Abstract
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a novel coronavirus causing coronavirus disease 2019 (COVID-19), has been expanding globally since late 2019. SARS-CoV-2, an RNA virus, has a genome sequence that can easily undergo mutation. Several mutated SARS-CoV-2 strains, including those with higher infectivity than others, have been reported. To reduce SARS-CoV-2 transmission, it is crucial to trace its infection sources. Here, we developed a simple, easy-to-use genotyping method to identify SARS-CoV-2 variants using a high-resolution melting (HRM) analysis.
Methods
We investigated five mutation sites, A23403G, G25563T, G26144T, T28144C, and G28882A, which are known strain determinants according to GISAID clades (L, S, V, G, GH, and GR).
Results
We first employed synthetic DNA fragments containing the five characteristic sites for HRM analysis. All sequences clearly differentiated wild-type from mutant viruses. We then confirmed that RNA fragments were suitable for HRM analysis following reverse transcription. Human saliva did not negatively affect the HRM analysis, which supports the absence of a matrix effect.
Conclusions
Our results indicate that this HRM-based genotyping method can identify SARS-CoV-2 variants. This novel assay platform potentially paves the way for accurate and rapid identification of SARS-CoV-2 infection sources.
Keywords: SARS-CoV-2, COVID-19, High resolution melting, Genotyping method, Real-time PCR, Virus mutation
1. Introduction
In December 2019, a novel coronavirus named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged from China and caused coronavirus disease 2019 (COVID-19) [[1], [2], [3]]. SARS-CoV-2 has been rapidly spreading worldwide, and the World Health Organization (WHO) declared COVID-19 a pandemic on March 11, 2020. One of the world's highest priority tasks is to prevent the breakneck spread of SARS-CoV-2 as early as possible.
SARS-CoV-2, a Betacoronavirus member, is an enveloped positive-sense, single-strand RNA virus with infectivity towards humans and mammals [4]. Although the SARS-CoV-2 evolutionary rate seemed to be slower than those of other RNA viruses, various SARS-CoV-2 variants have been reported worldwide [[5], [6], [7]]. Six SARS-CoV-2 clades have been identified based on GISAID (https://www.gisaid.org), a global initiative for data sharing on influenza viruses and latterly SARS-CoV-2. Emergent SARS-CoV-2 mutants can exhibit higher transmissibility and virulence. Indeed, a mutant strain, D614G [8], carries a spike amino acid mutation and possesses higher infectivity towards human cells and in animal models [9]. Therefore, the rapid genotyping of SARS-CoV-2, especially to detect the D614G mutant, is now essential for tracing its infection source and reducing its further transmission.
Real-time reverse transcription (RT) PCR is a commonly used indirect method for diagnosing COVID-19 infections worldwide [10,11]. Alternative indirect diagnostic methods for COVID-19 have been developed (e.g., RT-LAMP tests [12,13] and point-of-care tests [14,15]). These tests, including real-time RT-PCR, detect the presence of SARS-CoV-2 in samples, but sequence data cannot be acquired from these tests. Contrastingly, next-generation sequencing (NGS) tests can confirm viral sequences and thereby enable researchers to characterize SARS-CoV-2 strains and trace their infection pathways [16,17]. However, NGS analysis has long data-acquisition times and needs computational power. Therefore, simple and inexpensive tests to rapidly and reliably trace SARS-CoV-2 infections are needed.
High-resolution melting (HRM) analysis has been used to identify single-nucleotide polymorphisms in DNA sequences [18,19] and in bacterial strains [20,21]. Because double-stranded DNA shows characteristic thermo-dissociation behavior (i.e., a melting profile) depending on its GC content and base distribution, HRM can detect subtle differences at single-base resolution between two DNA sequences. Previous studies confirm that HRM analysis can successfully identify human immunodeficiency virus (HIV) variants [22,23], which in common with SARS-CoV-2, are RNA viruses. These facts suggest that HRM analysis could potentially be a general and versatile method for determining SARS-CoV-2 transmission. In this study, we developed a novel HRM assay for genotyping five SARS-CoV-2 mutant types. This assay enabled us to identify six types of viral clades, L, S, V, G, GH, and GR (Table 1 ).
Table 1.
SARS-CoV-2 types based on GISAID data.
| Type | Nucleotide mutation | Amino acid mutation |
|---|---|---|
| L | (NCBI Reference Sequence: NC_045512.2) | |
| S | C8782T, T28144C | NS8-L84S |
| V | G11083T, G26144T | NSP6-L37F, NS3-G251V |
| G | C241T, C3037T, A23403G | S-D614G |
| GH | C241T, C3037T, A23403G, G25563T | S-D614G, NS3-Q57H |
| GR | C241T, C3037T, A23403G, G28882A | S-D614G, N-G204R |
NS/NSP, Nonstructural proteins; S, Spike protein; N, Nucleocapsid protein.
2. Materials and methods
2.1. Synthesis of DNA fragments
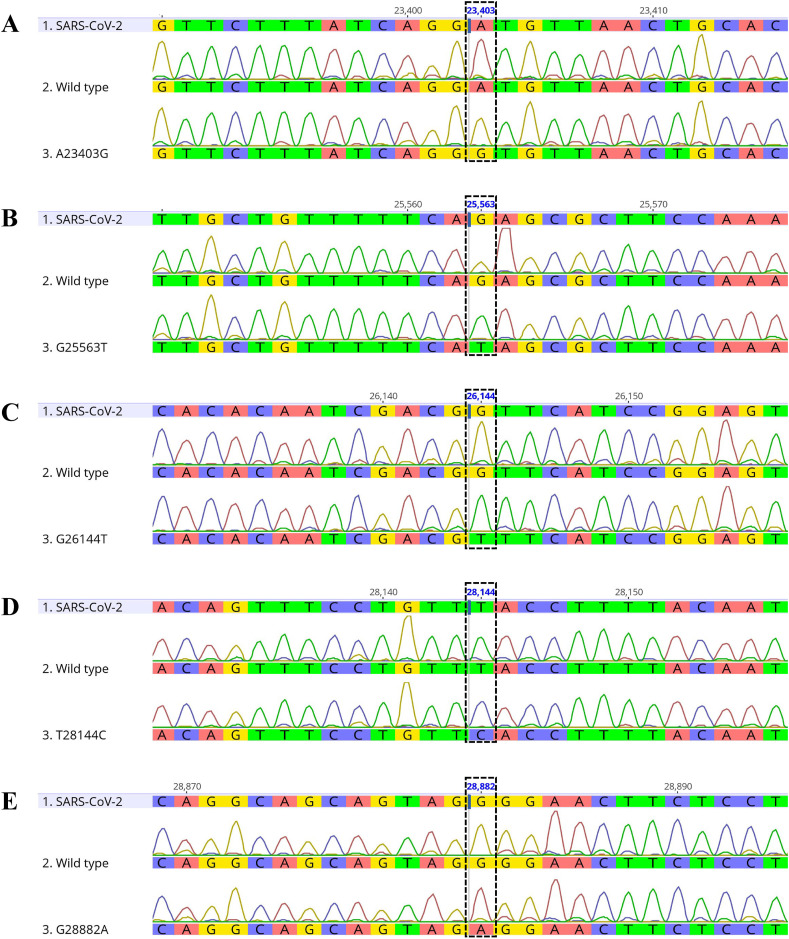
The SARS-CoV-2 sequence used herein was obtained from NCBI (NCBI Reference Sequence: NC_045512.2) and the GISAID database (www.gisaid.org). pMA vectors harboring a series of five wild-type DNA fragments (A23403, G25563, G26144, T28144, and G28882) and mutants (A23403G, G25563T, G26144T, T28144C, and G28882A) were obtained from Thermo Fisher Scientific (Waltham, MA, USA) (Fig. 1 ). Each fragment (400 or 500 bp in length) was used as a PCR amplification template.
Fig. 1.
DNA sequences of the target regions in the plasmid templates. Each target region was amplified using specific primers and the amplicon sequences were analyzed. The five mutational sites were A23403 (A), G25563 (B), G26144 (C), T28144 (D), and G28882 (E). Dashed rectangles indicate single point mutations (Table 1).
2.2. PCR amplification and sequence analysis
PCR was performed using a high-fidelity PCR enzyme (KOD FX neo; Toyobo Co., Ltd., Osaka, Japan) according to the manufacturer's instructions. The primer pairs used for amplifying SARS-CoV-2 genome sequences were M13 Forward (−20) 5′-GTAAAACGACGGCCAG-3′ and M13 Reverse 5′-CAGGAAACAGCTATGAC-3′ (Thermo Fisher Scientific). PCR amplifications were performed with an initial denaturation at 94 °C for 2 min followed by 35 cycles of denaturation at 98 °C for 10 s, annealing at 58 °C for 30 s, and extension at 68 °C for 30 s. PCR products were purified on spin columns (MinElute PCR Purification Kit; Qiagen GmbH, Hilden, Germany). After purification, the DNA fragments were sequenced (Eurofins Genomics KK, Tokyo, Japan), and the sequence data were analyzed using Geneious Prime (Biomatters Ltd., Auckland, New Zealand). The purified PCR products were used for HRM analysis and for in vitro T7 transcription.
2.3. HRM analysis
Normal human saliva, which was collected from a healthy donor, was purchased from Lee Biosolutions Inc. (Maryland Heights, MO, USA), and confirmed to be COVID-19 negative using a SARS-CoV-2 detection kit (SARS-CoV-2 Direct Detection RT-qPCR Kit; Takara Bio Inc., Shiga, Japan) according to the manufacturer's instructions. All reactions were performed in duplicate on a real-time PCR system (LightCycler 480 System; F. Hoffmann-La Roche Ltd., Basel, Switzerland).
Saliva was incubated at 95 °C for 5 min to inactivate endogenous deoxyribonuclease (DNase). Each DNA solution was added to a 4-fold volume of heat-inactivated saliva (or water). HRM analysis was performed using HRM reagents (LightCycler 480 High-Resolution Melting Master; F. Hoffmann-La Roche Ltd.) according to the manufacturer's instructions. The primer pairs used to PCR amplify A23403, G25563, G26144, T28144, and G28882 were as follows: A23403 forward 5′-CACCAGGAACAAATACTTCTAACC-3′ and A23403 reverse 5′- AACAGGGACTTCTGTGCAGTTAA-3′; G25563 forward 5′-GCGTTGCACTTCTTGCTGTTTT-3′ and G25563 reverse 5′-GCTAGTTGCCATCTCTTTTTGAGG-3′; G26144 forward 5′- GCCTGAAGAACATGTCCAAATT-3′ and G26144 reverse 5′-TGGATTAACAACTCCGGATGA-3′; T28144 forward 5′-CCCATTCAGTACATCGATATCGG-3′ and T28144 reverse 5′- CAATTTAGGTTCCTGGCAATTAA-3′; G28882 forward 5′-TCAACTCCAGGCAGCAGTAG-3′ and G28882 reverse 5′-GCCATTGCCAGCCATTCTA-3′. Each reaction mixture (20 μL) contained DNA solution (20 pg), 400 nmol/L of each primer, 1 × Master mix, and 1.5 mmol/L of MgCl2. All reactions were performed in duplicate on a real-time PCR system (LightCycler 96 System; F. Hoffmann-La Roche Ltd.). PCR amplification was performed with an initial denaturation at 95 °C for 5 min, followed by 40 cycles of denaturation at 95 °C for 10 s, annealing at 60 °C for 10 s, and extension at 72 °C for 20 s. After amplification, HRM was performed with denaturation at 95 °C for 60 s, cooling at 40 °C for 60 s, preheating at 65 °C for 1 s, and melting curve generation from 75 °C to 95 °C in 1 °C/s increments with 25 acquisitions. HRM curves were analyzed using Gene Scanning Software (F. Hoffmann-La Roche Ltd.) under default settings. Normalized melting curves, melting peaks (-dF/dT), and curve differences (Δ) were acquired by setting pre-melt and post-melt fluorescences as 100% and 0%, respectively. After HRM analysis, each PCR product (10 μL/lane) was separated by agarose gel electrophoresis, and the images were captured by the FAS-IV imaging system (Nippon Genetics Co, Ltd., Tokyo, Japan).
2.4. RNA fragment synthesis: in vitro T7 transcription
Because each synthetic DNA fragment possesses a 5′ T7 upstream promoter sequence, the SARS-CoV-2 RNA fragments were synthesized by in vitro T7 transcription (CUGA 7 in vitro Transcription Kit; Nippon Gene Co. Ltd.) according to the manufacturer's instructions. Synthetic RNA fragments were purified on spin columns (RNeasy Mini Kit, Qiagen). After purification, each eluent was treated with DNase I (RNase-Free DNase Set; Qiagen) and repurified on a new spin column. Synthetic single-stranded RNA fragments (250 ng/lane) were separated by non-denaturing agarose gel electrophoresis using RNA loading buffer and RNA ladder (DynaMarker RNA High for Electrophoresis; BioDynamics Laboratory Inc., Tokyo, Japan) according to the manufacturer's instructions. Images were captured by FAS-IV.
2.5. One-step RT-PCR HRM analysis
To remove the endogenous ribonuclease (RNase), human saliva was centrifuged using a centrifugal ultrafiltration (10 kDa) device (Ultrafree-MC; Sigma-Aldrich, St. Louis, MO, USA) (12,000×g, 30 min, room temperature). The flow-through fraction obtained was regarded RNase-free saliva.
RNA templates were treated with DNase (ezDNase; Thermo Fisher Scientific) according to the manufacturer's protocol. Briefly, the reaction mixture (5 μL) consisted of RNA solution (500 pg), 1 μL of RNase-free saliva (or water), 0.5 μL of ezDNase buffer, and 0.5 μL of DNase. The mixtures were reincubated at 37 °C for 5 min and 95 °C for 5 min. After DNase treatment, one-step RT-PCR HRM analysis was performed using a one-step reverse transcriptase kit (SuperScript IV One-Step RT-PCR System; Thermo Fisher Scientific) and LightCycler 480 High-Resolution Melting Master (F. Hoffmann-La Roche Ltd.). Each reaction mixture (20 μL) contained RNA solution (200 pg), 0.2 μL of SuperScript IV RT mix, 400 nmol/L of each primer, ten μL of master mix, and 1.5 mmol/L MgCl2. Reverse transcription was performed at 52 °C for 10 min, followed by 98 °C for 2 min. Real-time PCR and HRM were performed under the same conditions as used for the HRM analysis with DNA templates.
3. Results
3.1. HRM analysis on DNA templates
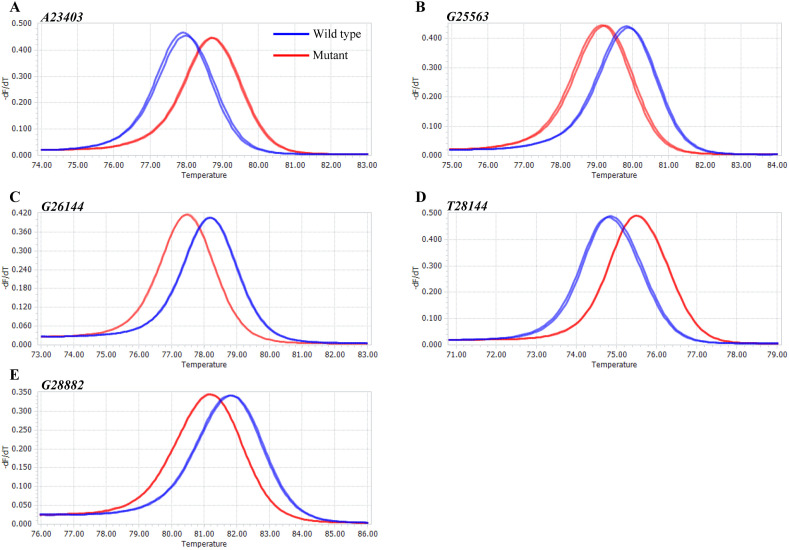
Before using the single-stranded RNA, we validated the HRM PCR conditions with double-stranded DNA. DNA templates were amplified from pre-prepared pMA vectors harboring five mutational sites (A23403G, G25563T, G26144T, T28144C, and G28882A; Table 1), before proceeding to HRM analysis. The HRM curve analysis showed that the characteristic melting peaks were distinguishable between wild-types and mutants at all five mutation sites (Fig. 2 ). Upon electrophoretic separation, a single correctly-sized DNA fragment was observed in each HRM reaction mixture (Fig. S1). These results indicate that our PCR conditions were suitable for HRM analysis.
Fig. 2.
DNA template melting peaks. -dF/dT plots for the five mutational sites were acquired using wild-type (blue) and mutant (red) DNA templates. (A) A23403, (B) G25563, (C) G26144, (D) T28144, and (E) G28882.
3.2. Effect of saliva on the HRM analysis
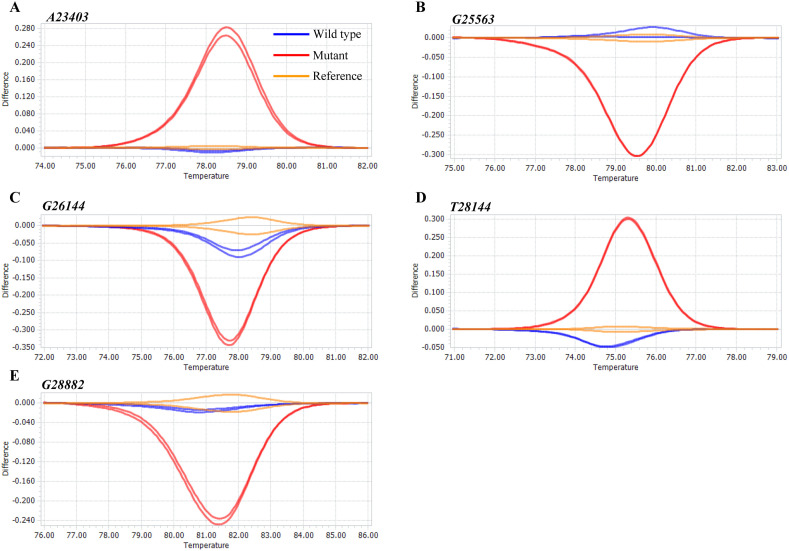
In clinical practice, saliva rather than nasopharyngeal swabs is used for SARS-CoV-2 infection testing [[24], [25], [26]]. Therefore, we determined the effect of saliva on the HRM analysis results using commercially available human saliva. A DNA fragment was spiked into this saliva at 1 × 107 copies/mL, and the spiked saliva was analyzed by HRM. DNA templates without saliva were used as references, and the normalized melting curves from the samples were subtracted by that of the respective reference to obtain difference plots for the five mutational sites (Fig. 3 ). The mutants’ plots were dissimilar to those of the references, whereas the wild-type plots were in agreement with those of the references. These results indicate that saliva in the HRM reaction mixture did not interfere with the amplification and melting processes.
Fig. 3.
Difference plots for DNA templates in the presence of human saliva. Each DNA fragment was added to human saliva at 107 copies/mL. DNA templates (wild-types) without saliva were used as the references. The difference plots indicate that wild-types (blue) and references (orange) were distinguishable from mutants (red). (A) A23403, (B) G25563, (C) G26144, (D) T28144, and (E) G28882.
3.3. One-step RT-PCR HRM analysis for RNA templates
We next confirmed the applicability of RNA templates for HRM analysis after reverse transcription. To prepare the RNA templates, in vitro T7 transcription was carried out using pMA vectors harboring five mutational sites in SARS-CoV-2. Upon electrophoretic separation, the transcripts were confirmed to be single, correctly-sized fragments (Fig. S2). Thus, our one-step RT-PCR HRM analysis involved reverse transcription and subsequent amplification of synthetic single-stranded RNA in a single tube.
In the one-step RT-PCR HRM analysis, single-stranded RNA was compared between the wild-types and mutants. Double-stranded DNA was used as the positive control reference for DNA amplification. Although there is no direct evidence that our reaction conditions resulted in reverse-transcribed single-stranded RNA production and PCR amplification of any resultant cDNA, the plots showing the differences between the wild-type sequences were in agreement with those of the references, implying that reverse transcription was successful during the one-step RT-PCR HRM (Fig. S3). Conversely, the mutants’ plots were distinctly different to those of the references. These results suggest that our one-step method identified wild-type and mutant SARS-CoV-2 viral RNAs, but the effect of saliva on the reverse transcription reaction was unclear. Therefore, we performed the one-step RT-PCR HRM analysis in the presence of RNase-free saliva (Fig. S4). For all five mutational sites, one-step RT-PCR HRM analysis distinguished the wild types from the mutants in the presence of a saliva matrix, indicating that saliva did not impact the assay.
4. Discussion
Since the WHO declared COVID-19 a pandemic on March 11, 2020, SARS-CoV-2 infection has rapidly spread worldwide. In Japan, COVID-19 patients are currently increasing daily, and healthcare is almost facing collapse. Recently, several COVID-19 vaccines have been developed and vaccination programs have just started. Although full deployment of such vaccines in Japan will take many months, epidemiological determinations and governmental mobility restrictions are still the standard approaches for suppressing viral transmission. In the current epidemiological situation, infection sources are identified based on contact tracing of COVID-19 cases and broad-range PCR testing of people. Conventional PCR testing targets consensus sequences within SARS-CoV-2 variants [10,27], but these tests do not detect variants possessing characteristics such as higher infectivity. It would be helpful, therefore, if PCR testing was able to detect SARS-CoV-2 variants, as this would allow healthcare professionals to trace not only close contacts but also know which SARS-CoV-2 variant is responsible, thereby providing more in-depth insight into infection sources and pathways. Our newly developed novel PCR-based HRM genotyping assay, which determines SARS-CoV-2 genome sequences, may be helpful in this respect.
NGS can identify novel strains based on the whole-genome sequence data it produces. This technique has characterized five mutants (S, V, G, GH, and GR; Table 1), among which G, GH, and GR mutants possess an identical mutation site (D614G) in the spike protein's coding region. These three mutated forms have higher infectivity and proliferative adaptability than the wild-type viruses [9], and are currently spreading worldwide. In mid to late December 2020, two more mutants emerged in the UK and South Africa (namely VOC-202012/01 and 501.V2, respectively) [28,29]. Based on the WHO's report, VOC-202012/01 seems to be 75% more transmissible than the previous circulating viruses, implying its future rapid transmission outside the UK [28]. Therefore, large-scale sequencing of SARS-CoV-2 viral genomes is needed for monitoring the infection status of VOC-202012/01, but whole-genome sequence analysis takes time and effort. In contrast, our HRM-based genotyping analysis identifies selected mutation sites, making it possible to detect SARS-CoV-2 variants rapidly and efficiently.
This analysis was also performed using serially diluted RNA templates corresponding to viral copy numbers from 104 to 1011 copies/mL in saliva. Among the five mutational sites, the successful copy number ranges were from 107 to 1011 copies/mL (for A23403, G26144, G28882) and from 105 to 1011 copies/mL (for G25563, T28144) (data not shown). Previous studies [30,31] have shown that saliva from SARS-CoV-2 patients contains about 104 to 108 virus copies per mL. Therefore, when used at > 107 copies per mL, the current HRM assay is applicable for the five mutational sites. Additional work is needed to improve the detection limit of HRM analysis. One option to improve it's the detection limit and specificity is to modify nested PCR by using two primer sets, which can improve real-time RT-PCR sensitivity towards SARS-CoV-2 [32,33].
We have not used HRM analysis on clinical samples yet, but it is reportedly a useful genotyping tool for HIV [22,23] which, like SARS-CoV-2, is also an RNA virus. Cousins et al. reported that, similarly to NGS, HRM analysis can capture HIV diversity [34]. Likewise, HRM analysis should be a useful tool for tracking the distribution of SARS-CoV-2 variants. Indeed, HRM analysis correctly identified the genomic RNA of GISAID clade S virus to be A23403, G25563, G26144, T28144C, and G28882 (Fig. S5).
Because HRM analysis is reflected in single-nucleotide polymorphisms, an unexpected mutation within the target regions would affect the melting temperature, by which each specific melting peak would be generated. It is necessary for both wild-type and mutants template as positive controls in order to identify SARS-CoV-2 variants using HRM analysis. Although sequence analyses are needed for samples with different nucleotide mutations, our HRM analysis can be applicable for high-throughput screening of the emerging unknown mutations.
In conclusion, we developed a novel method for genotyping SARS-CoV-2 variants using a real-time PCR system. This assay is faster and less costly than NGS analysis. Our results suggest that the current technique based on HRM analysis is a powerful high-throughput tool for determining SARS-CoV-2 clusters, but the technique requires future confirmation using samples from SARS-CoV-2-infected patients.
Authorship statement
All authors meet the ICMJE authorship criteria. All authors participated in interpreting study results. AA conducted all experiments in the manuscript and drafted the manuscript. YM performed the sequence analysis. YO planned the study and drafted the manuscript. HJ planned the study and supervised the project. All authors critically reviewed and approved the final version of the manuscript.
Declaration of competing interest
None.
Acknowledgments
This work was supported in part by Meijo University Research Project for Countermeasures Against COVID-19. We thank Edanz Group for editing a draft of this manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jiac.2021.06.007.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Coronaviridae Study Group of the International Committee on Taxonomy of V The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu J., Cui J., Qian Z., Wang Y., Zhang H., Duan Y., et al. On the origin and continuing evolution of SARS-CoV-2. Natl Sci Rev. 2020;7:1012–1023. doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rahimi A., Mirzazadeh A., Tavakolpour S. Genetics and genomics of SARS-CoV-2: a review of the literature with the special focus on genetic diversity and SARS-CoV-2 genome detection. Genomics. 2020 doi: 10.1016/j.ygeno.2020.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forster P., Forster L., Renfrew C., Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc Natl Acad Sci USA. 2020;117:9241–9243. doi: 10.1073/pnas.2004999117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hou Y.J., Chiba S., Halfmann P., Ehre C., Kuroda M., Dinnon K.H., et al. SARS-CoV-2 D614G variant exhibits enhanced replication ex vivo and earlier transmission in vivo. bioRxiv. 2020 doi: 10.1101/2020.09.28.317685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shirato K., Nao N., Katano H., Takayama I., Saito S., Kato F., et al. Development of genetic diagnostic methods for detection for novel coronavirus 2019 (nCoV-2019) in Japan. Jpn J Infect Dis. 2020;73:304–307. doi: 10.7883/yoken.JJID.2020.061. [DOI] [PubMed] [Google Scholar]
- 12.Lamb L.E., Bartolone S.N., Ward E., Chancellor M.B. Rapid detection of novel coronavirus/severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by reverse transcription-loop-mediated isothermal amplification. PloS One. 2020;15 doi: 10.1371/journal.pone.0234682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Augustine R., Hasan A., Das S., Ahmed R., Mori Y., Notomi T., et al. Loop-mediated isothermal amplification (LAMP): a rapid, sensitive, specific, and cost-effective point-of-care test for coronaviruses in the context of COVID-19 pandemic. Biology. 2020;9 doi: 10.3390/biology9080182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Assennato S.M., Ritchie A.V., Nadala C., Goel N., Tie C., Nadala L.M., et al. Performance evaluation of the SAMBA II SARS-CoV-2 test for point-of-care detection of SARS-CoV-2. J Clin Microbiol. 2020;59 doi: 10.1128/JCM.01262-20. e01262-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smithgall M.C., Scherberkova I., Whittier S., Green D.A. Comparison of Cepheid Xpert Xpress and Abbott ID Now to Roche cobas for the Rapid Detection of SARS-CoV-2. J Clin Virol. 2020;128 doi: 10.1016/j.jcv.2020.104428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hourdel V., Kwasiborski A., Baliere C., Matheus S., Batejat C.F., Manuguerra J.C., et al. Rapid genomic characterization of SARS-CoV-2 by direct amplicon-based sequencing through comparison of MinION and Illumina iSeq100TM system. Front Microbiol. 2020;11:571328. doi: 10.3389/fmicb.2020.571328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/s0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vossen R.H., Aten E., Roos A., den Dunnen J.T. High-resolution melting analysis (HRMA): more than just sequence variant screening. Hum Mutat. 2009;30:860–866. doi: 10.1002/humu.21019. [DOI] [PubMed] [Google Scholar]
- 19.Zhou L., Palais R.A., Paxton C.N., Geiersbach K.B., Wittwer C.T. Copy number assessment by competitive PCR with limiting deoxynucleotide triphosphates and high-resolution melting. Clin Chem. 2015;61:724–733. doi: 10.1373/clinchem.2014.236208. [DOI] [PubMed] [Google Scholar]
- 20.Nagai Y., Iwade Y., Hayakawa E., Nakano M., Sakai T., Mitarai S., et al. High resolution melting curve assay for rapid detection of drug-resistant mycobacterium tuberculosis. J Infect Chemother. 2013;19:1116–1125. doi: 10.1007/s10156-013-0636-3. [DOI] [PubMed] [Google Scholar]
- 21.Tamburro M., Ripabelli G. High resolution melting as a rapid, reliable, accurate and cost-effective emerging tool for genotyping pathogenic bacteria and enhancing molecular epidemiological surveillance: a comprehensive review of the literature. Ann Ig. 2017;29:293–316. doi: 10.7416/ai.2017.2153. [DOI] [PubMed] [Google Scholar]
- 22.Sacks D., Ledwaba J., Morris L., Hunt G.M. Rapid detection of common HIV-1 drug resistance mutations by use of high-resolution melting analysis and unlabeled probes. J Clin Microbiol. 2017;55:122–133. doi: 10.1128/JCM.01291-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Towler W.I., James M.M., Ray S.C., Wang L., Donnell D., Mwatha A., et al. Analysis of HIV diversity using a high-resolution melting assay. AIDS Res Hum Retrovir. 2010;26:913–918. doi: 10.1089/aid.2009.0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakanashi D., Asai N., Nakamura A., Miyazaki N., Kawamoto Y., Ohno T., et al. Comparative evaluation of nasopharyngeal swab and saliva specimens for the molecular detection of SARS-CoV-2 RNA in Japanese patients with COVID-19. J Infect Chemother. 2021;27:126–129. doi: 10.1016/j.jiac.2020.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeuchi Y., Furuchi M., Kamimoto A., Honda K., Matsumura H., Kobayashi R. Saliva-based PCR tests for SARS-CoV-2 detection. J Oral Sci. 2020;62:350–351. doi: 10.2334/josnusd.20-0267. [DOI] [PubMed] [Google Scholar]
- 26.To K.K., Tsang O.T., Yip C.C., Chan K.H., Wu T.C., Chan J.M., et al. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis. 2020;71:841–843. doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inoue G., Uchida K., Fukushima K., Uchiyama K., Nakazawa T., Aikawa J., et al. Experience of an orthopaedic surgery department early during the COVID-19 outbreak in Japan including real-time polymerase chain reaction assay results for SARS-CoV-2. Cureus. 2020;12:e11140. doi: 10.7759/cureus.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leung K., Shum M.H., Leung G.M., Lam T.T., Wu J.T. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Euro Surveill. 2021;26 doi: 10.2807/1560-7917.ES.2020.26.1.2002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. bioRxiv. 2020 doi: 10.1101/2020.12.21.20248640. [DOI] [Google Scholar]
- 30.To K.K.-W., Tsang O.T.-Y., Leung W.-S., Tam A.R., Wu T.-C., Lung D.C., et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20:565–574. doi: 10.1016/s1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu J., Guo J., Xu Y., Chen X. Viral dynamics of SARS-CoV-2 in saliva from infected patients. J Infect. 2020;81:e48–e50. doi: 10.1016/j.jinf.2020.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J., Cai K., Zhang R., He X., Shen X., Liu J., et al. Novel one-step single-tube nested quantitative real-time PCR assay for highly sensitive detection of SARS-CoV-2. Anal Chem. 2020;92:9399–9404. doi: 10.1021/acs.analchem.0c01884. [DOI] [PubMed] [Google Scholar]
- 33.Yip C.C., Sridhar S., Leung K.H., Ng A.C., Chan K.H., Chan J.F., et al. Development and evaluation of novel and highly sensitive single-tube nested real-time RT-PCR assays for SARS-CoV-2 detection. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21165674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cousins M.M., Ou S.S., Wawer M.J., Munshaw S., Swan D., Magaret C.A., et al. Comparison of a high-resolution melting assay to next-generation sequencing for analysis of HIV diversity. J Clin Microbiol. 2012;50:3054–3059. doi: 10.1128/JCM.01460-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.