Abstract
With the growing evidence on the variable human susceptibility against COVID-19, it is evident that some genetic loci modulate the severity of the infection. Recent studies have identified several loci associated with greater severity. More recently, a study has identified a 50 kb genomic segment introgressed from Neanderthal adding a risk for COVID-19, and this genomic segment is present among 16% and 50% people of European and South Asian descent, respectively. Our studies on ACE2 identified a haplotype present among 20% and 60% of European and South Asian populations, respectively, which appears to be responsible for the low case fatality rate among South Asian populations. This result was also consistent with the real-time infection rate and case fatality rate among various states of India. We readdressed this issue using both of the contrasting datasets and compared them with the real-time infection rates and case fatality rate in India. We found that the polymorphism present in the 50 kb introgressed genomic segment (rs10490770) did not show any significant correlation with the infection and case fatality rate in India.
Subject terms: Evolutionary genetics, Molecular evolution, Population genetics, Risk factors
Introduction
Since the beginning of COVID-19 pandemic, it has been observed that people with a different ethnic background and country or continent of origin have variable degrees of susceptibility1,2. Though there are a few well known factors for higher susceptibility, e.g. age and comorbidity3,4, the hospitalisation of younger healthy people has also been reported5. Recent genome wide association study has identified a gene cluster at chromosome 3 as well as the ABO gene at chromosome 9 associated with the severe risk factor for COVID-19 among Europeans6. Subsequently, the COVID-19 Host Genetics Initiative has corroborated this result7. The worldwide meta-analysis of the COVID-19 Host Genetics Initiative has identified 13 genetic loci associated with higher susceptibility or higher severity8.
Zeberg and Pääbo9 have identified a risk haplotype of 50 kb size introgressed from Neanderthals, which they called the ‘Neanderthal core haplotype’. This risk haplotype was found to be present with an allele frequency of 30% among South Asians, 8% in Europeans and 4% among admixed Americans. The peak carrier frequency was estimated among the Bangladeshi population, where 63% carried at least one copy of this haplotype. The study also cited twice the risk of mortality among people of Bangladeshi extraction living in the UK as opposed to the native population of Brittanic pedigree10.
Conversely, three of our studies on ACE2, the gateway of SARS-CoV-2, identified a haplotype, shared among South Asians and East Eurasians, likely protecting them from severe risk11–13. Additionally, the spatial distribution of this haplotype showed strong association with the low infection as well as low case fatality rate (CFR)13. To resolve this discrepancy between the two sets of findings and the associated claims, we have extracted a SNP (rs10490770) reported to be associated with the high risk for COVID-199, from our published and unpublished genome wide datasets (Supplementary Table S1), and looked for existing association with the state-wise COVID-19 data of India.
Materials and methods
Zeberg and Pääbo9 have mainly discussed about the SNP rs35044562. However, they reported 12 other SNPs present in the ‘Neanderthal core haplotype’ that are in high linkage disequilibrium (r2 > 0.98) (Supplementary Table S2). SNP rs10490770 showed high LD (r2 = 0.99) with the SNP rs35044562. The genome-wide genotype data by Illumina tagged rs2285666 and rs10490770 SNPs in their panel. Therefore, we searched the genotype datasets generated by this platform. The frequency data for both of the SNPs from various Indian populations were extracted using Plink 1.914, from 1000 genome project data phase 315, data published by the Estonian Biocentre16–19 and our newly genotyped samples for various Indian states and Bangladesh (Supplementary Table S1). In addition to our previous study13, more samples were added for the SNP rs2285666. The state-wise COVID-19 infection and CFR datasets were extracted from https://www.covid19india.org/. The regression estimations and plots were built by https://www.graphpad.com/quickcalcs/linear1/ and reverified by the Microsoft Excel regression calculations. We have also used Pearson’s correlation coefficient test20 to evaluate the effect of both the SNPs. The spatial distribution of both SNPs were drawn using the web tool available at https://www.datawrapper.de/.
Results and discussion
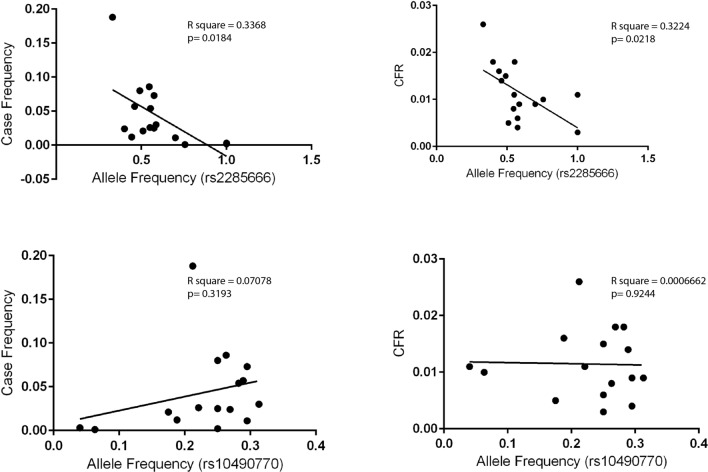
In contrast to the conclusions drawn by Zeberg and Pääbo9, our study on ACE2 identified a haplotype that is frequent among South Asians and East Eurasians11–13. This haplotype is derived by a polymorphism rs2285666 responsible for elevated expression of ACE2. We have found high inverse correlation of this haplotype with the state-wise cases as well as the case-fatality rate (CFR) among Indian populations13. This correlation was significant at various timelines of the pandemic in India (Table 1). We verified the statistical tests with the updated data up to December 2020 and found these data to be consistent with previous observations (Fig. 1 and Supplementary Fig. S1). Thus, it is likely that the ACE2 SNP rs2285666 has played a significant role in modulating the susceptibility to the disease among Indian populations.
Table 1.
Estimates of Pearson correlation coefficient for the rs2285666 and rs10490770 with the real-time COVID-19 cases as well as case fatality rate among Indian populations. The significant values are shown in bold letters.
| rs2285666 (T) | rs10490770 (C) | FreqMay2020 | FreqAug2020 | FreqDec2020 | CFRAug2020 | CFRAug2020 | ||
|---|---|---|---|---|---|---|---|---|
| rs2285666 |
Pearson correlation Sig. (2-tailed) |
1 |
− 0.428 0.098 |
− 0.588* 0.017 |
− 0.558* 0.025 |
− 0.580* 0.018 |
− 0.649** 0.007 |
− 0.568 0.022 |
| rs10490770 |
Pearson correlation Sig. (2-tailed) |
− 0.428 0.098 |
1 |
0.204 0.450 |
0.164 0.543 |
0.266 0.319 |
0.244 0.362 |
− 0.026 0.924 |
*Correlation is significant at the 0.05 level (2-tailed).
**Correlation is significant at the 0.01 level (2-tailed).
Figure 1.
The regression analyses of rs2285666 (ACE2) and rs10490770 (LZTFL1) with the COVID-19 cases as well as case fatality rate in various states of India (Supplementary Table S1).
In our search of the SNPs reported to be associated with high risk by Zeberg and Pääbo9, we found rs10490770 from genome-wide datasets17,18,21–23. We applied the same tests done for the ACE2 SNPs (Fig. 1). The state-wise frequency variation of this SNP did not show any association either with the number of cases or the CFR (Table 1 and Supplementary Fig. S1). We repeated these regression tests for the number of cases as well as the CFR data, obtained during all the three months. However, none of them showed any association with the rs10490770 (p > 0.3) (Table 1). It is interesting to note that this SNP (rs10490770) has been found to be associated with disease severity in the global data8. However, the lack of association for rs10490770 with COVID-19 cases or CFR in India is striking and suggests instead a complex susceptibility response among Indian populations. Along with the complex genetic structure11,24, socio-economic status25 and hygiene26 may have contributed to such a complex scenario. Furthermore, a detailed clinical and genome-wide association study on Indian COVID-19 patients would be useful to resolve this complexity.
Zeberg and Pääbo9 used the data of higher susceptibility to the disease among the Bangladeshi population living in UK10 to support their findings. By considering the effect of sex, age, socio-economic deprivation and region, this report found that people of Bangladeshi origin had double the risk of mortality as compared to people of British origin. However, the higher mortality rate for Bangladeshi population in the UK needs more detailed investigation on comorbidity, genetic admixture as well as local environment and socio-economic circumstances in their particular British context. More importantly, a similar trend had also been observed among admixed Americans, where some of the same qualifications may apply mutatis mutandis27–29. Furthermore, it is notable that among the Bangladeshi samples analysed by us, the tribal populations of Bangladesh showed almost three times less frequency of rs10490770 (Supplementary Table S1). This is likely due to the different population histories of the caste and tribal populations of Bangladesh30,31. The Tibeto-Burman speakers of Bangladesh show a closer genetic affinity with the East and Southeast Asian populations, whereas the Indo-European speaking caste populations incline with the Indian populations. Therefore, it is advised explicitly to differentiate between the caste and tribal populations while making any statement about Bangladeshi populations. Significantly, our data also show that the incidence of the allele rs2285666 has been found to occur in the highest frequency of 100% in Indian populations such as the Nishi and Kokborok (Tripuri), who represent Trans-Himalayan language communities (Supplementary Fig. S1 and Supplementary Table S1). As a linguistic phylum, the Trans-Himalayan language family is widespread in parts of eastern Eurasia and includes languages such as Tibetan, Burmese, Mandarin, Cantonese and Hokkien.
Thus, our extensive analyses on real-time data did not show any association of rs10490770 with the state-wise infection rates as well as CFRs, suggesting that the risk allele for COVID-19 in Europe does not play a significant role in COVID-19 severity in South Asia.
Supplementary Information
Acknowledgements
KT was supported by Council of Scientific and Industrial Research (CSIR) and J C Bose Fellowship from Science and Engineering Research Board (SERB), Department of Science and Technology, Government of India. PPS was supported by the CSIR-JRF fellowship from CSIR, India.
Author contributions
G.C., K.T., R.S. and P.S. conceived and designed this study. P.P.S., A.S., G.S., N.K., A.P., Pr.S., R.S., and P.S. collected the data for alleles and COVID-19. P.P.S., A.S., Pr.S., and G.C. analyzed the data. P.P.S., Pr.S., K.T., Gv.D. and G.C. wrote the manuscript from the inputs of other co-authors. All authors contributed to the article and approved the submitted version.
Data availability
All datasets generated for this study are included in the article/Supplementary Material.
Competing interests
The authors declare no competing interests.
Footnotes
The original online version of this Article was revised: In the original version of this Article, Kumarasamy Thangaraj was omitted as a corresponding author. Correspondence and request for materials should also be addressed to thangs@ccmb.res.in . In addition, the email address of the co-corresponding author Gyaneshwer Chaubey was incorrectly given as thangs@ccmb.res.in. Correspondence and request for materials should also be addressed to gyaneshwer.chaubey@bhu.ac.in .
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
8/5/2021
A Correction to this paper has been published: 10.1038/s41598-021-94864-4
Contributor Information
Kumarasamy Thangaraj, Email: thangs@ccmb.res.in.
Gyaneshwer Chaubey, Email: gyaneshwer.chaubey@bhu.ac.in.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-91711-4.
References
- 1.Mackey K, et al. Racial and ethnic disparities in COVID-19–related infections, hospitalizations, and deaths: A systematic review. Ann. Intern. Med. 2021;174:362–373. doi: 10.7326/M20-6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shelton JF, et al. Trans-ethnic analysis reveals genetic and non-genetic associations with COVID-19 susceptibility and severity. medRxiv. 2020 doi: 10.1101/2020.09.04.20188318. [DOI] [Google Scholar]
- 3.Alberca RW, de Oliveira LM, Branco ACCC, Pereira NZ, Sato MN. Obesity as a risk factor for COVID-19: An overview. Crit. Rev. Food Sci. Nutr. 2020;1:15. doi: 10.1080/10408398.2020.1775546. [DOI] [PubMed] [Google Scholar]
- 4.Fang L, Karakiulakis G, Roth M. Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection? Lancet Respir. Med. 2020;8:21. doi: 10.1016/S2213-2600(20)30116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Godri Pollitt KJ, et al. COVID-19 vulnerability: The potential impact of genetic susceptibility and airborne transmission. Hum. Genom. 2020;14:1–7. doi: 10.1186/s40246-020-00267-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellinghaus D, et al. Genomewide association study of severe Covid-19 with respiratory failure. N. Engl. J. Med. 2020 doi: 10.20944/preprints202007.0178.v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.COVID-19 Host Genetics Initiative The COVID-19 Host Genetics Initiative a global initiative to elucidate the role of host genetic factors in susceptibility and severity of the SARS-CoV-2 virus pandemic. Eur. J. Hum. Genet. 2020;28:715. doi: 10.1038/s41431-020-0636-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ganna A. Mapping the human genetic architecture of COVID-19 by worldwide meta-analysis. medRxiv. 2021 doi: 10.1101/2021.03.10.21252820. [DOI] [Google Scholar]
- 9.Zeberg H, Pääbo S. The major genetic risk factor for severe COVID-19 is inherited from Neanderthals. Nature. 2020;587:610–612. doi: 10.1038/s41586-020-2818-3. [DOI] [PubMed] [Google Scholar]
- 10.Public Health England. COVID-19: Review of disparities in risks and outcomes (2020). https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/908434/Disparities_in_the_risk_and_outcomes_of_COVID_August_2020_update.pdf
- 11.Srivastava A, et al. Most frequent South Asian haplotypes of ACE2 share identity by descent with East Eurasian populations. PLoS ONE. 2020;15:e0238255. doi: 10.1371/journal.pone.0238255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh KK, Chaubey G, Chen JY, Suravajhala P. Decoding SARS-CoV-2 hijacking of host mitochondria in pathogenesis of COVID-19. Am. J. Physiol. 2020 doi: 10.1152/ajpcell.00224.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Srivastava A, et al. Genetic association of ACE2 rs2285666 polymorphism with COVID-19 spatial distribution in India. Front. Genet. 2020;11:1163. doi: 10.3389/fgene.2020.564741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang CC, et al. Second-generation PLINK: Rising to the challenge of larger and richer data sets. Gigascience. 2015;4:S13742-015. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.1000 Genomes Project Consortium et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Estonian Biocentre Public_Data. https://evolbio.ut.ee/.
- 17.Chaubey G, et al. “Like sugar in milk”: Reconstructing the genetic history of the Parsi population. Genome Biol. 2017;18:110. doi: 10.1186/s13059-017-1244-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pathak AK, et al. The genetic ancestry of modern Indus valley populations from Northwest India. Am. J. Hum. Genet. 2018;103:918–929. doi: 10.1016/j.ajhg.2018.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tätte K, et al. The genetic legacy of continental scale admixture in Indian Austroasiatic speakers. Sci. Rep. 2019;9:3818. doi: 10.1038/s41598-019-40399-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benesty, J., Chen, J., Huang, Y., and Cohen, I. (2009). “Pearson correlation coefficient,” in Noise Reduction in Speech Processing, Springer Topics in Signal Processing, vol. 2, Berlin, Heidelberg: Springer. 10.1007/978-3-642-00296-0_5
- 21.Chaubey G, et al. Population genetic structure in Indian austroasiatic speakers: The role of landscape barriers and sex-specific admixture. Mol. Biol. Evol. 2011;28:1013–1024. doi: 10.1093/molbev/msq288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Metspalu M, et al. Shared and unique components of human population structure and genome-wide signals of positive selection in South Asia. Am. J. Hum. Genet. 2011;89:731–744. doi: 10.1016/j.ajhg.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamang R, et al. Reconstructing the demographic history of the Himalayan and adjoining populations. Hum. Genet. 2018;137:129–139. doi: 10.1007/s00439-018-1867-2. [DOI] [PubMed] [Google Scholar]
- 24.Metspalu M, Mondal M, Chaubey G. The genetic makings of South Asia. Genet. Hum. Orig. 2018;53:128–133. doi: 10.1016/j.gde.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 25.Rajkumar RP. The relationship between demographic, socioeconomic, and health-related parameters and the impact of COVID-19 on 24 regions in India: Exploratory cross-sectional study. JMIR Public Health Surveill. 2020;6:e23083. doi: 10.2196/23083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finlay BB, et al. The hygiene hypothesis, the COVID pandemic, and consequences for the human microbiome. Proc. Natl. Acad. Sci. USA. 2021;118:e2010217118. doi: 10.1073/pnas.2010217118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kullar R, et al. Racial disparity of coronavirus disease 2019 in African American communities. J. Infect. Dis. 2020;222:890–893. doi: 10.1093/infdis/jiaa372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yancy CW. COVID-19 and African Americans. JAMA. 2020;323:1891–1892. doi: 10.1001/jama.2020.6548. [DOI] [PubMed] [Google Scholar]
- 29.Hooper MW, Nápoles AM, Pérez-Stable EJ. COVID-19 and racial/ethnic disparities. JAMA. 2020;323:2466–2467. doi: 10.1001/jama.2020.8598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sultana GNN, Sharif MI, Asaduzzaman M, Chaubey G. Evaluating the genetic impact of South and Southeast Asia on the peopling of Bangladesh. Leg. Med. Tokyo Jpn. 2015;17:446–450. doi: 10.1016/j.legalmed.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 31.Gazi NN, et al. Genetic structure of Tibeto-Burman populations of Bangladesh: Evaluating the gene flow along the Sides of Bay-of-Bengal. PLoS ONE. 2013;8:e75064. doi: 10.1371/journal.pone.0075064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.