Abstract
SARS-CoV-2-induced COVID-19 is a serious pandemic of the 21st century, which has caused a devastating loss of lives and a global economic catastrophe. A successful vaccine against SARS-CoV-2 has suffered a delay due to lack of substantial knowledge about its mechanisms of action. Understanding the innate immune system against SARS-CoV-2 and the role of heat shock proteins' (HSP) inhibiting and resolution of inflammatory pathways may provide information to the low SARS-CoV-2 mortality rates in Africa. In addition, bats being a host to different viruses, including SARS-CoV-2 possess a well specialized IFN-innate antiviral inflammatory response, showing no signs of disease or pro-inflammatory cytokine storm. We discuss the molecular pathways in COVID-19 with a focus on innate immunity, inflammation, HSP responses, and suggest appropriate candidates for therapeutic targets and The contribution of the innate immune system to the efficacy of mRNA or vector based Corona immunizations.
Keywords: Innate immunity, COVID-19, Heat shock protein, NLRP3 inflammasome, Inflammation
Abbreviations: SARS, severe acute respiratory syndrome; COVID-19, coronavirus disease 2019; MERS-CoV, Middle East Respiratory Syndrome-corona virus; IFN, interferon; ACE2, angiotensin converting enzyme 2; RBD, receptor binding domains; MRRs, microbial recognition receptors; TLRs, Toll-like receptors; ARDS, acute respiratory distress syndrome; IRF3, interferon regulatory factor 3; NF-kB, nuclear factor kappa-light-chain-enhancer of activated B-cells; JAK-STAT, Janus kinase/signal transducers and activators of transcription; TNF-α, tumor necrosis factor-alpha; IL-6, interleukin 6; ASC, apoptosis-associated speck-like protein containing a caspase recruitment domain; LRR, leucine-rich repeat domain; HSPs, heat shock proteins; GSDMD, gasdermin D; COX-2, Cyclooxygenase-2; PG, prostaglandin; HSR, heat shock response; HSF-1, heat shock factor 1; IFN-γ, interferon gamma; NSAIDs, Non-steroid anti-inflammatory drugs.
1. Introduction
1.1. Coronavirus
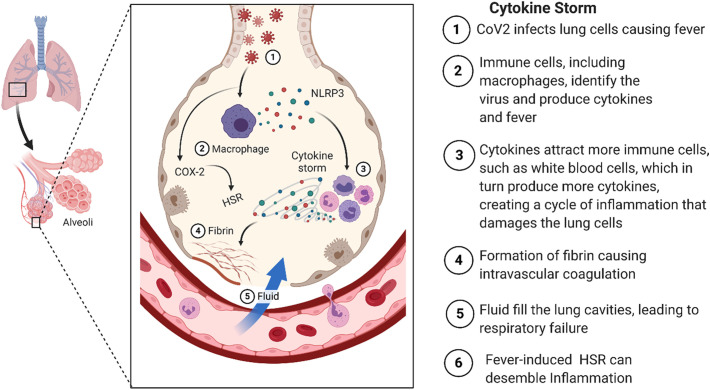
The first reported cases of the new coronavirus (CoV) occurred in Huanan seafood market in Wuhan City/China, where a small number of individuals were identified with pneumonia of unknown origin leading to a severe acute respiratory syndrome (SARS) (Zhu et al., 2019). Respiratory samples from these patients were analyzed and the etiological agent was identified as CoV. The novel CoV was named by the world health organization (WHO) as the novel pneumonia infectious disease, “coronavirus disease 2019 (COVID-19) or COVID-SARS-CoV-2” (Novel, 2020). The overall genomic sequence identity of SARS-CoV-2 showed a concordance of 96.2% to COV-RATG13 found in bats (Zhou et al., 2020b). According to global statistics, more than 164.284.766 people were infected between December 2019 and May 2021, with a mortality incidence of over 3.406.261 cases (https://coronavirus.jhu.edu). The clinical severity of COVID-19 infection may lead to severe respiratory failure, especially in the elderly and patients with pre-existing comorbidities such as hypertension, diabetes mellitus, coronary heart disease, and chronic obstructive lung disease (Polanco et al., 2014; Zhou et al., 2020a). Currently, humans affected by COVID-19 may suffer from dysregulated immune responses, resulting in excessive inflammation, known as cytokine storm (Weiss and Leibowitz, 2011) (Fig. 1 ). Previous severe cases of SARS-CoV and Middle East Respiratory Syndrome (MERS)-CoV showed high serum levels of several pro-inflammatory cytokines (CHIEN et al., 2006; Kim et al., 2016). Compared to the previous CoV infections, COVID-19 is highly contagious (Liu et al., 2020b), and the progression rate to SARS is quick in some of the cases. Although research has intensively focused on understanding the pathophysiology of COVID-19 infection, the specific molecular and biochemical host factors that derive severe lung pathology are not well understood yet. A retrospective analysis of adult patients suffering from COVID-19 SARS showed that high viral titer, increased inflammatory monocytes/macrophages, neutrophil infiltration, delayed interferon (IFN) response, and multiple organ failure, contribute to disease severity (Channappanavar et al., 2017; Gorla et al., 2018; Matthay et al., 2019; Zhou et al., 2020a).
Fig. 1.
Schematic representation of clinical features versus pathogenic inflammatory cytokine response in CoV-2 infections.
1.2. Infection
The spread of the novel coronavirus SARS-CoV-2 has caused a global emergency, which demands an immediate solution to reduce any further global threat to health, social life, and the economy.
Global strategies focus on controlling SARS-CoV-2 by suppressing transmission of the virus and therapeutic intervention. According to current evidences, it has been suggested that SARS-CoV-2 can spread from person to person. However, understanding how, when, and environmental settings favoring SARS-CoV-2 spread is crucial to the development of effective infection prevention and control measures. SARS-CoV-2 possible modes of transmission includes droplet, airborne, fomite, fecal-oral, blood borne, mother to child, and animal-to-human transmission. Transmission of SARS-CoV-2 can occur through direct close contact with infected people through saliva and respiratory secretions or their respiratory droplets, which are released when an infected person coughs, sneezes, talks or sings (Burke et al., 2020; Ghinai et al., 2020; Liu et al., 2020a). SARS-CoV-2 transmission can also occur by dissemination of droplet nuclei (aerosols) that remain infectious when suspended in air over long distances and time (Organization WH, 2014, Organization WH, 2020).
Microscopic aerosols generated from infected patients can evaporate and can be exhaled during normal breathing and talking. As a result, a susceptible person could inhale aerosols and become infected. The amount of SARS-CoV-2 in aerosol sufficient to cause infection in another person is yet known. However, studies have found SARS-CoV-2 RNA in air samples 3 h and 16 h after the induction of aerosols (Fears et al., 2020; Van Doremalen et al., 2020). Studies have also found SARS-CoV-2 RNA in air samples without aerosol induction in a health care setting (Chia et al., 2020; Guo et al., 2020; Liu et al., 2020c; Santarpia et al., 2020; Zhang et al., 2020). SARS-CoV-2 RNA are found to be viable from hours to days under a favorable condition (temperature and humidity) and the type of surface. Therefore, transmission may occur through direct contact with surfaces in the immediate environment contaminated with virus from an infected person.
At the moment, there are no published reports of transmission of SARS-CoV-2 through feces or urine, but recently, SARS-CoV-2 RNA has been detected in urine and feces of infected patients (Guan et al., 2020; Sun et al., 2020a; Wang et al., 2020b; Zheng et al., 2020). Also, some studies have reported detection of SARS-CoV-2 RNA in either plasma or serum, with complete replication in blood cells. However, the role of blood borne transmission remains uncertain. Low viral titers in plasma and serum suggest that the risk of transmission through this route may be low (Le Chang et al., 2020; Wang et al., 2020b).
The spike protein (S) of SARS-CoV-2 binds to the human receptor angiotensin converting enzyme 2 (ACE2) initiating the infection of host cells. ACE2 could therefore serve as a primary target for vaccines preventing viral entry into host cells (Li, 2016; Panda et al., 2020). The receptor binding domains (RBD) between SARS-CoV-1 and SARS-CoV-2 show a structural difference in S protein, and therefore it is not possible to use the available SARS-CoV-1 vaccine for the treatment of SARS-CoV-2 (Berry et al., 2004). In high-risk COVID-19 patients, the innate immune system lacks the possibility to reduce inflammation and prevent the cytokine storm (Sun et al., 2020b).
This review article summarizes the innate immunity, inflammation, and heat shock protein (HSPs) responses activated by SARS-CoV-2. The responses to SARS-CoV-2 infection in Africa and lessons learned from bats are discussed in the second section of this review. Finally, we discuss.
novel potential clinical studies and alternative treatments for COVID-19 patients and the innate immune response to the mRNA or vector based Corona immunizations.
2. Innate immune response to SARS-CoV-2
The evolutionary conserved innate immune system is the host's first defense line of action against viral infections (Netea et al., 2019). The innate immune system plays a role in the removal of virus-infected cells, leading to rapid coordinated adaptive immune response (Catanzaro et al., 2020).
In the mammalian hosts, microbial recognition receptors (MRRs) including Toll-like receptors (TLRs) and the nucleotide-binding oligomerization domain (NOD)-like receptor family proteins (NLRs) are involved in the detection of various microbes (Franchi et al., 2008; Franchi et al., 2009b; Sansonetti, 2006). The pattern recognition receptors (PRRs) assemble multiple proteins to form a complex called inflammasome (Man et al., 2017), which can induce membrane pore formation and proinflammatory cytokine overload leading to inflammatory cell death called pyroptosis (He et al., 2015; Man et al., 2017; Shi et al., 2015). Therefore, the response point between the host innate immune response and viral replication is a potential therapeutic target in viral infections by reducing excessive inflammation while retaining antiviral functions.
2.1. NLRP3 inflammasome pathogenesis in SARS-CoV-2 infection
Among pattern-recognition receptors, nucleotide-binding and oligomerization domain (NOD)-like receptors (NLRs) are unique cytosolic receptors, which constantly patrol for invading pathogens in the cytoplasm.
NLRP3 inflammasome is a well-studied activated inflammasome in many families of viruses. Full NLRP3 inflammasome activation requires two signaling pathways (Shrivastava et al., 2016). The primary, or priming signal can be initiated by TLRs and RIG-I-like receptors (RLRs) or by a protein receptor, which leads to the upregulation of pro-caspase-1 and pro-IL-1β and pro-IL-18 (Bauemfeind et al., 2009). The second signal pathway of NLRP3 inflammasome activation, involves pro-caspase-1 recruitment to NLRP3 and the subsequent production of mature caspase-1 and IL-1β and IL-18, the main stress signals associated with tissue damage or infection (Franchi et al., 2008). The complete mechanisms of NLRP3 inflammasome activation are still not fully understood. However, three diverse classes of stimuli are involved in the activation of NLRP3 inflammasome: the invading microbial pathogens and their products, including lipopolysaccharide, muramyl dipeptide, nucleic acids, and pore-forming toxins; the endogenous danger signals like extracellular ATP, urate crystals, hyaluronan, and fibrillar amyloid-β; and the crystalline environmental pollutants, such as alum adjuvant, and ultraviolet irradiation (Baral et al., 2014; Feldmeyer et al., 2007; Franchi et al., 2009a; Schroder and Tschopp, 2010; Sha et al., 2014).
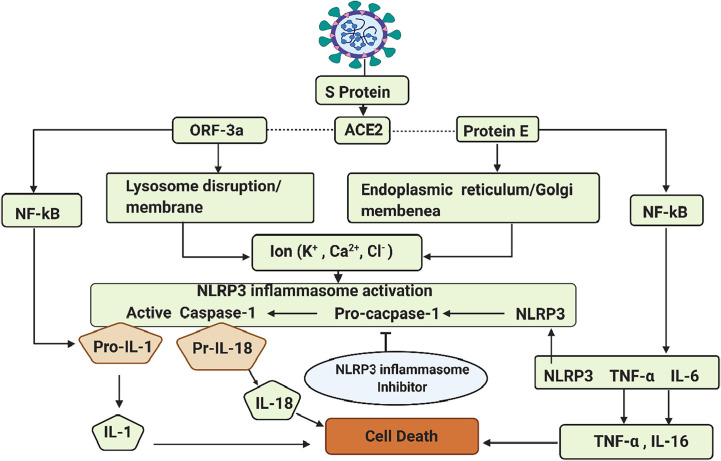
As previously mentioned, SARS-CoV-2 genome encodes S proteins that bind to the host cell receptor ACE2, which facilitates viral entry. The SARS-CoV-2 envelope (E) consists of a small hydrophobic protein-membrane (M) and nucleocapsid (N). These four SARS-CoV-2 structures are essential for viral assembly and infection (Weiss and Leibowitz, 2011). As described before, the initial binding of SARS-CoV-2 to the host cell is initiated between the S protein and the ACE2 receptor (Hoffmann et al., 2020; Patel and Verma, 2020). SARS-CoV-2 encodes three putative ion channels (IC): protein E, ORF-3a, and ORF-8a (Chan et al., 2020; Ramaiah and Arumugaswami, 2020; Wu et al., 2020a). The dominant proteins E and ORF3a have a (PDZ) binding motif and are involved in triggering cytokine storm and leading to cell death via the innate immune signaling sensor NLRP3 inflammasome (Nieto-Torres et al., 2015) (Fig. 2 ). Clinically this may result in increased pulmonary edema causing acute respiratory distress syndrome (ARDS) (Jimenez-Guardeño et al., 2014; Nieto-Torres et al., 2014; Torres et al., 2015). In addition, the E protein plays a principal role in several signaling mechanisms including the activation of interferon regulatory factor 3 (IRF3) and nuclear factor kappa-light-chain-enhancer of activated B-cells (NF-kB). IRF3 is known to mediate the secretion of type 1 interferon, which causes the activation of the Janus kinase/signal transducers and activators of transcription (JAK-STAT) pathway and the expression of interferon-stimulated genes. On the other hand, E protein triggers the activation of the NF-kB inflammatory signaling cascade and the interaction of its PDZ-binding motif (PBM) with inflammatory factors, such as tumor necrosis factor-alpha (TNF-α) and interleukin 6 (IL-6) (Wang et al., 2007). These changes form a calcium ion (Ca2+) channel in the endoplasmic reticulum/Golgi apparatus intermediate compartment membrane, and act as powerful stimuli activating the cytosolic innate immune NLRP3 inflammasome. Moreover, multiple cellular signaling events have been shown to activate NLRP3 at the membrane, leading to efflux of potassium (K+) or chloride ions (Cl−), and flux of Ca2+ (Di et al., 2018; Domingo-Fernández et al., 2017; Muñoz-Planillo et al., 2013; Perregaux and Gabel, 1994; Samways et al., 2014; Surprenant et al., 1996; Tang et al., 2017; Triantafilou et al., 2013) as well as lysosomal disruption, mitochondrial dysfunction, metabolic changes, and trans-Golgi disassembly (Swanson et al., 2019) (Fig. 2). Virally-induced activation of NLRP3 and downstream mediators often lead to pathological tissue injury during infection.
Fig. 2.
Schematic representation of SARS-CoV, after host receptor interaction with S viral protein [angiotensin-converting enzyme 2 (ACE2)]. SARS-CoV mediated NLRP3 inflammasome activation and downstream inflammatory cascades leading to inflammation and cell death via the open reading frame (ORF)-3a translate to disrupt lysosomal membrane and the disruption of endoplasmic reticulum/Golgi membrane by protein E. However, inflammasome activates nuclear factor kappa beta (NF-kB) and the production of active caspase 1 can then cleave to pro-inflammatory cytokines tumor necrotic factor alpha (TNF-α), interleukin 1 (IL1), interleukin 6 (IL6), and interleukin 18 (IL18) leading to inflammation and cell death.
The NLRP3 inflammasome assembles and activates caspase-1, inducing the inflammation-associated cell death process pyroptosis, and the maturation of the key pro-inflammatory cytokines IL-1β and IL-18, leading to the development of inflammatory responses (de Torre-Minguela et al., 2017). NLRP3 cleaves apoptosis-associated speck-like protein containing a caspase recruitment domain (ASC) via its N-terminal pyrin domain through hemophilic interactions, resulting in the formation of prion-like oligomerization (Verkhratsky and Pelegrín, 2014). The presence of various domains such as MHC class II transcription activator (CIITA), HET-E (incompatibility locus protein from Podospora anserina), and telomerase-associated protein located in the middle of the NLRP3 inflammasome complex, possess deoxynucleosidtriphosphate (dNTPs) activity that mediates downstream oligomerization (Lu et al., 2014; Ruland, 2014; Schmidt et al., 2016). The C-terminal leucine-rich repeat domain (LRR) associated with heat shock proteins (HSPs) and SGT1 is considered responsible for the regulation of NLRP3 inflammasome activity (Lo et al., 2013; Mayor et al., 2007). Once the NLRP3 inflammasome is activated, it triggers the auto-cleavage of pro-caspase-1, which acts as an activator to mediate the proteolytic processing of pro-IL-1β, pro-IL-18, and proapoptotic factor gasdermin D (GSDMD) (Shi et al., 2015). The GSDMD attaches and forms a pore on the host cells membrane, thereby facilitating the secretion of IL-1β/IL-18 and further causing pyroptosis (He et al., 2015). Accordingly, it is evident that SARS-CoV E protein and ORF-3a activate the NLRP3 inflammasome and establish host antiviral status (Zhao and Zhao, 2020). The NLRP3 mediated inflammasome response to SARS-CoV-2 may be a potential specific drug target for the treatment of SARS-CoV-2 disease. Ambient temperature, promoting the production of anti-inflammatory heat shock protein, may be a critical factor in the progression of SARS-CoV-2 treatment. The molecular involvement of HSPs has been shown to play a vital role in the resolution of the inflammatory pathway of COVID-19 infection (Heck et al., 2020).
2.2. Toll-like receptors pathogenesis in SARS-CoV infection
The TLRs are a group of innate immune receptors that are involved in activation of the innate immunity, regulation of the cytokine expression, indirect activation of the adaptive immune system, and recognition of the pathogen-associated molecular patterns (PAMPs).
(Birra et al., 2020; Debnath et al., 2020; Hedayat et al., 2011). Before SARS-CoV-2 pandemic, several studies have shown that TLR pathways are important in the pathogenesis of SARS-CoV and the Middle East respiratory syndrome (MERS) (Birra et al., 2020). The TLRs comprise of ten superfamily members and are divided into membranous and endosomal receptors. The TLRs are expressed on different immune cells including the dendritic cells, macrophages, natural killer cells, and the adaptive immune cells (T and B cells) (Angelopoulou et al., 2020). The TLRs have a wide range of recognition for both single-strand and double-strand DNA pathogens.
The signal transduction of TLRs involves two major pathways, the Myeloid differentiation primary response 88 (MyD88) and the Toll/interleukin-1 receptor (TIR)-domain containing adaptor inducing interferon-β (IFN-β) also known as toll like receptor adaptor molecule 1 (TRIF or TICAM1). The presence of tumor necrosis factor receptor–associated factor (TRAF) and IL-1 receptor-associated kinases (IRAK) proteins can initiate a downstream activation of nuclear factor-kB (NF-kB) and Interferon regulatory factor (IRF) and lead to the production of type 1 IFN and pro-inflammatory cytokines-interleukin-1 (IL-1), IL-6, tumor necrosis factor-α (TNF-α), and IL-12. The TLRs also play an indirect role in the adaptive immune system by modulating the expression of co-stimulatory molecules. The activation of TLRs by SARS-CoV-2 activates inflammasome and production of IL-1β and IL-6. Studies have reported that the long-term activation of inflammasomes has been the primary cause for the poor outcome in COVID-19 patients (de Rivero Vaccari et al., 2020). In addition, TLRs induce the activation of Janus kinase transducers (JAK/STAT) leading to macrophage activation syndrome. In a fashionable manner, TLRs mediate host cell signaling pathways and decrease the expression of IFN receptors and type 1 IFN production, which lead to systemic inflammatory response (Angelopoulou et al., 2020), being vital in the pathogenesis of CoVs. Several studies have been conducted to study the involvement of TLR members. These studies have shown that TLR3 act via the TIRF pathway to offer protection in SARS-CoV and MERS-CoV infections. A TLR3 mouse model has shown to activate IRF3 and NF-kB pathways, and the production of type 1 IFN and pro-inflammatory cytokines (Birra et al., 2020). Also, no reduction in the secretion of cytokines following coronavirus infection in TLR3 knock-out mice has been shown (Birra et al., 2020). TLR4 activates the same pathway as TLR3, but the TLR4 is vital in bacterial infections and activated by oxidized phospholipids found in most of the viral lung infections, also being confirmed for COVID-19.
Activated TLR4 pathway in the pulmonary phase of infection causes oxidative injury. TLR4 signaling pathway plays a role in the activation of neutrophil extracellular traps (NETs) and NET formation in COVID-19 has been shown to sustain inflammation, which can lead to bad outcome of COVID-19 patients (Cicco et al., 2020). The SARS-CoV-2 spike protein binds TLR1, 4, and 6, with a higher affinity for TLR4. TLR4 blocker could be administered as a therapeutic remedy for COVID-19 patients (Choudhury and Mukherjee, 2020). Studies have shown that TLR7 and TLR8 are highly expressed in the lung during SARS-CoV infection and could play a role in cytokine storm in SARS-CoV-1 (de Groot and Bontrop, 2020). Studies from whole genomic sequencing has revealed that TLR7 could be more involved in SARS-CoV-2 pathogenesis compared to SARS-CoV and MERS-CoV because SARS-CoV-2`s single stranded RNA could primarily bind to TLR7 (Van Der Made et al., 2020). TLR7 could be another candidate for triggering NET formation in COVID-19 patients, as the activation of TLR7/8 pathway induces a strong pro-inflammatory response in patients, resulting in acute lung injury. Therefore, it may have a dual role disease progression (Moreno-Eutimio et al., 2020; Veras et al., 2020).
Moreover, several TLR agonists have been administered to activate the innate immune cells and the production of various resisting factors in the lung epithelial cell. In this effort to reduce COVID-19 death, several clinical trials have evaluated the effect of anti-inflammatory drugs in COVID-19 patients, using CD24Fc conjugate to block TLR activation (Florindo et al., 2020). Also, the antagonistic effects of glycyrrhetinic acid against TLR4 has an anti-inflammatory effect in the lung of mice with acute respiratory distress syndrome, thereby protecting tissue destruction (Huang et al., 2020). It may also stimulate an anti-inflammatory activity downstream of the less active ACE2 and may also stimulate an anti-inflammatory activity downstream of the less active ACE2. Therefore, it could be a potential approach to control COVID-19 (Murck, 2020).
3. HSPs-mediated CoV-2 tolerance versus immunopathology
Heat shock proteins (HSPs) are intracellular proteins that act as molecular chaperones in protein folding and protein trafficking between intracellular compartments. They are increasingly expressed by oxidative stress, nutritional deficiencies, and radiation. However, HSPs are released into the extracellular environment by not fully known mechanism, but they act with numbers of innate immunological effect. One of the specific pre-existing comorbidities associated with severe COVID-19 infection includes a relative deficiency of heat shock response (HSR) (Heck et al., 2020). A lack of sufficient HSR has been reported to be the likely underlying etiology for the unfavorable prognosis in most of the chronic inflammatory diseases (Newsholme and de Bittencourt Jr, 2014), and is also suspected in groups with high risk for COVID-19 mortality (Heck et al., 2020).
The innate immune cells are highly sensitive to stimuli and rapidly recruit cells within minutes (neutrophils) to hours (monocyte/macrophages) to the site of injury. These rapid responses are orchestrated primarily by the expression of NF-kB, which derives inflammation during the early phase (Oeckinghaus and Ghosh, 2009). Cyclooxygenase-2 (COX-2) is an inducible protein responsible for the production of proinflammatory arachidonic acid-derived prostaglandin (PG) and other lipid mediators as well as vasoactive compounds that increase vascular permeability and facilitate the arrival and activation of inflammatory cells and tissue repair (Medzhitov, 2008). COX-2-derived PG-E2 induces fever by blocking thermosensory information at the preoptic area of the anterior hypothalamus, and thalamus, leading to the activation of coordinated sympathetic/parasympathetic heat-sparing mechanisms, resulting in an elevation of core temperature (Miragem and Homem de Bittencourt, 2017). The rise in temperature of approximately 2–3 °C triggers the heat shock response (HSR) (Singh and Hasday, 2013).
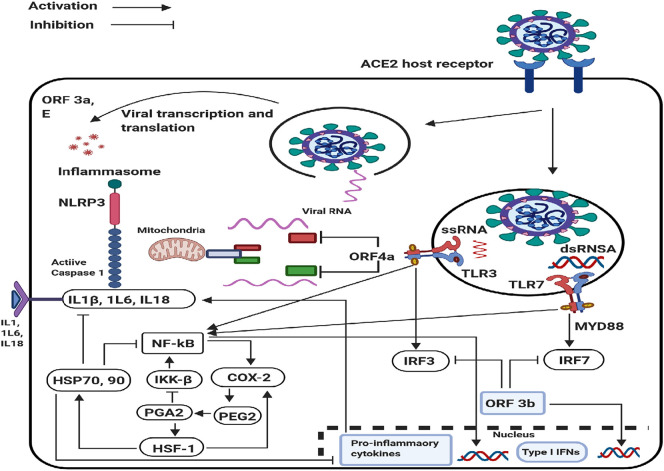
Structural changes in the plasma membrane including viral budding during the establishment of fever plays a direct role in heat shock factor 1 (HSF-1) activation (Anckar and Sistonen, 2011). The activation of HSF-1 regulates the transcription of HSPs, expression of cytokines, and early gene response (Chen et al., 2005), including the control of COX-2 transcription-induced production of PDE2 during the priming/action phase of inflammation (Gilroy et al., 1999). The HSR can disassemble acute inflammation by blocking NF-kB and other downstream pro-inflammatory signals (Gilroy et al., 1999; Newsholme and de Bittencourt Jr, 2014; Serhan, 2011). The production of HSP70 in response to HSF-1 activation is correlated with complex formation by NF-kB and its inhibitor (I-kB) to impede NF-kB translocation into the nucleus (Chen et al., 2005) (Fig. 3 ). The HSPs are anti-inflammatory chaperones that ameliorate a series of inflammatory conditions (de Bittencourt Jr et al., 2007; Ianaro et al., 2003), including after activation of the NLRP3 inflammasome. Persistent activation of NLRP3-dependent caspase-1-mediated cleavage of an RNA-binding protein can enhance both the expression and transcriptional activity of HSF-1 to promote a robust HSR (Newsholme and de Bittencourt Jr, 2014; Talwar et al., 2011; Wang et al., 2013).
Fig. 3.
Innate Immune signaling Pathways in SAR-CoV infection and Immune Evasion. Upon CoV infection, incoming double-stranded RNA(dsRNA) and genomic single-stranded RNA (ssRNA) are recognized by Toll-like receptor 3 (TLR3) and TLR7, respectively. The downstream signaling of these TLRs induces activation of nuclear factor-κB (NF-κB) to produce proinflammatory cytokines and phosphorylation of interferon regulatory factor 3 (IRF3) and IRF7 to drive type I interferon. NF-κB activation-induced COX-2 production, which in term induced fever and then triggers the response of heat shock factor 1 (HSF-1). The inhibitory of SARS-CoV-2-induced inflammation damage by heat shock protein (HSP70, 90) is via suppressing activation of NF-κB pathway in the cytosol NLRP3 inflammasome and suppression of pro-inflammatory cytokines in the nucleus.
Fatality after SARS-CoV-2 infection is thought to be due to the virus-induced elevated pro-inflammatory cytokine release, often called “cytokine storm” (Huang et al., 2005; Tisoncik et al., 2012). Alternatively, in those who present a robust HSR after infection, the anti-inflammatory activity of HSR may result in the inhibition/degradation of cytokines, preventing a cytokine storm (Tanaka et al., 2014). Although, several drugs targeting the anti-inflammatory and anti-apoptotic pathways in SARS-CoV-2 patients including chloroquine (Gao et al., 2020), hydroxychloroquine (Gautret et al., 2020), glucocorticoids (Wu et al., 2020b), remdesivir (Wang et al., 2020a), favipiravir (Cai et al., 2020), have been tested, but no significant benefit has been shown yet.
Although the mechanisms involved in the multiple beneficial effects of hyperthermic treatment in chronic inflammatory diseases have not been fully elucidated, it has been hypothesized that elevating core body temperature in humans between 38 and 39 °C can exert anti-inflammatory effects due to nitric oxide (NO)-based improvement of endothelial function as well as chronic NO-elicited HSP70 expression (Krause et al., 2015). In fact, HSPs can inhibit both NLRP3 inflammasome activation and caspase-1 activity in mouse macrophages (Levin et al., 2008). Therefore, purposefully increasing or maintaining core body temperature at fever range (38–39 °C) levels could activate the anti-inflammatory activity of the HSR and provide and alternative treatment.
4. Constitutive heat shock response and tolerance to CoV-2
Constitutive heat shock response is an evolutionary conserved innate stress response system. Under normal physiological conditions, it exist in low levels but their concentration can increase in many folds in response to a plethora of stimuli including thermal and nonthermal stimuli such as ischemia, iron overload, oxidants, and infections (Jäättelä, 1993; Jäättelä and Wissing, 1992; Villar et al., 1994). Stress-induced/chronic disease-induced HSPs accumulation is considered a powerful cytoprotective (Bakthisaran et al., 2015; Zhang et al., 1999). The ambient temperature in sub-Saharan Africa could be a potential stimulus of HSR and might play a significant role against SARS-CoV-2.
SARS-CoV pandemic in African countries is losing the perspective of Chad Wells and colleagues (Wells et al., 2020) who have estimated a total death toll of approximately 300,000 for the Democratic Republic of Congo alone. However, their prediction has been proven wrong because little is known about the dynamics of SARS-CoV-2 in African countries, including its infectiousness and the proportion of infected people who develop symptoms and the response of their innate immune cells. We are aware of the lack of test capacity in Africa and the quality of the collected data, however as the scientific evidence behind the low mortality rate of SARS-CoV-2 is lacking, we speculate that the high ambient temperature in sub-Saharan African countries could potentially stimulate the HSR and their anti-inflammatory effects to dissolve the inflammation caused by SARS-CoV-2 infection. Additionally, cases of COVID-19 outbreaks present a pattern of clustering in relatively cool and dry environments, just like the previous SARS-CoV-1 (Araujo and Naimi, 2020). According to COVID-19 weather models, warm and cold climates favors the spread of the virus, whereas arid and tropical climates are less favorable (Ma et al., 2020; Sajadi et al., 2020). However, this model is still uncertain across sub-Saharan Africa and South East Asia (Araujo and Naimi, 2020). Climate could help constrain SARS-CoV-2 (Araujo and Naimi, 2020; Bannister-Tyrrell et al., 2020; O'Reilly et al., 2020). The restoration of immunoinflammatory balance through hyperthermic treatment has been suggested (Cohen, 2020), and could be a promising treatment option in handling chronic auto-immune disease without the involvement of immunosuppressive approaches (Tukaj and Kaminski, 2019). Hyperthermia-induced heat stress has been shown to mitigate viral infections by a direct inhibition of pathogens, stimulation of both the innate and adaptive transcriptional genes of the immune system and the activation of regulatory processes that dissemble inflammation, and the prevention of cytokine storms that otherwise could cause excessive tissue damage (Evans et al., 2015). Heat intervention is one of the oldest forms of microbial control and today remains one of the most common methods for controlling and eradicating pathogens. Control temperatures at 60 °C for 30 min or 65 °C for 15 min or 80 °C for 1 min have been shown to reduced coronavirus infectious by at least 4 fold (Cohen, 2020). However, the therapeutic temperature, humidity, and time required to dissemble SAR-CoV-2 in vivo are yet to be determined.
Epidemiological evidence has suggested that frequent sauna bathing can reduce the risk of pneumonia (Kunutsor et al., 2017a), and decrease the incidence of respiratory viral infections (Ernst et al., 1990; Kunutsor et al., 2017b). Additionally, inhaling warmed and humidified air above 43 °C for 30 min can reduce viral shedding and relieve symptoms of the common cold (Tyrrell et al., 1989). The inhalation of hot air is significant for the immune system's first line of defense by direct inhibition or deactivation of virions in the ethmoidal sinus where they first lodge (Conti et al., 1999). Whole body heat application has also been proven to support the immune system's second line of defense dependent on the HSR pathway by mimicking the effects of fever (Schieber and Ayres, 2016). Higher temperatures at fever range can activate immune cells, promote cell membrane fluidity, and increase cell differentiation and activation of viral antigens, leading to rapid response to viral threats. The direct application of heat to the upper airways, at the first signs of infection, may further serve to inhibit or deactivate virions. In vitro exposure of cells to 45 °C for 20 min stimulates immune cells to release adequate HSPs and suppresses rhinovirus multiplication by more than 90% (Conti et al., 1999). The inhalation of steam with added essential oils such as Eucalyptus, peppermint, and lavender with anti-viral decongestant, may further assist in facilitating muco-ciliary clearance and reducing viral load as well as providing physical and psychological relief. This hypothesis necessitates re-evaluation and caution at the time while treating SARS-CoV-2 patients.
Currently no clinical study has been planned or designed using heat in the treatment of COVID-19, but heat has a long traditional use in this setting. Heat-based clinical protocols are needed to design future studies and inform clinical practice in order to minimize the risk of cross-infection during treatment as well as minimize the risks of treatment such as burns, cramps, dizziness and fainting, heat exhaustion, and heat stroke.
Interestingly, bats present an unequaled HSP-based anti-inflammatory HSR and do not show degenerative diseases nor cytokine storms (Ahn et al., 2019). Bat immune cells continue to suppress NLRP3 inflammasome activation in response to viral/bacterial and sterile stimuli (Ahn et al., 2019). The same occurs for different viruses, including MERS coronavirus, without influencing their ability to defeat viruses (Ahn et al., 2019). Constant inflammatory responses have been shown to correlate with HSP70-induced anti-viral interferon gamma (IFN-γ) production (Jacquemin et al., 2017) via IFNs regulate HSP expression, which further enhance the transcription rate of the heat shock gene and increasing the stability of mRNA coding for HSPs (Zhao et al., 2002). In bat, IFN-α can also enhance cyPG-induced HSP70 synthesis in virus infected cells (Pica et al., 1996), and IFN-γ-induced synthesis and release of HSP70 towards the extracellular exosomes pathway, which influence unaffected dendritic cells (Bausero et al., 2005). The HSR in the bats shows a constant rapid proteostasis response upon virus-induced ER stress, rapid resolution of inflammation to prevent tissue damage. Despite species differences, comparative physiology and similarity between bats' and human antiviral and anti-inflammatory protective pathways involving HSR can direct some clues on how to avoid or treat cytokines storm in COVID-19 patients.
5. Innate immune response to the mRNA or vector based Corona immunizations
The effort to protect the global population using the current vaccines may provide a possible path out of the pandemic and vaccines have been approved around the world. The goal of these vaccines is to induced and train the immune system to recognize a piece of SARS-CoV-2 antigen, targeting the spike protein, which the coronavirus uses in masking human cells. The approved vaccines developed by Pfizer/BioNTech and Moderna use mRNA and lipid nanoparticle (LNP) delivery technology, while the approved formulations by AstraZeneca, Johnson and Johnson and Gam-COVID-vac (Sputnik V) contain DNA within non-replicating recombinant adenovirus (AdV) vector systems (Baden et al., 2021; Logunov et al., 2021; Polack et al., 2020; Voysey et al., 2021). Both vaccines from Pfizer/BioNTech (BNT162b2) and Moderna (mRNA-1273) are mRNA vaccines, reporting a success of 90–95% efficacy against COVID-19 (Baden et al., 2021; Polack et al., 2020), while the AdV vaccines (ChAdOx1 nCoV-19) and Gam-COVID-vac (Sputnik V) have an average efficacy of 70% and 91% protection against COVID-19 (Logunov et al., 2021; Voysey et al., 2021). Little is only known about the vaccines mobilizing the immune response, the durability of protection, and how to further optimize against new variants. However, the vaccines have been reported to neutralize antibody and virus T cell after 2 to 4 weeks of injection (Sahin et al., 2020; Widge et al., 2021), and trigger the innate and adaptive response to stimulate adaptive immunity without inducing systemic inflammation that may cause severe side effects. The mRNA vaccines function as both immunogen (encoding the viral protein) and adjuvant (capable of activating the Th or Th2 response). Upon entry of the single-stranded RNA (ssRNA) or double-stranded RNA (dsRNA), these are recognized by various endosomal and cytosolic innate immune sensors. Endosomal Toll-like receptors (TLR3 and TLR7) are the main TLRs that binds dsRNA and stimulates the expression of inflammatory chemokines while components of the inflammasome such as MDA5, RIG-I, NOD2, and PKR bind to ssRNA and dsRNA in the cytosol leading to cellular activation and production of type I interferon and multiple inflammatory mediators (Pardi et al., 2018).
The lymph node nanoparticle (LNP) carrier helps to protect the mRNA for safe-target delivery to lymphatics, where it promotes protein translation to occur (Pardi et al., 2018). Once in the LN, the LNP is engulfed by dendritic cells (DCs), which subsequently produce and present the antigen to T cells for activation of the adaptive immune response. The AdV vaccines also contain inherent adjuvant properties, although these reside with the virus particle that encases the DNA encoding the immunogen. After the injection of AdV, dendritic cells and macrophages stimulate innate immune to engage multiple pattern-recognition receptors including, TLR9-induced type I interferon secretion in the infected lungs (Sayedahmed et al., 2020). Although, mRNA vaccines do not bind TLR9, both vaccines are involved in the production of type I interferon. Dendritic cells and other cells that are involved in the production of type I interferon take up the vaccine-derived nucleic acids encoding S protein, which can thereafter stimulate both antigenic and inflammatory signals to activate T cells in lymph nodes mobilizing the adaptive immunity against SARS-CoV-2. The mRNA and AdV vaccines promote intracellular production of S protein and innate immune responses, which prime both CD8+ and CD4+ T cells to differentiate into effector and memory subsets. Vaccine-driven production of type I interferon promotes differentiation of CD4+ and CD8+ effector T cells producing inflammatory and cytotoxic mediators, and CD4+ T follicular helper (TFH) cells, which promote B cell differentiation into antibody secreting plasma cells. This secondary inflammatory response may provide a short-term change to the innate cells like macrophages through a phenomenon called ‘trained immunity’ (Yao et al., 2018), and the activation of memory T cells and B cells from the initial injection that activate type I interferon, amplifies T cell memory and promote B cell differentiation and survival, thus suggesting that vaccine-associated inflammation can booster and promote the generation of long-term immunological memory.
6. Conclusions
This review was written at the time; global SARS-CoV-2 infections rate have exceeded 164 million with over 3 million deaths, increasingly spreading worldwide. The old and new information in this review provides hope and solutions for the treatment of COVID-19. Based on the current understanding, SARS-CoV-2 is primarily spread through contact and respiratory droplets. Under some circumstances airborne transmission may occur both indoor and out indoor crowded poorly ventilated settings elsewhere.
It could be hypothesized that TLRs have both harmful and beneficial effects in COVID-19 infection. According to the old data from SARS-CoV and MERS, could help in better understanding of the exact role of component of innate and adaptive immunity in COVID-19 infection. Only the TLR7/8 recognizes ssRNA of COVID-19, while TLR3, TLR4, and TLR6 could be involved in COVID-19 infection. The use of both antagonists and agonists, should be investigated to determine the therapeutic and harmful effects of TLR in SARS-CoV-2 infection. The stage of infection is also important in determining the type of TLRs involvement. In addition, the attenuation of excessive activation of inflammasomes and NET formation could also be a therapeutic target. Finally, bioinformatics studies could help in understanding of interactions of TLRs with proteins and RNA of COVID-19.
Recent research has demonstrated that, during certain pathogen including SARS-CoV-2 infections, NLRP3 is capable of detecting specific ligands, activate caspase-1, and induce the release of various pro-inflammatory cytokines with vital roles against viral infection (Komune et al., 2011). Some years back, efforts have been put into the investigation of the relationship between virus and NLRP3 inflammasome. Viral RNA, viroporin, and infectious viral particles activate the NLRP3 inflammasome (Chen and Ichinohe, 2015). Most RNA viral infection activates or inhibits NLRP3 inflammasome by regulating ion channels and ROS model. The K+ efflux plays a major role in NLRP3 activation, although, Ca2+ channel and ROS model remain controversial.
Presently bat research has gained attraction. In addition to flight, various biological traits make bats unique among mammals. Bodies of research such as those of the Bat1K consortium147, and technologies that use single-cell RNA sequencing, allow unbiased and deeper characterization of bats immune cell populations and their specific functions and pathways. Bat host defense immune tolerate and balance virus inflammation confers exceptional health. The key regulators and machinery used by bat in maintaining this homeostatic balance is a valuable lesson for controlling and combating viruses' numerous inflammatory diseases in humans.
We suggest that HSR, is an essential pathway for inflammation resolution. Finally, we propose that the use of HSR activators should be investigated, since they could potentially alleviate the COVID-19 complications. Although, constant inflammatory responses have been shown to correlate with HSP70-induced anti-viral interferon gamma (IFN-γ) production (Jacquemin et al., 2017) via IFNs regulate HSP expression, which further enhance the transcription rate of the heat shock gene and increasing the stability of mRNA coding for HSPs (Zhao et al., 2002).
The presently recommended newly developed vaccines against SARS-CoV-2 has proven successful, but it administration has been associated with autoimmune manifestations in some groups of predisposed individuals. It has been demonstrated that these vaccines do not pose prominent danger than natural infections themselves, also both patients and clinicians are concerned about the potential risk for relapse or worsening of autoimmune diseases mainly because of insufficient data. To avoid the associated vaccination risk should not lead to vaccine refusal furthermore, trials would clarify the underlying immunological mechanisms of the newly implemented vaccines/adjuvants in these population.
Authors contribution
The authors actively contributed and co-wrote the manuscript. JD and HS defined the content, the literature of the manuscript. JD prepared the figures.
Declaration of Competing Interest
All authors critically reviewed, no competing interest and approved the final version of this manuscript content for publication.
Acknowledgments
Acknowledgement
We would like to sincerely thank Dr. Thomas Ragnar Wood for his input during the writing of this manuscript.
References
- Ahn M., Anderson D.E., Zhang Q., Tan C.W., Lim B.L., Luko K., et al. Dampened NLRP3-mediated inflammation in bats and implications for a special viral reservoir host. Nat. Microbiol. 2019;4:789–799. doi: 10.1038/s41564-019-0371-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anckar J., Sistonen L. Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu. Rev. Biochem. 2011;80:1089–1115. doi: 10.1146/annurev-biochem-060809-095203. [DOI] [PubMed] [Google Scholar]
- Angelopoulou A., Alexandris N., Konstantinou E., Mesiakaris K., Zanidis C., Farsalinos K., et al. Imiquimod-a toll like receptor 7 agonist-is an ideal option for management of COVID 19. Environ. Res. 2020;188:109858. doi: 10.1016/j.envres.2020.109858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo M.B., Naimi B. Spread of SARS-CoV-2 Coronavirus likely to be constrained by climate. medRxiv. 2020 doi: 10.1101/2020.03.12.20034728. [DOI] [Google Scholar]
- Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakthisaran R., Tangirala R., Rao C.M. Small heat shock proteins: role in cellular functions and pathology. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics. 2015;1854:291–319. doi: 10.1016/j.bbapap.2014.12.019. [DOI] [PubMed] [Google Scholar]
- Bannister-Tyrrell M., Meyer A., Faverjon C., Cameron A. Preliminary evidence that higher temperatures are associated with lower incidence of COVID-19, for cases reported globally up to 29th February 2020. medRxiv. 2020 doi: 10.1101/2020.03.18.20036731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baral P., Batra S., Zemans R.L., Downey G.P., Jeyaseelan S. Divergent functions of Toll-like receptors during bacterial lung infections. Am. J. Respir. Crit. Care Med. 2014;190:722–732. doi: 10.1164/rccm.201406-1101PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauemfeind F., Horvath G., Stutz A., Alnemri E., MacDonald K., Speert D., et al. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J. Immunol. 2009;183:787–791. doi: 10.4049/jimmunol.0901363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bausero M.A., Gastpar R., Multhoff G., Asea A. Alternative mechanism by which IFN-γ enhances tumor recognition: active release of heat shock protein 72. J. Immunol. 2005;175:2900–2912. doi: 10.4049/jimmunol.175.5.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry J.D., Jones S., Drebot M.A., Andonov A., Sabara M., Yuan X.Y., et al. Development and characterisation of neutralising monoclonal antibody to the SARS-coronavirus. J. Virol. Methods. 2004;120:87–96. doi: 10.1016/j.jviromet.2004.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birra D., Benucci M., Landolfi L., Merchionda A., Loi G., Amato P., et al. COVID 19: a clue from innate immunity. Immunol. Res. 2020;68:161–168. doi: 10.1007/s12026-020-09137-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bittencourt Jr P.I.H., Lagranha D.J., Maslinkiewicz A., Senna S.M., Tavares A.M., Baldissera L.P., et al. LipoCardium: endothelium-directed cyclopentenone prostaglandin-based liposome formulation that completely reverses atherosclerotic lesions. Atherosclerosis. 2007;193:245–258. doi: 10.1016/j.atherosclerosis.2006.08.049. [DOI] [PubMed] [Google Scholar]
- Burke R.M., Midgley C.M., Dratch A., Fenstersheib M., Haupt T., Holshue M., et al. Active monitoring of persons exposed to patients with confirmed COVID-19—United States, January–February 2020. Morb. Mortal. Wkly Rep. 2020;69:245. doi: 10.15585/mmwr.mm6909e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q., Yang M., Liu D., Chen J., Shu D., Xia J., et al. Experimental treatment with favipiravir for COVID-19: an open-label control study. Engineering. 2020;6(10):1192–1198. doi: 10.1016/j.eng.2020.03.007. S2095809920300631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanzaro M., Fagiani F., Racchi M., Corsini E., Govoni S., Lanni C. Immune response in COVID-19: addressing a pharmacological challenge by targeting pathways triggered by SARS-CoV-2. Signal Trans. Target. Therapy. 2020;5:1–10. doi: 10.1038/s41392-020-0191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.-W., Kok K.-H., Zhu Z., Chu H., To K.K.-W., Yuan S., et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerging Microbes Infections. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fett C., Mack M., Ten Eyck P.P., Meyerholz D.K., Perlman S. Sex-based differences in susceptibility to severe acute respiratory syndrome coronavirus infection. J. Immunol. 2017;198:4046–4053. doi: 10.4049/jimmunol.1601896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.-W., Kuo H.-T., Wang S.-J., Lu T.-S., Yang R.-C. In vivo heat shock protein assembles with septic liver NF-κB/I-κB complex regulating NF-κB activity. Shock. 2005;24:232–238. doi: 10.1097/01.shk.0000174020.87439.f2. [DOI] [PubMed] [Google Scholar]
- Chen I.-Y., Ichinohe T. Response of host inflammasomes to viral infection. Trends Microbiol. 2015;23:55–63. doi: 10.1016/j.tim.2014.09.007. [DOI] [PubMed] [Google Scholar]
- Chia P.Y., Coleman K.K., Tan Y.K., Ong S.W.X., Gum M., Lau S.K., et al. Detection of air and surface contamination by SARS-CoV-2 in hospital rooms of infected patients. Nat. Commun. 2020;11:1–7. doi: 10.1038/s41467-020-16670-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHIEN J.Y., HSUEH P.R., CHENG W.C., YU CJ, YANG PC. Temporal changes in cytokine/chemokine profiles and pulmonary involvement in severe acute respiratory syndrome. Respirology. 2006;11:715–722. doi: 10.1111/j.1440-1843.2006.00942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury A., Mukherjee S. In silico studies on the comparative characterization of the interactions of SARS-CoV-2 spike glycoprotein with ACE-2 receptor homologs and human TLRs. J. Med. Virol. 2020;92:2105–2113. doi: 10.1002/jmv.25987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicco S., Cicco G., Racanelli V., Vacca A. Neutrophil extracellular traps (NETs) and damage-associated molecular patterns (DAMPs): two potential targets for COVID-19 treatment. Mediat. Inflamm. 2020;2020 doi: 10.1155/2020/7527953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M. Turning up the heat on COVID-19: heat as a therapeutic intervention. F1000Research. 2020;9:292. doi: 10.12688/f1000research.23299.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti C., De Marco A., Mastromarino P., Tomao P., Santoro M. Antiviral effect of hyperthermic treatment in rhinovirus infection. Antimicrob. Agents Chemother. 1999;43:822–829. doi: 10.1128/aac.43.4.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath M., Banerjee M., Berk M. Genetic gateways to COVID-19 infection: implications for risk, severity, and outcomes. FASEB J. 2020;34:8787–8795. doi: 10.1096/fj.202001115R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di A., Xiong S., Ye Z., Malireddi R.S., Kometani S., Zhong M., et al. The TWIK2 potassium efflux channel in macrophages mediates NLRP3 inflammasome-induced inflammation. Immunity. 2018;49:56–65.e4. doi: 10.1016/j.immuni.2018.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo-Fernández R., Coll R.C., Kearney J., Breit S., O’Neill L.A. The intracellular chloride channel proteins CLIC1 and CLIC4 induce IL-1β transcription and activate the NLRP3 inflammasome. J. Biol. Chem. 2017;292:12077–12087. doi: 10.1074/jbc.M117.797126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst E., Pecho E., Wirz P., Saradeth T. Regular sauna bathing and the incidence of common colds. Ann. Med. 1990;22:225–227. doi: 10.3109/07853899009148930. [DOI] [PubMed] [Google Scholar]
- Evans S.S., Repasky E.A., Fisher D.T. Fever and the thermal regulation of immunity: the immune system feels the heat. Nat. Rev. Immunol. 2015;15:335–349. doi: 10.1038/nri3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fears A.C., Klimstra W.B., Duprex P., Hartman A., Weaver S.C., Plante K.S., et al. Persistence of severe acute respiratory syndrome coronavirus 2 in aerosol suspensions. Emerg. Infect. Dis. 2020;26:2168. doi: 10.3201/eid2609.201806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmeyer L., Keller M., Niklaus G., Hohl D., Werner S., Beer H.-D. The inflammasome mediates UVB-induced activation and secretion of interleukin-1β by keratinocytes. Curr. Biol. 2007;17:1140–1145. doi: 10.1016/j.cub.2007.05.074. [DOI] [PubMed] [Google Scholar]
- Florindo H.F., Kleiner R., Vaskovich-Koubi D., Acúrcio R.C., Carreira B., Yeini E., et al. Immune-mediated approaches against COVID-19. Nat. Nanotechnol. 2020;15:630–645. doi: 10.1038/s41565-020-0732-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi L., Park J.H., Shaw M.H., Marina-Garcia N., Chen G., Kim Y.G., et al. Intracellular NOD-like receptors in innate immunity, infection and disease. Cell. Microbiol. 2008;10:1–8. doi: 10.1111/j.1462-5822.2007.01059.x. [DOI] [PubMed] [Google Scholar]
- Franchi L., Eigenbrod T., Muñoz-Planillo R., Nuñez G. The inflammasome: a caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat. Immunol. 2009;10:241–247. doi: 10.1038/ni.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi L., Warner N., Viani K., Nuñez G. Function of Nod-like receptors in microbial recognition and host defense. Immunol. Rev. 2009;227:106–128. doi: 10.1111/j.1600-065X.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Tian Z., Yang X. Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends. 2020;14(1):72–73. doi: 10.5582/bst.2020.01047. (Epub 2020 Feb 19) [DOI] [PubMed] [Google Scholar]
- Gautret P., Lagier J.-C., Parola P., Meddeb L., Mailhe M., Doudier B., et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents. 2020;105949 doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ghinai I., McPherson T.D., Hunter J.C., Kirking H.L., Christiansen D., Joshi K., et al. First known person-to-person transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the USA. Lancet. 2020;395:1137–1144. doi: 10.1016/S0140-6736(20)30607-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy D.W., Colville-Nash P., Willis D., Chivers J., Paul-Clark M., Willoughby D. Inducible cyclooxygenase may have anti-inflammatory properties. Nat. Med. 1999;5:698–701. doi: 10.1038/9550. [DOI] [PubMed] [Google Scholar]
- Gorla R., Erbel R., Eagle K.A., Bossone E. Systemic inflammatory response syndromes in the era of interventional cardiology. Vasc. Pharmacol. 2018;107:53–66. doi: 10.1016/j.vph.2018.04.003. [DOI] [PubMed] [Google Scholar]
- de Groot N.G., Bontrop R.E. Springer; 2020. COVID-19 Pandemic: Is a Gender-Defined Dosage Effect Responsible for the High Mortality Rate among Males? [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.-j., Ni Z.-y., Hu Y., Liang W.-h., Ou C.-q., He J.-x., et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z.-D., Wang Z.-Y., Zhang S.-F., Li X., Li L., Li C., et al. Aerosol and surface distribution of severe acute respiratory syndrome coronavirus 2 in hospital wards, Wuhan, China, 2020. Emerg. Infect. Dis. 2020;26:1586. doi: 10.3201/eid2607.200885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W.-t., Wan H., Hu L., Chen P., Wang X., Huang Z., et al. Gasdermin D is an executor of pyroptosis and required for interleukin-1β secretion. Cell Res. 2015;25:1285–1298. doi: 10.1038/cr.2015.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck T.G., Ludwig M.S., Frizzo M.N., Rasia-Filho A.A., Homem de Bittencourt Jr PI. Suppressed anti-inflammatory heat shock response in high-risk COVID-19 patients: lessons from basic research (inclusive bats), light on conceivable therapies. Clin. Sci. 2020;134:1991–2017. doi: 10.1042/CS20200596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedayat M., Netea M.G., Rezaei N. Targeting of Toll-like receptors: a decade of progress in combating infectious diseases. Lancet Infect. Dis. 2011;11:702–712. doi: 10.1016/S1473-3099(11)70099-8. [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. Epub 2020 Mar 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K.J., Su I.J., Theron M., Wu Y.C., Lai S.K., Liu C.C., et al. An interferon-γ-related cytokine storm in SARS patients. J. Med. Virol. 2005;75:185–194. doi: 10.1002/jmv.20255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianaro A., Maffia P., Cuzzocrea S., Mazzon E., Santoro M.G., Di Rosa M., et al. 2-Cyclopenten-1-one and prostaglandin J2 reduce restenosis after balloon angioplasty in rats: role of NF-κB. FEBS Lett. 2003;553:21–27. doi: 10.1016/s0014-5793(03)00873-1. [DOI] [PubMed] [Google Scholar]
- Jäättelä M. Overexpression of major heat shock protein hsp70 inhibits tumor necrosis factor-induced activation of phospholipase A2. J. Immunol. 1993;151:4286–4294. [PubMed] [Google Scholar]
- Jäättelä M., Wissing D. Emerging role of heat shock proteins in biology and medicine. Ann. Med. 1992;24:249–258. doi: 10.3109/07853899209149952. [DOI] [PubMed] [Google Scholar]
- Jacquemin C., Rambert J., Guillet S., Thiolat D., Boukhedouni N., Doutre M.S., et al. Heat shock protein 70 potentiates interferon alpha production by plasmacytoid dendritic cells: relevance for cutaneous lupus and vitiligo pathogenesis. Br. J. Dermatol. 2017;177:1367–1375. doi: 10.1111/bjd.15550. [DOI] [PubMed] [Google Scholar]
- Jimenez-Guardeño J.M., Nieto-Torres J.L., DeDiego M.L., Regla-Nava J.A., Fernandez-Delgado R., Castaño-Rodriguez C., et al. The PDZ-binding motif of severe acute respiratory syndrome coronavirus envelope protein is a determinant of viral pathogenesis. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E.S., Choe P.G., Park W.B., Oh H.S., Kim E.J., Nam E.Y., et al. Clinical progression and cytokine profiles of Middle East respiratory syndrome coronavirus infection. J. Korean Med. Sci. 2016;31:1717–1725. doi: 10.3346/jkms.2016.31.11.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komune N., Ichinohe T., Ito M., Yanagi Y. Measles virus V protein inhibits NLRP3 inflammasome-mediated interleukin-1β secretion. J. Virol. 2011;85:13019–13026. doi: 10.1128/JVI.05942-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M., Ludwig M.S., Heck T.G., Takahashi H.K. Heat shock proteins and heat therapy for type 2 diabetes: pros and cons. Curr. Opinion Clin. Nut. Metabolic Care. 2015;18:374–380. doi: 10.1097/MCO.0000000000000183. [DOI] [PubMed] [Google Scholar]
- Kunutsor S.K., Laukkanen T., Laukkanen J.A. Frequent sauna bathing may reduce the risk of pneumonia in middle-aged Caucasian men: the KIHD prospective cohort study. Respir. Med. 2017;132:161–163. doi: 10.1016/j.rmed.2017.10.018. [DOI] [PubMed] [Google Scholar]
- Kunutsor S.K., Laukkanen T., Laukkanen J.A. Sauna bathing reduces the risk of respiratory diseases: a long-term prospective cohort study. Eur. J. Epidemiol. 2017;32:1107–1111. doi: 10.1007/s10654-017-0311-6. [DOI] [PubMed] [Google Scholar]
- Le Chang L.Z., Gong H., Wang L., Wang L. Severe acute respiratory syndrome coronavirus 2 RNA detected in blood donations. Emerg. Infect. Dis. 2020;26:1631. doi: 10.3201/eid2607.200839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin T.C., Wickliffe K.E., Leppla S.H., Moayeri M. Heat shock inhibits caspase-1 activity while also preventing its inflammasome-mediated activation by anthrax lethal toxin. Cell. Microbiol. 2008;10:2434–2446. doi: 10.1111/j.1462-5822.2008.01220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, function, and evolution of coronavirus spike proteins. Ann Rev. Virology. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Liao X., Qian S., Yuan J., Wang F., Liu Y., et al. Community transmission of severe acute respiratory syndrome coronavirus 2, Shenzhen, China, 2020. Emerg. Infect. Dis. 2020;26:1320. doi: 10.3201/eid2606.200239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Ning Z., Chen Y., Guo M., Liu Y., Gali N.K., et al. Aerodynamic analysis of SARS-CoV-2 in two Wuhan hospitals. Nature. 2020;582:557–560. doi: 10.1038/s41586-020-2271-3. [DOI] [PubMed] [Google Scholar]
- Liu Y.-C., Kuo R.-L., Shih S.-R. COVID-19: the first documented coronavirus pandemic in history. Biom. J. 2020;43(4):328–333. doi: 10.1016/j.bj.2020.04.007. (Epub 2020 May 5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Y.-H., Huang Y.-W., Wu Y.-H., Tsai C.-S., Lin Y.-C., Mo S.-T., et al. Selective inhibition of the NLRP3 inflammasome by targeting to promyelocytic leukemia protein in mouse and human. Blood J, Am. Soc. Hematol. 2013;121:3185–3194. doi: 10.1182/blood-2012-05-432104. [DOI] [PubMed] [Google Scholar]
- Logunov D.Y., Dolzhikova I.V., Shcheblyakov D.V., Tukhvatulin A.I., Zubkova O.V., Dzharullaeva A.S., et al. Safety and efficacy of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine: an interim analysis of a randomised controlled phase 3 trial in Russia. Lancet. 2021;397:671–681. doi: 10.1016/S0140-6736(21)00234-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A., Magupalli V.G., Ruan J., Yin Q., Atianand M.K., Vos M.R., et al. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. 2014;156:1193–1206. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Zhao Y., Liu J., He X., Wang B., Fu S., et al. Effects of temperature variation and humidity on the mortality of COVID-19 in Wuhan. medRxiv. 2020 doi: 10.1101/2020.03.15.20036426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man S.M., Karki R., Kanneganti T.D. Molecular mechanisms and functions of pyroptosis, inflammatory caspases and inflammasomes in infectious diseases. Immunol. Rev. 2017;277:61–75. doi: 10.1111/imr.12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthay M.A., Zemans R.L., Zimmerman G.A., Arabi Y.M., Beitler J.R., Mercat A., et al. Acute respiratory distress syndrome (Primer) Nat. Rev. Disease Primers. 2019;5(1):18. doi: 10.1038/s41572-019-0069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor A., Martinon F., De Smedt T., Pétrilli V., Tschopp J. A crucial function of SGT1 and HSP90 in inflammasome activity links mammalian and plant innate immune responses. Nat. Immunol. 2007;8:497–503. doi: 10.1038/ni1459. [DOI] [PubMed] [Google Scholar]
- Medzhitov R. Origin and physiological roles of inflammation. Nature. 2008;454:428–435. doi: 10.1038/nature07201. [DOI] [PubMed] [Google Scholar]
- Miragem A.A., Homem de Bittencourt P.I. Nitric oxide-heat shock protein axis in menopausal hot flushes: neglected metabolic issues of chronic inflammatory diseases associated with deranged heat shock response. Hum. Reprod. Update. 2017;23:600–628. doi: 10.1093/humupd/dmx020. [DOI] [PubMed] [Google Scholar]
- Moreno-Eutimio M.A., López-Macías C., Pastelin-Palacios R. Bioinformatic analysis and identification of single-stranded RNA sequences recognized by TLR7/8 in the SARS-CoV-2, SARS-CoV, and MERS-CoV genomes. Microbes Infect. 2020;22:226–229. doi: 10.1016/j.micinf.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Planillo R., Kuffa P., Martınez-Coló n G., Smith B., Rajendiran T., Núñez G. K+ efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity. 2013;38:1142–1153. doi: 10.1016/j.immuni.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murck H. Symptomatic protective action of glycyrrhizin (Licorice) in Covid-19 infection? Front. Immunol. 2020;11:1239. doi: 10.3389/fimmu.2020.01239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea M.G., Schlitzer A., Placek K., Joosten L.A., Schultze J.L. Innate and adaptive immune memory: an evolutionary continuum in the host’s response to pathogens. Cell Host Microbe. 2019;25:13–26. doi: 10.1016/j.chom.2018.12.006. [DOI] [PubMed] [Google Scholar]
- Newsholme P., de Bittencourt Jr P.I.H. The fat cell senescence hypothesis: a mechanism responsible for abrogating the resolution of inflammation in chronic disease. Curr. Opinion Clin. Nut. Metabolic Care. 2014;17:295–305. doi: 10.1097/MCO.0000000000000077. [DOI] [PubMed] [Google Scholar]
- Nieto-Torres J., Verdiá-Bá guena C., Jimenez-Guardeño J.M., Regla-Nava J.A., Castaño-Rodriguez C., Fernandez-Delgado R., et al. Severe acute respiratory syndrome coronavirus E protein transports calcium ions and activates the NLRP3 inflammasome. Virology. 2015;485:330–339. doi: 10.1016/j.virol.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Torres J.L., DeDiego M.L., Verdiá-Báguena C., Jimenez-Guardeño J.M., Regla-Nava J.A., Fernandez-Delgado R., et al. Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novel CPERE The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. Zhonghua liu xing bing xue za zhi=Zhonghua liuxingbingxue zazhi. 2020;41:145. doi: 10.3760/cma.j.issn.0254-6450.2020.02.003. [DOI] [PubMed] [Google Scholar]
- Oeckinghaus A., Ghosh S. The NF-κB family of transcription factors and its regulation. Cold Spring Harb. Perspect. Biol. 2009;1:a000034. doi: 10.1101/cshperspect.a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly K.M., Auzenbergs M., Jafari Y., Liu Y., Flasche S., Lowe R. Effective transmission across the globe: the role of climate in COVID-19 mitigation strategies. Lancet Planet. Health. 2020;4 doi: 10.1016/S2542-5196(20)30106-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organization WH . World Health Organization; 2014. Infection Prevention and Control of Epidemic-and Pandemic-Prone Acute Respiratory Infections in Health Care. [PubMed] [Google Scholar]
- Organization WH . World Health Organization; 2020. Advice on the Use of Masks in the Context of COVID-19: Interim Guidance, 6 April 2020. [Google Scholar]
- Panda P.K., Arul M.N., Patel P., Verma S.K., Luo W., Rubahn H.-G., et al. Structure-based drug designing and immunoinformatics approach for SARS-CoV-2. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abb8097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines—a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A.B., Verma A. COVID-19 and angiotensin-converting enzyme inhibitors and angiotensin receptor blockers: what is the evidence? Jama. 2020;323:1769–1770. doi: 10.1001/jama.2020.4812. [DOI] [PubMed] [Google Scholar]
- Perregaux D., Gabel C.A. Interleukin-1 beta maturation and release in response to ATP and nigericin. Evidence that potassium depletion mediated by these agents is a necessary and common feature of their activity. J. Biol. Chem. 1994;269:15195–15203. [PubMed] [Google Scholar]
- Pica F., Rossi A., Santirocco N., Palamara A., Garaci E., Santoro M. Effect of combined αIFN and prostaglandin A1 treatment on vesicular stomatitis virus replication and heat shock protein synthesis in epithelial cells. Antivir. Res. 1996;29:187–198. doi: 10.1016/0166-3542(95)00834-9. [DOI] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polanco C., Castañón-González J., Samaniego J., Arabi Y.M., Arifi A.A., Balkhy H.H., Najm H., Aldawood A.S., Ghabashi A., et al. Clinical course and outcomes of critically ill patients with Middle East respiratory syndrome coronavirus infection. Ann. Intern. Med. 2014;160:389–397. doi: 10.7326/M13-2486. [DOI] [PubMed] [Google Scholar]
- Ramaiah A., Arumugaswami V. Insights into cross-species evolution of novel human coronavirus 2019-nCoV and defining immune determinants for vaccine development. bioRxiv. 2020 doi: 10.1101/2020.01.29.925867. [DOI] [Google Scholar]
- de Rivero Vaccari J.C., Dietrich W.D., Keane R.W., de Rivero Vaccari J.P. The inflammasome in times of COVID-19. Front. Immunol. 2020;11:2474. doi: 10.3389/fimmu.2020.583373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruland J. Inflammasome: putting the pieces together. Cell. 2014;156:1127–1129. doi: 10.1016/j.cell.2014.02.038. [DOI] [PubMed] [Google Scholar]
- Sahin U., Muik A., Derhovanessian E., Vogler I., Kranz L.M., Vormehr M., et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH 1 T cell responses. Nature. 2020;586:594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- Sajadi M.M., Habibzadeh P., Vintzileos A., Shokouhi S., Miralles-Wilhelm F., Amoroso A., et al. March 5, 2020. Latitude Analysis to Predict Potential Spread and Seasonality for COVID-19. (Available at SSRN 3550308) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samways D.S.K., Li Z., Egan T.M. Principles and properties of ion flow in P2X receptors. Front. Cell. Neurosci. 2014;8:6. doi: 10.3389/fncel.2014.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansonetti P.J. The innate signaling of dangers and the dangers of innate signaling. Nat. Immunol. 2006;7:1237–1242. doi: 10.1038/ni1420. [DOI] [PubMed] [Google Scholar]
- Santarpia J., Rivera D., Herrera V., Morwitzer M., Creager H., Santarpia G., et al. Aerosol and surface transmission potential of SARS-CoV-2. medRxiv. 2020;3 (Preprint published online June) [Google Scholar]
- Sayedahmed E.E., Elkashif A., Alhashimi M., Sambhara S., Mittal S.K. Adenoviral vector-based vaccine platforms for developing the next generation of influenza vaccines. Vaccines. 2020;8:574. doi: 10.3390/vaccines8040574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieber A.M.P., Ayres J.S. Thermoregulation as a disease tolerance defense strategy. Pathogens Disease. 2016;74 doi: 10.1093/femspd/ftw106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt F.I., Lu A., Chen J.W., Ruan J., Tang C., Wu H., et al. A single domain antibody fragment that recognizes the adaptor ASC defines the role of ASC domains in inflammasome assembly. J. Exp. Med. 2016;213:771–790. doi: 10.1084/jem.20151790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder K., Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- Serhan C.N. The resolution of inflammation: the devil in the flask and in the details. FASEB J. 2011;25:1441–1448. doi: 10.1096/fj.11-0502ufm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha W., Mitoma H., Hanabuchi S., Bao M., Weng L., Sugimoto N., et al. Human NLRP3 inflammasome senses multiple types of bacterial RNAs. Proc. Natl. Acad. Sci. 2014;111:16059–16064. doi: 10.1073/pnas.1412487111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Zhao Y., Wang K., Shi X., Wang Y., Huang H., et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- Shrivastava G., León-Juárez M., García-Cordero J., Meza-Sánchez D.E., Cedillo-Barrón L. Inflammasomes and its importance in viral infections. Immunol. Res. 2016;64:1101–1117. doi: 10.1007/s12026-016-8873-z. [DOI] [PubMed] [Google Scholar]
- Singh I.S., Hasday J.D. Fever, hyperthermia and the heat shock response. Int. J. Hyperth. 2013;29:423–435. doi: 10.3109/02656736.2013.808766. [DOI] [PubMed] [Google Scholar]
- Sun J., Zhu A., Li H., Zheng K., Zhuang Z., Chen Z., et al. Isolation of infectious SARS-CoV-2 from urine of a COVID-19 patient. Emerging Microbes Infections. 2020;9:991–993. doi: 10.1080/22221751.2020.1760144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Wang T., Cai D., Hu Z., Liao H., Zhi L., et al. Cytokine storm intervention in the early stages of COVID-19 pneumonia. Cytokine Growth Factor Rev. 2020;53:38–42. doi: 10.1016/j.cytogfr.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surprenant A., Rassendren F., Kawashima E., North R.A., Buell G. The cytolytic P2Z receptor for extracellular ATP identified as a P2X receptor (P2X7) Science. 1996;272:735–738. doi: 10.1126/science.272.5262.735. [DOI] [PubMed] [Google Scholar]
- Swanson K.V., Deng M., Ting J.P.-Y. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat. Rev. Immunol. 2019;19:477–489. doi: 10.1038/s41577-019-0165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talwar S., Jin J., Carroll B., Liu A., Gillespie M.B., Palanisamy V. Caspase-mediated cleavage of RNA-binding protein HuR regulates c-Myc protein expression after hypoxic stress. J. Biol. Chem. 2011;286:32333–32343. doi: 10.1074/jbc.M111.255927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Shibazaki A., Ono R., Kaisho T. HSP70 mediates degradation of the p65 subunit of nuclear factor κB to inhibit inflammatory signaling. Sci. Signal. 2014;7 doi: 10.1126/scisignal.2005533. [DOI] [PubMed] [Google Scholar]
- Tang T., Lang X., Xu C., Wang X., Gong T., Yang Y., et al. CLICs-dependent chloride efflux is an essential and proximal upstream event for NLRP3 inflammasome activation. Nat. Commun. 2017;8:1–12. doi: 10.1038/s41467-017-00227-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisoncik J.R., Korth M.J., Simmons C.P., Farrar J., Martin T.R., Katze M.G. Into the eye of the cytokine storm. Microbiol. Mol. Biol. Rev. 2012;76:16–32. doi: 10.1128/MMBR.05015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Torre-Minguela C., Mesa del Castillo P., Pelegrín P. The NLRP3 and pyrin inflammasomes: implications in the pathophysiology of autoinflammatory diseases. Front. Immunol. 2017;8:43. doi: 10.3389/fimmu.2017.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres J., Surya W., Li Y., Liu D.X. Protein-protein interactions of viroporins in coronaviruses and paramyxoviruses: new targets for antivirals? Viruses. 2015;7:2858–2883. doi: 10.3390/v7062750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triantafilou K., Hughes T.R., Triantafilou M., Morgan B.P. The complement membrane attack complex triggers intracellular Ca2+ fluxes leading to NLRP3 inflammasome activation. J. Cell Sci. 2013;126:2903–2913. doi: 10.1242/jcs.124388. [DOI] [PubMed] [Google Scholar]
- Tukaj S., Kaminski M. Heat shock proteins in the therapy of autoimmune diseases: too simple to be true? Cell Stress Chaperones. 2019;24:475–479. doi: 10.1007/s12192-019-01000-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrrell D., Barrow I., Arthur J. Local hyperthermia benefits natural and experimental common colds. Br. Med. J. 1989;298:1280–1283. doi: 10.1136/bmj.298.6683.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Made C.I., Simons A., Schuurs-Hoeijmakers J., Van Den Heuvel G., Mantere T., Kersten S., et al. Presence of genetic variants among young men with severe COVID-19. Jama. 2020;324:663–673. doi: 10.1001/jama.2020.13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020;382:1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veras F.P., Pontelli M.C., Silva C.M., Toller-Kawahisa J.E., de Lima M., Nascimento D.C., et al. SARS-CoV-2–triggered neutrophil extracellular traps mediate COVID-19 pathology. J. Exp. Med. 2020;217 doi: 10.1084/jem.20201129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkhratsky D.B., Pelegrín P. 2014. Apoptosis-Associated Speck-Like Protein. [Google Scholar]
- Villar J., Ribeiro S.P., Mullen J., Kuliszewski M., Post M., Slutsky A.S. Induction of the heat shock response reduces mortality rate and organ damage in a sepsis-induced acute lung injury model. Crit. Care Med. 1994;22:914–921. [PubMed] [Google Scholar]
- Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Guo Y., Chu H., Guan Y., Bi J., Wang B. Multiple functions of the RNA-binding protein HuR in cancer progression, treatment responses and prognosis. Int. J. Mol. Sci. 2013;14:10015–10041. doi: 10.3390/ijms140510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Ye L., Ye L., Li B., Gao B., Zeng Y., et al. Up-regulation of IL-6 and TNF-α induced by SARS-coronavirus spike protein in murine macrophages via NF-κB pathway. Virus Res. 2007;128:1–8. doi: 10.1016/j.virusres.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., et al. Detection of SARS-CoV-2 in different types of clinical specimens. Jama. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S.R., Leibowitz J.L. Elsevier; 2011. Coronavirus Pathogenesis. Advances in Virus Research; pp. 85–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells C.R., Stearns J.K., Lutumba P., Galvani A.P. COVID-19 on the African continent. Lancet Infect. Dis. 2020;20(12):1368–1370. doi: 10.1016/S1473-3099(20)30374-1. Epub 2020 May 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widge A.T., Rouphael N.G., Jackson L.A., Anderson E.J., Roberts P.C., Makhene M., et al. Durability of responses after SARS-CoV-2 mRNA-1273 vaccination. N. Engl. J. Med. 2021;384:80–82. doi: 10.1056/NEJMc2032195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27(3):325–328. doi: 10.1016/j.chom.2020.02.001. (Epub 2020 Feb 7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Chen X., Cai Y., Zhou X., Xu S., Huang H., et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern. Med. 2020;180(7):934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Jeyanathan M., Haddadi S., Barra N.G., Vaseghi-Shanjani M., Damjanovic D., et al. Induction of autonomous memory alveolar macrophages requires T cell help and is critical to trained immunity. Cell. 2018;175:1634–1650.e17. doi: 10.1016/j.cell.2018.09.042. [DOI] [PubMed] [Google Scholar]
- Zhang H., Kim Y., Slutsky A. Springer; 1999. Role of Heat Shock Proteins in Cytoprotection. Anaesthesia, Pain, Intensive Care and Emergency Medicine—APICE; pp. 561–570. [Google Scholar]
- Zhang Y., Zhang J., Chen Y., Luo B., Yuan Y., Huang F., et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through potently downregulating MHC-I. biorxiv. 2020 doi: 10.1101/2020.05.24.111823. [DOI] [Google Scholar]
- Zhao C., Zhao W. NLRP3 inflammasome-a key player in host-virus interactions. Front. Immunol. 2020;11:211. doi: 10.3389/fimmu.2020.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M., Tang D., Lechpammer S., Hoffman A., Asea A., Stevenson M.A., et al. Double-stranded RNA-dependent protein kinase (pkr) is essential for thermotolerance, accumulation of HSP70, and stabilization of ARE-containing HSP70 mRNA during stress. J. Biol. Chem. 2002;277:44539–44547. doi: 10.1074/jbc.M208408200. [DOI] [PubMed] [Google Scholar]
- Zheng S., Fan J., Yu F., Feng B., Lou B., Zou Q., et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January–March 2020: retrospective cohort study. Bmj. 2020;369 doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. Epub 2020 Mar 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. (Epub 2020 Jan 24) [DOI] [PMC free article] [PubMed] [Google Scholar]