Abstract
Background
New SARS-CoV-2 variants with increased transmissibility, like B.1.1.7, first detected in England or B.1.351, first detected in South Africa, have caused considerable concern worldwide. In order to contain the spread of these lineages, it is of utmost importance to have rapid, sensitive and high-throughput detection methods at hand.
Methods
A set of RT-qPCR assays was modified for a diagnostic SARS-CoV-2 multiplex assay including detection of the del-HV69/70 and N501Y mutations on the cobas6800 platform. Analytical sensitivity was assessed for both wild-type SARS-CoV-2 and B.1.1.7 lineage by serial dilution. For clinical performance, a total of 176 clinical samples were subjected to the test and results compared to a commercial manual typing-PCR assay and next generation sequencing as gold standard.
Results
The multiplex assay was highly sensitive for detection of SARS-CoV-2 RNA in clinical samples, with an LoD of 6.16 cp/ml (CI: 4.00–8.31). LoDs were slightly higher for detection of the HV69/70 deletion (85.92, CI: 61–194.41) and the N501Y SNP (105.99 cp/ml, CI: 81.59 – 183.66). A total of 176 clinical samples were tested with the assay, including 50 samples containing SARS-CoV-2 of the B.1.1.7 lineage, one containing B.1.351 and 85 non-B.1.1.7/B.1.351 lineage, of which three also harbored a HV69/70 deletion. All were correctly identified by the multiplex assay.
Conclusion
We describe here a highly sensitive, fully automated multiplex PCR assay for the simultaneous detection of the del-HV69/70 and N501Y mutations that can distinguish between B.1.1.7 and other lineages. The assay allows for high-throughput screening for currently relevant variants in clinical samples prior to sequencing.
Keywords: SARS-CoV-2, N501Y, Del-HV69/70, Variants, RT-qPCR, High-throughput
Abbreviations: LOD, limit of detection; SNP, single nucleotide polymorphism; RBD, receptor binding domain; IC, internal control; IVD, in-vitro diagnostic; RFI, relative fluorescence increase; CI, confidence interval; NGS, next generation sequencing
1. Introduction
A number of novel SARS-CoV-2 variants that emerged in fall 2020, some of which show increased transmissibility and possible immune escape, are increasingly attracting worldwide attention [1, 2].
The B.1.1.7 lineage (VOC202012/01, 501Y.V1) first emerged in southern England and is notable for an unusually high number of mutations with no direct common ancestor compared to previous sequences [3, 4]. In particular, non-synonymous spike-gene mutations such as the N501Y SNP (single nucleotide polymorphism) may lead to changes in receptor binding properties [5]. Another variant of concern, B.1.351 (501Y.V2) first described in south Africa, are characterized by the same spike-gene mutation, along with E484K and others [6]. Both have largely replaced previously circulating lineages in their respective areas of origin, indicating improved host adaptation and immune evasion.
As a result of increased awareness in public health and science institutions, there has been a rapidly growing demand for whole genome sequencing worldwide in order to recognize and map the spread of these emerging variants. In this context, the speed and scalability of fully automated RT-PCR can be highly beneficial to pre-screen samples for relevant mutations.
The aim of this study was to create and validate a high-throughput first-line screening assay with the built-in ability to discriminate between relevant SARS-CoV-2 variants, most importantly B.1.1.7, on a fully automated sample-to-result PCR-platform [7]. Inhouse assays have previously been used successfully for SARS-CoV-2 detection and diagnostics with this system via its open mode (Cobas Omni Utility Channel) [8].
2. Multiplex assay setup
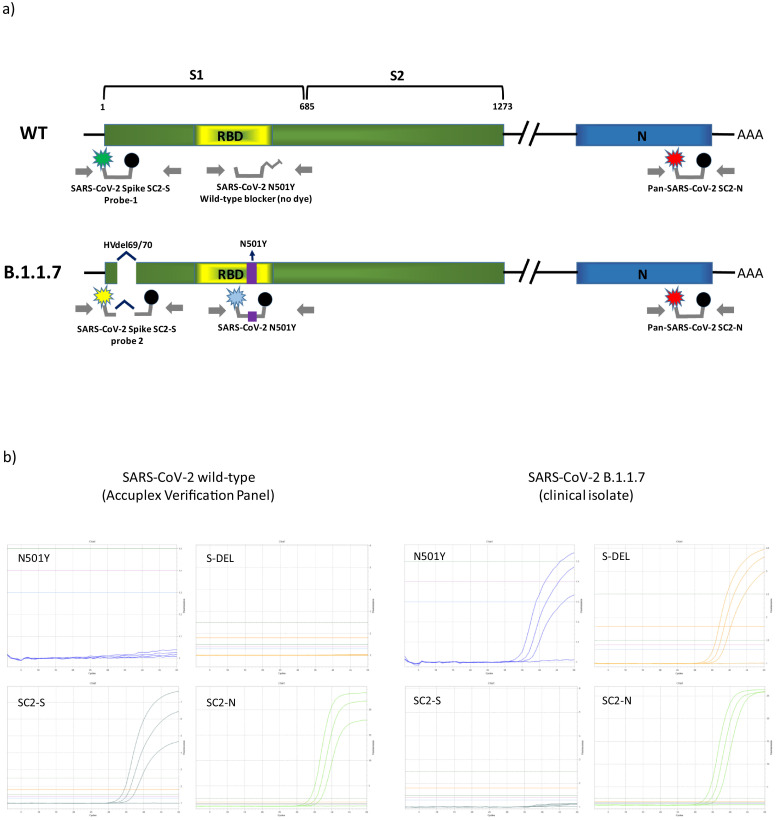
The basic rationale of the multiplex assay was to combine two highly sensitive diagnostic SARS-CoV-2 PCR assays with additional assays to simultaneously detect relevant mutations. The SC2-assay by the US CDC (N-gene, SC2-N) was modified to serve as Pan-SCoV2 target [9]. A publicly available diagnostic S-gene assay by Zhen et al. serves as second target (SC2-S) [10], while also featuring a drop-out phenomenon in the presence of a HV69/70 deletion due to probe location. An additional Taqman-probe (Probe-2, S-Del) was created to match the mutated sequence and allow differentiation for wild-type and del-HV69/70. An additional assay was designed (using Beacon designer and PrimerQuest software) and integrated into the multiplex to detect the N501Y SNP. See Fig. 1 for a schematic overview of the PCRs and their target regions.
Fig. 1.
a) Schematic overview of the primers and probes used in the novel multiplex assay and their target regions within the SARS-CoV-2 lineages (wildtype (WT) and B.1.1.7). SARS-CoV-2 Spike SC2 PCR targets the spike gene (S1) of both lineages, with probe-1 detecting the wildtype sequence and probe-2 being specific for the HV69/70 deletion present in the B.1.1.7 lineage. The SARS-CoV-2 N501YPCR targeting the receptor binding domain (RBD) is specifically designed for detecting the N501Y SNP. Both lineages are detected by the third PCR targeting the N-gene (N). b) Exemplary amplification curves of the SCOV2_VAR_UCT for low concentration SARS-CoV-2 wild-type and B.1.1.7 as seen in the Utility Channel optimization software.
Primers and Probes were added to MMX-R2 to form the MMX-R2 mastermix (see supplementary Table 1) and loaded into cobas omni utility channel cassettes, according to instructions by the manufacturer. Final concentrations of primers and probes are indicated in Table 1 . The utility channel run protocol is outlined in Table 2 .
Table 1.
Primers and probes were custom made by Integrated DNA Technologies (Coralville, USA), Ella Biotech GmbH (Martinsried, Germany) and biomers.net GmbH (Ulm, Germany). Final concentrations of oligonucleotides indicated above refer to concentrations within the final reaction mix. OMe-X, 2′O-methyl-RNA base. BMN-Q620, proprietary dark-quencher by biomers.net, can likely be replaced with e.g. BHQ2. +X, Locked nucleic acid (LNA) base. *Probe-2 of the SARS-CoV-2 Spike assay is not part of Zhen et al.’s original publication and was created as part of this study.
| Assay | Primer/Probe | Sequence (5′ - 3′) | Conc. [nM] | Inclusivity | Ref. |
|---|---|---|---|---|---|
|
Pan-SARS-CoV-2 SC2-N |
Fwd: Rev: Probe: |
5´-CTG CAG ATT TGG ATG ATT TCT (OMe-C)C −3′ 5´- CCT TGT GTG GTC TGC ATG AGT T(Ome-U)A G −3´ 5´- Atto620- ATT GCA ACA (BMN-Q620)ATC CAT GAG CAG TGC TGA CTC -BMN-Q620 −3′ |
400 400 100 |
SARS-CoV-2 any lineage |
[9] |
|
SARS-CoV-2 Spike SC2-S |
Fwd: Rev: Probe-1: Probe-2: |
5´- TCA ACT CAG GAC TTG TTC T(OMe-U)A C −3′ 5´- TGG TAG GAC AGG GTT AT(Ome-C) AAA C −3´ 5´- Fam- TGG TCC CAG (ZEN)AGA CAT GTA TAG CAT -Iowa Black NFQ −3′ 5´- YakYellow- TGG TCC CAG A(+G)A T(+A)G C(+A)T -BHQ1 |
400 400 75 75 |
SARS-CoV-2 Probe-1: HV69/70 WT Probe-2: del-HV69/70 |
[10]* |
| SARS-CoV-2 Spike N501Y | Fwd: Rev: Probe: Blocker: |
5´- CCG GTA GCA CAC CTT G(OMe-U)A AT −3′ 5´- AGT TGC TGG TGC ATG TA(OMe-G) AA −3´ 5´- Atto390- CC(+A) (+A)CC CAC (+T)(+T)(+A) TGG T(+G) -BHQ1 −3′ 5´- CC(+A) (+A)CC CAC (+T)(+A)(+A) TGG T(+G) -C3-Spacer −3′ |
400 400 150 150 |
Spike N501Y | [This study] |
Table 2.
Run protocol for the SCOV2_VAR_UCT assay. Configuration was done with cobas omni Utility Channel software according to instructions by the manufacturer. Material type used was Swab, 400µL of sample is used for NA extraction. RFI (relative fluorescence increase) is used as threshold for automatic calling for results.
| Software settings | |||||
| Sample type | Swab (400 µL) | ||||
| Channels | 1: N501Y | 2: SC2-S | 3: S-DEL | 4: SC2-N | 5: IC |
| RFI | 1.25 | 1.5 | 1.8 | 1.4 | 1.5 |
| PCR cycling conditions | |||||
| UNG incubation | Pre-PCR step | 1st measurement | 2nd measurement | Cooling | |
| No. of cycles | Predefined | 1 | 5 | 45 | Predefined |
| No. of steps | 3 | 2 | 2 | ||
| Temperature | 55 °C; 60 °C; 65 °C | 95 °C; 55 °C | 91 °C; 58 °C | ||
| Hold time | 120 s; 360 s; 240 s | 5 s; 30 s | 5 s; 25 s | ||
| Data acquisition | None | End of each cycle | End of each cycle | ||
The cobas 6800/8800 internal control (IC) is a spike-in (packaged) RNA target, which is automatically added by the system during extraction. MMRX-R2-reagent already contains the internal control assay by default; the respective sequences are not disclosed by the manufacturer. The IC acts as a full process control in the same way as in commercial cobas
6800/8800 IVD tests manufactured by Roche.
3. Evaluation of sensitivity, specificity and clinical performance
For analytical performance evaluation, quantified SARS-COV-2 reference material (SeraCare Accuplex SARS-CoV-2, FluA/B and RSV Verification Panel, Milford, USA) and a patient sample containing B.1.1.7 lineage (according to SARS CoV-2 whole genome sequencing) were used to prepare dilution series in pooled negative patient samples. Absolute quantification of the latter was carried out using the same assay with preexisting linearity data and the Accuplex verification panel as reference on the cobas6800 system [11]. 2-fold dilution series (8 repeats per dilution step) were used to determine lower limit of detection. Probit analysis was carried out using medCalc software (Ostend, Belgium)
To evaluate clinical performance and specificity of differentiation, a set of 136 samples RT-PCR positive for SARS-CoV-2 was subjected to the SCOV2_VAR_UCT. Of these samples, 50 were predetermined as B.1.1.7 lineage by whole genome sequencing. A single B.1.351 sample was simulated by diluting a nucleic acid extract of a patient sample containing NGS-confirmed B.1.351 lineage in UTM. The remaining 85 SARS-CoV-2 samples were constituted of 22 different lineages (allocated as of 25/03/21) with B.1.177.81, B.1.177 and B.1.221 being most abundant. A complete list of the detected lineages can be found in supplementary Table 2. Additionally, all samples were tested with a set of commercial assays to confirm the relevant mutations (TIB MOL, VirSNiP Spike N501Y and Del69/70, Berlin, Germany). A further 40 SARS-CoV-2 negative samples (by cobas SARS-CoV-2 IVD test), and a cross-reactivity panel containing various respiratory pathogens (supplementary Table 4) were tested with the assay to assess overall specificity.
Overall analytic LoD (95% chance of detection) was determined as 6.16 cp/ml (CI: 4.00 – 8.31) (SC2-S LoD: 12.79; CI: 8.50 – 63.67. SC2-N LoD: 9.01, CI 6.37 – 32.13). LoD for a B.1.1.7 lineage sample showed variance less than one log-step (LoD: 27.99, CI: 14.42 – 41.58; S-DEL LoD: 85.92, CI: 61 – 194.41; SC2-N LoD: 27.99; CI: 14.42 – 41.58). Limit for successful detection of an N501Y SNP was 105.99 cp/ml (CI: 81.59 – 183.66), indicating that the N501Y SNP-assay is less sensitive than the two diagnostic-grade SARS-CoV-2 assays. All detection limits are based on manufacturer values (cp/ml) for the Accuplex verification panel as stated above.
Of the 136 SARS-CoV-2 positive sample-set, all were correctly identified by the SCOV2_VAR_UCT. Median of SC2-N for the entire positive-set was 18.60 (IQR 14.63 – 21.80). 50 B1.1.7 lineage samples were classified as positive for N501Y and del-HV69/70. One was detected as N501Y positive and del-HV69/70 negative (B.1.351). 3 of the 85 non-B.1.1.7 samples (B.1.258 lineage) were positive for del-HV69/70, but not N501Y, which was confirmed by the TIB MOL reference assays (Table 3 ). This is in line with existing data showing sporadic occurrences of del-HV69/70 mutations within the endemic SARS-CoV-2 population [12].
Table 3.
Clinical samples (UTM based) were predetermined positive or negative in routine diagnostics (commercial and inhouse methods) and checked for the Del-69/70 and N501Y spike-gene mutations using commercial VirSNiP assays by TIB MOL (Berlin, Germany). Lineages were assigned based on whole genome sequencing. All samples and mutations were correctly detected by the SCOV2_VAR assay.
| SCOV2_VAR_UCT | |||||
|---|---|---|---|---|---|
| Target-1: N501Y |
Target-2: SC2-S |
Target-3: S-Del |
Target-4: SC2-N |
Total | |
|
SARS-CoV-2 positive Non-B.1.1.7, HV69/70 WT |
0/82 | 82/82 | 0/82 | 82/82 | 82 |
| Non-B.1.1.7, Del-HV69/70 | 0/3 | 0/3 | 3/3 | 3/3 | 3 |
| B.1.1.7 lineage | 50/50 | 0/50 | 50/50 | 50/50 | 50 |
| B.1.351 lineage* | 1/1 | 1/1 | 0/1 | 1/1 | 1 |
| SARS-CoV-2 negative | 0/40 | 0/40 | 0/40 | 0/40 | 40 |
| Total Samples | 176 | ||||
4. Discussion and conclusion
Diagnostic labs in Europe are increasingly confronted with demands for rapid differentiation of SARS-CoV-2 isolates due to concerns about potentially higher transmissibility and immune escape with recently emerged lineages such as B.1.1.7 and B1.351 [1, 6]. Previous reports have demonstrated how inclusivity issues of commercial assays such as the “TaqPath RT-PCR COVID-19 kit” (Thermo Fischer Scientific, USA) can coincidentally be used to pre-screen for B.1.1.7 lineage variants because of a drop-out phenomenon in one of the targets associated with del-HV69/70 [12]. However, this method runs the risk of misinterpreting sporadic HV69/70 deletions, of which we found three within the B.1.1.7 negative set. Mutations like N501Y have independently occurred multiple times in areas with high incidence rates, implying advantages in an environment of high background host-immunity. Their relevance may further grow in the context of ongoing vaccination campaigns. Commercial solutions for SARS-CoV-2 SNP detection such as the TIB MOL VirSNiP-kits are in the process of entering the diagnostics market; however, they mostly consist of manual protocols, thus limiting their suitability for large scale application. The SCOV2_VAR_UCT is able to detect both the del-HV69/70 and N501Y mutations, relevant for B.1.1.7, B1.351 and P.1 and further adaptions can be implemented to also pick up new emerging SNPs if necessary. It is self-evident that such methods cannot be a replacement for whole genome sequencing; they do, however, represent a valuable asset to quickly detect clusters and help better direct NGS capacities.
It has to be noted, that the SARS-CoV-2 Spike-gene is a variable region and further mutations are likely to emerge over time, potentially impairing the two spike-gene assays in this multiplex in the future. It is advisable to check contemporary SARS-CoV-2 sequences for mismatches with the provided sequences before using the assay. Furthermore, while SARS-CoV-2 P1 lineage (first detected in Brazil) should be identifiable as suspicious (N501Y positive, del-HV69/70 negative) by the multiplex, we were unable to test this claim as there were no samples available. For B.1.351, only a single sample was tested due to the very limited availability of this lineage. These represent limitations of this study.
In conclusion, the SCOV2_VAR_UCT multiplex presented in this study combines highly sensitive SARS-CoV-2 detection with relatively reliable identification of the B.1.1.7 lineage by detecting two hallmark mutations. Furthermore, other lineages featuring the N501Y SNP, but lacking the deletion, (i.e., B.1.351 and P.1) may also be detected as abnormal for further investigation, though further validation is necessary regarding these two VOCs. It can thus be used either as a secondary assay for pre-screening prior to sequencing, or as a first-line diagnostic assay. Using the cobas6800/8800 automated systems, this setup can be employed for high-throughput screening for B.1.1.7 and other variants, requiring minimal hands-on time.
Author contribution
ML, SP, DN, FO, NF and MA conceptualized and supervised the study. DN and MG performed the experiments. DN, ML, SP, NF, FO, and MA wrote and edited the manuscript. All authors agreed to the publication of the final manuscript.
Declaration of Competing Interest
ML received speaker honoraria and related travel expenses from Roche Diagnostics.
All other authors declare no conflict of interest.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jcv.2021.104894.
Appendix. Supplementary materials
References
- 1.Kirby T. New variant of SARS-CoV-2 in UK causes surge of COVID-19. Lancet Respir. Med. 2021;9:e20–e21. doi: 10.1016/S2213-2600(21)00005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang P., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., Graham B.S., Mascola J.R., Chang J.Y., Yin M.T., Sobieszczyk M., Kyratsous C.A., Shapiro L., Sheng Z., Nair M.S., Huang Y., Ho D.D. Increased resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7 to antibody neutralization. bioRxiv. 2021 doi: 10.1101/2021.01.25.428137:2021.01.25.428137. [DOI] [PubMed] [Google Scholar]
- 3.Volz E., Mishra S., Chand M., Barrett J.C., Johnson R., Geidelberg L., Hinsley W.R., Laydon D.J., Dabrera G., Á O'Toole, Amato R., Ragonnet-Cronin M., Harrison I., Jackson B., Ariani C.V., Boyd O., Loman N.J., McCrone J.T., Gonçalves S., Jorgensen D., Myers R., Hill V., Jackson D.K., Gaythorpe K., Groves N., Sillitoe J., Kwiatkowski D.P., Flaxman S., Ratmann O., Bhatt S., Hopkins S., Gandy A., Rambaut A., Ferguson N.M. Transmission of SARS-CoV-2 Lineage B.1.1.7 in England: insights from linking epidemiological and genetic data. medRxiv. 2021 doi: 10.1101/2020.12.30.20249034:2020.12.30.20249034. [DOI] [Google Scholar]
- 4.Kemp S., Harvey W., Lytras S., Carabelli A., Robertson D., Gupta R. Recurrent emergence and transmission of a SARS-CoV-2 Spike deletion H69/V70. bioRxiv. 2021 doi: 10.1101/2020.12.14.422555:2020.12.14.422555. [DOI] [Google Scholar]
- 5.Luan B., Wang H., Huynh T. Molecular Mechanism of the N501Y mutation for enhanced binding between SARS-CoV-2’s spike protein and human ACE2 receptor. bioRxiv. 2021 doi: 10.1101/2021.01.04.425316:2021.01.04.425316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N., Mlisana K., von Gottberg A., Walaza S., Allam M., Ismail A., Mohale T., Glass A.J., Engelbrecht S., Van Zyl G., Preiser W., Petruccione F., Sigal A., Hardie D., Marais G., Hsiao M., Korsman S., Davies M.-.A., Tyers L., Mudau I., York D., Maslo C., Goedhals D., Abrahams S., Laguda-Akingba O., Alisoltani-Dehkordi A., Godzik A., Wibmer C.K., Sewell B.T., Lourenço J., Alcantara L.C.J., Pond S.L.K., Weaver S., Martin D., Lessells R.J., Bhiman J.N., Williamson C., de Oliveira T. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv. 2020 doi: 10.1101/2020.12.21.20248640:2020.12.21.20248640. [DOI] [Google Scholar]
- 7.Poljak M., Korva M., Knap Gašper N., Fujs Komloš K., Sagadin M., Uršič T., Avšič Županc T., Petrovec M. Clinical evaluation of the cobas SARS-CoV-2 test and a diagnostic platform switch during 48 h in the midst of the COVID-19 pandemic. J. Clin. Microbiol. 2020 doi: 10.1128/jcm.00599-20:JCM.00599-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfefferle S., Reucher S., Nörz D., Lütgehetmann M. Evaluation of a quantitative RT-PCR assay for the detection of the emerging coronavirus SARS-CoV-2 using a high throughput system. Eurosurveillance. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.9.2000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.CDC . CDC Website; 2020. Research Use Only CDC Influenza SARS-CoV-2 (Flu SC2) Multiplex Assay Real-Time RT-PCR Primers and Probes. [Google Scholar]
- 10.Zhen W., Berry G.J. Development of a New multiplex real-time RT-PCR assay for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) detection. J. Mol. Diagn.: JMD. 2020;22:1367–1372. doi: 10.1016/j.jmoldx.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nörz D., Frontzek A., Eigner U., Oestereich L., Fischer N., Aepfelbacher M., Pfefferle S., Lütgehetmann M. Pushing beyond specifications: evaluation of linearity and clinical performance of a fully automated SARS-CoV-2 RT-PCR assay for reliable quantification in blood and other materials outside recommendations. medRxiv. 2020 doi: 10.1101/2020.05.28.20115469:2020.05.28.20115469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bal A., Destras G., Gaymard A., Stefic K., Marlet J., Eymieux S., Regue H., Semanas Q., d'Aubarede C., Billaud G., Laurent F., Gonzalez C., Mekki Y., Valette M., Bouscambert M., Gaudy-Graffin C., Lina B., Morfin F., Josset L., Group CO-DHS Two-step strategy for the identification of SARS-CoV-2 variant of concern 202012/01 and other variants with spike deletion H69-V70, France, August to December 2020. Euro Surveill. 2021;26 doi: 10.2807/1560-7917.ES.2021.26.3.2100008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.