Abstract
Abstract
Folding and processing of proteins in the endoplasmic reticulum (ER) are major impediments in the production and secretion of proteins from Pichia pastoris (Komagataella sp.). Overexpression of recombinant genes can overwhelm the innate secretory machinery of the P. pastoris cell, and incorrectly folded proteins may accumulate inside the ER. To restore proper protein folding, the cell naturally triggers an unfolded protein response (UPR) pathway, which upregulates the expression of genes coding for chaperones and other folding-assisting proteins (e.g., Kar2p, Pdi1, Ero1p) via the transcription activator Hac1p. Unfolded/misfolded proteins that cannot be repaired are degraded via the ER-associated degradation (ERAD) pathway, which decreases productivity. Co-expression of selected UPR genes, along with the recombinant gene of interest, is a common approach to enhance the production of properly folded, secreted proteins. Such an approach, however, is not always successful and sometimes, protein productivity decreases because of an unbalanced UPR. This review summarizes successful chaperone co-expression strategies in P. pastoris that are specifically related to overproduction of foreign proteins and the UPR. In addition, it illustrates possible negative effects on the cell’s physiology and productivity resulting from genetic engineering of the UPR pathway. We have focused on Pichia’s potential for commercial production of valuable proteins and we aim to optimize molecular designs so that production strains can be tailored to suit a specific heterologous product.
Key points
• Chaperones co-expressed with recombinant genes affect productivity in P. pastoris.
• Enhanced UPR may impair strain physiology and promote protein degradation.
• Gene copy number of the target gene and the chaperone determine the secretion rate.
Keywords: Pichia pastoris, Productivity of recombinant protein production, Folding and secretion, Unfolded protein response (UPR), Chaperone, Co-expression strategy
Introduction
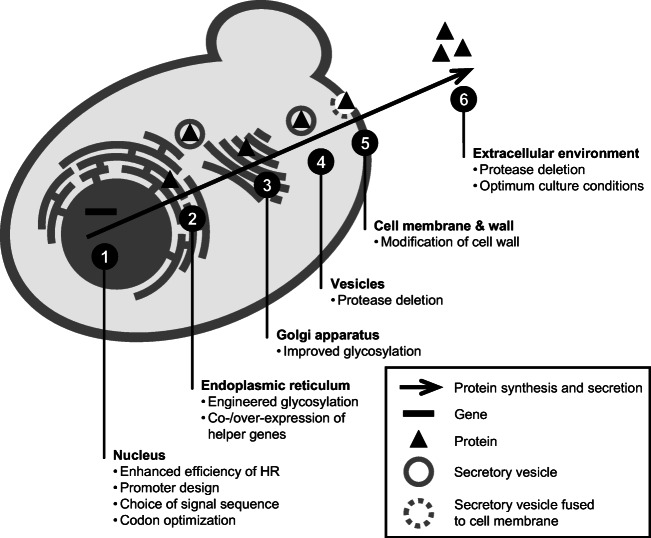
The methylotrophic yeast Pichia pastoris (Komagataella phaffii) is an established platform for applied research, specifically for the biotechnological production of a wide range of recombinant proteins. These include various intracellular, membrane and surface-displayed proteins and, most importantly, recombinant proteins that are secreted in large quantities (Cereghino and Cregg 2000; Daly and Hearn 2005; Gasser et al. 2013; Emmerstorfer et al. 2014; Spohner et al. 2015). The ability of P. pastoris to efficiently secrete recombinant proteins of unparalleled high quality (i.e., correctly folded, and post-translationally modified, without contamination from other proteins) makes this yeast an appropriate host for the industrial production of biopharmaceuticals or commercially valuable enzymes. Protein secretion is a multistep process, involving various cellular compartments (Fig. 1). After the transcription of its recombinant gene in the nucleus, the protein is synthesized, folded, and post-translationally modified in the endoplasmic reticulum (ER). From there, it is translocated in COPII vesicles to the Golgi apparatus (Antonny and Schekman 2001), where the post-translational modifications are finalized. The protein is then packed and shipped in a vesicle towards its destination, which, in the case of proteins intended for secretion, is towards the cell membrane. The vesicles fuse with the cell membrane and the protein is finally released to the extracellular environment (Puxbaum et al. 2015). It was found that in P. pastoris, recombinant proteins aimed for secretion, localized in the ER, are inherited during cell division to buds, from whence exocytosis of these soluble proteins predominantly occurs (Puxbaum et al. 2016).
Fig. 1.
Approaches to enhance recombinant protein secretion in P. pastoris. The production and secretion of recombinant protein can be enhanced by different approaches, aimed at different stages of the recombinant protein’s production and secretion. By improving the rate of homologous recombination (HR), the integration of (multiple) expression cassettes is enhanced. The expression level of the heterologous gene is determined by the promoter used, and processing and secretion of the protein can be improved by its codon optimization and the choice of a suitable secretion signal sequence, respectively. Correct glycosylation can be ensured in glycoengineered production strains, and folding or building of disulfide bridges might be enhanced by co-expressing chaperone or other helper genes. The intracellular proteolytic degradation of the recombinant proteins can be avoided by deletion of genes encoding proteases. The release of the proteins to the extracellular environment may be enhanced by modifications of the cell membrane and cell wall. Stability of the secreted protein in the extracellular environment is preserved by the choice of appropriate cultivation conditions (pH, temperature) and can be improved by the deletion of genes encoding secreted proteases
Information about secretory mechanisms in P. pastoris are still mainly based on knowledge derived from the model yeast S. cerevisiae. Nevertheless, information about the P. pastoris cell factory has advanced over the last decade due to whole genome characterization (De Schutter et al. 2009), available omics analyses (Zahrl et al. 2017), and the development of novel tools facilitating genomic engineering, such as CRISPR-Cas9 technology (Weninger et al. 2016; Raschmanová et al. 2018; Weninger et al. 2018). Based on genomic comparisons of different yeast species and mammals, it was shown that some patterns of P. pastoris’s secretory pathway resemble those of mammalian cells rather than those of S. cerevisiae (Delic et al. 2013). For example, structural organization of the Golgi compartment differs in S. cerevisiae and P. pastoris; the Golgi apparatus in P. pastoris is arranged in stacks and embedded in a ribosome-excluding matrix, which is similar to mammalian and plant cells (Rossanese et al. 1999; Mogelsvang et al. 2003). Also, some patterns of response to ER stress observed in P. pastoris resemble those of mammalian cells (Graf et al. 2008). These results indicate that valid conclusions for P. pastoris cannot be generally drawn from the model species S. cerevisiae. More intensive basic research on the secretory pathway and its bottlenecks in P. pastoris is needed, to effectively optimize the production/secretion of recombinant proteins by this host.
Because of the complex character of the secretory pathway, optimization of productivity of secreted proteins is challenging and often requires a combinatorial approach. The right optimization strategy seems not to be generally predictable, even for proteins of similar structures and properties (Obst et al. 2017), so, unfortunately, it must be designed for each protein specifically. Production/secretion might be generally optimized at various levels: the expression cassette (promoter engineering, secretion signal sequences, codon optimization etc.), the host strain (co-/overexpression of chaperone genes or genes of other folding-assisting proteins, co-expression of transcription- and translation-enhancing elements, disruption of protease genes, modification of cell wall properties etc.), the cultivation conditions (pH, temperature), and the bioprocess strategy (specific growth rate of biomass etc.) (Marx et al. 2006; Emmerstorfer et al. 2014; Looser et al. 2015; Barrero et al. 2018; Gidijala et al. 2018; Zepeda et al. 2018; Duan et al. 2019; Fischer and Glieder 2019; Liu et al. 2019; Naranjo et al. 2019) (Fig. 1).
An extensive effort has been mounted to increase protein secretion by co-expression of different folding factor genes involved in the UPR pathway. However, this strategy has not been successful in all cases and rather, has been applied on an ad hoc basis. In this review, we have analyzed published data on co-expression strategies, with the aim of identifying the best strategy to enhance recombinant protein production/secretion in P. pastoris. Importantly, we also point out the undesirable effects on strain physiology and production, potentially resulting from the co-expression of folding factor genes, i.e., an unbalancing of the UPR pathway by its genetic engineering.
Secretion bottlenecks and unfolded protein response (UPR) in P. pastoris
Proteins intended for secretion enter the lumen of the ER through the Sec61 protein-translocation channel (Marsalek et al. 2019). Integral membrane proteins (except for peroxisomal and mitochondrial membrane proteins) also enter the secretory pathway, starting in the ER (Emmerstorfer et al. 2014). In the lumen of the ER, post-translational modifications and folding take place. Correct folding of the proteins is ensured by folding-assisting proteins such as chaperones or foldases (Zimmermann et al. 2011; Delic et al. 2013), and only correctly folded proteins may leave the ER and proceed through the secretory pathway. The formation of disulfide bonds (Damasceno et al. 2012), protein folding (Helenius et al. 1992), and/or the transport of folded proteins out of the ER (Love et al. 2012) are considered to be the rate-limiting steps of the secretory pathway in P. pastoris, as previously shown for recombinant human serum albumin (Shen et al. 2012; Puxbaum et al. 2016) and Rhizopus chinensis lipase (Sha et al. 2013b), or suggested for bovine lactoferrin (Sun et al. 2019), penicillin-G-acylase (Borčinová et al. 2020), or peptidoglycan recognition protein (Yang et al. 2016). Inappropriate cultivation conditions or high levels of production of recombinant proteins (Gasser et al. 2007a), especially those that are surface-displayed, as well as membrane or complex secreted proteins, may overwhelm the folding capacity of the ER, where-upon misfolded/unfolded proteins begin to accumulate in the lumen of the ER. These proteins cause stress to the cell and trigger the UPR (Graf et al. 2008), a signaling cascade aimed at reducing the level of incorrectly folded proteins in the ER, and thus eliminating the stress. The UPR results in upregulation of the expression of genes encoding chaperones and foldases, proteins ensuring correct post-translational modifications, and genes encoding proteins involved in protein translocation and ER quality control (Gasser et al. 2007a; Vogl et al. 2014). At the same time, the expression of many genes involved in protein synthesis is downregulated (Vogl et al. 2014). If the proteins fail to fold correctly, they are translocated back to the cytosol, ubiquitinated, and degraded by the ER-associated degradation (ERAD) pathway (Xie and Ng 2010). Upregulation of ERAD may also be a way to decrease the protein load on the ER if the secretory capacity of the cell is exceeded (Zahrl et al. 2018).
The regulatory mechanisms of UPR were first studied and extensively described in S. cerevisiae (Cox and Walter 1996). The key components of the UPR pathway are the following: the kinase/RNase Ire1p, the transcription factor Hac1p, and the chaperone Kar2p, which is a yeast homologue of the mammalian BiP (Casagrande et al. 2000). The Kar2p chaperone resides in the lumen of the ER and, under non-stress conditions, associates with the luminal domain of the monomeric Ire1p. As soon as unfolded proteins occur in the lumen of the ER, Kar2p dissociates from the luminal domain of Ire1p to assist with proper folding (Sidrauski and Walter 1997; Okamura et al. 2000). When Kar2p unbinds, Ire1p assembles into dimers, which results in its phosphorylation and activation of the RNase function of the cytosolic domain of Ire1p (Papa et al. 2003; Kimata et al. 2007). In S. cerevisiae, it was demonstrated that besides Kar2p dissociation, there is an additional mechanism of Ire1p activation, based on a direct interaction of unfolded proteins with clustered Ire1p (Kimata et al. 2004, 2007). The RNase domain of Ire1p then non-conventionally (i.e., spliceosome-independent) splices the HAC1 pre-mRNA (HAC1u mRNA) into its mature form (HAC1i mRNA) (Cox and Walter 1996). The HAC1 pre-mRNA is targeted to Ire1p via a stem-loop structure within the 3′ UTR of the pre-mRNA (Aragon et al. 2009; Kohno 2010). After the excision of the intron from the HAC1 mRNA, the two exons are joined by tRNA ligase, encoded by the RLG1 gene (Sidrauski et al. 1996). The mature HAC1i mRNA is translated to the protein Hac1p, which is translocated to the nucleus where it acts as a transcription activator recognizing the so-called UPRE (unfolded protein response element) sequence, and initiates the transcription of UPR-associated genes in the nucleus (Travers et al. 2000). Besides genes of ER chaperones and proteins involved in folding in P. pastoris, Hac1p also induces genes encoding cytosolic chaperones, and genes involved in translation, ribosome biogenesis, organelle biosynthesis, intracellular membrane expansion, protein glycosylation, and translocation (Graf et al. 2008; Guerfal et al. 2010). The UPR was also shown to play an important role in regulating lipid metabolism in P. pastoris (Zhang et al. 2016; Adelantado et al. 2017), and to affect the cytosolic redox balance, because redox processes in the ER are counterbalanced by redox processes in the cytosol (Delic et al. 2012). Imbalanced redox processes enhance the likely development of folding-related diseases (e.g., Alzheimer’s or Parkinson’s).
UPR regulation by Ire1 and Hac1 is highly phylogenetically conserved in eukaryotes and is the main pathway that responds to ER-stress (Bernales et al. 2006; Ron and Walter 2007). Nevertheless, there are variations in the molecular mechanism, and the physiological and stress-responsive roles of the UPR between different yeast species (Hernández-Elvira et al. 2018). The differences in UPR between P. pastoris and S. cerevisiae include the sequence of UPRE (Mori et al. 1996; Guerfal et al. 2010), the regulation of HAC1 mRNA splicing (Guerfal et al. 2010; Baumann et al. 2011; Fauzee et al. 2020; Raschmanová et al., in preparation), the length of the HAC1 intron (Mori et al. 2000; Guerfal et al. 2010), and the role of UPR genes in inositol biosynthesis (Chang et al. 2004; Raschmanová et al., in preparation). Recently, it was reported that the basal level of ER stress (i.e., without external stressing stimuli) in P. pastoris is higher than in S. cerevisiae, likely due to the enhanced passage of endogenous N-glycosylated proteins through the ER and the secretory pathway (Fauzee et al. 2020). It becomes evident that information about the UPR cannot be solely adopted from S. cerevisiae, and more basic research in this area is needed for P. pastoris in order to engineer the UPR pathway effectively in terms of increasing productivity. Moreover, general knowledge on chaperones involved in membrane protein folding is limited in yeasts (Emmerstorfer et al. 2014).
In P. pastoris, intracellular retention or aggregation of recombinant proteins, or even their intracellular degradation, was observed, accompanied by the upregulation of UPR and/or ERAD (Table 1). Intracellular degradation of the protein may account for up to 60% of the total product (Pfeffer et al. 2011). Recombinant proteins triggering the UPR are of different types, including various secreted proteins: antibody fragments (Gasser et al. 2006, 2007a; Khatri et al. 2011; Pfeffer et al. 2011; Delic et al. 2012; Pfeffer et al. 2012), human interleukin (Zhong et al. 2014), human serum albumin (Aw et al. 2017), different secreted enzymes (Resina et al. 2007; Tawde and Freimuth 2012; Lin et al. 2013; Sha et al. 2013a; Raschmanová et al. 2019), membrane proteins (Vogl et al. 2014), and enhanced green fluorescent protein (EGFP) (Liu et al. 2014). For example, human serum albumin (HSA) is considered to be a well-secreted protein by P. pastoris, as grams per liter of secreted HSA can be obtained (Kobayashi et al. 2000), while heterodimeric antibody fragments are typically produced in only milligrams per liter (Gasser et al. 2006). Yet, both were shown to upregulate the UPR (Table 1). To assign secretion capability (good vs. poor) of the recombinant protein, a combination of several characteristics should be considered (Raschmanová et al. 2019): titer (e.g., grams of secreted protein per liter), specific productivity (e.g., grams of secreted protein per liter and per gram of biomass), intracellular protein accumulation/degradation, and physiological state of the cells (e.g., compromised growth, proportion of non-viable cells). However, all these characteristics are rarely assessed and described in the available literature. Typically therefore, the distinction between a well and poorly secreted protein is based solely on the titer achieved, i.e., extracellular protein concentration.
Table 1.
ER-stress during production of recombinant proteins in P. pastoris
| Recombinant protein | Promoter | Intracellular accumulation/degradation | ER-stress | References |
|---|---|---|---|---|
| Secreted proteins | ||||
| Antibody Fab fragment | PGAP | Accumulation | Expression of KAR2, PDI1, ROT2, ERO1, calnexin, SEC31, SEC53 ↑ | (Gasser et al. 2006; Gasser et al. 2007a) |
| Antibody Fab3H6 fragment | PGAP | Degradation | Expression of KAR2 ↑ 3.5-fold, LHS1 ↑ 1.6-fold, proteolytic activity ↑ by more than 20% | (Pfeffer et al. 2011; Pfeffer et al. 2012) |
| Anti-HIV antibody 2F5 Fab fragment | PGAP | Not analyzed | Expression of KAR2, ERO1, PDI1, HAC1 ↑ 2–3-fold | (Delic et al. 2012) |
| Single-chain antibody fragment (scFv) | PAOX1 | Not analyzed | Expression of KAR2 ↑ app. 1.6-fold, PDI unchanged | (Khatri et al. 2011) |
| Hepatitis B virus surface antigen (8 copies) | PAOX1 | Potential degradation (ERAD) | Content of Pdi, ERAD proteins ↑ | (Vanz et al. 2012) |
| Anti-CD3 immunotoxin | PAOX1 or PGAP | No, but slow secretion | Content of Kar2p ↑ 1.5–3-fold | (Liu et al. 2005) |
| Synovial sarcoma X break point 2 | PAOX1 | Accumulation | Expression of KAR2 ↑ 2.3–3.5-fold | (Huang et al. 2010) |
| Human interleukin-10 (different copy number: 1, 5, 10; different temperature: 20°C or 30°C) | PAOX1 | Accumulation (both immature and mature protein) |
20°C: Expression of HAC1 ↑ app. 1.5-fold, KAR2 ↑ app. 1.9-fold, ERO1 ↑ app. 2.4-fold (compared to 30°C) 5-copy strain: Expression of HAC1 ↑ app. 2.5-fold, KAR2 ↑ app. 2.5-fold, ERO1 ↑ app. 3.8-fold Higher level of ER-phagy at 30°C than at 20°C |
(Zhong et al. 2014) |
| Human serum albumin, nine different clones (all single copy) | PAOX1 | Not analyzed | Expression of HAC1, KAR2, and PDI ↑, ↓ or unchanged, depending on the clone | (Aw et al. 2017) |
| Rabies virus glycoprotein (2–8 copies) | PAOX1 | Accumulation | Expression of HAC1 ↑ up to 3-fold (8-copy strain), PDI1 ↑ up to 2.8-fold (7-copy strain), KAR2 ↑ up to 2.8-fold (7-copy strain), ERAD genes HRD1 and CDC48 unchanged | (Ben Azoun et al. 2016a) |
| Rabies virus glycoprotein (1–8 copies) | PGAP | Accumulation | Expression of HAC1 ↑ up to 5.1-fold, PDI1 ↑ up to 4.8-fold, KAR2 ↑ up to 5.1-fold, HRD1 ↑ up to 2.3-fold, CDC48 ↑ up to 1.5-fold (highest for 8-copy strain) | (Ben Azoun et al. 2016b) |
| Rabies virus glycoprotein (1, 2, 3, 5, or 10 copies) | PAOX1 | Degradation | Expression of HAC1 ↑ up to 4.3, IRE1 ↑ up to 3.6-fold, PDI1 ↑ up to 3.1-fold, KAR2 ↑ up to 3.6-fold, HRD1 ↑ up to 7.5-fold, CDC48 ↑ up to 6.1-fold (highest for 10-copy strain) | (Ben Azoun et al. 2017) |
| Porcine insulin precursor (PIP) | PAOX1 | Not analyzed | 6-copy strain: Expression of KAR2 ↑ 1.68-fold, PDI1 ↑ 1.43-fold. 18-copy strain: Expression of KAR2 ↑ 5.78-fold, PDI1 ↑ 2.14-fold | (Zhu et al. 2011) |
| Insulin precursor | PAOX1 | Accumulation less than 10% | Amount of UPR- and ERAD-proteins (Kar2p, Pdi) ↓ | (Vanz et al. 2014) |
| Human lysozyme (nine mutational variants with different stability) | PAOX1 | Accumulation (the lower stability, the higher amount) |
Expression of HAC1 ↑ up to 6-fold, KAR2 ↑ up to 7.5-fold, PDI1 ↑ up to 5-fold, DER1 ↑ up to 1.8-fold, HRD3 ↑ up to 1.6-fold, SEC61 ↑ up to 1.2-fold The lower stability, the higher increase |
(Whyteside et al. 2011) |
| Human lysozyme (variant prone to intracellular aggregation I56T and misfolded but secretable variant T70N) | PAOX1 | Aggregation (20–30% in case of T70N, up to 60% in case of I56T) | Expression of KAR2 and PDI1 ↑ 2-fold transiently | (Hesketh et al. 2013) |
| Human trypsinogen | PAOX1 or PGAP | Accumulation | Content of Kar2p ↑ up to 4-fold | |
| Human trypsinogen | PAOX1 | Not analyzed | Expression of PDI1, HAC1, ERO1, etc. ↑ | |
| Human trypsinogen (1, 2, or 3 copies) | PAOX1 | Not analyzed |
Expression of HAC1, KAR2, PDI, ERAD genes ↑ 3-copy strain: Expression of HAC1 ↑ 3–4-fold higher than in 1-copy strain |
|
| Porcine trypsinogen | PGAP | Not analyzed | >1-copy strain: Expression of KAR2, ERO1, PDI1, HAC1 ↑ 2–3-fold | (Delic et al. 2012) |
| Prolyl endopeptidase | PAOX1 | Degradation | Expression of HAC1 ↑ up to app. 4.7-fold, KAR2 ↑ up to app. 5-fold, and PDI1 ↑ up to 5.7-fold | (Wang et al. 2017) |
| Enhanced green fluorescent protein (1–6 copies) | PAOX1 | Accumulation (in strains with 4 and 5 copies) | Expression of KAR2 ↑ app. 1.5–5.5-fold, PDI ↑ app. 1.5–4-fold. The highest increase in 4-copy strain | (Liu et al. 2014) |
| Rhizopus oryzae lipase | PFLD1 | Not analyzed |
Shake flasks: Expression of KAR2 ↑ app. 4-fold, PDI ↑ app. 5-fold Bioreactors: Expression of KAR2 ↑ from 0 to 2.5 fmol mg−1 total RNA, PDI ↑ from 0 to 2 fmol mg−1 total RNA |
(Resina et al. 2007) |
| Rhizopus chinensis lipase (1, 3, 5, or 6 copies) | PAOX1 | No | Expression of ERO1 ↑ up to 1.7-fold, PDI1 ↑ up to 3.7-fold (highest for 6-copy strain) | (Sha et al. 2013a) |
| Arabidopsis modular cellulases AtGH9C1 (C1) and AtGH9C2 (C2) and their truncated versions | PAOX1 | Accumulation (truncated versions) | Expression of KAR2 ↑ 5–15-fold, PDI1 ↑ 3–6-fold | (Tawde and Freimuth 2012) |
| Xylanase A from Bacillus halodurans (1 or 4 copies) | PAOX1 | Not analyzed specifically (only total intracellular protein), probably no degradation |
1-copy strain: Expression of HAC1 ↑ app. 1.5-fold, ERO1 unchanged, KAR2 ↓ app. 0.5-fold, CNE1 ↓ app. 0.4-fold, levels of proteins involved in folding and stress response ↓ 4-copy strain: Expression of HAC1 ↑ 2.2-fold, KAR2 ↑ 1.9-fold, ERO1 ↑ 1.5-fold, CNE1 ↑ 1.9-fold, chaperone content ↑ |
(Lin et al. 2013) |
| Escherichia coli penicillin G acylase (EcPGA), Candida antarctica lipase B (CaLB), Thermomyces lanuginosus xylanase A (TlXynA) | PAOX1 | Accumulation in case of EcPGA (50–70%) | Activity of PKAR2 ↑ (60% cells producing EcPGA, 35% cells producing CaLB, and 30% cells producing TlXynA), expression of KAR2 ↑ up to 5.6-fold in case of EcPGA production | (Raschmanová et al. 2019) |
| Membrane proteins | ||||
| Alternative oxidase from P. pastoris (PpAodp), human CMP-Sia transporter (HsCstp), copper transporter Ctr3 from S. cerevisiae (ScCtr3p), all linked to GFP | PAOX1 | --- | Expression of genes involved in stress response (PpAodp, HsCstp, ScCtr3p) and protein folding (PpAodp) ↑ | (Vogl et al. 2014) |
It seems that the secretion is not predominantly determined by the origin of the protein, in the sense of being naturally secreted or cytosolic; typically, intracellular proteins can also be successfully secreted by P. pastoris, e.g., human catalase (0.55 g per liter) (Shi et al. 2007). Rather, the ease of secretion and UPR upregulation seem to be the result of combined effects of the strength of expression (i.e., promoter), gene copy number, protein thermostability, and cultivation conditions used. In all the cases listed in Table 1, the heterologous genes were expressed from strong promoters, either the constitutive glyceraldehyde phosphate dehydrogenase (GAP) promoter, or the methanol inducible alcohol oxidase 1 (AOX1) promoter. Generally, the higher the copy number of the heterologous gene, the more pronounced was the UPR (Table 1). In the study performed by Love et al. (2012), increasing the copy number of genes expressed from PAOX1 led to decreased rates of secretion for three proteins with different folding complexities: EGFP, and glycosylated and aglycosylated versions of a human Fc fragment. Nevertheless, there are also proteins whose secretion increases with high-copy number (Huang et al. 2017). The relationship between protein thermostability, secretion, and UPR/ERAD was studied (Whyteside et al. 2011). These authors showed that the production of mutationally destabilized variants of human lysozyme led to higher UPR and ERAD levels, and the protein was retained intracellularly, i.e., poorly secreted and targeted for degradation, more so than the stable variant of lysozyme (Whyteside et al. 2011). Cultivation conditions such as specific growth rate of biomass, temperature, or osmolarity regulate the UPR. An increased specific growth rate of biomass upregulated the UPR, while proteolytic degradation of secretory proteins (ERAD) was downregulated (Rebnegger et al. 2014). Reduction of the cultivation temperature from 30 to 20°C upregulated UPR (Zhong et al. 2014), which probably led to a more rapid processing of the recombinant product in the ER, decreased levels of immature forms of the protein, and increased product yield (Zhong et al. 2014).
Enhancing protein secretion by overexpression of UPR genes
A possible strategy to enhance production and secretion of a recombinant protein is to co-express a chaperone gene or other genes involved in the UPR, assuming that the co-expressed partner will assist and ensure correct protein folding. Nevertheless, upregulation of the UPR is beneficial only in the cases where protein folding, rather than its passage through the secretory pathway, becomes rate-limiting (Love et al. 2012). In P. pastoris, increased expression or secretion of many different recombinant proteins resulted from co-expression of the following: the ER-chaperone Kar2p or protein disulfide isomerase Pdi1 (Inan et al. 2006; Damasceno et al. 2007; Sallada et al. 2019), enzymes involved in the ER redox control and oxidative stress such as Ero1, Gpx1, Aha1, or Ypt6 (Sha et al. 2013c; Ben Azoun et al. 2016a; Huangfu et al. 2016; Sallada et al. 2019), the UPR transcription factor Hac1p (Guerfal et al. 2010; Vogl et al. 2014; Li et al. 2015; Krainer et al. 2016; Huang et al. 2017; Han et al. 2020; Liu et al. 2020), the kinase/RNase Ire1p (Yu et al. 2020), or new co-chaperones (Huangfu et al. 2016) (Table 2). Glycosylation activity was also increased (Moon et al. 2015) or product homogeneity and processing of the secretion α-factor were improved (Guerfal et al. 2010). Recently, three novel folding factors, Mpd1p (member of the PDI family), Pdi2p (protein disulfide isomerase), and Sil1p (nucleotide exchange factor for the ER lumenal Hsp70 chaperone Kar2p), were characterized and their genes co-expressed in P. pastoris (Duan et al. 2019). In this work, only Sil1p improved the specific extracellular activity and the secretion ratio of one out of three recombinant proteins tested (Duan et al. 2019).
Table 2.
Examples of co-expression strategies involving UPR genes to improve production/secretion of recombinant proteins in P. pastoris
| Recombinant protein (secreted, if not stated otherwise) | Co-expressed helper gene | Promoter for recombinant/helper gene expression | Production/secretion-related effect(s) | References |
|---|---|---|---|---|
| Human parathyroid hormone | Sc*1PDI1 | PAOX1/PAOX1 | Secretion ↑ app. 3-fold | (Vad et al. 2005) |
| Antibody Fab fragment | ScHAC1i | PAOX1 or PGAP/PGAP | Secretion ↑ 1.3-fold | (Gasser et al. 2006) |
| Antibody Fab fragment | ScPDI | PAOX1 or PGAP/PGAP | Secretion ↑ 1.9-fold | (Gasser et al. 2006) |
| Antibody Fab fragment | ScPDI1, ScERO1, ScKAR2, or ScHAC1 | PGAP/PGAP | Productivity ↑ 1.7-fold (PDI1), ↑ 1.4-fold (ERO1), 1.5-fold (KAR2), 1.5-fold (HAC1) | (Gasser et al. 2007b) |
| A33 single-chain antibody fragment | Pp*2KAR2 and/or PpPDI | PAOX1/PAOX1 | Secretion ↑ 3-fold (KAR2), no effect (PDI), no effect (KAR2 and PDI) | (Damasceno et al. 2007) |
| Interleukin-2-human serum albumin fusion protein | PDI1, KAR2, ERO1 | PAOX1/PGAP | Secretion level ↑ 2.2-fold (PDI1), 1.9-fold (KAR2), 2.3-fold (ERO1) | (Guan et al. 2016) |
| Interleukin-1 receptor antagonist-human serum albumin fusion protein (high-copy number) | PpPDI, PpKAR2 | PAOX1/PAOX1 | Yield ↑ 2.4–3.7-fold (different copy numbers of PDI1), ↓ (different copy numbers of KAR2) | (Shen et al. 2012) |
| Human granulocyte-colony stimulating factor | ScKAR2 and/or ScPDI | PAOX1/PGAP | Activity ↑ 5.6-fold (ScKAR2), 4-fold (ScPDI), 6.5-fold (ScKAR2 and ScPDI) | (Zhang et al. 2006) |
|
Mouse interferon-γ, human interferon-β, human thrombomodulin, human erythropoietin (all surface-displayed proteins) mIL*3-10, Trypanosoma cruzi trans-sialidase protein Adenosine A2A receptor (membrane protein) |
PpHAC1i | PAOX1/PAOX1 or PGAP |
PAOX1-expressed HAC1i: Expression of human thrombomodulin ↑ 1.9-fold, mIL-10 ↑ 2.2-fold, Trypanosoma cruzi trans-sialidase ↑ 2.1-fold, other proteins ↓, homogeneity and processing of the α-mating factor of adenosine A2A receptor improved PGAP-expressed HAC1i: Little or no improvement of production |
(Guerfal et al. 2010) |
| Human CMP-Sia transporter (HsCstp), copper transporter Ctr3 from S. cerevisiae (ScCtr3p), rice (Oryza sativa) CMP-Sia transporter (OsCstp), human copper transporter Ctr1 (HsCtr1p), all linked to GFP (membrane proteins) | PpHAC1i | PAOX1/PAOX1 | Expression of ScCtr3p unchanged, HsCstp ↑ 2.1-fold, HsCtr1p ↑ 1.7-fold, OsCstp ↑ 1.5-fold | (Vogl et al. 2014) |
| Necator americanus secretory protein (different copy numbers) | PpPDI1 | PAOX1/PAOX1 | Secretion ↑ app. 4–8-fold | (Inan et al. 2006) |
| Rabies virus glycoprotein | PDI1, ERO1, GPX1, GLR1, or YAP1 | PAOX1/PGAP | Level ↑ up to 9.6-fold (PDI1), ↑ app. 3-fold (ERO1), ↑ 8.2-fold (GPX1), unchanged (GLR1, YAP1) | (Ben Azoun et al. 2016a) |
| Rabies virus glycoprotein | PDI1, ERO1, GPX1, GLR1, or YAP1 | PGAP/PGAP | Expression ↑ up to 15-fold (PDI1), ↑ 4-fold (ERO1), ↑ 9-fold (GPX1), ↑ 1.7-fold (GLR1), unchanged (YAP1) | (Ben Azoun et al. 2016b) |
| Porcine peptidoglycan recognition protein (low-, medium-, high-copy) | PDI1 and/or KAR2 | PAOX1/PGAP | Amount in medium-copy strain ↑ up to app. 2.8-fold (PDI1), high-copy strain ↑ up to app. 5-fold (PDI1), unchanged in low-copy strain (PDI1). Amount unchanged or ↓ (KAR2) | (Yang et al. 2016) |
| Hydrophobin HFBI (1–3-copies) | PpKAR2, PpPDI1, PpERO1 | PAOX1/PAOX1 |
Expression in 1-copy strain ↑ 14-fold (KAR2), insignificant change (PDI1, ERO1) Expression in 2-copy strain ↑ 9.8-fold (KAR2), insignificant change (PDI1, ERO1) Expression in 3-copy strain ↑ 22-fold (KAR2), ↑ 7.8-fold (PDI1), 30-fold (ERO1) |
(Sallada et al. 2019) |
| Bovine lactoferrin (2 copies) | PpHAC1i |
PAOX1/PGAP PAOX1/P0547*4 |
Yield ↓ by 20.9% Yield ↑ by 109.5% |
(Sun et al. 2019) |
| Human lysozyme (4 copies) | PpHAC1i | PAOX1/PAOX1 | Lysozyme activity ↑ by 21.3% | (Liu et al. 2020) |
| Human lysozyme (6 copies) | PpKAR2, PpERO1, PpPDI1 | PAOX1/PGAP | Activity ↑ (PpERO1, PpPDI1, PpERO1+PpPDI1), ↓ (PpKAR2) | (He et al. 2020) |
| Porcine trypsinogen | PpPDI1 or PpERO1 | PGAP/PGAP | Titer ↑ 2-fold (PpPDI1), unchanged (PpERO1) | (Delic et al. 2012) |
| Rhizopus oryzae lipase | ScHAC1 | PFLD1/PGAP | Extracellular activity ↑ 1.5-fold | (Resina et al. 2007) |
| Rhizopus oryzae lipase | ScHAC1 | PFLD1/PGAP | Specific productivity ↑ 3-fold | (Resina et al. 2009) |
| Rhizopus chinensis lipase | PpERO1 and PpPDI1 (simultaneously) | PAOX1/PAOX1 | Enzyme yield ↑ by 30% | (Sha et al. 2013c) |
| Candida antarctica lipase B | PpKAR2 | PAOX1/PAOX1 | Activity ↓ 0.7-fold | (Samuel et al. 2013) |
| Rhizomucor miehei lipase (2 or 4 copies) | PpPDI, PpERO1 | PAOX1/PAOX1 |
Activity in 2-copy strain unchanged (PDI1), ↓ by app. 33% (ERO1) Activity in 4-copy strain ↑ 2-fold (PDI1), ↓ by app. 12% (ERO1) |
(Huang et al. 2020) |
| Lipase MAS1 from marine Streptomyces sp. | PpPDI, PpHAC1, PpKAR2 | PAOX1/PAOX1 | Activity ↑ 1.7-fold (PDI), ↑ slightly (HAC1, KAR2) | (Lan et al. 2016) |
| α-glucosidase from Aspergillus niger | PpPDI1 | PAOX1/PAOX1 | Concentration unchanged or ↓ | (Liu et al. 2013) |
| Xylanase A from Bacillus halodurans (1 or 4 copies) | HAC1 | PAOX1/PAOX1 | Amount ↑ 1.4-fold (4-copy strain), unchanged (1-copy strain) | (Lin et al. 2013) |
| Phytase from Citrobacter amalonaticus | HAC1i | PAOX1 (engineered)/PAOX1 | Concentration ↑ 1.4-fold | (Li et al. 2015) |
| Horseradish peroxidase | HAC1i | PAOX1/PAOX1 or PHTX1 (bidirectional) | Specific activity ↑ | (Krainer et al. 2016) |
| β-glucuronidase | AHA1, SBA1, SIS1, YPT6 | PAOX1/PGAP | Specific activity ↑ 1.9-fold (AHA1), ↑ 1.6-fold (SBA1), ↑ 1.4-fold (SIS1), ↑ 1.8-fold (YPT6), ↑ 2.3 (AHA1 with YPT6), altered glycosylation | (Huangfu et al. 2016) |
| Endo-β-1,4-xylanase | AHA1, YPT6 | PAOX1/PGAP | Specific activity ↑ 2-fold (AHA1), ↑ app. 2.5-fold (YPT6) | (Huangfu et al. 2016) |
| Phospholipase C-Y from Bacillus cereus | HAC1, PDI, KAR2 | PAOX1/PAOX1 (attenuated) | Titer ↑ 6.2-fold (HAC1), unchanged (PDI, KAR2) | (Elena et al. 2016) |
| Raw-starch hydrolyzing α-amylase | HAC1i | PAOX1/PAOX1 or PGAP | Concentration ↑ up to 7.2-fold (6 copies of HAC1i from PAOX1), ↑ 12.1-fold (+ other 17 copies of HAC1i from PGAP) | (Huang et al. 2017) |
| Starmerella bombicola lactone esterase | HAC1i | PAOX1/PAOX1 | Concentration ↑ 1.8-fold | (De Waele et al. 2018) |
| β-galactosidase from A. oryzae (Lac), β-mannanase from Bacillus (Man), glucose oxidase from A. niger (Gox) (all codon-optimized for P. pastoris) | HAC1 homologues: PpHAC1, ScHAC1, Tr*5HAC1 or Hs*6XBP1 | PAOX1/PAOX1 |
Specific activity of Lac ↑ by 75% (PpHAC1), ↑ by 57% (ScHAC1), ↑ by 81% (TrHAC1), ↓ by 62% (HsXBP1) Specific activity of Man ↑ by 8% (TrHAC1), ↑ by 49% (HsXBP1), ↓ by 3% (PpHAC1), ↓ by 41% (ScHAC1) Specific activity of Gox ↑ by 13% (PpHAC1), ↑ by 10% (ScHAC1), ↑ by 5% (HsXBP1), ↓ by 3% (TrHAC1) |
(Bankefa et al. 2018) |
| Yeast-enhanced green fluorescent protein (yEGFP), β-galactosidase (Gal), cephalosporin C acylase (SECA) | PDI1, KAR2, HAC1, MPD1, PDI2, SIL1 | PAOX1/PAOX1 |
Specific extracellular fluorescence of yEGFP ↑ by 26% (PDI1), ↑ by 14% (KAR2), ↑ by 99% (HAC1), ↓ (others), secretion ratio*7 unchanged (all) Extracellular production of Gal ↑ slightly (PDI2, KAR2), ↓ dramatically (others) Extracellular production of SECA ↑ 3-fold (SIL1, HAC1), ↓ or unchanged (others), secretion ratio ↑ 2.7-fold (SIL1) and 3.2-fold (HAC1) |
(Duan et al. 2019) |
| Pseudomonas aeruginosa elastase | HAC1i | PAOX1/PAOX1 | Activity ↑ 1.8–3.9-fold. Negligible effect on N-glycosylation | (Han et al. 2020) |
| Zobellia κ-carrageenase | KAR2, ERO1, PDI, YAP1, AHA1, YPT6, PRX1, RPN4, IRE1 | PAOX1/PAOX1 | Enzymatic activity unchanged (KAR2, PDI), ↑ 1.24–1.35-fold (all others) | (Yu et al. 2020) |
*1Saccharomyces cerevisiae, *2Pichia pastoris, *3mouse interleukin, *4novel methanol-inducible promoter (Xu et al. 2018), *5Trichoderma reesei, *6Homo sapiens, *7secreted to total protein amount
In the vast majority of published works, the helper gene, as well as the target gene of interest, was expressed from the classic strong Pichia promoters, GAP or AOX1 (Table 2). When co-expressing 1, 2, 4, 6, 8, or 11 copies of HAC1 from the AOX1 promoter and additional 4, 6, 9, 10, 13, 17 copies of HAC1 from the GAP promoter along with the raw-starch hydrolyzing enzyme, α-amylase, the best improvement of product concentration was reached with 6 copies of HAC1 expressed from the AOX1 promoter and 17 copies of HAC1 expressed from the GAP promoter (Huang et al. 2017). In another work, the effect of HAC1 overexpression on heterologous protein levels was stronger when HAC1 was expressed from the inducible AOX1 promoter than from the constitutive GAP promoter (Guerfal et al. 2010). As shown recently, it might also be beneficial to examine alternative promoters. The yield of bovine lactoferrin was improved by 109.5% by HAC1i expressed from a novel methanol-inducible promoter P0547, while it decreased when using the GAP promoter (Sun et al. 2019). Recently, the UPR-inducible PDI1 promoter, whose strength was found to be equivalent to 20–25% of the GAP promoter and 4.5–5% of the AOX1 promoter, was used for moderate expression of the Candida antarctica lipase B gene (Prattipati et al. 2020).
Improved expression/secretion was also affected by the copy number of the recombinant gene (Lin et al. 2013; Yang et al. 2016; Sallada et al. 2019; Huang et al. 2020), as well as of the co-expressed helper gene (Yang et al. 2016; Huang et al. 2017). For example, while the amount of secreted xylanase A from Bacillus halodurans increased 1.4-fold in a 4-copy strain by the co-expression of HAC1, it was not changed in a co-expressing strain containing only one copy of the xylanase A gene (Lin et al. 2013). A similar trend was observed for the production of secreted Rhizomucor miehei lipase; overexpression of PDI1 led to enhanced activity (2-fold) in a 4-copy strain, whereas activity in the strain carrying two copies of the lipase gene remained unchanged (Huang et al. 2020). In a P. pastoris strain producing hydrophobin HFBI, co-expression of KAR2 increased expression of hydrophobin 14-fold in a 1-copy strain, 9.8-fold in a 2-copy strain, and 22-fold in a 3-copy strain (Sallada et al. 2019). Co-expression of other helper genes, PDI1 and ERO1, only increased the expression of hydrophobin in the 3-copy strain (7.8-fold and 30-fold, respectively) (Sallada et al. 2019). Another example was the co-expression of PDI1 in P. pastoris strains containing low-, medium-, and high-copy numbers of the porcine peptidoglycan recognition protein gene (Yang et al. 2016). Improvements in the amount of secreted product were more significant the higher the copy number, i.e., unchanged, 2.8-fold higher, and 5-fold higher in the low-, medium-, and high-copy strains, respectively (Yang et al. 2016). These results indicate that co-expression of helper UPR genes is particularly helpful, or more pronounced, in strains containing higher copy numbers of the heterologous gene. In the end, this can lead to higher secretion by strains containing a high-copy number of the heterologous gene than by the low-copy strains, which originally, i.e., without the co-expressed chaperone, secreted more product (Yang et al. 2016). The co-expression of multiple copies of the chaperone genes improved the secretion of porcine peptidoglycan recognition protein (high-copy strain) (Yang et al. 2016) or α-amylase from Geobacillus sp. (Huang et al. 2017).
In addition, the origin (i.e., the homologue used) of the co-expressed helper gene is important for the extent of its effect on recombinant protein secretion (Bankefa et al. 2018). While the specific activity of β-galactosidase produced with P. pastoris was the most improved by co-expression of HAC1 from Trichoderma reesei (by 81%), in the case of β-mannanase, the best co-expression partner was the HAC1 homologue from Homo sapiens (improvement of 49%), and for glucose oxidase, the co-expression of P. pastoris HAC1 worked the best (improvement of 13%). These results indicate that the native Hac1p or its homologue from a closely related species does not necessarily have to be the best option generally for enhancing the secretion of any protein (Bankefa et al. 2018).
An alternative strategy, based on regulating/engineering the UPR, which may improve protein secretion in P. pastoris, is inhibition of the proteasome, including ERAD (Pfeffer et al. 2012). However, recent research revealed that the disruption of proteasomal and ERAD components did not increase the secretion of an antibody fragment produced by P. pastoris and the authors proposed that the protein was probably degraded prior to entering the secretory pathway (Zahrl et al. 2018). Another approach enhancing recombinant protein production might be deletion of certain chaperones; in S. cerevisiae, deletion of CNE1, encoding the yeast homologue of mammalian calnexin and calreticulin, increased the production of human transferrin receptor (Prinz et al. 2003). In another review, the strategy of improving the production of recombinant G-protein coupled receptors (GPCR) in yeasts by addition of GPCR-specific ligands or chemical chaperones, such as DMSO, histidine, or glycerol, was discussed (Emmerstorfer et al. 2014). These chemical chaperones are involved in, e.g., gene regulation, modulating ER/Golgi transport, cell wall integrity, membrane permeability, stabilizing protein conformation, or supposedly acting as antioxidants (Emmerstorfer et al. 2014). Other engineering approaches to improve secretion by P. pastoris are reviewed elsewhere (Ahmad et al. 2014; Puxbaum et al. 2015; Fischer and Glieder 2019).
Pitfalls of engineering the UPR
It is apparent from the published studies that the effect of co-expressed factors is product-specific (Table 2); in some cases, the production/secretion of the recombinant proteins was unchanged (Damasceno et al. 2007; Delic et al. 2012; Liu et al. 2013; Vogl et al. 2014; Ben Azoun et al. 2016a; Ben Azoun et al. 2016b; Elena et al. 2016; Duan et al. 2019), and sometimes it was even reduced (Liu et al. 2013; Yang et al. 2016; Bankefa et al. 2018; Duan et al. 2019; Sun et al. 2019). For example, the secretion of A33 single-chain antibody fragment was increased by KAR2 co-expression but was not changed by the co-expression of PDI or simultaneous co-expression of KAR2 and PDI (Damasceno et al. 2007). In contrast, PDI co-expression increased secretion levels of an antibody Fab fragment (Gasser et al. 2006), Necator americanus secretory protein (different copy numbers) (Inan et al. 2006), or porcine trypsinogen (Delic et al. 2012).
A decrease in protein secretion in P. pastoris was reported for different membrane- and surface-displayed proteins after co-expression of PpHAC1i from PAOX1 (Guerfal et al. 2010), α-glucosidase from Aspergillus niger after co-expression of PpPDI1 from PAOX1 (Liu et al. 2013), bovine lactoferrin after co-expression of PpHAC1i from PGAP (by 20.9%) (Sun et al. 2019), porcine peptidoglycan recognition protein after co-expression of KAR2 from PGAP (Yang et al. 2016), or Candida antarctica lipase B after co-expression of KAR2 from PAOX1 (0.7-fold) (Samuel et al. 2013). These negative effects might be attributed to the use of a strong promoter for the co-expression of the UPR gene, which induces the UPR to an inappropriately high level and results in elevated ERAD, re-translocation of the protein to the cytosol and its subsequent degradation (Guerfal et al. 2010; Liu et al. 2013). The overexpression of KAR2 increased the intracellular insoluble fraction of a recombinant peptidoglycan recognition protein, and the prolonged retention of the protein in the ER probably led to its degradation via ERAD (Yang et al. 2016). Moreover, excess Kar2p molecules in the ER, caused by KAR2 overexpression, might — even in the presence of unfolded proteins — lead to sustained association of Kar2p with Ire1p, and thus prevent activation of Ire1p and subsequent upregulation of the UPR (Samuel et al. 2013). The efficiency of UPR regulation is also determined by the source of the overexpressed HAC1 (Bankefa et al. 2018); the specific activity of β-galactosidase from A. oryzae was decreased in the case of co-expression of the Homo sapiens homologue of HAC1 from PAOX1 (by 62%), and the specific activity of β-mannanase from Bacillus was decreased (by 41%) after co-expression of the S. cerevisiae homologue of HAC1 from PAOX1 (Bankefa et al. 2018). In the case of β-mannanase, it was shown that overexpression of ScHAC1 had little or even a negative effect on the expression of chaperones, compared to the HAC1 homologue from Homo sapiens, which also increased the specific β-mannanase activity (Bankefa et al. 2018).
It is important to keep in mind that overexpression of the UPR genes affects the UPR balance and other cellular processes. Overexpression of PDI1 in P. pastoris producing an antibody fragment (Fab) enhanced the secretion rate of Fab, but did not reduce the UPR stress (Gasser et al. 2007a). In addition, the constitutive expression of HAC1 activated ERAD (Guerfal et al. 2010). Prolonged activation of the UPR can result in so-called ER-phagy, when parts of the ER are removed to relieve the ER stress and remove the misfolded proteins (Kruse et al. 2006). In addition, a sustained activation of UPR can impair cellular growth, as reported for different yeasts (Cox et al. 1993; Kawahara et al. 1997; Chawla et al. 2011; Cheon et al. 2011; Miyazaki et al. 2013; Moon et al. 2015). In P. pastoris, slower growth was observed in the case of co-expression of HAC1 in strains producing xylanase A from Bacillus halodurans or human lysozyme (Lin et al. 2013; Liu et al. 2020), of PDI1 in a strain producing α-glucosidase from Aspergillus niger (Liu et al. 2013), or of ERO1 in a strain producing Rhizomucor miehei lipase (Huang et al. 2020). Other authors reported a decreased (by 27%) maximum specific growth rate (μmax) of a P. pastoris strain producing β-galactosidase, as a result of the co-expression of KAR2, but a comparable final cell density (Duan et al. 2019). Co-expression of PDI1 increased the final cell concentration by 35% but did not affect the μmax of that strain. The growth rate of a P. pastoris strain producing cephalosporin C acylase was not affected by the co-expression of PDI1, but was decreased by the co-expression of folding factors HAC1, KAR2, MPD1, PDI2, and SIL1, with SIL1 having the most detrimental effect: μmax was decreased by 39% (Duan et al. 2019). Nevertheless, in other works, no negative effect of overexpression of PDI1 and/or KAR2 on the growth of cells was observed (Damasceno et al. 2007; Guan et al. 2016), and the co-expression of HAC1i was even reported to enhance cellular growth (Han et al. 2020). These results suggest that the effect of the co-expressed gene on a strain’s physiology and growth has to be determined individually for each product. In this context, it is important to note that it is not quite correct to evaluate the effect of the co-expressed helper gene on protein production/secretion only by comparing protein concentrations or activities. Knowing that co-expression might influence the strain’s growth characteristics, it is essential to also assess the biomass growth. To evaluate the effect of the co-expression strategy, protein to biomass yields (mass of protein produced per mass of biomass) or specific productivities (mass of protein produced per mass of biomass per hour) should be compared, instead of only protein mass (mass of protein produced) or concentrations (mass of protein per liter). This is, however, usually not taken into account (Table 2).
Outcomes: Recommendations for co-expression strategies
The correct folding and rate of secretion of a recombinant protein are affected by the strength of expression of its gene, gene copy number (Love et al. 2012), thermostability of the protein (Whyteside et al. 2011), and cultivation conditions (Rebnegger et al. 2014; Zhong et al. 2014). It seems that the combination of these effects can outweigh the effect of the protein’s origin (cytosolic vs. secreted) and character with respect to its folding and secretion complexity. Low gene copy number (Love et al. 2012; Yang et al. 2016), increased thermostability of the protein (Whyteside et al. 2011), and decreased cultivation temperature (Zhong et al. 2014) can enhance the folding and secretion rate by alleviating the UPR.
Based on the currently available information, it seems that the effect of a co-expressed folding partner on recombinant protein secretion cannot be predicted a priori. The most suitable folding partner must be verified experimentally for each individual product. According to the literature search summarized in Table 2, the most frequently used co-expression partner genes employed to promote recombinant protein secretion in P. pastoris were as follows: HAC1 encoding a transcription factor of UPR genes, PDI1 encoding a protein disulfide isomerase, and KAR2 encoding an ER chaperone. In the case of HAC1, the use of different promoters (PGAP, PAOX1, P0547, and PHTX1) for its expression, different copy numbers, or different homologues was investigated, which makes HAC1 the best so far described co-expression partner in P. pastoris. As summarized in Table 2, overexpression of yeast HAC1 (i.e., the homologue from P. pastoris or S. cerevisiae) enhanced recombinant protein production/secretion in approx. 60% of reported cases (as reported in the literature) and co-expression of PDI1 and KAR2 improved protein production/secretion in approx. 73% and 53% of the published cases, respectively. However, such a broad brush view should be taken with care: The number of publications describing overexpression of PDI1 and KAR2 was lower than those reporting HAC1 co-expression (Table 2). We acknowledge that the number of unpublished results, either negative or positive, is uncertain. Nevertheless, this purely statistical view should be helpful given the wide scientific interest in the UPR-topic.
There were only a few studies where the effects of HAC1, PDI1, and KAR2 co-expression were compared for the same product. For the antibody Fab fragment and lipase from marine Streptomyces sp., PDI1 co-expression resulted in a greater increase in secreted product than HAC1 and KAR2 co-expression (Gasser et al. 2007b; Lan et al. 2016), while the secreted amount of yeast-enhanced green fluorescent protein (yEGFP) was increased the most significantly by HAC1 co-expression (Duan et al. 2019). The extracellular production of cephalosporin C acylase was improved by HAC1 co-expression, but not by PDI or KAR2 (Duan et al. 2019). Due to the low number of studies comparing the effect of HAC1, PDI1, and KAR2 co-expression, it is not possible to draw general conclusions about which co-expression partner would be the most suitable for any particular recombinant protein. Additionally, it cannot be concluded whether constitutive or inducible expression of the co-expressed helper gene would be more suitable, as both were shown to result in improved, but also unchanged or reduced secretion of recombinant proteins. There is a lack of literature describing the effect of chaperone gene co-expression on the production of membrane and surface-displayed proteins; only Hac1p was tested as a helper, and this improved the production of only some proteins (Guerfal et al. 2010; Vogl et al. 2014). It might be beneficial to employ promoters alternative to PGAP and PAOX1 for expression of the helper gene, including weak to moderate promoters for a better fine-tuning of the UPR. Failed co-expression strategies were, in some cases, attributed to the UPR having been upregulated to inappropriately high levels by the co-expression of the UPR genes from strong GAP or AOX1 promoters, which might have resulted in increased ERAD (Guerfal et al. 2010; Liu et al. 2013). It is also necessary to note that there might be many failed co-expression experiments in P. pastoris that were never published, but which might actually shed more light on the UPR mechanism and expression fine-tuning.
The literature search for co-expression strategies that employ a UPR-involved gene to enhance recombinant protein production in P. pastoris led to the following recommendations:
Consider the copy number of the heterologous gene of interest. Folding stress can be reduced, thus secretion enhanced, by reducing the copy number of the heterologous gene (Love et al. 2012, Yang et al. 2016). However, co-expression of a helper UPR gene might reverse this trend, resulting in more enhanced secretion in strains with a higher copy number of the heterologous gene than in low-copy number strains (Yang et al. 2016).
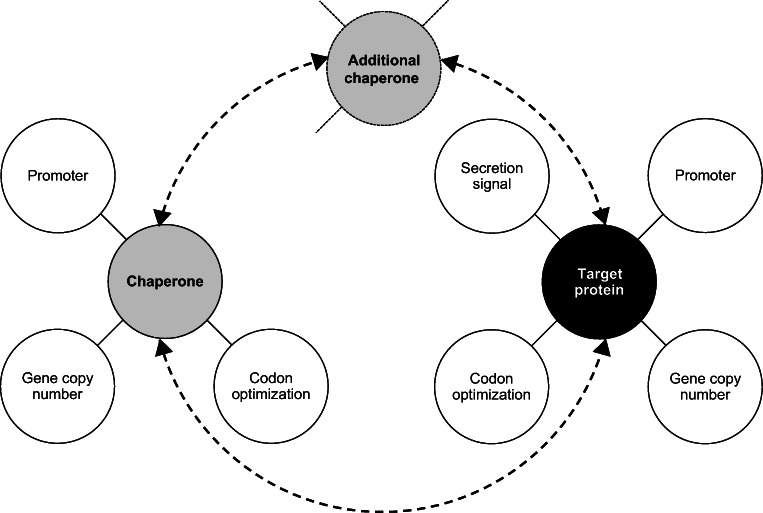
Use a combinatorial approach to optimize the co-expression strategy. Try different co-expression helper genes and promoters (also weak ones) for their expression, different copy numbers of the helper gene, different homologues of the helper gene, simultaneous co-expression of multiple helper genes etc. (Fig. 2).
If an extensive combinatorial approach is not feasible, as a minimum we recommend examining several different co-expression partners; this might improve the chances of an unknown bottleneck in protein processing in the ER being overcome. We suggest the co-expression of the following: (1) HAC1i as the transcription factor upregulating the entire UPR, thus also increasing the expression of genes of chaperones, foldases, and others; (2) PDI and/or ERO1, which are involved in the formation of disulfide bonds and oxidative stress in the ER; and (3) the ER lumenal chaperone KAR2 that assists in correct protein folding.
For “one-shot” scenarios, when testing of several different co-expression partners is not feasible, we suggest using HAC1i as a co-expression partner, since it was shown to improve the secretion of different types of proteins including antibody fragments, transporter proteins, lysozyme, a broad range of hydrolytic enzymes, and enhanced the production of a surface-displayed protein (Table 2). However, if the protein of interest is rich in disulfide bridges (Sha et al. 2013c; Guan et al. 2016), the co-expression of PDI1 or ERO1 might be preferred (Gasser et al. 2007b; Guan et al. 2016).
Along with protein titer/productivity, we recommend assessing the effect of the co-expressed helper gene on the strain’s physiology and growth of the production strain (Raschmanová et al. 2019), as these, and thus overall productivity and robustness of a bioprocess may be impaired. A negative effect on biomass growth was reported for all of the three most frequently used co-expression partners, Hac1p, Pdi1, and Kar2p. When evaluating protein production/secretion, it is reasonable to calculate the specific productivity (mass of product produced per mass of biomass per hour), which reflects the effect of the co-expressed gene on product secretion, as well as on biomass growth. For difficult-to-secrete proteins, it is useful to assess the proportion of secreted as well as intracellularly retained protein (Duan et al. 2019; Borčinová et al. 2020), and calculate a secretion ratio, i.e., the ratio of secreted to total protein, as a relevant characteristic diffentiating between the effect of the co-expressed gene on total production, versus its secretion (Duan et al. 2019).
Fig. 2.
Combinatorial map of a co-expression strategy for genes of the target protein together with chaperones in P. pastoris. Co-expression of a chaperone gene is a possible method to enhance production/secretion of a target recombinant protein, in addition to the choice of appropriate promoter and secretion signal sequences, codon optimization, and optimized copy number of the gene. Various chaperone genes with codon-optimized sequences and optimized gene copy numbers should be considered and tested with promoters of different strengths (strong, moderate, weak). The promoter used to control expression of the target gene might be different from those used with chaperones. The recombinant protein also needs a secretion signal, but not the chaperon which acts within the cell
Conclusions and outlook
A commonly used strategy to boost folding and protein processing in the ER, and thus to overcome secretory bottlenecks in P. pastoris, is the overexpression of genes encoding proteins involved in the UPR, such as the transcription activator of UPR genes, Hac1p, or chaperones and foldases, e.g., Kar2p, Pdi1, or Ero1p. In this review, we comprehensively analyzed the successes and failures of such co-expression strategies in P. pastoris. Currently, as basic research about the UPR in P. pastoris is limited and no general instructions that guarantee enhanced protein secretion can be followed, it is necessary to design and optimize a co-expression strategy for each individual product, since different proteins may benefit from different levels of UPR activity. Nevertheless, we have summarized recommendations on the best practices for co-expression strategies. In terms of future prospects for recombinant protein production and secretion, the application of novel folding-factors and promoters weaker than the classic PGAP and PAOX1 for their co-expression could promote folding and secretion of diverse recombinant proteins that require fine-tuning of the UPR.
Acknowledgements
The authors wish to thank John Brooker for English proofreading.
Author contribution
The idea of describing some rationale behind construction of Pichia strains has driven Hana Raschmanová’s curiosity since the beginning of her PhD thesis, jointly supervised by Karin Kovar, Zdeněk Knejzlík, and Karel Melzoch. Hana Raschmanová performed an in-depth literature search and data analysis that was amended and critically revised by Astrid Weninger and Karin Kovar.
Upon invitation from the AMAB journal (by Prof. Alexander Steinbüchel), all authors jointly shaped the texts to create a conceptual framework for a better understanding of the rationale behind strain construction.
Funding
This work was supported by specific university research (MSMT No 21-SVV/2018) and the Swiss Government Excellence Scholarship ESKAS-Nr: 2016.0162.
Availability of data and material
Not applicable.
Code availability
Not applicable.
Declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Adelantado N, Tarazona P, Grillitsch K, García-Ortega X, Monforte S, Valero F, Feussner I, Daum G, Ferrer P. The effect of hypoxia on the lipidome of recombinant Pichia pastoris. Microb Cell Factories. 2017;16:86. doi: 10.1186/s12934-017-0699-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad M, Hirz M, Pichler H, Schwab H. Protein expression in Pichia pastoris: recent achievements and perspectives for heterologous protein production. Appl Microbiol Biotechnol. 2014;98:5301–5317. doi: 10.1007/s00253-014-5732-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonny B, Schekman R. ER export: public transportation by the COPII coach. Curr Opin Cell Biol. 2001;13:438–443. doi: 10.1016/s0955-0674(00)00234-9. [DOI] [PubMed] [Google Scholar]
- Aragon T, van Anken E, Pincus D, Serafimova IM, Korennykh AV, Rubio CA, Walter P. Messenger RNA targeting to endoplasmic reticulum stress signalling sites. Nature. 2009;457:736–7U9. doi: 10.1038/nature07641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aw R, Barton GR, Leak DJ. Insights into the prevalence and underlying causes of clonal variation through transcriptomic analysis in Pichia pastoris. Appl Microbiol Biotechnol. 2017;101:5045–5058. doi: 10.1007/s00253-017-8317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankefa OE, Wang M, Zhu T, Li Y. Hac1p homologues from higher eukaryotes can improve the secretion of heterologous proteins in the yeast Pichia pastoris. Biotechnol Lett. 2018;40:1149–1156. doi: 10.1007/s10529-018-2571-y. [DOI] [PubMed] [Google Scholar]
- Barrero JJ, Casler JC, Valero F, Ferrer P, Glick BS. An improved secretion signal enhances the secretion of model proteins from Pichia pastoris. Microb Cell Factories. 2018;17:161. doi: 10.1186/s12934-018-1009-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann K, Dato L, Graf AB, Frascotti G, Dragosits M, Porro D, Mattanovich D, Ferrer P, Branduardi P. The impact of oxygen on the transcriptome of recombinant S. cerevisiae and P. pastoris - a comparative analysis. BMC Genomics. 2011;12:218. doi: 10.1186/1471-2164-12-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Azoun S, Belhaj AE, Göngrich R, Gasser B, Kallel H. Molecular optimization of rabies virus glycoprotein expression in Pichia pastoris. Microb Biotechnol. 2016;9:355–368. doi: 10.1111/1751-7915.12350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Azoun S, Belhaj AE, Kallel H. Rabies virus glycoprotein enhanced expression in Pichia pastoris using the constitutive GAP promoter. Biochem Eng J. 2016;113:77–85. doi: 10.1016/j.bej.2016.05.013. [DOI] [Google Scholar]
- Ben Azoun S, Ben Zakour M, Sghaier S, Kallel H. Expression of rabies virus glycoprotein in the methylotrophic yeast Pichia pastoris. Biotechnol Appl Biochem. 2017;64:50–61. doi: 10.1002/bab.1471. [DOI] [PubMed] [Google Scholar]
- Bernales S, Papa FR, Walter P. Intracellular signaling by the unfolded protein response. Annu Rev Cell Dev Biol. 2006;22:487–508. doi: 10.1146/annurev.cellbio.21.122303.120200. [DOI] [PubMed] [Google Scholar]
- Borčinová M, Raschmanová H, Zamora I, Looser V, Marešová H, Hirsch S, Kyslík P, Kovar K. Production and secretion dynamics of prokaryotic Penicillin G acylase in Pichia pastoris. Appl Microbiol Biotechnol. 2020;104:5787–5800. doi: 10.1007/s00253-020-10669-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casagrande R, Stern P, Diehn M, Shamu C, Osario M, Zú M, Brown PO, Ploegh H. Degradation of proteins from the ER of S. cerevisiae requires an intact unfolded protein response pathway. Mol Cell. 2000;5(4):729–735. doi: 10.1016/s1097-2765(00)80251-8. [DOI] [PubMed] [Google Scholar]
- Cereghino JL, Cregg JM. Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol Rev. 2000;24:45–66. doi: 10.1111/j.1574-6976.2000.tb00532.x. [DOI] [PubMed] [Google Scholar]
- Chang HJ, Jesch SA, Gaspar ML, Henry SA. Role of the unfolded protein response pathway in secretory stress and regulation of INO1 expression in Saccharomyces cerevisiae. Genetics. 2004;168:1899–1913. doi: 10.1534/genetics.104.032961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla A, Chakrabarti S, Ghosh G, Niwa M. Attenuation of yeast UPR is essential for survival and is mediated by IRE1 kinase. J Cell Biol. 2011;193:41–50. doi: 10.1083/jcb.201008071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheon SA, Jung KW, Chen YL, Heitman J, Bahn YS, Kang HA. Unique evolution of the UPR pathway with a novel bZIP transcription factor, HxL1, for controlling pathogenicity of Cryptococcus neoformans. PLoS Pathog. 2011;7:e1002177. doi: 10.1371/journal.ppat.1002177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-A. [DOI] [PubMed] [Google Scholar]
- Daly R, Hearn MT. Expression of heterologous proteins in Pichia pastoris: a useful experimental tool in protein engineering and production. J Mol Recognit. 2005;18:119–138. doi: 10.1002/jmr.687. [DOI] [PubMed] [Google Scholar]
- Damasceno LM, Anderson KA, Ritter G, Cregg JM, Old LJ, Batt CA. Cooverexpression of chaperones for enhanced secretion of a single-chain antibody fragment in Pichia pastoris. Appl Microbiol Biotechnol. 2007;74:381–389. doi: 10.1007/s00253-006-0652-7. [DOI] [PubMed] [Google Scholar]
- Damasceno LM, Huang CJ, Batt CA. Protein secretion in Pichia pastoris and advances in protein production. Appl Microbiol Biotechnol. 2012;93:31–39. doi: 10.1007/s00253-011-3654-z. [DOI] [PubMed] [Google Scholar]
- De Schutter K, Lin YC, Tiels P, Van Hecke A, Glinka S, Weber-Lehmann J, Rouze P, Van de Peer Y, Callewaert N. Genome sequence of the recombinant protein production host Pichia pastoris. Nat Biotechnol. 2009;27:561–566. doi: 10.1038/nbt.1544. [DOI] [PubMed] [Google Scholar]
- De Waele S, Vandenberghe I, Laukens B, Planckaert S, Verweire S, Van Bogaert INA, Soetaert W, Devreese B, Ciesielska K. Optimized expression of the Starmerella bombicola lactone esterase in Pichia pastoris through temperature adaptation, codon-optimization and co-expression with HAC1. Protein Expr Purif. 2018;143:62–70. doi: 10.1016/j.pep.2017.10.016. [DOI] [PubMed] [Google Scholar]
- Delic M, Rebnegger C, Wanka F, Puxbaum V, Haberhauer-Troyer C, Hann S, Kollensperger G, Mattanovich D, Gasser B. Oxidative protein folding and unfolded protein response elicit differing redox regulation in endoplasmic reticulum and cytosol of yeast. Free Radic Biol Med. 2012;52:2000–2012. doi: 10.1016/j.freeradbiomed.2012.02.048. [DOI] [PubMed] [Google Scholar]
- Delic M, Valli M, Graf AB, Pfeffer M, Mattanovich D, Gasser B. The secretory pathway: exploring yeast diversity. FEMS Microbiol Rev. 2013;37:872–914. doi: 10.1111/1574-6976.12020. [DOI] [PubMed] [Google Scholar]
- Duan G, Ding L, Wei D, Zhou H, Chu J, Zhang S, Qian J. Screening endogenous signal peptides and protein folding factors to promote the secretory expression of heterologous proteins in Pichia pastoris. J Biotechnol. 2019;306:193–202. doi: 10.1016/j.jbiotec.2019.06.297. [DOI] [PubMed] [Google Scholar]
- Elena C, Ravasi P, Cerminati S, Peiru S, Castelli ME, Menzella HG. Pichia pastoris engineering for the production of a modified phospholipase C. Process Biochem. 2016;51:1935–1944. doi: 10.1016/j.procbio.2016.08.022. [DOI] [Google Scholar]
- Emmerstorfer A, Wriessnegger T, Hirz M, Pichler H. Overexpression of membrane proteins from higher eukaryotes in yeasts. Appl Microbiol Biotechnol. 2014;98(18):7671–7698. doi: 10.1007/s00253-014-5948-4. [DOI] [PubMed] [Google Scholar]
- Fauzee YNBM, Taniguchi N, Ishiwata-Kimata Y, Takagi H, Kimata Y. The unfolded protein response in Pichia pastoris without external stressing stimuli. FEMS Yeast Res. 2020;7:foaa053. doi: 10.1093/femsyr/foaa053. [DOI] [PubMed] [Google Scholar]
- Fischer JE, Glieder A. Current advances in engineering tools for Pichia pastoris. Curr Opin Biotechnol. 2019;59:175–181. doi: 10.1016/j.copbio.2019.06.002. [DOI] [PubMed] [Google Scholar]
- Gasser B, Maurer M, Gach J, Kunert R, Mattanovich D. Engineering of Pichia pastoris for improved production of antibody fragments. Biotechnol Bioeng. 2006;94:353–361. doi: 10.1002/bit.20851. [DOI] [PubMed] [Google Scholar]
- Gasser B, Maurer M, Rautio J, Sauer M, Bhattacharyya A, Saloheimo M, Penttila M, Mattanovich D. Monitoring of transcriptional regulation in Pichia pastoris under protein production conditions. BMC Genomics. 2007;8:18. doi: 10.1186/1471-2164-8-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser B, Sauer M, Maurer M, Stadlmayr G, Mattanovich D. Transcriptomics-based identification of novel factors enhancing heterologous protein secretion in yeasts. Appl Environ Microbiol. 2007;73:6499–6507. doi: 10.1128/AEM.01196-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser B, Prielhofer R, Marx H, Maurer M, Nocon J, Steiger M, Puxbaum V, Sauer M, Mattanovich D. Pichia pastoris: protein production host and model organism for biomedical research. Future Microbiol. 2013;8:191–208. doi: 10.2217/fmb.12.133. [DOI] [PubMed] [Google Scholar]
- Gidijala L, Uthoff S, Kampen SJ, Steinbüchel A, Verhaert RMD. Presence of protein production enhancers results in significantly higher methanol-induced protein production in Pichia pastoris. Microb Cell Factories. 2018;17:112. doi: 10.1186/s12934-018-0961-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf A, Gasser B, Dragosits M, Sauer M, Leparc GG, Tuchler T, Kreil DP, Mattanovich D. Novel insights into the unfolded protein response using Pichia pastoris specific DNA microarrays. BMC Genomics. 2008;9:390. doi: 10.1186/1471-2164-9-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan B, Chen F, Su S, Duan Z, Chen Y, Li H, Jin J. Effects of co-overexpression of secretion helper factors on the secretion of a HSA fusion protein (IL2-HSA) in Pichia pastoris. Yeast. 2016;33:587–600. doi: 10.1002/yea.3183. [DOI] [PubMed] [Google Scholar]
- Guerfal M, Ryckaert S, Jacobs PP, Ameloot P, Van Craenenbroeck K, Derycke R, Callewaert N. The HAC1 gene from Pichia pastoris: characterization and effect of its overexpression on the production of secreted, surface displayed and membrane proteins. Microb Cell Factories. 2010;9:49. doi: 10.1186/1475-2859-9-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M, Wang W, Zhou J, Gong X, Xu C, Li Y, Li Q. Activation of the unfolded protein response via co-expression of the HAC1i gene enhances expression of recombinant elastase in Pichia pastoris. Biotechnol Bioprocess Eng. 2020;25:302–307. doi: 10.1007/s12257-019-0381-2. [DOI] [Google Scholar]
- He H, Wu S, Mei M, Ning J, Li C, Ma L, Zhang G, Yi L. A combinational strategy for effective heterologous production of functional human lysozyme in Pichia pastoris. Front Bioeng Biotechnol. 2020;8:118. doi: 10.3389/fbioe.2020.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A, Marquardt T, Braakman I. The endoplasmic reticulum as a protein-folding compartment. Trends Cell Biol. 1992;2:227–231. doi: 10.1016/0962-8924(92)90309-B. [DOI] [PubMed] [Google Scholar]
- Hernández-Elvira M, Torres-Quiroz F, Escamilla-Ayala A, Domínguez-Martin E, Escalante R, Kawasaki L, Ongay-Larios L, Coria R. The unfolded protein response pathway in the yeast Kluyveromyces lactis. A comparative view among yeast species. Cells. 2018;7:106. doi: 10.3390/cells7080106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesketh AR, Castrillo JI, Sawyer T, Archer DB, Oliver SG. Investigating the physiological response of Pichia (Komagataella) pastoris GS115 to the heterologous expression of misfolded proteins using chemostat cultures. Appl Microbiol Biotechnol. 2013;97:9747–9762. doi: 10.1007/s00253-013-5186-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CJ, Anderson KA, Damasceno LM, Ritter G, Old LJ, Batt CA. Improved secretion of the cancer-testis antigen SSX2 in Pichia pastoris by deletion of its nuclear localization signal. Appl Microbiol Biotechnol. 2010;86:243–253. doi: 10.1007/s00253-009-2275-2. [DOI] [PubMed] [Google Scholar]
- Huang M, Gao Y, Zhou X, Zhang Y, Cai M. Regulating unfolded protein response activator HAC1p for production of thermostable raw-starch hydrolyzing Α-amylase in Pichia pastoris. Bioprocess Biosyst Eng. 2017;40:341–350. doi: 10.1007/s00449-016-1701-y. [DOI] [PubMed] [Google Scholar]
- Huang J, Zhao Q, Chen L, Zhang C, Bu W, Zhang X, Zhang K, Yang Z. Improved production of recombinant Rhizomucor miehei lipase by coexpressing protein folding chaperones in Pichia pastoris, which triggered ER stress. Bioengineered. 2020;11:375–385. doi: 10.1080/21655979.2020.1738127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu J, Xu Y, Li C, Li J. Overexpressing target helper genes enhances secretion and glycosylation of recombinant proteins in Pichia pastoris under simulated microgravity. J Ind Microbiol Biotechnol. 2016;43:1429–1439. doi: 10.1007/s10295-016-1817-8. [DOI] [PubMed] [Google Scholar]
- Inan M, Aryasomayajula D, Sinha J, Meagher MM. Enhancement of protein secretion in Pichia pastoris by overexpression of protein disulfide isomerase. Biotechnol Bioeng. 2006;93:771–778. doi: 10.1002/bit.20762. [DOI] [PubMed] [Google Scholar]
- Kawahara T, Yanagi H, Yura T, Mori K. Endoplasmic reticulum stress-induced mRNA splicing permits synthesis of transcription factor Hac1p/Ern4p that activates the unfolded protein response. Mol Biol Cell. 1997;8:1845–1862. doi: 10.1091/mbc.8.10.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri NK, Gocke D, Trentmann O, Neubauer P, Hoffmann F. Single-chain antibody fragment production in Pichia pastoris: benefits of prolonged pre-induction glycerol feeding. Biotechnol J. 2011;6:452–462. doi: 10.1002/biot.201000193. [DOI] [PubMed] [Google Scholar]
- Kimata Y, Oikawa D, Shimizu Y, Ishiwata-Kimata Y, Kohno K. A role for BiP as an adjustor for the endoplasmic reticulum stress-sensing protein Ire1. J Cell Biol. 2004;167:445–456. doi: 10.1083/jcb.200405153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata Y, Ishiwata-Kimata Y, Ito T, Hirata A, Suzuki T, Oikawa D, Takeuchi M, Kohno K. Two regulatory steps of ER-stress sensor Ire1 involving its cluster formation and interaction with unfolded proteins. J Cell Biol. 2007;179:75–86. doi: 10.1083/jcb.200704166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Kuwae S, Ohya T, Ohda T, Ohyama M, Ohi H, Tomomitsu K, Ohmura T. High-level expression of recombinant human serum albumin from the methylotrophic yeast Pichia pastoris with minimal protease production and activation. J Biosci Bioeng. 2000;89:55–61. doi: 10.1016/S1389-1723(00)88050-0. [DOI] [PubMed] [Google Scholar]
- Kohno K. Stress-sensing mechanisms in the unfolded protein response: similarities and differences between yeast and mammals. J Biochem. 2010;147:27–33. doi: 10.1093/jb/mvp196. [DOI] [PubMed] [Google Scholar]
- Krainer FW, Gerstmann MA, Darnhofer B, Birner-Gruenberger R, Glieder A. Biotechnological advances towards an enhanced peroxidase production in Pichia pastoris. J Biotechnol. 2016;233:181–189. doi: 10.1016/j.jbiotec.2016.07.012. [DOI] [PubMed] [Google Scholar]
- Kruse KB, Brodsky JL, McCracken AA. Autophagy: an ER protein quality control process. Autophagy. 2006;2:135–137. doi: 10.4161/auto.2.2.2388. [DOI] [PubMed] [Google Scholar]
- Lan D, Qu M, Yang B, Wang Y. Enhancing production of lipase MAS1 from marine Streptomyces sp. strain in Pichia pastoris by chaperones co-expression. Electron J Biotechnol. 2016;22:62–67. doi: 10.1016/j.ejbt.2016.06.003. [DOI] [Google Scholar]
- Li C, Lin Y, Zheng X, Pang N, Liao X, Liu X, Huang Y, Liang S. Combined strategies for improving expression of Citrobacter amalonaticus phytase in Pichia pastoris. BMC Biotechnol. 2015;15:88. doi: 10.1186/s12896-015-0204-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin XQ, Liang SL, Han SY, Zheng SP, Ye YR, Lin Y. Quantitative iTRAQ LC-MS/MS proteomics reveals the cellular response to heterologous protein overexpression and the regulation of HAC1 in Pichia pastoris. J Proteome. 2013;91:58–72. doi: 10.1016/j.jprot.2013.06.031. [DOI] [PubMed] [Google Scholar]
- Liu YY, Woo JH, Neville DM., Jr Overexpression of an anti-CD3 immunotoxin increases expression and secretion of molecular chaperone BiP/Kar2p by Pichia pastoris. Appl Environ Microbiol. 2005;71:5332–5340. doi: 10.1128/AEM.71.9.5332-5340.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wu D, Wu J, Chen J. Optimization of the production of Aspergillus niger alpha-glucosidase expressed in Pichia pastoris. World J Microbiol Biotechnol. 2013;29:533–540. doi: 10.1007/s11274-012-1207-y. [DOI] [PubMed] [Google Scholar]
- Liu H, Qin Y, Huang Y, Chen Y, Cong P, He Z. Direct evaluation of the effect of gene dosage on secretion of protein from yeast Pichia pastoris by expressing EGFP. J Microbiol Biotechnol. 2014;24:144–151. doi: 10.4014/jmb.1308.08010. [DOI] [PubMed] [Google Scholar]
- Liu WC, Inwood S, Gong T, Sharma A, Yu LY, Zhu P. Fed-batch high-cell-density fermentation strategies for Pichia pastoris growth and production. Crit Rev Biotechnol. 2019;39:258–271. doi: 10.1080/07388551.2018.1554620. [DOI] [PubMed] [Google Scholar]
- Liu J, Han Q, Cheng Q, Chen Y, Wang R, Li X, Liu Y, Yan D. Efficient expression of human lysozyme through the increased gene dosage and co-expression of transcription factor Hac1p in Pichia pastoris. Curr Microbiol. 2020;77:846–854. doi: 10.1007/s00284-019-01872-9. [DOI] [PubMed] [Google Scholar]
- Looser V, Bruhlmann B, Bumbak F, Stenger C, Costa M, Camattari A, Fotiadis D, Kovar K. Cultivation strategies to enhance productivity of Pichia pastoris: a review. Biotechnol Adv. 2015;33:1177–1193. doi: 10.1016/j.biotechadv.2015.05.008. [DOI] [PubMed] [Google Scholar]
- Love KR, Politano TJ, Panagiotou V, Jiang B, Stadheim TA, Love JC. Systematic single-cell analysis of Pichia pastoris reveals secretory capacity limits productivity. PLoS One. 2012;7:e37915. doi: 10.1371/journal.pone.0037915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsalek L, Puxbaum V, Buchetics M, Mattanovich D, Gasser B. Disruption of vacuolar protein sorting components of the HOPS complex leads to enhanced secretion of recombinant proteins in Pichia pastoris. Microb Cell Factories. 2019;18:119. doi: 10.1186/s12934-019-1155-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx H, Sauer M, Resina D, Vai M, Porro D, Valero F, Ferrer P, Mattanovich D. Cloning, disruption and protein secretory phenotype of the GAS1 homologue of Pichia pastoris. FEMS Microbiol Lett. 2006;264:40–47. doi: 10.1111/j.1574-6968.2006.00427.x. [DOI] [PubMed] [Google Scholar]
- Miyazaki T, Nakayama H, Nagayoshi Y, Kakeya H, Kohno S. Dissection of Ire1 functions reveals stress response mechanisms uniquely evolved in Candida glabrata. PLoS Pathog. 2013;9:20. doi: 10.1371/journal.ppat.1003160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogelsvang S, Gomez-Ospina N, Soderholm J, Glick BS, Staehelin LA. Tomographic evidence for continuous turnover of Golgi cisternae in Pichia pastoris. Mol Biol Cell. 2003;14:2277–2291. doi: 10.1091/mbc.E02-10-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon HY, Cheon SA, Kim H, Agaphonov MO, Kwon O, Oh DB, Kim JY, Kang HA. Hansenula polymorpha Hac1p is critical to protein N-glycosylation activity modulation, as revealed by functional and transcriptomic analyses. Appl Environ Microbiol. 2015;81:6982–6993. doi: 10.1128/aem.01440-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Kawahara T, Yoshida H, Yanagi H, Yura T. Signalling from endoplasmic reticulum to nucleus: transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells. 1996;1:803–817. doi: 10.1046/j.1365-2443.1996.d01-274.x. [DOI] [PubMed] [Google Scholar]
- Mori K, Ogawa N, Kawahara T, Yanagi H, Yura T. mRNA splicing-mediated C-terminal replacement of transcription factor Hac1p is required for efficient activation of the unfolded protein response. Proc Natl Acad Sci U S A. 2000;97:4660–4665. doi: 10.1073/pnas.050010197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naranjo CA, Jivan AD, Vo MN, de Sa Campos KH, Deyarmin JS, Hekman RM, Uribe C, Hang A, Her K, Fong MM, Choi JJ, Chou C, Rabara TR, Myers G, Moua P, Thor D, Risser DD, Vierra CA, Franz AH, Lin-Cereghino J, Lin-Cereghino GP. Characterization of the role of BGS13 in the secretory mechanism of Pichia pastoris. Appl Environ Microbiol. 2019;24:e01615–e01619. doi: 10.1128/aem.01615-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obst U, Lu TK, Sieber V. A modular toolkit for generating Pichia pastoris secretion libraries. ACS Synth Biol. 2017;6:1016–1025. doi: 10.1021/acssynbio.6b00337. [DOI] [PubMed] [Google Scholar]
- Okamura K, Kimata Y, Higashio H, Tsuru A, Kohno K. Dissociation of Kar2p/BiP from an ER sensory molecule, Ire1p, triggers the unfolded protein response in yeast. Biochem Biophys Res Commun. 2000;279:445–450. doi: 10.1006/bbrc.2000.3987. [DOI] [PubMed] [Google Scholar]
- Papa FR, Zhang C, Shokat K, Walter P. Bypassing a kinase activity with an ATP-competitive drug. Science. 2003;302:1533–1537. doi: 10.1126/science.1090031. [DOI] [PubMed] [Google Scholar]
- Pfeffer M, Maurer M, Kollensperger G, Hann S, Graf AB, Mattanovich D. Modeling and measuring intracellular fluxes of secreted recombinant protein in Pichia pastoris with a novel 34S labeling procedure. Microb Cell Factories. 2011;10:47. doi: 10.1186/1475-2859-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer M, Maurer M, Stadlmann J, Grass J, Delic M, Altmann F, Mattanovich D. Intracellular interactome of secreted antibody Fab fragment in Pichia pastoris reveals its routes of secretion and degradation. Appl Microbiol Biotechnol. 2012;93:2503–2512. doi: 10.1007/s00253-012-3933-3. [DOI] [PubMed] [Google Scholar]
- Prattipati M, Ramakrishnan K, Sankaranarayanan M. Pichia pastoris protein disulfide isomerase (PDI1) promoter for heterologous protein production and its sequence characterization. Enzym Microb Technol. 2020;140:109633. doi: 10.1016/j.enzmictec.2020.109633. [DOI] [PubMed] [Google Scholar]
- Prinz B, Stahl U, Lang C. Intracellular transport of a heterologousmembrane protein, the human transferrin receptor, in Saccharomyces cerevisiae. Int Microbiol. 2003;6:49–55. doi: 10.1007/s10123-003-0100-9. [DOI] [PubMed] [Google Scholar]
- Puxbaum V, Mattanovich D, Gasser B. Quo vadis? The challenges of recombinant protein folding and secretion in Pichia pastoris. Appl Microbiol Biotechnol. 2015;99:2925–2938. doi: 10.1007/s00253-015-6470-z. [DOI] [PubMed] [Google Scholar]
- Puxbaum V, Gasser B, Mattanovich D. The bud tip is the cellular hot spot of protein secretion in yeasts. Appl Microbiol Biotechnol. 2016;100:8159–8168. doi: 10.1007/s00253-016-7674-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschmanová H, Weninger A, Glieder A, Kovar K, Vogl T. Implementing CRISPR-Cas technologies in conventional and non-conventional yeasts: current state and future prospects. Biotechnol Adv. 2018;36:641–665. doi: 10.1016/j.biotechadv.2018.01.006. [DOI] [PubMed] [Google Scholar]
- Raschmanová H, Zamora I, Borčinová M, Meier P, Weninger A, Mächler D, Glieder A, Melzoch K, Knejzlík Z, Kovar K. Single-cell approach to monitor the unfolded protein response during biotechnological processes with Pichia pastoris. Front Microbiol. 2019;10:335. doi: 10.3389/fmicb.2019.00335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebnegger C, Graf AB, Valli M, Steiger MG, Gasser B, Maurer M, Mattanovich D. In Pichia pastoris, growth rate regulates protein synthesis and secretion, mating and stress response. Biotechnol J. 2014;9:511–525. doi: 10.1002/biot.201300334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resina D, Bollok M, Khatri NK, Valero F, Neubauer P, Ferrer P. Transcriptional response of P. pastoris in fed-batch cultivations to Rhizopus oryzae lipase production reveals UPR induction. Microb Cell Factories. 2007;6:11. doi: 10.1186/1475-2859-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resina D, Maurer M, Cos O, Arnau C, Carnicer M, Marx H, Gasser B, Valero F, Mattanovich D, Ferrer P. Engineering of bottlenecks in Rhizopus oryzae lipase production in Pichia pastoris using the nitrogen source-regulated FLD1 promoter. New Biotechnol. 2009;25:396–403. doi: 10.1016/j.nbt.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Rossanese OW, Soderholm J, Bevis BJ, Sears IB, O’Connor J, Williamson EK, Glick BS. Golgi structure correlates with transitional endoplasmic reticulum organization in Pichia pastoris and Saccharomyces cerevisiae. J Cell Biol. 1999;145:69–81. doi: 10.1083/jcb.145.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallada ND, Harkins LE, Berger BW. Effect of gene copy number and chaperone coexpression on recombinant hydrophobin HFBI biosurfactant production in Pichia pastoris. Biotechnol Bioeng. 2019;116:2029–2040. doi: 10.1002/bit.26982. [DOI] [PubMed] [Google Scholar]
- Samuel P, Prasanna Vadhana AK, Kamatchi R, Antony A, Meenakshisundaram S. Effect of molecular chaperones on the expression of Candida antarctica lipase B in Pichia pastoris. Microbiol Res. 2013;168:615–620. doi: 10.1016/j.micres.2013.06.007. [DOI] [PubMed] [Google Scholar]
- Sha C, Yu XW, Li F, Xu Y. Impact of gene dosage on the production of lipase from Rhizopus chinensis CCTCC M201021 in Pichia pastoris. Appl Biochem Biotechnol. 2013;169:1160–1172. doi: 10.1007/s12010-012-0050-9. [DOI] [PubMed] [Google Scholar]
- Sha C, Yu XW, Lin NX, Zhang M, Xu Y. Enhancement of lipase r27RCL production in Pichia pastoris by regulating gene dosage and co-expression with chaperone protein disulfide isomerase. Enzym Microb Technol. 2013;53:438–443. doi: 10.1016/j.enzmictec.2013.09.009. [DOI] [PubMed] [Google Scholar]
- Sha C, Yu XW, Zhang M, Xu Y. Efficient secretion of lipase r27RCL in Pichia pastoris by enhancing the disulfide bond formation pathway in the endoplasmic reticulum. J Ind Microbiol Biotechnol. 2013;40:1241–1249. doi: 10.1007/s10295-013-1328-9. [DOI] [PubMed] [Google Scholar]
- Shen Q, Wu M, Wang HB, Naranmandura H, Chen SQ. The effect of gene copy number and co-expression of chaperone on production of albumin fusion proteins in Pichia pastoris. Appl Microbiol Biotechnol. 2012;96:763–772. doi: 10.1007/s00253-012-4337-0. [DOI] [PubMed] [Google Scholar]
- Shi X-L, Feng M-Q, Shi J, Shi Z-H, Zhong J, Zhou P. High-level expression and purification of recombinant human catalase in Pichia pastoris. Protein Expr Purif. 2007;54:24–29. doi: 10.1016/j.pep.2007.02.008. [DOI] [PubMed] [Google Scholar]
- Sidrauski C, Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–1039. doi: 10.1016/S0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- Sidrauski C, Cox JS, Walter P. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell. 1996;87:405–413. doi: 10.1016/S0092-8674(00)81361-6. [DOI] [PubMed] [Google Scholar]
- Spohner SC, Müller H, Quitmann H, Czermak P. Expression of enzymes for the usage in food and feed industry with Pichia pastoris. J Biotechnol. 2015;202:118–134. doi: 10.1016/j.jbiotec.2015.01.027. [DOI] [PubMed] [Google Scholar]
- Sun J, Jiang J, Zhai X, Zhu S, Qu Z, Yuan W, Wang Z, Wei C. Coexpression of Kex2 endoproteinase and Hac1 transcription factor to improve the secretory expression of bovine lactoferrin in Pichia pastoris. Biotechnol Bioprocess Eng. 2019;24:934–941. doi: 10.1007/s12257-019-0176-5. [DOI] [Google Scholar]
- Tawde MD, Freimuth P. Toxic misfolding of Arabidopsis cellulases in the secretory pathway of Pichia pastoris. Protein Expr Purif. 2012;85:211–217. doi: 10.1016/j.pep.2012.08.009. [DOI] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/S0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Vad R, Nafstad E, Dahl LA, Gabrielsen OS. Engineering of a Pichia pastoris expression system for secretion of high amounts of intact human parathyroid hormone. J Biotechnol. 2005;116:251–260. doi: 10.1016/j.jbiotec.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Vanz AL, Lunsdorf H, Adnan A, Nimtz M, Gurramkonda C, Khanna N, Rinas U. Physiological response of Pichia pastoris GS115 to methanol-induced high level production of the Hepatitis B surface antigen: catabolic adaptation, stress responses, and autophagic processes. Microb Cell Factories. 2012;11:103. doi: 10.1186/1475-2859-11-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanz AL, Nimtz M, Rinas U. Decrease of UPR- and ERAD-related proteins in Pichia pastoris during methanol-induced secretory insulin precursor production in controlled fed-batch cultures. Microb Cell Factories. 2014;13:23. doi: 10.1186/1475-2859-13-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogl T, Thallinger GG, Zellnig G, Drew D, Cregg JM, Glieder A, Freigassner M. Towards improved membrane protein production in Pichia pastoris: general and specific transcriptional response to membrane protein overexpression. New Biotechnol. 2014;31:538–552. doi: 10.1016/j.nbt.2014.02.009. [DOI] [PubMed] [Google Scholar]
- Wang XD, Jiang T, Yu XW, Xu Y. Effects of UPR and ERAD pathway on the prolyl endopeptidase production in Pichia pastoris by controlling of nitrogen source. J Ind Microbiol Biotechnol. 2017;44:1053–1063. doi: 10.1007/s10295-017-1938-8. [DOI] [PubMed] [Google Scholar]
- Weninger A, Hatzl AM, Schmid C, Vogl T, Glieder A. Combinatorial optimization of CRISPR/Cas9 expression enables precision genome engineering in the methylotrophic yeast Pichia pastoris. J Biotechnol. 2016;235:139–149. doi: 10.1016/j.jbiotec.2016.03.027. [DOI] [PubMed] [Google Scholar]
- Weninger A, Fischer JE, Raschmanová H, Kniely C, Vogl T, Glieder A. Expanding the CRISPR/Cas9 toolkit for Pichia pastoris with efficient donor integration and alternative resistance markers. J Cell Biochem. 2018;119:3183–3198. doi: 10.1002/jcb.26474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyteside G, Alcocer MJ, Kumita JR, Dobson CM, Lazarou M, Pleass RJ, Archer DB. Native-state stability determines the extent of degradation relative to secretion of protein variants from Pichia pastoris. PLoS One. 2011;6:e22692. doi: 10.1371/journal.pone.0022692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Ng DTW. ERAD substrate recognition in budding yeast. Semin Cell Dev Biol. 2010;21:533–539. doi: 10.1016/j.semcdb.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Xu N, Zhu J, Zhu Q, Xing Y, Cai M, Jiang T, Zhou M, Zhang Y. Identification and characterization of novel promoters for recombinant protein production in yeast Pichia pastoris. Yeast. 2018;35:379–385. doi: 10.1002/yea.3301. [DOI] [PubMed] [Google Scholar]
- Yang J, Lu Z, Chen J, Chu P, Cheng Q, Liu J, Ming F, Huang C, Xiao A, Cai H, Zhang L. Effect of cooperation of chaperones and gene dosage on the expression of porcine PGLYRP-1 in Pichia pastoris. Appl Microbiol Biotechnol. 2016;100:5453–5465. doi: 10.1007/s00253-016-7372-4. [DOI] [PubMed] [Google Scholar]
- Yu Y, Liu Z, Chen M, Yang M, Li L, Mou H. Enhancing the expression of recombinant κ-carrageenase in Pichia pastoris using dual promoters, co-expressing chaperones and transcription factors. Biocatal Biotransfor. 2020;38(10):1–10. doi: 10.1080/10242422.2019.1655001. [DOI] [Google Scholar]
- Zahrl RJ, Peña DA, Mattanovich D, Gasser B. Systems biotechnology for protein production in Pichia pastoris. FEMS Yeast Res. 2017;17:fox068. doi: 10.1093/femsyr/fox068. [DOI] [PubMed] [Google Scholar]
- Zahrl RJ, Mattanovich D, Gasser B. The impact of ERAD on recombinant protein secretion in Pichia pastoris (Syn Komagataella spp.) Microbiology (Reading) 2018;164:453–463. doi: 10.1099/mic.0.000630. [DOI] [PubMed] [Google Scholar]
- Zepeda AB, Pessoa A, Farías JG. Carbon metabolism influenced for promoters and temperature used in the heterologous protein production using Pichia pastoris yeast. Braz J Microbiol. 2018;49:119–127. doi: 10.1016/j.bjm.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Zhao HL, Xue C, Xiong XH, Yao XQ, Li XY, Chen HP, Liu ZM. Enhanced secretion of heterologous proteins in Pichia pastoris following overexpression of Saccharomyces cerevisiae chaperone proteins. Biotechnol Prog. 2006;22:1090–1095. doi: 10.1021/bp060019r. [DOI] [PubMed] [Google Scholar]
- Zhang M, Yu Q, Liang C, Zhang B, Li M. Lipid homeostasis is involved in plasma membrane and endoplasmic reticulum stress in Pichia pastoris. Biochem Biophys Res Commun. 2016;478:777–783. doi: 10.1016/j.bbrc.2016.08.024. [DOI] [PubMed] [Google Scholar]
- Zhong YJ, Yang L, Guo YG, Fang F, Wang D, Li R, Jiang M, Kang WY, Ma JJ, Sun J, Xiao WH. High-temperature cultivation of recombinant Pichia pastoris increases endoplasmic reticulum stress and decreases production of human interleukin-10. Microb Cell Factories. 2014;13:10. doi: 10.1186/s12934-014-0163-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Guo M, Zhuang Y, Chu J, Zhang S. Understanding the effect of foreign gene dosage on the physiology of Pichia pastoris by transcriptional analysis of key genes. Appl Microbiol Biotechnol. 2011;89:1127–1135. doi: 10.1007/s00253-010-2944-1. [DOI] [PubMed] [Google Scholar]
- Zimmermann R, Eyrisch S, Ahmad M, Helms V. Protein translocation across the ER membrane. Biochim Biophys Acta Biomembr. 2011;1808:912–924. doi: 10.1016/j.bbamem.2010.06.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.
Not applicable.