Abstract
Mycobacterium tuberculosis (Mtb) is an obligate human pathogen that can adapt to the various nutrients available during its life cycle. However, in the nutritionally stringent environment of the macrophage phagolysosome, Mtb relies mainly on cholesterol. In previous studies, we demonstrated that Mtb can accumulate and utilize cholesterol as the sole carbon source. However, a growing body of evidence suggests that a lipid-rich environment may have a much broader impact on the pathogenesis of Mtb infection than previously thought. Therefore, we applied high-resolution transcriptome profiling and the construction of various mutants to explore in detail the global effect of cholesterol on the tubercle bacillus metabolism. The results allow re-establishing the complete list of genes potentially involved in cholesterol breakdown. Moreover, we identified the modulatory effect of vitamin B12 on Mtb transcriptome and the novel function of cobalamin in cholesterol metabolite dissipation which explains the probable role of B12 in Mtb virulence. Finally, we demonstrate that a key role of cholesterol in mycobacterial metabolism is not only providing carbon and energy but involves also a transcriptome remodeling program that helps in developing tolerance to the unfavorable host cell environment far before specific stress-inducing phagosomal signals occur.
Subject terms: Microbial genetics, Pathogens, Microbiology, Bacteria, Bacterial pathogenesis
Introduction
Compared with other bacteria, Mycobacterium tuberculosis (Mtb) is capable of simultaneously consuming multiple carbon sources to augment growth1. However, numerous studies have established that in the nutritionally stringent environment of macrophage phagolysosome Mtb relies on fatty acids and cholesterol2. In our previous studies, we demonstrated that Mtb can accumulate and utilize cholesterol as the sole carbon source. Moreover, functional cholesterol degradation pathway is required for Mtb to multiply in human macrophages3,4. Other studies have demonstrated that Mtb employs a cholesterol‐dependent pathway to infect host cells, induces a foamy macrophage phenotype, and extracts cholesterol from caseous granuloma or serum-derived lipoproteins5,6. The ability to catabolize cholesterol is crucial during the early and late stages of Mtb infection7–12. Therefore, a thorough study on the cholesterol fate inside a mycobacterial cell is indispensable in understanding the tubercle bacilli survival success. Unfortunately, the process of cholesterol breakdown is complex, and an exceptionally large number of genes are annotated as involved in Mtb lipid metabolism13, thus greatly complicating the identification of genes involved specifically in cholesterol degradation. Therefore, robust categorization through transcriptomics may greatly help delineate genes tightly involved in this pathway. Through microarray-based transcriptional profiling, Nesbitt et al. identified over 200 Mtb genes that are regulated by cholesterol in a standard growth medium9. However, along with the emergence of RNA-Seq, an RNA sequencing technique that has an advantage over microarrays because it can produce perfectly quantified transcriptomes with a higher resolution, the growing body of evidence suggests that the lipid-rich environment may have a much broader impact on the Mtb transcriptome than previously thought. The use of RNA-Seq has allowed for the delineation of the transcriptional background of Mtb grown in even-length fatty acids14 or a mixture of lipids15. Although cholesterol is the key constituent of the Mtb lipid environment during pathogenesis, a complete transcriptomic landscape of the tubercle bacillus utilizing cholesterol as a sole carbon source is still lacking. Therefore, in this study, we applied RNA-Seq and constructed and analyzed specific mutants to precisely describe Mtb metabolic adaptations during growth on cholesterol as the only carbon and energy source and explain how cholesterol may facilitate the establishment of persistent infection.
Results and discussion
Differential gene expression of Mycobacterium tuberculosis H37Rv in the presence of cholesterol
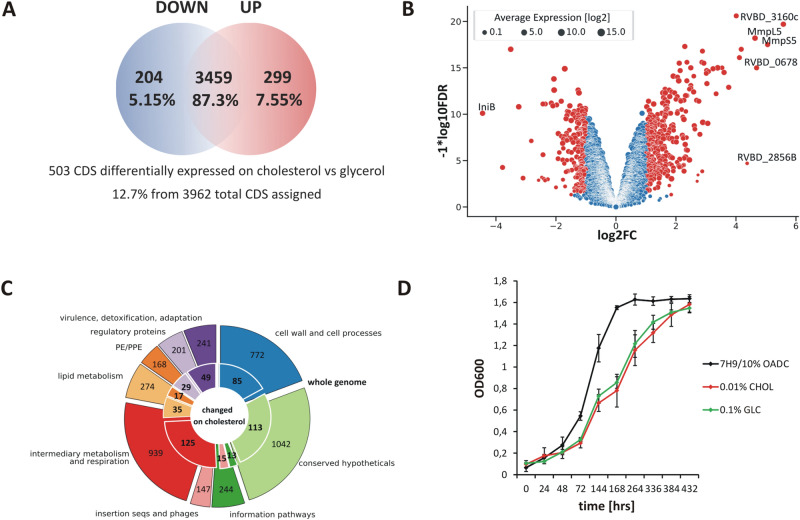
With a single change in providing cholesterol instead of glycerol as the sole carbon source the expression of more than 500 M. tuberculosis (Mtb) genes changed (Fig. 1A, Dataset S1). Genes demonstrating a change in expression with a false discovery rate (FDR) of <0.05 and a log2 fold change greater than an absolute value of 1 (changing two times or more) were considered differentially expressed (DE) in the study (Fig. 1B). Among the 3962 CDSs assigned, 204 genes with decreased (5.15%) and 299 with increased (7.55%) expression were identified (Fig. 1A). Interestingly, the vast majority of DE genes are not functionally related to lipid metabolism (Fig. 1C). The pool of DE genes was significantly enriched with operon members of more than 35 Mtb transcription factors (Dataset S2). We did not observe any difference in Mtb growth rate between the two main carbon sources discussed - glycerol and cholesterol (Fig. 1D), however, compared to the rich medium, Mtb demonstrates an extended Lag phase on both defined carbon sources which probably results from the need for complex metabolic adaptation (Fig. 1D). Conversely, the growth of propionate metabolism regulator (PrpR) mutant - ∆prpR that was used to study the transcriptional effect of methylcitrate cycle (MCC) inhibition is significantly retarded on cholesterol (Fig. 3A). Supplementation of the growth medium with the vitamin B12 that was used to analyze the influence of B12 on the Mtb transcriptome does not influence the bacterial growth rate (data not shown).
Figure 1.
Summary of RNA sequencing results and growth characteristics of M. tuberculosis grown on cholesterol as the sole carbon source versus glycerol used as a control. (A) Venn diagram. A total of 503 differentially expressed CDSs were found. Among the 3962 CDSs assigned, 204 genes with decreased and 299 with increased expression were identified in the presence of cholesterol as the sole carbon source (B) Volcano plot. The negative log of false discovery rate (FDR) (base 10) is plotted on the Y-axis, and the log of the fold change (FC) (base 2) is plotted on the X-axis. The red points represent transcripts that are differentially expressed on cholesterol (FC > 1 and FDR < 0.05). The blue points represent transcripts that are not differentially expressed. The log average expression level (base 10) correlates with the size of individual points. (C) Donut plot. Protein functional categories were assigned to each transcript carrying information for protein synthesis. The number of genes with a change in expression observed for the individual categories (inner part of the chart) is plotted against the total number of transcripts encoding proteins belonging to the given category (outer part of the chart). Functional categories were assigned according to Mycobrowser (www.mycobrowser.epfl.ch, former Tuberculist). (D) Growth rate characteristics of the Mtb wild-type strain on 7H9/10% OADC medium and mineral medium supplemented with 0.01% cholesterol or 0.1% glycerol.
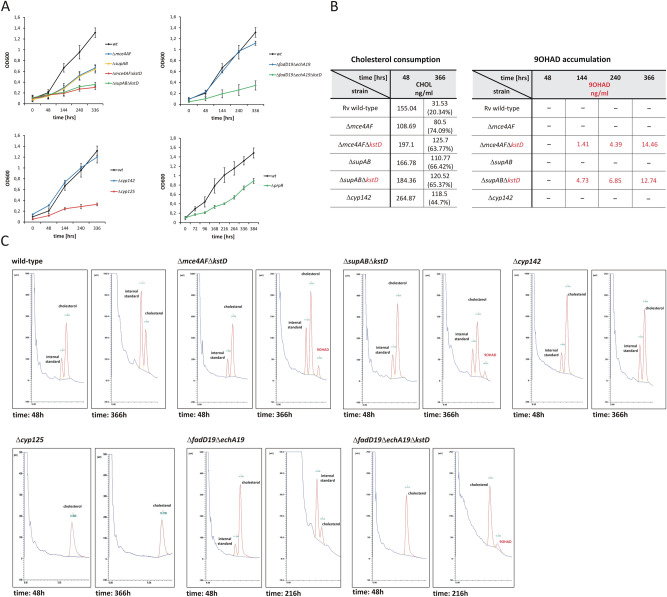
Figure 3.
Phenotype of the Mtb mutants constructed in this study. (A) Growth rate analysis of the Mtb wild-type strain (wt) and Mtb mutants described in this study on mineral medium supplemented with 0.01% cholesterol. (B) Quantitative analysis of cholesterol bioconversion and 9-hydroxy-4-androstene-3,17-dione (9OHAD) accumulation by the M. tuberculosis wild-type strain and the Mtb mutants ∆mce4AF, ∆mce4AF∆kstD, ∆supAB, ∆supAB∆kstD, Δcyp142 growing in mineral medium supplemented with cholesterol as the sole source of carbon and energy, monitored by gas chromatography. Data concerning 9OHAD accumulation are marked red. The analysis was done in the selected time points. Equal amounts of the internal standard (4-androstene-3,11,17-trione) were added to each sample. The amounts of cholesterol and 9OHAD in the sample (nanograms per milliliter of the bacterial culture) were calculated from the ratio between the internal standard and the cholesterol and 9OHAD peaks. After adding cholesterol to the bacterial culture a 48-hour incubation was conducted for better cholesterol solubilization. Therefore, % of the initial amount of cholesterol in the sample was calculated considering the value measured at 48 hours as 100% (C) Chromatograms demonstrating cholesterol consumption and 9-hydroxy-4-androstene-3,17-dione (9OHAD) accumulation in M. tuberculosis wild-type strain and ∆mce4AF∆kstD, ∆supAB∆kstD, Δcyp142, Δcyp125, ∆fadD19∆echA19, ∆fadD19∆echA19∆kstD mutant cells. The selected time points are shown in the figure. Each time, the quantitative analysis was conducted, an equal amount of the internal standard (4-androstene-3,11,17-trione) was added to each sample.
Transcriptional profile of cholesterol uptake and degradation
Through microarray-based transcriptional profiling, Nesbitt et al. identified over 200 Mtb genes that may be regulated by cholesterol9. These genes include regulons of TetR family repressors KstR1 and KstR216,17 situated in the Cho-region9 as well as genes that belong to the regulon of Mce3R18 or SigE19. However, cholesterol-induced changes were analyzed in a standard medium containing other carbon sources. Therefore, in our experiment, to determine the clear effect of cholesterol on the tubercle bacillus transcriptome, we applied a more accurate technique, RNA-Seq, to analyze the transcriptome of Mtb growing on cholesterol as the sole carbon source. The data were related to the results obtained on a minimal medium supplemented with glycerol. Additionally, the analysis was corrected for transcriptional change that is induced in standard 7H9/10% OADC medium.
The study confirmed the cholesterol-induced up-regulation of only 26 of the 83 Cho-region genes. Specifically, the results showed that only 32 of 71 KstR1 genes, including kstR1 (Rv3574), were up-regulated, and the observed induction was strictly (excluding hsaE) cholesterol-specific (Fig. 2A, Dataset S1). Thirteen of 44 KstR1 genes previously classified as induced by cholesterol9 did not change their expression. Almost all KstR2 genes responded to cholesterol; however, the response was not specific because the same up-regulation was also observed in the 7H9/10% OADC medium. Therefore, the metabolite of A & B sterol ring degradation20 is surely not the only chemical inducer alleviating KstR2 repressor binding, and KstR2-regulated genes are probably also involved in β-oxidation of fatty acids. Despite being described as unique to cholesterol metabolism21 and involved in host-Mtb interactions22,23, the Mce3R regulon did not respond specifically to cholesterol, except Rv1936 encoding putative monooxygenase. Similarly, none of the 16 genes of the SigE regulon changed its expression in our experiment.
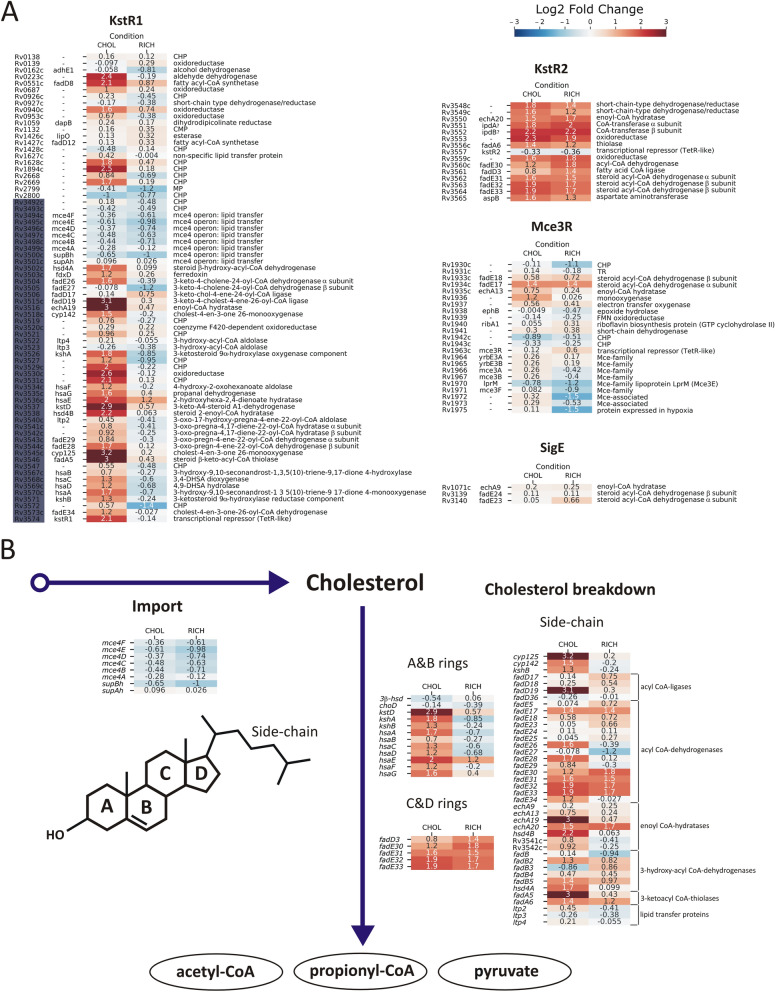
Figure 2.
Differential gene expression of cholesterol import and breakdown genes. DGE analysis was performed comparing conditions of Mtb grown in glycerol vs cholesterol (CHOL) or glycerol vs 7H9/10% OADC medium (RICH). The results are presented as a heatmap of the log2-fold change in expression. See the color key for log2FC values. (A) Gene expression within KstR1, KstR2, MceR, and SigE regulon genes that were previously assigned as cholesterol-induced/associated. Genes that belong to the Cho-region9 are marked dark blue (B) DGE analysis of a functional compilation of genes that were previously classified in various studies28 as induced by cholesterol and involved in cholesterol import and breakdown.
Several studies have demonstrated that the mce4 locus of the KstR1 regulon is involved in ATP-dependent cholesterol import7,24,25. Neither genes encoding putative permease supA (yrbE4A) and supB (yrbE4B) nor probable substrate-binding proteins (mce4A-mce4F)26 of mce4 regulon were up-regulated by cholesterol in the described culture conditions (Fig. 2B, Fig. 5A). To date, prolonged hypoxia is the only reported condition that induces the whole mce4 locus, suggesting its special role during persistence27. The essentiality of the permease component for the in vitro Mtb growth on cholesterol was previously determined7; however, the significance of the remaining genes of the mce4 locus has never been assessed by Mtb knockout studies. Therefore, we constructed two Mtb mutants in which permease component genes (supA-supB) and accessory mce4 genes (mce4A-mce4F) were deleted separately (Fig. S1A) to confirm their joint action in cholesterol transport. Both growth and cholesterol consumption were significantly impaired in the ∆mce4AF multiple mutant (Fig. 3A, B). However, 25% of the initial cholesterol content in the culture medium was still successfully metabolized (Fig. 3B). To confirm this observation, the ∆mce4AF mutant was subjected to deletion of the 3-ketosteroid Δ1-dehydrogenase gene (kstD) (Fig. S1B). During growth on cholesterol, the ∆kstD mutant accumulates the cholesterol degradation intermediate 9-hydroxy-4-androstene-3,17-dione (9OHAD)3. Therefore, the presence of 9OHAD in the lipid extract of ∆mce4AF∆kstD could confirm that cholesterol was successfully transported into the mutant cells. Chromatographic analysis revealed that after 144 hours of incubation on cholesterol the ∆mce4AF∆kstD strain starts accumulating 9OHAD (Fig. 3B, C), thereby confirming that the deletion of mce4A-mce4F only partially inhibits cholesterol transport. Similarly, analysis of the ∆supAB double mutant demonstrated that despite impaired growth it still metabolizes approximately 33% of the added cholesterol (Fig. 3A, B), which was then confirmed by the accumulation of 9OHAD in the ∆supAB strain with additional deletion in kstD gene - ∆supAB∆kstD (Fig. S1B, Fig. 3B, C). Our data demonstrate that all mce4 genes are jointly required for optimal cholesterol transport; however, cholesterol does not influence their expression. Assuming SupA/B is the core cholesterol transporter, the incomplete inhibition of cholesterol transport in the ∆supAB mutant may strongly suggest the presence of another (less efficient in the described growth conditions) system of sterol uptake in tubercle bacilli.
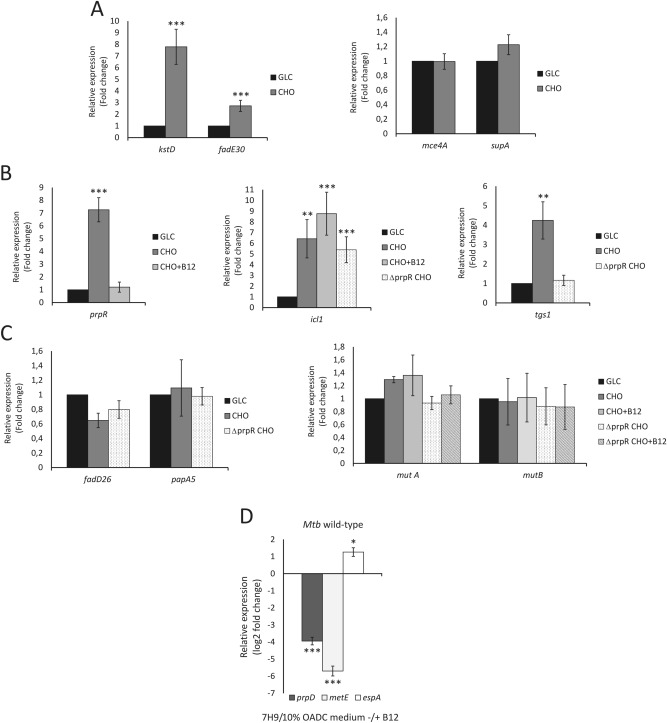
Figure 5.
qRT-PCR validation of the expression of the selected genes in various growth conditions. Selected genes represent the main metabolic pathways discussed in this study. (A) Differential expression of the representative cholesterol breakdown (kstD, fadE30) and import (mce4, supA) genes. (B) Differential expression of the representative genes of Mtb central metabolism (prpR, icl1) and TAG synthesis gene tgs-1. (C) Differential expression of the representative genes of Mtb virulence lipid synthesis (fadD26, papA5) and the methylmalonyl pathway (MMP) (mutA, mutB). (D) Change in expression of PrpR operon genes (represented here by prpD) in response to vitamin B12 in Mtb wild-type growing in standard 7H9/10% OADC medium. The expression of metE was used as an example of confirmed B12-dependent regulation of gene expression. qRT-PCR analysis of espA (representative of the EspR regulon) expression in medium with or without B12 supplementation confirms the discussed influence of vit. B12 on the expression of ESX-1 associated genes. GLC Mtb growing on glycerol, CHO Mtb growing on cholesterol, CHO+B12 Mtb growing on cholesterol and vitamin B12, ∆prpR CHO ∆prpR mutant growing on cholesterol, ∆prpR CHO+B12 ∆prpR mutant growing on cholesterol and vitamin B12. Statistical significance: one, two and three stars denote p values ≤0.05, ≤0.01 and ≤0.001, respectively (Student’s t-test).
Apparently, many genes within regulons previously described as regulated by cholesterol have shown no transcriptional response to this molecule. However, grouping them by functional categories revealed that almost every step of cholesterol breakdown was covered by significantly up-regulated genes (Fig. 2B). The expression of all ten genes required for sequential oxidation of the A & B ring carbon framework28 was cholesterol-induced in our study (Fig. 2B, Fig. 5A) and the induction was clearly cholesterol-specific. Prior to A & B ring degradation, cholesterol must be oxidized and isomerized to cholest-4-ene-3-one. The two genes encoding the enzymes able to catalyze this step - cholesterol oxidase ChoD and hydroxysteroid dehydrogenase 3β-HSD, are not cholesterol-regulated, and our previous study demonstrated that both are dispensable for cholesterol degradation in Mtb29. Since initial oxidation is an essential step of cholesterol breakdown, other proteins with this function must occur among mycobacterial dehydrogenases. Among the five genes that have been proposed to be involved in C & D ring metabolism28, four were up-regulated, although this change was not cholesterol-specific (Fig. 2B). In contrast to the findings of previous studies9, fadD3, encoding a probable acyl-CoA ligase introducing substrate for C & D ring metabolism, was not regulated by cholesterol.
The Mtb genome encodes 20 cytochrome P450 enzymes (Cyp)13 that have been proposed to be responsible for the initial step of cholesterol side-chain degradation. Cholesterol up-regulated four cyp genes (cyp125, cyp136, cyp138 and cyp142), with the strongest increase observed for cyp125 (Fig. 2B, Dataset S1). Cyp125, together with KshB reductase (also up-regulated), is responsible for the initiation of cholesterol side-chain β-oxidation in vitro however, is considered dispensable for cholesterol breakdown in Mtb H37Rv30 and Cyp142 is proposed as the functional backup for Cyp12531. Since cyp142 was also moderately induced in our study, we decided to re-establish the involvement of both cytochromes in cholesterol degradation by the construction and analysis of ∆cyp125 and ∆cyp142 mutant (Fig. S1C). Surprisingly, our study demonstrates that along with its high level of cholesterol-dependent induction, cyp125 is essential for Mtb growth on cholesterol. The ∆cyp125 mutant did not grow on cholesterol (Fig. 3A) and was unable to metabolize it as a sole carbon source (Fig. 3C). Conversely, the deletion of cyp142 did not disturb the growth on cholesterol (Fig. 3A). Cyp142 is, therefore, unable to provide compensatory activity for Cyp125; however, its cholesterol-dependent induction and slightly delayed cholesterol degradation in ∆cyp142 mutant (Fig. 3B, C) may suggest unresolved cooperation of both proteins in cholesterol breakdown.
Among the 36 acyl-CoA ligase genes in Mtb, four (fadD3, fadD17, fadD18, and fadD19) were originally reported to be up-regulated on cholesterol9. In our study, only fadD8 and fadD19 showed cholesterol-dependent induction (Fig. 2A, B, Dataset S1). Both genes belong to the KstR1 regulon; however, only for FadD19 function in the first step of cholesterol side-chain thioesterification was demonstrated32. Within 34 genes of probable acyl CoA-dehydrogenases that may be involved in the first step of side-chain β-oxidation, only fadE26, fadE28 and fadE34 induction was cholesterol-specific (Fig. 2A, B).
Of the 21 genes annotated as (S)–specific enoyl–CoA hydratases involved in the second step of β-oxidation13, only echA19 was specifically up-regulated on cholesterol (Fig. 2B, Dataset S1). In the Mtb genome, echA19 is adjacent to the acyl-CoA ligase gene fadD19, which is also highly induced on cholesterol. Both genes are also up-regulated during infection of human macrophages or in artificial granuloma33,34. Therefore, EchA19 is considered the enzyme catalyzing hydration in the 1st cycle of cholesterol side-chain degradation, just after thioesterification by FadD19. However, our previous study on M. smegmatis demonstrated that FadD19 is required for the biodegradation of branched sterol side-chains but not straight ones, such as cholesterol, and EchA19 is not essential for the degradation of either substrate35. Since no experimental data were available in Mtb, we constructed an H37Rv mutant with a deletion within the fadD19/echA19 locus (Fig. S1A). Strikingly, also in this species, phenotypic analysis of ΔfadD19ΔechA19 double mutant demonstrated that both genes are dispensable for growth on cholesterol (Fig. 3A) and that the constructed strain consumes cholesterol as the wild-type Mtb (Fig. 3C). To further confirm undisturbed cholesterol side-chain degradation, the ΔfadD19ΔechA19 strain was subjected to deletion of the abovementioned kstD gene (Fig. S1B). The presence of the 9OHAD intermediate in lipid samples of the ΔfadD19ΔechA19ΔkstD triple mutant growing on cholesterol finally demonstrated that cholesterol side-chain metabolism in Mtb devoid of echA19 and fadD19 genes was unaffected (Fig. 3C). These results indicate that despite significant induction by cholesterol, both genes may not be directly involved in cholesterol breakdown or other Mtb proteins within a large group of acyl-CoA ligases and enoyl-CoA hydratases may complement their essential enzymatic functions. hsd4B is the only cholesterol-induced gene annotated as coding for the probable (R)-enoyl-CoA hydratase that catalyzes hydration in the 2nd round of side-chain β-oxidation (Fig. 2B).
Within three up-regulated β-hydroxy-acyl-CoA dehydrogenase genes (fadB2, fadB5, hsd4A), only hsd4A is required for growth on cholesterol, and it is located in the cholesterol regulon36. Among the 6 Mtb 3-ketoacyl-CoA thiolase genes, only fadA5 was specifically up-regulated on cholesterol (Fig. 2B). Highly induced fadA5 encodes thiolase of confirmed function, which is required for growth on cholesterol and up-regulated in macrophages and lungs9,33,37. The last carbon-carbon bond between the side-chain and the D ring is cleaved by aldolase, and the Ltp2 protein was demonstrated to conduct this step38. ltp2, together with two other homologous genes, ltp3 and ltp4, was previously reported as cholesterol-regulated9; however, our study does not confirm this regulation (Fig. 2B).
Flux of cholesterol catabolism metabolites: novel regulatory functions of PrpR (Rv1129) and vitamin B12 in mycobacterial virulence
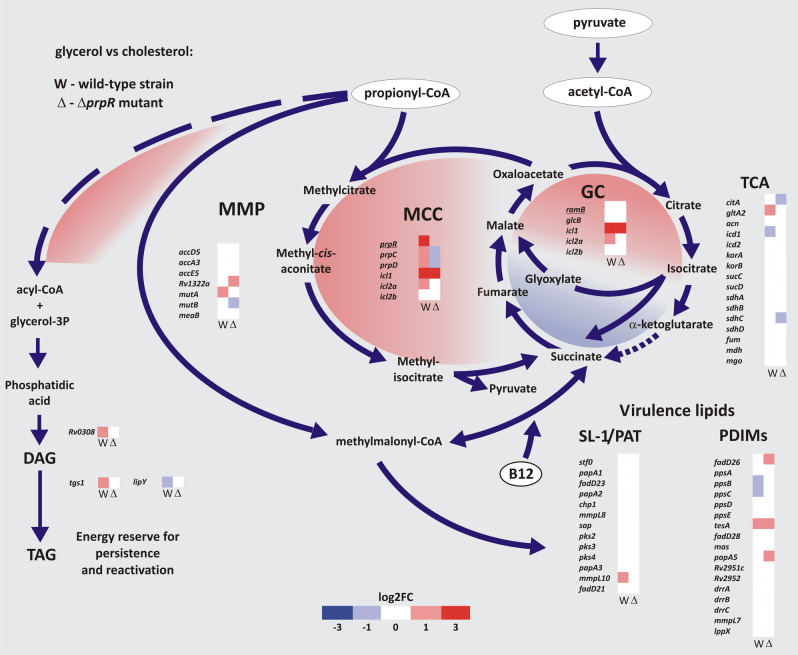
Cholesterol consumption increases the cellular abundance of primary metabolites, mainly propionyl but also acetyl-CoA39. Due to the potential toxicity, their effective amelioration is a prerequisite for the ability to grow on this carbon source. We analyzed the transcriptional response of Mtb intermediary metabolism to delineate the contribution of different pathways in channeling cholesterol degradation metabolites (Fig. 4). Cholesterol utilization leads to substantial induction of the methylcitrate cycle (MCC), which is manifested by the massive increase in prpR (Rv1129c) propionate regulator gene expression along with prpD/C (Rv1130, Rv1131), which are directly controlled by this transcription factor (Figs. 4, 5B). We also observed induction of gltA2 (Rv0896), an essential citrate synthase gene36 conducting the entry reaction of the tricarboxylic acid cycle (TCA), with concomitant repression of icd1 (Rv3339c), which encodes isocitrate dehydrogenase, and strong induction of the isocitrate/methylisocitrate lyase (ICL/MCL) gene icl1 (Rv0467). This finding confirms that in addition to the induction of MCC (propionyl-CoA disposal), Mtb cholesterol consumption induces the bypass of the oxidative part of TCA in favor of increased flux through the glyoxylate shunt (GS) for effective and carbon-saving acetyl-CoA disposal. The demonstrated changes clearly reflect those observed in the transcriptome of M. bovis BCG infecting human THP-1 cells40. Considering propionate the main metabolite of cholesterol breakdown, we decided to investigate the role of the key propionate disposal regulator PrpR in Mtb cholesterol metabolism by comparing the transcriptome of the Mtb wild-type and ΔprpR mutant growing on cholesterol (Fig. 4, Dataset S1, Dataset S2). Same as for the Mtb wild-type transcriptome, for DGE analysis the ΔprpR mutant RNA-Seq data were related to the results obtained on a minimal medium supplemented with glycerol and the analysis was corrected for transcriptional change that is induced in standard 7H9/10% OADC medium. PrpR deletion caused an obvious decrease in prpC/D expression, but the icl1 level was still very high in the ΔprpR strain (Figs. 4, 5B). Although PrpR is a confirmed regulator of icl1 during growth on propionate41, our results demonstrated that on cholesterol, which is not only a source of propionate but also acetate, icl1 expression can be controlled jointly by regulators associated with MCC and/or GS. Conversely, we have shown that the second ICL/MCL gene, icl2a (aceA) (Rv1915), previously considered a pseudogene42, is also induced on cholesterol, but its up-regulation seems to be solely PrpR dependent (Fig. 4). Induction of icl2a was not observed on carbon sources whose metabolism does not generate propionate14,42. There are also no reports on the interaction between Icl2a and RamB, an acetate metabolism regulator, as was demonstrated for Icl143. Icl2a may therefore be an enzyme associated mainly with propionate metabolism induced when the propionate level rises above a certain threshold, whereas Icl1 seems to be designated primarily to GS. Indeed, Icl1 possesses a 10-fold higher affinity to isocitrate than Icl2a42. We have also demonstrated that PrpR may control the cholesterol-induced TCA-GS transition through the regulation of gltA2, citA, and icd1 expression (Fig. 4).
Figure 4.
Transcriptional response of Mtb intermediary metabolism in channeling cholesterol degradation metabolites when cholesterol is utilized as a sole carbon source. DGE analysis of the main pathways of acetyl-CoA, propionyl-CoA, and pyruvate amelioration in Mtb and ∆prpR mutant. The two datasets represent DGE analysis comparing growth in glycerol vs cholesterol of the Mtb wild-type strain (W) and ΔprpR mutant (∆). Transcriptional changes that were not cholesterol-specific (were present also in standard 7H9/10% OADC medium) were excluded from the analysis. See the color key for log2FC values. TCA tricarboxylic acid cycle, GC glyoxylate cycle, MCC methylcitrate cycle, MMP methylmalonyl pathway. Up-regulated and down-regulated pathways are shown in red and blue, respectively.
Functional MCC is required for in vitro growth of Mtb on cholesterol44 or in murine macrophages45. However, it has been found to be dispensable during infection in mice45, suggesting activation of other pathways of propionate disposal. During the Mtb lifecycle, carboxylated propionyl-CoA can also be incorporated into methyl-branched, cell wall virulence lipids (PDIM, SL-1/PAT)46,47. Importantly, this anabolic pathway not only ameliorates the toxic excess of propionate but also dissipates reducing equivalents that arise during lipid β-oxidation45,48. Our study did not show clear cholesterol-induced transcriptional changes within PDIM or SL-1/PAT metabolism (Figs. 4, 5C). It confirms that virulence lipid synthesis is largely posttranscriptionally regulated48 and depends simply on common precursor availability47. Interestingly, we have shown that inhibition of MCC during growth on cholesterol through prpR deletion increases the expression of the WhiB3 (Rv3416) redox sensor directly modulating virulence lipid synthesis in response to reductive stress48. This finding suggests an interplay between MCC and virulence lipid synthesis in cholesterol metabolite amelioration. Surprisingly, we also noticed a cholesterol-induced increase in the expression of the triacylglycerol (TAG) synthesis genes tgs-1 (Rv3130c) and Rv0308 with concomitant down-regulation of the LipY lipase gene, which is involved in TAG hydrolysis upon nutrient starvation. The shift in tgs-1 expression on cholesterol was not observed in the ∆prpR mutant (Figs. 4, 5B). Since tgs-1 is a member of the DosR/S/T regulon, synthesis of TAG, which acts as an energy reserve for reactivation, has been strictly associated with entering dormancy to date49,50. However, it appears that in a cholesterol-rich environment, Mtb can initiate storage lipid synthesis without induction through DosR/S/T, even in aerated in vitro culture, and this process may be PrpR-dependent.
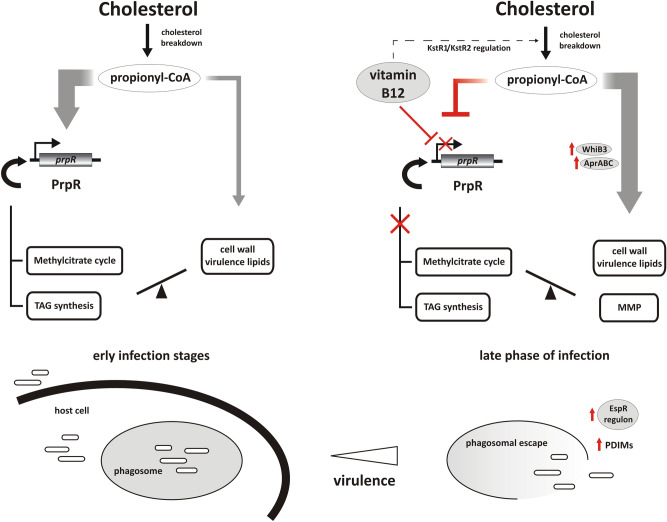
In addition to the MCC, the generation of the TCA intermediate from propionyl-CoA can also be carried out by the methylmalonyl pathway (MMP), provided that vitamin B12, a cofactor of methylmalonyl-CoA mutase (as adenosylcobalamin), is supplied exogenously51. Despite being functional in Mtb, MMP did not respond transcriptionally to cholesterol even after MCC inhibition and/or vitamin B12 supplementation (Figs. 4, 5C). Therefore, the expression of MMP genes is rather continuous in Mtb, and the functionality of the whole pathway is regulated solely through B12 availability. It was previously suggested that in the presence of B12, both MCC and MMP cooperate to provide optimal growth on propionate51. However, our study revealed that providing Mtb with vitamin B12 not only renders MMP functional but also strongly decreases the expression of the prpR and prpDC regulon, thus inhibiting MCC. The observed effect of B12 is independent of the utilized carbon source (Figs. 4, 5B, D). RNA-Seq analysis of the Mtb wild-type strain growing in standard 7H9/10% OADC medium with or without vitamin B12 supplementation (Dataset S1) revealed that among the entire Mtb transcriptome, genes of the PrpR operon undergo the strongest repression on medium supplemented with B12 together with closely located metE encoding methionine synthase, whose regulation is a confirmed example of B12-sensing riboswitch in Mtb52 (Fig. 5D, Dataset S1). Whether MCC genes are regulated through the same mechanism requires further study. Previous studies demonstrated that an Mtb mutant with impaired B12 import reveals no impact on the early establishment of infection but a profound effect on the maintenance of chronic disease53. Therefore, both the B12-dependent and B12-independent pathways of cholesterol metabolite dissipation are essential for maintaining infection; however, we can speculate that they operate in a specific sequence imposed by the identified interaction between PrpR and B12 (Fig. 6). We propose that at the early infection stage, intensive cholesterol consumption highly increases the expression of PrpR. PrpR directs carbon flux through the MCC and GC and induces TAG biosynthesis. It is uncertain whether B12 is continuously available in Mtb cell during infection or becomes available at some distinct stage. Nevertheless, under an unclear stimulus, which may be a change in internal redox potential and/or pH54, B12 inhibits the expression of PrpR and pathways controlled by this regulator, and this change directs most of the propionate flux to virulence lipid synthesis. In addition, B12-unlocked MMP allows for efficient conversion of some part of carboxylated propionate into succinate, thereby bypassing down-regulated MCC and the need for anaplerosis via GC (Fig. 6). Moreover, B12 strongly up-regulated the AprA regulator, inducer of PDIM biosynthesis (Dataset S1, Fig. 6). Induction of the aprABC locus, a modulator of macrophage phagosome adaptation, has been described to date as pH-driven only55. An increase in the amount of cell wall virulence lipids at the late phase of infection facilitates phagosomal escape and host cell exit, allowing Mtb dissemination56. Indeed, the gene pool that significantly changed its expression in response to B12 also includes members of the EspR regulon (Dataset S2, Figs. 5D, 6) that are directly involved in phagosomal escape and work synergistically with PDIM to help M. tuberculosis escape the host cell56. Interestingly, in addition to affecting the route of intermediary metabolite amelioration, B12 may also control their accumulation rate by down-regulating the expression of KstR2 and KstR1 genes (cholesterol metabolism) and by regulation of fatty acid metabolism (MabR, FasR regulon genes) (Fig. 6, Dataset S2). Overall, we noticed a B12-induced transcriptional change in many Mtb regulons involved in survival under various stresses, cell wall integrity, antibiotic resistance and tolerance to toxic compounds, cell division or regulation of latency (Dataset S2). Therefore, vitamin B12 may not only be an enzyme cofactor but also an important regulator coordinating the shift in Mtb metabolism and virulence in response to the variable environment inside the host.
Figure 6.
The role of vitamin B12 and PrpR joint action in the channeling of propionyl-CoA and virulent phenotype induction during Mtb infection. See the text for description.
Cholesterol induces DosR/S/T regulon
DosR regulates the expression of more than 50 genes involved in the transition of actively replicating Mtb to dormant bacilli upon gradual depletion of oxygen inside granuloma57 and resumption of growth upon return to normoxia58. Induction of tgs-1, a member of the DosR/S/T regulon, in cholesterol medium prompted us to analyze the cholesterol-induced response of the other Dos genes. Our study revealed that cholesterol in the Mtb growth environment is a potent inducer of the Dos response, even in actively replicating cells, much earlier than specific hypoxia symptoms. 36 of 51 Dos genes showed significantly up-regulated expression (Fig. S2). Importantly, tgs-1 is the only Dos gene whose induction on cholesterol is inhibited in response to prpR deletion, thereby confirming that TAG synthesis is PrpR- rather than DosR-regulated on this carbon source. Although previous transcriptomic studies mention some Dos genes as induced during Mtb growth in the lipid environment14,15, the effect of cholesterol is the strongest reported so far.
Cholesterol supports the development of Mtb drug tolerance and prevents the inhibition of own metabolism
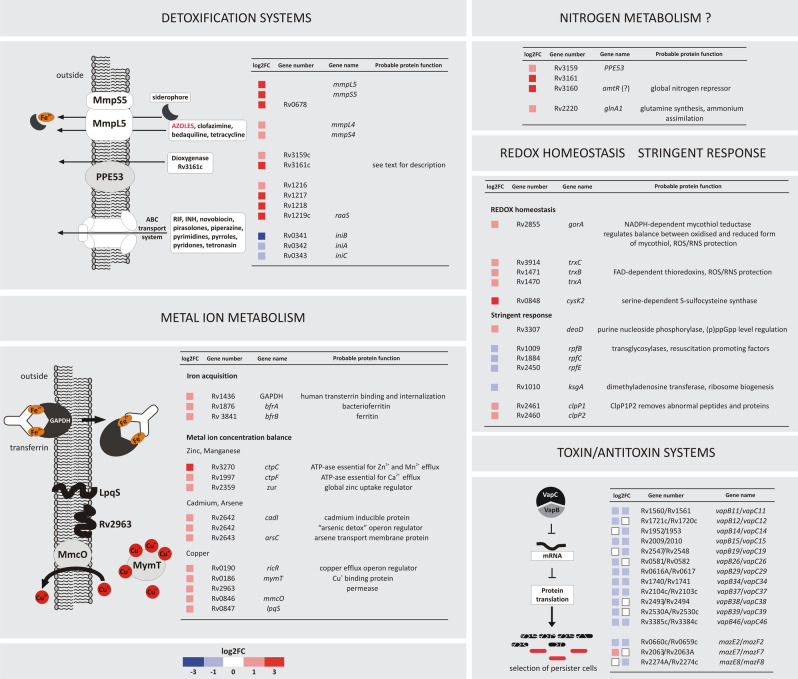
The majority of Mtb genes with the highest (≥ 4 log FC) change in expression on cholesterol encode efflux pumps conferring resistance to numerous antibiotics. Cholesterol greatly increased the expression of MmpS5/MmpL5 multidrug efflux pump components (Fig. 7). Together with mmpS4/L4, they are the only members of the mmpS/L family up-regulated on cholesterol and have been linked to the iron acquisition mechanism59. In addition to resistance to various antibiotics, MmpS5-L5 efflux determines Mtb resistance to azoles that target cholesterol metabolism by inhibiting cytochrome P450 (Cyp) monooxygenases60,61. Some Mtb Cyps, including Cyp125 and Cyp142, have already been demonstrated to bind azole drugs62. Up-regulation of MmpS5-L5 during growth on cholesterol may, therefore, secure continuity of cholesterol metabolism in two ways: by providing iron for many heme-containing catabolic enzymes and/or by effluxing heme-binding inhibitors. Cholesterol also up-regulates the Rv1216c-Rv1218c operon encoding the main ATP-dependent efflux pump conferring Mtb resistance to many chemically unrelated antibiotics63 (Fig. 7). The operon is up-regulated together with its regulatory protein RaaS (Rv1219c), which plays an important role in mycobacterial long-term survival in vitro and in vivo64. The activity of the mentioned efflux systems is strictly associated with the ability to survive within macrophages during latency. These systems were previously reported as co-induced in the presence of drugs or during growth in a mixture of various lipids15,65. However, as demonstrated here, the sole presence of cholesterol is a sufficient condition for such induction. Interestingly, cholesterol not only up-regulates Mtb efflux systems. The iniBAC operon, which encodes a pump-like efflux mechanism conferring tolerance to multiple anti-TB drugs66, was significantly down-regulated in this study (Fig. 7). iniBAC is highly induced in response to the inhibition of cell wall biosynthesis67; however, conditions that down-regulate iniBAC are, to date, largely unknown. Cholesterol affects not only Mtb drug efflux but also detoxification systems, such as the Rv3161c-Rv3159c gene cluster encoding a putative TetR regulator, probable dioxygenase, and outer membrane protein (Fig. 7). It is induced by various aromatic compounds, such as thioridazine and triclosan68,69. The operon was also reported to be induced in a lipid-rich environment15; however, again our results demonstrate that cholesterol is a sufficient inducer. Rv3160, the most cholesterol-up-regulated gene in the present study, encodes a protein controlling the whole set of metabolic events during active infection, dormancy, or resuscitation of dormant “nonculturable” Mtb70–72. Rv3160 is also considered a homolog of C. glutamicum AmtR, a global nitrogen metabolism regulator controlling ammonium uptake and assimilation73. Although this pathway in Mtb may be GlnR- rather than AmtR-regulated we have also observed, despite providing a nitrogen source in the culture media, cholesterol-dependent up-regulation of glnA1 (Rv2220), which encodes glutamine synthetase essential for ammonium assimilation and an important determinant of Mtb pathogenesis74.
Figure 7.
Examples of Mtb metabolic pathways that are modulated in a cholesterol-rich environment. Changes in the expression of the selected representative genes are shown. See the color key for log2FC values.
Metal ion metabolism
Mtb has developed numerous mechanisms for maintaining cellular metal ion balance, thereby preventing intoxication inside the phagosome75. Here, we demonstrated that growth on cholesterol up-regulates the expression of many metal ion efflux/uptake systems and their regulators (Fig. 7). Many of them, such as the copper-responsive regulon RicR or “arsenic detox” operon, are essential for Mtb infection and in vivo growth, regulate drug susceptibility and are described as new virulence factors76,77. We have also noticed that despite providing iron in the culture medium, cholesterol specifically up-regulates genes of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Rv1436) and iron storage protein ferritin (bfrB) (Rv3841), although their up-regulation has been previously only associated with iron depletion78. Both GAPDH and ferritin promote the most efficient and essential for virulence, siderophore-independent iron acquisition system that allows binding and internalization of the whole human transferrin78. Cholesterol-dependent GAPDH induction may thus secure iron for the sterol degradative pathway.
Cholesterol modulates the expression of Mtb persistence-associated genes
We noticed a cholesterol-induced change in the expression of many genes seemingly unrelated in terms of supposed function. However, a closer look suggests their common contribution in providing a metabolic background for Mtb persistence and long-term dormancy. Cholesterol substantially up-regulates genes encoding mycothiol-dependent redox mechanism (Fig. 7), which is first-line defence against reactive oxygen and nitrogen species inside the host cell79. These genes determine entering persistence and are overexpressed in various dormancy models80,81. Importantly, the mycothiol switch controls Mtb cholesterol catabolism in response to the redox state inside the host macrophage54. Long-term survival of nonreplicating Mtb is ensured by the coordinated shutdown of active metabolism through the stringent response mediated by hyperphosphorylated guanosine ((p)ppGpp)82. Cholesterol induces the expression of purine nucleoside phosphorylase DeoD (Rv3307), which is thought to be involved in the Rel-dependent regulation of (p)ppGpp levels83 (Fig. 7). Its expression correlates with dormancy and is considered among the top 100% persistence targets (TBSGC, www.webtb.org). Strikingly, we observed also cholesterol-induced change in expression of genes encoding resuscitation-promoting factors (Rpfs) coordinating the transition between Mtb nonreplicating persistence and active growth84. Mtb grown in cholesterol down-regulates three (rpfBCE) of five Rpf genes as well as cotranscribed and coregulated ksgA (Rv1010)85. KsgA methyltransferase is involved in ribosome biogenesis; thus, its down-regulation affects translation and growth86. Intriguingly, one of the most represented groups of Mtb genes that changes expression in response to cholesterol is a toxin-antitoxin (TA) system. Among 15 TA pairs with altered expression, 13 are represented by the VapBC family. It is expected that in response to various stressors, VapC activity generates the stochastic expression of many proteins interfering with key cellular processes, ultimately leading to the emergence of persister cells87,88. Therefore, in this paper, we have demonstrated a link between cholesterol in the Mtb growth environment and a specific metabolic program that may facilitate entering dormancy, resumption of growth upon exposure to ambient O2 levels and selection of persister cells.
Conclusions
Cholesterol is considered essential for Mtb to maintain a chronic infection; however, the molecular basis of this phenomenon is largely unknown. In this study, we delineated the transcriptomic landscape of tubercle bacilli that utilize cholesterol as the sole carbon source, which provided insights on the contribution of cholesterol to establishing persistent infection. The data allowed us to verify the list of cholesterol breakdown genes that are truly cholesterol-induced. We further proposed a new function of vitamin B12 in directing cholesterol metabolites which, together with analysis of B12-induced changes in Mtb transcriptome, is the first attempt to the identification of the role of cobalamin in mycobacterial virulence. Apparently many changes in Mtb transcriptome that were previously ascribed to the lipid-rich environment are in fact induced by cholesterol. We demonstrated that the essential role of cholesterol in Mtb intracellular survival is not only coupled to central carbon metabolism and energy production but also to the ability to induce a transcriptomic program that promotes persistence. Therefore, cholesterol may facilitate the development of Mtb tolerance to unfavorable host cell environment far before specific stress-inducing phagosomal signals occur. The direct mechanism of this transcriptome remodeling program demands further study.
Methods
Bacterial strains and culture conditions
The Mycobacterium tuberculosis H37Rv wild-type strain and derived mutants (described below) were maintained on Middlebrook 7H10 agar and 7H9 broth (Becton-Dickinson) supplemented with 10% OADC enrichment (Becton-Dickinson). For RNA sequencing, the Mtb wild-type and ∆prpR mutant were grown on defined carbon sources in minimal medium (0.5 g/liter asparagine, 1 g/liter KH2PO4, 2.5 g/liter Na2HPO4, 50 mg/liter ferric ammonium citrate, 0.5 g/liter MgSO4∙7H2O, 0.5 mg/liter CaCl2, and 0.1 mg/liter ZnSO4) containing either 0.1% glycerol (vol/vol) or 0.01% cholesterol (wt/vol)7. RNA sequencing data resulting from a comparison of the expression profile of glycerol vs cholesterol were also related to the results obtained for bacteria growing in standard 7H9/10% OADC medium, which is referred to as a “rich medium”. For the study on the influence of vitamin B12 on the Mtb transcriptome, bacteria were grown in 7H9/10% OADC medium with or without vitamin B12 (Cyanocob(III)alamin) (Sigma Aldrich) supplementation (10 μg/ml). Each bacterial culture was set in triplicate and grown to an optical density of OD600 0.6. Then, 100 ml of each culture was spun down and subjected directly to total RNA isolation. Minimal medium supplemented with 0.01% cholesterol (wt/vol) was also used in cholesterol uptake and biotransformation experiments.
Library preparation and sequencing
For total RNA sequencing, RNA was isolated using TRIzol LS reagent (Invitrogen) as described previously89. Cells were lysed by bead beating with the MP FastPrep system (MP Biomedicals) using lysing matrix B. DNA contamination of the RNA samples was removed by DNase I turbo (Invitrogen) digestion according to the manufacturer’s protocol. The quantity and integrity of the DNase I-treated RNA was assessed using an Agilent 2100 BioAnalyzer according to the manufacturer's instructions (Agilent RNA 6000 Nano Kit). The Illumina-compatible RNA/cDNA sequencing libraries were prepared essentially as described earlier90. For total RNA sequencing, RNA samples were purified using AMPure XP magnetic beads (Becton Dickinson) with a 2.2:1 bead-to-RNA ratio. Two micrograms of the purified total RNA was used per library. The ribosomal RNA was removed with the Ribo-Zero Bacteria rRNA Removal Kit (Illumina) according to the manufacturer’s protocol. Ribodepleted RNA was used to generate cDNA libraries with the KAPA Stranded RNA-Seq kit (Roche) according to the detailed instructions provided by the manufacturer. Thirteen amplification cycles were performed on the initial cDNA libraries to obtain the final sequencing libraries. The Illumina True Seq v2 indexing system was used as a barcoding system for multiplex sequencing. The quality and quantity of the resulting libraries were assessed with the help of an Agilent 2100 BioAnalyzer fitted with a DNA 1000 chip. Before assembling the sequencing run, the libraries were additionally quantified by qPCR with the NEBNext Library Quant Kit for Illumina (New England Biolabs). The NextSeq500 System (Illumina) with the NextSeq 500/550 Mid Output v2 sequencing kit (150 cycles) (Illumina) was used to sequence RNA-Seq libraries, ensuring between 5 and 10 million paired-end reads per sample.
RNA-Seq data analysis
Demultiplexed high-throughput RNA sequencing results were processed with a series of software and bioinformatic scripts as previously described90. Briefly, after the initial removal of sequencing adapters with Cutadapt v. 1.9.191, the sequencing reads were quality trimmed with the windowed adaptive trimming tool Sickle, which allowed for a minimum quality of 30% and a minimal read length of 20 bp. Reads of sufficient quality and length were next aligned to the M. tuberculosis H37Rv genome (NC_018143.2) using the Bowtie2 short read aligner92. SAMtools93 software suite was used for aligned data handling, conversion, and indexing. Gene-mapped reads or read pairs were counted according to appropriate genomic features (coding DNA sequences - CDSs) with the help of the HTSeq 0.6.1 python module94. Integrative Genomics Viewer (IGV) was used to visualize sequencing results in the genomic context95. The gene expression matrix file was generated by merging individual HTSeq output files, with the gene name treated as an index. The resulting file was submitted to the online Degust RNA-Seq analysis platform, which was used to estimate the differential expression. Default parameters were used during the analysis, with voom/limma algorithms set for differential gene expression estimation (http://degust.erc.monash.edu/)96. The statistical analysis of differential gene expression (DGE) was performed by Degust and is provided as the false discovery rate (FDR) value. To identify significant changes in gene expression, genes with an FDR of <0.05 and a log2 fold change greater than an absolute value of 1 (changing two times or more) were considered differentially expressed in the current study. The whole procedure from bacterial culture to cDNA library sequencing was performed in three independent replicates for each strain/condition studied, and the obtained RNA-Seq data were averaged during Degust analysis. The RNA-seq related data have been deposited in NCBI's Gene Expression Omnibus (GEO) database and are accessible through GEO Series accession number GSE175812 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE175812).
qRT-PCR validation of selected genes
To validate the RNA-Seq results, a qRT-PCR analysis was performed for selected genes. SuperScript III First-Strand Synthesis SuperMix (Invitrogen) was used for reverse transcription of relevant RNA samples according to the manufacturer’s instructions. Subsequently, 1 μl of cDNA (equivalent to 50 ng of RNA) was used in the qRT-PCR experiment. qRT-PCR was performed using Maxima SYBR Green qPCR master mix (Thermo Scientific) and a 7900HT real-time PCR system (Applied Biosystems). To verify the specificity and identity of the PCR products generated, a melting curve analysis was performed at the end of each PCR.
Constructions of gene replacement vectors
Standard molecular biology protocols were used for all cloning procedures97. All PCR products were obtained using thermostable AccuPrime Pfx DNA polymerase (Invitrogen). They were initially cloned into a pJET1.2/blunt vector (Thermo Scientific), followed by sequencing and digestion with the appropriate restriction enzymes, and then they were cloned into the final vectors. To facilitate subcloning, certain restriction enzyme recognition sites were incorporated into the primer sequences (see Table S1). To create an unmarked deletion of the cyp125, cyp142, or multigenic regions, supA-supB (yrbE4A – yrbE4B), mce4A-mce4F, and fadD19-echA19, suicidal recombination delivery vectors based on p2NIL98 were constructed (see Table S2). In the case of cyp125 and cyp142, each vector carried the fragment upstream of the gene together with the 5′ end of the gene (GR1-GR2) cloned next to the 3′ end of the gene and its downstream fragment (GR3-GR4) cloned into p2NIL to generate p2Ncyp125Tb and p2Ncyp142Tb vectors, respectively. The GR1-GR2 and GR3-GR4 fragment lengths as well as the PCR primers used for their amplification on M. tuberculosis chromosomal DNA are listed in Table S1 and Table S2. For the deletion of the two-gene supA-supB region, the 5′ end of supA and the fragment upstream of a gene (GR1-GR2) were cloned next to the 3′ end of supB and its downstream fragment (GR3-GR4) to generate a p2NsupTb vector. For the deletion of the six-gene mce4A-mce4F region, the 5′ end of a gene together with the upstream sequence (GR1-GR2) was cloned next to the 3′ end of mce4F together with the sequence downstream of a gene (GR3-GR4) to generate the p2Nmce4Tb vector. For simultaneous deletion of the fadD19 and echA19 genes, the suicidal vector carried the fragment downstream of the fadD19 together with the 3′ end of the gene (GR1-GR2) was cloned next to the 3′ end of the echA19 and its downstream fragment (GR3-GR4) to generate the p2NfadechTb vector. All GR1-GR2 and GR3-GR4 fragments in the gene replacement vectors were ligated out of frame, resulting in the expression of nonfunctional proteins.
Disruption of genes by homologous recombination
The plasmid DNA of p2Ncyp125Tb, p2Ncyp142Tb, p2NsupTb, p2Nmce4Tb and p2NfadechTb suicide delivery vectors was UV-treated and electroporated into wild-type M. tuberculosis competent cells. The two-step homologous recombination protocol of Parish and Stoker98 was used to introduce unmarked deletions into the above-described genes or gene regions. The genotypes of selected double-crossover mutants ∆cyp125, ∆cyp142, ∆supAB, ∆mce4AF, and ∆fadD19∆echA19 were confirmed by PCR and Southern blot hybridization on a chromosomal DNA template digested with BglII, ClaI, BamHI, PstI, and ClaI (Fig. S1). Hybridization probes were generated by PCR on the appropriate gene replacement vector DNA as the template. Probe labeling, hybridization, and signal detection were performed using the AlkPhos Direct labeling and detection system (GE Healthcare) according to the manufacturer’s instructions. For the construction of the ∆supAB∆kstD, ∆mce4AF∆kstD, and ∆fadD19∆echA19∆kstD mutants, ∆supAB, ∆mce4AF, ∆fadD19∆echA19 strains were used as the recipients of a kstD gene replacement vector. Disruption of kstD by homologous recombination and genotype verification were performed as described elsewhere3. The construction of the M. tuberculosis ∆prpR mutant was described previously41.
Cholesterol biotransformation
Wild-type M. tuberculosis and the constructed mutant strains were cultured in minimal medium supplemented with 0.01% cholesterol7. To follow the process of cholesterol biotransformation and detect intermediates, five-milliliter culture samples were withdrawn from the culture at 24 h intervals and extracted three times with an equal volume of chloroform. The extracts were dried under vacuum, the residue was dissolved in 0.5 ml of acetone, and the isolated steroids were analyzed by gas chromatography as previously described99. To quantify the cholesterol and 9-hydroxy-4-androstene-3,17-dione (9OHAD), equal amounts of the internal standard 4-androstene-3,11,17-trione (Sigma) were added to each sample.
Supplementary information
Acknowledgements
This work was supported by the Statutory Fund of Institute of Medical Biology of the Polish Academy of Sciences and by the National Science Centre of Poland grants given to J.P. [2012/05/N/NZ2/00622] and A.M. [2015/19/D/NZ6/03011].
Author contributions
J.P. Conceptualization, Investigation, Formal analysis, Visualization, Writing—Original Draft, Funding acquisition A.B. Conceptualization, Investigation, Validation A.M. Investigation, Funding acquisition P.P. Software, Formal analysis, Data Curation A.R-G. Investigation D.S. Investigation J.Z-C. Writing—Review & Editing J.D. Supervision, Project administration, Validation, Funding acquisition.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jakub Pawełczyk and Anna Brzostek.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-91812-0.
References
- 1.de Carvalho LP, et al. Metabolomics of Mycobacterium tuberculosis reveals compartmentalized co-catabolism of carbon substrates. Chem. Biol. 2010;17:1122–31. doi: 10.1016/j.chembiol.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 2.Wilburn KM, Fieweger RA, VanderVen BC. Cholesterol and fatty acids grease the wheels of Mycobacterium tuberculosis pathogenesis. Pathog. Dis. 2018;76:fty021. doi: 10.1093/femspd/fty021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brzostek A, Pawelczyk J, Rumijowska-Galewicz A, Dziadek B, Dziadek J. Mycobacterium tuberculosis is able to accumulate and utilize cholesterol. J. Bacteriol. 2009;191:6584–91. doi: 10.1128/JB.00488-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brzezinska M, et al. The role of 3-ketosteroid 1(2)-dehydrogenase in the pathogenicity of Mycobacterium tuberculosis. BMC Microbiol. 2013;13:43. doi: 10.1186/1471-2180-13-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muñoz S, Rivas-Santiago B, Enciso JA. Mycobacterium tuberculosis entry into mast cells through cholesterol-rich membrane microdomains. Scand. J. Immunol. 2009;70:256–63. doi: 10.1111/j.1365-3083.2009.02295.x. [DOI] [PubMed] [Google Scholar]
- 6.Peyron P, et al. Foamy macrophages from tuberculous patients' granulomas constitute a nutrient-rich reservoir for M. tuberculosis persistence. PLoS Pathog. 2008;4:e1000204. doi: 10.1371/journal.ppat.1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pandey AK, Sassetti CM. Mycobacterial persistence requires the utilization of host cholesterol. Proc. Natl. Acad. Sci. USA. 2008;105:4376–4380. doi: 10.1073/pnas.0711159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu Y, et al. 3-Ketosteroid 9alpha-hydroxylase is an essential factor in the pathogenesis of Mycobacterium tuberculosis. Mol. Microbiol. 2010;75:107–21. doi: 10.1111/j.1365-2958.2009.06957.x. [DOI] [PubMed] [Google Scholar]
- 9.Nesbitt NM, et al. A thiolase of Mycobacterium tuberculosis is required for virulence and production of androstenedione and androstadienedione from cholesterol. Infect. Immun. 2010;78:275–82. doi: 10.1128/IAI.00893-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang JC, Harik NS, Liao RP, Sherman DR. Identification of Mycobacterial genes that alter growth and pathology in macrophages and in mice. J. Infect. Dis. 2007;196:788–95. doi: 10.1086/520089. [DOI] [PubMed] [Google Scholar]
- 11.VanderVen BC, et al. Novel inhibitors of cholesterol degradation in Mycobacterium tuberculosis reveal how the bacterium’s metabolism is constrained by the intracellular environment. PLoS Pathog. 2015;11:e1004679. doi: 10.1371/journal.ppat.1004679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yam KC, et al. Studies of a ring-cleaving dioxygenase illuminate the role of cholesterol metabolism in the pathogenesis of Mycobacterium tuberculosis. PLoS Pathog. 2009;5:e1000344. doi: 10.1371/journal.ppat.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole ST, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–44. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 14.Rodríguez JG, et al. Global adaptation to a lipid environment triggers the dormancy-related phenotype of Mycobacterium tuberculosis. mBio. 2014;5:e01125. doi: 10.1128/mBio.01125-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aguilar-Ayala DA, et al. The transcriptome of Mycobacterium tuberculosis in a lipid-rich dormancy model through RNAseq analysis. Sci. Rep. 2017;7:17665. doi: 10.1038/s41598-017-17751-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kendall SL, et al. A highly conserved transcriptional repressor controls a large regulon involved in lipid degradation in Mycobacterium smegmatis and Mycobacterium tuberculosis. Mol. Microbiol. 2007;65:684–99. doi: 10.1111/j.1365-2958.2007.05827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kendall SL, et al. Cholesterol utilization in mycobacteria is controlled by two TetR-type transcriptional regulators: KstR and KstR2. Microbiology. 2010;156:1362–71. doi: 10.1099/mic.0.034538-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De La Paz Santangelo M, et al. Mce3R, a TetR-type transcriptional repressor, controls the expression of a regulon involved in lipid metabolism in Mycobacterium tuberculosis. Microbiology. 2009;155:2245–55. doi: 10.1099/mic.0.027086-0. [DOI] [PubMed] [Google Scholar]
- 19.Fontan PA, et al. Mycobacterium tuberculosis sigma factor E regulon modulates the host inflammatory response. J. Infect. Dis. 2008;198:877–85. doi: 10.1086/591098. [DOI] [PubMed] [Google Scholar]
- 20.Casabon I, Crowe AM, Liu J, Eltis LD. FadD3 is an acyl-CoA synthetase that initiates catabolism of cholesterol rings C and D in actinobacteria. Mol. Microbiol. 2013;87:269–83. doi: 10.1111/mmi.12095. [DOI] [PubMed] [Google Scholar]
- 21.Wipperman MF, Yang M, Thomas ST, Sampson NS. Shrinking the FadE proteome of Mycobacterium tuberculosis: insights into cholesterol metabolism through identification of an α2β2 heterotetrameric acyl coenzyme A dehydrogenase family. J. Bacteriol. 2013;195:4331–41. doi: 10.1128/JB.00502-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Senaratne RH, et al. Mycobacterium tuberculosis strains disrupted in mce3 and mce4 operons are attenuated in mice. J. Med. Microbiol. 2008;57:164–70. doi: 10.1099/jmm.0.47454-0. [DOI] [PubMed] [Google Scholar]
- 23.Gioffre A, et al. Mutation in mce operons attenuates Mycobacterium tuberculosis virulence. Microbes Infect. 2005;7:325–34. doi: 10.1016/j.micinf.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 24.Mohn WW, et al. The actinobacterial mce4 locus encodes a steroid transporter. J. Biol. Chem. 2008;283:35368–74. doi: 10.1074/jbc.M805496200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh P, et al. PDIM and SL1 accumulation in Mycobacterium tuberculosis is associated with mce4A expression. Gene. 2018;642:178–187. doi: 10.1016/j.gene.2017.09.062. [DOI] [PubMed] [Google Scholar]
- 26.Casali N, Riley LW. A phylogenomic analysis of the Actinomycetales mce operons. BMC Genomics. 2007;8:60. doi: 10.1186/1471-2164-8-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rathor N, et al. Expression profile of mce4 operon of Mycobacterium tuberculosis following environmental stress. Int. J. Mycobacteriol. 2016;5:328–332. doi: 10.1016/j.ijmyco.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 28.Wipperman MF, Sampson NS, Thomas ST. Pathogen roid rage: cholesterol utilization by Mycobacterium tuberculosis. Crit. Rev. Biochem. Mol. Biol. 2014;49:269–93. doi: 10.3109/10409238.2014.895700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brzostek A, Rumijowska-Galewicz A, Dziadek B, Wojcik EA, Dziadek J. ChoD and HsdD can be dispensable for cholesterol degradation in mycobacteria. J. Steroid Biochem. Mol. Biol. 2013;134:1–7. doi: 10.1016/j.jsbmb.2012.09.028. [DOI] [PubMed] [Google Scholar]
- 30.Capyk JK, et al. Mycobacterial cytochrome P450 125 (Cyp125) catalyzes the terminal hydroxylation of C27 steroids. J. Biol. Chem. 2009;284:35534–42. doi: 10.1074/jbc.M109.072132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnston JB, Ouellet H, Ortiz De Montellano PR. Functional redundancy of steroid C26-monooxygenase activity in Mycobacterium tuberculosis revealed by biochemical and genetic analyses. J. Biol. Chem. 2010;285:36352–60. doi: 10.1074/jbc.M110.161117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Casabon I, Swain K, Crowe AM, Eltis LD, Mohn WW. Actinobacterial acyl coenzyme A synthetases involved in steroid side-chain catabolism. J. Bacteriol. 2014;196:579–87. doi: 10.1128/JB.01012-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dubnau E, Fontan P, Manganelli R, Soares-Appel S, Smith I. Mycobacterium tuberculosis genes induced during infection of human macrophages. Infect. Immun. 2002;70:2787–95. doi: 10.1128/IAI.70.6.2787-2795.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karakousis PC, et al. Dormancy phenotype displayed by extracellular Mycobacterium tuberculosis within artificial granulomas in mice. J. Exp. Med. 2004;200:647–57. doi: 10.1084/jem.20040646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wrońska N, et al. The role of fadD19 and echA19 in sterol side chain degradation by Mycobacterium smegmatis. Molecules. 2016;21:598. doi: 10.3390/molecules21050598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Griffin JE, et al. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 2011;7:e1002251. doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dubnau E, Chan J, Mohan VP, Smith I. Responses of mycobacterium tuberculosis to growth in the mouse lung. Infect. Immun. 2005;73:3754–7. doi: 10.1128/IAI.73.6.3754-3757.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gilbert S, Hood L, Seah SYK. Characterization of an aldolase involved in cholesterol side chain degradation in Mycobacterium tuberculosis. J. Bacteriol. 2017;200:e00512–17. doi: 10.1128/JB.00512-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang X, Nesbitt NM, Dubnau E, Smith I, Sampson NS. Cholesterol metabolism increases the metabolic pool of propionate in Mycobacterium tuberculosis. Biochemistry. 2009;48:3819–21. doi: 10.1021/bi9005418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rienksma RA, et al. Comprehensive insights into transcriptional adaptation of intracellular mycobacteria by microbe-enriched dual RNA sequencing. BMC Genomics. 2015;16:34. doi: 10.1186/s12864-014-1197-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Masiewicz P, Brzostek A, Wolański M, Dziadek J, Zakrzewska-Czerwińska J. A novel role of the PrpR as a transcription factor involved in the regulation of methylcitrate pathway in Mycobacterium tuberculosis. PLoS One. 2012;7:e43651. doi: 10.1371/journal.pone.0043651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Höner Zu Bentrup K, Miczak A, Swenson DL, Russell DG. Characterization of activity and expression of isocitrate lyase in Mycobacterium avium and Mycobacterium tuberculosis. J. Bacteriol. 1999;181:7161–7. doi: 10.1128/JB.181.23.7161-7167.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Micklinghoff JC, et al. Role of the transcriptional regulator RamB (Rv0465c) in the control of the glyoxylate cycle in Mycobacterium tuberculosis. J. Bacteriol. 2009;191:7260–9. doi: 10.1128/JB.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Griffin JE, et al. Cholesterol catabolism by Mycobacterium tuberculosis requires transcriptional and metabolic adaptations. Chem. Biol. 2012;19:218–27. doi: 10.1016/j.chembiol.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muñoz-Elías EJ, Upton AM, Cherian J, McKinney JD. Role of the methylcitrate cycle in Mycobacterium tuberculosis metabolism, intracellular growth, and virulence. Mol. Microbiol. 2006;60:1109–22. doi: 10.1111/j.1365-2958.2006.05155.x. [DOI] [PubMed] [Google Scholar]
- 46.Rainwater DL, Kolattukudy PE. Synthesis of mycocerosic acids from methylmalonyl coenzyme A by cell-free extracts of Mycobacterium tuberculosis var. bovis BCG. J. Biol. Chem. 1983;258:2979–85. doi: 10.1016/S0021-9258(18)32817-5. [DOI] [PubMed] [Google Scholar]
- 47.Jain M, et al. Lipidomics reveals control of Mycobacterium tuberculosis virulence lipids via metabolic coupling. Proc. Natl. Acad. Sci. U S A. 2007;104:5133–8. doi: 10.1073/pnas.0610634104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh A, et al. Mycobacterium tuberculosis WhiB3 maintains redox homeostasis by regulating virulence lipid anabolism to modulate macrophage response. PLoS Pathog. 2009;5:e1000545. doi: 10.1371/journal.ppat.1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Daniel J, et al. Induction of a novel class of diacylglycerol acyltransferases and triacylglycerol accumulation in Mycobacterium tuberculosis as it goes into a dormancy-like state in culture. J. Bacteriol. 2004;186:5017–5030. doi: 10.1128/JB.186.15.5017-5030.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deb C, et al. A novel in vitro multiple-stress dormancy model for Mycobacterium tuberculosis generates a lipid-loaded, drug-tolerant, dormant pathogen. PLoS ONE. 2009;4:e6077. doi: 10.1371/journal.pone.0006077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Savvi S, et al. Functional characterization of a vitamin B12-dependent methylmalonyl pathway in Mycobacterium tuberculosis: implications for propionate metabolism during growth on fatty acids. J. Bacteriol. 2008;190:3886–95. doi: 10.1128/JB.01767-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Warner DF, Savvi S, Mizrahi V, Dawes SS. A riboswitch regulates expression of the coenzyme B12-independent methionine synthase in Mycobacterium tuberculosis: implications for differential methionine synthase function in strains H37Rv and CDC1551. J. Bacteriol. 2007;189:3655–9. doi: 10.1128/JB.00040-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Domenech P, Kobayashi H, LeVier K, Walker GC, Barry CE., 3rd BacA, an ABC transporter involved in maintenance of chronic murine infections with Mycobacterium tuberculosis. J. Bacteriol. 2009;191:477–85. doi: 10.1128/JB.01132-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lu R, et al. Catabolism of the cholesterol side chain in Mycobacterium tuberculosis Is controlled by a redox-sensitive thiol switch. ACS Infect. Dis. 2017;3:666–675. doi: 10.1021/acsinfecdis.7b00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abramovitch RB, Rohde KH, Hsu FF, Russell DG. aprABC: a Mycobacterium tuberculosis complex-specific locus that modulates pH-driven adaptation to the macrophage phagosome. Mol. Microbiol. 2011;80:678–94. doi: 10.1111/j.1365-2958.2011.07601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Quigley J, et al. The cell wall lipid PDIM contributes to phagosomal escape and host cell exit of Mycobacterium tuberculosis. mBio. 2017;8:e00148–17. doi: 10.1128/mBio.00148-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park HD, et al. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol. Microbiol. 2003;48:833–843. doi: 10.1046/j.1365-2958.2003.03474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leistikow RL, et al. The Mycobacterium tuberculosis DosR regulon assists in metabolic homeostasis and enables rapid recovery from nonrespiring dormancy. J. Bacteriol. 2010;192:1662–1670. doi: 10.1128/JB.00926-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jones CM, et al. Self-poisoning of Mycobacterium tuberculosis by interrupting siderophore recycling. Proc. Natl. Acad. Sci. U S A. 2014;111:1945–1950. doi: 10.1073/pnas.1311402111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hartkoorn RC, Uplekar S, Cole ST. Cross-resistance between clofazimine and bedaquiline through upregulation of MmpL5 in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2014;58:2979–2981. doi: 10.1128/AAC.00037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Milano A, et al. Azole resistance in Mycobacterium tuberculosis is mediated by the MmpS5-MmpL5 efflux system. Tuberculosis (Edinb) 2009;89:84–90. doi: 10.1016/j.tube.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 62.Ahmad Z, et al. Antimycobacterial activity of econazole against multidrug-resistant strains of Mycobacterium tuberculosis. Int. J. Antimicrob. Agents. 2006;28:543–544. doi: 10.1016/j.ijantimicag.2006.07.028. [DOI] [PubMed] [Google Scholar]
- 63.Balganesh M, et al. Rv1218c, an ABC transporter of Mycobacterium tuberculosis with implications in drug discovery. Antimicrob. Agents Chemother. 2010;54:5167–72. doi: 10.1128/AAC.00610-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Turapov O, et al. Antimicrobial treatment improves mycobacterial survival in non-permissive growth conditions. Antimicrob. Agents Chemother. 2014;58:2798–2806. doi: 10.1128/AAC.02774-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boshoff HIM, et al. The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism: Novel insights into drug mechanisms of action. J. Biol. Chem. 2004;279:40174–84. doi: 10.1074/jbc.M406796200. [DOI] [PubMed] [Google Scholar]
- 66.Colangeli R, et al. The Mycobacterium tuberculosis iniA gene is essential for activity of an efflux pump that confers drug tolerance to both isoniazid and ethambutol. Mol. Microbiol. 2005;55:1829–1840. doi: 10.1111/j.1365-2958.2005.04510.x. [DOI] [PubMed] [Google Scholar]
- 67.Alland D, Steyn AJ, Weisbrod T, Aldrich K, Jacobs WR., Jr Characterization of the Mycobacterium tuberculosis iniBAC promoter, a promoter that responds to cell wall biosynthesis inhibition. J. Bacteriol. 2000;182:1802–11. doi: 10.1128/JB.182.7.1802-1811.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Betts JC, et al. Signature gene expression profiles discriminate between isoniazid-, thiolactomycin-, and triclosan-treated Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2003;47:2903–13. doi: 10.1128/AAC.47.9.2903-2913.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dutta NK, Mehra S, Kaushal D. A Mycobacterium tuberculosis sigma factor network responds to cell-envelope damage by the promising anti-mycobacterial thioridazine. PLoS One. 2010;5:e10069. doi: 10.1371/journal.pone.0010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ramos JL, et al. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 2005;69:326–56. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Abhishek S, et al. Transcriptional profile of Mycobacterium tuberculosis in an in vitro model of intraocular tuberculosis. Front. Cell Infect. Microbiol. 2018;8:330. doi: 10.3389/fcimb.2018.00330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Salina EG, et al. Resuscitation of dormant "non-culturable" Mycobacterium tuberculosis is characterized by immediate transcriptional burst. Front. Cell Infect. Microbiol. 2019;9:272. doi: 10.3389/fcimb.2019.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Harper C, Hayward D, Wiid I, van Helden P. Regulation of nitrogen metabolism in Mycobacterium tuberculosis: a comparison with mechanisms in Corynebacterium glutamicum and Streptomyces coelicolor. IUBMB Life. 2008;60:643–50. doi: 10.1002/iub.100. [DOI] [PubMed] [Google Scholar]
- 74.Tullius MV, Harth G, Horwitz MA. Glutamine synthetase GlnA1 is essential for growth of Mycobacterium tuberculosis in human THP-1 macrophages and guinea pigs. Infect. Immun. 2003;71:3927–3936. doi: 10.1128/IAI.71.7.3927-3936.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Neyrolles O, Wolschendorf F, Mitra A, Niederweis M. Mycobacteria, metals, and the macrophage. Immunol. Rev. 2015;264:249–63. doi: 10.1111/imr.12265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shi X, et al. The copper-responsive RicR regulon contributes to Mycobacterium tuberculosis virulence. mBio. 2014;5:e00876–13. doi: 10.1128/mBio.00876-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang M, et al. Cd(II)-binding transcriptional regulator interacts with isoniazid and regulates drug susceptibility in mycobacteria. J. Biochem. 2020;24:mvaa086. doi: 10.1093/jb/mvaa086. [DOI] [PubMed] [Google Scholar]
- 78.Boradia VM, et al. Mycobacterium tuberculosis acquires iron by cell-surface sequestration and internalization of human holo-transferrin. Nat. Commun. 2014;5:4730. doi: 10.1038/ncomms5730. [DOI] [PubMed] [Google Scholar]
- 79.Hayward D, Wiid I, van Helden P. Differential expression of mycothiol pathway genes: are they affected by antituberculosis drugs? IUBMB Life. 2004;56:131–8. doi: 10.1080/15216540410001674012. [DOI] [PubMed] [Google Scholar]
- 80.Torrey HL, Keren I, Via LE, Lee JS, Lewis K. High persister mutants in Mycobacterium tuberculosis. PLoS One. 2016;11:e0155127. doi: 10.1371/journal.pone.0155127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Voskuil MI, et al. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 2003;198:705–713. doi: 10.1084/jem.20030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Primm TP, et al. The stringent response of Mycobacterium tuberculosis is required for long-term survival. J. Bacteriol. 2000;182:4889–98. doi: 10.1128/JB.182.17.4889-4898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ducati RG, Santos DS, Basso LA. Substrate specificity and kinetic mechanism of purine nucleoside phosphorylase from Mycobacterium tuberculosis. Arch. Biochem. Biophys. 2009;486:155–64. doi: 10.1016/j.abb.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 84.Gengenbacher M, Kaufmann SH. Mycobacterium tuberculosis: success through dormancy. FEMS Microbiol. Rev. 2012;36:514–32. doi: 10.1111/j.1574-6976.2012.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schwenk S, Moores A, Nobeli I, McHugh TD, Arnvig KB. Cell-wall synthesis and ribosome maturation are co-regulated by an RNA switch in Mycobacterium tuberculosis. Nucleic Acids Res. 2018;46:5837–5849. doi: 10.1093/nar/gky226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.O'Farrell HC, Rife JP. Staphylococcus aureus and Escherichia coli have disparate dependences on KsgA for growth and ribosome biogenesis. BMC Microbiol. 2012;12:244. doi: 10.1186/1471-2180-12-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ahidjo BA, et al. VapC toxins from Mycobacterium tuberculosis are ribonucleases that differentially inhibit growth and are neutralized by cognate VapB antitoxins. PLoS One. 2011;6:e21738. doi: 10.1371/journal.pone.0021738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Winther K, Tree JJ, Tollervey D, Gerdes K. VapCs of Mycobacterium tuberculosis cleave RNAs essential for translation. Nucleic Acids Res. Nov. 2016;44:9860–9871. doi: 10.1093/nar/gkw781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pawelczyk J, et al. AccD6, a key carboxyltransferase essential for mycolic acid synthesis in Mycobacterium tuberculosis, is dispensable in a nonpathogenic strain. J Bacteriol. 2011;193:6960–72. doi: 10.1128/JB.05638-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Płociński P, et al. Proteomic and transcriptomic experiments reveal an essential role of RNA degradosome complexes in shaping the transcriptome of Mycobacterium tuberculosis. Nucleic Acids Res. 2019;47:5892–5905. doi: 10.1093/nar/gkz251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 92.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li H, et al. The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Robinson JT, et al. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Powell D. 2015. R. Degust: interactive RNA-seq analysis. [DOI]
- 97.Sambrook J, Russell D, editors. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 98.Parish T, Stoker NG. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology. 2000;146:1969–1975. doi: 10.1099/00221287-146-8-1969. [DOI] [PubMed] [Google Scholar]
- 99.Rumijowska A, Lisowska K, Ziolkowski A, Sedlaczek L. Transformation of sterols by Mycobacterium vaccae: Effect of lecithin on the permeability of cell envelopes to sterols. World J. Microbiol. Biotechnol. 1997;13:89–95. doi: 10.1007/BF02770813. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Powell D. 2015. R. Degust: interactive RNA-seq analysis. [DOI]