Abstract
The nuclear pore complex (NPC) is the massive protein assembly that regulates transport of macromolecules between the nucleus and the cytoplasm. Recent breakthroughs have provided major insights into the structure of the NPC in different eukaryotes, revealing a previously unsuspected diversity of NPC architectures. In parallel, the NPC has been shown to be a key player in regulating essential nuclear processes such as chromatin organization, gene expression, or DNA repair. However, our knowledge of the NPC structure has not been able to address the molecular mechanisms underlying its regulatory roles. We discuss potential explanations, including the coexistence of alternative NPC architectures with specific functional roles.
Keywords: Nuclear pore complex, nucleoporin, structure, NPC, nuclear transport, gene regulation
The Nuclear Pore Complex Regulates Trafficking In and Out of the Nucleus
The nucleus is without a doubt the quintessential eukaryotic organelle. The nucleus of a cell is defined by the presence of a double lipid membrane, called the nuclear envelope (NE; see Glossary), that surrounds and encloses the chromatin. The flux of genetic information as defined by the central dogma of molecular biology (DNA makes RNA makes protein) is divided into two main steps, transcription and translation, that are physically segregated by the NE. To overcome the NE barrier and ensure a fast, constant, and regulated exchange of macromolecules between the nucleus (where transcription takes place) and the cytoplasm (where translation happens), eukaryotic cells developed a remarkable molecular machine, a massive protein complex called the nuclear pore complex (NPC). The NPC is a large (~100 nm wide and ~40nm high) eight-fold symmetrical assembly composed of more than 550 copies of ~30 different proteins called nucleoporins or Nups [1–3]. The NPC core scaffold coats the NE membrane, shaping and stabilizing it to form a central channel of ~40–60 nm that traverses the NE and acts as the sole communication hub between the inside of the nucleus and the cytoplasm. The central channel of the NPC is filled by a mixture of transport factors and their cargoes together with intrinsically disordered Nup domains rich in phenylalanine-glycine (FG) repeats [4, 5], that collectively make up the so-called central transporter [6]; while the transporter and mechanism of transport are not our main focus here, we address some interesting recent insights in Box 1. We will, rather, summarize our knowledge of the NPC structure across organisms, and discuss the implications of those findings in the context of our understanding of the multiple roles the NPC plays as a platform for regulation of nuclear processes.
Text Box 1: The Nuclear Pore Complex (NPC) Central Transporter.
The central transporter is the mixture of proteins forming the permeability barrier that regulates transport and maintains the NPC’s selectivity. A significant part of the central transporter is formed by FG repeat nucleoporin (Nup) domains (see main text) [122]. Only cargo proteins containing specific signals can be recognized by transport factors that then, through specific interactions with these FG repeats, traverse the NPC with their cargoes [123–125]. Approximately one-third of all Nups contain these FG regions, and their close-packed anchoring along the walls of the central channel form a high-density polymer brush. Although the biophysical nature and molecular mechanism of nuclear transport remains a topic of heated debate, there is now some consensus that FG repeats function as a highly dynamic and fluid “phase” in vivo [124, 126, 127], with FG repeats being intrinsically disordered regions that are either weakly cohesive or non-cohesive and form a highly mobile tethered solution in the central transporter that excludes non-specific macromolecules while allowing the rapid passage of transport factors. Crucially, the bulk of this selective ‘phase’ is not made of FG repeat regions; instead, roughly three-quarters is actually comprised of a heterogeneous mixture of transport factors and their cognate cargos [1, 128].
Cryo-electron microscopy (cryo-EM) studies have discerned few hints about the organization for the central transporter, but this is not surprising as the central transporter is highly heterogeneous, extraordinarily dynamic and is in significant part disordered. However a few studies did resolve the central transporter as an hourglass shaped density that is connected through thin bridges to the inner ring region in the NPC equator [1, 6, 129]. The position of these bridges coincides with the predicted emanating points for the Nic96 complex FG domains (Nsp1, Nup57 and Nup49; Figure 1), strongly suggesting that the bridges are formed by these FG regions emanating as plumes from the inner ring to form the transporter [1]. This observation constitutes the first hint into the organization of the central transporter, but to unravel its full dynamic organization and mechanism will likely require numerous alternative and complementary approaches. For example, groundbreaking advances have been made by the use of atomic force microscopy, a method that imaged the shape and stiffness of the X. laevis central transporter [130] and, when used in its high-speed mode, to observe the time-resolved dynamics of its components [131].
The Nuclear Pore Complex is a Key Regulatory Platform for Nuclear Processes
Although the NPC’s paramount function is to mediate transport between the nucleus and the cytoplasm, research in recent years is also highlighting the importance of the NPC as a regulatory platform. The nuclear side of the NE could be envisioned as a surface in which NPCs would appear as prominent reference points where molecular machineries can be recruited, anchored and coordinated. This dual role for NPCs as both communication and regulation hubs was suggested by Gunter Blobel [7] and expanded upon by a multitude of subsequent studies (reviewed in detail in [8–11]). NPCs have been shown to organize chromatin and regulate gene expression by partially overlapping mechanisms that can be broadly grouped into: i) modulation of chromatin state and architecture [12–14]; ii) posttranslational modification of chromatin-associated proteins [15–18] or Nups themselves [19]; and iii) tethering of specific transcription factors to control gene positioning by recruitment of their cognate DNA binding sites [20–22]. As well as these roles, the NPC is also involved in maintaining genome integrity; upon the appearance of certain DNA lesions, DNA damage response components are actively recruited to peripheral regions of the NPC for repair of the DNA insults [23–25].
Structure of the Nuclear Pore Complex: One Size Does Not Fit All
Despite a remarkable amount of effort, we are still far from a clear mechanistic understanding for most of the NPC’s functional roles that we just summarized. As “structure determines function” in biology, one of the key pieces of information that was missing to solve this puzzle was a complete and comprehensive description of the structure of the NPC at the molecular detail. Once we know the arrangement of each Nup and subcomplex, we should be able to start making sense of the accumulated functional data by placing it all in a proper structural context. But the sheer size of the NPC, its complexity, and the presence of a significant amount of disordered proteins in its composition have made the structural characterization of the NPC a formidable challenge. Only the combination of groundbreaking technical and methodological advances in different areas, including biochemistry, proteomics, integrative methods, crystallography, and cryo-electron microscopy (cryo-EM), have finally allowed the field to tackle this problem and gain a higher resolution view of the structure of the NPC in a variety of organisms (Figure 1).
Figure 1. Structural diversity of nuclear pore complexes (NPCs) across eukaryotes.
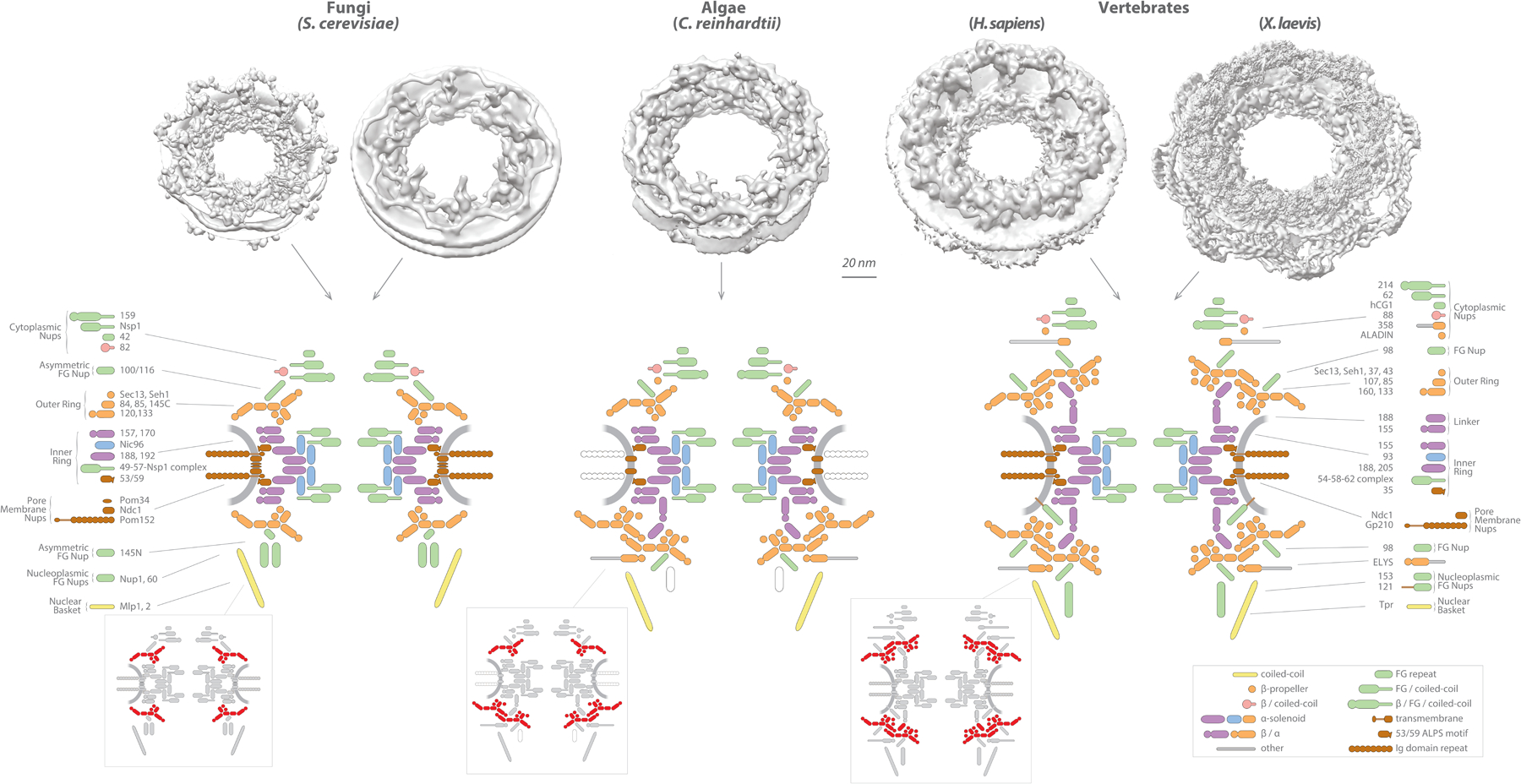
Top row, NPC structures for the indicated organisms solved by integrative methods (left S. cerevisiae PDBDEV_00000010–12 [1]), FIB-cryo-ET (right S. cerevisiae EMD-10198 [31], and C. reinhardtii EMD-4355 [32]), cryo-ET (H. sapiens EMD-3103 [47]), or cryo-ET and single particle analysis (X. laevis, kindly shared by Gaoxingyu Huang and Yigong Xi [57, 80]). Middle row, diagrams reflecting the known composition and arrangement of the different nucleoporins (Nups) and NPC modules in S. cerevisiae (left), C. reindhardtii (middle) and H. sapiens (right). The identity of each Nup is shown to the left in S. cerevisiae or to the right of the H. sapiens diagram. Empty, non-colored shapes indicate a non-identified component. Grey arches: nuclear envelope (NE). Bottom row, NPC diagrams showing the outer ring components in red to highlight their alternative arrangement in the different organisms. Bottom right corner, box depicting the protein folds and domains found in Nups. β / α indicates a β-propeller followed by an α-solenoid.
To build an NPC, most Nups assemble into biochemically stable subcomplexes that in combination form eight identical protomer units called spokes. These “spokes” were first described by EM [26], and although the terminology now is established we may have to consider more accurate descriptions as more is revealed about the NPC’s fine structure. Each of the eight spokes are attached to the NE membrane, and connect radially to each other to form concentric rings: the outer, inner, and membrane rings. Each ring runs parallel to the equatorial plane of the NPC, with a single membrane ring protruding towards the NE lumen at the point of junction between the outer and inner NE membranes, two inner rings that delineate the central channel, and two outer rings sandwiching the inner rings and stabilizing the NE membrane curvature as it enters the NPC. The outer rings also serve as anchoring platforms for asymmetric Nups and cofactors that form the cytoplasmic messenger RNA (mRNA) export machinery and the nuclear basket [1, 27–29]. All these major modules of the NPC are connected by an extensive network of flexible connectors [30], intrinsically disordered Nup domains containing short linear motifs (SliMs, see Glosary and below for detailed explanation), that act as flexible cables tying together the more rigid modules to generate a strong yet flexible assembly [1].
Several models for the structure of the NPC in different organisms are now available [1, 28, 29, 31, 32]. Altogether, they have revealed two key insights. The first insight is that the inner rings of all analyzed NPCs share an overall conserved arrangement. However, the second insight is that different organisms show significant variability in the more peripheral regions of the NPC; a diversity of NPC architectures is thus generated by gain or loss of components, as well as different combinations and stoichiometries of conserved building modules. As a consequence, we can now safely state that there is no such a thing as “the” structure of the NPC, but instead there is a common structural bauplan from which a - yet-uncharacterized - variety of structures arose through evolution to adapt to each organism’s biological needs and constraints [1, 33–37]. Using the model organism Saccharomyces (baker’s yeast) as our initial reference for the NPC, we will summarize what is known currently about the structure of the different NPC modules, starting from the conserved core scaffold and then working our way outwards to the more variable peripheral regions, discussing their alternative architectures and their possible biological relevance.
The inner ring
The inner rings form the structural heart of the NPC, and as such its most conserved module. The inner rings are roughly symmetric with respect to the equator of the NPC, with two superposed symmetric and laterally offset rings, one located in the cytoplasmic side and one on the nuclear side. Equatorially, the inner ring extends from the NE membrane all the way to the central channel, serving both as a framework to shape and stabilize the membrane and as the anchor point for much of the rest of the NPC’s core scaffold, as well as for most of the mass of FG Nups that form the central transporter (Figure 1).
The main Nup components of the inner ring scaffold are relatively large proteins (>90 KDa) that are formed by either alpha-helical solenoid domains or a specific combination of N-terminal beta-propeller and C-terminal alpha-helical solenoid [38]. The alpha-helical solenoid and beta-propeller / alpha-solenoid signature, also found in the outer ring Nups (see below) [39], is only found in components of other membrane coating complexes - such as clathrin/adaptin, COPI, and COPII – and tethering complexes – IFT, SEA, and HOPS/CORVET. Such architectural similarities between molecular machineries devoted to molding membranes led to the proposal of a common evolutionary origin for the NPC and these coating complexes in an ancestral protocoatomer complex, from which they inherited and retained structural and mechanistic signatures [27, 38–40].
The closest inner ring components to the NE membrane are a double pair of paralogous proteins, the beta-propeller/alpha-solenoid Nups, Nup157 and Nup170, and the flexible connector containing Nups, Nup53 and Nup59. All these Nups contain membrane binding motifs (MBMs) [41–43], that appear to anchor them to the membrane around the point of insertion of the transmembrane domains of Pom152, the main component of the membrane ring. Nup157 and Nup170 are oriented perpendicular to the equatorial plane of the NPC, while Nup53 and Nup59 extend from the membrane into the central channel, connecting with other major Nups of the inner ring and also serving as bridges between adjacent spokes [1, 28, 29] that are also connected through relatively small interaction surfaces between opposing Nup170 beta-propellers [1]. Further from the NE membrane, and adjacent to the surfaces of Nup157 and Nup170 facing the central channel, four copies of Nic96 form a diagonally oriented column across the spoke that serves as the keystone of the NPC [1, 28, 29], holding the large alpha-solenoids of Nup192 and Nup188, bracing Nup170 and Nup157, and orienting four coiled-coil trimeric Nsp1-Nup57-Nup49 complexes [44, 45] (Figure 1). These coiled-coils enclose the central transport channel and serve as projection points for the FG repeat domains of Nsp1-Nup57-Nup49 that represent ~4 MDa worth of mass in the central transporter. The flexible connector domains of Nup53/Nup59 and Nup145N/Nup116/Nup100 thread through the whole inner ring, interconnecting its components and tying it to the outer rings and mRNA export machineries on both nuclear and cytoplasmic sides of the NPC [1].
Although the overall architecture of the inner ring was found to be quite conserved in the organisms studied so far, clear variability has been observed in the degree of compaction, overall dimensions, and manner of connection to the outer rings [1, 28, 29, 31, 32]. One of the most obvious differences is the presence of additional copies of a beta-propeller/alpha solenoid Nup157/Nup170 ortholog, termed Nup155, that form upward facing pillars, running roughly perpendicular to the NPC’s equatorial plane, connecting the inner rings to the outer rings in the vertebrate NPC [46, 47]. These connection pillars are missing in the S. cerevisiae NPC [1, 31] and appear to be present only on the nuclear side of the algae Chlamydomonas reinhardtii NPC [32]. Although the functional reason for the presence of these pillars is not clear, one can speculate that their role could be related to the difference in height of the NE between these organisms (vertebrate ~40 nm; algae and yeast ~25 nm) and by the presence of staggered double rings in both vertebrate outer rings and in the algae nuclear outer ring (see below) [32, 46, 47], which might require additional spacers to span this extra width and maintain NPC integrity.
Another difference is that the diameter of the human [48], algae [32], and yeast [31] NPCs appears significantly wider when analyzed by in situ cryo-electron tomography (ET) (central channel ~60 nm) than in isolated vertebrates or yeast NPCs (~40 nm) [1, 47]. Apparently, this difference in diameter does not seem to involve major rearrangements of the inner ring and spokes architecture [31, 32, 48]. Such ranges in diameters have been observed in vertebrate NPCs using other methods [49, 50], collectively leading to a speculation that these different conformations might reflect a continuum of dynamic dilation/constriction that the NPCs could adopt in response to physiological events [51, 52]. Indeed, a groundbreaking study recently showed that transmission of mechanical force from the cytoskeleton to the NE results in stretching of the NPCs, changing their permeability and allowing nuclear import of a mechanosensitive transcriptional activator, YAP [53]. This malleability of the NPC central channel diameter may be facilitated by the fact that the lateral contacts between spokes at the inner ring are limited to a few, small interaction surfaces, as discussed above. Such thin connections should allow some degree of sliding and opening of the spokes to adapt to mechanical stress [54], facilitate passage of large cargoes or to coordinate responses to extracellular stimuli by changes in the NPC permeability barrier [53].
The membrane ring
The membrane ring runs parallel to the equatorial plane of the NPC and is formed by integral membrane Nups with varying numbers of transmembrane regions [27] (Figure 1). Until recently, no indication of its arrangement was available from cryo-EM analyses [46, 47]. However, studies in fungi [1, 55, 56] and Xenopus oocytes [57] have provided a first glance at the surprisingly diverse arrangement of this part of the NPC. In yeast, the main component of the membrane ring, Pom152, forms homo-dimeric interactions through their elongated pearl-on-string luminal domains that generate arches connecting adjacent spokes within the NE lumen [1, 55]. The wider part of these arches coincides with the boundaries between spokes, delineating what may be the transport pathway for integral membrane proteins [1]. The remaining components of the membrane ring (Pom34 and Ndc1) are structurally not so well defined, although they seem to help establish multiple connections between the NE membrane and the major Nups of the inner ring [1, 58].
A recent analysis performed in X. laevis oocyte NEs identified a structural feature that most likely corresponds to the vertebrate membrane ring [57] (Figure 1). Unexpectedly, it is not a diaphanous feature, but a bulky density formed by multiple copies of an elongated protomer (reminiscent of the Pom152 luminal domain shape [55, 56]) that arrange to form a two-winged symmetric subunit in each spoke. Wing-to-wing connections are established between spokes to form the so called “bumper domains”, suggested to help cushion neighboring NPCs when they get close to each other in the highly crowded oocyte NE (~60 NPCs per square micron) [57]. The most likely scenario is that the elongated protomers are mainly formed by the luminal domain of vertebrate Nup210, although the presence of other proteins could not be discarded. Nup210/Gp210 is the largest component of the vertebrate membrane ring, appears structurally homologous to yeast Pom152 and, although it is not present in some metazoan cell lines [59, 60], it has been shown to be a key regulator of gene expression and cell differentiation processes [61]. Thus, confirming that Nup210 is the protomer forming the membrane ring in X. laevis oocytes would be vital to start placing these functional data into a structural context and to arrive at a mechanistic understanding of how changes in NPC composition can define cellular fate.
The NPC outer rings and associated peripheral machineries
The conserved building block of the NPC outer rings is the so-called Y-complex, owing its name to its characteristic elongated Y-shape [62, 63]. Based on the organisms characterized so far, this complex has a conserved heterohexameric core that delineates the Y, and a variable number of additional components that add up to nine members in the vertebrate complex [46, 64–67]. Most of the conserved components are either beta-propeller/alpha solenoid proteins (Nup133, Nup120 [68–71]) or COPII-like alpha-solenoid Nups (Nup145C, Nup85, and Nup84 [72–74]), adding to the overwhelming evidence for a common evolutionary origin between the NPC and membrane coating complexes in an ancestral protocoatomer [39] (reviewed in detail in [34, 75]). The Y-complex contacts the NE membrane through MBMs located at the tips of the complex, within the beta-propellers of Nup120 and Nup133 [1, 42, 46, 68, 76, 77], and it arranges in a head-to-tail fashion to form the outer rings on both sides of the NPC [27, 64, 71, 77].
However, the number of Y-complex copies on each outer ring have been shown to vary from organism to organism (Figure 1). In the human NPC, two staggered rings of eight Y-complexes each form both cytoplasmic and nuclear outer rings, adding up to 32 copies of the complex per NPC [46, 78], while in yeast, only one octameric ring per side (16 Y-complexes per NPC) has been observed [1, 27]. Remarkably, an algae cryo-EM NPC map indicates an in-between composition, with a yeast-like cytoplasmic single ring arrangement and a vertebrate-like nuclear double staggered ring (24 Y-complexes per NPC) [32]. An even more complex arrangement has been recently suggested for the organization of the outer rings of the fission yeast Schizosaccharomyces pombe [79]; using immunoelectron and fluorescence microscopy methods, Asakawa et al. observed that components of the Y-complex are asymmetrically distributed between the nuclear and cytoplasmic outer rings, resulting in an arrangement of fragmented Y-complexes that would strongly deviate from the canonical view of how the outer rings are organized [79].
Whenever a double Y-complex ring has been identified, eight beta-propeller / alpha-solenoid Nup155 pillars have been shown to connect it to the inner ring [32, 46, 47] and additional components have been proposed as required to stabilize the arrangement. First, the vertebrate-specific Nup358/RanBP2 is present in the cytoplasmic outer ring, where it forms a “clamp” around the stem of the double Y-complex ring [47, 80]. This Nup358 clamp helps stabilize the arrangement as gene silencing of Nup358 leads to disassembly of the outer Y-complex ring [47]. Second, question-mark-shaped densities, tentatively assigned to Nup188 based on cross-linking and mass spectrometry data [28, 29, 46], are present in each spoke connecting the double Y-complex rings on both sides of the NPC. However, a higher resolution cryo-EM map of the X. laevis cytoplasmic outer ring indicated instead that these are formed by two copies of Nup205 [80]. It is thus an open question whether this is a conserved or a differential feature between nuclear and cytoplasmic spokes, whether the number of outer rings is diagnostic for the path of NPC evolution between organisms, as proposed [33], or if it is yet another feature that can be regulated to meet differential demands on NPC functionality.
The modules that control RNA remodeling and export in the NPC, the cytoplasmic export platform and the nuclear basket, are anchored to the outer rings, though also make connections with the inner rings [1]. Mechanistically, the export of RNA is largely distinct from that of other cargoes. First, RNAs are packaged into export competent ribonucleoproteins that dock to the nuclear basket. These are then chaperoned across the NPC by specific transport factors. Finally, they are remodeled and released by the export platform on the cytoplasmic face of the NPC [81–83]. Both the basket and export platform seem to have a major attachment point in the short arm and hub of the most internal Y-complex [1, 84], where they likely establish contacts with the Y-complex backbone and through short linear motifs located in flexible connectors, as shown in the case of a thermophilic fungus [85]. The core scaffold of the cytoplasmic RNA export platform is the Nup82 complex, formed by a complex arrangement of heterotrimeric coiled-coil bundle subunits [84, 86]. Variability in the Nup82 complex components between yeast and vertebrates suggest that the detailed architecture of the complex may vary between species and may be a somewhat malleable structure, but cryo-EM maps agree in positioning the Nup82 complex as a hook-like structure protruding into the central channel of the NPC [1, 31, 32, 46, 80, 84], where it recruits the RNA helicase Dbp5 and associated cofactors to disassemble the messenger ribonucleoproteins in the last step of nuclear export [82, 87].
Although it is known that its major component is the large coiled-coil protein Tpr in vertebrates [88, 89] and its homologs Mlp1/Mlp2 in Saccharomyces [90, 91], the arrangement and orientation of the nuclear basket is still poorly defined, probably due to the intrinsic flexibility and potential heterogeneity of Tpr/Mlp and its other components, including the observation that not all NPCs seem to possess a nuclear basket [90, 92, 93]. In situ focused ion beam and cryo-ET of Chlamydomonas cells have revealed filament-like structures attached to the nuclear side of the NPC [94] that would be reminiscent of our traditional picture of the nuclear basket, but their nature has not been verified and it is not clear if such an arrangement would be present in other type of cells. Instead, we still rely on much older work to give us some idea concerning the organization of these structures, such as high resolution scanning EM images of both yeast and vertebrates which showed the nuclear basket as being made of eight filaments attached to the nucleoplasmic ring to conjoin on a distal basket ring [95, 96].
Flexible connectors
It has been estimated that approximately 60% of the NPC mass comes from the relatively large alpha-solenoid and beta-propeller containing Nups and coiled-coil bundles that from the core scaffold of the outer and inner rings, and they are the components that generate most of the observed density in the published NPC cryo-EM structures [1, 3]. However, there are much less noticeable NPC elements that nevertheless perform the crucial role of tying together all these large NPC modules. Firstly identified in Ed Hurt’s laboratory [30], they have been called SLiMs or flexible connectors, and they are extended, intrinsically disordered domains within certain Nups that contain one or more short interaction sequences. These sequences establish specific, yet not extensive interactions with folded domains of large Nups and subcomplexes [97]. Taken together, evidence indicates that the flexible connector containing Nups have been shown to form an extensive network of “cables” that extend vertically, from the cytoplasmic face of the NPC through the inner ring and all the way to the nuclear side, and horizontally, extending from the NE membrane, traversing the inner ring and connecting to the central transporter, towards which some of them project their associated FG regions, generating a flexible, yet stress-resilient structure [1, 30, 85, 98].
It is actually quite common to find FG domains associated to flexible connectors within Nups, and it has been suggested that both domains may have a common evolutionary origin [99], and that they could act synergistically to modulate their interaction with folded Nups and karyopherins during NPC biogenesis and to maintain the mature NPC stability [100, 101]. Several of the SLiMs have been crystallized in interactions with their Nup partners [29], and some have been shown to only interact with assembled NPC subcomplexes, mainly formed by coiled-coil bundles [30, 44, 85], strengthening the idea that they might be important for coordination during NPC biogenesis. In summary, the flexible connectors perform an analogous role to that of the suspender cables in a bridge, tying together the major modules of the structure, to provide flexibility and strength to the structure.
Can we understand the NPC regulatory roles in the light of its structure?
While in this short review we can only provide the briefest synopsis, we hope to have shown the reader that in the last few years the field has come a long way in its efforts to dissect the architecture of the NPC. Nevertheless, the variabilities that have been observed clearly indicate that we may still be far from grasping the real structural and functional diversity of the NPC across eukaryotes, and even within individual cells. The picture that is emerging is that of a relatively conserved core to which peripheral modules can be plugged in and out, potentially combining into a multitude of possibilities, similar to the popular Lego toy suite. Such a “plug and play” character for its peripheral regions might explain some of the regulatory mechanisms we discussed above for the NPC, as changes in those modules would imply changes in their associated interactomes and functional purposes. However, the situation appears more nuanced, as recent studies are challenging the idea that the NPC regulatory roles could be solely related to its more variable and less conserved outer peripheral regions, and rather show that components buried in the core structure of the NPC, allegedly quite inaccessible to interaction, play key roles in gene expression regulation.
The most remarkable cases are the metazoan inner ring components Nup155 and Nup93, and their fungal homologs Nup170 and Nic96, respectively (Figure 1). Although both proteins are buried within the structure of the inner ring, they have been shown to have conserved roles in recruitment of heterochromatin to the NPC. In Saccharomyces, Nup170 was shown to recruit the silencing factor Sir4 and promote transcriptional repression of subtelomeric regions at the NPC [102]; its metazoan homolog, Nup155, was also shown to interact with the histone deacetylase HDAC4 [103] and to be required for chromatin association to the NPC in an RNA interference screen looking for factors required for chromatin detachment in meiotic Drosophila oocytes [12]. Similarly, a pair of recent studies demonstrated that Drosophila Nup93 [104] and its fission yeast homolog Npp106 [105] are physically associated with heterochromatin and are involved in gene silencing. Human Nup93 has also been shown to participate in binding superenhancers [106] and in tethering and repressing the HOXA gene cluster [16], suggesting a highly conserved role for Nup93 in recruitment and silencing of chromatin at the NPC. Altogether, these studies strongly indicate that two Nups buried at the core of the NPC spoke, where the available structures show that they act as true structural keystones and anchor point for many other Nups, are nevertheless able to establish physical interactions with bulky silencing protein complexes and heterochromatin domains within the NPC.
How could these two apparently conflicting possibilities be reconciled into a coherent structure/function model for the NPC? Paradoxically, there is plenty of evidence suggesting that other Nups and NPC modules may restrict heterochromatin access to the inner ring Nups or even serve to recruit actively transcribed genes. A negative regulatory loop in which Nup62 can repress the observed Nup155 chromatin tethering activity has been described in Drosophila [12], and the presence of the nuclear basket seems to be required for exclusion of heterochromatin and maintenance of open chromatin architecture at NPCs, as suggested by studies in fission yeast [105] and virus-infected human cells [107–109]. Recent reports describe the association of euchromatin and active genes with outer ring complex components in the NPC [21, 104], supporting previous studies that showed recruitment of actively transcribed genes [61, 110] and a role for the NPC as a key platform for transcriptional memory and tethering of inducible genes [111, 112]. We are thus facing apparently contradictory evidence indicating that, on the one hand, within a relatively compact, inaccessible NPC structure, several of its inner components are nevertheless directly involved in recruiting heterochromatin and promoting gene silencing, while on the other hand, peripheral components recruit euchromatin, to prevent access of heterochromatin to the NPC, and thus support expression of active genes.
Based on our knowledge of NPC biology, we speculate on several non-mutually exclusive scenarios that could help explain this apparent conundrum: first, the remarkable flexibility of the NPC core scaffold (discussed above) might facilitate openings in the structure that would expose certain components to interaction with co-factors and other assemblies; second, Nups could be having “out-of-the-NPC” functions, as several NPC components have been shown to perform intranuclear regulatory roles independently of their association with the NPC [17, 113–117], although the vast majority of inner ring Nups in a cell are devoted to build NPCs [2], and it is as part of the mature NPC that they perform most of their regulatory functions [17, 104, 106]; finally, a third explanation that would fit with the available evidence would be the coexistence of different types of NPCs or Nup assemblies with alternative architectures and differential functionality within the same cell types.
This type of scenario arose for example from the work of Lapetina et al. [118], that during dissection of the Nup170 and silencing factors interaction network in Saccharomyces identified what they called the Snup, an assembly formed by core Nups (mainly members of the inner and outer ring) that is physically distinct from a mature NPC and does not contain FG, flexible connector or nuclear basket Nups. The authors suggest that the Snup, and not the mature NPC, would be responsible for interacting with silencing factors like Sir4 and drive recruitment of subtelomeric chromatin [118]. Although more detailed work is needed to investigate the exact nature of the Snup, this study together with ones previously mentioned, raise the interesting possibility that alternative NPC architectures with distinct functional roles might coexist in the same cell. Similarly to the architectural variability observed between divergent eukaryotes (Figure 1), an unidentified diversity of NPC architectures might be present within a single cell or be assembled in response to certain physiological cues. Examples of this are already present in the literature, where NPCs lacking a nuclear basket were detected around the nucleolus in Saccharomyces cells [92, 93], distinct NPC architectures were found in the Tetrahymena somatic macronucleus and germline micronucleus [36], and changes in Nup stoichiometry were described across human cell types [3, 119, 120].
Concluding Remarks
Far from the traditional, somewhat monolithic picture of a ring that we have seen for decades in textbooks, technical and methodological advances in the structural characterization of the NPC are revealing a beautifully intricate architecture with a significant degree of both compositional and conformational flexibility [1, 28, 29, 31, 32, 57, 80]. To explain the malleable character of the functional architecture of the NPC, we have previously used the analogy of a suspension bridge [1]: both the NPC and a suspension bridge are structures that create a stable passageway for a large and continuous flow of traffic that passes through them at all times; both are formed by firm anchors to a substrate (NE or bedrock respectively), relatively rigid modules connected by hinges and a network of cables tying all the modules together. This type of architecture produces a flexible yet resilient assembly, that could accommodate many types of stresses without suffering damage to its integrity. We could now even expand this analogy, as, like bridges, the NPC seems to have been shaped into diverse incarnations and variations that accommodate the differing needs of a variety of organisms.
How the observed alternative NPC architectures relate to the different biology of these organisms is still an open question, as is the question of how these alternative NPC architectures were shaped during eukaryotic evolution [33, 34] (see Outstanding Questions). Moreover, recent advances in the functional characterization of the NPC are helping to underscore its key role as a nuclear regulatory platform, independently from its primary role as regulator of transport between the nucleus and the cytoplasm. The NPC is directly involved in regulating a large variety of pathways and physiological processes, such as chromatin architecture or transcriptional control and memory [9, 10, 121]. It is thus hard to imagine how a protein assembly could support such a variety of functional roles without undergoing significant changes in both its composition and/or structure. Similarly to the diverse NPC architectures identified across organisms, we expect that a still unidentified variety of NPCs might exist even within tissues and cell types. If confirmed, this exciting possibility would open vast new avenues of research that would require the development of novel approaches and methodologies to explore the diverse and dynamic nature of NPCs.
Outstanding Questions Box.
How is the nuclear pore complex (NPC) 8-fold symmetry defined? What are the structural determinants that define the number of modules that form each NPC?
What are the molecular determinants that define the oligomerization state of an NPC outer rings? What is the functional reason behind a single or double outer ring arrangement?
How do nucleoporins (Nups), that do not have any obvious DNA binding motifs, interact with chromatin for its regulation? If it is through adaptor proteins, which are they and how is their interaction with the NPC modulated?
Do all NPCs transport the same type of cargo? Are there specialized NPCs devoted to organizing scaffolds for gene regulation?
Do all NPCs have a membrane ring? How does the composition of the vertebrate NPC membrane ring define its ability to regulate gene expression and cell fate determination?
Does the Snup complex represent a different type of mature NPC, an NPC biogenesis precursor or a completely different assembly?
Highlights.
The nuclear pore complex (NPC) is a large protein assembly that regulates macromolecular transport between the nucleus and the cytoplasm and acts as a regulatory platform for many other essential nuclear processes
The NPC structural characterization has been challenging, but recent technical and methodological advances are pushing our understanding of the NPC architecture
Structural analyses of the NPC in different organisms revealed that, although there is a common bauplan, a significant degree of variability is observed in the peripheral modules that build these NPCs
The multiple regulatory roles shown for the NPC suggest an even greater degree of NPC architectural diversity that may have not yet been unveiled
Acknowledgments
We would like to apologize to the colleagues whose work has not been included in this review solely due to space limitations. We thank Gaoxingyu Huang and Yigong Xi for kindly sharing the X. laevis cryo-EM image in Figure 1 and Ilona Nudelman for her help with Figure 1. The authors are supported by NSF grant NSF-1818129 (J.F-M.) and NIH grants R01 GM112108, R01 CA228351 and P41 GM109824 (M.P.R.).
Glossary
- Atomic force microscopy
technique that enables the imaging of a sample’s surface and the analysis of its mechanical properties by the use of a sharp tip about 10–20 nm in diameter attached to a cantilever
- Cryo-electron microscopy (Cryo-EM)
imaging technique that analyzes the shape and dimensions of a frozen-hydrated sample using a transmission electron microscope under cryogenic conditions
- Cryo-electron tomography (Cryo-ET)
cryo-EM method where the samples are imaged as they are tilted at defined angles relative to the electron beam, to obtain a series of 2D images that are then computationally combined into a three dimensional reconstruction of the sample
- Focused ion beam (FIB)
microscopy technique based on the use of a focused beam of ions to ablate layers of material from a biological sample, carving it into a thin layer with nanometer precision; it is usually applied to frozen-hydrated specimens and combined with cryo-electron microscopy and tomographic methods
- Integrative method
strategy for determining the structure of biological systems based on data produced by multiple experimental and theoretical methods that are combined into a single, coherent structural calculation; the combination of several complementary methods to solve a structure is also called a hybrid approach
- Membrane Binding Motif (MBM)
short amino acid sequences that directly interact with the nuclear envelope (NE) membrane; they are usually amphipathic alpha-helices that insert their hydrophobic region into one face of the membrane lipid bilayer
- Nuclear Envelope (NE)
double lipid membrane, contiguous to the endoplasmic reticulum, that surrounds, delimits, and shapes the nucleus in eukaryotic cells
- Nuclear Pore Complex (NPC)
large protein complex that creates a channel through the nuclear envelope and mediates transport of macromolecules between the nucleus and the cytoplasm
- Nucleoporin (Nup)
protein constituent of the NPC, defined as a protein that spends most of its life in the NPC forming a stable part of its structure
- Phenylalanine-glycine (FG) repeats
intrinsically disordered Nup domains rich in phenylalanine-glycine (FG) short linear motifs separated by very hydrophilic spacer residues
- Polymer brush
mass of polymer chains tethered to a solid substrate with a density higher than the polymer radius of gyration
- Short Linear Motifs (SLiMs)
intrinsically disordered domains within certain Nups that contain one or more short interaction sequences; these interaction sequences establish specific, yet not extensive interactions with folded domains of large Nups and NPC subcomplexes
- Y-complex
NPC module that forms the outer rings; it is formed by 6–9 Nups and adopts a characteristic Y-shape
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kim SJ et al. (2018) Integrative structure and functional anatomy of a nuclear pore complex. Nature 555 (7697), 475–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rout MP et al. , The yeast nuclear pore complex: composition, architecture, and transport mechanism, The Journal of Cell Biology, 2000, pp. 635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ori A et al. (2013) Cell type-specific nuclear pores: a case in point for context-dependent stoichiometry of molecular machines. Mol Syst Biol 9, 648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wente SR and Rout MP, The Nuclear Pore Complex and Nuclear Transport, Cold Spring Harbor Perspectives in Biology, 2010, pp. a000562–a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sakiyama Y et al. (2017) Structural dynamics of the nuclear pore complex. Semin Cell Dev Biol 68, 27–33. [DOI] [PubMed] [Google Scholar]
- 6.Akey CW, Interactions and structure of the nuclear pore complex revealed by cryo-electron microscopy, The Journal of Cell Biology, 1989, pp. 955–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blobel G (1985) Gene gating: a hypothesis. Proc Natl Acad Sci U S A 82 (24), 8527–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D’Angelo MA (2018) Nuclear pore complexes as hubs for gene regulation. Nucleus 9 (1), 142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuhn TM and Capelson M (2019) Nuclear Pore Proteins in Regulation of Chromatin State. Cells 8 (11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ptak C et al. (2014) The multifunctional nuclear pore complex: a platform for controlling gene expression. Curr Opin Cell Biol 28C, 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun J et al. (2019) The Nuclear Pore Complex in Cell Type-Specific Chromatin Structure and Gene Regulation. Trends Genet 35 (8), 579–588. [DOI] [PubMed] [Google Scholar]
- 12.Breuer M and Ohkura H (2015) A negative loop within the nuclear pore complex controls global chromatin organization. Genes Dev 29 (17), 1789–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuhn TM et al. (2019) Chromatin targeting of nuclear pore proteins induces chromatin decondensation. J Cell Biol 218 (9), 2945–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pascual-Garcia P et al. (2017) Metazoan Nuclear Pores Provide a Scaffold for Poised Genes and Mediate Induced Enhancer-Promoter Contacts. Mol Cell 66 (1), 63–76 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fiserova J et al. (2017) Chromatin organization at the nuclear periphery as revealed by image analysis of structured illumination microscopy data. J Cell Sci 130 (12), 2066–2077. [DOI] [PubMed] [Google Scholar]
- 16.Labade AS et al. (2016) HOXA repression is mediated by nucleoporin Nup93 assisted by its interactors Nup188 and Nup205. Epigenetics Chromatin 9, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Light WH et al. (2013) A conserved role for human Nup98 in altering chromatin structure and promoting epigenetic transcriptional memory. PLoS Biol 11 (3), e1001524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saik NO et al. (2020) Recruitment of an Activated Gene to the Yeast Nuclear Pore Complex Requires Sumoylation. Front Genet 11, 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar A et al. (2018) Daughter-cell-specific modulation of nuclear pore complexes controls cell cycle entry during asymmetric division. Nat Cell Biol 20 (4), 432–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brickner DG et al. (2019) The Role of Transcription Factors and Nuclear Pore Proteins in Controlling the Spatial Organization of the Yeast Genome. Dev Cell 49 (6), 936–947 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Z et al. (2019) Nucleoporin Seh1 Interacts with Olig2/Brd7 to Promote Oligodendrocyte Differentiation and Myelination. Neuron 102 (3), 587–601 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raices M and D’Angelo MA (2017) Nuclear pore complexes and regulation of gene expression. Curr Opin Cell Biol 46, 26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aguilera P et al. (2020) The nuclear pore complex prevents sister chromatid recombination during replicative senescence. Nat Commun 11 (1), 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Folz H et al. (2019) SUMOylation of the nuclear pore complex basket is involved in sensing cellular stresses. J Cell Sci 132 (7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horigome C et al. (2019) Ribosomal RNA gene repeats associate with the nuclear pore complex for maintenance after DNA damage. PLoS Genet 15 (4), e1008103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Unwin PN and Milligan RA (1982) A large particle associated with the perimeter of the nuclear pore complex. J Cell Biol 93 (1), 63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alber F et al. , The molecular architecture of the nuclear pore complex, Nature, 2007, pp. 695–701. [DOI] [PubMed] [Google Scholar]
- 28.Kosinski J et al. , Molecular architecture of the inner ring scaffold of the human nuclear pore complex., Science, American Association for the Advancement of Science, 2016, pp. 363–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin DH et al. , Architecture of the symmetric core of the nuclear pore, Science, 2016, pp. aaf1015–aaf1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fischer J et al. , Linker Nups connect the nuclear pore complex inner ring with the outer ring and transport channel, Nat Struct Mol Biol, Nature Publishing Group, 2015, pp. 1–10. [DOI] [PubMed] [Google Scholar]
- 31.Allegretti M et al. (2020) In-cell architecture of the nuclear pore and snapshots of its turnover. Nature. [DOI] [PubMed] [Google Scholar]
- 32.Mosalaganti S et al. (2018) In situ architecture of the algal nuclear pore complex. Nat Commun 9 (1), 2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beck M et al. (2018) From the resolution revolution to evolution: structural insights into the evolutionary relationships between vesicle coats and the nuclear pore. Curr Opin Struct Biol 52, 32–40. [DOI] [PubMed] [Google Scholar]
- 34.Field MC and Rout MP (2019) Pore timing: the evolutionary origins of the nucleus and nuclear pore complex. F1000Res 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holzer G and Antonin W (2018) Nuclear Pore Complexes: Global Conservation and Local Variation. Curr Biol 28 (11), R674–R677. [DOI] [PubMed] [Google Scholar]
- 36.Iwamoto M et al. (2017) Compositionally distinct nuclear pore complexes of functionally distinct dimorphic nuclei in the ciliate Tetrahymena. J Cell Sci 130 (10), 1822–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shav-Tal Y and Tripathi T (2018) Yeast and Human Nuclear Pore Complexes: Not So Similar After All. Trends Cell Biol 28 (8), 589–591. [DOI] [PubMed] [Google Scholar]
- 38.Devos D et al. , Simple fold composition and modular architecture of the nuclear pore complex, Proc Natl Acad Sci USA, 2006, pp. 2172–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Devos D et al. , Components of coated vesicles and nuclear pore complexes share a common molecular architecture, Plos Biol, 2004, p. e380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rout MP and Field MC (2017) The Evolution of Organellar Coat Complexes and Organization of the Eukaryotic Cell. Annu Rev Biochem 86, 637–657. [DOI] [PubMed] [Google Scholar]
- 41.De Magistris P et al. (2018) A self-inhibitory interaction within Nup155 and membrane binding are required for nuclear pore complex formation. J Cell Sci 131 (1). [DOI] [PubMed] [Google Scholar]
- 42.Drin G et al. (2007) A general amphipathic alpha-helical motif for sensing membrane curvature. Nat Struct Mol Biol 14 (2), 138–46. [DOI] [PubMed] [Google Scholar]
- 43.Marelli M et al. , A link between the synthesis of nucleoporins and the biogenesis of the nuclear envelope, The Journal of Cell Biology, 2001, pp. 709–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stuwe T et al. , Architecture of the fungal nuclear pore inner ring complex., Science, American Association for the Advancement of Science, 2015, pp. 56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chug H et al. (2015) Crystal structure of the metazoan Nup62*Nup58*Nup54 nucleoporin complex. Science 350 (6256), 106–10. [DOI] [PubMed] [Google Scholar]
- 46.Bui KH et al. (2013) Integrated structural analysis of the human nuclear pore complex scaffold. Cell 155 (6), 1233–43. [DOI] [PubMed] [Google Scholar]
- 47.von Appen A et al. (2015) In situ structural analysis of the human nuclear pore complex. Nature 526 (7571), 140–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mahamid J et al. (2016) Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science 351 (6276), 969–72. [DOI] [PubMed] [Google Scholar]
- 49.Liashkovich I et al. (2011) Exceptional structural and mechanical flexibility of the nuclear pore complex. J Cell Physiol 226 (3), 675–82. [DOI] [PubMed] [Google Scholar]
- 50.Feldherr CM and Akin D (1990) The permeability of the nuclear envelope in dividing and nondividing cell cultures. J Cell Biol 111 (1), 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Denais CM et al. (2016) Nuclear envelope rupture and repair during cancer cell migration. Science 352 (6283), 353–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raab M et al. (2016) ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science 352 (6283), 359–62. [DOI] [PubMed] [Google Scholar]
- 53.Elosegui-Artola A et al. (2017) Force Triggers YAP Nuclear Entry by Regulating Transport across Nuclear Pores. Cell 171 (6), 1397–1410 e14. [DOI] [PubMed] [Google Scholar]
- 54.Thaller DJ and Patrick Lusk C (2018) Fantastic nuclear envelope herniations and where to find them. Biochem Soc Trans 46 (4), 877–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Upla P et al. (2017) Molecular Architecture of the Major Membrane Ring Component of the Nuclear Pore Complex. Structure 25 (3), 434–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hao Q et al. (2018) Electron microscopy of Chaetomium pom152 shows the assembly of ten-bead string. Cell Discov 4, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y et al. (2020) Molecular architecture of the luminal ring of the Xenopus laevis nuclear pore complex. Cell Res 30 (6), 532–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Onischenko E et al. , Role of the Ndc1 interaction network in yeast nuclear pore complex assembly and maintenance, The Journal of Cell Biology, 2009, pp. 475–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Olsson M et al. (2004) Limited expression of nuclear pore membrane glycoprotein 210 in cell lines and tissues suggests cell-type specific nuclear pores in metazoans. Exp Cell Res 292 (2), 359–70. [DOI] [PubMed] [Google Scholar]
- 60.D’Angelo MA et al. , A Change in Nuclear Pore Complex Composition Regulates Cell Differentiation, Developmental Cell, 2012, pp. 446–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raices M et al. (2017) Nuclear Pores Regulate Muscle Development and Maintenance by Assembling a Localized Mef2C Complex. Dev Cell 41 (5), 540–554 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lutzmann M et al. , Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins, EMBO J, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Siniossoglou S et al. , Structure and assembly of the Nup84p complex, The Journal of Cell Biology, 2000, pp. 41–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fernandez-Martinez J et al. , Structure-function mapping of a heptameric module in the nuclear pore complex., The Journal of Cell Biology, Rockefeller Univ Press, 2012, pp. 419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kelley K et al. (2015) Atomic structure of the Y complex of the nuclear pore. Nat Struct Mol Biol 22 (5), 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kampmann M and Blobel G (2009) Three-dimensional structure and flexibility of a membrane-coating module of the nuclear pore complex. Nat Struct Mol Biol 16 (7), 782–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Loïodice I et al. , The entire Nup107–160 complex, including three new members, is targeted as one entity to kinetochores in mitosis, Molecular biology of …, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim SJ et al. (2014) Integrative structure-function mapping of the nucleoporin Nup133 suggests a conserved mechanism for membrane anchoring of the nuclear pore complex. Mol Cell Proteomics 13 (11), 2911–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Whittle JR and Schwartz TU (2009) Architectural nucleoporins Nup157/170 and Nup133 are structurally related and descend from a second ancestral element. J Biol Chem 284 (41), 28442–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Leksa NC et al. (2009) The structure of the scaffold nucleoporin Nup120 reveals a new and unexpected domain architecture. Structure 17 (8), 1082–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seo H-S et al. , Structural and functional analysis of Nup120 suggests ring formation of the Nup84 complex., Proceedings of the National Academy of Sciences, National Acad Sciences, 2009, pp. 14281–14286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brohawn SG et al. (2008) Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science 322 (5906), 1369–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brohawn SG and Schwartz TU (2009) Molecular architecture of the Nup84-Nup145CSec13 edge element in the nuclear pore complex lattice. Nat Struct Mol Biol 16 (11), 1173–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hsia KC et al. (2007) Architecture of a coat for the nuclear pore membrane. Cell 131 (7), 1313–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leksa NC and Schwartz TU (2010) Membrane-coating lattice scaffolds in the nuclear pore and vesicle coats: commonalities, differences, challenges. Nucleus 1 (4), 314–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shi Y et al. , Structural characterization by cross-linking reveals the detailed architecture of a coatomer-related heptameric module from the nuclear pore complex, Mol Cell Proteomics, American Society for Biochemistry and Molecular Biology, 2014, p. mcp.M114.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nordeen SA et al. (2020) Yeast Nup84-Nup133 complex structure details flexibility and reveals conservation of the membrane anchoring ALPS motif. Nat Commun 11 (1), 6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ori A et al. (2014) The use of targeted proteomics to determine the stoichiometry of large macromolecular assemblies. Methods Cell Biol 122, 117–46. [DOI] [PubMed] [Google Scholar]
- 79.Asakawa H et al. (2019) Asymmetrical localization of Nup107–160 subcomplex components within the nuclear pore complex in fission yeast. PLoS Genet 15 (6), e1008061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huang G et al. (2020) Structure of the cytoplasmic ring of the Xenopus laevis nuclear pore complex by cryo-electron microscopy single particle analysis. Cell Res 30 (6), 520–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Heinrich S et al. (2017) Temporal and spatial regulation of mRNA export: Single particle RNA-imaging provides new tools and insights. Bioessays 39 (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Folkmann AW et al. (2011) Dbp5, Gle1-IP6 and Nup159: a working model for mRNP export. Nucleus 2 (6), 540–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Oeffinger M and Zenklusen D (2012) To the pore and through the pore: a story of mRNA export kinetics. Biochim Biophys Acta 1819 (6), 494–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fernandez-Martinez J et al. , Structure and Function of the Nuclear Pore Complex Cytoplasmic mRNA Export Platform, Cell, Elsevier, 2016, pp. 1215–1228.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Teimer R et al. (2017) A short linear motif in scaffold Nup145C connects Y-complex with pre-assembled outer ring Nup82 complex. Nat Commun 8 (1), 1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gaik M et al. , Structural basis for assembly and function of the Nup82 complex in the nuclear pore scaffold, The Journal of Cell Biology, 2015, pp. 283–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xie Y and Ren Y (2019) Mechanisms of nuclear mRNA export: A structural perspective. Traffic 20 (11), 829–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Frosst P et al. (2002) Tpr is localized within the nuclear basket of the pore complex and has a role in nuclear protein export. J Cell Biol 156 (4), 617–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Krull S et al. (2004) Nucleoporins as components of the nuclear pore complex core structure and Tpr as the architectural element of the nuclear basket. Mol Biol Cell 15 (9), 4261–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Strambio-de-Castillia C et al. , Proteins connecting the nuclear pore complex with the nuclear interior, The Journal of Cell Biology, 1999, pp. 839–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kosova B et al. (2000) Mlp2p, a component of nuclear pore attached intranuclear filaments, associates with nic96p. J Biol Chem 275 (1), 343–50. [DOI] [PubMed] [Google Scholar]
- 92.Galy V et al. (2004) Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 116 (1), 63–73. [DOI] [PubMed] [Google Scholar]
- 93.Niepel M et al. (2005) The nuclear pore complex-associated protein, Mlp2p, binds to the yeast spindle pole body and promotes its efficient assembly. J Cell Biol 170 (2), 225–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Albert S et al. (2017) Proteasomes tether to two distinct sites at the nuclear pore complex. Proc Natl Acad Sci U S A 114 (52), 13726–13731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ris H and Malecki M (1993) High-resolution field emission scanning electron microscope imaging of internal cell structures after Epon extraction from sections: a new approach to correlative ultrastructural and immunocytochemical studies. J Struct Biol 111 (2), 148–57. [DOI] [PubMed] [Google Scholar]
- 96.Goldberg MW and Allen TD (1996) The nuclear pore complex and lamina: three-dimensional structures and interactions determined by field emission in-lens scanning electron microscopy. J Mol Biol 257 (4), 848–65. [DOI] [PubMed] [Google Scholar]
- 97.Beck M and Hurt E (2017) The nuclear pore complex: understanding its function through structural insight. Nat Rev Mol Cell Biol 18 (2), 73–89. [DOI] [PubMed] [Google Scholar]
- 98.Amlacher S et al. , Insight into Structure and Assembly of the Nuclear Pore Complex by Utilizing the Genome of a Eukaryotic Thermophile, Cell, Elsevier Inc., 2011, pp. 277–289. [DOI] [PubMed] [Google Scholar]
- 99.Hayama R et al. (2017) The nuclear pore complex core scaffold and permeability barrier: variations of a common theme. Curr Opin Cell Biol 46, 110–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Blus BJ et al. (2019) Allosteric modulation of nucleoporin assemblies by intrinsically disordered regions. Sci Adv 5 (11), eaax1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Onischenko E et al. (2017) Natively Unfolded FG Repeats Stabilize the Structure of the Nuclear Pore Complex. Cell 171 (4), 904–917 e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Van de Vosse DW et al. (2013) A role for the nucleoporin Nup170p in chromatin structure and gene silencing. Cell 152 (5), 969–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kehat I et al. (2011) Modulation of chromatin position and gene expression by HDAC4 interaction with nucleoporins. J Cell Biol 193 (1), 21–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gozalo A et al. (2020) Core Components of the Nuclear Pore Bind Distinct States of Chromatin and Contribute to Polycomb Repression. Mol Cell 77 (1), 67–81 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Iglesias N et al. (2020) Native Chromatin Proteomics Reveals a Role for Specific Nucleoporins in Heterochromatin Organization and Maintenance. Mol Cell 77 (1), 51–66 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ibarra A et al. (2016) Nucleoporin-mediated regulation of cell identity genes. Genes Dev 30 (20), 2253–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Krull S et al. (2010) Protein Tpr is required for establishing nuclear pore-associated zones of heterochromatin exclusion. EMBO J 29 (10), 1659–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lelek M et al. (2015) Chromatin organization at the nuclear pore favours HIV replication. Nat Commun 6, 6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wong RW et al. (2015) Impact of Nucleoporin-Mediated Chromatin Localization and Nuclear Architecture on HIV Integration Site Selection. J Virol 89 (19), 9702–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dultz E et al. (2016) Global reorganization of budding yeast chromosome conformation in different physiological conditions. J Cell Biol 212 (3), 321–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ahmed S et al. (2010) DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat Cell Biol 12 (2), 111–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Light WH et al. (2010) Interaction of a DNA zip code with the nuclear pore complex promotes H2A.Z incorporation and INO1 transcriptional memory. Mol Cell 40 (1), 112–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Capelson M et al. , Chromatin-Bound Nuclear Pore Components Regulate Gene Expression in Higher Eukaryotes, Cell, 2010, pp. 372–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Franks TM et al. (2017) Nup98 recruits the Wdr82-Set1A/COMPASS complex to promoters to regulate H3K4 trimethylation in hematopoietic progenitor cells. Genes Dev 31 (22), 2222–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Juhlen R and Fahrenkrog B (2018) Moonlighting nuclear pore proteins: tissue-specific nucleoporin function in health and disease. Histochem Cell Biol 150 (6), 593–605. [DOI] [PubMed] [Google Scholar]
- 116.Pascual-Garcia P et al. (2014) Nucleoporin Nup98 associates with Trx/MLL and NSL histone-modifying complexes and regulates Hox gene expression. Cell Rep 9 (2), 433–42. [DOI] [PubMed] [Google Scholar]
- 117.Ptak C and Wozniak RW, ScienceDirect Nucleoporins and chromatin metabolism, Curr Opin Cell Biol, Elsevier Ltd, 2016, pp. 153–160. [DOI] [PubMed] [Google Scholar]
- 118.Lapetina DL et al. (2017) Yeast silencing factor Sir4 and a subset of nucleoporins form a complex distinct from nuclear pore complexes. J Cell Biol 216 (10), 3145–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Coyne AN et al. (2020) G4C2 Repeat RNA Initiates a POM121-Mediated Reduction in Specific Nucleoporins in C9orf72 ALS/FTD. Neuron 107 (6), 1124–1140 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kinoshita Y et al. (2012) Nuclear distributions of NUP62 and NUP214 suggest architectural diversity and spatial patterning among nuclear pore complexes. PLoS One 7 (4), e36137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gomar-Alba M and Mendoza M (2019) Modulation of Cell Identity by Modification of Nuclear Pore Complexes. Front Genet 10, 1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Terry LJ and Wente SR, Flexible Gates: Dynamic Topologies and Functions for FG Nucleoporins in Nucleocytoplasmic Transport, Eukaryotic Cell, 2009, pp. 1814–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Peters R (2009) Translocation through the nuclear pore: Kaps pave the way. Bioessays 31 (4), 466–77. [DOI] [PubMed] [Google Scholar]
- 124.Aramburu IV and Lemke EA (2017) Floppy but not sloppy: Interaction mechanism of FG-nucleoporins and nuclear transport receptors. Semin Cell Dev Biol 68, 34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jovanovic-Talisman T and Zilman A (2017) Protein Transport by the Nuclear Pore Complex: Simple Biophysics of a Complex Biomachine. Biophys J 113 (1), 6–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lemke EA (2016) The Multiple Faces of Disordered Nucleoporins. J Mol Biol 428 (10 Pt A), 2011–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rout MP et al. (2003) Virtual gating and nuclear transport: the hole picture. Trends Cell Biol 13 (12), 622–8. [DOI] [PubMed] [Google Scholar]
- 128.Paradise A et al. (2007) Significant proportions of nuclear transport proteins with reduced intracellular mobilities resolved by fluorescence correlation spectroscopy. J Mol Biol 365 (1), 50–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Akey CW and Radermacher M, Architecture of the Xenopus nuclear pore complex revealed by three-dimensional cryo-electron microscopy, The Journal of Cell Biology, 1993, pp. 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bestembayeva A et al. (2015) Nanoscale stiffness topography reveals structure and mechanics of the transport barrier in intact nuclear pore complexes. Nat Nanotechnol 10 (1), 60–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sakiyama Y et al. (2016) Spatiotemporal dynamics of the nuclear pore complex transport barrier resolved by high-speed atomic force microscopy. Nat Nanotechnol 11 (8), 719–23. [DOI] [PubMed] [Google Scholar]


