or HDL was via syringe pump at a rate of 1 mL/min at concentrations of 0.05 mg/mL (0,1 µM) and 0.132 mg/mL, respectively. The S protein concentration hereby was chosen to be among the low range of that typically used for protein-lipid interactions by surface sensitive techniques [10], [27], [28], [29], [30]. The HDL concentration was chosen to be fully comparable to previous studies [14], [16], [17]. After 5 h of incubation in all cases, the bilayers were rinsed with buffer and re-characterized in all three contrasts. The fittings of the final bilayers were based on the initial bilayer characterizations, keeping the head-group thickness the same but allowing the tail region thickness and SLD to change in some cases. All bilayers also required an additional layer on top corresponding to remaining bound spike protein and/or HDL. Due to restricted beamtime availability, experiments were not repeated. However, NR is a highly reproducible technique with the variations within different replicates typically being among the fit erros [31].
2.6. ATR-FTIR to study the S protein interacting with model membranes
The silicon block used for the ATR-FTIR (Thermo Nicolet spectrophotometer) was cleaned via bath sonication in 2% Hellmanex solution, ethanol and MilliQ water, rinsing between each solvent change and then exposed to ozone for 30 min before use. All tubing was cleaned with 2% Hellmanex, ethanol and plenty of MilliQ water. The sample environment was maintained at 37 °C via a water bath. The lipids, S protein and HDL were all introduced slowly to the cell by hand via syringe injection. All injections and washes were done in D2O or d-TBS to minimize the signal from the OH stretches from H2O. Continuous measurements were taken to follow the incubation of the S protein and/or HDL with the lipid membranes. Experiments were performed in duplicates.
3. Results and discussions
Supported lipid bilayers were formed via the vesicle fusion on silicon surfaces with a composition of 80:20 mol% dDMPC:dcholesterol at 37 °C. Cholesterol was added since it was shown to be determinant for the fusion of SARS-CoV to cellular membranes [19] and since it reduces the capacity of HDL to exchange lipids [16]. NR profiles were in three isotopic contrasts at 37 °C (Supporting Information Figure S1). Such cholesterol-containing model membranes have a lipid core and head-group thickness of 34.0 ± 0.5 Å and 7 ± 1 Å, respectively, with a mean molecular area of 58 ± 2 Å2 , in agreement with previous reports [16].
Three independent model membranes were then exposed to either 0.05 mg/mL (0,1 µM) S protein in h-TBS (50 mM Tris, 150 mM NaCl, pH 7.4 in H2O), 0.13 mg/mL HDL, or to a freshly prepared mixture of 0.13 mg/mL HDL and 0.05 mg/mL S protein. The HDL was a sample pooled from three healthy male volunteers obtained via the blood bank at Malmö University Hospital and purified as described previously [14]. The samples were allowed to incubate for 5 h at 37 °C, and reflectivities were measured at different times during equilibration (Fig. 1 and Fig. 2 ).
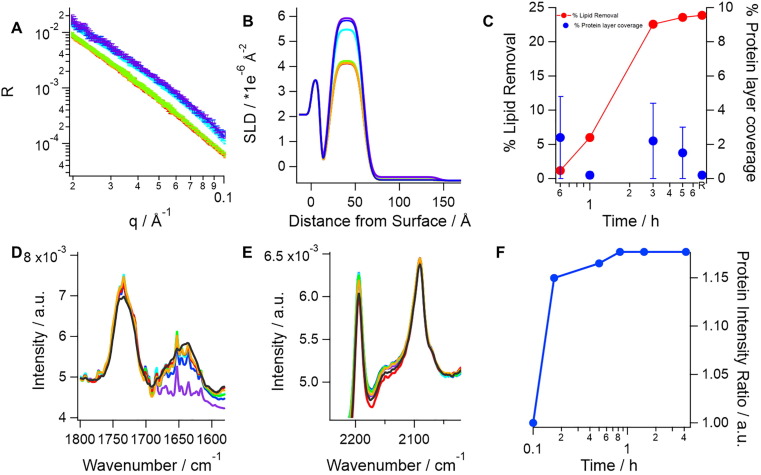
Fig. 1.
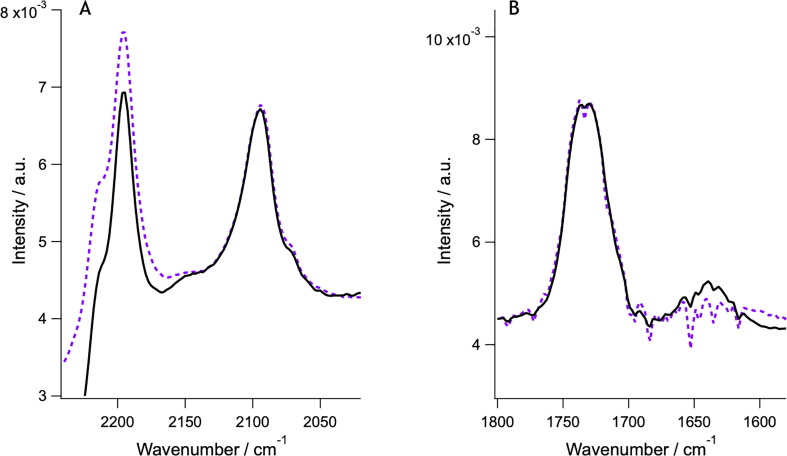
SARS-CoV-2 spike protein (0.05 mg/mL or 0,1 µM) incubation with model membranes composed of saturated lipids and cholesterol lead to lipid removal, which is accompanied by protein adsorption on top of the lipid membrane. The top panel, A shows neutron reflection profiles (markers) and best fits (lines) in h-TBS at 37 °C upon incubation time of 0 min (purple), 40 min (blue), 1 h (cyan), 3 h (green), 5 h (orange) and upon rinsing with excess h-TBS (red). Corresponding scattering length density (SLD) profiles for best fits to the neutron reflection data are shown in B. Apparent lipid removal as calculated from a change in the SLD (red) of the lipid tail layer as well as additional protein layer coverage (blue) are shown in C. Protein binding was corroborated by ATR-FTIR (bottom panel). In this case, a model membrane composed of dDMPC and hcholesterol is formed and pre-characterized in d-TBS (purple) and a broad amide I peak (~1640 cm−1, D) appears as soon as the protein is introduced in the cell (blue, 10 min; cyan, 30 min; green, 50 min; red, 90 min; black, 4 h). A small decrease in the C O stretching (D) and the CD2 symmetric and asymmetric stretch (E) bands (occurring at ~ 1734, ~2090 and ~ 2194 cm−1, respectively) suggests that at least some dDMPC was removed. The change in amide I peak intensity over time (right) shows that protein binding occurs within the first 10 min of incubation (F). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
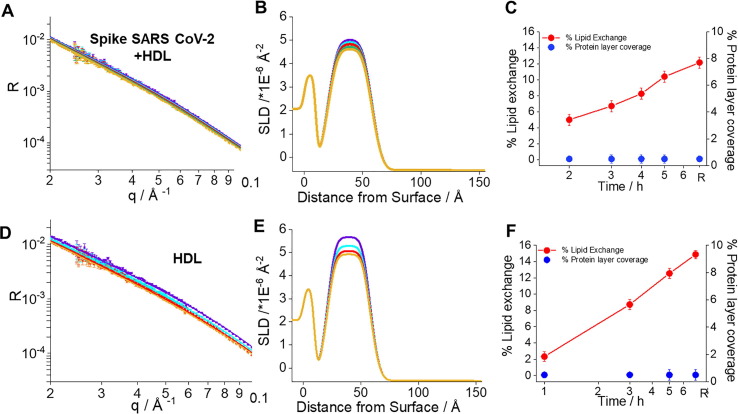
Fig. 2.
Neutron Reflection profiles including best fits (A,D), corresponding scattering length density profiles (B,E) and kinetics of interaction for the model membranes (C,F) exposed to 0.05 mg/mL or 0.1 µM S freshly mixed with 0.132 mg/mL HDL (top) in h-TBS at 37 °C and upon 2 h (purple), 3 h (cyan), 4 h (green), 5 h (dark green) incubation time and upon rinsing with excess h-TBS (orange), and 0.132 mg/mL HDL (bottom) at time 1 h (purple), 3 h (cyan), 5 h (green), and upon rinsing with excess h-TBS (orange). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Incubation with S protein induced a progressive decrease in reflectivity over time, with most of the changes occurring between the third and fifth incubation hour (Fig. 1). Decreased reflectivity at this contrast implies a loss of deuterated material adsorbed at the interface - which can only occur as a result of lipid removal by S protein. The lipid removal occurs faster during the first 3 h, after which it reaches a steady state at roughly under 25% removal of the original membrane.
A similar behaviour (although attenuated) was observed upon incubation with HDL (Fig. 2, lower panel). This is due to deuterated lipid removal by HDL and replacement by hydrogenated lipids [14], [16], [17]. The extent of the reflectivity decrease upon incubation with both S protein and HDL was slightly reduced over time (Fig. 2, upper panel) indicating both that S protein did indeed affect how HDL exchanged lipids, and that HDL compensated for the lipid removal by the S protein.
Unambiguous determination of the extent of lipid removal and deposition by NR can only be achieved upon simultaneous data analysis of 2 or ideally 3 isotopic contrasts. Here all datasets were measured upon equilibrium in h-TBS, D2O based buffer (d-TBS) and a mixture of 62% h-TBS and 38% d-TBS to match the silicon substrate SLD (Supporting Information Figure SI2). The extents of lipid deposition and lipid removal by S protein, HDL or their mixture to model membranes are shown in Fig. 3 . S protein incubation with model membranes leads to a loss of 28% of the lipids present in the membrane. Here, 28% lipid removal is determined by the simultaneous analysis of three contrast NR data, which give a more accurate value to the one estimated from a single contrast in Fig. 2. This is accompanied by an additional adsorbed layer, which should correspond to the membrane-associated S protein. The total adsorbed amount of S protein at the steady state condition is very small, and within the sensitivity range of NR. However, the NR kinetic data show that the adsorbed amount of S protein (fixed to a layer thickness of 80 Å) seems higher at first but then decreases over time, although the error is high due to the overall low layer-coverage. This may suggest that once S protein becomes coated by lipids, it detaches from the membrane. To demonstrate definitively that lipid loss is accompanied by S protein adsorption, an independent experiment was performed using ATR-FTIR. For this study, a model membrane made of dDMPC and hydrogenous cholesterol (hcholesterol) was formed in D2O, characterized in d-TBS and exposed to 0.05 mg/mL S in d-TBS. The data (Fig. 1, bottom panel) clearly show the rapid S protein adsorption that is associated with the amide 1 band at 1640 cm−1. This was accompanied by a small decrease in the C O stretch band at 1734 cm−1, and both the symmetric and asymmetric C-D stretch bands at 2090 and 2194 cm−1, respectively. This demonstrates that at least some dDMPC is removed by S protein.
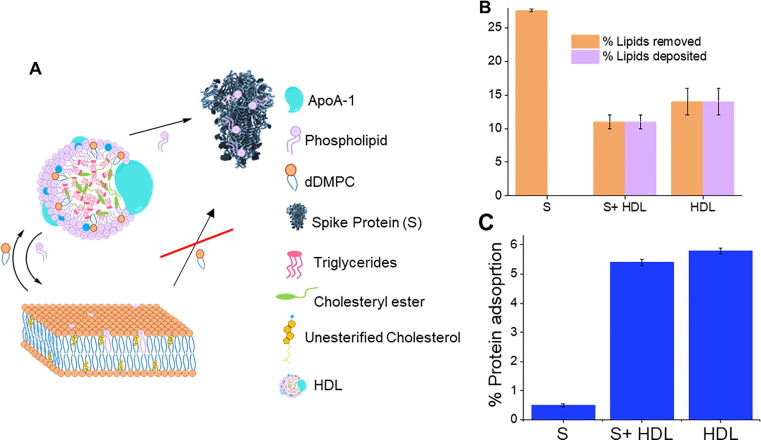
Fig. 3.
Schematics (not to scale) for the dynamics of lipid exchange between high density lipoprotein (HDL), SARS-CoV-2 spike protein (S) and model membranes composed of saturated lipids and cholesterol (A). Extent of lipid removal/deposition (B) and adsorbed protein/lipoprotein layer coverage (C) upon 5 h of incubation of either HDL, SARS-CoV-2 spike protein or co-incubation of both SARS-CoV-2 spike protein and HDL. HDL removes and deposits lipids from model membranes. S protein removes lipids from model membranes. Co-incubation of HDL and S protein leads to reduced lipid removal and deposition at model membranes, suggesting that S removes lipids from HDL rather than the membrane.
A separate experiment was performed with membranes composed of only dDMPC exposed to the S protein (Fig. 4 ). In this case, minimal lipid removal and very little protein binding took place as measured by ATR-FTIR. This suggests that cholesterol is crucial for S protein binding to model membranes, and for its ability to remove lipids. Indeed, it is well accepted that the membrane composition plays a key role in the behaviour of fusion proteins and their ability to enter the cell [32]. In particular, increased levels of cholesterol in human plasma membranes correlate with stronger binding affinity for the fusion peptide of SARS-CoV [33] and, thus, with increased rates of SARS-CoV infection [19]. Recent results showed that the S protein in SARS-CoV-2 binds cholesterol and HDL [1], and here cholesterol is shown to be essential for significant S protein binding to model membranes and lipid removal from membranes. ATR-FTIR studies on dDMPC and hcholesterol containing membranes (Fig. 1) confirm that some dDMPC may be removed by the S protein but, in any case, the symmetric and asymmetric C-D stretch bands did not decrease by 28% as expected from the NR results for the S protein against a dDMPC-dcholesterol model membrane (Fig. 3 and Supporting Information Figure S2). Thus, it seems likely that cholesterol is the main component removed by S protein from the membrane. Additional NR experiments could clarify whether or not both phospholipids and cholesterol are removed equally: for example, by using membranes containing different combinations of hydrogenated or deuterated membrane components, the specific removal of either cholesterol or phospholipids could be determined as done previously for ApoE-based reconstituted HDL [34]. We attempted to carry out ATR-FTIR experiments with hDMPC and dcholesterol model membranes, but the CD2 bands were, in this case, too weak to draw any conclusions from the data (probably due to the low cholesterol molar content used in these experiments). Finally, the coverage of the pristine membranes was ~ 85%. Despite the presence of some defects on the membrane, no S protein incorporation on the core of the bilayer or translocation across the membrane took place. This suggests that the S protein interacts mainly via electrostatic interactions with the headgroup regions of the model membrane. The fact that the presence of cholesterol is needed for S protein binding and lipid removal (Fig. 4) is in line with previous results for the envelope protein of the Dengue virus, which also forms a trimer and triggers the fusion of the Dengue virus to the host membrane. In particular, the presence of negative lipids or cholesterol was found to facilitate the binding of the envelope protein to the model membranes by NR and atomistic molecular dynamic simulations due to strengthened electrostatic interactions or indirectly via regulation of the headgroup interdistance across the membrane, respectively [10]. Moreover, an ATR-FTIR experiment was performed using the soluble domain of the cytochrome P450 reductase (POR) from Sorghum bicolor [35] against a dDMPC and hcholesterol model membrane (Supporting Information Figure S3) in order to demonstrate that the lipid removal by S protein is specific to it. The results show that POR binds to the model membrane (a clear amide band appears quickly after POR injection) but no observable lipid removal took place (the symmetric and asymmetric CD2 bands remain constant over the time of the experiment).
Fig. 4.
ATR-FTIR data for dDMPC model membranes (no cholesterol was present) before (purple) and after S protein incubation (black). No significant changes in the CD2 asymmetric stretch (~2194 cm−1) bands take place (A) and minimal appearance of the amide I peak (~1640 cm−1) occurs even after two hours of incubation (B). There is a change in the baseline that affects the signal for the C O stretching and CD2 symmetric bands (occurring at ~ 1734 and ~ 2090 cm−1 respectively). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Model membrane incubation with HDL leads to similar extent of lipid deposition and removal (Fig. 3). It was noted previously that more lipid molecules are removed than deposited by HDL [14], [16], [17]. The results here could be a peculiarity of this pooled HDL sample, which may contain altered proportions of apolipoprotein variants having for example altered affinity for lipid exchange [34]. Membrane co-incubation of both HDL and S protein maintains this trend (Fig. 3), with the total proportion of lipids deposited and removed being slightly lower than for HDL incubation alone (significantly different for p = 0.1). Assuming no interaction between S protein and HDL, the same level of lipid deposition by HDL would be expected, while a greater extent of lipid removal should occur as a result of the collective action of both S protein and HDL.
Indeed, the surface coverage for the additional layer seen on top of the model membrane (which corresponds to either membrane associated S protein, HDL, or their mixture) is not the sum of the additional layer’s coverage derived for the independent S protein and HDL experiments, but is actually lower than for the HDL alone (significant, p = 0.05). Thus, the observed effect for the amount of lipid removed and deposited must be a consequence of S protein binding to HDL in solution. This could lead to lipid removal from HDL by S protein, which in turn affects how HDL exchanges lipids. Indeed, SARS-CoV-2 infection changes the lipodomic profile in serum, resulting in a reduced size of HDL particles [36]. The ratio of the ApoA to total protein remained the same, which suggests that the lipid content per particle is affected. This leads to an increase in the proportion of triglyceride rich lipoproteins [36].
Earlier, it was shown that S protein binds fatty acids with a preference for the essential linoleic acid [6]. The lipids in the HDL surface are mainly composed of phospholipids and sphingolipids [23], [37] besides free cholesterol. Lipidomics have not detected free fatty acids but this does not exclude the possibility that they are present in HDL. Overall, it is likely that the S protein interacts with HDL and that cholesterol and other lipids are removed because of that interaction. Additional studies are required to understand these processes, and further determine how the HDL composition and structure is changed.
Finally, HDL from individuals suffering from lipid disorders causes a higher risk for the development of atherosclerosis [38], [39], and contains different associated lipids and proteins [40] that may impact HDL function and S protein affinity for it. S protein-induced HDL remodelling could have major impact on the ability of HDL to remove lipids and therefore on the metabolism of lipids. Further studies are required to understand the processes by which S protein interacts with HDL, and how the HDL composition and structure is changed. Indeed, an increase in the average size of LDL particles has been seen in Covid-19 patients, also consistent with the triglyceride rich composition [36]. However, it is not known whether it is the lipid loss due to interaction with the S protein that leads to such change in LDL particle size although this would seem quite likely.
4. Conclusions
In this paper, the effect of co-incubation of HDL pooled from three healthy adult male donors with the Spike SARS-CoV-2 protein on the capacity for HDL to exchange lipids from model membranes was studied using NR and ATR-FTIR. For comparison, parallel experiments were performed with HDL or the Spike protein alone at the model membrane. The hypothesis was that Spike interactions with HDL might affect the capacity of HDL to exchange lipids from model membranes, since there is a correlation in the clinics between HDL levels and the severity of the Covid-19 disease [3], [4], [20]. Our studies suggest that the SARS-CoV-2 spike protein binds to model membranes and removes lipids, and potentially the process is facilitated by the presence of cholesterol. Recently, the SARS-CoV-2 spike protein was found to bind cholesterol and HDL [1]. Upon co-incubation of both the SARS-CoV-2 spike protein and HDL on model membranes, slightly lower levels of lipid removal and deposition take place as compared to incubation with HDL alone [14], [16], [17] Collectively, the facts strongly suggest that S protein binds HDL, and that this binding leads to remodelling of HDL structure and composition. More experiments are needed to establish whether cholesterol and lipids are taken up by the SARS-CoV-2 spike protein, and which lipids the SARS-CoV-2 spike protein removes from HDL. Studies to explore how co-incubation or pre-exposure of HDL to the S protein affect cholesterol efflux should be performed in the future. The present studies using lipoproteins extracted from individuals with different serum lipid profiles may help elucidate why some individuals are more seriously afflicted by Covid-19 than others.
CRediT authorship contribution statement
Yubexi Correa: Methodology, Data curation, Visualization, Writing - review & editing. Sarah Waldie: Methodology, Data curation, Writing - review & editing. Michel Thépaut: Methodology, Writing - review & editing. Samantha Micciula: Data curation, Writing - review & editing. Martine Moulin: Methodology, Writing - review & editing. Franck Fieschi: Methodology, Writing - review & editing. Harald Pichler: Methodology, Writing - review & editing. V. Trevor Forsyth: Conceptualization, Writing - review & editing. Michael Haertlein: Conceptualization, Writing - review & editing. Marité Cárdenas: Conceptualization, Methodology, Writing - original draft, Supervision, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
MC, YC and SW acknowledges support from the Swedish Research Council grant numbers 2018-03990 and 2018-04833 . IBS acknowledges integration into the Interdisciplinary Research Institute of Grenoble (IRIG, CEA). VTF acknowledges support from the UK Engineering and Physical Sciences Council (EPSRC) which funded the creation of the Deuteration Laboratory within the Life Sciences Group at the ILL [GR/R99393/01 and EP/C015452/1]. The soluble domain of the POR was a kind gift by Dr. Rita del Giudice. Thanks to Dr. Federica Sebastiani and Dr. Nicolo Paracini for support during ATR-FTIR experiments. We thank the ILL for granted beamtime with DOI: 10.5291/ILL-DATA.DIR-212.
Footnotes
HDL, high density lipoprotein; S, SARS-CoV-2 Spike protein; ACAT, acyl-CoA:cholesterol acyltransferase; dDMPC, 1,2-dimyristoyl-D54-3-sn-glycerophosphatidylcholine, dcholesterol, perdeuterated cholesterol; hcholesterol, hydrogenated cholesterol; NR, neutron reflection; ATR-FTIR, Attenuated Total Reflection –Fourier Transform Infrared Spectroscopy; h-Tris, Tris pH 8, 150 mM NaCl made in H2O; d-Tris, Tris pH 8, 150 mM NaCl made in D2O; cmSi-Tris, Tris pH 8, 150 mM NaCl made in 58:32 v% H2O: D2O; ApoE, apolipoprotein E; SLD, scattering length density.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jcis.2021.06.056.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Y. Peng, L. Wan, C. Fan, P. Zhang, X. Wang, J. Sun, Y. Zhang, Q. Yan, J. Gong, H. Yang, X. Yang, H. Li, Y. Wang, Y. Zong, F. Yin, X. Yang, H. Zhong, Y. Cao, C. Wei, Cholesterol Metabolism--Impacts on SARS-CoV-2 Infection Prognosis, medRxiv, 2020.
- 2.Wang G., Zhang Q., Zhao X., Dong H., Wu C., Wu F., Yu B., Lv J., Zhang S., Wu G., Wu S., Wang X., Wu Y., Zhong Y. Low high-density lipoprotein level is correlated with the severity of COVID-19 patients: an observational study. Lipids Health Dis. 2020;19(1):204. doi: 10.1186/s12944-020-01382-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun J.T., Chen Z., Nie P., Ge H., Shen L., Yang F., Qu X.L., Ying X.Y., Zhou Y., Wang W., Zhang M., Pu J. Lipid Profile Features and Their Associations With Disease Severity and Mortality in Patients With COVID-19. Front. Cardiovascular Med. 2020;7(290) doi: 10.3389/fcvm.2020.584987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanaka S., De Tymowski C., Assadi M., Zappella N., Jean-Baptiste S., Robert T., Peoc'h K., Lortat-Jacob B., Fontaine L., Bouzid D., Tran-Dinh A., Tashk P., Meilhac O., Montravers P. Lipoprotein concentrations over time in the intensive care unit COVID-19 patients: Results from the ApoCOVID study. PLoS ONE. 2020;15(9) doi: 10.1371/journal.pone.0239573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ke Z., Oton J., Qu K., Cortese M., Zila V., McKeane L., Nakane T., Zivanov J., Neufeldt C.J., Cerikan B., Lu J.M., Peukes J., Xiong X., Kräusslich H.-G., Scheres S.H.W., Bartenschlager R., Briggs J.A.G. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature. 2020 doi: 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toelzer C., Gupta K., Yadav S.K.N., Borucu U., Davidson A.D., Kavanagh Williamson M., Shoemark D.K., Garzoni F., Staufer O., Milligan R., Capin J., Mulholland A.J., Spatz J., Fitzgerald D., Berger I., Schaffitzel C. Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein. Science. 2020;370:725–730. doi: 10.1126/science.abd3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang S., Li W., Hui H., Tiwari S.K., Zhang Q., Croker B.A., Rawlings S., Smith D., Carlin A.F., Rana T.M. Cholesterol 25-Hydroxylase inhibits SARS-CoV-2 and other coronaviruses by depleting membrane cholesterol. EMBO J. 2020;39(21) doi: 10.15252/embj.2020106057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trinick T.R., Duly E.B. In: Encyclopedia of Human Nutrition (Third Edition) Caballero B., editor. Academic Press; Waltham: 2013. Hyperlipidemia: Overview; pp. 442–452. [Google Scholar]
- 9.Carro A.C., Damonte E.B. Requirement of cholesterol in the viral envelope for dengue virus infection. Virus Res. 2013;174(1–2):78–87. doi: 10.1016/j.virusres.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 10.Vanegas J.M., Heinrich F., Rogers D.M., Carson B.D., La Bauve S., Vernon B.C., Akgun B., Satija S., Zheng A., Kielian M., Rempe S.B., Kent M.S. Insertion of dengue E into lipid bilayers studied by neutron reflectivity and molecular dynamics simulations. Biochim. Biophys. Acta Biomembr. 2018;1860(5):1216–1230. doi: 10.1016/j.bbamem.2018.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kitchens R.L., Thompson P.A., Munford R.S., O'Keefe G.E. Acute inflammation and infection maintain circulating phospholipid levels and enhance lipopolysaccharide binding to plasma lipoproteins. J. Lipid Res. 2003;44(12):2339–2348. doi: 10.1194/jlr.M300228-JLR200. [DOI] [PubMed] [Google Scholar]
- 12.Barlage S., Fröhlich D., Böttcher A., Jauhiainen M., Müller H.P., Noetzel F., Rothe G., Schütt C., Linke R.P., Lackner K.J., Ehnholm C., Schmitz G. ApoE-containing high density lipoproteins and phospholipid transfer protein activity increase in patients with a systemic inflammatory response. J. Lipid Res. 2001;42(2):281–290. [PubMed] [Google Scholar]
- 13.Chang C., Dong R., Miyazaki A., Sakashita N., Zhang Y., Liu J., Guo M., Li B.L., Chang T.Y. Human acyl-CoA:cholesterol acyltransferase (ACAT) and its potential as a target for pharmaceutical intervention against atherosclerosis. Acta Biochim. Biophy. Sin. 2006;38(3):151–156. doi: 10.1111/j.1745-7270.2006.00154.x. [DOI] [PubMed] [Google Scholar]
- 14.Browning K.L., Lind T.K., Maric S., Malekkhaiat-Häffner S., Fredrikson G.N., Bengtsson E., Malmsten M., Cárdenas M. Human lipoproteins at model cell membranes: effect of lipoprotein class on lipid exchange. Sci. Rep. 2017;7(1):7478. doi: 10.1038/s41598-017-07505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plochberger B., Sych T., Weber F., Novacek J., Axmann M., Stangl H., Sezgin E. Lipoprotein particles interact with membranes and transfer their cargo without receptors. Biochemistry. 2020;59(45):4421–4428. doi: 10.1021/acs.biochem.0c00748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.S. Waldie, F. Sebastiani, K. Browning, S. Maric, T.K. Lind, N. Yepuri, T.A. Darwish, M. Moulin, G. Strohmeier, H. Pichler, M.W.A. Skoda, A. Maestro, M. Haertlein, V.T. Forsyth, E. Bengtsson, M. Malmsten, M. Cárdenas, Lipoprotein ability to exchange and remove lipids from model membranes as a function of fatty acid saturation and presence of cholesterol, Biochim. et Biophys. Acta (BBA) – Mol. Cell Biol. Lipids 1865(10) (2020) 158769. [DOI] [PubMed]
- 17.Browning K.L., Lind T.K., Maric S., Barker R.D., Cárdenas M., Malmsten M. Effect of bilayer charge on lipoprotein lipid exchange. Colloids Surf., B. 2018;168:117–125. doi: 10.1016/j.colsurfb.2018.01.043. [DOI] [PubMed] [Google Scholar]
- 18.Moulin M., Strohmeier G.A., Hirz M., Thompson K.C., Rennie A.R., Campbell R.A., Pichler H., Maric S., Forsyth V.T., Haertlein M. Perdeuteration of cholesterol for neutron scattering applications using recombinant Pichia pastoris. Chem. Phys. Lipids. 2018;212:80–87. doi: 10.1016/j.chemphyslip.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 19.Meher G., Bhattacharjya S., Chakraborty H. Membrane cholesterol modulates oligomeric status and peptide-membrane interaction of severe acute respiratory syndrome coronavirus fusion peptide. J. Phys. Chem. B. 2019;123(50):10654–10662. doi: 10.1021/acs.jpcb.9b08455. [DOI] [PubMed] [Google Scholar]
- 20.Radenkovic D., Chawla S., Pirro M., Sahebkar A., Banach M. Cholesterol in relation to COVID-19: should we care about It? J. Clin. Med. 2020;9(6):1909. doi: 10.3390/jcm9061909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.M. Thépaut, J. Luczkowiak, C. Vivès, N. Labiod, I. Bally, F. Lasala, Y. Grimoire, D. Fenel, S. Sattin, N. Thielens, G. Schoehn, A. Bernardi, R. Delgado, F. Fieschi, DC/L-SIGN recognition of spike glycoprotein promotes SARS-CoV-2 trans-infection and can be inhibited by a glycomimetic antagonist, bioRxiv (2020) 2020.08.09.242917. [DOI] [PMC free article] [PubMed]
- 23.Maric S., Lind T.K., Raida M.R., Bengtsson E., Fredrikson G.N., Rogers S., Moulin M., Haertlein M., Forsyth V.T., Wenk M.R., Pomorski T.G., Arnebrant T., Lund R., Cárdenas M. Time-resolved small-angle neutron scattering as a probe for the dynamics of lipid exchange between human lipoproteins and naturally derived membranes. Sci. Rep. 2019;9(1):7591. doi: 10.1038/s41598-019-43713-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waldie S., Lind T.K., Browning K., Moulin M., Haertlein M., Forsyth V.T., Luchini A., Strohmeier G.A., Pichler H., Maric S., Cárdenas M. Localization of cholesterol within supported lipid bilayers made of a natural extract of tailor-deuterated phosphatidylcholine. Langmuir. 2018;34(1):472–479. doi: 10.1021/acs.langmuir.7b02716. [DOI] [PubMed] [Google Scholar]
- 25.Cubitt R., Fragneto G. D17: the new reflectometer at the ILL. Appl. Phys. A. 2002;74(1):s329–s331. [Google Scholar]
- 26.Nelson A. Co-refinement of multiple-contrast neutron/X-ray reflectivity data using MOTOFIT. J. Appl. Crystallogr. 2006;39(2):273–276. [Google Scholar]
- 27.Hellstrand E., Grey M., Ainalem M.L., Ankner J., Forsyth V.T., Fragneto G., Haertlein M., Dauvergne M.T., Nilsson H., Brundin P., Linse S., Nylander T., Sparr E. Adsorption of alpha-synuclein to supported lipid bilayers: positioning and role of electrostatics. ACS Chem. Neurosci. 2013;4(10):1339–1351. doi: 10.1021/cn400066t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shenoy S., Shekhar P., Heinrich F., Daou M.C., Gericke A., Ross A.H., Losche M. Membrane association of the PTEN tumor suppressor: molecular details of the protein-membrane complex from SPR binding studies and neutron reflection. PLoS ONE. 2012;7(4) doi: 10.1371/journal.pone.0032591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delhom R., Nelson A., Laux V., Haertlein M., Knecht W., Fragneto G., Wacklin-Knecht H.P. The antifungal mechanism of amphotericin B elucidated in ergosterol and cholesterol-containing membranes using neutron reflectometry. Nanomaterials (Basel) 2020;10(12) doi: 10.3390/nano10122439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.T.K. Lind, L. Darre, C. Domene, Z. Urbanczyk-Lipkowska, M. Cardenas, H.P. Wacklin, Antimicrobial peptide dendrimer interacts with phosphocholine membranes in a fluidity dependent manner: A neutron reflection study combined with molecular dynamics simulations, Biochim. Biophys. Acta 1848(10 Pt A) (2015) 2075–2084. [DOI] [PubMed]
- 31.Åkesson A., Lind T., Ehrlich N., Stamou D., Wacklin H., Cárdenas M. Composition and structure of mixed phospholipid supported bilayers formed by POPC and DPPC. Soft Matter. 2012;8(20):5658–5665. [Google Scholar]
- 32.Kočar E., Režen T., Rozman D. Cholesterol, lipoproteins, and COVID-19: Basic concepts and clinical applications. Biochim. et Biophys. Acta (BBA) – Mol. Cell Biol. Lipids. 2021;1866(2) doi: 10.1016/j.bbalip.2020.158849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abu-Farha M., Thanaraj T.A., Qaddoumi M.G., Hashem A., Abubaker J., Al-Mulla F. The role of lipid metabolism in COVID-19 virus infection and as a drug target. Int. J. Mol. Sci. 2020;21(10):3544. doi: 10.3390/ijms21103544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waldie S., Sebastiani F., Moulin M., Del Giudice R., Paracini N., Roosen-Runge F., Gerelli Y., Prevost S., Voss J.C., Darwish T.A., Yepuri N., Pichler H., Maric S., Forsyth V.T., Haertlein M., Cardenas M. ApoE and ApoE Nascent-Like HDL Particles at Model Cellular Membranes: Effect of Protein Isoform and Membrane Composition. Front Chem. 2021;9 doi: 10.3389/fchem.2021.630152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laursen T., Borch J., Knudsen C., Bavishi K., Torta F., Martens H.J., Silvestro D., Hatzakis N.S., Wenk M.R., Dafforn T.R., Olsen C.E., Motawia M.S., Hamberger B., Moller B.L., Bassard J.E. Characterization of a dynamic metabolon producing the defense compound dhurrin in sorghum. Science. 2016;354(6314):890–893. doi: 10.1126/science.aag2347. [DOI] [PubMed] [Google Scholar]
- 36.C. Bruzzone, M. Bizkarguenaga, R. Gil-Redondo, T. Diercks, E. Arana, A. García de Vicuña, M. Seco, A. Bosch, A. Palazón, I. San Juan, A. Laín, J. Gil-Martínez, G. Bernardo-Seisdedos, D. Fernández-Ramos, F. Lopitz-Otsoa, N. Embade, S. Lu, J.M. Mato, O. Millet, SARS-CoV-2 Infection Dysregulates the Metabolomic and Lipidomic Profiles of Serum, iScience 23(10) (2020) 101645. [DOI] [PMC free article] [PubMed]
- 37.Yetukuri L., Söderlund S., Koivuniemi A., Seppänen-Laakso T., Niemelä P.S., Hyvönen M., Taskinen M.-R., Vattulainen I., Jauhiainen M., Orešič M. Composition and lipid spatial distribution of HDL particles in subjects with low and high HDL-cholesterol[S] J. Lipid Res. 2010;51(8):2341–2351. doi: 10.1194/jlr.M006494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Camont L., Lhomme M., Rached F., Le Goff W., Nègre-Salvayre A., Salvayre R., Calzada C., Lagarde M., Chapman M.J., Kontush A. Small, dense high-density lipoprotein-3 particles are enriched in negatively charged phospholipids: relevance to cellular cholesterol efflux, antioxidative, antithrombotic, anti-inflammatory, and antiapoptotic functionalities. Arterioscler. Thromb. Vasc. Biol. 2013;33(12):2715–2723. doi: 10.1161/ATVBAHA.113.301468. [DOI] [PubMed] [Google Scholar]
- 39.M. Ståhlman, B. Fagerberg, M. Adiels, K. Ekroos, J.M. Chapman, A. Kontush, J. Borén, Dyslipidemia, but not hyperglycemia and insulin resistance, is associated with marked alterations in the HDL lipidome in type 2 diabetic subjects in the DIWA cohort: Impact on small HDL particles, Biochim. et Biophys. Acta (BBA) – Mol. Cell Biol. Lipids 1831(11) (2013) 1609–1617. [DOI] [PubMed]
- 40.Martin S.S., Jones S.R., Toth P.P. High-density lipoprotein subfractions: current views and clinical practice applications. Trends Endocrinol. Metab. 2014;25(7):329–336. doi: 10.1016/j.tem.2014.05.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.