Abstract
Objectives
There is an urgent need to ameliorate the possibilities of transporting reconstituted mRNA vaccines from the centralized preparation centres to the vaccination sites to improve the efficiency of the vaccination campaign against coronavirus disease 2019 (COVID-19). We have analysed the integrity of the Pfizer–BioNTech and Moderna vaccines under different movement conditions to provide information that may improve the distribution of vaccines to the target population.
Methods
Syringes of reconstituted Pfizer–BioNTech or Moderna COVID-19 vaccines were prepared in a laminar flow chamber to be subjected to a stability analysis in order to evaluate the impact of movement on mRNA integrity. RNA integrity was checked by the lack of RNA peaks under the original mRNA peak in the electropherogram resulting from potential fragments from RNA degradation. Samples were then exposed for 180 min at room temperature (21 ± 1°C, 55 ± 10% humidity) under different movement conditions.
Results
We report that the integrity of the mRNA in the reconstituted COVID-19 vaccines after continuous moderate movement at room temperature is maintained for at least 3 hours, with values of fluorescence units (FU) under the original mRNA peak of 0.38 ± 0.06 in the Pfizer–BioNTech vaccine and 0.96 ± 1.18 FU in the Moderna vaccine, equal to the values obtained without movement (0.36 ± 0.08 FU in the Pfizer–BioNTech and 1.12 ± 0.19 FU in the Moderna). In contrast, the integrity of these vaccines exposed to repeated Vortex shaking was significantly impaired (p < 0.001) with values under the original mRNA peak of 1.34 ± 0.31 FU for the Pfizer–BioNTech and 5.03 ± 1.16 FU for the Moderna samples.
Conclusions
The stability of these reconstituted vaccines reported here may improve the efficiency of the ground transportation and distribution of the vaccines, which may lead to shorter and more homogeneous vaccinations in cities and rural areas.
Keywords: COVID-19, SARS-CoV2, Transportation, Vaccination, Stability
Graphical abstract
Introduction
The Pfizer–BioNTech and Moderna vaccines are composed of mRNA, and the main difference between them lies in the lipid nanoparticles that protect the mRNA integrity and facilitate cell entry. Due to this difference, the Pfizer–BioNTech vaccine requires storage in freezers at between –90°C and –60°C, whereas the Moderna vaccine should be stored between –25°C and –15°C [1]. Pfizer–BioNTech vaccine reception in hospitals entails their reconstitution and re-storage at temperatures between 2°C and 8°C with an expiration of 5 days, while the expiration at room temperature is only 2 h. The Moderna vaccine can be stored refrigerated at 2–8°C for 30 days and at 8–25°C for up to 12 h before use. When the first dose is withdrawn from the vial, both vaccines should be held between 2°C and 25°C and discarded 6 h later.
It is recommended that reconstituted vials of the Pfizer–BioNTech and Moderna vaccines should not be transported to avoid unnecessary movement that could alter the mRNA integrity; this represents a major limitation for the rapid distribution of the vaccines [1,2]. However, no information is currently available about the consequences of movement to the integrity of the reconstituted vaccines. We have now analysed the integrity of the Pfizer–BioNTech and Moderna vaccines under different movement conditions, and we provide novel information crucial to improving the distribution of the vaccines to the target population.
Materials and methods
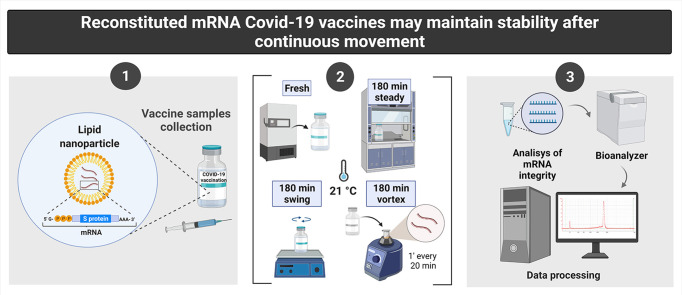
The Hospital del Mar (Barcelona, Spain) acted as a reference hospital and the Pharmacy department as vaccine distributor. Several vials and syringes of Pfizer–BioNTech and Moderna COVID-19 vaccines were returned to the pharmacy department by the vaccination teams after exceeding the expiration time or for potential microbiological contamination, such as vials falling to the ground. For this analysis we selected only the vials and syringes with loss of guarantee of lack of microbiological contamination. Syringes of reconstituted Pfizer–BioNTech or Moderna COVID-19 vaccines (0.5–2 mL) were prepared in a laminar flow chamber to be subjected to a stability analysis to evaluate the impact of movement conditions on mRNA integrity.
For this purpose, samples were exposed for 180 min at room temperature (21 ± 1°C, 55 ± 10% humidity) under different conditions. A first group (n = 12 from Pfizer–BioNTech and n = 17 from Moderna) remained without movement during 180 min to mimic the manufacturer's vaccine conditions for use after reconstitution. A second group (n = 10 from Pfizer–BioNTech and n = 16 from Moderna) was exposed to continuous movement during 180 min in a swing shaker (Ovan SW-3DE) at 40 rpm with an inclination angle of ±8° to mimic the movement that may occur in ground transportation under the most unfavourable road conditions. A third group (n = 8 from Pfizer–BioNTech and n = 14 from Moderna) was exposed to 1 min Vortex (Scientific Industries SI™ Vortex-Genie™ 2) at 3200 rpm every 20 min for 180 min, a massive shaking reported to impair RNA integrity [3]. A small amount (2 μL) of the fresh samples (n = 12 from Pfizer–BioNTech and n = 23 from Moderna) was extracted before this period under sterile conditions and immediately analysed to compare the integrity of the samples before and after exposure to these conditions.
Microfluidic measurements to analyse mRNA integrity were performed using Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, USA) with the RNA 6000 Nano LabChip kit and the assay Eukaryote total RNA Nano (Genomics Core Facility, University Pompeu Fabra). Results were generated and analysed with the Bioanalyzer 2100 Expert Software (Version B.0210.SI764) and manual integration combined with smear analysis was used to define regions following the Bioanalyzer user guide. This technique has been validated for the evaluation of RNA integrity with high accuracy and precision [4], and it is widely used to determine RNA integrity [5].
One-way ANOVA (group as between-subjects factor) was used, followed by multiple-group comparisons (Newman–Keuls) when the main variable was significant (p < 0.05) using the Statistical Package for Social Science program SPSS 20.0. Results are expressed as means ± SEMs. The study achieved a power of 99% with our sample size of 8–23 per group.
Results
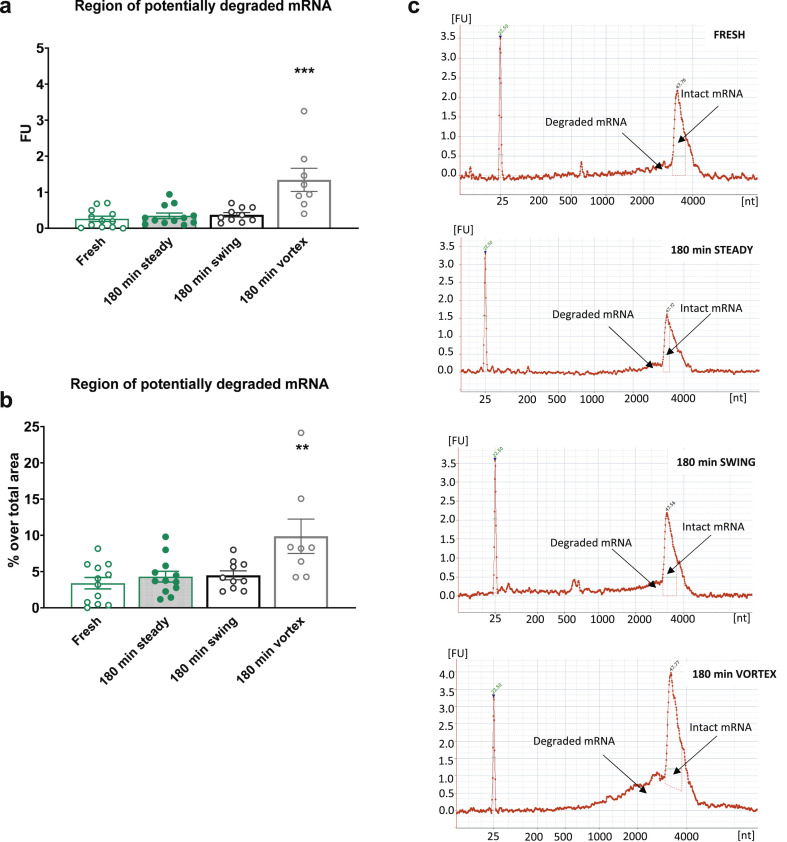
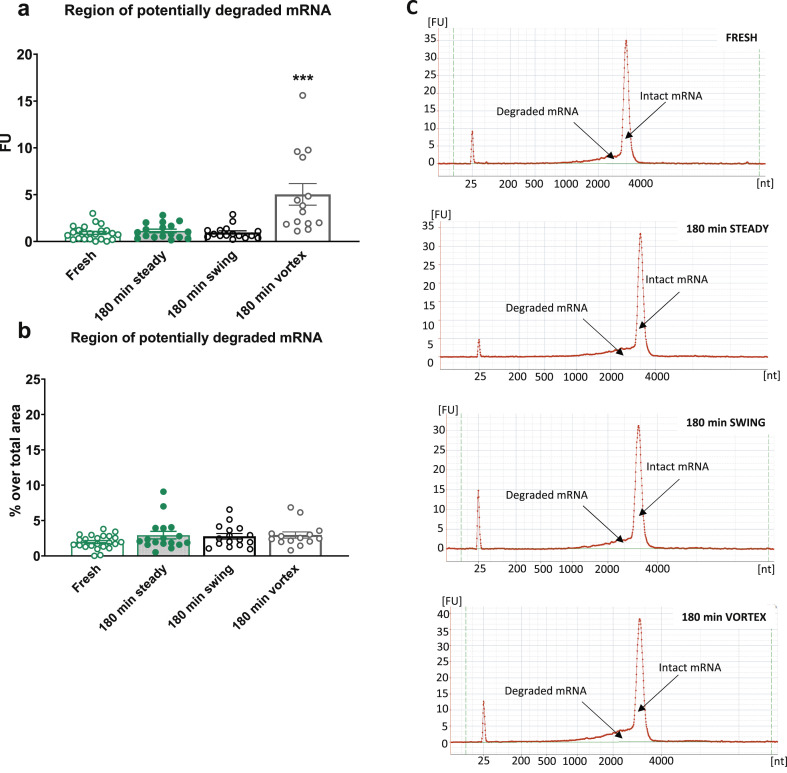
Our results revealed a negligible mRNA degradation (non-significant) in the Pfizer–BioNTech and Moderna samples exposed to room temperature for 180 min with the swing shaker (Pfizer–BioNTech 0.38 ± 0.06 fluorescence units (FU), Moderna 0.96 ± 1.18 FU) or without (Pfizer–BioNTech 0.36 ± 0.08 FU, Moderna 1.12 ± 0.19 FU) when compared to the fresh samples (Pfizer–BioNTech 0.26 ± 0.07 FU, Moderna 0.91 ± 0.16 FU). Indeed, the RNA fractions areas under the original mRNA peak were similar in these three experimental groups with very low FU values (Fig. 1, Fig. 2 a) corresponding to <5% of the original mRNA degradation for both Pfizer–BioNTech and Moderna samples (Fig. 1, Fig. 2b). Any possible product of the original mRNA degradation should be identified in these lower-molecular-weight fractions [4].
Fig. 1.
Analysis of the integrity of mRNA in reconstituted Pfizer–BioNTech coronavirus disease 2019 (COVID-19) vaccines. (a) Measurement of the region of potentially degraded mRNA in fluorescence units (FU) of the samples analysed immediately (fresh) or after 180 min without movement (180 min steady), with continuous swing shaker movement (180 min swing), or with Vortex shaking (180 min Vortex). Individual values of FU with the means ± SEMs are represented; one-way ANOVA, F (3,38) = 13.39, p < 0.001, Newman–Keus (N–K) post hoc test ∗∗∗p < 0.001 Vortex versus the remaining groups (n = 8–12). (b) Percentage over the region's total area of potentially degraded mRNA of the samples analysed immediately or after 180 min without movement, with continuous swing shaker movement, or with Vortex shaking. Individual values of FU percentages with the means ± SEMs are represented; one-way ANOVA, F (3,38) = 5.772, p < 0.01, N–K post hoc test ∗∗p < 0.01 Vortex versus the remaining groups (n = 8–12). (c) Representative electropherograms expressed in FU of RNA integrity for different mRNA samples, detailing the regions that indicate potentially degraded and intact mRNA peaks in the samples analysed immediately, or after 180 min without movement, with continuous swing shaker movement, or with Vortex shaking.
Fig. 2.
Analysis of the integrity of mRNA in reconstituted Moderna coronavirus disease 2019 (COVID-19) vaccines. (a) Measurement of the region of potentially degraded mRNA in fluorescence units (FU) of the samples analysed immediately (fresh) or after 180 min without movement (180 min steady), with continuous swing shaker movement (180 min swing), or with Vortex shaking (180 min Vortex). Individual values of FU with the means ± SEMs are represented; one-way ANOVA, F (3,66) = 14.77, p < 0.001, Newman–Keus (N–K) post hoc test ∗∗∗p < 0.001 Vortex versus the remaining groups (n = 14–23). (b) Percentage over the region's total area of potentially degraded mRNA of the samples analysed immediately or after 180 min without movement, with continuous swing shaker movement, or with Vortex shaking. Individual FU percentage values with the means ± SEMs are represented; one-way ANOVA, F (3,66) = 1.745, not significant (n = 14–23). (c) Representative electropherograms expressed in FU of RNA integrity for different mRNA samples detailing the regions that indicate potentially degraded and intact mRNA peaks in the samples analysed immediately or after 180 min without movement, with continuous swing shaker movement, or with Vortex shaking.
By contrast, the integrity of the mRNA in Pfizer–BioNTech and Moderna samples exposed to Vortex shaking was significantly impaired. Indeed, the RNA fractions area under the original mRNA peak was 1.34 ± 0.31 FU (p < 0.001) for the Pfizer–BioNTech samples (Fig. 1a): 9.87 ± 2.36% of original mRNA degradation (p < 0.01) (Fig. 1b). RNA fractions area under the original mRNA peak of the Moderna samples after Vortex was 5.03 ± 1.16 FU (p < 0.001) (Fig. 2a), corresponding to only 2.95 ± 0.45% of mRNA degradation (not significant) (Fig. 2b) due to the higher RNA fraction of the peak of non-degraded mRNA in the Moderna (147.26 ± 17.80 FU) than in the Pfizer–BioNTech (10.92 ± 1.00 FU) samples.
Discussion
A continued moderate movement at room temperature during 180 min of the reconstituted Pfizer–BioNTech and Moderna vaccines does not impair the quality of the mRNA since this quality was similar without movement and did not differ from the original quality of the fresh samples. As expected, the mRNA in these reconstituted vaccines may be degraded under conditions reported to impair mRNA integrity [3], although the degradation after this massive shaking was only moderate, mainly with the Moderna vaccine. Therefore, exposure of the Pfizer–BioNTech and Moderna COVID-19 reconstituted vaccines to continuous movement mimicking ground transportation during 180 min at room temperature does not impair mRNA quality. Such a period of 180 min could guarantee ground transportation over 180–300 km in road conditions of Europe and North America. This distance would ensure the distribution of the vaccine after reconstitution to the institutions that do not have the appropriate facilities to do it. The permanence of the mRNA's integrity under these experimental conditions suggests that the Pfizer–BioNTech and Moderna reconstituted vaccines, subsequently fractionated in syringes in pharmacy departments, could be more easily distributed than initially expected.
Some limitations of this study must be addressed. The total number of Moderna and Pfizer–BioNTech samples available was small, since few samples were returned to the pharmacy department due to potential loss of microbiological traceability. This scarcity leads to a discrepant number of samples for the various arms to prioritize the groups with a lower range of changes induced by the independent variable (fresh, 180 min steady, and swing) to optimize the detection of any possible modification due to this variable. Furthermore, no information is currently available about the possible direct consequences of RNA stability variation on alteration of vaccine efficacy. However, prevention of the degradation of the active component of the medication, the mRNA in these vaccines, represents a mandatory requirement.
In conclusion, the stability of Pfizer–BioNTech and Moderna reconstituted vaccines after continuous movement at room temperature may improve the efficiency in the administration of the vaccines, which may lead to shorter and more homogeneous vaccination in cities and rural areas. The possibility of preserving the mRNA after medium-distance ground transportation of reconstituted and fractionated syringes of both vaccines may optimize the distribution.
Author contributions
All the authors contributed to the acquisition, analysis, and interpretation of data and to the critical revision of the manuscript. RM and SG are responsible for conceptualizing the research idea and design, making the final interpretation of the statistical analyses, and writing the first draft and revision of the manuscript. EMG is responsible for the selection, realization and interpretation of statistical tests/analyses, and participated in writing the manuscript. OF participated in conceptualizing the research idea, making the primary interpretation of the statistical analyses, and in writing the manuscript.
Transparency declaration
RM received in the past 36 months grants or contracts from Aelis, Angelini, Boehringer Ingelheim, Camurus, Esteve, GlaxoSmithKline, Grünenthal, Lundbeck, Pharmaleads, Sanofi, Spherium and Uriach. SG received in past 36 months consulting fees from Pfizer and has been Advisor for the Spanish Medicine Agency. Funding: #ICREA Acadèmia2015 to RM.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Acknowledgements
We are grateful to the Genomics Core Facility of University Pompeu Fabra for assistance with the mRNA integrity analysis. We thank R. Martín and J. Jancyte for their technical support. Figures with drawings are created with BioRender.com.
Editor: L. Kaiser
References
- 1.COVID-19 Vaccines . 2020. FDA US food drug.https://www.fda.gov/emergency-preparedness-and-response/coronavirus-disease-2019-covid-19/covid-19-vaccines [Google Scholar]
- 2.Ministry of Health . 2020. COVID-19 vaccine storage handl guid – pfizer handl guid – pfizer–BioNTech mod COVID-19 vaccines. [Google Scholar]
- 3.Phua K.K.L., Nair S.K., Leong K.W. Messenger RNA (mRNA) nanoparticle tumour vaccination. Nanoscale. 2014;6:7715–7729. doi: 10.1039/c4nr01346h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sodowich B.I., Fadl I., Burns C. Method validation of in vitro RNA transcript analysis on the Agilent 2100 Bioanalyzer. Electrophoresis. 2007;28:2368–2378. doi: 10.1002/elps.200600673. [DOI] [PubMed] [Google Scholar]
- 5.Technical overviews: bioanalyzer RNA analysis. Agil Technol; 2020. https://www.agilent.com/en/product/automated-electrophoresis/bioanalyzer-systems/bioanalyzer-rna-kits-reagents/bioanalyzer-rna-analysis-228256#literature [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.