Abstract
We studied a group of tick-associated viruses with characteristics of members of the family Iflaviridae, a family of viruses frequently found in arthropods. Our aim was to gain insight into the evolutionary dynamics of this group of viruses, which may be linked to the biology of ticks. We explored assembled RNA-Seq data sets for different species of ticks. We identified members of five different iflavirus species, four of them novel, and discovered nine new genome sequences, including variants. Five variants represented a virus species associated with Ixodes ricinus. Unexpectedly, a sequence found in the Ixodes scapularis cell line ISE6 was nearly identical to the sequences of I. ricinus variants, suggesting a contamination of this cell line by I. ricinus material. Analysing patterns of substitutions between these variants, we detected a strong excess of synonymous mutations, suggesting evolution under strong positive selection. The phylogenies of the viruses and of their tick hosts were not congruent, suggesting recurrent host changes across tick genera during their evolution. Overall, our work constitutes a step in the understanding of the interactions between this family of viruses and ticks.
Introduction
Ticks are blood-feeding parasites, which makes of them a "hub" for a number of microorganisms (bacterial pathogens and symbionts, viruses, protozoa) that potentially interact with ticks and their vertebrate hosts. The discovery of new RNA viruses has rapidly increased in recent years, and they appear to be far more prevalent and diverse than previously expected, especially in invertebrates [1, 2]. High-throughput transcriptome sequencing (or RNA-Seq) has proven to be the tool of choice for characterizing the genomes of these viruses in many types of arthropods [2, 3], including ticks [4, 5], as this technique does not require known sequences to be targeted, while allowing the retrieval of contigs that may contain full viral genome sequences, given a sufficient coverage and sufficient prevalence of the virus in the analysed samples. The Iflaviridae are a family of RNA viruses within the order Picornavirales. They are non-enveloped and have a positive-sense, single-stranded non-segmented genome of ~9-11 kb [6]. This family of viruses has been shown to have a privileged association with insects, being detected in a growing number of species belonging to several orders. Iflavirus infections can range from asymptomatic to severe, causing developmental anomalies (e.g., deformed wing virus in honey bees) or death [6]. Iflaviruses have also been detected in parasitic mites (subphylum Chelicerata) [7]. Recently, genomes of viruses belonging to the family Iflaviridae have been detected in pools of tick species [2], in an endemic Australian tick species (Ixodes holocyclus iflavirus [IhIV]) [8], in a tick species associated with marine birds, Ixodes uriae [9, 10] (Gerbovitch virus), and in the ISE6 tick cell line (Ixodes scapularis iflavirus [ISIV]) [11]. Overall, members of the family Iflaviridae are among the most common viruses that have been identified in different species of ticks, although many aspects of their biology are still unknown, including their mode of transmission, their effect on the fitness of the tick host, and the phylogenetic relationships among members of this family. For this reason, we decided to explore the many transcriptome data sets that have been published for different tick species, searching for new sequences of members of the family Iflaviridae. We identified nine new genome sequences representing five different species of tick-associated iflaviruses, including several variants of the same virus species in Ixodes ricinus, and studied the evolution and phylogenetic relationships of this group.
Materials and methods
Search for iflavirus sequences
We searched for sequences matching iflaviruses in public tick transcriptome assemblies (available from TSA, GenBank, as of August 2020). For each transcriptome assembly, we searched for matches using tblastn [12], using the polyprotein sequence of ISIV (accession number BBD75427) as a query and RNA-Seq contigs (nucleotide sequences) as targets. Of note, several of these transcriptomes were produced by our research group for a phylogenetic study of hard ticks [13] or for expression profiling studies of I. ricinus (BioProject accession numbers PRJEB40724 and PRJNA662253). We used low-stringency matching criteria (e-value threshold 1-e3) to ensure that even relatively distantly related sequences could be found. Since new genome sequences identified in our study were retrieved from transcriptome data, they represent uncultivated virus genomes (UViGs). Genomes were defined as representing novel species if their sequence identity to members of existing species was less than 90%, according to the species delineation criteria of the International Committee on Taxonomy of Viruses (ICTV) for iflaviruses (https://talk.ictvonline.org/ictv-reports/ictv_online_report/positive-sense-rna-viruses/w/iflaviridae). We also performed a BLASTp search on the nr database, selecting all of the matches described as being associated with ticks and adding them to our data set. This approach ensured that our study included all of the tick-associated iflavirus genome sequences published to date (as of September 2020).
Phylogeny methods
Based on a recent phylogenetic study of iflaviruses [14], we included two outgroups, one associated with the mite Tetranychus truncatus (representing the closest outgroup to tick-associated iflaviruses) and the other associated with Apis mellifera (deformed wing virus [DWV]). We aligned amino acid sequences of all iflavirus polyprotein sequences using MUSCLE in MEGA X [15]. This alignment was filtered using Gblocks [16] to exclude poorly aligned regions and gaps. The filtered alignment comprised 1,149 amino acid positions. A phylogenetic ML tree was constructed using IQ-TREE [17]. The best model of substitution was determined using Model Finder [18], and branch support was assessed using 1000 ultrafast bootstrap replicates [19]. A graphical representation of the consensus ML tree was made using ITOL [20].
Test of congruence between host and virus phylogenies
To test the congruence between virus and host phylogenies, we used the cophylogeny testing tool Jane 4 [21]. The host phylogeny was derived from a previous transcriptome-based phylogenetic study of hard ticks, using a large number of nuclear markers [13]. Three iflavirus genomes, obtained from pools of different species of ticks, could not be included in this analysis because it was impossible to assign a host species to these iflaviruses. We also collapsed branches containing virus sequences that were nearly identical in the virus phylogeny, since they can be considered to represent the same species. This resulted not only in eliminating two iflavirus sequences associated with I. holocyclus but also another group of six sequences, five of which were found in I. ricinus and one of which was found in a cell line of I. scapularis (see “Results”). The name given to this group was IricIV-ISIV.
Estimation of genetic distances and evolutionary rates
We estimated genetic distances and evolutionary rates within two ensembles of sequences, one including ISIV and I. ricinus variants and the other including two variants associated with I. holocyclus, that grouped closely in the phylogenetic analysis. We first estimated genetic distances (number of substitutions per site) at both the nucleotide level and the amino acid level, using the maximum composite likelihood model [22] and the Poisson correction model [23], respectively, with the complete deletion option (all gaps excluded). This analysis was performed in MEGA X [15]. We then determined the ratios of non-synonymous to synonymous substitutions (dN/dS) using Codeml [24] with the one-ratio model for the ISIV + I. ricinus group and the pairwise estimate of dN/dS for the two variants found in I. holocyclus.
Virus presence across different populations and stages of Ixodes ricinus
The abundance of virus genomes was assessed by mapping RNA-Seq reads to the genome sequence of the iflavirus IricIV-1. Because a very low abundance of a virus can be caused by library cross-contamination (index hopping), a library was considered positive only if the read count per million was above one, following an approach used in a recent study [10].
Results
Iflavirus sequences identified
We found five variants of a novel iflavirus associated with Ixodes ricinus (Tables 1 and 2). These sequences were associated with different tissues or stages of the tick life cycle (in the GIDG transcriptome assembly) and with ticks from different geographical regions. All viral genome sequences obtained from I. ricinus were highly similar to the ISIV sequence obtained from a cell line of I. scapularis (~98% amino acid identity, while higher variation was detected at the nucleotide level, as shown below). Although the ISIV sequence was found in the transcriptome of a cell line from I. scapularis, there was no match with this sequence in the three available de novo assemblies obtained for three independently sequenced transcriptomes of I. scapularis (two of which were obtained from large collections of wild ticks, with a total of ~200 males and females from three locations for the GGIX TSA assembly). Novel iflaviruses were also identified in Ixodes frontalis (IfronIV), Ixodes vespertilionis (IvespIV), and Hyalomma dromedarii (HydromIV) (Table 2). For the first two of these viruses, the retrieved sequences were incomplete (5' partial), and the sequenced region of the ORF comprised 2,437, and 1,945 amino acids, representing ~81% and ~65% of the complete genome, respectively. In the case of HydromIV, the match with the ISIV sequence was in two frames, suggesting a frameshift. This frameshift could be authentic, especially if this sequence corresponds to an endogenous viral element (EVE), given that EVEs do not necessarily maintain open reading frames [25]. A detailed analysis of reads mapping to the region of the frameshift showed that it was located in a homopolymeric (poly-A) region and that the reads were polymorphic in that region, most containing a deletion of one A compared to the contig. The corrected sequence, based on the majority of reads, contained an intact ORF and no frameshift. We therefore corrected this contig, and all subsequent analysis of this sequence was based on the corrected sequence. Additionally, a 5' partial sequence was also found in a transcriptome assembly of I. holocyclus (2,039 amino acids, IhIV-2). This sequence was similar but not identical to that of the first iflavirus genome (IhIV) identified for that tick species. The new sequences were deposited in the GenBank database (accession numbers in Table 1) with metadata specifying the source of these viruses, the assembly methods used, and the quality of the sequence data (following the guidelines for the Minimum Information about an Uncultivated Virus Genome, or MIUVIG [26]).
Table 1.
List of iflavirus sequences found in tick transcriptomes (with tblastn or blastp, using the ISIV polyprotein sequence as a query). Columns: Species, Tissues (HEM: haematocytes, MG: midgut, MT: Malpighian tubules, OV: ovaries, SG: salivary glands, SYN: synganglion, WB: whole bodies) followed by details on the stages or conditions between parentheses, when available, Location of the sampling (or source of the strains), Accessions: GenBank accessions, protein and nucleotide, and if available, related TSA or BioProject accession between parentheses, Publication (or authors of the sequences), Percent identity with ISIV -% id at the amino acid level of the first hsp (tblastn)- and query range of the match. Lines in bold correspond to the nine iflavirus genome sequences newly discovered in the present study. For HydromIV, the sequence used was a corrected contig sequence
| Species | Tissues and conditions | Location | Accession numbers | Publication | Identity | Match range(s) | Virus name |
|---|---|---|---|---|---|---|---|
| Amblyomma americanum | WB (wild questing ticks) | NY and Connecticut, USA | ASU47553.1, KX774633.1 | Tokarz et al. 2018 | 65.8% | 1142-2838 | Lone star tick dicistrovirus |
| Haemaphysalis flava | WB | Japan | BBK20270.1, LC483655.1 | Kobayashi et al. 2020 | 45.6% | 63-2937 | HflFV |
| Hyalomma asiaticum | WB | China | APG77501.1, KX883729.1 | Shi et al. 2016 | 48.4% | 379-2990 | Bole Hyalomma asiaticum |
| Hyalomma dromedarii | SG, wild ticks | Tunisia | BK012003 (GFGI01) | Bensaoud et al. 2018 | 40.0% | 1-1880 and 2000-2989 | HydromIV |
| Ixodes frontalis | WB | Carquefou, France | QPI13027.1, MT008333 (PRJNA528282) | Charrier et al. 2019 | 41.1% | 560-2989 | IfronIV |
| Ixodes holocyclus | SG,WB,MG | QLD and NSW, Australia | AQZ42314.1, KY020412.1 (GIBQ01) | O'Brien et al. 2018 | 62.2% | 1-2989 | IhIV |
| Ixodes holocyclus | WB | NSW, Australia | QPI13026.1, MT008332 (PRJNA528282) | Charrier et al. 2019 | 65.4% | 913-2989 | IhIV-2 |
| Ixodes ricinus | SYN (wild unfed ticks) | Chizé, France | QPI13029.1, MT008330.1 (PRJEB40724) | Rispe et al (unpub.) | 97.7% | 1-2991 | IricIV-1 |
| Ixodes ricinus | SG, OV, MT (feeding adult females) | Neuchâtel (Switzerland), lab strain | QPI13030.1, MT008331 (PRJNA662253) | Daveu et al (unpub.) | 98.4% | 1-2991 | IricIV-2 |
| Ixodes ricinus | WB (various stages) | Czech Republic | BK012002 (GIDG01012278) | Vechtova et al. (unpub.) | 98.1% | 1-2991 | IricIV-3 |
| Ixodes ricinus | WB (wild nymphs) | Switzerland | QPI13031.1, MT050463 (PRJNA662080) | Rispe et al (unpub.) | 98.3% | 6-2991 | IricIV-4 |
| Ixodes ricinus | WB (wild nymphs) | Nancy, France | QPI13032.1, MT050464 (PRJNA662080) | Rispe et al (unpub.) | 98.2% | 1-2991 | IricIV-5 |
| Ixodes scapularis | Cell line ISE6 | - | BBD75427.1, LC094426.1 | Nakao et al. 2017 | 100.0% | 1-2991 | ISIV |
| Ixodes uriae | WB | Antartic peninsula | QIS88066.1, MT025175.1 | Wille et al. 2020 | 42.0% | 383-2991 | Gerbovich virus |
| Ixodes verspertilionis | WB | Maine-et-Loire, France | QPI13028, MT008334 (PRJNA528282) | Charrier et al. 2019 | 68.2% | 1016-2989 | IvespIV |
| Pool of tick species | WB | China | YP_009336552.1, NC_032764.1 | Shi et al. 2016 | 38.6% | 381-2991 | Ht-V1 |
| Pool of tick species | WB | China | YP_009336542.1, NC_032758.1 | Shi et al. 2016 | 44.3% | 391-2991 | Ht-V2 |
| Pool of tick species | WB | China | YP_009336533.1, NC_032751.1 | Shi et al. 2016 | 46.1% | 63-2989 | Ht-V3 |
Table 2.
Percentage of amino acid identity between iflavirus polyprotein sequences, for each pair of sequences included in our study. In bold, sequences newly identified in this work. Values shaded in gray indicates identity above 90%, defining sequences that belong to the same species based on species delineation criteria of iflaviruses.
All other assemblies obtained from multiple genera of hard and soft ticks gave negative results. Finally, other sequences were found after a BLASTp search against the GenBank protein database (nr), which allowed the inclusion of sequences from viruses associated with Amblyomma americanum (lone star tick dicistrovirus), Haemaphysalis flava (HfFV), Hyalomma asiaticum (Bole Hyalomma virus), and Ixodes holocyclus (IhIV), Ixodes uriae (Gerbovich virus) as well as viruses from pools of tick species (Table 1).
Genome organization
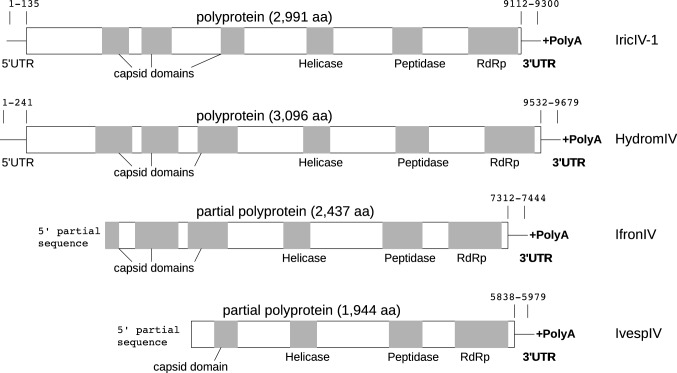
We analyzed the genome organization of the four sequences we consider possible representatives of novel iflavirus species (Fig. 1). Two of them (for IricIV and HydromIV) appear to contain a complete ORF and both 5' and 3' UTRs. These sequences contain conserved domains, including three capsid domains, followed by a helicase, a peptidase, and an RNA-dependant RNA polymerase (RdRp) domain. Two other sequences (from IfronIV and IvespIV) were incomplete at the 5' end, lacking the 5' UTR and at least part of the capsid domains, while the other domains and the 3' UTRs were present.
Fig. 1.
Genome organization of the four novel tick-associated iflaviruses discovered in this study, respectively found in Ixodes ricinus (IricIV-1), Hyalomma dromedarii (HydromIV), Ixodes frontalis (IfronIV), and Ixodes vespertilionis (IvespIV). ORFs were predicted and domains were searched with Interproscan [34]. Predicted domains are shown as grey bars.
Phylogeny
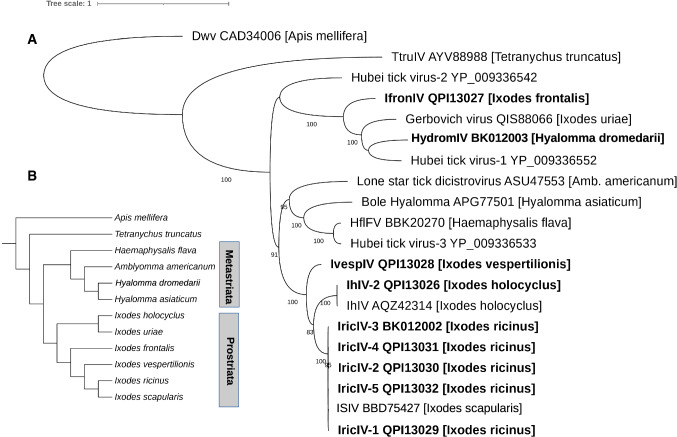
The iflavirus genome sequences included in our work are described in Table 1, including nine newly identified sequences from the present study (Fig. 2). The best-fit model (BIC criteria) was LG+G4. An ML tree (consensus from 1,000 bootstrap trees) showed that the five variants found in I. ricinus and the ISIV sequence grouped closely together (see below for details on genetic distances). This group therefore represents variants of the same virus species. This was also the case of the two variants of I. holocyclus. Another iflavirus sequence, found in I. vespertilionis, formed a sister group with the two groups above. Two other iflavirus genomes, found in I. frontalis and H. dromedarii, respectively, grouped with Ht-V1 [2]. The latter sequence was obtained from a pool of tick species that contained members of several species of the Metastriata (non-Ixodes hard ticks) or soft tick species (Argasidae), but no members of the genus Ixodes [2]. Of note, a sequence found in A. americanum in a study by Tokarz et al. [5] corresponds to an iflavirus, although it was named "dicistrovirus" in that publication.
Fig. 2.
A. Maximum-likelihood phylogenetic tree of tick-associated iflaviruses. The tree was based on the amino acid sequence of iflavirus sequences found in the transcriptomes of different hard tick species (Acari; Parasitiformes; Ixodida; Ixodidae) and rooted with two outgroups, a honey bee iflavirus (deformed wing virus [DWV]) and an iflavirus associated with Tetranychus truncatus (Acari; Acariformes; Prostigmata), TtruIV. The host taxon is indicated in brackets. Details related to each taxon are given in Table 1. Taxon names in bold correspond to the nine iflavirus sequences discovered in the present study. Bootstrap support is indicated at the nodes. B. Expected topology of the phylogenetic tree of arthropod hosts of iflaviruses included in this study, based on reference 13 and on the delimitation of the two groups recognized within the family Ixodidae (Prostriata and Metastriata).
Test of cophylogeny
A test of cophylogeny was performed using Jane 4, allowing us to depict several possible coevolutionary scenarios, all of which had the same scoring and differed very little in structure (we present one of them in Fig. 3). For all scenarios, there was an initial event of cophylogeny due to the fact that all iflavirus genomes associated with ticks form a monophyletic clade, but after this conserved ancestral node, all scenarios included four instances of host switching. An additional anomaly was a "failure to diverge" between viruses associated with I. ricinus and I. scapularis (see “Discussion”).
Fig. 3.
Test of the congruence between the phylogenies of tick-associated iflaviruses and of tick hosts, using Jane 4. This tree contains only part of the sequences analysed in Fig. 2 (three virus genomes found in pools of tick species could not be included). Iric-ISIV represents iflavirus sequences found either in I. ricinus or in a cell line of I. scapularis (these sequences being nearly identical). Open circles at the nodes indicate cophylogeny, HS and solid colored circles, red or yellow, indicate a host shift, FD indicates a failure to diverge
Distances among iflavirus genome sequences and rates
For the group comprising variants found in I. ricinus and the ISIV sequence, the estimated pairwise distances varied between 0.010 and 0.078 at the nucleotide level, suggesting that these sequences are closely related but not identical, whereas amino acid distances were lower, ranging between 0.004 and 0.024 (Table 3). Consistently, the estimated ratio of non-synonymous to synonymous substitutions was low and well below one (with the one-ratio model, dN/dS = 0.024). The sequences of the two variants found in I. holocyclus were extremely similar but not identical (105 differences over 6,117 nucleotide positions, but only five differences at the amino acid level). For this pair of variants, the estimated pairwise distances were 0.018 (nucleotide level) and 0.002 (amino acid level), respectively, whereas the estimated ratio of non-synonymous to synonymous substitutions was also very low (pairwise ratio, dN/dS = 0.014).
Table 3.
Pairwise distances among complete genomes of iflavirus associated with Ixodes ricinus (five variants, IricIV-1 to 5, first identified in the present study) or the I. scapularis cell line ISE6 (ISIV). Number of base substitutions per site (Maximum composite likelihood model, 8,976 positions) followed by number of amino acid substitutions per site (Poisson correction model, 2,991 positions). Analyses were conducted with MegaX
| IricIV-1 | IricIV-2 | IricIV-3 | IricIV-4 | IricIV-5 | ISIV | |
|---|---|---|---|---|---|---|
| IricIV-1 | ||||||
| IricIV-2 | 0.059 / 0.015 | |||||
| IricIV-3 | 0.078 / 0.020 | 0.064 / 0.015 | ||||
| IricIV-4 | 0.060 / 0.016 | 0.010 / 0.004 | 0.066 / 0.016 | |||
| IricIV-5 | 0.076 / 0.019 | 0.065 / 0.015 | 0.028 / 0.006 | 0.066 / 0.017 | ||
| ISIV | 0.077 / 0.024 | 0.049 / 0.016 | 0.065 / 0.019 | 0.050 / 0.017 | 0.062 / 0.019 |
Evaluation of the prevalence of iflaviruses in different populations and stages of I. ricinus
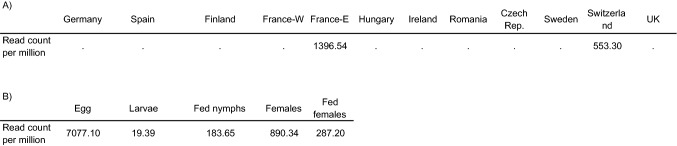
We detected the presence of the iflavirus associated with I. ricinus in only two out of 12 individual field populations: those from northwestern France and Switzerland. The virus abundance was null or very low in the remaining populations (i.e., below our detection threshold). In a second data set corresponding to a lab strain, the virus was detected in all life stages and conditions, ranging from a low abundance in larvae to a maximum abundance in the egg stage (Table 4), where reads assigned to the virus represented around 0.7% of all reads sequenced.
Table 4.
Relative abundance of an iflavirus associated with I. ricinus, in different field populations and life-stages. Abundance was assessed by read counts per million in different RNA-Seq libraries, comparing A) different field populations (each library was obtained from a pool of 50 nymphs), data from BioProject PRJNA662080 B) different life-stages and conditions, for a lab-reared strain, data from BioProject PRJNA595586. A dot indicates a negative library (i.e. less than the one read per million threshold), where the virus is presumed absent from the pool of individuals
Discussion
Communities of microorganisms associated with ticks are receiving special attention in the context of increasing concern about tick-transmitted diseases [27]. In recent years, there has been a growing interest in viruses associated with ticks, and iflavirus-like viruses have emerged as some of the most common viruses in ticks [2, 5, 8, 11, 14]. Our study, based on the exploration of assembled transcriptomes, allowed us to discover nine new iflavirus genome sequences in ticks, representing five different species of tick-associated iflaviruses. We then performed a phylogenetic analysis that included these new genome sequences as well as previously published sequences. This analysis shows that the iflavirus genome sequences associated with ticks are closely grouped together and form a monophyletic clade. This suggests an ancient association between this virus subgroup and ticks, which could mean relatively infrequent host switches of the virus between major groups of arthropods. It is noteworthy, however, that two iflavirus-like genome sequences have been described recently in members of the genus Antricola [28], a genus of neotropical soft ticks that have a peculiar biology, being associated with hot bat caves and feeding partially on bat guano instead of having an exclusive vertebrate blood diet. These two sequences (not included in our phylogeny) grouped closely with insect iflaviruses and not with sequences associated with ticks, showing that there has been more than one infection of ticks by viruses of this family. Within ticks, the virus phylogeny and the tick phylogeny were not congruent, i.e., there was no strict co-cladogenesis (Figs. 2 and 3). One monophyletic group comprised sequences found in I. ricinus, I. holocyclus, and I. vespertilionis, suggesting that they form an "Ixodes" subclade among the iflaviruses associated with ticks. However, the virus sequence found in I. frontalis did not group with this ensemble and was closer to iflavirus genomes found in ticks of other genera. Also, the closer grouping of I. holocyclus and I. ricinus was inconsistent with the phylogeny of this genus [13, 29], since I. holocyclus, a species belonging to the Australasian subgroup of Ixodes, is phylogenetically distant from I. ricinus and I. vespertilionis. Although the data for iflaviruses in genera other than Ixodes is still relatively scarce, we finally note that the sequences associated with two species of the genus Hyalomma did not group together. In addition, a formal test of cophylogeny (Fig. 3) suggested several events of switching of the virus between tick species or genera over evolutionary time. A more precise picture of these dynamics will require a denser data set including more iflavirus genomes and a larger sample of tick species.
RNA viruses have relatively fast-evolving sequences, which was also suggested here by the relatively low sequence identity among genomes associated with different tick species. There was, however, a striking exception, concerning the sequence found in a cell line of I. scapularis (ISIV), which formed a tight cluster in the ML phylogenetic tree with five different variants identified in I. ricinus. In fact, these sequences were nearly identical at the amino acid level (>98% pairwise identity of protein sequences). Therefore, ISIV and the I. ricinus iflavirus sequences represent different strains of the same virus – a puzzling result given that the tick host species I. scapularis and I. ricinus have evolved on two different continents (eastern North America and Europe) and are separated by several million years of divergence. The geographic distance and the scarcity of animal movements (birds would be the only shared potential tick hosts) between western Europe and North America leaves little chance of a recent natural inter-species contamination through shared tick hosts. We thus propose that a contamination could explain this anomaly. Indeed, ISIV was found in the transcriptome of a cell line of I. scapularis (ISE6), but we failed to detect it in any other assembled transcriptomes of this species, including a data set comprising as many as 200 adult ticks from several locations [30]. In addition, another study assessed the presence of viral sequences in I. scapularis after surveying several pools of wild samples (a total of > 1,100 individuals, combining nymphs and adults) from different locations – New York and Connecticut, USA – but did not report any iflavirus-like sequence [5]. Of note, using the same methods, this study did identify an iflavirus genome in Amblyomma americanum (although it was named "dicistrovirus", we demonstrated that this sequence groups with members of the family Iflaviridae). By contrast, the five I. ricinus variants found in the present study were obtained from several independent samples of wild ticks (or from one lab strain initially derived from wild ticks), which strongly suggests that the I. ricinus iflavirus variants discovered in the present study correspond to strains of iflaviruses that naturally infect this species. Based on the above, we argue that the presence of the virus in the I. scapularis cell line is best explained by a contamination from a virus associated in the field with I. ricinus. Such contamination could have occurred due to the establishment or maintenance of cell lines from the two species in a same lab, a possibility that was discussed previously for other viruses and cell lines of I. scapularis used in a study by Alberdi et al. [31].
Much remains to be known about the evolutionary dynamics of the association between iflaviruses and tick species. For example, the effect of iflaviruses on the fitness of their tick hosts is unknown, as there are no reports, to our knowledge, of evident symptoms in ticks infected by these viruses. A second aspect to explore is the possibility that some of the tick-associated iflaviruses could have been incorporated into the tick genome, becoming endogenous viral elements (EVEs). The initial observation of a frameshift in the contig identified for Hyalomma dromedarii could suggest a scenario of incorporation into the host genome, followed by pseudogenization. However, further examination of the reads led us to correct the sequence, which in fact contains an intact ORF. For HflFV, RT-PCR and PCR analysis of DNA and RNA showed that the iflavirus sequence was only amplified from RNA, and thus that it was not derived from an EVE, but this test remains to be performed for other tick-associated iflaviruses. Other related points that need to be explored are the prevalence and modes of transmission of the virus in each of the tick species studied here. In one tick species, I. ricinus, the RNA-Seq data set allowed us to evaluate patterns of prevalence in different life stages or different field populations. For example, the high abundance of the virus in a library obtained from a pool of eggs suggests that the virus can be transmitted vertically. More experiments, based on testing of mothers and their egg mass, will be needed to determine whether this is the exclusive mode of transmission. Another data set for I. ricinus, based on twelve field populations (pools of nymphs) showed that the virus was only present in a minority of populations. Based on this observation, the negative results obtained for most species (i.e., all transcriptomes of 12 species of the Metastriata and three Ornithodoros spp.) should not be taken as evidence of absence of this family of viruses from the entire species. Our study supports the hypothesis of an ancient association and relatively widespread presence of iflavirus genomes in tick species. We suggest that a more systematic use of RNA-Seq, based on large pools of wild individuals, would maximize the chance of detecting sequences of tick-associated viruses, even in cases of low prevalence [32]. More generally, this would enhance progress of the description of the virome associated with ticks, comprising viruses that are pathogenic for their vertebrate hosts, including humans [33].
Acknowledgements
We thank O. Rais, M. Voordouw, and Franck Boué, who provided part of the material used for transcriptome sequencing of I. ricinus. We acknowledge the support of the Genotoul bioinformatics platform Toulouse Midi-Pyrénées (Bioinfo Genotoul) for bioinformatic analyses. Thanks to Sara Moutailler (ANSES) for helpful comments on the manuscript.
Author contributions
Conceptualization and methodology: Claude Rispe and Olivier Plantard. Resources and funding acquisition: Claude Rispe, Caroline Hervet, Davide Sassera, Aaron Jex, Jean-Marc Aury, Karine Labadie, and Olivier Plantard. Formal analysis and data curation: Claude Rispe, Romain Daveu, and Louane Sigrist. The original draft of the manuscript was written by Claude Rispe and Romain Daveu, and all authors commented on previous versions of the manuscript. All authors read and agreed to the final version of the manuscript.
Funding
This work was supported by the Genoscope, the Commissariat à l’Energie Atomique et aux Energies Alternatives (CEA) and France Génomique (ANR-10-INBS-09-08), by the Italian Ministry of Education, University and Research (MIUR): Dipartimenti di Eccellenza Program (2018–2022) – Dept. of Biology and Biotechnology "L. Spallanzani", University of Pavia (to D.S.), and by a grant from the Institut Carnot France Futur Elevage (XENOBIOTICK project).
Declarations
Conflict of interest
The authors declare no conflict of interest.
Availability of data and material
Genome sequences of iflaviruses have been submitted to the GenBank database.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Romain Daveu, Email: romain.daveu@inrae.fr.
Caroline Hervet, Email: caroline.hervet@oniris-nantes.fr.
Louane Sigrist, Email: louane_sigrist@hotmail.fr.
Davide Sassera, Email: davide.sassera@unipv.it.
Aaron Jex, Email: jex.a@wehi.edu.au.
Karine Labadie, Email: Karine.Labadie@genoscope.cns.fr.
Jean-Marc Aury, Email: jmAury@genoscope.cns.fr.
Olivier Plantard, Email: olivier.plantard@oniris-nantes.fr.
Claude Rispe, Email: claude.rispe@inrae.fr.
References
- 1.Li C-X, Shi M, Tian J-H, et al. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. Elife. 2015;4:e05378. doi: 10.7554/eLife.05378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi M, Lin X-D, Tian J-H, et al. Redefining the invertebrate RNA virosphere. Nature. 2016;540:539–543. doi: 10.1038/nature20167. [DOI] [PubMed] [Google Scholar]
- 3.Liu S, Chen Y, Bonning BC. RNA virus discovery in insects. Curr Opin Insect Sci. 2015;8:54–61. doi: 10.1016/j.cois.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 4.Harvey E, Rose K, Eden J-S, et al. Extensive diversity of RNA viruses in Australian ticks. J Virol. 2019 doi: 10.1128/JVI.01358-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tokarz R, Sameroff S, Tagliafierro T, et al. Identification of novel viruses in Amblyomma americanum, Dermacentor variabilis, and Ixodes scapularis Ticks. Sphere. 2018 doi: 10.1128/mSphere.00614-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valles SM, Chen Y, Firth AE, et al. ICTV virus taxonomy profile: iflaviridae. J Gen Virol. 2017;98:527–528. doi: 10.1099/jgv.0.000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Miranda JR, Genersch E. Deformed wing virus. J Invertebr Pathol. 2010;103:S48–S61. doi: 10.1016/j.jip.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 8.O’Brien CA, Hall-Mendelin S, Hobson-Peters J, et al. Discovery of a novel iflavirus sequence in the eastern paralysis tick Ixodes holocyclus. Arch Virol. 2018;163:2451–2457. doi: 10.1007/s00705-018-3868-9. [DOI] [PubMed] [Google Scholar]
- 9.Wille M, Harvey E, Shi M, et al. Sustained virome diversity in Antarctic penguins and their ticks: geographical connectedness and no evidence for low pathogen pressure. bioRxiv. 2019 doi: 10.1101/2019.12.11.873513. [DOI] [Google Scholar]
- 10.Pettersson JH-O, Ellström P, Ling J, et al. Circumpolar diversification of the Ixodes uriae tick virome. PLoS Pathog. 2020;16:e1008759. doi: 10.1371/journal.ppat.1008759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakao R, Matsuno K, Qiu Y, et al. Putative RNA viral sequences detected in an Ixodes scapularis-derived cell line. Ticks Tick Borne Dis. 2017;8:103–111. doi: 10.1016/j.ttbdis.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 12.Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charrier NP, Hermouet A, Hervet C, et al. A transcriptome-based phylogenetic study of hard ticks (Ixodidae) Sci Rep. 2019 doi: 10.1038/s41598-019-49641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobayashi D, Murota K, Itokawa K, et al. RNA virome analysis of questing ticks from Hokuriku District, Japan, and the evolutionary dynamics of tick-borne phleboviruses. Ticks Tick Borne Dis. 2020;11:101364. doi: 10.1016/j.ttbdis.2019.101364. [DOI] [PubMed] [Google Scholar]
- 15.Kumar S, Stecher G, Li M, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 17.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalyaanamoorthy S, Minh BQ, Wong TKF, et al. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoang DT, Chernomor O, von Haeseler A, et al. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Letunic I, Bork P. Interactive tree of life (iTOL) v4: recent updates and new developments. Nucl Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conow C, Fielder D, Ovadia Y, Libeskind-Hadas R. Jane: a new tool for the cophylogeny reconstruction problem. Algorithms Mol Biol. 2010;5:16. doi: 10.1186/1748-7188-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zuckerkandl E, Pauling L. Molecules as documents of evolutionary history. J Theor Biol. 1965;8:357–366. doi: 10.1016/0022-5193(65)90083-4. [DOI] [PubMed] [Google Scholar]
- 24.Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Bioinformatics. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 25.Katzourakis A, Gifford RJ. Endogenous viral elements in animal genomes. PLoS Genet. 2010;6:e1001191. doi: 10.1371/journal.pgen.1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roux S, Adriaenssens EM, Dutilh BE, et al. Minimum Information about an uncultivated virus genome (MIUViG) Nat Biotechnol. 2019;37:29–37. doi: 10.1038/nbt.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Díaz-Sánchez S, Estrada-Peña A, Cabezas-Cruz A, de la Fuente J. Evolutionary insights into the tick hologenome. Trends Parasitol. 2019;35:725–737. doi: 10.1016/j.pt.2019.06.014. [DOI] [PubMed] [Google Scholar]
- 28.Blomström A-L, Luz HR, Öhlund P, et al. Novel viruses found in Antricola ticks collected in Bat Caves in the Western Amazonia of Brazil. Viruses. 2019 doi: 10.3390/v12010048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mans BJ, Featherston J, Kvas M, et al. Argasid and ixodid systematics: Implications for soft tick evolution and systematics, with a new argasid species list. Ticks Tick Borne Dis. 2019;10:219–240. doi: 10.1016/j.ttbdis.2018.09.010. [DOI] [PubMed] [Google Scholar]
- 30.Josek T, Walden KKO, Allan BF, et al. A foreleg transcriptome for Ixodes scapularis ticks: candidates for chemoreceptors and binding proteins that might be expressed in the sensory Haller’s organ. Ticks Tick Borne Dis. 2018;9:1317–1327. doi: 10.1016/j.ttbdis.2018.05.013. [DOI] [PubMed] [Google Scholar]
- 31.Alberdi MP, Dalby MJ, Rodriguez-Andres J, et al. Detection and identification of putative bacterial endosymbionts and endogenous viruses in tick cell lines. Ticks Tick Borne Dis. 2012;3:137–146. doi: 10.1016/j.ttbdis.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moutailler S, Popovici I, Devillers E, et al. Diversity of viruses in Ixodes ricinus, and characterization of a neurotropic strain of Eyach virus. New Microbes New Infect. 2016;11:71–81. doi: 10.1016/j.nmni.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vandegrift KJ, Kapoor A. The ecology of new constituents of the tick virome and their relevance to public health. Viruses. 2019 doi: 10.3390/v11060529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones P, Binns D, Chang H-Y, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]