Abstract
SARS-CoV-2 has caused a worldwide epidemic of coronavirus disease 19 (COVID-19). Antibody drugs present an effective weapon for tens of millions of COVID-19 patients. Antibodies disrupting the interactions between the receptor-binding domain (RBD) of SARS-CoV-2 S protein and the angiotensin converting enzyme 2 (ACE2) effectively block SARS-CoV-2 cell entry into host cells. In order to rapidly develop more potent neutralizing antibodies, we utilized virtual scanning mutageneses and molecular dynamics simulations to optimize the antibody of P2B-2F6 isolated from single B cells of SARS-CoV-2 infected patients. Two potent P2B-2F6 mutants, namely H:V106R and H:V106R/H:P107Y, were found to possess higher binding affinities with the RBD domain of SARS-CoV-2 than others. Polar interactions are preferred near 106 and 107 paratope residues of the heavy chain. The mutations also increase the hydrogen-bonding network formed between the antibody and the RBD. Notably, the optimized antibodies possess potential neutralizing activity against the alarming SARS-CoV-2 variant of N501Y. This study provides insights into structure-based optimization of antibodies with higher affinity to the antigen. We hope that our proposed antibody mutants could contribute to the development of improved therapies against COVID-19.
Keywords: Antibody, SARS-CoV-2, COVID-19, Scanning mutageneses, Molecular dynamics simulation
1. Introduction
The epidemic of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spreads at an alarming rate and has led to a global pandemic [1]. With its first severe outbreak in late 2019, SARS-CoV-2 has been identified to cause an acute respiratory distress syndrome (ARDS) called coronavirus disease 2019 (COVID-19) [[2], [3], [4]]. SARS-CoV-2 is highly contagious, and transmission of COVID-19 occurs mainly via airborne droplets and fecal-oral route [5]. As of May 6, 2021, SARS-CoV-2 has infected over 155,665,000 people worldwide with a death toll of over 3,250,000 [6]. The fatality rate becomes higher in elderly patients and patients with comorbidities [4]. Specific SARS-CoV-2 drugs are in urgent need clinically. Although some therapeutic options have been approved by FDA, their efficacies are yet to be validated [[7], [8], [9]].
Antibodies have remarkable potential for therapeutic and prophylactic applications against SARS-CoV-2, and various antibody treatments are currently being explored [10,11]. SARS-CoV-2 is a positive-stranded RNA virus belonging to the β genus coronavirus, which includes SARS-CoV, bat SARS-like CoV, and MERS-CoV [12]. The surface spike glycoprotein (S protein) mediating the infection of target cells is the major antigen of coronaviruses. Angiotensin-converting enzyme 2 (ACE2) has been identified as the receptor for SARS-CoV-2 as well as SARS-CoV [[13], [14], [15]]. The receptor-binding domain (RBD) of the S protein binds with ACE2 to trigger cell membrane fusion and thus facilitate SARS-CoV-2 entry into host cells [16]. This indicates that disruption of the RBD and ACE2 interaction could block SARS-CoV-2 cell entry. RBD is immunogenic and can elicit potent neutralizing antibodies that inhibit RBD binding with ACE2. Several studies have already demonstrated that RBD-targeted antibodies of SARS-CoV and MERS-CoV constitute a major component of protective immunity against viral infection in humans [17,18]. Although the RBD sequence of SARS-CoV-2 and that of SARS-CoV share ~73% identity, they lead to distinctive immunological responses [12]. Therefore, antibodies specifically targeting the RBD of SARS-CoV-2 with maximum efficacy are desired to treat the tens of millions of patient with COVID-19.
The constant emergence of SARS-CoV-2 variants is another significant concern. Recently, new SARS-CoV-2 variants such as B.1.1.7, B.1.351, and P.1 lineages are detected in many parts of the world [[19], [20], [21]]. These variants have an unusually large number of genetic changes, including the noteworthy spike protein mutation N501Y, one of the key residues in RBD domain interacting with ACE2 [19]. Experimental data suggests mutation N501Y increases the binding affinity of the spike protein with human and murine ACE2 [20]. It is estimated that the spread of SARS-CoV-2 N501Y variant is 70% faster than previous strains, indicating a much higher infectivity [22]. The mutation might affect the immune recognition of current antibodies and/or vaccines targeting the RBD domain.
P2B-2F6 is the first reported antibody isolated from a SARS-CoV-2 infected patient with atomic-level structural characterization of its interactions with RBD [23]. N501 of RBD is spatially distant from its interacting interface with P2B-2F6, and thereby the newly identified mutation N501Y is unlikely to affect the neutralizing activity of P2B-2F6. Hence, P2B-2F6 was selected for further development. Although it neutralizes live SARS-CoV-2 with IC50 value of 0.41 μg/ml, its binding affinity with SARS-CoV-2 RBD (Kd, 5.14 nM) is lower than ACE2, other SARS-CoV-2 antibodies such as P2C-1F11, and chronic HIV-1 infection antibodies [[24], [25], [26], [27]]. In general, the binding affinity of antibody with SARS-CoV-2 RBD reflects the capability of competition with ACE2. Therefore, this study aims to computationally optimize the antibody of P2B-2F6 by increasing the binding affinities based on the recently released crystal structure of P2B-2F6 in complex with the RBD domain of SARS-CoV-2 (PDB code: 7BWJ). Two potent P2B-2F6 mutants of H:V106R and H:V106 R/H:P107Y were found to have higher binding affinities with the RBD domain of SARS-CoV-2. This study provides a potential guidance in structure-based optimization of antibodies with higher affinities to the antigen. We hope that our proposed antibody mutants could be more efficient to combat SARS-CoV-2 as well as the more infectious N501Y variant.
2. Methods
Mutants Design. The crystal structure of P2B-2F6 binding with SARS-CoV-2 RBD was obtained from the Protein Data Bank (PDB ID: 7BWJ) [23]. The complex structure was prepared at pH 7.4 using CHARMm force field. Scanning mutagenesis [28] in protein complex was performed in the Protein Design module of Discovery Studio 4.1. The paratope residues of S30, S31, Y33, H54, I103, V104, V105, V106, P107 in the heavy chain (H) and G31, Y32 in the light chain (L) of P2B-2F6 were mutated into 20 standard amino acid types to screen out mutations that stabilize the binding with RBD. For each mutant, the difference in the free energy of binding between the wild type and mutated structure was calculated. The total free energy, ΔGtot, of bound or unbound state were calculated as the following weighted sum of energy terms:
| ΔGtot(T) = aEvdW + bΔGelec(T) - cTSsc + ΔGnp |
where a, b, and c are empirical scaling parameters. The mutation energy < −0.5 kcal/mol, between −0.5 and 0.5 kcal/mol and >0.5 kcal/mol respectively indicate the mutation stabilizes, neutralizes and destabilizes the antibody binding with SARS-CoV-2 RBD.
Representative single mutants with higher mutation energy were subject to molecular dynamics (MD) simulations and the binding free energy of the antibody with RBD was confirmed to be strengthened by H:V106R, H:V104Y and L:G31L mutations of P2B-2F6. Finally, double mutations were screened using scanning mutagenesis in Discovery Studio 4.1 at the crucial paratope residues of L:G31 and H:V104-P107 of P2B-2F6 antibody. Representative double mutants with higher mutation energy were subject to another round of MD simulations to optimize the complex structures.
Building of Mutant Complex Structures. The complex structures of potent P2B-2F6 mutants binding with SARS-CoV-2 RBD were built using the crystal structure 7BWJ as the template in the Build Mutants module of Discovery Studio 4.1. The disulfide bridges between cysteine residues were constructed according to the template. High level of sampling was performed to optimize the built structures. Five models of each mutant were obtained and the model with the lowest total energy score was used for subsequent simulations.
Molecular Dynamics (MD) Simulations in Water. All MD simulations were performed in Amber 12 [29]. The complex structures of P2B-2F6 and its mutants binding with SARS-CoV-2 RBD were prepared. Hydrogens were added using the Leap module and the protonation states of the titratable residues were set using the program PROPKA [30]. Amber ff12SB force field [31] was assigned for the proteins. The complexes were solvated in a truncated octahedral box of TIP3P water molecules with 10.0 Å buffer along each dimension [32]. Cl− counterions were added to neutralize the systems.
For each system, energy minimization and MD simulations were performed using the PMEMD module in Amber 12. A two-step energy minimization process was carried out to relieve bad contacts. At the first step, water molecules and counterions were relaxed by restraining the complex with a harmonic constant of 2.0 kcal/mol·Å−2 based on the steepest descent method. At the second step, the restraint was removed to allow all of the atoms to move freely using the conjugate gradient algorithm. After that, each system was gently heated from 0 to 300 K in 300 ps at constant volume with a harmonic constant of 10.0 kcal/mol·Å−2 and then equilibrated at 300 K and 1 bar constant pressure for 500 ps. Finally, a 100 ns MD simulation was performed with a time step of 2 fs. During the simulation, all bonds involving hydrogen atoms were constrained using the SHAKE algorithm [33]. The non-bonded cutoff was set to 10.0 Å, and electrostatic interactions were calculated using the particle-mesh Ewald method (PME) [34,35]. The temperature was controlled using the Langevin thermostat method [36].
MM/GBSA Method. Compared with MM/PBSA, MM/GBSA is more suitable for binding free energy comparisons [37]. Therefore, MM/GBSA of the antibodies binding with SARS-CoV-2 RBD in each system was calculated in Amber 12. A total of 100 snapshots from the 80–100 ns equilibrated dynamics trajectory with a time interval of 200 ps were extracted, and the MM/GBSA calculation was performed on each snapshot. The binding free energy (ΔG) was computed according to the following equations [37]:
| ΔG = <ΔGgas> + <ΔGsolv> − <TΔS> | (1) |
| ΔGgas = ΔGelec + ΔGvdW + ΔGint | (2) |
| ΔGsol = ΔGGB + ΔGnp | (3) |
| ΔGnp = γ × SASA + β | (4) |
where < … > indicates an average of the energy term. ΔGgas and ΔGsolv represent the vacuum and solvation binding free energies, respectively. –TΔS is the entropic contribution, which is not considered in the relative free energy analysis. ΔGgas is composed of intermolecular electrostatic energy (ΔGelec), van der Waals energy (ΔGvdW), and internal energy (ΔGint). ΔGsolv includes the electrostatic solvation energy (ΔGGB) and the nonpolar solvation energy (ΔGnp). γ is the surface tension proportionality constant and β is the offset value. The solvent accessible surface area (SASA) was estimated by the MSMS algorithm with a probe radius of 1.4 Å.
Data analysis. The cpptraj analysis module within Amber 12 was used for the calculations of root mean square deviation (RMSD), root-mean-square fluctuation (RMSF) and hydrogen bond analysis, etc. T-test was used to compare the difference significance between the wild-type and mutant antibodies in Graphpad Prism 5.0. The structures were viewed in PyMOL 1.7.4.
Aggregation propensity calculation. Hydrophobicity is the most important factor leading to antibody aggregation. We compared the aggregation propensity of WT P2B-2F6 and the optimized mutants by identifying hydrophobic surface patches in the CDR regions using MOE 2020 software. Zeta values of the antibodies were also calculated at 300 K and pH 7.4 in MOE 2020.
3. Results
3.1. Structural basis of P2B-2F6 binding with SARS-CoV-2 RBD
The SARS-CoV-2 neutralizing antibody of P2B-2F6 was isolated and characterized from single B cells of a SARS-CoV-2 infected patient. The crystal structure shows that P2B-2F6 binds with the RBD of SARS-CoV-2 through hydrophobic and hydrophilic interactions primarily using the heavy chain (Fig. 1 A). The paratope consists 12 heavy chain residues (Y27, S28, S30, S31, and Y33 of HCDR1 [IMGT number: Y28, S29, S31, S35, Y37]; H54 of HCDR2 [IMGT number: H58]; I103-P107 and R112 of HCDR3 [IMGT number: I111, V111A, V111B, V111C, P112D, and R113]) and 4 light chain residues (G31-Y34 of LCDR1 [IMGT number: G35-Y38]) interacting with R346, K444-L452 and V483-S494 of RBD (Fig. 1B).
Fig. 1.
Structures of SARS-CoV-2 RBD binding with P2B-2F6 and ACE2. (A) Overall structure of P2B-2F6 Fab binding with SARS-CoV-2 RBD. N501 of RBD is shown in sphere. (B) Detailed binding interactions between P2B-2F6 and SARS-CoV-2 RBD. The crucial residues of SARS-CoV-2 RBD, P2B-2F6 light chain and heavy chain are respectively shown in blue, green and magenta sticks. (C) Overall structure of ACE2 binding with SARS-CoV-2 RBD. (D) The population of crucial residues of RBD interacting with ACE2 analyzed from the crystal structures (PDB codes: 6LZG, 6M0J, 7A98, 7DF4 and 7KNB) in PDBsum server.
The structure of RBD binding with ACE2 (Fig. 1C) and the contacting residues on RBD were analyzed using PDBsum server (PDB codes: 6LZG, 6M0J, 7A98, 7DF4 and 7KNB) (Fig. 1D). The most important domain of RBD (residues K417 to Y505) interacts with H1 helix (residues S19 to W48) of ACE2 mainly through hydrogen bonds, non-bonded contacts and salt bridges. The binding site of RBD with P2B-2F6 is partially overlapped with that of ACE2, and the overlapped interacting residues include G446, Y449, F490 and S494 (Fig. 1BCD). Therefore, P2B-2F6 competes with ACE2 to bind RBD [23], indicating that blocking the RBD and ACE2 interaction is a useful surrogate for neutralization.
N501Y mutation has been reported to accelerate the spread of SARS-CoV-2 recently [20]. Although N501 of RBD forms essential interaction with ACE2 [16], it is spatially distant from the binding site of P2B-2F6 (Fig. 1AD). The calculated mutation energy of N501Y of RBD binding with P2B-2F6 is 0.01 kcal/mol, confirming that N501Y mutation does not affect their interaction. This indicates that the N501Y variant of SARS-CoV-2 would not escape the immune effect of P2B-2F6.
3.2. P2B-2F6 single mutation scanning
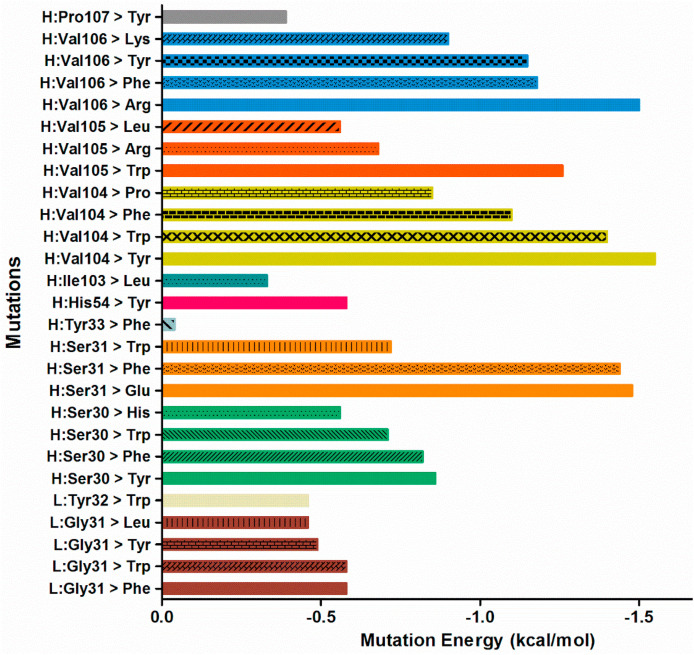
Computational biology is widely used in antibody design. To obtain antibodies with more potent neutralizing activity against SARS-CoV-2, scanning mutageneses in P2B-2F6 complex binding with RBD were performed. The paratope residues of S30, S31, Y33, H54, I103, V104, V105, V106, P107 in the heavy chain (H) and G31, Y32 in the light chain (L) were mutated into each of the 20 standard amino acids for individual assessment. The single mutations of the paratope residues with top ranking mutation energies were summarized in Fig. 2 . The mutation energies of H:V104Y and H:V106R were greater than −1.5 kcal/mol, while those of H:S31F, H:S31E, H:V104W, H:V104F, H:V105W, H:V106F and H:V106Y were greater than −1.0 kcal/mol, indicating that these mutations might strengthen the binding of the antibody with RBD domain of SARS-CoV-2.
Fig. 2.
The mutation energy of P2B-2F6 mutants binding with SARS-CoV-2 RBD calculated by single mutation scanning. The mutation energy < −0.5 kcal/mol, between −0.5 and 0.5 kcal/mol and >0.5 kcal/mol respectively indicate the mutation stabilizes, neutralizes and destabilizes the antibody binding with SARS-CoV-2 RBD.
3.3. P2B-2F6 single mutation optimization
To further optimize the complex structures of P2B-2F6 mutants with RBD, MD simulations were carried out for a series of representative mutant systems. RMSDs of the heavy atoms were calculated in reference to the first structure through the entire MD simulations to evaluate the equilibrium of the systems (Supplementary Fig. S1). All systems reached equilibrium after 20 ns of MD simulations, and the last 20 ns trajectories of all systems were very stable, with the mean RMSD values < 2.0 Å (Supplementary Table S1). The RMSFs of Cα atoms near the given mutated residues were analyzed comparing to the WT P2B-2F6 (Supplementary Fig. S2). L:G31F and L:G31L mutations caused large fluctuations near the mutation site. The conformation changes may lead to rearrangement of the residues, which would affect the interactions of the flaps with SARS-CoV-2 RBD.
MM/PBSA and MM/GBSA are arguably very popular methods for binding free energy prediction since they are more accurate than most scoring functions and less computationally demanding than alchemical free energy methods in biomolecular studies such as protein−protein interaction [38,39]. MM/GBSA method is more suitable for binding free energy comparisons, so it was employed to compare the affinity of RBD binding with the wild-type (WT) and mutant antibodies. To obtain a good conformational sampling, a total of 100 conformations extracted from the last 20 ns trajectories of MD simulations were used to calculate binding free energies and separate free energy components.
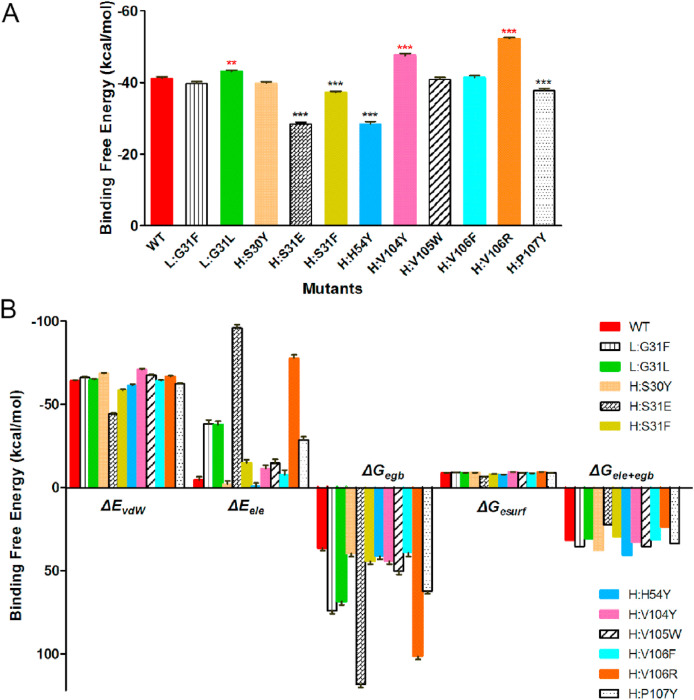
It is found that the MM/GBSA binding free energies of H:V106R, H:V104Y, and L:G31L mutants of P2B-2F6 with RBD are strengthened by 11.07, 6.40, and 1.84 kcal/mol, respectively, relative to that of WT P2B-2F6 (Fig. 3 A and Supplementary Table S2), indicating stronger binding of these P2B-2F6 mutants. The binding free energies are decomposed into van der Waals interactions (ΔE vdW), electrostatics interactions (ΔE ele), polar solvation free energies (ΔG egb), and non-polar solvation free energies (ΔG esurf). The increase of van der Waals interactions induced by H:V104Y and polar interactions (ΔG ele + egb) induced by H:V106R are the main contributors to the enhanced binding of the mutated P2B-2F6 with RBD (Fig. 3B). Binding free energy decomposition of the residues at the mutation sites showed that H:Y30, H:F31, H:F106 and H:R106 lead to their binding free energies with SARS-CoV-2 RBD significantly increase in comparison with that of WT P2B-2F6, while the H:E31 and H:Y54 mutations lead to the binding free energies decrease. (Supplementary Fig. S3). The binding free energy changes of the mutated residues are not completely consistent with the overall binding free energy changes of the antibody mutants, which may be because of binding free energy changes of other associated residues caused by conformational rearrangement.
Fig. 3.
The binding free energies (A) and separate free energy components (B) of P2B-2F6 single mutants binding with SARS-CoV-2 RBD calculated by MM/GBSA method.
3.4. P2B-2F6 double mutations design
Based on the screening results of single mutations, we evaluated double mutations of the crucial paratope residues L:G31 and H:V104-P107 of P2B-2F6 antibody. Scanning mutageneses suggested that a series of double mutations could stabilize the binding of P2B-2F6 with SARS-CoV-2 RBD (Fig. 4 A). Among them, the mutation energies of H:V106R/H:P107Y and H:V104Y/H:V106W are higher than others, with the values < −3.0 kcal/mol. Thereby, H:V106R/H:P107Y, H:V104Y/H:V106W, and H:V104Y/H:V106R (considering the mutation effect of H:V104Y and H:V106R, Fig. 3) mutants of P2B-2F6 were selected for optimization by MD simulations.
Fig. 4.
P2B-2F6 double mutation design. (A) The mutation energy of P2B-2F6 mutants binding with SARS-CoV-2 RBD calculated by double mutations scanning. The binding free energies (B) and separate free energy components (C) of P2B-2F6 double mutants binding with SARS-CoV-2 RBD calculated by MM/GBSA method. Structural characteristics of the potent antibody mutants.
The MM/GBSA binding free energies of H:V106R/H:P107Y, H:V104Y/H:V106W, and H:V104Y/H:V106R mutants with RBD are 13.59, 4.80, and 4.59 kcal/mol, respectively, higher than WT P2B-2F6 (Fig. 4B, Supplementary Table S2). The free energy decomposition demonstrates that the enhanced binding ability of the mutant antibody to RBD is mainly induced by the increase of polar interactions (ΔG ele + egb) for H:V106 R/H:P107Y and H:V104Y/H:V106R mutants, and van der Waals interactions for H:V104Y/H:V106W mutant (Fig. 4C). Binding free energy decomposition of the residues at the mutation sites showed that H:R106, H:W106 and H:Y107 mutation residues in the double mutants of P2B-2F6 possess higher binding free energies with SARS-CoV-2 RBD than that of WT P2B-2F6 (Supplementary Fig. S3). Among them, the binding energy of H:R106 increases most significantly in H:V106 R/H:P107Y double mutant.
From computational scanning mutageneses and MD simulations, we found the single mutation H:V106R and the double mutations H:V106R/H:P107Y of P2B-2F6 resulted in higher binding affinities with the RBD domain of SARS-CoV-2 than others. To reveal changes in interactions between the antibody and RBD of SARS-CoV-2 upon mutations, hydrogen bonding and structural analyses were performed after MD simulations.
Hydrogen bond analysis reveals that RBD forms hydrogen bonds with Y27, S30, S31, H54, R112, Y53 in the heavy chain and N33, Y34 in the light chain of WT P2B-2F6 (Table 1 ). Mutations of H:V106R and H:V106R/H:P107Y produce an obvious impact on the hydrogen-bonding network formed by the antibody and RBD of SARS-CoV-2. The H:V106R mutation leads to additional hydrogen bonds formation between the side chains of H:R106 and E484 of RBD (Table 1). For the H:V106R/H:P107Y double mutant, the phenolic hydroxyl group of H:Y107 donates a hydrogen bond to the amide oxygen of T470 on RBD with a high occupancy of 88.12% (Table 1). Interactions between RBD and the antibodies around the mutation sites are shown (Fig. 5 ). Formation of additional hydrogen bonds contributes to the increased binding affinity between the antibody and antigen, consistent with the significant increase of electrostatics interactions (ΔE ele) in energy decomposition (Fig. 4B).
Table 1.
Main hydrogen-bonding interactions involved in antibodies binding with the RBD domain of SARS-CoV-2.
| Hydrogen Bonda | Occupancyb |
||
|---|---|---|---|
| WT | H:V106R | H:V106 R/H:P107Y | |
| O@G447···O-H@H:Y27 | 86.14 | 87.13 | 93.07 |
| O@H:S31⋯N-H@Y449 | 70.30 | 66.34 | 85.15 |
| OD1@N450···NE2-HE2@H:H54 | 67.33 | 44.55 | 69.31 |
| O@H:S30···ND2-HD21@N450 | 64.36 | 59.41 | 70.30 |
| OE2@E484···NH1-HH12@H:R112 | 61.39 | 55.45 | 94.06 |
| OE2@E484···NH2-HH22@H:R112 | 54.46 | 31.68 | 33.66 |
| OE1@E484···NH2-HH22@H:R112 | 27.72 | 30.69 | 38.61 |
| OE1@E484···NH1-HH12@H:R112 | 18.81 | 41.58 | |
| O@E484···ND2-HD22@L:N33 | 43.56 | 53.47 | 8.91 |
| O-H@ L:Y34⋯N-H@E484 | 27.72 | 16.83 | 20.79 |
| OD1@N450···NE2-HE2@H:Y53 | 21.78 | 11.88 | |
| OE2@E484···NH2-HH21@H:R106 | 44.55 | 4.95 | |
| OE1@E484···NH2-HH21@H:R106 | 53.47 | 86.14 | |
| OE1@E484···NE-HE@H:R106 | 38.61 | 13.86 | |
| OE2@E484···NE-HE@H:R106 | 37.62 | 65.35 | |
| O@T470···O-H@H:Y107 | 88.12 | ||
Residues of the antibody are shown in bold.
The hydrogen bonds are determined by an acceptor···donor distance of <3.5 Å and an acceptor···H−donor angle of >120°.
Occupancy is defined as the percentage of simulation time that a specific hydrogen bond exists.
Fig. 5.
Binding interactions around residue 106 in the heavy chain of P2B-2F6 WT (A) and H:V106R (B), H:V106R/H:P107Y (C) mutants with SARS-CoV-2 RBD. SARS-CoV-2 RBD is shown in green. P2B-2F6, H:V106R and H:V106R/H:P107Y mutants are shown in cyan, purple and yellow, respectively. Crucial residue Y34 in the light chain is labeled L:Y34, other labeled residues of the antibody are all in the heavy chain (H).
Aggregation propensity is an important developability metric in protein-based biotherapeutics, as aggregation can occur with a concomitant significant loss of drug efficacy and higher rate of immunogenic response [40]. There are many properties that affect protein aggregation, including hydrophobicity, charge, propensity to form β-sheets and α-helical structures. Hydrophobicity is thought to be of major importance during protein aggregation [41]. Therefore, we predicted the aggregation propensity of WT P2B-2F6 and the optimized mutants of H:V106R and H:V106R/H:P107Y by identifying hydrophobic surface patches in the CDR regions. The results indicate that H:V106R mutation of P2B-2F6 might reduce aggregation but H:V106R/H:P107Y double mutations might increase aggregation (Fig. 6 , Table 2 ). We also calculated Zeta values of the WT and optimized antibodies (Table 2). The Zeta values of H:V106R and H:V106R/H:P107Y mutants are greater than WT P2B-2F6, suggesting that the optimized antibodies are more stable than WT P2B-2F6.
Fig. 6.
The hydrophobic patches in the CDR regions of WT P2B-2F6 (A), H:V106R (B) and H:V106R/H:P107Y (C) mutants. The heavy chain and light chain of the antibodies are shown in purple and green ribbons, respectively. The patches are colored in orange.
Table 2.
The calculated CDR hydrophobic patch areas and Zeta values of WT P2B-2F6 and V106R, V106R/P107Y mutants.
| Antibodies | CDR Hydrophobic Patch Area (Å2) | Zeta (pH 7.4) |
|---|---|---|
| WT P2B-2F6 | 160 | 14.08 |
| H:V106R | 120 | 17.74 |
| H:V106R/H:P107Y | 180 | 17.74 |
3.5. The potential of the optimized antibodies against SARS-CoV-2 spike variants
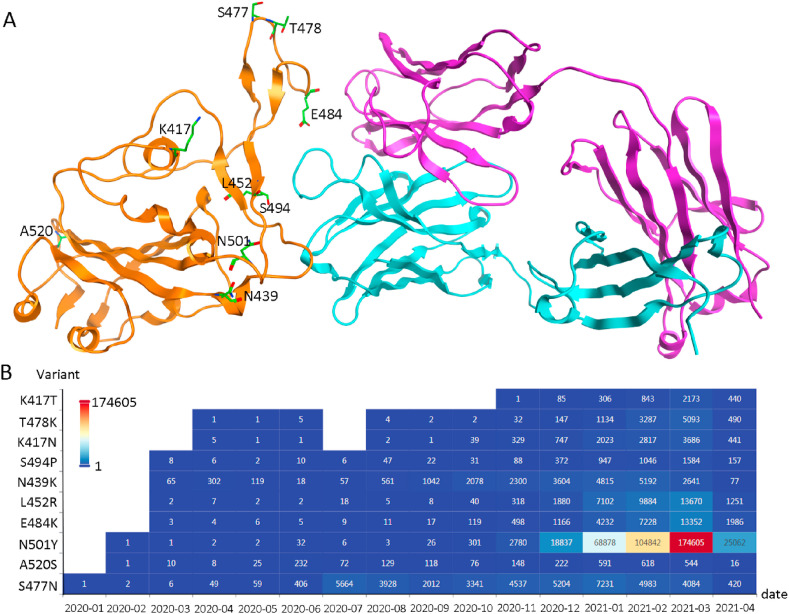
Mutations in the spike protein's RBD region could potentially impact disease severity and treatment strategies. The top 10 RBD region mutations (data from CovMT, a COVID-19 virus mutation tracker system [42]) were structurally analyzed (Fig. 7 ). Among them, E484, S494, L452, S477, T478, N501, and N439 are either in direct contact with P2B-2F6 or in close proximity to the interaction site. We calculated the mutation energy (ΔΔG) between these SARS-CoV-2 spike mutants and the WT or optimized P2B-2F6 antibodies (Table 3 ). The results indicate the binding affinity of WT P2B-2F6 remains almost unchanged with most single spike protein mutants except for E484K and L452R. On the other hand, the optimized antibodies of H:V106R and H:V106R/H:P107Y have increased binding affinities with most single spike protein mutants including the highest frequency mutant of N501Y. However, the E484K spike protein mutation decreased its binding affinity with both the WT and optimized P2B-2F6 antibodies. Some SARS-CoV-2 variants possess combination mutations such as E484K/N501Y and K417N/E484K/N501Y [42]. We also calculated the mutation energy between the combination spike mutants and the antibodies (Table 3). Unfortunately, the results indicate that neither the WT nor optimized P2B-2F6 antibodies have potential neutralizing activity against these variants. Comparatively, the mutation energies of E484K, E484K/N501Y and K417N/E484K/N501Y binding with H:V106R/H:P107Y are smaller than WT P2B-2F6.
Fig. 7.
The top 10 identified RBD region mutations (data from CovMT). (A). The structural locations of the top 10 RBD region mutations. SARS-CoV-2 RBD, the heavy chain and light chain of P2B-2F6 are shown in orange, purple and cyan ribbons, respectively. The top 10 mutations are shown in green sticks. (B). The isolate counts of the top 10 RBD mutations updated by 2021-04-23.
Table 3.
Mutation energy (ΔΔG) between SARS-CoV-2 spike mutants and the WT/optimized P2B-2F6 antibodies.
| Spike Mutants | Impact on binding strength |
||
|---|---|---|---|
| WT P2B-2F6 | H:V106R | H:V106 R/H:P107Y | |
| E484Ka | Decrease | Decrease | Decrease |
| S494P | Neutral | Increase | Increase |
| L452R | Decrease | Increase | Increase |
| S477N | Neutral | Increase | Increase |
| T478K | Neutral | Increase | Increase |
| N501Y | Neutral | Increase | Increase |
| N439K | Neutral | Increase | Increase |
| E484K/N501Yb | Decrease | Decrease | Decrease |
| K417N/E484K/N501Yc | Decrease | Decrease | Decrease |
Increase in binding strength equates to the mutation energy ΔΔG < −0.5 kcal/mol.
Neutral in binding strength equates to the mutation energy −0.5 < ΔΔG <0.5 kcal/mol.
Decrease in binding strength equates to the mutation energy ΔΔG >0.5 kcal/mol.
a. The mutation energy: H:V106R/H:P107Y (1.16 kcal/mol) < WT P2B-2F6 (2.57 kcal/mol) < H:V106R (2.67 kcal/mol). b. The mutation energy: H:V106R/H:P107Y (1.47 kcal/mol) < WT P2B-2F6 (2.50 kcal/mol) < H:V106R (3.40 kcal/mol). c. The mutation energy: H:V106R/H:P107Y (0.75 kcal/mol) < WT P2B-2F6 (2.02 kcal/mol) < H:V106R (2.75 kcal/mol).
4. Discussion
SARS-CoV-2 poses a serious global health emergency. It is reported that ~20% of COVID-19 patients develop serious symptoms such as acute respiratory distress, severe pneumonia, sepsis, and even death [4]. Although vaccines are effective in preventing virus infection in susceptible people, antibody drugs with advantage of precise targeting and rapid onset of action present an effective alternative for the treatment of COVID-19. SARS-CoV-2 encodes four main structural proteins, spike (S), envelope (E), membrane (M), and nucleocapsid (N) [43]. S protein binds to the host cell receptor and is responsible for the host-viral cell fusion, making it the most promising target for neutralizing antibodies [17]. The discovery of anti-S protein antibodies is progressing rapidly. Many potent neutralizing antibodies have been identified from convalescent human donors, immunized animal models, and re-engineering [23,[44], [45], [46], [47], [48], [49]], with several human clinical trials in progress.
The antibody of P2B-2F6 is isolated from single B cells of SARS-CoV-2 infected patients. It is competitive with ACE2 by binding to the RBD of SARS-CoV-2 and blocks the pathogen cell entry [23]. To obtain antibodies with more potent neutralizing activity against SARS-CoV-2, scanning mutageneses of the paratope residues of P2B-2F6 and MD simulations were performed. We found that two potent P2B-2F6 mutants, H:V106R and H:V106R/H:P107Y, produce higher binding affinities with the RBD domain of SARS-CoV-2 than others. These mutations changed the hydrogen-bonding network formed by the antibody and S protein of SARS-CoV-2. The enhanced binding ability between the mutant antibodies and RBD is mainly induced by the increase of electrostatic interactions and polar solvation free energy. In general, competition with ACE2 predicts antibody potency, while the binding affinity with SARS-CoV-2 RBD reflects certain capability of competition. Therefore, we propose that H:V106R and H:V106R/H:P107Y mutants of P2B-2F6 might have more potent neutralizing activity against SARS-CoV-2 than P2B-2F6.
SARS-CoV-2 is constantly evolving to stabilize its genome. New SARS-CoV-2 variants such as B.1.1.7, B.1.351, and P.1 lineages are emerging and spreading rapidly in many parts of the world [[19], [20], [21]]. The most alarming evolution is the newly detected spike mutation N501Y, common among B.1.1.7, B.1.351 and P.1 lineages, which has increased the infectivity of SARS-CoV-2 [19,50]. N501, one of the key contact residues of RBD with ACE2, has no direct interaction with P2B-2F6. Although N501Y mutation increases the binding affinity of SARS-CoV-2 RBD with host ACE2 [20], it has little effect on the binding of antibodies investigated in this study. Therefore, we speculate that the optimized antibodies also have high neutralizing activity against N501Y variant of SARS-CoV-2. However, for other concerning mutations of E484K, E484K/N501Y and K417N/E484K/N501Y observed in some SARS-CoV-2 variants [20,21], the neutralizing activity of both the WT antibody and the optimized mutants might be significantly decreased due to the loss of interactions from E484 and the probable appearance of electrostatic repulsion with H:R112 and H:R106 of the antibodies when this glutamate mutates to lysine. However, these analyses need further in vitro and in vivo validations before the antibodies can be used against SARS-CoV-2.
ChloeRees-Spear et al. designed a series of spike mutations to identify potential escape variants. They found that some of the tested antibodies [51] lost or exhibited less potency against the RBD-mutated pseudotyped virus, including K417V, KVG444-446TST, L452K, LF455-6YL, TEI470-2NVP, and S494D [50]. From the binding interactions of P2B-2F6 with SARS-CoV-2, we speculate that the K417V, KVG444-446TST, LF455-6YL and TEI470-2NVP mutations have little effect on the neutralizing activity of P2B-2F6 and the optimized antibodies, but the L452K and S494D mutated pseudotyped virus might reduce the neutralizing activity of the antibodies. Fortunately, these variants of SARS-CoV-2 have not yet emerged in real world.
Owing to the fast development of artificial intelligence technology, computer-aided drug discovery has been widely applied in the development of anti-SARS-CoV-2 drugs [[52], [53], [54], [55]]. A number of different approaches to guide the rational engineering of antibody binding are available. However, most approaches are of limited accuracy when applied to antibody design, and have largely been limited to analyzing single point mutations [[56], [57], [58]]. This study proposed a computational approach for the optimization of antibodies for COVID-19 via combined virtual scanning mutageneses and MD simulations (Fig. 8 ). Firstly, the paratope residues from the binding interactions between antibody and RBD are analyzed. Then the single mutations that could strengthen the binding of the antibody with RBD are identified through scanning mutageneses. Such single mutations are subsequently evaluated by MD simulations and MM/GBSA calculations. Double-site scanning mutageneses are performed afterwards to predict double mutations that could strengthen the binding of the antibody with RBD, which are eventually subjected to MD simulations and MM/GBSA calculations to obtain the final suggested double mutations. Combination of scanning mutageneses and MD simulations help reduce the probability of false positive antibody mutants.
Fig. 8.
Computational workflow for identifying possible mutations that enhance the binding of antibody with SARS-CoV-2. Ab: antibody. ΔΔG = ΔGmutation – ΔGWT.
Most of the predictions made by our approach are consistent with mmCSM-AB [59], a recently developed benchmarked method using graph-based signatures and atomic interaction information in analyzing mutations on antigen binding affinity, confirming that our methodology is of reliability (Supplementary Table S3). Unlike previous studies that rely on researchers’ expertise to select possible mutations [[60], [61], [62]], this workflow can be easily performed on high-performance clusters or supercomputers without such expertise. Moreover, our designed workflow can also be applied to optimizing other protein drugs. However, this methodology is based on 3D structures of protein-protein complexes. For those proteins without solved structures, homology modeling and molecular docking should be done first.
5. Conclusions
The simplest and most direct approach to combating SARS-CoV-2 is antibodies or vaccines. Antibodies possessing specific binding affinities with SARS-CoV-2 RBD are promising therapeutics for COVID-19. Integrating virtual scanning mutageneses with MD simulations, we improved the binding affinity of P2B-2F6 with RBD by mutating single and/or double paratope residues. The mutants of H:V106R and H:V106R/H:P107Y might have more potent neutralizing activity against both wild type SARS-CoV-2 as well as the alarming variant of N501Y. This precise structural matching method should be considered in future antibody design protocols.
Declaration of competing interest
The authors declare no conflicts of interest with respect to this work.
Acknowledgments
We acknowledge the National Natural Science Foundation of China (81973527).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2021.104550.
Author contributions
J. Chen and P.Cao conceived the presented idea, J.Chen, F.Wu and X.Sun designed the research; J.Chen, F.Wu and W.Kong performed research; X.Cai and J.Yang helped in analyzing data; J.Chen, D.Lin, X.Sun and P.Cao drafted and revised the manuscript. All authors read and approved the final manuscript. J.Chen and F.Wu contributed equally to this work.
Data availability statement
The data that support the findings of this study are available on request from the corresponding author.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Cheng Z.J., Shan J. Novel coronavirus: where we are and what we know. Infection. 2019;48(2):155–163. doi: 10.1007/s15010-020-01401-y. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. China novel coronavirus investigating and research team. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang C.L., Wang Y.M., Li X.W., Ren L.L., Zhao J.P., Hu Y., Zhang L., Fan G.H., Xu J.Y., Gu X.Y., Cheng Z.S., Yu T., Xia J., Wei Y., Wu W.J., Xie X.L., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J.G., Wang G.F., Jiang R.M., Gao Z.C., Jin Q., Wang J.W., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese center for disease control and prevention. J. Am. Med. Assoc. 2020;323(13):1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 5.Zhang W., Du R.H., Li B., Zheng X.S., Yang X.L., Hu B., Wang Y.Y., Xiao G.F., Yan B., Shi Z.L., Zhou P. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg. Microb. Infect. 2020;9(1):386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization Coronavirus disease 2019 (COVID-19) situation reports. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports Available at: [PubMed]
- 7.Li G.D., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug Discov. 2020;19(3):149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 8.Gao J.J., Tian Z.X., Yang X. Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci Trends. 2020;14(1):72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 9.Luo H., Tang Q.L., Shang Y.X., Liang S.B., Yang M., Robinson N., Liu J.P. Can Chinese medicine Be used for prevention of corona virus disease 2019 (COVID-19)? A review of historical classics, research evidence and current prevention programs. Chin. J. Integr. Med. 2020;26(4):243–250. doi: 10.1007/s11655-020-3192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klasse P.J., Moore J.P. Antibodies to SARS-CoV-2 and their potential for therapeutic passive immunization. Elife. 2020;9 doi: 10.7554/eLife.57877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gavor E., Choong Y.K., Er S.Y., Sivaraman H., Sivaraman J. Structural basis of SARS-CoV-2 and SARS-CoV antibody interactions. Trends Immunol. 2020;41(11):1006–1022. doi: 10.1016/j.it.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li W.H., Zhang C.S., Sui J.H., Kuhn J.H., Moore M.J., Luo S., Wong S.K., Huang I.C., Xu K.M., Vasilieva N., Murakami A., He Y.Q., Marasco W.A., Guan Y., Choe H., Farzan M. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005;24(8):1634–1643. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lan J., Ge J.W., Yu J.F., Shan S.S., Zhou H., Fan S.L., Zhang Q., Shi X.L., Wang Q.S., Zhang L.Q., Wang X.Q. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 17.Du L.Y., He Y.X., Zhou Y.S., Liu S.W., Zheng B.J., Jiang S.B. The spike protein of SARS-CoV--a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7(3):226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu J.Y., Jia W.X., Wang P.F., Zhang S.Y., Shi X.L., Wang X.Q., Zhang L.Q. Antibodies and vaccines against Middle East respiratory syndrome coronavirus. Emerg. Microb. Infect. 2019;8(1):841–856. doi: 10.1080/22221751.2019.1624482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi B., Choudhary M.C., Regan J., Sparks J.A., Padera R.F., Qiu X., Solomon I.H., Kuo H.H., Boucau J., Bowman K., Adhikari U.D., Winkler M.L., Mueller A.A., Hsu T.Y., Desjardins M., Baden L.R., Chan B.T., Walker B.D., Lichterfeld M., Brigl M., Kwon D.S., Kanjilal S., Richardson E.T., Jonsson A.H., Alter G., Barczak A.K., Hanage W.P., Yu X.G., Gaiha G.D., Seaman M.S., Cernadas M., Li J.Z. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N. Engl. J. Med. 2020;383(23):2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., King N.P., Veesler D., Bloom J.D. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182(5):1295–1310. doi: 10.1016/j.cell.2020.08.012. e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N., et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv. 2020 doi: 10.1101/2020.12.21.20248640. [DOI] [Google Scholar]
- 22.Gu H.J., Chen Q., Yang G., He L., Fan H., Deng Y.Q., Wang Y.X., Teng Y., Zhao Z.P., Cui Y.J., Li Y.C., Li X.F., Li J.F., Zhang N.N., Yang X.L., Chen S.L., Guo Y., Zhao G.Y., Wang X.L., Luo D.Y., Wang H., Yang X., Li Y., Han G.C., He Y.X., Zhou X.J., Geng S.S., Sheng X.L., Jiang S.B., Sun S.H., Qin C.F., Zhou Y.S. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369(6511):1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ju B., Zhang Q., Ge J.W., Wang R.K., Sun J., Ge X.Y., Yu J.Z., Shan S.S., Zhou B., Song S., Tang X., Yu J.F., Lan J., Yuan J., Wang H.Y., Zhao J.J., Zhang S.Y., Wang Y.C., Shi X.L., Liu L., Zhao J.C., Wang X.Q., Zhang Z., Zhang L.Q. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584(7819):115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 24.Cao Y.L., Su B., Guo X.H., Sun W.J., Deng Y.Q., Bao L.L., Zhu Q.Y., Zhang X., Zheng Y.H., Geng C.Y., Chai X.R., He R.S., Li X.F., Lv Q., Zhu H., Deng W., Xu Y.F., Wang Y.J., Qiao L.X., Tan Y.F., Song Y.L., Wang G.P., Du X.X., Gao N., Liu J.N., Xiao J.Y., Su X.D., Du Z.M., Feng Y.M., Qin C., Qin C.F., Jin R.H., Xie X.S. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients' B cells. Cell. 2020;182(1):73–84. doi: 10.1016/j.cell.2020.05.025. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Y., Wang F.R., Shen C.G., Peng W.Y., Li D.L., Zhao C., Li Z.H., Li S.H., Bi Y.H., Yang Y., Gong Y.H., Xiao H.X., Fan Z., Tan S.G., Wu G.Z., Tan W.J., Lu X.C., Fan C.F., Wang Q.H., Liu Y.X., Zhang C., Qi J.X., Gao G.F., Gao F., Liu L. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368(6496):1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu X.L., Zhou T.Q., Zhu J., Zhang B.S., Georgiev I., Wang C., Chen X.J., Longo N.S., Louder M., McKee K., O'Dell S., Perfetto S., Schmidt S.D., Shi W., Wu L., Yang Y.P., Yang Z.Y., Yang Z.J., Zhang Z.H., Bonsignori M., Crump J.A., Kapiga S.H., Sam N.E., Haynes B.F., Simek M., Burton D.R., Koff W.C., Doria-Rose N.A., Connors M., NISC Comparative Sequencing Program. Mullikin J.C., Nabel G.J., Roederer M., Shapiro L., Kwong P.D., Mascola J.R. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333(6049):1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao H.X., Lynch R., Zhou T.Q., Gao F., Alam S.M., Boyd S.D., Fire A.Z., Roskin K.M., Schramm C.A., Zhang Z., Zhu J., Shapiro L., NISC Comparative Sequencing Program. Mullikin J.C., Gnanakaran S., Hraber P., Wiehe K., Kelsoe G., Yang G., Xia S.M., Montefiori D.C., Parks R., Lloyd K.E., Scearce R.M., Soderberg K.A., Cohen M., Kamanga G., Louder M.K., Tran L.M., Chen Y., Cai F.P., Chen S.R., Moquin S., Du X.L., Joyce M.G., Srivatsan S., Zhang B.S., Zheng A.Q., Shaw G.M., Hahn B.H., Kepler T.B., Korber B.T., Kwong P.D., Mascola J.R., Haynes B.F. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496(7446):469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spassov V.Z., Yan L. pH-selective mutagenesis of protein-protein interfaces: in silico design of therapeutic antibodies with prolonged half-life. Proteins. 2013;81(4):704–714. doi: 10.1002/prot.24230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Case D.A., Darden T.A., Cheatham T.E., Simmerling C.L., Roberts B.P. University of California; San Francisco: 2012. Amber 12. [Google Scholar]
- 30.Li H., Robertson A.D., Jensen J.H. Very fast empirical prediction and rationalization of protein pKa values. Proteins. 2005;61(4):704–721. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- 31.Ponder J.W., Case D.A. Force fields for protein simulations. Adv. Protein Chem. 2003;66:27–85. doi: 10.1016/s0065-3233(03)66002-x. [DOI] [PubMed] [Google Scholar]
- 32.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–930. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 33.Ryckaert J.P., Ciccotti G., Berendsen H.J. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of N-alkanes. J. Comput. Phys. 1977;23:327–341. doi: 10.1016/0021-9991(77)90098-5. [DOI] [Google Scholar]
- 34.Darden T., York D., Pedersen L. Particle mesh Ewald: an N·log (N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- 35.Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. doi: 10.1063/1.470117. [DOI] [Google Scholar]
- 36.Izaguirre J.A., Catarello D.P., Wozniak J.M., Skeel R.D. Langevin stabilization of molecular dynamics. J. Chem. Phys. 2001;114:2090–2098. doi: 10.1063/1.1332996. [DOI] [Google Scholar]
- 37.Sun H.Y., Li Y.Y., Tian S., Xu L., Hou T.J. Assessing the performance of MM/PBSA and MM/GBSA methods. 4. Accuracies of MM/PBSA and MM/GBSA methodologies evaluated by various simulation protocols using PDBbind data set. Phys. Chem. Chem. Phys. 2014;16(31):16719–16729. doi: 10.1039/c4cp01388c. [DOI] [PubMed] [Google Scholar]
- 38.Wang E.C., Sun H.Y., Wang J.M., Wang Z., Liu H., Zhang J.Z.H., Hou T.J. End-point binding free energy calculation with MM/PBSA and MM/GBSA: strategies and applications in drug design. Chem. Rev. 2019;119(16):9478–9508. doi: 10.1021/acs.chemrev.9b00055. [DOI] [PubMed] [Google Scholar]
- 39.Wang E.C., Weng G.Q., Sun H.Y., Du H.Y., Zhu F., Chen F., Wang Z., Hou T.J. Assessing the performance of the MM/PBSA and MM/GBSA methods. 10. Impacts of enhanced sampling and variable dielectric model on protein-protein Interactions. Phys. Chem. Chem. Phys. 2019;21(35):18958–18969. doi: 10.1039/c9cp04096j. [DOI] [PubMed] [Google Scholar]
- 40.Lauer T.M., Agrawal N.J., Chennamsetty N., Egodage K., Helk B., Trout B.L. Developability index: a rapid in silico tool for the screening of antibody aggregation propensity. J Pharm Sci. 2012;101(1):102–115. doi: 10.1002/jps.22758. [DOI] [PubMed] [Google Scholar]
- 41.Bowerman C.J., Ryan D.M., Nissan D.A., Nilsson B.L. The effect of increasing hydrophobicity on the self-assembly of amphipathic beta-sheet peptides. Mol. Biosyst. 2009;5(9):1058–1069. doi: 10.1039/b904439f. [DOI] [PubMed] [Google Scholar]
- 42.Alam I., Radovanovic A., Incitti R., Kamau A.A., Alarawi M., Azhar E.I., Gojobori T. CovMT: an interactive SARS-CoV-2 mutation tracker, with a focus on critical variants. Lancet Infect. Dis. 2021;21(5):602. doi: 10.1016/S1473-3099(21)00078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Srinivasan S., Cui H., Gao Z., Liu M., Lu S., Mkandawire W., Narykov O., Sun M., Korkin D. Structural genomics of SARS-CoV-2 indicates evolutionary conserved functional regions of viral proteins. Viruses. 2020;12(4):360. doi: 10.3390/v12040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., Peter A., Guarino B., Spreafico R., Cameroni E., Case J.B., Chen R.E., Havenar-Daughton C., Snell G., Telenti A., Virgin H.W., Lanzavecchia A., Diamond M.S., Fink K., Veesler D., Corti D. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583(7815):290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 45.Padoan A., Sciacovelli L., Basso D., Negrini D., Zuin S., Cosma C., Faggian D., Matricardi P., Plebani M. IgA-Ab response to spike glycoprotein of SARS-CoV-2 in patients with COVID-19: a longitudinal study. Clin. Chim. Acta. 2020;507:164–166. doi: 10.1016/j.cca.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., Marrama D., de Silva A.M., Frazier A., Carlin A.F., Greenbaum J.A., Peters B., Krammer F., Smith D.M., Crotty S., Sette A. Targets of T Cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(7):1489–1501. doi: 10.1016/j.cell.2020.05.015. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang C.Y., Li W.T., Drabek D., Okba N.M.A., van Haperen R., Osterhaus A.D.M.E., van Kuppeveld F.J.M., Haagmans B.L., Grosveld F., Bosch B.J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11(1):2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chi X.J., Liu X.Y., Wang C.H., Zhang X.H., Li X., Hou J.H., Ren L.L., Jin Q., Wang J.W., Yang W. Humanized single domain antibodies neutralize SARS-CoV-2 by targeting the spike receptor binding domain. Nat. Commun. 2020;11(1):4528. doi: 10.1038/s41467-020-18387-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lei C.H., Qian K.W., Li T., Zhang S., Fu W.Y., Ding M., Hu S. Neutralization of SARS-CoV-2 spike pseudotyped virus by recombinant ACE2-Ig. Nat. Commun. 2020;11(1):2070. doi: 10.1038/s41467-020-16048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rees-Spear C., Muir L., Griffith S.A., Heaney J., Aldon Y., Snitselaar J.L., Thomas P., Graham C., Seow J., Lee N., Rosa A., Roustan C., Houlihan C.F., Sanders R.W., Gupta R.K., Cherepanov P., Stauss H.J., Nastouli E. SAFER Investigators, Doores KJ, van Gils MJ, McCoy LE. The effect of spike mutations on SARS-CoV-2 neutralization. Cell Rep. 2021;34(12) doi: 10.1016/j.celrep.2021.108890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., Bentlage A.E.H., van Haaren M.M., Guerra D., Burger J.A., Schermer E.E., Verheul K.D., van der Velde N., van der Kooi A., van Schooten J., van Breemen M.J., Bijl T.P.L., Sliepen K., Aartse A., Derking R., Bontjer I., Kootstra N.A., Wiersinga W.J., Vidarsson G., Haagmans B.L., Ward A.B., de Bree G.J., Sanders R.W., van Gils M.J. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369(6504):643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 53.Han Y., Král P. Computational design of ACE2-based peptide inhibitors of SARS-CoV-2. ACS Nano. 2020;14(4):5143–5147. doi: 10.1021/acsnano.0c02857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oany A.R., Mia M., Pervin T., Junaid M., Hosen S.M.Z., Moni M.A. Design of novel viral attachment inhibitors of the spike glycoprotein (S) of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) through virtual screening and dynamics. Int. J. Antimicrob. Agents. 2020;56(6) doi: 10.1016/j.ijantimicag.2020.106177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baig A.M. Computing the effects of SARS-CoV-2 on respiration regulatory mechanisms in COVID-19. ACS Chem. Neurosci. 2020;11(16):2416–2421. doi: 10.1021/acschemneuro.0c00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pahari S., Li G., Murthy A.K., Liang S., Fragoza R., Yu H., Alexov E. SAAMBE-3D: predicting effect of mutations on protein-protein interactions. Int. J. Mol. Sci. 2020;21(7):2563. doi: 10.3390/ijms21072563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang N., Chen Y., Lu H., Zhao F., Alvarez R.V., Goncearenco A., Panchenko A.R., Li M. MutaBind2: predicting the impacts of single and multiple mutations on protein-protein interactions. iScience. 2020;23(3) doi: 10.1016/j.isci.2020.100939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.MyungY, Rodrigues C.H.M., Ascher D.B., Pires D.E.V. mCSM-AB2: guiding rational antibody design using graph-based signatures. Bioinformatics. 2020;36:1453–1459. doi: 10.1093/bioinformatics/btz779. [DOI] [PubMed] [Google Scholar]
- 59.Myung Y., Pires D.E.V., Ascher D.B. mmCSM-AB: guiding rational antibody engineering through multiple point mutations. Nucleic Acids Res. 2020;48(W1):W125–W131. doi: 10.1093/nar/gkaa389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Y.J., Liu M.Y., Gao J.L. Enhanced receptor binding of SARS-CoV-2 through networks of hydrogen-bonding and hydrophobic interactions. Proc. Natl. Acad. Sci. U. S. A. 2020;117(25):13967–13974. doi: 10.1073/pnas.2008209117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luan B.Q., Xu G.X., Feng M., Cong L., Zhou R.H. Combined computational-experimental approach to explore the molecular mechanism of SaCas9 with a broadened DNA targeting range. J. Am. Chem. Soc. 2019;141(16):6545–6552. doi: 10.1021/jacs.8b13144. [DOI] [PubMed] [Google Scholar]
- 62.Zhu L.M., Gao B., Yuan S.L., Zhu S.Y. Scorpion toxins: positive selection at a distal site modulates functional evolution at a bioactive site. Mol. Biol. Evol. 2019;36(2):365–375. doi: 10.1093/molbev/msy2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author.