Supplemental Digital Content is available in the text.
Background.
There is an unmet need for noninvasive tools for diagnosis of rejection after kidney transplantation. The aim of this study was to determine the discriminative value of a combined cellular and molecular biomarker platform in urine for the detection of rejection.
Methods.
First, microRNA (miR) molecules were screened in transplant biopsies and urine sediments of patients with acute rejection and patients without rejection and stable graft function. Second, the expression of 15 selected miRs was quantified in an independent set of 115 urine sediments of patients with rejection and 55 urine sediments of patients without histological signs of rejection on protocol biopsy. Additionally, CXCL-9 and CXCL-10 protein levels were quantified in the urine supernatant.
Results.
Levels of miR-155-5p (5.7-fold), miR-126-3p (4.2-fold), miR-21-5p (3.7-fold), miR-25-3p (2.5-fold), and miR-615-3p (0.4-fold) were significantly different between rejection and no-rejection urine sediments. CXCL-9 and CXCL-10 levels were significantly elevated in urine from recipients with rejection. In a multivariable model (sensitivity: 89.1%, specificity: 75.6%, area under the curve: 0.94, P < 0.001), miR-155-5p, miR-615-3p, and CXCL-9 levels were independent predictors of rejection. Stratified 10-fold cross validation of the model resulted in an area under the curve of 0.92.
Conclusions.
A combined urinary microRNA and chemokine profile discriminates kidney transplant rejection from stable graft conditions.
INTRODUCTION
Despite the introduction of powerful immunosuppressive agents, the risk of acute rejection within the first year after kidney transplantation is still 15%.1 The gold standard to diagnose rejection is the histological evaluation of a transplant biopsy. However, limitations of this procedure including its invasiveness urge the identification of noninvasive biomarkers of rejection. Since single biomarkers may not provide sufficient sensitivity and specificity, we hypothesize that a cross-platform approach is optimal for the noninvasive detection of rejection.2
MicroRNAs (miRs) are a class of small noncoding RNA molecules of approximately 22 nucleotides in length. These molecules can influence gene expression by affecting degradation of mRNA transcripts and/or inhibition of protein translation.3 Several research groups published on alterations in miR expression in kidney allografts at the time of acute rejection.4-7 Besides their presence in the graft tissue, miR levels can be determined in the urine sediment, which contains cells shed from the kidney parenchyma and graft-infiltrating leukocytes.8 Lorenzen et al9 reported decreased levels of miR-210 in total (uncentrifuged) urine samples at time of cellular rejection compared to stable graft function conditions. MiR levels have been investigated in the urine sediments from recipients with interstitial fibrosis and tubular atrophy (IF/TA).10,11
Chemokines are important in the recruitment of leukocytes to sites of inflammation, which is an early event in the rejection process. CXCL-9 (monokine induced by interferon-gamma, MIG) and CXCL-10 (interferon-inducible protein 10, IP-10) bind receptor CXCR3, which is restricted to activated T-lymphocytes.12 Increased numbers of CXCR3-positive T cells have been observed in inflammatory regions of grafts with cellular rejection.13 Furthermore, increased urinary levels of CXCL-9 and CXCL-10 have been observed in renal transplant recipients with acute rejection.14-16
The aim of this study was to identify a combined urinary signature, enabling the noninvasive detection of rejection after kidney transplantation. Hereto, levels of miRs and chemokines in urine samples were analyzed. We show that a combined urinary miR and chemokine profile identifies kidney transplant rejection.
MATERIALS AND METHODS
Patients and Sample Collection
The study was approved by the Medical Ethics Committee of the Leiden University Medical Center, The Netherlands. Urine sediments and supernatants, collected from kidney transplant recipients between 2007 and 2015 and stored at −20°C, were studied. Before storage, urine samples (maximum of 50 mL) were centrifuged at 3000 rpm for 10 min to spin down the urine sediment. After washing the sediment with 900 µL of PBS and centrifugation at 14 000 rpm, 50 µL of RNAlater (Qiagen, Venlo, The Netherlands) was added to preserve the RNA. An overview of patients and samples studied is depicted in a flow chart (Figure 1).
FIGURE 1.
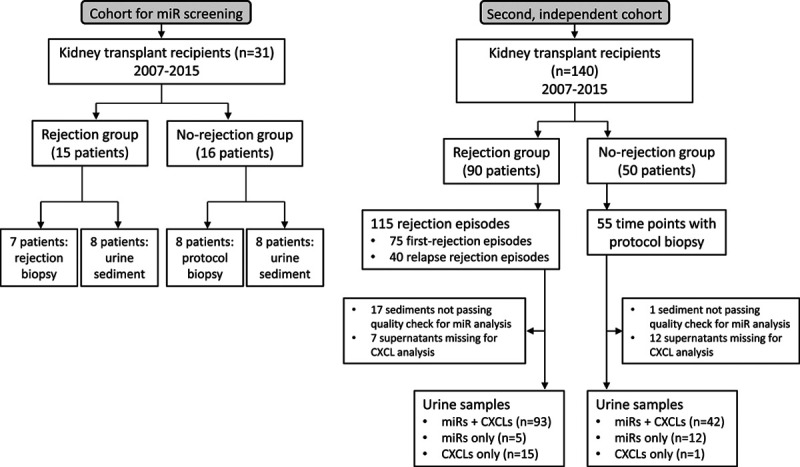
Flowchart of patients and samples studied. The study for miR screening has been depicted to the left and the validation phase in the second, independent cohort has been depicted to the right. miR, microRNA.
Biopsy Cohort for miR Screening
Fresh–frozen biopsies from 7 patients with cellular rejection (71.4% Banff IA and 28.6% Banff IIA; 163 ± 30 µmol/L) and 8 patients with stable graft function (123 ± 22 µmol/L) without histological signs of rejection in their 6-mo protocol biopsy were selected for miR profiling.
Urine Sediment Cohort for miR Screening
A set of 16 urine sediments, including 8 urine sediments from recipients at the moment of (biopsy-supported) cellular rejection (66.7% Banff IA and 33.3% Banff IIA; 178 ± 64 µmol/L) and 8 urine sediments from transplant recipients with stable graft function (126 ± 24 µmol/L; 3 were biopsy-supported), were selected for miR profiling.
Independent Cohort of Urine Samples
An independent cohort of kidney transplant recipients was selected based on medical files and histology reports. Inclusion criteria for the rejection group were acute graft dysfunction with suspicion of rejection and the availability of a transplant biopsy and urine sample both taken before antirejection treatment. Severity scores were assigned to the biopsies according to Banff criteria.17 Antibody-mediated rejection (AMR) was characterized by the presence of typical lesions in the tissue and/or C4d positivity in peritubular capillaries with presence of donor-specific antibodies. This group was further distinguished into AMR only or mixed AMR + T-cell–mediated rejection (TCMR).
Inclusion criteria for the no-rejection group were the availability of a urine sample and a protocol biopsy, showing the absence of morphologic abnormalities indicative of active rejection. In the rejection group, 115 urine sediments were collected from 90 recipients having rejection, of which 75 samples were taken at time of a first rejection episode and 40 samples during a relapse rejection episode. In the no-rejection group, 55 urine sediments were selected from 50 recipients at hospitalization for a protocol biopsy, performed in a time window of 7 d before and 7 d after urine sampling. From 5 recipients, 2 urine sediments were collected on 2 separate protocol biopsy moments.
To investigate urinary miR expression at time of BK viral nephropathy (BKVN) and CMV infection, 5 urine sediments were collected from 5 recipients (Leiden University Medical Center) with a biopsy proven BKVN. In addition, RNA samples from 8 urine sediments of recipients with a biopsy proven BKVN and 8 urine sediments of recipients with an active CMV infection were obtained from the Radboud university medical center (RUMC), Nijmegen, The Netherlands. Patients in the BKVN group showed BK positivity in blood plasma by PCR (3.2 ± 3.5 × 106 copies/L) and positive simian virus-40 immunostaining in the transplant biopsy. Patients in the CMV group had a positive CMV-PCR (≥106 copies/L) in peripheral blood and clinical signs of CMV infection.
RNA Extraction From Biopsies and Urine Sediments
On average, ten 10-µm sections per biopsy were cut for RNA extraction. Sectioned tissue was kept on dry ice, placed in 300 µL of ML buffer (Nucleospin miRNA kit; Macherey-Nagel, Düren, Germany), and stored at −20 oC until further use. From the RNAlater-impregnated urine sediments, small and large RNA were isolated with the Nucleospin miRNA kit, following the manufacturer’s protocol. The RNA from each spin column was diluted in 50 µL of RNase-free water. RNA samples from the RUMC Nijmegen were derived from urine sediments using the miRVANA miRNA Isolation Kit (Thermo Fisher Scientific, Waltham, MA).
MiR Screening in Biopsy and Urine Sediment Cohort
Real-time PCR (RT-PCR) was performed to screen the expression of approximately 750 conserved human miRs in a set of renal biopsies and urine sediments. Global miR expression analysis for the 15 kidney transplant biopsies was performed using human miRNOME v3 panels I+II (Exiqon, Vedbaek, Denmark) on a CFX384 RT-PCR instrument (Biorad, Hercules, CA). MiR expression profiling in 16 urine sediments was performed using human miRNOME v2 panels (Exiqon). To obtain sufficient material per sample for screening, the total RNA eluates were evaporated in a SpeedVac to a volume of 10 µL and reverse transcribed into cDNA using the Universal cDNA synthesis kit II (Exiqon). Heating steps were performed on a T100 thermal cycler (Biorad). Undiluted cDNA was added to SYBR green mix (Biorad). Each well of the miRNOME panel contained a dried down LNA primer set for a 10-µL RT-PCR reaction. Forty-five cycles of PCR were performed on a CFX384 instrument (Biorad).
MiR Quantification in Urinary Sediments of Independent Cohort
In the independent cohort of urine sediments, 15 miRs were selected for quantification by single-assay RT-qPCRs. Hereto, 4 µL of RNA template was reverse transcribed into cDNA using the miRCURY LNA Universal RT miR PCR kit (Exiqon). UniSp6 template was added as a spike-in to control for cDNA synthesis efficiency. Afterward, 4 µL of diluted cDNA (1:20) was used in a 10-µL real-time qPCR reaction using miR-specific LNA enhanced primer sets (Exiqon). Real-time qPCR (40 cycles) was performed on the ViiA 7 Real-Time PCR System (ThermoFisher Scientific) with settings according to the manufacturer’s PCR protocol (miRCURY, Exiqon). PCR assays were performed in duplicate. RNA from human liver, kidney, spleen, and synthetic human reference RNA was included as positive controls for interplate calibration of duplicates. Samples with aberrant amplification of UniSp6 and/or of 2 or more reference miRs (based on outlier analysis in the whole group) were excluded from the analysis, as well as duplicates with a coefficient of variation >10%.
Protein Quantification in Urine Supernatant
Urinary protein levels of CXCL-9 and CXCL-10 were quantified in a multiplex format using a Bio-plex cytokine assay (Biorad) according to the manufacturer’s instructions. Before the assay, BSA (Sigma) was added to urine supernatant to a final concentration of 0.5%. Briefly, 50 µL of a working solution of magnetic, colored beads (10× stock beads), coated with CXCL-9- and CXCL-10-specific antibodies, was added to a 96-well filter plate. After washing of the filter plate, 50 µL of urine supernatant or diluted standards were added to each well. After incubation and several washing procedures, 12.5 µL of biotinylated detection antibody (10×) solution was added to each well. After the second incubation and washing of the filter plate, 25 µL of streptavidin-PE (100×) was added. After a final incubation and filter plate washing, constituents were drawn into a flow-based Bio-plex suspension array system (Biorad) for the identification and quantification of fluorescent signals. Standard curves derived from a recombinant protein standard were obtained to quantify concentrations of the analytes. For each sample, 1 single run was performed. No normalization for urine creatinine values was performed, since urine protein concentrations correlate well with creatinine-normalized protein concentrations.18
Statistical Analyses
MiR expression data were normalized using a global median normalization strategy.19,20 Hereto, median expression levels of all significantly expressed miRs (ie, Cq < 35) of a given sample were calculated. For the preliminary screening phase only miRs having a significant expression in at least 8 samples (50%) were included in the Mann–Whitney rank sum test (MWU) analysis. Additionally, corrections for multiple comparisons by false discovery rates were applied.
For the independent urine sediment cohort, miR expression levels were normalized using the geometric mean21 of 3 miRs (namely miR-30c-5p, miR-423-3p, and miR-744-5p) that were selected from the urine sediment miRNA expression profiling data based on: relatively low coefficient of variation (SD/mean) between the sediments, sufficiently high miR expression in the independent cohort (median Cq values of 21.7, 24.9, and 26.8, respectively), and a high extent of intercorrelation (r = 0.81–0.85). Relative miR expression levels and absolute urine concentrations of CXCL-9 and CXCL-10 were log10 transformed before statistical testing. Nonparametric Spearman’s correlation was used to estimate the relationship between analytes. For clinical characteristics of samples and recipients, nominal variables were compared using the Chi-square test. Continuous variables were assessed for normal distribution and were compared by using the MWU test. Post hoc Bonferroni corrections were performed to correct for multiple testing of cases versus controls in the independent cohort. Differences were considered significant at P < 0.05.
To determine a urine signature distinctive for rejection, univariate analysis was performed for each parameter that was significantly different between the rejection and no-rejection group. The 10 most significant variables from univariate analysis were included in a multivariable logistic analysis. The number of variables was limited to prevent overfitting of the model. The forward Wald method was used to create the optimal model. The model was internally verified by a stratified 10-fold cross validation. Hereto, the cohort was randomly partitioned 10 times into a training set (90%) and test set (10%), and the validation results were combined over the rounds. IBM SPSS statistics version 24 and Microsoft Excel (Office for Windows 10) were used for statistical analyses.
RESULTS
MiR Expression Screening in Biopsies and Urine Sediments
A total of 263 ± 26 miRs were significantly expressed in the biopsy specimens of the screening set. In Figure S1A, SDC, http://links.lww.com/TXD/A336, interplate corrected miR expression levels with median expression values are shown for each rejection and control biopsy specimen. From the 254 miRs that were included in the MWU analysis 20 showed a significantly different expression between the rejection and no-rejection group at a P < 0.01 (Table S1, SDC, http://links.lww.com/TXD/A336). Among these, 13 were underexpressed in rejection biopsies compared to normal biopsies, and 7 miRs were overexpressed, including miR-155-5p, miR-142-3p, miR-21-5p, miR-142-5p, and miR-223-3p showing the highest fold difference between groups.
In the urine sediments, 542 ± 53 miRs were significantly expressed. In Figure S1B, SDC, http://links.lww.com/TXD/A336, miR levels are shown for each sample. A total of 568 miRs were included in the MWU analysis, of which 2 (miR-92b-3p and miR-296-3p) showed a significantly elevated level in the urine sediments of recipients with rejection (P < 0.01). At a P value of <0.05, an additional set of 29 miRs were differentially expressed between rejection and no-rejection sediments (Table S2, SDC, http://links.lww.com/TXD/A336).
Fifteen miRs of interest (Table S3, SDC, http://links.lww.com/TXD/A336) were selected for quantification in an independent cohort of urine sediments based on the results of the preliminary screening and on literature data as described below. Three increased miRs (miR-92b-5p, miR-296-3p, and miR-25-3p) and 2 decreased miRs (miR-203a and miR-224-5p) were selected for validation based on fold change between rejection and no-rejection sediments and their absolute expression level. Four miRNAs appearing in the list from both biopsy and urine screening (miR-149-5p, miR-141-3p, miR-615-3p, and miR-126-3p) were also analyzed further. Levels of miR-210-3p, found to be decreased in urine during acute kidney transplant rejection,22 were also studied in the independent cohort. The increased relative expression of miR-155-5p, miR-142-3p, miR-21-5p, miR-142-5p, and miR-223-3p observed in the rejection biopsies confirms the results of previous miR studies in biopsies.23,24 Although these miRs were not differentially expressed between groups in our urine sediment miR profiling data, we selected them for quantification in the urine sediment cohort.
MiR Quantification in Urine Sediments of the Independent Cohort
Demographic and clinical characteristics of the transplant recipients and urine samples are shown in Table 1. Based on the UniSp6 internal control and on reference miR values, 98 rejection samples and 54 no-rejection urine sediments passed quality control. Results of miR quantification are shown in Table 2. MiR-296-3p and miR-92b were excluded for further analysis because of low correlation between duplicate measurements, probably as a result of their relatively low expression (median Cq of 32.0 and 31.4, respectively). Five miRs showed the lowest significance for differences between groups (P < 0.001, Table 2). Of these, 4 miRs were significantly higher expressed in the rejection sediments than in no-rejection sediments, namely miR-155-5p (5.7-fold), miR-126-3p (4.2-fold), miR-21-5p (3.7-fold), and miR-25-3p (2.5-fold) (see Figure 2). In contrast, miR-615-3p was significantly lower expressed (0.4-fold) in the rejection sediments. A strong positive correlation was found between miR-25-3p and miR-126-3p expression (ρ = 0.78, P < 0.001). A negative correlation was found between miR-615-3p and miR-21-5p (ρ = −0.40, P < 0.001).
TABLE 1.
Demographics and clinical characteristics of kidney transplant recipients and urine samples
| Rejection | No-rejection | P | |
|---|---|---|---|
| Transplant recipients (number) | 90 | 50 | |
| Underlying disease | 0.798 | ||
| Vascular diseasea | 22.5% | 17.4% | |
| Diabetes | 11.2% | 6.5% | |
| Nephropathyb | 15.7% | 8.7% | |
| Nephrotic syndrome/FSGS | 5.6% | 8.7% | |
| Congenital diseasec | 21.3% | 30.4% | |
| Glomerulonephritisd | 12.4% | 15.2% | |
| Vasculitise | 2.2% | 2.2% | |
| Interstitial/pyelonephritis | 5.6% | 4.3% | |
| Other (trauma, tumor) | 3.4% | 6.5% | |
| Recipient gender (M/F) | 48.8%/51.2% | 72.0%/28.0% | 0.008* |
| Recipient age (med, min–max), y | 49.0 (20.0–75.0) | 58.0 (20.0–75.0) | 0.001* |
| Donor age (median, min–max), y | 50.5 (17.0–79.0) | 57.0 (13.0–75.0) | 0.087 |
| Donor type (living/deceased) | 56.7%/43.3% | 74.0%/26.0% | 0.042* |
| hPRA (>5%/≤5%) | 30.0%/70.0% | 4.0%/96.0% | <0.001* |
| HLA-A and HLA-B mismatches, 0/1/2/3/4 (%) | 10.0/16.7/34.4/26.7/12.2 | 10.0/14.0/28.0/32.0/16.0 | 0.875 |
| HLA-DR mismatches, 0/1/2 (%) | 15.6/61.1/23.3 | 24.0/36.0/40.0 | 0.017* |
| DGF (yes/no) | 32.2%/67.8% | 10.0%/90.0% | 0.003* |
| Induction therapy (anti-IL2RA/anti-CD52) | 93.0%/7.0% | 86.0%/14.0% | 0.229 |
| Primary transplant (yes/no) | 85.6%/14.4% | 98.0%/2.0% | 0.019* |
| Urine samples (number) | 115 | 55 | |
| Time interval Tx to sampling | 0.010* | ||
| ≤1 mo | 29.6% | 12.7% | |
| 1–3 mo | 13.0% | 5.5% | |
| >3 mo | 57.4% | 81.8% | |
| Rejection severity | - | - | |
| Borderline | 6.1% | ||
| Banff IA | 25.2% | ||
| Banff IB | 21.7% | ||
| Banff IIA | 19.1% | ||
| Banff IIB | 6.1% | ||
| Banff III | 2.6% | ||
| AMR | 13.0% | ||
| Mixed AMR/TCMR | 6.1% | ||
| Serum creatinine at sampling, µmol/L | 252 ± 209 | 117 ± 32 | <0.001* |
| CKD-EPI eGFR at sampling, mL/min/1.73 m2 | 29.7 ± 13.8 | 60.3 ± 19.4 | <0.001* |
| Urinary protein, mg/24 h | 864 ± 116 | 247 ± 120 | <0.001* |
| Urine protein to creatine ratio, mg/mmol | 98.7 ± 121.6 | 22.3 ± 11.5 | <0.001* |
a(Malignant) Hypertension and/or renal vascular disease.
bIgA nephropathy, membranous nephropathy, and not otherwise specified causes of nephropathy.
cPolycystic kidney disease, congenital renal dysplasia, and Alport’s syndrome.
dMPGN, lupus glomerulonephritis, and not otherwise specified causes of glomerulonephritis.
eHenoch–Schönlein purpura and Wegener’s granulomatosis.
* P < 0.05. Differences for underlying disease, recipient gender, donor type, hPRA, HLA mismatches, DGF, primary transplant, and sampling time were tested by chi-square. Differences in recipient age, donor age, serum creatinine, eGFR, and urinary protein excretion were tested by the Mann–Whitney U test. Difference in induction therapy was tested by the Fisher exact test.
AMR, antibody-mediated rejection; DGF, delayed graft function; F, female; hPRA, historical PRA; M, male; TCMR, T-cell–mediated rejection; Tx, transplantation.
TABLE 2.
Results of RT-PCR of miRs in urine sedimentsa
| Rejection group | No-rejection group | |||
|---|---|---|---|---|
| Relative expression | Relative expression | FC | P b | |
| miR-21-5p | 73.8 (2.20–1748.2) | 20.0 (2.79–231.4) | 3.7 | <0.001 |
| miR-25-3p | 5.51 (0.73–213.3) | 2.24 (0.29–25.4) | 2.5 | <0.001 |
| miR-126-3p | 0.20 (0.01–19.1) | 0.05 (0.00–1.19) | 4.2 | <0.001 |
| miR-142-5p | 0.47 (0.04–8.94) | 0.50 (0.04–7.10) | 0.9 | 0.374 |
| miR-149-5p | 0.19 (0.00–1.96) | 0.14 (0.01–1.76) | 1.4 | 0.034 |
| miR-155-5p | 0.74 (0.04–171.4) | 0.13 (0.01–1.60) | 5.7 | <0.001 |
| miR-210-3p | 2.07 (0.05–54.6) | 1.76 (0.21–29.5) | 1.2 | 0.920 |
| miR-615-3p | 0.01 (0.00–0.88) | 0.03 (0.00–1.39) | 0.4 | <0.001 |
| miR-203a | 20.6 (0.04–502.2) | 14.4 (0.75–495.3) | 1.4 | 0.506 |
| miR-224 | 0.04 (0.00–33.5) | 0.06 (0.00–0.74) | 0.7 | 0.090 |
| miR-141-3p | 3.38 (0.07–49.4) | 2.37 (0.23–47.5) | 1.4 | 0.210 |
| miR-142-3p | 17.2 (0.57–257.1) | 12.3 (0.66–165.4) | 1.4 | 0.172 |
| miR-223-3p | 52.6 (1.93–2041.7) | 27.0 (0.64–521.9) | 1.9 | 0.004 |
aResults are presented as median relative (min-max).
bGroup sizes were 98 rejections and 54 no-rejections. Differences were tested by Mann–Whitney U tests. MiRs with significant P according to a post hoc Bonferroni correction of the alpha level (0.05/13) are shown in bold.
FC, fold change; miR, microRNA.
FIGURE 2.
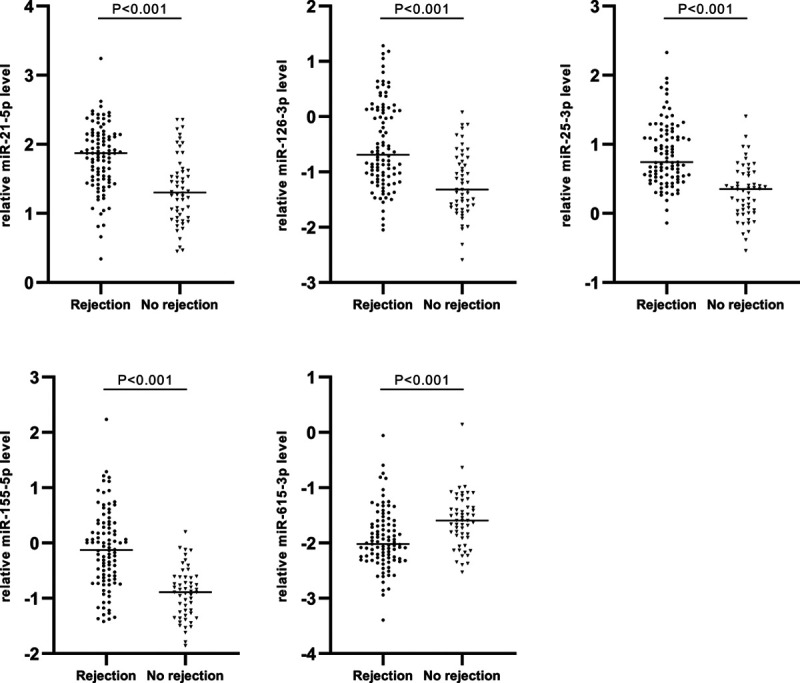
Differentially expressed miRNAs between rejection (n = 98) and no-rejection (n = 54) urine sediments. Scatter plots of relative miR expression levels are shown on a logarithmic scale. Medians are presented by horizontal lines. Differences were tested by Mann–Whitney U tests. miR, microRNA.
Quantification of Urine Proteins
CXCL-9 and CXCL-10 concentrations were determined in the urine supernatant by Luminex assays. A median CXCL-9 concentration of 1808.1 pg/mL (22.7–77 863.9) was observed in the rejection samples, which was 8-fold higher than the median CXCL-9 concentration in the no-rejection group (215.7 pg/mL, 34.6–2400.0) (P < 0.001). The median CXCL-10 concentration in the rejection samples (1227.8 pg/mL, 12.0–135962.2) was 10-fold higher compared to the no-rejection samples (115.5 pg/mL, 17.1–7429.0) (P < 0.001) (Figure 3). A strong correlation was found between urine concentrations of CXCL-9 and CXCL-10 (Figure 3; ρ = 0.82, P < 0.001).
FIGURE 3.
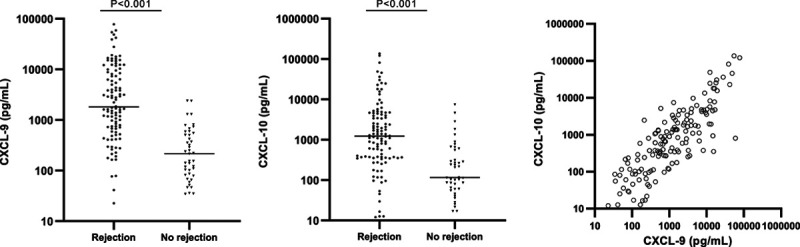
Absolute concentrations of urinary CXCL-9 and CXCL-10 in urine supernatants of the rejection (n = 108) and no-rejection groups (n = 43). Scatter plots of absolute concentrations are shown on a log-scale. Horizontal lines represent the median levels. Differences were tested by Mann–Whitney U tests.
Identifying a Urinary Signature of Rejection
To determine a set of urinary analytes that can identify active kidney rejection, logistic regression analysis was performed. The 5 miRs of interest (miR-21-5p, miR-25-3p, miR-126-3p, miR-155-5p, and miR-615-3p) were significant predictors of rejection in a univariate regression analysis (Table 3). The same was true for CXCL-9 and CXCL-10 protein concentrations (Table 3). In univariate analysis, recipient gender and age at transplantation, donor age, donor type, historical PRA, the occurrence of delayed graft function, and a previous kidney transplantation were also significantly associated with rejection. A multivariable logistic regression model was built by combining the most significant variables. In this model, miR-155-5p and miR-615-3p in urine sediments and CXCL-9 levels in urine supernatant were independent predictors of rejection. Together with recipient age, this model could distinguish urine samples from recipients with rejection and those without rejection with a sensitivity of 89.1% and specificity of 75.6% (Table 4) and an area under the curve of 0.94 in the receiver operating characteristic curve (Figure 4A). Finally, we performed a stratified 10-fold cross validation of the model, which resulted in an area under the curve of 0.92 for the receiver operating characteristic curve (Table 4; Figure 4B).
TABLE 3.
Univariate and multivariable analysis of markers and clinical characteristics
| Parametera | Univariate logistic regressionb | Multivariable logistic regression | ||
|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | |
| Recipient gender (M) | 0.28 (0.14–0.57) | <0.001* | — | |
| Recipient age at Tx | 0.96 (0.93–0.98) | 0.001* | 0.94 (0.89–0.99) | 0.014 |
| Donor age | 0.97 (0.95–0.99) | 0.037 | ||
| Donor type (deceased) | 2.05 (1.02–4.13) | 0.044 | ||
| hPRA (>5%) | 11.12 (2.56–48.26) | 0.001* | — | |
| HLA-A and -B mismatches | ||||
| 1 | 0.82 (0.23–2.91) | 0.76 | ||
| 2 | 1.29 (0.42–4.01) | 0.66 | ||
| 3 | 0.87 (0.28–2.71) | 0.80 | ||
| 4 | 0.72 (0.20–2.58) | 0.61 | ||
| HLA-DR mismatches | ||||
| 1 | 2.10 (0.91–4.83) | 0.083 | ||
| 2 | 0.93 (0.38–2.27) | 0.88 | ||
| DGF | 4.20 (1.54–11.44) | 0.005* | — | |
| Anti-IL2RA induction | 0.40 (0.13–1.24) | 0.11 | ||
| Primary transplantation | 0.12 (0.02–0.89) | 0.038 | ||
| miR-21-5p | 7.02 (3.16–15.60) | <0.001* | — | |
| miR-25-3p | 29.34 (8.60–100.09) | <0.001* | — | |
| miR-126-3p | 4.58 (2.43–8.63) | <0.001* | — | |
| miR-155-5p | 9.91 (4.41–22.26) | <0.001* | 4.36 (1.18–16.18) | 0.028 |
| miR-615-3p | 0.28 (0.14–0.57) | <0.001* | 0.12 (0.03–0.47) | 0.002 |
| CXCL-9 | 10.91 (4.78–24.91) | <0.001* | 6.72 (2.21–20.45) | 0.001 |
| CXCL-10 | 4.14 (2.32–7.38) | <0.001 | NA | |
| Days post-Tx | ||||
| 181–730 | 0.41 (0.19–0.88) | 0.378 | ||
| >730 | 0.28 (0.12–0.69) | 0.005 | ||
aThe following group sizes (rejection/no-rejection) applied to the logistic regression tests; for the miRs: 98/54, for CXCL-9: 108/43, and for the combined models: 92/41.
bTo prevent overfitting the multivariable model, a maximum of 10 of the most significant parameters from univariate (marked with *) were entered in the model. CXCL-10 was not included since it highly correlates with CXCL-9.
CI, confidence interval; DGF, delayed graft function; F, female; hPRA, historical PRA; M, male; miR, microRNA; OR, odds ratio; Tx, transplantation.
TABLE 4.
Predictive value parameters for separate markers and combined modelsa
| Variable | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | ROC AUC | 95% CI |
|---|---|---|---|---|---|---|
| miR-155-5p | 84.7 | 56.6 | 78.3 | 66.7 | 0.82 | 0.75–0.89 |
| miR-615-3p | 88.7 | 24.1 | 67.7 | 54.2 | 0.30 | 0.21–0.39 |
| CXCL-9 | 90.7 | 58.1 | 84.5 | 71.4 | 0.86 | 0.80–0.93 |
| Model miRs/CXCL-9 | 89.1 | 78.0 | 90.1 | 76.2 | 0.92 | 0.87–0.97 |
| Model plus rec.age | 89.1 | 75.6 | 89.1 | 75.6 | 0.94 | 0.90–0.98 |
| Stratified 10-fold | — | — | — | — | 0.92 | 0.87–0.97 |
Crossvalidation.
aThe following group sizes (rejection/no-rejection) applied to the logistic regression tests; for the miRs: 98/54, for CXCL-9: 108/43, and for the combined models: 92/41. The model was internally verified by a stratified 10-fold crossvalidation.
AUC, area under the curve. CI, confidence interval; miR, microRNA; NPV, negative predictive value; PPV, positive predictive value; ROC, receiver operating characteristic.
FIGURE 4.
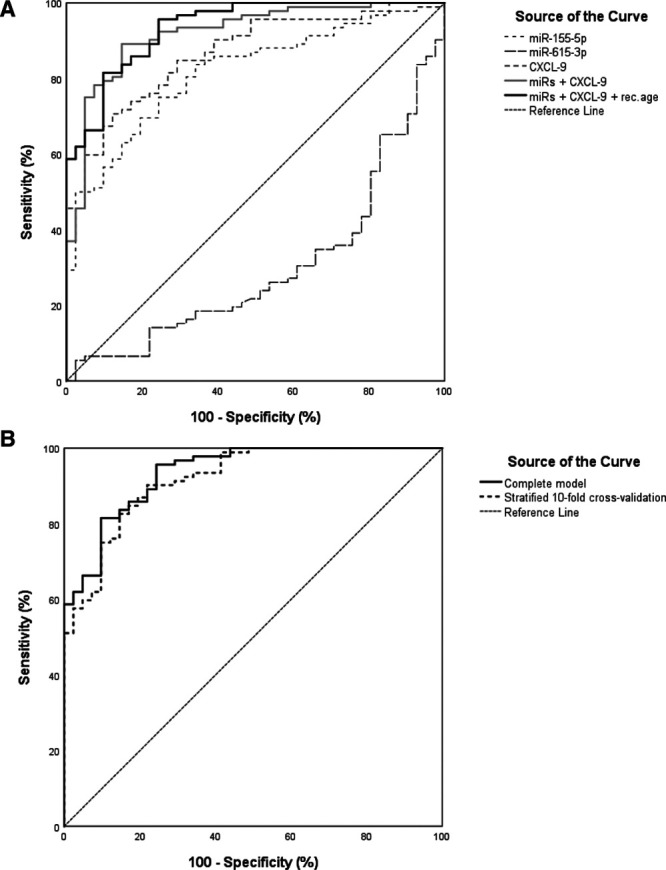
Receiver operating characteristic (ROC) curves of markers and combined models for distinction of rejection. A, ROC curves are shown for each individual miR and for CXCL-9, which were independent predictors in the combined miR/CXCL-9 model (gray line) and the combined miR/CXCL-9/recipient age model (black line). B, ROC curves for the complete model (dashed line) from (A) and for the stratified 10-fold cross validated model (solid line). The following group sizes (rejection/no rejection) applied to the ROC curves; for the miRs: 98/54, for CXCL-9: 108/43, and for the models: 92/41. miR, microRNA.
Markers in Relation to Transplant Function, Time Point, and Rejection Severity
The total study cohort was stratified according to the different stages of kidney function at the time of sampling (ie, eGFR ≤15, 15–29, 30–60, ≥60 mL/min), and we tested if the levels of key markers (miR-155-5p, miR-615-3p, and CXCL-9) differed between these categories. Significant differences between eGFR categories were especially found for CXCL-9 levels (Figure 5), partly due to case-control differences. We further investigated if the marker levels might be dependent on proteinuria and eGFR. When comparing only patients with similar urinary protein levels (≤0.25 mg/24 h) or eGFR (40–60 mL/min/1.73 m2), miRNAs and CXCL-9 still differed between the rejection and no-rejection group (Figure S2A, SDC, http://links.lww.com/TXD/A336), suggesting that differences between groups could not be explained by differences in proteinuria or graft function. Furthermore, none of the markers significantly correlated with eGFR levels in the no-rejection group (Figure S2B, SDC, http://links.lww.com/TXD/A336), whereas in the rejection group only CXCL-9 showed weak correlation with eGFR (ρ = −0.39, P < 0.001). Finally, as urinary flow rate may influence CXCL-9 concentrations, we correlated urine protein concentrations with creatinine-normalized concentrations. This correlation was high (ρ = 0.97, P < 0.0001; Figure S2C, SDC, http://links.lww.com/TXD/A336) showing minimal effect of urinary output on chemokine levels assessed.
FIGURE 5.
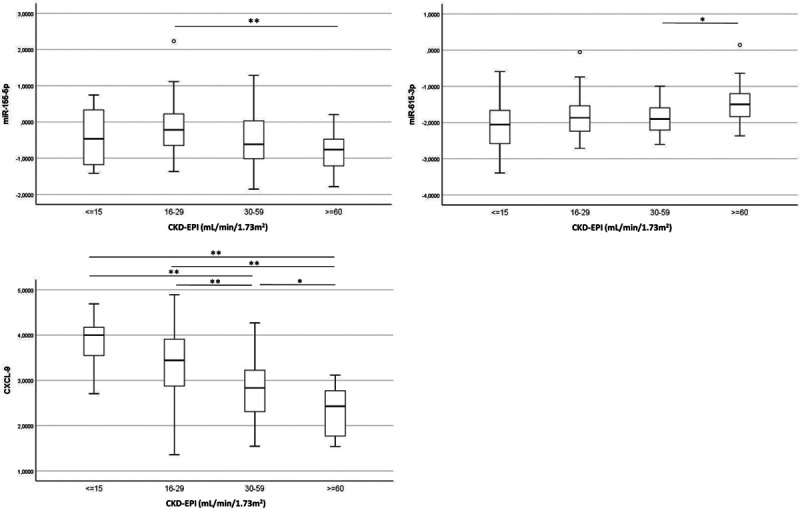
Association between markers and transplant function. MiR- and CXCL-9 levels were calculated according to eGFR category. Box plots show 50% of the observations with the whiskers representing variability outside the upper and lower quartiles. Group sizes (eGFR ≤ 15, 16–29, 30–59, ≥60) were as follows; for the miRs: 14/44/69/21 and for CXCL-9: 17/46/67/18. Differences between categories were tested by Mann–Whitney U tests and corrected for multiple testing. *P < 0.05; **P < 0.005. miR, microRNA.
When the total group of samples was split according to sampling time, differences in key markers between the rejection and no-rejection group at both an early period (≤1 mo) and later period (>3 mo) posttransplant showed a similar trend as seen for the whole groups (Figure 6).
FIGURE 6.
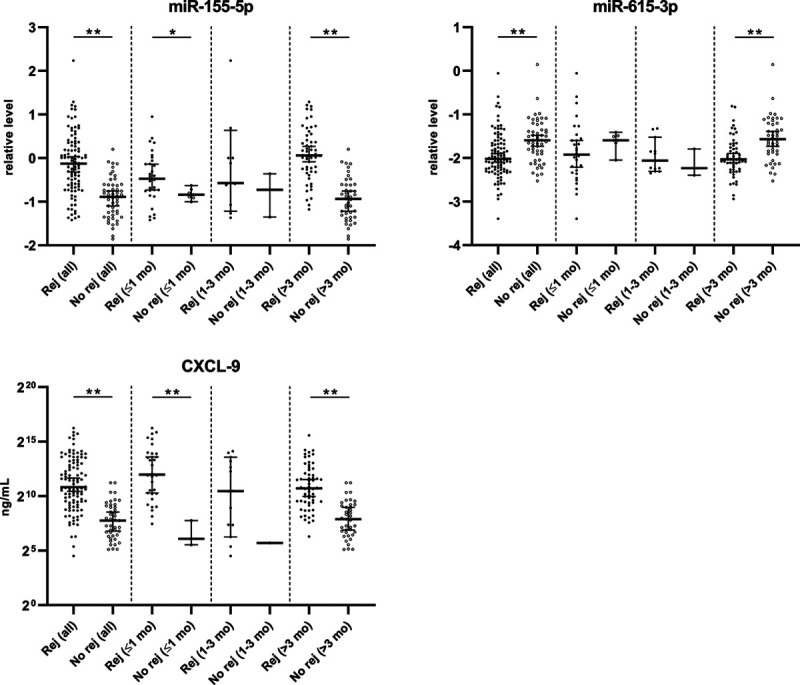
MiR- and CXCL-9 levels in the rejection and no-rejection groups, according to timing after transplantation (≤1; 1–3; >3 mo). Medians with 95% confidence interval are shown. Rej: rejection group, No rej: no-rejection group. *P < 0.05. **P < 0.005. The 1- to 3-mo category was not tested for statistical difference between groups because of low sample size in the no-rejection group. miR, microRNA.
When the rejection group was split into TCMR only, AMR only or mixed AMR/TCMR, the markers reached almost significance level (P between 0.080 and 0.099) with respect to difference between TCMR and AMR. In contrast, mixed AMR/TCMR samples showed a higher similarity to the TCMR samples (P > 0.4; Figure S3, SDC, http://links.lww.com/TXD/A336).
MiR Expression in Urine of Recipients With BKVN and CMV
We investigated the potential of miR-21-5p, miR-25-3p, miR-126-3p, miR-155-5p, and miR-615-3p to distinguish rejection from BKVN and from CMV. Only miR-615-3p levels were significantly higher in urine sediments from recipients with BKVN compared to urine sediments from recipients with rejection (P < 0.001) (Figure S4, SDC, http://links.lww.com/TXD/A336). No significant differences in miR levels were detected between rejection and CMV infection.
DISCUSSION
Urine is an accessible source for biomarker research as its molecular composition may reflect intrarenal events. We demonstrated that a combined measurement of urinary miR-155-5p and miR-615-3p, together with CXCL-9 levels can distinguish the presence of active rejection with high accuracy.
We presented a n miR signature in individual urine sediments from kidney transplant recipients, using a n RT-PCR platform that enables the simultaneous screening of >700 miRs. Although we did not use paired biopsy tissue and urine samples for the initial miR screening, 3 miRs were identified as significantly downregulated in both graft tissue and urine sediments of recipients with rejection, including miR-615-3p, miR-149-5p, and miR-141-3p. Interestingly, although miR-216-3p levels were significantly decreased in the rejecting graft, a significant increase was observed in the urine sediments of recipients with rejection.
In the second stage, we quantified in an independent cohort of urine sediments a set of miRs, which had been selected on the basis of the preliminary miR screening in biopsies and urine samples and of previous results from literature. We observed a significant increase of miR-21-5p, miR-25-3p, miR-155-5p, and miR-126-5p and a decrease of miR-615-3p in urine sediments of recipients with rejection compared to urine sediments from recipients without rejection. Distinction between rejection and infection after kidney transplantation by the use of urinary biomarkers has been notoriously difficult.25-28 Here, in urine sediments of recipients with BKV viral nephropathy, only miR-615-3p showed significant difference when compared to the rejection group. While this indicates that active rejection may be discernible from BKVN to some extent, the set of BKVN and CMV samples was rather limited.
Our findings in both urine and graft tissue confirm previous observations of increased expression of miR-155-5p, miR-142-3p, miR-142-5p, miR-21-5p, and miR-223-3p and decreased expression of miR-615-3p during acute rejection.7,22,24,29 Increased levels of miR-155-5p, miR-223-3p, and miR-142-5p probably indirectly reflect the presence of graft-infiltrating immune cells, since it was previously shown that these miRs strongly correlate with intragraft CD3 (T cell) and CD20 (B cell) mRNA levels.5 In support of this notion, miR-155-5p levels in the sediment were found to correlate significantly with levels of CXCL-9 and CXCL-10 (data not shown), which both are chemoattractants of T cells. Furthermore, the difference we detected between TCMR and AMR for several urinary markers was bigger than that between TCMR and mixed TCMR/AMR. Although the former comparison did not reach significance, this observation may suggest that the biomarker profile in the urine reflects cellular immune mechanisms in the graft.
Urinary miRs were previously investigated by Lorenzen et al who demonstrated a significant downregulation of miR-210 in urine samples of recipients with acute cellular rejection with a diagnostic sensitivity of 74% and specificity of 52% ( area under the curve 0.7). We could not confirm a decrease in the level of this miR in urine sediments at time of rejection. However, in their study, miR expression was investigated in whole urine samples instead of the cellular fraction alone. Furthermore, RNA pools of samples, rather than individual specimens, were studied by miR expression profiling, and an exogenous spike-in was added to urine samples for subsequent qPCR data normalization (cel-miR-39). We preferred to select highly expressed, relatively stable miRs from our urine sediment profiling data set to normalize expression data from the independent set of urine sediments, since adequate reference miRs for studies in urine are currently unavailable. Millán et al29 showed a significantly increased expression of miR-155-5p and miR-142-3p in urine cell pellets of recipients with acute rejection. Although both miRs were significantly upregulated in rejecting allografts in our study, we could only confirm an increased expression of miR-155-5p in urine sediments.
We found increased levels of both CXCL-9 and CXCL-10 in urine samples of recipients during a rejection episode, which is consistent with findings by other groups.14,15,29 A multivariable model containing miR-155-5p, miR-615-3p, and CXCL-9 as independent variables, together with recipient age, could be used to noninvasively distinguish the presence of active rejection. The use of a combination of biomarkers resulted in a model with a better diagnostic performance compared to the measurement of a single biomarker (Figure 4; Table 4).
One remark concerning the methodology of the current study is that urines were not collected immediately at the bed side, resulting in a variable time among the samples between collection from the patient and processing, up to several hours maximally. It may be possible that the particular urinary analytes were affected by incubation time of the sample. It is also worth mentioning that our study has a case-control design, and that several demographic variables were significantly different between the rejection and no-rejection groups. Regardless of this, after performing a multivariable analysis to correct for these demographic variables, several variables in the urine were found to be independently associated with rejection. Relatively many of the rejection samples had been taken within the first month posttransplant, whereas for the no-rejection group relatively many samples had been taken beyond 3 mo. But nevertheless, the key markers showed differences between cases and controls both in samples taken early and in samples taken later after transplantation. Obviously, to validate the findings from the current study prospective analysis of the biomarkers is further needed, including assessment of positive and negative predictive values of the biomarker-based tests. This will be challenging30 given the fact that incidence of rejection is lower nowadays. Our case group consisted of samples taken at time of graft dysfunction. Nevertheless, to rule-out dependence of marker expression on eGFR and urinary protein levels, we did verify that levels of the markers of interest still differed between subgroups of rejection and no-rejection that were similar in clinical graft function parameters. Still, a clinical biomarker gains utility when it would identify rejection before changes in graft function and before clinical diagnosis. Therefore, it is useful to set up a longitudinal study and to test such markers in urine samples from patients with subclinical rejection and from rejecting patients taken before the biopsy. In view of the current results, it would also be interesting to investigate if miRNA and chemokine levels normalize after successful treatment.
In conclusion, this retrospective cross-sectional study provides evidence that miR-155p-5p and miR-615-3p levels, together with CXCL-9 concentrations, in the urine identify the presence of active rejection in the kidney transplant.
ACKNOWLEDGMENTS
The authors thank the team of technicians and PhD students of the Transplant Immunology research group who were involved in processing and freezing the clinical samples. The authors also thank Prof. Dr Benedicte de Winter (University of Antwerp, Belgium) for a critical review of the manuscript.
Supplementary Material
Footnotes
Published online 10 June, 2021.
E.M.G. evaluated patient data, performed experiments, participated in data analyses, carried out statistical analyses, and wrote the paper. J.D.H.A. performed qPCR experiments. E.v.B. performed Luminex experiments. G.W.H. assisted in statistical analyses. H.W.D.F. coordinated collection of patient data. I.B. evaluated morphology in the transplant biopsies. S.H. participated in data analyses and critically assessed the paper. M.v.d.V. collected urine samples and patient data for the CMV/BK group. L.B.H. coordinated data collection for the CMV/BK group. M.J.K.M. coordinated collection of patient data. K.L. participated in data analysis and critically assessed the paper. F.H.J.C. participated in research design and critically assessed the paper. M.E. participated in research design and data analysis, carried out statistical analyses, and wrote parts of the paper.
The authors declare no conflicts of interest.
This work was supported by the young fellowship programme of the European Renal Association—European Dialysis and Transplant Association (ERA-EDTA), granted to E.M.G. (application number 425), and by the Roche Organ Transplant Research Foundation (# 119749307 to M.E. and F.H.J.C.).
Data in this article may be shared.
This work was supported by the young fellowship programme of the European Renal Association—European Dialysis and Transplant Association (ERA-EDTA), granted to E.G. (application number 425), and by the Roche Organ Transplant Research Foundation (# 119749307 to M.E. and F.C.).
Supplemental digital content (SDC) is available for this article. Direct URL citations appear in the printed text, and links to the digital files are provided in the HTML text of this article on the journal’s Web site (www.transplantjournal.com).
REFERENCES
- 1.Nankivell BJ, Alexander SI. Rejection of the kidney allograft. N Engl J Med. 2010; 363:1451–1462. [DOI] [PubMed] [Google Scholar]
- 2.Heidt S, Eikmans M, Roelen DL, et al. Novel strategies for immunological monitoring of kidney transplant recipients: from microRNA to alloantibodies. Clin Transpl 2013:257–267. [PubMed] [Google Scholar]
- 3.Lorenzen JM, Thum T. Circulating and urinary microRNAs in kidney disease. Clin J Am Soc Nephrol. 2012; 7:1528–1533. [DOI] [PubMed] [Google Scholar]
- 4.Wilflingseder J, Regele H, Perco P, et al. miRNA profiling discriminates types of rejection and injury in human renal allografts. Transplantation. 2013; 95:835–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anglicheau D, Sharma VK, Ding R, et al. MicroRNA expression profiles predictive of human renal allograft status. Proc Natl Acad Sci U S A. 2009; 106:5330–5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sui W, Dai Y, Huang Y, et al. Microarray analysis of MicroRNA expression in acute rejection after renal transplantation. Transpl Immunol. 2008; 19:81–85. [DOI] [PubMed] [Google Scholar]
- 7.Vitalone MJ, Sigdel TK, Salomonis N, et al. Transcriptional perturbations in graft rejection. Transplantation. 2015; 99:1882–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van de Vrie M, Deegens JK, Eikmans M, et al. Urinary microRNA as biomarker in renal transplantation. Am J Transplant. 2017; 17:1160–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lorenzen JM, Volkmann I, Fiedler J, et al. Urinary miR-210 as a mediator of acute T-cell mediated rejection in renal allograft recipients. Am J Transplant. 2011; 11:2221–2227. [DOI] [PubMed] [Google Scholar]
- 10.Scian MJ, Maluf DG, David KG, et al. MicroRNA profiles in allograft tissues and paired urines associate with chronic allograft dysfunction with IF/TA. Am J Transplant. 2011; 11:2110–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maluf DG, Dumur CI, Suh JL, et al. The urine microRNA profile may help monitor post-transplant renal graft function. Kidney Int. 2014; 85:439–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baggiolini M. Chemokines and leukocyte traffic. Nature. 1998; 392:565–568. [DOI] [PubMed] [Google Scholar]
- 13.Panzer U, Reinking RR, Steinmetz OM, et al. CXCR3 and CCR5 positive T-cell recruitment in acute human renal allograft rejection. Transplantation. 2004; 78:1341–1350. [DOI] [PubMed] [Google Scholar]
- 14.Rabant M, Amrouche L, Lebreton X, et al. Urinary C-X-C motif chemokine 10 independently improves the noninvasive diagnosis of antibody-mediated kidney allograft rejection. J Am Soc Nephrol. 2015; 26:2840–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rabant M, Amrouche L, Morin L, et al. Early low urinary CXCL9 and CXCL10 might predict immunological quiescence in clinically and histologically stable kidney recipients. Am J Transplant. 2016; 16:1868–1881. [DOI] [PubMed] [Google Scholar]
- 16.Hricik DE, Nickerson P, Formica RN, et al. ; CTOT-01 consortium. Multicenter validation of urinary CXCL9 as a risk-stratifying biomarker for kidney transplant injury. Am J Transplant. 2013; 13:2634–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haas M, Loupy A, Lefaucheur C, et al. The Banff 2017 kidney meeting report: revised diagnostic criteria for chronic active T cell-mediated rejection, antibody-mediated rejection, and prospects for integrative endpoints for next-generation clinical trials. Am J Transplant. 2018; 18:293–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kooiman J, van de Peppel WR, Sijpkens YW, et al. No increase in kidney injury molecule-1 and neutrophil gelatinase-associated lipocalin excretion following intravenous contrast enhanced-CT. Eur Radiol. 2015; 25:1926–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mestdagh P, Van Vlierberghe P, De Weer A, et al. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009; 10:R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet. 2012; 13:358–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple control genes. Genome Biol. 2003; 3:0034.0031–0034.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X, Dong C, Jiang Z, et al. MicroRNA-10b downregulation mediates acute rejection of renal allografts by derepressing BCL2L11. Exp Cell Res. 2015; 333:155–163. [DOI] [PubMed] [Google Scholar]
- 23.Van Aelst LN, Summer G, Li S, et al. RNA profiling in human and murine transplanted hearts: identification and validation of therapeutic targets for acute cardiac and renal allograft rejection. Am J Transplant. 2016; 16:99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soltaninejad E, Nicknam MH, Nafar M, et al. Differential expression of microRNAs in renal transplant patients with acute T-cell mediated rejection. Transpl Immunol. 2015; 33:1–6. [DOI] [PubMed] [Google Scholar]
- 25.Jackson JA, Kim EJ, Begley B, et al. Urinary chemokines CXCL9 and CXCL10 are noninvasive markers of renal allograft rejection and BK viral infection. Am J Transplant. 2011; 11:2228–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Ham SM, Heutinck KM, Jorritsma T, et al. Urinary granzyme A mRNA is a biomarker to diagnose subclinical and acute cellular rejection in kidney transplant recipients. Kidney Int. 2010; 78:1033–1040. [DOI] [PubMed] [Google Scholar]
- 27.Øzbay A, Tørring C, Olsen R, et al. Transcriptional profiles in urine during acute rejection, bacteriuria, CMV infection and stable graft function after renal transplantation. Scand J Immunol. 2009; 69:357–365. [DOI] [PubMed] [Google Scholar]
- 28.Yannaraki M, Rebibou JM, Ducloux D, et al. Urinary cytotoxic molecular markers for a noninvasive diagnosis in acute renal transplant rejection. Transpl Int. 2006; 19:759–768. [DOI] [PubMed] [Google Scholar]
- 29.Millán O, Budde K, Sommerer C, et al. Urinary miR-155-5p and CXCL10 as prognostic and predictive biomarkers of rejection, graft outcome and treatment response in kidney transplantation. Br J Clin Pharmacol. 2017; 83:2636–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Willis JC, Lord GM. Immune biomarkers: the promises and pitfalls of personalized medicine. Nat Rev Immunol. 2015; 15:323–329. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


