Abstract
Background
The first COVID-19 vaccines are being distributed to the general population. However, the shortage of doses is slowing down the goal of reaching herd immunity. The aim of the study was to verify whether previously SARS-CoV-2 infected subjects, a considerable portion of the population, should receive the same vaccination treatment of seronegative individuals.
Methods
Health-professionals either recovered from COVID-19 or never infected by SARS-CoV-2 were serologically tested at different time-points right before, and several days after, vaccination.
Results
Previously infected individuals showed humoral immune responses, 21 days after the first dose, that was approximately 10-folds higher than the seronegative group 21 days after the second dose. Seropositivity persists for at least 11 months.
Conclusion
During a shortage of COVID-19 vaccine doses, previously SARS-CoV-2 infected individuals should be dispensed from the vaccination campaign. When dose availability returns to normality, injection of a single dose for seropositive individuals should be considered.
Keywords: COVID-19, Serological test, mRNA vaccine, Roche Anti-SARS-CoV-2-S, Immune response, Vaccination
1. Introduction
The coronavirus disease (COVID-19) is a pandemic threatening the health and economy of the world’s population. Exceptional research efforts led to the rapid development of vaccines, which are now starting to be distributed to the general population. The BioNTech/Pfizer vaccine BNT162b2 Comirnaty was the first to be approved, in both Europe and US, showing a remarkable 95% efficacy [1], [2]. Unlike conventional vaccines, Comirnaty is a lipid nanoparticle-formulated, nucleoside-modified RNA (modRNA) encoding the SARS-CoV-2 full-length spike-protein (S-protein) [1], locked in the pre-fusion conformation [3]. Comirnaty represents the first large-scale mRNA vaccination campaign thus, data regarding the serological response in the general population are scarce [4], [5]. Furthermore, this vaccine was never thoroughly tested on individuals previously infected by SARS-CoV-2 and only preliminary information is available on its effect on this relatively large portion of the population [6], [7].
Most importantly, due to the shortage of vaccine doses, scientists are wondering for how long antibodies against SARS-CoV-2 persist in previously infected individuals and whether they should be administered with two, one or no vaccination doses.
The aim of this study was 1) to evaluate if, and for how long, previously infected subjects retain a degree of immunity (i.e., immunological memory), and 2) to compare immune response mounting upon vaccination in naturally seropositive and seronegative individuals.
2. Methods
2.1. Covidiagnostix study
From January 2021 till mid-February 2021, the IRCCS Ospedale San Raffaele (OSR), Milan, Italy underwent the COVID-19 vaccination campaign. Healthcare professionals were offered the Comirnaty vaccine. Among them, 3511 agreed to participate in a serological monitoring study (COVIDIAGNOSTIX) approved by the Institutional Ethical Review Board (CE:199/INT/2020).
A serological test right before the first vaccination dose (T0) was implemented to discriminate between naturally SARS-CoV-2 seropositive and seronegative individuals.
2.2. Inclusion criteria
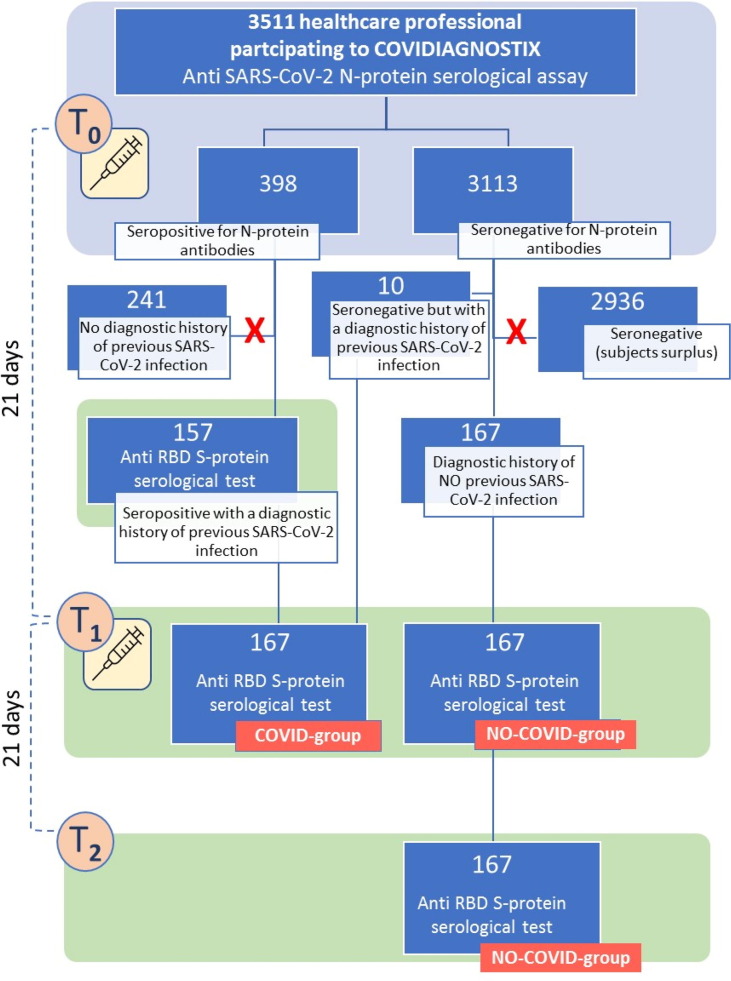
All seropositive individuals at T0, with also a diagnostic history of previous COVID-19 (n = 157) allowing the dating of the disease (i.e., positive RT-PCR swab test and/or positive serological during the COVID-19 period), as well as 10 seronegative individuals, yet having a diagnostic history of previous COVID-19, were included in the study (Scheme 1 ). This group was called the “COVID-group”. An equal number (n = 167) of randomly selected (alphabetical order) seronegative individuals with a diagnostic history of no previous COVID-19 (i.e., negative RT-PCR swab test and/or negative serological test during the COVID-19 period), matching the COVID-group for gender distribution, were also included in the study (Scheme 1). This group was called the “NO-COVID-group”.
Scheme 1.
Study design scheme. Light blue background is associated with serological test targeted anti-N-protein antibodies. Light green background is associated with serological test targeted anti-S-protein antibodies.
2.3. Study procedures
Blood samples, collected as described elsewhere [8], [9] into clot-activator BD vacutainer tubes (cat# 369032) without a separator (Becton, Dickinson and Company, NJ, US), were withdrawn at T0, right before receiving the first vaccination dose, and time-1 (T1), 21 days later, before the injection of the second dose. For the 167 seronegative subjects, a third blood sample was withdrawn at time-2 (T2), 21 days after the second vaccination dose (Scheme 1).
At T0, all serum samples were tested for the presence of SARS-CoV-2 nucleocapsid-protein (N-protein) specific antibodies using the Roche electrochemiluminescence immunoassay (ECLIA) Elecsys Anti-SARS-CoV-2 test (Ref# 09203095190), on a COBAS-601-platform (Roche Diagnostic, Basel, Switzerland), targeted on total immunoglobulins (IgTot) against the N-protein. The test was chosen because of its specificity near 100% (manufacturers’ suggested cutoff: 1.0 U/mL) as described in several studies [10], [11]. Thanks to an instrumental query, upon a positive result, same samples were further tested on the same platform with the Roche SARS-CoV-2-S test (Ref# 09289275190), targeted on IgTot against the receptor binding domain (RBD) of the viral S-protein.
At T1, serum samples of both groups were tested for the presence antibodies specific for the S-protein RBD using the Anti-SARS-CoV-2-S assay. The latter has a signal interval ranging from 0.4 to 250 U/ml. Above 250 U/mL the instrument automatically performs a 1:10 dilution that further extends the upper limit to 2500 U/mL. As reported in the manufacturer’s datasheet, the positivity cutoff is set at 0.8 U/mL.
The NO-COVID-group was further tested at T2 with the Anti-SARS-CoV-2-S test.
2.4. Covid-19 diagnostic data
From the beginning of the COVID-19 pandemic, thanks to a follow up institutional program, RT-PCR swab tests were performed routinely but also whenever a healthcare professional showed symptoms consistent with COVID-19. Samples were taken from back of the throat and analyzed using the Tib-Molbiol’s 2019-nCoV Real-Time Reverse-Transcription PCR Kit (cat# 61011896) on a Roche Cobas Z480 thermocycler (Roche Diagnostic, Basel, Switzerland). RNA purification was performed using the Roche Magna pure system (cat# A42352) [12]. Additionally, during May 2020, OSR health professionals were subjected to a serological evaluation using the LIAISON SARS-CoV-2 S1/S2 IgG chemiluminescence immunoassay (CLIA) (Ref# 311450) targeted on IgG specific for the viral S-protein.
2.5. Statistical analysis
Statistical analyses were performed with the software Excel (Microsoft, Redmond, WA, USA). Median and, when possible, average ± standard deviation (SD) were quoted. Comparisons of the quantitative antibody titers were performed by a two-tailed, unequal variances t-test (Welch test). Differences were considered statistically significant if the p-value was lower than 0.05.
3. Results
3.1. Covid-group
Of the 167 health professionals (40.7 ± 11.1 years) belonging to the COVID-group, 96 were females (41.5 ± 11.0 years) and 71 were males (39.5 ± 11.2 years). At T0, 157 were seropositive for the N-protein (supplementary Table 1S) and were further tested for the presence of antibodies against the S-protein RBD (Fig. 1 , panel A). They showed a median value at the Anti-SARS-CoV-2-S test of 110 U/mL, with one subject above the upper 2500 U/mL instrumental limit and one subject below the 0.4 U/mL lower instrumental limit (Fig. 1, panel A). By excluding these two extreme values the arithmetic mean, and its corresponding standard deviation, were 224.6 ± 345.0 U/mL. In addition to the subject showing a titer below the 0.4 U/mL instrumental limit, two subjects were below the 0.8 U/mL instrumental cutoff limit (0.6 and 0.7 U/mL respectively), resulting “negative” for the presence of antibodies specific for the S-protein RBD. The 10 subjects (7 females 38.4 ± 12.6 years and 3 males 36.3 ± 7.2 years) negative at the T0 N-protein serological screening and assigned to the COVID-group because of their proved diagnostic history of COVID-19, were missing the T0 S-protein RBD titer, which was measured, according to an instrumental query, only for the N-protein positive subjects. Seven of them had a positive RT-PCR test in March/April 2020 (9–10 months before vaccination) and experienced COVID-19 with very light symptoms (2–3 days of fever, ~37.5C°). The remaining three were completely asymptomatic and discovered to have recovered from the disease through the institutional serological test in May 2020. Thus, they experienced COVID-19 more than 8 months before vaccination.
Fig. 1.
Serological results of the subjects involved in the study. COVID-group: Panel A: anti-S-protein RBD assay results at T0 (n = 157 black dots) and T1 (n = 167, red dots). Panel B: 19 samples, exceeding 2500 U/mL (upper instrumental limit) at T1, were diluted with pre-pandemic serum to bring the signal within the instrumental range. Dilution factor back-calculated values are shown. NO-COVID-group: Panel C: anti-S-protein RBD assay results at T1 (n = 167, red dots) and T2 (n = 167, blue dots). Panel D: 13 samples, exceeding 2500 U/mL (upper instrumental limit) at T2, were diluted with pre-pandemic serum to bring the signal within the instrumental range. Dilution factor back-calculated values are shown. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
At T1, 165 subjects (98.8%) showed an antibody titer above the 2500 U/mL upper instrumental limit (Fig. 1, panel A). The remaining 2 subjects showed antibody titers increased with respect to T0 (<0.4, 40 and 547, 755 U/mL for T0 and T1, respectively). It must be noted that one of them was the only subject showing an Anti-S-protein RBD result at T0 below the instrumental limit (<0.4 U/mL). The 10 subjects that, at T0, showed no presence of anti N-protein antibodies but were included in the COVID-group because of their COVID-19 diagnostic history, as well as the two subjects “negative”, at T0, for the presence of S-protein RBD specific antibodies, all showed titers at T1 above the instrumental limit. To better inquire into the 98.8% of values above the 2500 U/mL instrumental limit, we diluted (1:50) 19 randomly chosen samples (accounting for >10% of the total samples above the limit) with a single pool of human pre-pandemic serum to bring the instrumental response within the instrumental range (Fig. 1, panel B). After adjusting for the dilution factor, the 19 samples showed a median value of 22,650 U/mL (the arithmetic mean, and its corresponding standard deviation were 30589 ± 26893 U/mL), consistent to what previously observed in a study, including 51 participants, using the same Roche instrumentation [13] (Table 2S). Thus, the median anti-S titer have increased 205-fold with respect to pre-vaccine levels. Two of the diluted samples were from subjects negative at the T0 N-protein serological screening and showed titers of 37,820 and 12,280 U/mL.
3.1.1. COVID-group: Time-interval between disease and first vaccination dose
The documented diagnostic history of COVID-19, available at the OSR database, showed that the longer time-intervals between a positive swab test and the first vaccination dose were 11 months (1 subject), 10 months (11 subjects) and 9 months (6 subjects). Ninety-one subjects showed a positive serological test in May 2020, thus they had experienced COVID-19 at least 8 months before T0. The remaining 58 health professionals had time-intervals ranging from 1 to 7 months. Among the 109 subjects (65.2%) showing the longer time-intervals (8 to 11 months), all of them (100%) showed antibody titers at T1 > 2500 U/mL.
3.2. No-Covid-group
The NO-COVID-group was formed by randomly selecting 167 subjects, matching the COVID-group for gender distribution (96 females aged 42.8 ± 9.3 years, and 71 males aged 42.6 ± 11.1 years), showing both a negative result at the T0 N-protein serological assay (supplementary Table 1S) and a negative diagnostic history of COVID-19. For the latter reasons, anti-S-protein RBD antibody were also assumed to absent in these subjects. At T1 no subjects showed an anti-S-protein RBD titer above the 2500 U/mL and only one showed a titer still below the 0.8 U/mL cutoff limit (Fig. 1, panel C). The median value was 44.1 U/mL (arithmetic mean, and corresponding standard deviation, equal to 112.8 ± 269.9 U/mL). At T2, 21 days after the second dose, all of the 167 subjects, belonging to the NO-COVID-group, showed antibody titers above the 0.8 U/mL cutoff limit and 58 of them (34.7%) were above the 2500 U/mL upper instrumental limit (Fig. 1, panel C). Their median value was 1806 U/mL. As for the COVID-group, we diluted a number (n = 13) of randomly chosen samples (>2500 U/mL), with human pre-pandemic serum. In contrast to the COVID-group, as for the COVID-group, a 1:50 dilution was applied to bring the instrumental response within the instrumental range (Fig. 1, panel D). The 13 samples showed a median value of 3290 U/mL (arithmetic mean, and corresponding standard deviation, equal to 4328 ± 2920 U/mL), (Table 2S), consistent to what previously observed by Manisty et al. [13].
3.3. Covid-group vs No-Covid-group
At T1, 98.8% of the individuals included in the COVID-group were above the 2500 U/mL upper instrumental limit whereas none of the NO-COVID was. At T2 only 34.7% of the latter group was above that limit. By comparing the instrumental signal outcomes, the T1 diluted samples of the COVID-group showed an averaged value (31240 ± 28089 U/mL) statistically significantly (p-value < 0.0001) higher (>7-folds) than that of the NO-COVID-group at T2 (4328 ± 2920 U/mL). It must be noted that, in contrast with the COVID-group, only 34.7% of the NO-COVID-group were above the 2500 U/mL upper instrumental limit at T2, thus the 4328 ± 2920 U/mL (obtained from diluted samples) largely overestimated the average of the entire group.
4. Discussion
The comparison between the humoral immune responses upon Comirnaty vaccination in naturally seropositive and seronegative individuals showed different behaviors. Those with a past SARS-CoV-2 infection showed antibody titers, 21 days after the first vaccination dose, exceeding the upper limit of detection in almost 99% of the cases. This was consistent with previous studies [7], [13], one of them using the same Roche instrumentation [13], showing that in subjects with a previous SARS-CoV-2 infection, the anti-S titers (three weeks after vaccination) increase approximately 200-fold with respect to pre-vaccine levels. In contrast, none of the seronegative individuals was above the upper instrumental limit 21 days after the first vaccination dose, whereas, 21 days after the second dose, 34.7% of them was above the 2500 U/mL upper limit. Similar results were obtained by Manisty et al. [13]. Further investigation of the samples showed that seropositive subjects, 21 days after the first vaccination dose, have antibody titers approximately one order of magnitude higher than their seronegative counterpart 21 days after the second dose.
Interestingly, the 10 subjects with no presence of anti N-protein antibodies, yet included in the COVID-group, and two of the three subjects showing very low anti-S RBD antibody titers at T0 (below the 0.8 U/mL instrumental cutoff limit), also exhibited titers at T1 above the 2500 U/mL instrumental limit. The 10 subjects with a documented COVID-19 diagnostic history, yet negative at the anti N-protein test, might be considered as false negative tests. However, the Elecsys Anti-SARS-CoV-2 test has a very high specificity and the lack of N-protein antibodies could also be related to a physiological decay. Unfortunately, the anti-S titers, which could have been useful to discriminate between the two above hypotheses, were not available for these subjects. We might speculate that, the fact that all of them experienced the disease at least 8 months before vaccination and had either asymptomatic or very light disease course, could be consistent with a physiological antibody decay as shown in previous studies [14]. Yet, dilution of two of these samples showed that their titers were comparable with those displaying a clear anti-N seropositivity at T0. Thus, it seems that even very low antibody levels are sufficient to induce the observed strong humoral immune response, upon the first vaccination dose, in individuals previously infected by SARS-CoV-2.
Furthermore, available diagnostic data showed that all of the subjects with the longer disease-to-vaccination time-intervals (8–11 months), representing 65.2% of the COVID-group, showed antibody titers at T1 above the 2500 U/mL upper instrumental limit. Thus, immunological memory (the ability of the immune system to specifically recognize an antigen that has been previously encountered and initiate a corresponding immune response), seems to last for at least 11 months, much longer than the six months previously hypothesized [15]. Noteworthy, the first Italian autochthonous case was on February 21st, 2020, meaning that 11 months was the largest disease-to-vaccination time-interval possibly available for this study.
In summary, our data showed that in previously SARS-CoV-2 infected individuals immunological memory lasts for at least 11 months and that the first dose of the Comirnaty vaccine acts in these individuals as a boost approximately 10-fold higher than it does the second vaccine dose in seronegative subjects. Based on this data, it is plausible to think that certain measures can be adopted to improve the COVID-19 vaccination campaign. For instance, in case of a critical scarcity of doses, vaccination of previously infected individuals can be safely postponed, whereas, in case of a full vaccine doses availability, we suggest that previously infected individuals should be injected with a single vaccine dose only.
Funding
This project was supported by Ministry of Health of Italy, “Bando Ricerca COVID-19”; project number: COVID-2020-12371619; project title: COVIDIAGNOSTIX - Health Technology Assessment in Covid serological diagnostics.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.vaccine.2021.06.020.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N Engl J Med. 2020;383(27):2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mulligan M.J., Lyke K.E., Kitchin N., Absalon J., Gurtman A., Lockhart S., et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020;586(7830):589–593. doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- 3.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines-a new era in vaccinology. Nat Rev Drug Discov. 2018;17(4):261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iavarone C, O’hagan DT, Yu D, Delahaye NF, Ulmer JB. Mechanism of action of mRNA-based vaccines. Expert Rev Vacc 2017;16(9):871–81. [DOI] [PubMed]
- 6.Bradley T, Grundberg E, Study C, Selvarangan R, Banerjee D, Beldon B, et al. Antibody responses boosted in seropositive healthcare workers after single dose of SARS-CoV-2 mRNA vaccine. medRxiv. 10.1101/2021.02.03.21251078. [DOI]
- 7.Krammer F, Srivastava K, Team P, Simon V, Alshammary H, Amoako A, et al. Antibody responses in seropositive persons after a single dose of SARS-CoV-2 mRNA vaccine. N Engl J Med. 2021;Mar 10. 10.1101/2021.01.29.21250653. [DOI] [PMC free article] [PubMed]
- 8.Ferrari D., Strollo M., Vidali M., Motta A., Pontillo M., Locatelli M. Biochemical, immunochemical and serology analytes validation of the lithium heparin BD Barricor blood collection tube on a highly automated Roche COBAS8000 instrument. Acta Biomed. 2020;91(1):47–55. doi: 10.23750/abm.v91i1.9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrari D., Manca M., Banfi G., Locatelli M. Alcohol and illicit drugs in drivers involved in road traffic crashes in the Milan area. A comparison with normal traffic reveals the possible inadequacy of current cut-off limits. Forensic Sci Int. 2018;282:127–132. doi: 10.1016/j.forsciint.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 10.Perkmann T., Perkmann-Nagele N., Breyer M.K., Breyer-Kohansal R., Burghuber O.C., Hartl S., et al. Side-by-Side Comparison of Three Fully Automated SARS-CoV-2 Antibody Assays with a Focus on Specificity. Clin Chem. 2020;11(1405):1413. doi: 10.1093/clinchem/hvaa198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kittel M., Muth M.C., Zahn I., Roth H.J., Thiaucourt M., Gerhards C., et al. Clinical evaluation of commercial automated SARS-CoV-2 immunoassays. Int J Infect Dis. 2021;103:590–596. doi: 10.1016/j.ijid.2020.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrari D., Motta A., Strollo M., Banfi G., Locatelli M. Routine blood tests as a potential diagnostic tool for COVID-19. Clin Chem Lab Med. 2020;58(7):1095–1099. doi: 10.1515/cclm-2020-0398. [DOI] [PubMed] [Google Scholar]
- 13.Manisty C, Otter AD, Treibel TA, McKnight I, Altmann DM, Brooks T, et al. Antibody response to first BNT162b2 dose in previously SARS-CoV-2-infected individuals. Lancet 2021:1057–8. 10.1016/S0140-6736. [DOI] [PMC free article] [PubMed]
- 14.Long Q.X., Tang X.J., Shi Q.L., Li Q., Deng H.J., Yuan J., et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med. 2020;26(8):1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 15.L’Huillier AG, Meyer B, Andrey DO, Arm-Vernez I, Baggio S, Didierlaurent A, et al. Antibody persistence in the first six months following SARS-CoV-2 infection among hospital workers: a prospective longitudinal study. Clin Microbiol Infect. 2021. 10.1016/j.cmi.2021.01.005. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.