Abstract
Methionine metabolism arises as a key target to elucidate the molecular adaptations underlying animal longevity due to the negative association between longevity and methionine content. The present study follows a comparative approach to analyse plasma methionine metabolic profile using a LC-MS/MS platform from 11 mammalian species with a longevity ranging from 3.5 to 120 years. Our findings demonstrate the existence of a species-specific plasma profile for methionine metabolism associated with longevity characterised by: i) reduced methionine, cystathionine and choline; ii) increased non-polar amino acids; iii) reduced succinate and malate; and iv) increased carnitine. Our results support the existence of plasma longevity features that might respond to an optimised energetic metabolism and intracellular structures found in long-lived species.
Subject terms: Metabolomics, Ageing
Mota-Martorell and colleagues use a comparative metabolomics approach to examine plasma metabolite levels associated with methionine metabolism in 11 mammalian species. They identify species specific plasma profiles indicative of a link between lifetime longevity and methionine metabolism.
Introduction
All living organisms use the same 20 amino acids for protein synthesis. However, it seems that the minimum number to build functional proteins is around ten1. Indeed, the first proteins were built with the available amino acids generated upon the early earth via abiotic processes2. This abiotic set probably comprised the amino acids alanine (A), aspartate (D), glutamate (E), glycine (G), isoleucine (I), leucine (L), proline (P), serine (S), threonine (T) and valine (V)2. These early amino acids were encoded adaptively by the early genetic code. The evolutionary transition from this initial genetic code to a genetic code where all 20 amino acids are fixated seems to be linked to the adaptations demanded by the appearance of oxygen in the biosphere during early evolution3,4. Thus, amino acids with increased redox properties, such as arginine (R), asparagine (N), cysteine (C), glutamine (Q), histidine (H), lysine (K), methionine (M), phenylalanine (F), tyrosine (Y) and tryptophan (W) were fixated into the new genetic code as adaption to preserve aerobic life3.
Interestingly, the protein compositional content of the sulfur amino acids methionine and cysteine is species-specific and is associated with animal longevity. Thus, long-lived animal species show the lower methionine5–7,8 and cysteine9 protein content, surely as adaptive response4,10 to the low rate of endogenous damage and highly resistant macromolecular components also present in longevous species11,12,10. Reinforcing these observations, the free tissue methionine content is also lower in diverse long-lived animal species13,14; and the pro-longevity effects of nutritional (methionine restriction, MetR)15,11 and pharmacological (metformin)16 interventions are mediated by changes in methionine metabolism. In addition to its role in several intracellular processes, methionine is the core of a complex metabolic network which can be divided in three parts: methionine cycle, the transsulfuration pathway, and polyamine biosynthesis17,18 (for details see Fig. 1). Significantly, manipulation of each of these branches affects longevity in diverse experimental animal models15. Consequently, available findings point to the metabolism of methionine as a key target to study the molecular adaptive mechanisms underlying differences in animal longevity.
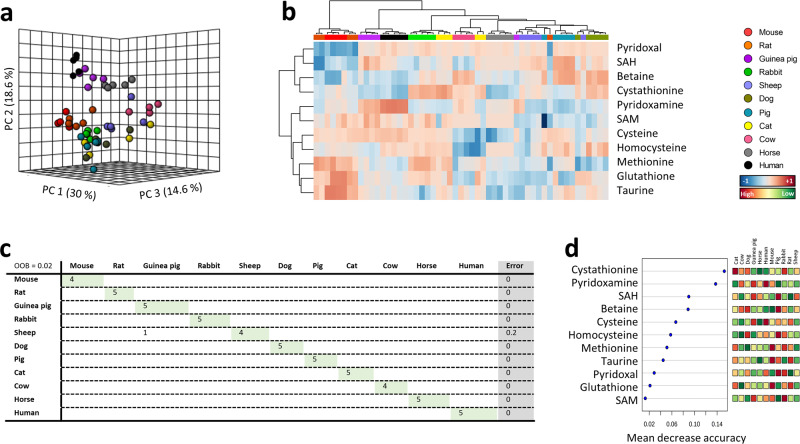
Fig. 1. Multivariate statistics reveals a species-specific methionine plasma profile.
a Principal component analyses (PCA) representation of methionine-related metabolites. X: Principal component 1 (PC1); Y: Principal component 2 (PC2); Z: Principal component 3 (PC3). b Hierarchical clustering of individual animal samples according to metabolite abundance. c Random forest (RF) classification algorithm. d VIP scores for RF. Data values are obtained from four (mice and cows), five (rats, rabbits, guinea pigs, sheep, dogs, pigs, cats and horses) or six (humans) specimens.
The purpose of this study was to investigate the methionine metabolic phenotype of long-lived mammalian species. Specifically, we have designed a study to detect and quantify a panel of metabolites including 35 different molecular species in plasma of eleven mammalian species showing more than one order of magnitude of difference in longevity, from 3.5 years in mice to 120 years in humans. The metabolites detected and quantified were: (1) methionine and its related metabolites, including the intermediates of the transmethylation pathway S-adenosylmethionine (SAM), S-adenosylhomocysteine (SAH) and homocysteine; betaine as metabolites involved in the regeneration of methionine plasma levels; the intermediates of the transsulfuration pathway cysteine and cystathionine; taurine and glutathione as downstream metabolites of the transsulfuration pathway; and vitamin B6 metabolites pyridoxal and pyridoxamine, as cofactors of the transsulfuration enzymes; (2) additional amino acids including eight non-polar amino acids (alanine, glycine, leucine, isoleucine, phenylalanine, proline, tryptophan and valine), four polar uncharged amino acids (asparagine, serine, threonine and tyrosine), two polar negatively charged amino acid (aspartate and glutamate) and two polar positively charged amino acids (arginine and histidine); (3) TCA cycle metabolites, including pyruvate, citrate, α-ketoglutarate, succinate, fumarate and malate; and (4) methionine-derived lipid intermediates, such as choline and carnitine. The plasma metabolites profile was determined using a LC-MS/MS platform to define specific phenotypic profiles associated with animal longevity.
Results
Multivariate statistics reveals a species-specific methionine-related metabolites plasma profile
In order to determine whether plasma methionine and its related metabolites concentration differed among mammals, multivariate statistics were applied. Non-supervised principal component analysis (PCA) suggested the existence of a species-specific methionine-related metabolites plasma profile (Fig. 1a), capable to explain 63.2% of sample variability. A hierarchical clustering of the samples represented by a heat map revealed a good sample clusterization according to animal species (Fig. 1b). Accordingly, the performance of a Random forest (RF) classification algorithm revealed a species overall classification error <0.1 (Fig. 1c), being cystathionine and pyridoxamine the variables with the highest contribution to classification accuracy (Fig. 1d). These data suggest that RF is a good model to identify specimens of specific mammalian species according to its plasma methionine metabolism profile.
Methionine and cystathionine plasma content are decreased in long-lived animals
The specific changes in plasma methionine and its related metabolites content across animal longevity was evaluated (Fig. 2a, Supplementary Tables 2 and 3). Specifically, methionine plasma content was decreased in long-lived animals, as well as the sulphur-containing metabolites cystathionine, taurine and glutathione. However, transmethylation metabolites, such as SAM, SAH and betaine, as well as the sulphur-containing metabolites homocysteine and cysteine, remained unchanged across animal longevity. Vitamin B6 intermediate metabolites pyridoxal and pyridoxamine, in turn, were increased in long-lived animals (Fig. 2b).
Fig. 2. Plasma methionine-related metabolites and its correlation with animal longevity.
Individual plasma correlation of the metabolites with animal longevity involved in the transmethylation and transsulfuration pathways (a) and vitamin B6 metabolites (b). Dashed lines refer to reactions in which more than one enzyme is involved. Grey-shaded boxes refer to non-detected metabolites. Pearson correlation was performed. Minimum signification level was set at p < 0.05. All metabolites were log-transformed in order to accomplish the assumptions of normality. Data values are obtained from four (mice and cows), five (rats, rabbits, guinea pigs, sheep, dogs, pigs, cats and horses) or six (humans) specimens. Enzyme codes refer to: MS methionine synthase, BHMT betaine-homocysteine S-methyltransferase, MAT methionine adenosyltransferase, MTs methyltransferases, AHCYL adenosylhomocysteinase-like 1, CBS cystathionine-β-synthase, CTH cystathionine-γ-lyase, PDXK pyridoxal kinase, PDXP pyridoxal phosphatase.
Plasma amino acid profile predicts animal species
Since plasma methionine is associated to animal longevity, we hypothesized the possibility that other amino acids could also be involved in the achievement of increased longevity. Specifically, we have been able to unambiguously detect 16 additional amino acids apart from cysteine and methionine. In order to determine whether plasma amino acid content defines animal species, multivariate statistics were applied. Non-supervised PCA suggested the existence of a different plasma amino acid profile within animal species (Fig. 3a), capable to explain 71.6% of sample variability. A hierarchical clustering of the samples represented by a heat map confirmed the existence of a species-specific plasma amino acid profile (Fig. 3b). Accordingly, RF classification algorithm revealed a species overall classification error of 0.11, being cow the specimens with the highest classification error (50% of cow specimens were not properly classified according to its amino acids plasma profile) (Fig. 3c), being serine the metabolite with the highest contribution to classification accuracy (Fig. 3d). These data suggest that RF is a good model to identify specimens of specific mammalian species according to its methionine metabolism plasma profile.
Fig. 3. Multivariate statistics reveals a species-specific amino acids plasma profile.
a Principal component analyses (PCA) representation of amino acids. X: Principal component 1 (PC1); Y: Principal component 2 (PC2); Z: Principal component 3 (PC3). b Hierarchical clustering of individual animal samples according to metabolite abundance. c RF classification algorithm. d VIP scores for RF. Data values are obtained from four (mice and cows), five (rats, rabbits, guinea pigs, sheep, dogs, pigs, cats and horses) or six (humans) specimens.
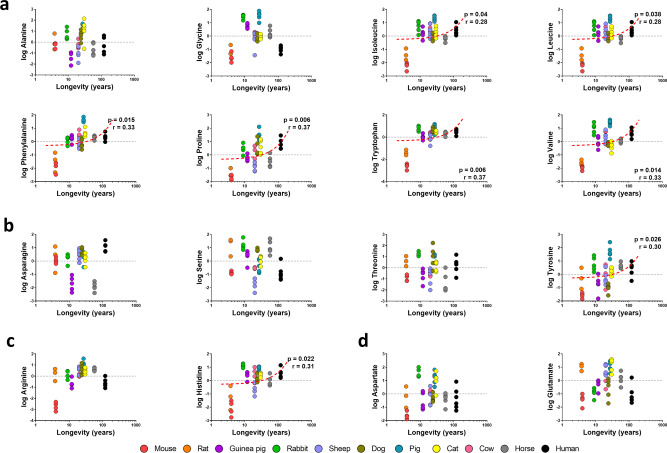
The specific changes in plasma amino acid content across animal longevity were also evaluated (Fig. 4, Supplementary Tables 2 and 3). Among the 16 additionally detected amino acid (methionine and cysteine amino acids are not included), we have found a global increase of non-polar amino acids plasma levels in long-lived animals, such as isoleucine, leucine, phenylalanine, proline, tryptophan and valine (Fig. 4a). Plasma content of specific polar amino acids, such as asparagine, tyrosine and histidine, was also increased in long-lived animals, unlike serine and glutamate, which were decreased (Fig. 4b–d).
Fig. 4. Individual plasma concentration of the detected amino acids.
a Non-polar amino acids. b Polar uncharged amino acids. c Positively charged amino acids. d Negatively charged amino acids. Pearson correlation between plasma metabolites and animal longevity was performed A–D. Minimum signification level was set at p < 0.05. All metabolites were log-transformed in order to accomplish the assumptions of normality. Data values are obtained from four (mice and cows), five (rats, rabbits, guinea pigs, sheep, dogs, pigs, cats and horses) or six (humans) specimens.
Succinate and malate plasma content are decreased in long-lived animals
Considering that amino acids can be metabolised into tricarboxylic acid (TCA) cycle intermediates, we have analysed the plasma changes for TCA cycle metabolites. Specifically, we had been able to unambiguously detect six intermediates, including pyruvate, citrate, α-ketoglutarate, succinate, fumarate and malate. In order to determine whether animal species have a specific plasma profile for TCA cycle intermediates, multivariate statistics were applied using the plasma concentration of the mentioned metabolites. A hierarchical clustering of the samples represented by a heat map revealed the existence of a shared TCA cycle intermediates plasma profile within animal species (Fig. 5a, b).
Fig. 5. Multivariate statistics reveals a shared TCA cycle intermediates plasma profile.
a Hierarchical clustering of individual animal samples according to metabolite abundance. b Hierarchical clustering animal species according to average metabolite abundance. c Individual plasma correlation of the metabolites with animal longevity involved TCA cycle. Dashed lines refer to reactions in which more than one enzyme is involved. Pearson correlation between plasma metabolites and animal longevity was performed. Minimum signification level was set at p < 0.05. All metabolites were log-transformed in order to accomplish the assumptions of normality. Data values are obtained from four (mice and cows), five (rats, rabbits, guinea pigs, sheep, dogs, pigs, cats and horses) or six (humans) specimens. Enzyme codes refer to: SDH or Cx II succinate dehydrogenase, FH fumarate hydratase.
The specific changes in plasma intermediates of TCA cycle across animal longevity were also evaluated (Fig. 5c, Supplementary Tables 2 and 3). Among the six detected metabolites, only succinate and malate were associated with animal longevity, being decreased in long-lived animals.
Amino acids and TCA cycle intermediates are bidirectionally related: the amino acids carbon skeleton can be used to synthetize TCA cycle intermediates and vice versa. Therefore, we have estimated the conversion of specific amino acids into the measured TCA intermediates across animal longevity (Supplementary Fig. 1d). In fact, long-lived animals were found to have an increased ratio of pyruvate/glycine and fumarate/aspartate. Positive correlation between fumarate and aspartate was also found (Supplementary Fig. 1e).
Methionine-derived lipid intermediates in long-lived animals
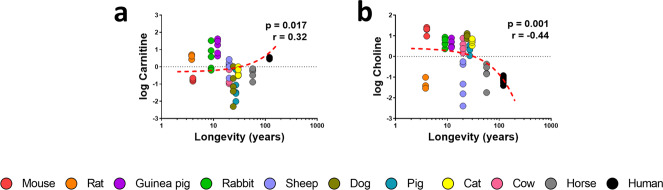
Methionine metabolism participates in the biosynthesis of lipid intermediates, such as choline and carnitine (Supplementary Tables 2 and 3). In long-lived animals, plasma carnitine was increased (Fig. 6a, b), whereas choline decreased (Fig. 6c).
Fig. 6. Methionine-derived lipid intermediates and its correlation with animal longevity.
Individual plasma concentration of lipid intermediates carnitine (a) and choline (b). Pearson correlation between plasma metabolites and animal longevity was performed. Minimum signification level was set at p < 0.05. All metabolites were log-transformed in order to accomplish the assumptions of normality. Data values are obtained from four (mice and cows), five (rats, rabbits, guinea pigs, sheep, dogs, pigs, cats and horses) or six (humans) specimens.
Pyridoxamine and succinate correlate with longevity after controlling for phylogenetic relationships
Animal species are evolutionarily related, and closely related species often have similar traits due to inheritance from a common ancestor. Most of statistical analysis, such as linear regression, assume the independence of the data, which might not be accomplished from data obtained from these close-related species. In order to find associations between longevity and plasma methionine metabolites, amino acids and TCA cycle intermediates, we have applied a phylogenetic comparative method, such as phylogenetically generalised least squares (PGLS) regression. A phylogenetic tree allowing to evolutionary relate the species in our study was inferred and is constructed in Fig. 7a.
Fig. 7. Plasma metabolites changes across mammalian longevity after correcting for phylogenetic relationships.
a Phylogenetic tree with a timescale of million years ago. b Phylogenetic generalized least squares (PGLS) regression between longevity and plasma content of pyridoxamine and succinate. Minimum signification level was set at p < 0.05. All metabolites were log-transformed in order to accomplish the assumptions of normality. Data values are obtained from four (mice and cows), five (rats, rabbits, guinea pigs, sheep, dogs, pigs, cats and horses) or six (humans) specimens.
First of all, under the assumption of a Brownian motion model of evolution (a branching, random walk of trait values from an ancestral value at the root to the tips of the tree19), we have estimated the Pagels λ. It allows to measure phylogenetic signal and indicates the relative extent to which a traits’ correlation among close relatives match a Brownian motion model of trait evolution. Pagels λ range from 0 to 1, where λ = 1 indicate that trait similarities between species are influenced by phylogenetic relationships; λ = 0 indicate that trait similarities between species are independent of phylogenetic relationships; and 0 < λ < 1 indicate different levels of phylogenetic signal. According to the estimated λ value, we have classified the measured traits according to its association degree with phylogenetic relationships (Supplementary Table 3): (1) independent (λ = 0), representing 60% of the analysed metabolites, including all the TCA cycle intermediates; (2) low dependence (λ < 0.6): methionine, taurine, pyridoxamine, glutamate and phenylalanine; (3) mild dependence (λ > 0.6): histidine, proline and tryptophan; and strong dependence (λ = 1): cystathionine, homocysteine, aspartate, tyrosine, choline. Finally, we have applied a PGLS regression, which revealed that only pyridoxamine (p = 0.045, r = −0.66) and succinate (p = 0.044, r = −0.69) (Fig. 7b, Supplementary Table 3) plasma content were positively correlated with animal longevity after controlling for phylogenetic relationships.
Discussion
Blood bathes cells and tissues, and serves as body’s major metabolite carrier20. Cells release molecules to bloodstream as a consequence of their active metabolism, which will be either excreted or transported, and up taken by cells located in different tissues and organs, according to their specific metabolic needs21. Therefore, plasma (blood’s liquid fraction) metabolome is modulated by cells and tissues, and its composition represents a global metabolic state from a living organism. Hence, plasma metabolome is the result of the metabolites that diffuse to bloodstream from each tissue as a consequence of its specific metabolism.
The achievement of a long-living phenotype requires the modulation of intracellular pathways which, in turn, regulate intracellular processes14,22,23 that converge in synthetizing resistant intracellular structures, and limiting endogenous damage production12. Consequently, a globally optimized metabolic state is achieved and expressed as specific transcriptomic24–26,19,27,28,29, proteomic30,31, lipidomic32–34,35 and metabolomic36,37,13,38 profile for each tissue38,39,40. In the present study, we have identified the methionine-related metabolites as clue molecules defining a species-specific plasma metabolic profile. Furthermore, our model suggests that using these metabolites it is possible to identify the species of a mammalian specimen, being plasma cystathionine and pyridoxamine, along with the amino acid serine, the highest predictors.
Reduced methionine content had been widely associated with extended longevity. Accordingly, previous studies had reported reduced DNA methionine-encoded5, as well as decreased tissue6,7,8 and plasma13 content in long-lived species. Furthermore, MetR leads to an extended longevity in different experimental models11,15,10. However, few studies have evaluated global changes in plasma13,41 and tissue14 methionine metabolism intermediates associated to animal longevity. Globally, these studies suggested that species longevity is associated with reduced sulphur-containing metabolites, and accompanied by decreased methionine plasma content13,41. Accordingly, our study revealed reduced steady-state plasma levels of methionine and cystathionine in long-lived mammals.
Transsulfuration, which starts with the metabolization of homocysteine into cystathionine via cystathionine-β-synthase, has been widely associated with animal longevity. Specifically, transsulfuration is enhanced in long-lived flies17 and mice42, in comparison to their non-longevous counterparts. The longevity effects of transsulfuration have been attributed to the generation of molecules with antioxidant properties, such as hydrogen sulfide (H2S)43–45,46, glutathione and taurine47,48. However, our study reveals that plasma transsulfuration intermediates or products remain unchanged (homocysteine and cysteine) across species, and even decreased (cystathionine, glutathione and taurine) in long-lived mammalian species. Under the presumption that “less is more” in long-lived species, we suggest that the steady-state levels of these intermediates are not increased since long-lived species produce less intracellular damage and are structurally built upon more resistant molecules. Therefore, these long-lived species do not need to synthetize a vast amount of antioxidant and can re-direct the energy saved to maintain other vital cellular functions. Altogether, these data support the existence of basal metabolic differences that are associated with interspecies longevity, such as decreased methionine and cystathionine plasma levels.
Proteins are essential structural components, but also the effectors of intracellular functions. Long-lived species have evolved by optimising protein composition to ensure proper protein structure, function and turnover. Synthesising more resistant proteins49 and enhancing autophagy50,51,52 to efficiently clear altered and non-functional proteins results in a negative correlation between global protein turnover and species longevity53. Accordingly, our data reveal a globally increased plasma free amino acid pool in long-lived species. Furthermore, the decreased serine plasma content, which can provide one-carbon units to regenerate methionine from homocysteine under methionine depleted conditions54, as we reported in the present study, supports the role of methionine metabolism as determinants of animal species and longevity.
Mitochondrion structural and functional adaptations associated with longevity had been widely described, and include decreased membrane unsaturation12, lower permeability23,55 and modulation of mitochondrial dynamics56. Nonetheless, quantitative and qualitative complex I (Cx I) adaptations, such as reduced total content12, and Cx I specific subunits23 that support its assembly57, resulting in reduced reactive oxygen species58–60,57,11,12, constitutes a hallmark of longevity.
Electron transport chain is feed from electron donors derived from the TCA cycle, such as NADH2 and FADH2, which transfer electrons to Cx I and Cx II, respectively. As expected, TCA cycle modulations have also been associated with animal longevity, and include the upregulation of TCA cycle genes61,62,27,63,64, which might result in an optimised energetic metabolism. In contrast with these data, our results revealed reduced basal content of succinate and malate in plasma from long-lived species, supporting the existence of metabolic adaptations associated to species longevity, different from those specific individual adaptations associated to specimen longevity. Succinate is the substrate of Cx II, and the only TCA cycle intermediate known to trigger organismal functions regulating the immune system65. We suggest that since long-lived species have an optimised metabolism with more efficient Cx I, they rely less in succinate and Cx II activity to enter electrons and generate ATP, therefore, they have reduced steady levels of succinate compared to short-lived ones. Accordingly, increased plasma levels of malate in aged individuals are associated with higher cardiovascular risk 36.
Methionine metabolism and TCA cycle are connected with lipid metabolism through choline and carnitine, respectively. Choline is synthetized from betaine, and is the biosynthetic precursor of phosphocholines (PC), which can be metabolised into lysoPC or catabolised into phosphoserine by exchanging choline by serine groups66. Previous studies in long-lived species had already reported the existence of a specific plasma PC profile in centenarians67,35, as well as a reduction of PC content in centenarians67 and the exceptionally long-living naked-mole rat (NMR)41. Our results revealed reduced plasma choline in long-lived species, which might result in a reduced PC biosynthesis, although its reduced plasma content as a consequence of increased PC biosynthesis cannot be discarded.
Carnitine acts as a shuttle essential to transport fatty acids from the cytosol to the mitochondrial matrix, where β-oxidation occurs68. When combined with fatty acids, acyl-carnitines are formed. Previous studies revealed that carnitine plasma content is reduced in NMR compared to mice41. Our data revealed higher plasma carnitine content in long-lived species, which might suggest an improved β-oxidation. Accordingly, previous proteomic results revealed a shift in mitochondrial metabolism from mitochondrial respiration to fatty acid metabolism for energy production in NMRs in comparison to short-lived guinea pigs30. Supporting our results, recent studies found an association between reduced serum carnitine and human frailty69. Furthermore, carnitine intake has been demonstrated to have neuroprotective effects 70,68.
Comparative studies across species with different lifespan are a powerful source of information to identify mechanisms linked to extended longevity39,38,14,23. However, due to species evolution, interspecies studies have to deal with the problem of non-independence of the data due to evolutionary relationships71. Therefore, it is important to elucidate whether a specific trait correlates with longevity differences, or alternatively, these differences arise because of the data similarity, by applying statistical phylogenetic comparative methods, such as PGLS32,72–74,14,23,28,75. In the present study, PGLS revealed that pyridoxamine and succinate plasma content changes associated with longevity were not due interspecies relationships. Therefore, these results support the involvement of sulphur-containing metabolites biosynthesis and energetic metabolism in determining species longevity.
When performing interspecies studies, it has been a matter of controversy whether to correct or not the observed correlations for species body size and/or metabolic rate76,77. Although it has been suggested that there is a positive correlation between longevity and body size, up to date, it has not been attributed a physiological mechanism to body size that contributes to the ageing process. Besides, there are several species that scape this rule of thumb, such as humans, and live longer than what is expected for their body size78. Accordingly, many exceptions exist to the Pearl’s rate of living theory of aging79 that suggests the existence of an association between species-specific metabolic rate and longevity. Hence, we believe that correcting our data for a correlation that is not shared for all living species, or at least for one of the species included in our analysis, would lead to misleading results. Besides, as stated by Barja76, correcting physiological associations using mathematical concepts would erase biologically meaningful information regarding species longevity.
Globally, the obtained results reveal the existence of a species-specific plasma methionine metabolic profile. Furthermore, this metabolic profile is an optimised feature of species longevity, characterised by (1) reduced plasma levels of sulphur-containing metabolites methionine and cystathionine and TCA cycle intermediates succinate and malate; (2) increased amino acids; (3) reduced choline; and (4) increased carnitine in long-lived species.
It is important to remark that although plasma composition reflects organismal metabolic status, we cannot disregard the possibility of the existence of controlled mechanisms regulating plasma composition, such as metabolite transports, which might differ across tissue and organs. Furthermore, intracellular function of the described metabolites has already been described, but a lack of knowledge is found regarding plasmatic function of plasma metabolites. Therefore, more studies focused on elucidating the function of the plasmatic profiles associated to longevity is needed.
Apart from lack-of-knowledge limitations, our work also has some technical limitations. First, the fact that although metabolome offers a wide amount of information, we cannot rule out the fact that plasma metabolome is highly affected by internal factors, such as tissue metabolism, or mixture of plasma with other biological fluids during animal sacrifice; and external factors, such as diet or circadian rhythms80,81. Nevertheless, we believe that these changes might be a reflection of individual or specimen-specific plasma metabolome modulations within a specie, and that the obtained plasma profile in our work reveals species-specific organismal metabolic adaptations associated to mammalian longevity. Besides, another limitation of our work is that we have only included a set of eleven mammalian species, which represents a minor percentage of living species. Therefore, it would be interesting to increase the number of analysed species, and also include species with similar body size but different longevities, such as birds and rodents.
The present work describes a species-specific longevity profile, which is the result of a whole organismal metabolic reorganisation that exert a “big effect” on longevity, but needs to be differentiated from other metabolic adaptations exerting a “small effect” on longevity, as suggested previously22. The later, represent the individual metabolic adaptations resulting from genetic modulations or nutritional interventions that determine whether a specimen from a specific specie becomes long-lived on not. Most of the efforts have been directed to study the longevity metabolomic determinants of long-lived specimens13,82,41, vertebrate83,42 and invertebrate long-lived mutants84,17, or its modulation by nutritional interventions increasing longevity85,86,87. The results obtained from these studies are of big value since allow to describe strategies that would be feasible to apply to humans in order to improve our health span. However, these results only represent the “small effect” determinants of longevity, and might resemble, such as decreased Cx I23,57,6 and/or reduced intracellular damage6,88,8, or differ, such as plasma metabolic profile82,83,13,69,41,87, from those obtained when comparing species of different longevities, as described previously. In the recent years, the number of interspecies studies have increased, since they provide the basal metabolic status of each species and to compare it with other species, which is essential to unravel the mechanisms underlying an improved longevity and to properly design strategies to improve human health span.
Methods
Chemicals
Unless otherwise specified, all reagents were from Sigma-Aldrich, and of the highest purity available.
Samples
Mammalian species included in the study were male adult specimens with an age representing their 15–30% of their longevity. The recorded values for longevity (in years) were: mouse (Mus musculus, n = 4), 4; rat (Rattus norvegicus, n = 5), 3.8; rabbit (Oryctolagus cuniculus, n = 5), 9; guinea pig (Cavia porcellus, n = 5), 12; sheep (Ovis aries, n = 5), 20; cow (Bos taurus, n = 4), 20; dog (Canis lupus, n = 5), 24; pig (Sus scrofa, n = 5), 27; cat (Felis catus, n = 5), 30; horse (Equus caballus, n = 5), 57; and human (Homo sapiens, n = 6), 120. Longevity values chosen for dog and sheep samples are approximated, and do not correspond to a specific breed. Rodent and rabbit specimens were obtained from rodent husbandries and blood samples were obtained after sacrifice by decapitation. For humans, blood samples were acquired from healthy adult individuals by venipuncture. The animal care protocols were approved by the Animal Experimentation Ethics Committee of the University of Lleida, and all relevant ethical regulations were complied. Human protocols were approved by the Committee for Ethics in Clinical Research of the Hospital Universitari Arnau de Vilanova, in accordance with the Declaration of Helsinki. All subjects were fully informed of the aims and scope of the research and signed an informed consent. Blood samples from rodent and rabbit specimens, and subjects, were obtained in the morning after fasting (8–12 h). Blood samples were centrifuged to separate plasma fraction, which was immediately frozen in liquid nitrogen and stored at −80 °C before 4 h until analyses. For dog, cat, sheep, pig, cow, and horse specimens, blood samples were acquired by venipuncture in the morning after fasting (8–12 h), and after centrifugation, plasma samples were obtained and preserved at −80 °C. Blood acquisition and processing to obtain plasma was performed by Charles Rivers Laboratories France Iffa-Credo (France).
Sample processing
Plasma metabolites extraction was performed based on the methodology previously described (Method 189). Briefly, 10 µL of plasma were added to 30 µL of cold methanol containing 1 μg/mL of Phe-13C as internal standard and 1 μM BHT as antioxidant. Then, samples were incubated at room temperature for 15 min and centrifuged at 12,000 × g for 3 min. Finally, the supernatant was filtrated through a 0.22-μm organic diameter filter (CLS8169, Sigma, Madrid, Spain) and transferred to Agilent (Barcelona, Spain) vials with glass inserts for further analysis.
Sulphur-containing metabolites were extracted on the bases of the methodology previously described (Method 290). Briefly, 2 µL of 5% DTT diluted in methanol (m/v) were added to 10 µL of plasma. The resulting solution was vortexed for 1 min and allowed to stand at room temperature for 10 min. For protein precipitation, 40 µL of acetonitrile containing 0.1% formic acid (v/v), 0.05% trifluoroacetic acid (v/v) and 1 µg/mL of Phe-13C as internal standard was added to the sample, and the solution was vortexed for 2 min. Then, samples were incubated at room temperature for 15 min and centrifuged at 12,000 × g for 3 min. Finally, the supernatant was filtrated through a 0.22-μm organic diameter filter (CLS8169, Sigma, Madrid, Spain) and transferred to Agilent (Barcelona, Spain) vials with glass inserts for further analysis.
Analysis conditions
The individual conditions for the detection and quantification of plasma metabolites are listed in Supplementary Table 1. For non-sulphur-containing metabolites, 2 µL of extracted sample was injected based on the method described (Method 189). Chromatographic separation was achieved on a reversed-phase column (Zorbax SB-Aq 2.1 × 50 mm, 1.8 µm particle size, Agilent Technologies, Barcelona, Spain) equipped with a pre-column (Zorba-SB-C8 Rapid Resolution Cartridge, 2.1 × 30 mm, 3.5 µm particle size, Agilent Technologies, Barcelona, Spain) with a column temperature of 60 °C. The flow rate was 0.6 mL/min during 19 min. Solvent A was composed of water containing 0.2 % acetic acid (v/v) and solvent B was composed of methanol containing 0.2 % acetic acid (v/v). The gradient started at 2 % of solvent B and increased to 98 % B in 13 min and held for 6 min. Post-time was established in 5 min. Electrospray ionization was performed in both positive and negative ion mode (depending on the target metabolite) using N2 at a pressure of 50 psi for the nebulizer with a flow of 12 L/min and a temperature of 325 °C, respectively.
For sulphur-containing metabolites, 10 µL of extracted sample was injected based on the method described (Method 290). Chromatographic separation was achieved on a reversed-phase Supelcosil LC-CN column (Supelco of 4.6 × 250 mm, 5 µm particle size, Sigma, Madrid, Spain) with a column temperature of 30 °C. The flow rate was maintained at 0.5 mL/min during 10 min using a mobile phase of 10:90 acetonitrile/water with 0.1% formic acid (v/v). Electrospray ionization was performed in both positive and negative ion mode (depending on the target metabolite) using N2 at a pressure of 50 psi for the nebulizer with a flow of 12 L/min and a temperature of 325 °C, respectively.
Data were collected using the MassHunter Data Analysis Software (Agilent Technologies, CA, USA). Samples were decoded and randomized before injection. Metabolite extraction quality controls (plasma samples with internal Phe-13C) were injected every ten samples. Peak determination and peak area integration were carried out with MassHunter Quantitative Analyses (Agilent Technologies, CA, USA).
Equipment
The analysis was performed through liquid chromatography coupled to a hybrid mass spectrometer with electrospray ionization and a triple quadrupole mass analyser. The liquid chromatography system was an ultra-performance liquid chromatography model 1290 coupled to LC-ESI-QqQ-MS/MS model 6420 both from Agilent Technologies (Barcelona, Spain).
Statistics and reproducibility
Prior to statistical analyses, data were pre-treated (auto-scaled and log-transformed). Multivariate statistics was performed using Metaboanalyst software91, and include PCA, hierarchical clustering analysis represented by a heat map, and RF used as a classification algorithm of animal species according to its plasma metabolome. Univariate statistics (Pearson correlation, Pearson correlation matrix, linear models and PGLS regression) were performed using RStudio (v1.1.453). Correlation functions were included in the packages Hmisc92 and corrplot93, and plotted with ggplot294. Linear regression was plotted using GrapPhad Prism (v8.0.1). PGLS regression functions were included in the package caper95, and used to analyse the correlation of individual metabolites with animal longevity after controlling for phylogenetic relationships, defined by a phylogeny. The phylogenetic tree was constructed using taxa names as described previously96. To ensure data reproducibility, 4–6 independent specimens for each species was included in the analyses.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
This work was supported by the Spanish Ministry of Science, Innovation and Universities (RTI2018-099200-B-I00), and the Generalitat of Catalonia (Agency for Management of University and Research Grants (2017SGR696) and Department of Health (SLT002/16/00250)) to R.P. This study has been co-financed by FEDER funds from the European Union (“A way to build Europe”). IRBLleida is a CERCA Programme/Generalitat of Catalonia. M.J. is a ‘Serra Hunter’ Fellow. N.M.M. received a predoctoral fellowship from the Generalitat of Catalonia (AGAUR, ref 2018FI_B2_00104).
Author contributions
R.P designed the study. N.M.M., M.J. and R.B. performed experimental work. N.M.M, M.J. and R.P. analysed the data. R.P. supervised the design and data interpretation. The manuscript was written by N.M.M, M.J. and R.P. and edited by R.P. All authors discussed the results and commented on the manuscript.
Data availability
All data generated or analysed during this study are included in this published article and its supplementary information files (Supplementary Data 1)
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Loredana Quadro and Luke R. Grinham. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-021-02254-3.
References
- 1.Fan K, Wang W. What is the minimum number of letters required to fold a protein? J. Mol. Biol. 2003;328:921–926. doi: 10.1016/S0022-2836(03)00324-3. [DOI] [PubMed] [Google Scholar]
- 2.Longo LM, Blaber M. Protein design at the interface of the pre-biotic and biotic worlds. Arch. Biochem. Biophys. 2012;526:16–21. doi: 10.1016/j.abb.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 3.Granold M, Hajieva P, Toşa MI, Irimie F-D, Moosmann B. Modern diversification of the amino acid repertoire driven by oxygen. Proc. Natl Acad. Sci. 2018;115:41–46. doi: 10.1073/pnas.1717100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moosmann B, Schindeldecker M, Hajieva P. Cysteine, glutathione and a new genetic code: biochemical adaptations of the primordial cells that spread into open water and survived biospheric oxygenation. Biol. Chem. 2020;401:213–231. doi: 10.1515/hsz-2019-0232. [DOI] [PubMed] [Google Scholar]
- 5.Aledo JC, Li Y, de Magalhães JP, Ruíz-Camacho M, Pérez-Claros JA. Mitochondrially encoded methionine is inversely related to longevity in mammals. Aging Cell. 2011;10:198–207. doi: 10.1111/j.1474-9726.2010.00657.x. [DOI] [PubMed] [Google Scholar]
- 6.Pamplona R, et al. Protein and lipid oxidative damage and complex I content are lower in the brain of budgerigar and canaries than in mice. Relat. Aging Rate. Age. 2005;27:267–280. doi: 10.1007/s11357-005-4562-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Portero-Otín M, Requena JR, Bellmunt MJ, Ayala V, Pamplona R. Protein nonenzymatic modifications and proteasome activity in skeletal muscle from the short-lived rat and long-lived pigeon. Exp. Gerontol. 2004;39:1527–1535. doi: 10.1016/j.exger.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 8.Ruiz MC, et al. Protein methionine content and MDA-lysine adducts are inversely related to maximum life span in the heart of mammals. Mech. Ageing Dev. 2005;126:1106–1114. doi: 10.1016/j.mad.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 9.Moosmann B. Respiratory chain cysteine and methionine usage indicate a causal role for thiyl radicals in aging. Exp. Gerontol. 2011;46:164–169. doi: 10.1016/j.exger.2010.08.034. [DOI] [PubMed] [Google Scholar]
- 10.Pamplona R, Barja G. An evolutionary comparative scan for longevity-related oxidative stress resistance mechanisms in homeotherms. Biogerontology. 2011;12:409–435. doi: 10.1007/s10522-011-9348-1. [DOI] [PubMed] [Google Scholar]
- 11.Pamplona R, Barja G. Mitochondrial oxidative stress, aging and caloric restriction: The protein and methionine connection. Biochim. Biophys. Acta. 2006;1757:496–508. doi: 10.1016/j.bbabio.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 12.Pamplona R, Barja G. Highly resistant macromolecular components and low rate of generation of endogenous damage: Two key traits of longevity. Ageing Res. Rev. 2007;6:189–210. doi: 10.1016/j.arr.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 13.Lewis KN, Rubinstein ND, Buffenstein R. A window into extreme longevity; the circulating metabolomic signature of the naked mole-rat, a mammal that shows negligible senescence. GeroScience. 2018;40:105–121. doi: 10.1007/s11357-018-0014-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mota-Martorell, N. et al. Gene expression and regulatory factors of the mechanistic target of rapamycin (mTOR) complex 1 predict mammalian longevity. GeroScience. 42, 1157–173 (2020a). [DOI] [PMC free article] [PubMed]
- 15.McIsaac RS, Lewis KN, Gibney PA, Buffenstein R. From yeast to human: exploring the comparative biology of methionine restriction in extending eukaryotic life span. Ann. N. Y. Acad. Sci. 2016;1363:155–170. doi: 10.1111/nyas.13032. [DOI] [PubMed] [Google Scholar]
- 16.Cabreiro F, et al. Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism. Cell. 2013;153:228–239. doi: 10.1016/j.cell.2013.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parkhitko, A. A., Jouandin, P., Mohr, S. E. & Perrimon, N. Methionine metabolism and methyltransferases in the regulation of aging and lifespan extension across species. Aging Cell18, e13034 (2019). [DOI] [PMC free article] [PubMed]
- 18.Sanderson SM, Gao X, Dai Z, Locasale JW. Methionine metabolism in health and cancer: a nexus of diet and precision medicine. Nat. Rev. Cancer. 2019;19:625–637. doi: 10.1038/s41568-019-0187-8. [DOI] [PubMed] [Google Scholar]
- 19.Inglés M, et al. Centenarians overexpress pluripotency-related genes. J. Gerontol. Ser. A. 2019;74:1391–1395. doi: 10.1093/gerona/gly168. [DOI] [PubMed] [Google Scholar]
- 20.Suarez-Diez M, et al. Plasma and serum metabolite association networks: comparability within and between studies using NMR and MS profiling. J. Proteome Res. 2017;16:2547–2559. doi: 10.1021/acs.jproteome.7b00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Psychogios N, et al. The human serum metabolome. PLoS One. 2011;6:e16957. doi: 10.1371/journal.pone.0016957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barja G. Towards a unified mechanistic theory of aging. Exp. Gerontol. 2019;124:110627. doi: 10.1016/j.exger.2019.05.016. [DOI] [PubMed] [Google Scholar]
- 23.Mota-Martorell, N. et al. Low abundance of NDUFV2 and NDUFS4 subunits of the hydrophilic complex I domain and VDAC1 predicts mammalian longevity. Redox Biol. 34, 101539 (2020b). [DOI] [PMC free article] [PubMed]
- 24.Borrás C, Serna E, Gambini J, Inglés M, Vina J. Centenarians maintain miRNA biogenesis pathway while it is impaired in octogenarians. Mech. Ageing Dev. 2017;168:54–57. doi: 10.1016/j.mad.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 25.Fushan AA, et al. Gene expression defines natural changes in mammalian lifespan. Aging Cell. 2015;14:352–365. doi: 10.1111/acel.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gambini J, et al. Identificación de polimorfismos de nucleótido simple en centenarios. Rev. Esp. Geriatr. Gerontol. 2016;51:146–149. doi: 10.1016/j.regg.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 27.Ma S, et al. Cell culture-based profiling across mammals reveals DNA repair and metabolism as determinants of species longevity. Elife. 2016;5:1–25. doi: 10.7554/eLife.19130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muntané G, et al. Biological processes modulating longevity across primates: a phylogenetic genome-phenome analysis. Mol. Biol. Evol. 2018;35:1990–2004. doi: 10.1093/molbev/msy105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Passtoors WM, et al. Transcriptional profiling of human familial longevity indicates a role for ASF1A and IL7R. PLoS One. 2012;7:e27759. doi: 10.1371/journal.pone.0027759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heinze I, et al. Species comparison of liver proteomes reveals links to naked mole-rat longevity and human aging. BMC Biol. 2018;16:82. doi: 10.1186/s12915-018-0547-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lehallier B, et al. Undulating changes in human plasma proteome profiles across the lifespan. Nat. Med. 2019;25:1843–1850. doi: 10.1038/s41591-019-0673-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jové M, et al. Plasma long-chain free fatty acids predict mammalian longevity. Sci. Rep. 2013;3:3346. doi: 10.1038/srep03346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jové M, et al. A stress-resistant lipidomicsignature confers extreme longevity to humans. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2017;72:30–37. doi: 10.1093/gerona/glw048. [DOI] [PubMed] [Google Scholar]
- 34.Mota-Martorell N, Pradas I, Jové M, Naudí A, Pamplona R. Biosíntesis de novo de glicerofosfolípidos y longevidad. Rev. Esp. Geriatr. Gerontol. 2019;54:88–93. doi: 10.1016/j.regg.2018.05.006. [DOI] [PubMed] [Google Scholar]
- 35.Pradas I, et al. Exceptional human longevity is associated with a specific plasma phenotype of ether lipids. Redox Biol. 2019;21:101127. doi: 10.1016/j.redox.2019.101127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng S, et al. Distinct metabolomic signatures are associated with longevity in humans. Nat. Commun. 2015;6:6791. doi: 10.1038/ncomms7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Houtkooper RH, Williams RW, Auwerx J. Metabolic networks of longevity. Cell. 2010;142:9–14. doi: 10.1016/j.cell.2010.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma S, et al. Organization of the mammalian metabolome according to organ function, lineage specialization and longevity. Cell Metab. 2015;22:332–343. doi: 10.1016/j.cmet.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bozek K, et al. Lipidome determinants of maximal lifespan in mammals. Sci. Rep. 2017;7:1–5. doi: 10.1038/s41598-017-00037-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sahm A, et al. Higher gene expression stability during aging in long-lived giant mole-rats than in short-lived rats. Aging. 2018;10:3938–3956. doi: 10.18632/aging.101683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Viltard M, et al. The metabolomic signature of extreme longevity: naked mole rats versus mice. Aging. 2019;11:4783–4800. doi: 10.18632/aging.102116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vitvitsky V, Martinov M, Ataullakhanov F, Miller RA, Banerjee R. Sulfur-based redox alterations in long-lived Snell dwarf mice. Mech. Ageing Dev. 2013;134:321–330. doi: 10.1016/j.mad.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hine C, et al. Endogenous hydrogen sulfide production is essential for dietary restriction benefits. Cell. 2015;160:132–144. doi: 10.1016/j.cell.2014.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller DL, Roth MB. Hydrogen sulfide increases thermotolerance and lifespan in Caenorhabditis elegans. Proc. Natl Acad. Sci. 2007;104:20618–20622. doi: 10.1073/pnas.0710191104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ng LT, et al. Lifespan and healthspan benefits of exogenous H2S in C. elegans are independent from effects downstream of eat-2 mutation. Npj Aging Mech. Dis. 2020;6:6. doi: 10.1038/s41514-020-0044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei Y, Kenyon C. Roles for ROS and hydrogen sulfide in the longevity response to germline loss in Caenorhabditis elegans. Proc. Natl Acad. Sci. 2016;113:E2832–E2841. doi: 10.1073/pnas.1524727113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ito T, et al. Tissue depletion of taurine accelerates skeletal muscle senescence and leads to early death in mice. PLoS One. 2014;9:e107409. doi: 10.1371/journal.pone.0107409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamori, Y. et al. Taurine as the nutritionalfactor for the longevity of the japanese revealed by a world-wide epidemiological survey. Adv Exp Med Biol.643, 13–25 (2009). [DOI] [PubMed]
- 49.Pickering AM, Lehr M, Kohler WJ, Han ML, Miller RA. Fibroblasts from longer-lived species of primates, rodents, bats, carnivores, and birds resist protein damage. J. Gerontol. Ser. A. 2015;70:791–799. doi: 10.1093/gerona/glu115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pickering AM, Lehr M, Miller RA. Lifespan of mice and primates correlates with immunoproteasome expression. J. Clin. Investig. 2015;125:2059–2068. doi: 10.1172/JCI80514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pride H, et al. Long-lived species have improved proteostasis compared to phylogenetically-related shorter-lived species. Biochem. Biophys. Res. Commun. 2015;457:669–675. doi: 10.1016/j.bbrc.2015.01.046. [DOI] [PubMed] [Google Scholar]
- 52.Rodriguez KA, et al. Determinants of rodent longevity in the chaperone-protein degradation network. Cell Stress Chaperones. 2016;21:453–466. doi: 10.1007/s12192-016-0672-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swovick K, et al. Cross-species comparison of proteome turnover kinetics. Mol. Cell. Proteom. 2018;17:580–591. doi: 10.1074/mcp.RA117.000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maddocks ODK, Labuschagne CF, Adams PD, Vousden KH. Serine metabolism supports the methionine cycle and DNA/RNA methylation through de novo ATP synthesis in cancer cells. Mol. Cell. 2016;61:210–221. doi: 10.1016/j.molcel.2015.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou B, et al. Mitochondrial permeability uncouples elevated autophagy and lifespan extension. Cell. 2019;177:299–314.e16. doi: 10.1016/j.cell.2019.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharma, A., Smith, H.J., Yao, P., and Mair, W.B. Causal roles of mitochondrial dynamics in longevity and healthy aging. EMBO Rep. 20, e48395 (2019). [DOI] [PMC free article] [PubMed]
- 57.Miwa S, et al. Low abundance of the matrix arm of complex I in mitochondria predicts longevity in mice. Nat. Commun. 2014;5:1–12. doi: 10.1038/ncomms4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barja G, Herrero A. Localization at complex I and mechanism of the higher free radical production of brain nonsynaptic mitochondria in the short-lived rat than in the longevous pigeon. J. Bioenerg. Biomembr. 1998;30:235–243. doi: 10.1023/A:1020592719405. [DOI] [PubMed] [Google Scholar]
- 59.Herrero A, Barja G. Sites and mechanisms responsible for the low rate of free radical production of heart mitochondria in the long-lived pigeon. Mech. Ageing Dev. 1997;98:95–111. doi: 10.1016/S0047-6374(97)00076-6. [DOI] [PubMed] [Google Scholar]
- 60.Herrero A, Barja G. H2O2 production of heart mitochondria and aging rate are slower in canaries and parakeets than in mice: sites of free radical generation and mechanisms involved. Mech. Ageing Dev. 1998;103:133–146. doi: 10.1016/S0047-6374(98)00035-9. [DOI] [PubMed] [Google Scholar]
- 61.Amador-Noguez D, Yagi K, Venable S, Darlington G. Gene expression profile of long-lived Ames dwarf mice and Little mice. Aging Cell. 2004;3:423–441. doi: 10.1111/j.1474-9728.2004.00125.x. [DOI] [PubMed] [Google Scholar]
- 62.Kamei Y, et al. GABA metabolism pathway genes, UGA1 and GAD1, regulate replicative lifespan in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2011;407:185–190. doi: 10.1016/j.bbrc.2011.02.136. [DOI] [PubMed] [Google Scholar]
- 63.Wang J, Jiang JC, Jazwinski SM. Gene regulatory changes in yeast during life extension by nutrient limitation. Exp. Gerontol. 2010;45:621–631. doi: 10.1016/j.exger.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yanai H, Budovsky A, Barzilay T, Tacutu R, Fraifeld VE. Wide-scale comparative analysis of longevity genes and interventions. Aging Cell. 2017;16:1267–1275. doi: 10.1111/acel.12659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martínez-Reyes I, Chandel NS. Mitochondrial TCA cycle metabolites control physiology and disease. Nat. Commun. 2020;11:102. doi: 10.1038/s41467-019-13668-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Z, Vance DE. Thematic review series: glycerolipids, phosphatidylcholine and choline homeostasis. J. Lipid Res. 2008;49:1187–1194. doi: 10.1194/jlr.R700019-JLR200. [DOI] [PubMed] [Google Scholar]
- 67.Collino S, et al. Metabolic signatures of extreme longevity in northern italian centenarians reveal a complex remodeling of lipids, amino acids, and gut microbiota metabolism. PLoS One. 2013;8:1–12. doi: 10.1371/annotation/5fb9fa6f-4889-4407-8430-6dfc7ecdfbdd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Calabrese V, et al. Cellular stress responses, hormetic phytochemicals and vitagenes in aging and longevity. Biochim. Biophys. Acta. 2012;1822:753–783. doi: 10.1016/j.bbadis.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 69.Rattray NJW, et al. Metabolic dysregulation in vitamin E and carnitine shuttle energy mechanisms associate with human frailty. Nat. Commun. 2019;10:5027. doi: 10.1038/s41467-019-12716-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Burks S, Raymick J, Robinson B, Hanig J, Sarkar S. Neuroprotective effects of acetyl-l-carnitine (ALC) in a chronic MPTP-induced Parkinson’s disease mouse model: endothelial and microglial effects. Neurosci. Lett. 2019;703:86–95. doi: 10.1016/j.neulet.2019.03.015. [DOI] [PubMed] [Google Scholar]
- 71.Cooper N, Thomas GH, FitzJohn RG. Shedding light on the ‘dark side’ of phylogenetic comparative methods. Methods Ecol. Evol. 2016;7:693–699. doi: 10.1111/2041-210X.12533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lindborg CM, Propert KJ, Pignolo RJ. Conservation of pro-longevity genes among mammals. Mech. Ageing Dev. 2015;146–148:23–27. doi: 10.1016/j.mad.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mayne B, Berry O, Davies C, Farley J, Jarman S. A genomic predictor of lifespan in vertebrates. Sci. Rep. 2019;9:17866. doi: 10.1038/s41598-019-54447-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Minias P, Podlaszczuk P. Longevity is associated with relative brain size in birds. Ecol. Evol. 2017;7:3558–3566. doi: 10.1002/ece3.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wilkinson GS, Adams DM. Recurrent evolution of extreme longevity in bats. Biol. Lett. 2019;15:20180860. doi: 10.1098/rsbl.2018.0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Barja G. Correlations with longevity and body size: to correct or not correct? J. Gerontol. Ser. A. 2014;69:1096–1098. doi: 10.1093/gerona/glu020. [DOI] [PubMed] [Google Scholar]
- 77.Speakman JR. Correlations between physiology and lifespan—two widely ignored problems with comparative studies. Aging Cell. 2005;4:167–175. doi: 10.1111/j.1474-9726.2005.00162.x. [DOI] [PubMed] [Google Scholar]
- 78.Szekely P, Korem Y, Moran U, Mayo A, Alon U. The mass-longevity triangle: pareto optimality and the geometry of life-history trait space. PLOS Comput. Biol. 2015;11:e1004524. doi: 10.1371/journal.pcbi.1004524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pearl, R. The Rate of Living (University Press London, 1928).
- 80.Olsen T, et al. Effects of dietary methionine and cysteine restriction on plasma biomarkers, serum fibroblast growth factor 21, and adipose tissue gene expression in women with overweight or obesity: a double-blind randomized controlled pilot study. J. Transl. Med. 2020;18:122. doi: 10.1186/s12967-020-02288-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang H, et al. Establishment of the circadian metabolic phenotype strategy in spontaneously hypertensive rats: a dynamic metabolomics study. J. Transl. Med. 2020;18:38. doi: 10.1186/s12967-020-02222-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.García-Cañaveras JC, Donato MT, Castell JV, Lahoz A. Targeted profiling of circulating and hepatic bile acids in human, mouse, and rat using a UPLC-MRM-MS-validated method. J. Lipid Res. 2012;53:2231–2241. doi: 10.1194/jlr.D028803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hoffman JM, Lyu Y, Pletcher SD, Promislow DEL. Proteomics and metabolomics in ageing research: from biomarkers to systems biology. Essays Biochem. 2017;61:379–388. doi: 10.1042/EBC20160083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gubina N, et al. Essential physiological differences characterize short- and long-lived strains of Drosophila melanogaster. J. Gerontol. Ser. A. 2019;74:1835–1843. doi: 10.1093/gerona/gly143. [DOI] [PubMed] [Google Scholar]
- 85.Jové M, et al. Caloric restriction reveals a metabolomic and lipidomic signature in liver of male mice. Aging Cell. 2014;13:828–837. doi: 10.1111/acel.12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pradas I, et al. Effects of aging and methionine restriction on rat kidney metabolome. Metabolites. 2019;9:280. doi: 10.3390/metabo9110280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Walters RO, et al. Sarcosine is uniquely modulated by aging and dietary restriction in rodents and humans. Cell Rep. 2018;25:663–676.e6. doi: 10.1016/j.celrep.2018.09.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gomez A, et al. Cysteine dietary supplementation reverses the decrease in mitochondrial ROS production at complex I induced by methionine restriction. J. Bioenerg. Biomembr. 2015;47:199–208. doi: 10.1007/s10863-015-9608-x. [DOI] [PubMed] [Google Scholar]
- 89.Cabré R, et al. Specific metabolomics adaptations define a differential regional vulnerability in the adult human cerebral cortex. Front. Mol. Neurosci. 2016;9:138. doi: 10.3389/fnmol.2016.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu Y, et al. Development of a matrix-based candidate reference material of total homocysteine in human serum. Anal. Bioanal. Chem. 2017;409:3329–3335. doi: 10.1007/s00216-017-0272-3. [DOI] [PubMed] [Google Scholar]
- 91.Chong, J., Wishart, D.S. & Xia, J. Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr. Protoc. Bioinforma. 68. e86 (2019) [DOI] [PubMed]
- 92.Frank, E. Harrell Jr, with contributions from Charles Dupont and many others. Hmisc: Harrell Miscellaneous. R package version 4.5-0. https://CRAN.R-project.org/package=Hmisc (2021).
- 93.Wei, T. & Simko, V. R package “corrplot”: Visualization of a Correlation Matrix (Version 0.88)https://github.com/taiyun/corrplot (2017).
- 94.Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer International Publishing, 2016).
- 95.Orme, D. et al. Comparative analyses of phylogenetics and evolution in R. (2018).
- 96.Kumar S, Stecher G, Suleski M, Hedges SB. TimeTree: a resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 2017;34:1812–1819. doi: 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files (Supplementary Data 1)