Abstract
The COVID-19 outbreak has catastrophically affected both public health system and world economy. Swift diagnosis of the positive cases will help in providing proper medical attention to the infected individuals and will also aid in effective tracing of their contacts to break the chain of transmission. Blending Artificial Intelligence (AI) with chest X-ray images and incorporating these models in a smartphone can be handy for the accelerated diagnosis of COVID-19. In this study, publicly available datasets of chest X-ray images have been utilized for training and testing of five pre-trained Convolutional Neural Network (CNN) models namely VGG16, MobileNetV2, Xception, NASNetMobile and InceptionResNetV2. Prior to the training of the selected models, the number of images in COVID-19 category has been increased employing traditional augmentation and Generative Adversarial Network (GAN). The performance of the five pre-trained CNN models utilizing the images generated with the two strategies has been compared. In the case of models trained using augmented images, Xception (98%) and MobileNetV2 (97.9%) turned out to be the ones with highest validation accuracy. Xception (98.1%) and VGG16 (98.6%) emerged as models with the highest validation accuracy in the models trained with synthetic GAN images. The best performing models have been further deployed in a smartphone and evaluated. The overall results suggest that VGG16 and Xception, trained with the synthetic images created using GAN, performed better compared to models trained with augmented images. Among these two models VGG16 produced an encouraging Diagnostic Odd Ratio (DOR) with higher positive likelihood and lower negative likelihood for the prediction of COVID-19.
Keywords: Convolutional Neural Network, Deep learning, COVID-19, Chest X-rays, GAN, Smartphone application
1. Introduction
The world is fighting against Corona Virus Disease 2019 (COVID-19) and till date, it has infected 92,983,900 people around the globe and claimed 2,009,781 lives (WHO, 2020). As the disease is fast spreading, the need for rapid diagnosis is of utmost importance during these crucial times. Generally, Reverse Transcription-Polymerase Chain Reaction (RT-PCR) tests are performed to detect the presence of SARS-CoV-2, the virus responsible for COVID-19 (FDA, 2020). Nevertheless, RT-PCR testing is expensive, time consuming, and suffers from low detection rate and sensitivity (Ozturk et al., 2020, Ucar and Korkmaz, 2020, Wang and Wong, 2020). Consequently, radiological imaging such as X-rays and Computed Tomography (CT) scans are being used in conjunction with PCR tests in order to improve diagnosis (Ozturk et al., 2020). However, due to the disparity in pathology and potential chances of human fatigue, there are chances of errors in the manual interpretation of such medical images (Shen, Wu, & Suk, 2017).
In this sense, computer-aided technological interventions, making use of Machine Learning (ML) techniques, have shown to be a fast and reliable alternative for medical image interpretation. In particular, Deep Learning (DL) approaches, implemented with Convolutional Neural Networks (CNNs), have proven to be useful in the detection of bone fractures (Guan, Zhang, Yao, Wang, & Wang, 2020), ostopenia and osteoporosis (Zhang, Yu, & Ning, 2020), bone age (Chen, Li, Zhang, Lu, & Liu, 2020), breast cancer (Ribili et al., 2018, Tsochatzidis et al., 2019, Cao et al., 2019, Abdelhafiz et al., 2019, Agarwal et al., 2020) and pneumonia (Rajpurkar et al., 2017, Wang et al., 2017, Al Mubarok et al., 2019, Islam et al., 2019, Liang and Zheng, 2020). These studies clearly show the usefulness of DL techniques, and in particular CNNs, for image-based medical diagnosis.
Detection of COVID-19 from chest X-ray images has also been addressed through various CNN models. However, in the early phase of the pandemic, studies based on such methods were carried out with a very limited number of chest X-ray images (Hemdan et al., 2020, Sethy et al., 2020), which resulted in model overfitting and less than satisfactory performance. To overcome this limitation, subsequent investigations have been made using augmentation approaches. Ucar and Korkmaz (2020) improved the discrimination rate, between normal, pneumonia and COVID-19 images, from 76.4% to 98.3% with offline augmented images from the COVIDx dataset. Similarly, Farooq (2020), using the same dataset, achieved a 96.23% accuracy on the three classes of images; while Minaee, Kafieh, and Sonka (2020) augmented the number of COVID-19 images from 84 to 450, and reported a 92.2% level of precision.
Loey, Smarandache, and Khalifa (2020) used an alternative approach to solve the issue of dataset shortage by employing Generative Adversarial Network (GAN). With this strategy, the number of images was increased from 270 to 8100 for training the Alexnet, GoogleNet and ResNet18 models. Three case scenarios were tested making use of X-ray images to discriminate COVID-19 class from different combinations of normal, bacterial pneumonia and viral pneumonia classes. Alexnet and ResNet18 achieved 100% testing accuracy for COVID-19 but reported a lower detection accuracy for normal and other pneumonia classes. In another study, Waheed et al. (2020) used GAN for generating synthetic X-ray images of normal and COVID-19 categories. The study reported an improvement of 10% in accuracy and 21% in sensitivity when using GAN generated images over original images. Although there is a reported improvement in performance with GAN generated images, further exploration is required with different CNN architectures along with comparison of diagnostic ability of the trained model using augmented and synthetic images. Table 1 provides a comprehensive summary of the different CNN models, hybrid models, datasets and methods proposed for this problem.
Table 1.
Details of previous studies carried out for the identification of COVID-19 using chest X-ray images employing deep learning models (For studies that have utilized more than 1 models, the one with the highest accuracy has been furnished).
| Work | Number of chest X-rays | Architecture | Accuracy (%) |
|---|---|---|---|
| Hemdan and Shouman (2020) | 25: COVID-19 + 25:Normal | VGG16 | 90 |
| Farooq (2020) | 68: COVID-19 + 1591: Pneumonia + 1203: Normal | COVID-ResNet | 96.23 |
| Loey et al. (2020) | 69: COVID-19 + 79: Normal + 158: Pneumonia | Googlenet | 81.5 |
| Ucar and Korkmaz (2020) | 76: COVID-19 + 1583: Normal + 4290:Pneumonia | Deep Bayes - SqeezeNet | 98.3 |
| Ezzat, Hassanien, and Ella (2020) | 99: COVID-19 + 104: Normal + 80: Pneumonia + 23: Others | GSA-DenseNet121-COVID-19 | 98.38 |
| Sethy et al. (2020) | 127: COVID-19 + 127: Normal + 127: Pneumonia | ResNet50 + SVM | 95.38 |
| Ozturk et al. (2020) | 125: COVID-19 + 500: Pneumonia + 500: Normal | DarkCovidNet | 87.02 |
| Ismael and Şengür (2021) | 180: COVID-19 + 200: Normal | ResNet50 + SVM | 94.7 |
| Luz, Silva, Silva, Silva, and Moreira (2020) | 183: COVID-19 + 8066: Normal + 5521: Pneumonia | EfficientNetB3 | 93.9 |
| Minaee et al. (2020) | 184: COVID-19 + 5000: Non-COVID-19 | SqueezeNet | 92.2 |
| Apostolopoulos and Mpesiana (2020) | 224:COVID-19 + 700:Pneumonia + 504:Normal | VGG19 | 93.48 |
| Brunese, Mercaldo, Reginelli, and Santone (2020) | 250: COVID-19 + 3520: Normal + 500: Pneumonia + 2753: Others | VGG16 | 98 |
| Jain, Mittal, Thakur, and Mittal (2020) | 250: COVID-19 + 315: Normal + 650: Pneumonia | ResNet50 & ResNet101 | 97.77 |
| Toğaçar, Ergen, and Cömert (2020) | 295: COVID-19 + 65: Normal + 98: Pneumonia | Features from combined MobileNetV2 & SqueezeNet | 99.2 |
| Mahmud, Rahman, and Fattah (2020) | 305: COVID-19 + 305: Normal + 610: Pneumonia | CovXNet | 90.2 |
| Narin and Kaya (2020) | 341: COVID-19 + 4265: Pneumonia + 2800: Normal | ResNet50 | 99. 7 |
| Afshar, Heidarian, Naderkhani, Oikonomou, and Plataniotis (2020) | 358: COVID-19 + 5538: Pneumonia + 8066: Normal | COVID-CAPS | 98.3 |
| Wang (2020) | 358: COVID-19 + 5538: Pneumonia + 8066: Normal | COVID-Net | 93.3 |
| Waheed et al. (2020) | 403: COVID-19 + 721: Normal | VGG16 | 95 |
| Karthik, Menaka, and Hariharan (2020) | 558: COVID-19 + 10,434: Normal + 4273: Pneumonia | CSDB CNN + DFL | 97.94 |
Many studies using the above-mentioned deep CNN-based approach need a laptop, PC or cloud based services with significant computing resources. This restricts the usage of such a powerful COVID-19 diagnosis tool to a hospital-type environment and does not provide easy access to all patients. In lower and middle income countries, with inadequate medical facilities, this restriction hinders the effective use of AI based diagnosis (Panth & Acharya, 2015). Implementation of these tools in widely available platforms such as smartphones or tablets can aid in the effective diagnosis even for people living in rural areas (Anderson, Henner, & Burkey, 2013). In such a scenario, a patient can remotely share X-ray images to the physician for AI supported diagnosis. This AI-supported non-contact telemedicine can eliminate the need for the patient to visit the hospital (Panth & Acharya, 2015), which is especially relevant in the COVID-19 scenario for patient and doctor safety. Despite the development of various CNN-based models for COVID-19 diagnosis, the feasibility of implementation of such a model in smartphones or tablets has not been explored widely.
This study proposes to develop a smartphone-based edge computing application for the effective diagnosis of COVID-19 using CNN models. Traditionally, cloud-based computing services are used in case of requirement of higher computing resources. The speed of the cloud based services are also dependent on the network traffic, resource availability, etc., and hence may not be able to provide real-time diagnosis (Hartmann, Hashmi, & Imran, 2019). In contrast, the edge computing technology can operate with low computing resources and does not need cloud-based services. In order to develop the smartphone application, five models which are popular and least explored for COVID diagnosis namely VGG16, Xception MobileNetV2, NASNetMobile and InceptionResNetV2 have been utilized in this study. The performance of the five selected CNN models using the augmented and GAN generated synthetic images have been compared. The best models have been identified using the analysis of selected performances metrics. Finally, a preliminary study has been conducted with the developed smartphone-based application integrating these best models for the screening of X-ray images.
This paper is organized in such a way that Section 2 covers materials and methods which include the details of utilized original dataset, techniques employed to increase the dataset and modifications performed in the selected CNN models for the diagnosis of COVID-19. The encouraging results along with shortcomings and scope for improvement have been discussed in Section 3 followed by concluding remarks in Section 4.
2. Materials and methods
2.1. Dataset
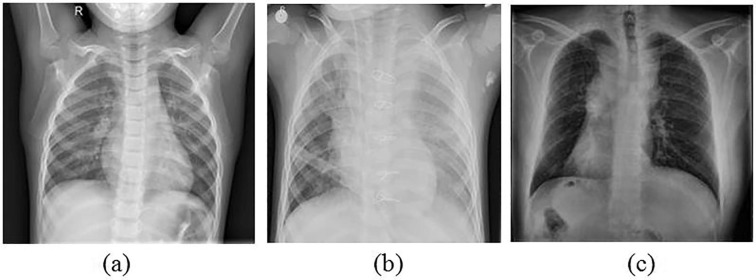
The dataset used for this study primarily consists of X-ray images categorized as normal, pneumonia and COVID-19. The images for the normal and pneumonia categories have been obtained from the Kaggle chest X-ray pneumonia dataset (Kermany et al., 2018, Mooney, 2018) and for COVID-19 category, the images have been obtained from the publicly available COVID chest X-ray image dataset (Cohen & Morrison, 2020). A total of 598 suitable COVID-19 X-ray images have been chosen from this dataset to be used in this study. Sample X-ray images for all the three categories are shown in Fig. 1 . In order to increase the number of images for COVID-19, two strategies have been followed namely, augmentation and GAN for generating synthetic images. These two strategies have been discussed in detail in the following sub-sections.
Fig. 1.
Sample X-ray images of category (a) Normal; (b) Pneumonia and (c) COVID-19.
2.1.1. Augmentation of images
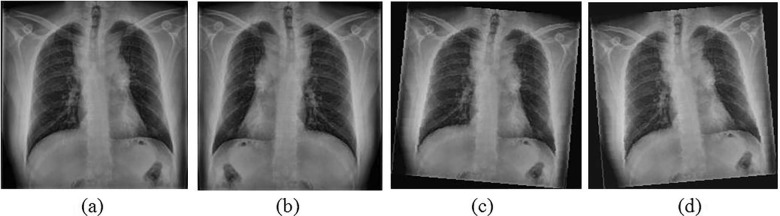
While applying augmentation, the images will be distorted in several ways to introduce uncertainties. This will generate new images with significant dissimilarities from the original images. This process not only increases dataset size but also improves the generalization ability of the CNN (Arvidsson, Overgaard, Astrom, & Heyden, 2019). These distortions can be introduced using numerous transformation operations and addition of noise to the image. In this study, flipping operation has been carried out on each image for generating one set of the mirrored COVID-19 images. In addition, another two sets of images have been created by applying a random rotation of a maximum of 5˚ to either sides. Subsequently, a random intensity value within the range of 20 for each pixel has been added in every image to manipulate the brightness. With these augmentation steps, the number of images increased from 598 to 2344 images. The intensity and rotation values have been selected based on several trials. Sample augmented images of COVID-19 are shown in Fig. 2 .
Fig. 2.
Augmentation applied on sample image (a) Original; (b) Flipped; (c) Rotated left and altered intensity values and (d) Rotated right and altered intensity values.
2.1.2. Generation of synthetic images using GAN
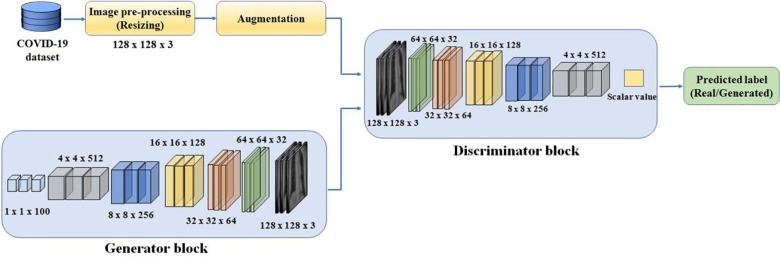
In the second strategy, GAN has been used to generate new synthetic images of COVID-19. GAN consists of a set of CNN layers with each forming the part of the discriminative and generative blocks (Goodfellow, Pouget-Abadie, Mirza, Xu, & Warde-Farley, 2014). The generative block generates images from the random noise as input and discriminative block predicts whether the generated images are real or synthetic based on the input of training data i.e. chest X-ray images of COVID-19.
In this study, a GAN architecture has been developed to generate synthetic X-ray images of dimension 128 × 128 × 3 as shown in Fig. 3 . A separate augmentation has also been performed prior to the delivery of images to the discriminator block to improve the variations in the generated images. The proposed architecture is best suited for generating the images of above dimension with the hardware that has limited computing resources (8 GB RAM with NVIDIA GTX 1050 4 GB GPU).
Fig. 3.
Developed GAN for the generation of synthetic images for COVID-19.
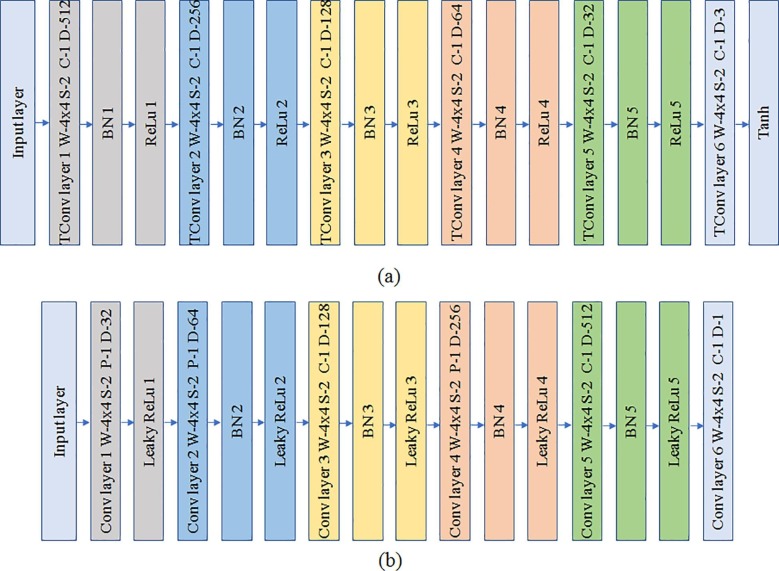
The generator block consists of six transposed convolution layers with a constant filter size of 4 × 4. Rectified Linear unit (ReLu) has been used as the activation function followed by batch normalization for the first five transposed convolution layers whereas, Tanh (hyperbolic tangent function) has been used for the last layer as shown in Fig. 4 . The number of filters for each layer is in the order of 2(4+n) where n ranges from 5 to 1 in a decreasing manner for the first five layers. The last layer has 3 filters corresponding to the 3 color channels of the output image. The input layer accepts an input array of 1 × 1 × 100 random values which is then passed through the transposed convolution layers for generating an output image of dimension 128 × 128 × 3. The discriminator block consists of six convolution layers with leaky ReLu for activation followed by batch normalization for each convolution layer as depicted in Fig. 4. The dimension of filter is a constant value of 4 × 4 similar to the generator block. The number of filters are also similar to the generator but in the increasing order which increases the depth.
Fig. 4.
Architecture of (a) Generator; (b) Discriminator.
Several hyperparameters for generator and discriminator have been configured for training the GAN architecture. The learning rates for the generator and the discriminator have been set to 0.0002 and 0.0001 respectively. For both the blocks, gradient decay factor and squared gradient decay factor have been set to 0.5 and 0.999 respectively. A mini-batch size of 128 has been set for training the generator and discriminator with the validation mini-batch size of 64. As the number of X-ray images for COVID-19 are low, training process has been carried for 1000 epochs with validation performed for every 100 epochs to visualize the generated images during training. The objective is to minimize the negative log likelihood of the generator, during the training process, in order to maximize the predicted probability to real images by the discriminator. Hence the loss function for the generator and discriminator with a mini-batch size m of 128 is given by (Goodfellow et al., 2014) as
| (1) |
| (2) |
Where and are the predicted probability for the real and generated synthetic images by the discriminator.
The weight and bias parameters will be updated during each iteration using the Adams optimizer with the estimated gradients and back propagation. Upon completion of the training process, synthetic images are generated and the number of images under COVID-19 category has been increased from 598 to 2598. The details of number of images in the datasets pertaining to normal & pneumonia categories and those that are created using augmentation & synthetic COVID-19 X-ray images using GAN have been consolidated and furnished in Table 2 .
Table 2.
Dataset utilized for the study.
| Categories | Dataset |
|---|---|
| Normal | 1304 |
| Pneumonia | 3804 |
| Augmented COVID-19 | 2344 |
| Synthetic COVID-19 | 2598 |
The next section discusses the utilized deep learning models and modification of the top layers for tuning the pre-trained models in the process of classification of normal, pneumonia and COVID-19 X-ray images.
2.2. Deep learning models for classification
The use of CNN in medical diagnosis is gaining momentum due to its ability in learning complex feature patterns inevitably making it an effective tool for assisting the physicians. A typical CNN consists of systematically architected stack of layers such as convolutions, ReLu, pooling, batch normalization or other customized layers for capturing the significant feature elements. The learnt features have been built from a simple to complex patterns through these series of convolution layers. It will then be fed to the classification layer that typically consists of fully connected neurons that predict the category to which the image belongs to. As these CNNs are data hungry models, an astonishingly large amount of data is required for training these models. With the limited availability of data using X-ray images, transfer learning approach has been adapted for this study. In transfer learning approach, the previously trained models with large dataset similar to Imagenet have optimized weights and bias parameters. Surprisingly, these pre-trained models have shown to produce unexpected higher performance with relatively limited dataset when configured for other image classification problems. In this study, five pre-trained models namely NASNetMobile, InceptionResNetV2, VGG16, Xception and MobileNetV2 (Chollet, 2017, Sandler et al., 2018, Simonyan and Zisserman, 2015, Szegedy et al., 2017, Zoph et al., 2018) have been used for the development of an AI based tool for the diagnosis of COVID-19 using X-ray images.
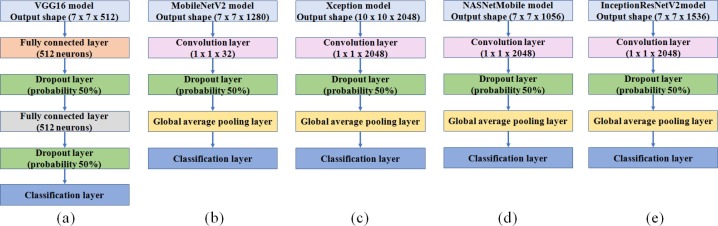
As these pre-trained models are primarily optimized for the recognition of 1000 categories of objects in the Imagenet dataset, a few layers have to be reformed and retrained for the diagnosis using X-ray images. The modification carried out for the five selected pre-trained models are shown in Fig. 5 .
Fig. 5.
Modified CNN models (a) VGG16; (b) MobileNetV2; (c) Xception (d) NASNetMobile and (e) InceptionResNetV2.
In VGG16, two hidden layers each with 512 neurons have been added after several trials of training. In the other modified models, a convolution, dropout and global average pooling layers have been added on the top of the pre-trained models. The features obtained from the convolution layers are directly provided to the classification layers as these architectures have been designed to avoid the need for the several fully connected layer since it exploits the computing resources. The learning rate for the introduced top layers has been set to 0.01 as learnable parameters of these layers are optimized for the classification. The training process has been carried out with 80% of the dataset for training and 20% for validation. The training for the augmented COVID-19 dataset and the generated synthetic images of COVID-19 has been performed separately.
2.3. Experiments
All the training and validation processes for transfer learning have been done in the Google Colab cloud service. The initial experiments have been performed independently with the augmented and synthetic images created using GAN in all the five selected models. The validation accuracy obtained for all the five models have been compared and misclassification among the three categories of X-ray images have been evaluated using the confusion matrix. Further, the performance of the models have been assessed with the performance metrics such as True Positive Rate (TPR), True Negative Rate (TNR), False Positive Rate (FPR), False Negative Rate (FNR), Positive Prediction Value (PPV), Negative Prediction Value (NPV), Positive likelihood (PL), Negative likelihood (NL) and Diagnostic Odd Ratio (DOR) estimated using the following equations (Tharwat, 2018):
| (3) |
| (4) |
| (5) |
| (6) |
| (7) |
| (8) |
| (9) |
| (10) |
| (11) |
TPR and TNR provides an insight on how well the models predict the positive and negative images pertaining to a particular category. The higher FPR or FNR poses risk of false prediction that significantly will affect the model’s performance. PPV provides the ratio of True Positive TP for a category to the total positive prediction for that particular category. NPV is similar to PPV but it considers negatively predicted images for a category. PL and NL can provide a measure on the probability for the occurrence of pneumonia, healthy or COVID-19, if the diagnosis provided by the models is positive or negative for that particular category. DOR is a measure to compare the discriminative capacity of these five different models which will assist in the selection of the best model for diagnosis of COVID-19, pneumonia or normal categories.
Based on the assessment, top two best models each for augmented and synthetic COVID-19 images have been selected and further assessment has been carried out using a separate test dataset with a smartphone application. The performance has been analyzed using several metrics such as memory requirement, prediction time, accuracy based on the trial diagnosis. The discussion on the results with the some of the prominent shortcomings and scope for improvement will be discussed in the following section.
3. Results and discussion
The trained models have been evaluated with the validation dataset and resulting accuracy for each model has been furnished in Table 3 . In the case of models trained using synthetic COVID-19 images, VGG16 and Xception emerged to be the top two performing models with highest validation accuracies of 98.6% and 98.1% respectively. Whereas, Xception and MobileNetV2 reported to be the top two performing models with highest accuracies of 98% and 97.9% respectively, in models trained using augmented COVID-19 images. Further analysis of the models using the performance metrics have been discussed in the subsequent sub-sections.
Table 3.
Validation accuracy for the five different models with different dataset.
| Models | Validation accuracy |
|
|---|---|---|
| Augmented COVID −19 | Synthetic COVID-19 | |
| VGG16 | 97.1% | 98.6% |
| MobileNetV2 | 97.9% | 87.9% |
| Xception | 98% | 98.1% |
| NASNetmobile | 93.5% | 92.7% |
| InceptionResNetV2 | 97.1% | 97.1% |
3.1. Performance of the models using augmented images
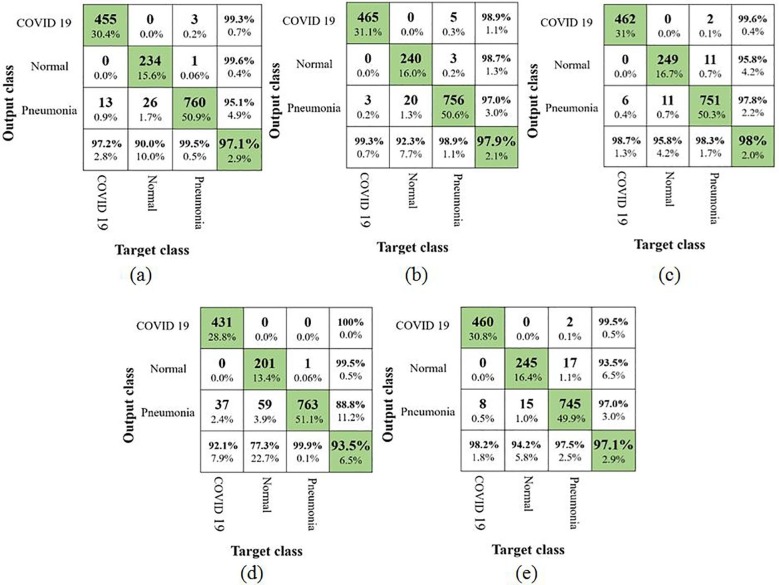
At first, the results of the five models using augmented COVID-19 images have been analyzed using confusion matrix. This will provide an inference on the misclassification of the X-ray images from a particular category to the other category as shown in Fig. 6 . Based on the observation in the confusion matrix, it can be inferred that X-ray images belonging to the normal category have been widely misclassified. It has to be also noted that, the NASNetMobile model had a poor performance as it obtained a TPR of 77.3%. The remaining 22.7% of the X-ray images have been falsely predicted to the pneumonia category. Earlier studies carried out by Loey et al. and Ucar et al. also reported a similar lower results for the normal category using AlexNet, GoogLeNet and ResNet18. In one of the other studies by Waheed et al, the authors have completely neglected pneumonia category and hence they reported a higher accuracy for the detection of normal category from COVID-19. The best TPR of 95.8% for normal category has been reported by Xception in this study. Considering the main intention of this study, i.e. identifying the X-ray images of COVID-19, MobileNetV2 reported the best TPR value of 99.3% followed by Xception with 98.7%. NASNetMobile produced the lowest TPR value of 92.1% and this reveals many false negatives of the X-ray images infected with COVID-19. The confusion matrices of all the five models also reveal that the X-ray images from COVID-19 category have been falsely misclassified as pneumonia but none of them have been predicted to be normal.
Fig. 6.
Confusion matrix using augmented COVID-19 images (a) VGG16; (b) MobileNetV2; (c) Xception; (d) NASNetMobile and (e) InceptionResNetV2.
This suggests that, even though the models were able to learn the distinct patterns that differentiated the images in normal and COVID-19 categories, they failed to some extent in differentiating the characteristics of images in pneumonia category. A few of the other performance metrics estimated using the confusion matrix are furnished in the Table 4 .
Table 4.
Performance metrics for the five models trained with augmented COVID-19 image.
| Models | Class | TPR | TNR | FPR | FNR | PPV | NPV | PL | NL | DOR |
|---|---|---|---|---|---|---|---|---|---|---|
| VGG16 | COVID-19 | 97.2% | 99.7% | 0.3% | 2.8% | 99.3% | 0.7% | 324 | 0.0278 | 11654.6 |
| Pneumonia | 99.5% | 94.6% | 5.4% | 0.5% | 95.1% | 4.9% | 18.55 | 0.0056 | 3312.5 | |
| Normal | 90% | 99.9% | 0.1% | 10% | 99.6% | 0.4% | 1000 | 0.1 | 10,000 | |
| MobileNetV2 | COVID-19 | 99.3% | 99.5% | 0.5% | 0.7% | 98.9% | 1.1% | 202.75 | 0.0065 | 31192.3 |
| Pneumonia | 98.9% | 96.8% | 3.2% | 1.1% | 97% | 3% | 31.31 | 0.0108 | 2899.1 | |
| Normal | 92.3% | 99.7% | 0.3% | 7.7% | 98.7% | 1.3% | 369.20 | 0.0771 | 4788.6 | |
| Xception | COVID-19 Pneumonia Normal |
98.7% 98.3% 95.8% |
99.8% 97.7% 99.1% |
0.2% 2.3% 0.9% |
1.3% 1.7% 4.2% |
99.6% 97.8% 95.8% |
0.4% 2.2% 4.2% |
493.55 42 106.40 |
0.0130 0.0174 0.0427 |
37965.4 2400 2491.8 |
| NASNetmobile | COVID-19 Pneumonia Normal |
92.1% 99.9% 77.3% |
100% 86.8% 99.9% |
0% 13% 0.1% |
7.9% 0.1% 22.7% |
100% 88.8% 99.5% |
0% 11.2% 0.5% |
∞ 7.57 393.3 |
0 1.6120 0.253 |
∞ 4.7 1554.3 |
| InceptionResNet V2 |
COVID-19 Pneumonia Normal |
98.2% 97.5% 94.2% |
99.8% 96.8% 98.6% |
0.2% 3.2% 1.4% |
1.8% 2.5% 5.8% |
99.5% 97% 93.5% |
0.5% 3% 6.5% |
491.45 30.89 68.28 |
0.0171 0.0243 0.0585 |
28739.8 1271.2 1167.2 |
According to Table 4, the TNR values for the pneumonia category is lower compared to the other two categories in all the five models. This suggests that a significant number of X-ray images belonging to the pneumonia category have been falsely predicted to the other categories. Similarly, the X-ray images from other categories have been falsely predicted to pneumonia. The models Xception, InceptionResNetV2 and MobileNetV2 reported higher TNR values of 97.7%, 96.8% and 96.8% respectively, which shows a lower misclassification compared to the other two models. The PPV values for the COVID-19 category evidently show that all the five models are performing significantly better with an average value of 99.46% for the detection of COVID-19.
Further analysis has been carried out using the positive likelihood PL, negative likelihood NL and DOR that can provide an insight on the odds of the existence of particular category, if the models predict an X-ray image to this category. When the values of PL for a category is close to infinity it suggests a strong odd for it to be actually true. Contrastingly, NL values should be close to zero which increases the odds of a category to be negative, if the models predict negative for that particular category. According to the literature, if the PL value is greater than 10 and NL is less than 0.1, then the prediction of the diagnostic model has better discriminative ability (Glas et al., 2003, Šimundić, 2009). All the models employed in this study have significantly higher PL values and lower NL values for prediction of the COVID-19 category X-ray images and this satisfies the conditions stated in the above literature. Further, DOR value can be estimated using the PL and NL values which can then be compared among these models for identifying the best models. From Table 3 it is evident that the DOR value for the detection of COVID-19 ranges from 11654.6 to ∞. The higher values depict a strong discriminative ability that differentiates the X-ray images with COVID-19 symptoms to normal and pneumonia categories. VGG16, MobileNetV2, Xception and InceptionResNetV2 produced the strongest discriminative ability in all the three categories which is evident from the DOR values.
3.2. Performance of the models using synthetic GAN images
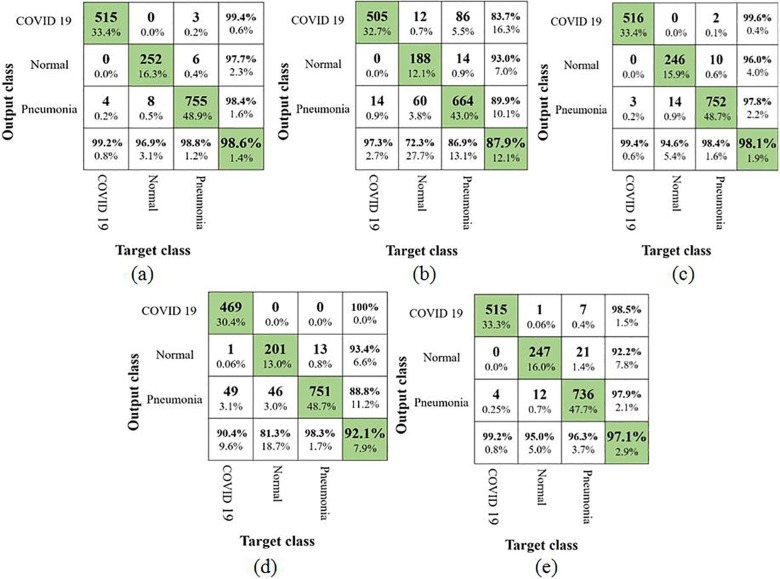
In the second part of this study, an analysis similar to augmented COVID-19 images have been performed in the generated synthetic COVID-19 images with the aid of confusion matrices (as shown in Fig. 7 ) and various other performance metrics. VGG16 and Xception showed an improved performance (i.e. from 97.1% to 98.6% for VGG16 and 98% to 98.1% for Xception)whereas MobileNetV2 and NasNetMobile depicted a drop in performance (i.e. from 97.9% to 87.9% for MobileNetV2 and from 93.5% to 92.1% for NasNetMobile). The accuracy (97.1%) of InceptionResNetV2 remained unchanged.
Fig. 7.
Confusion matrix using synthetic COVID-19 images (a) VGG16; (b) MobileNetV2; (c) Xception; (d) NASNetMobile and (e) InceptionResNetV2.
It was surprising to observe that the classification accuracy using MobileNetV2 dropped significantly mainly due to increase in the misclassification of X-ray images belonging to normal and pneumonia categories. Especially many images of pneumonia category have been misclassified to COVID-19 category in contrast to the results of model trained with augmented COVID-19 images. This shows that the model has lost its discriminative ability significantly for the normal and pneumonia categories.
In the case of VGG16, TPR value for COVID-19 category improved to 99.2% from 97.2% and similarly TPR value for normal category increased from 90% to 96.9%. The TPR value using Xception model also revealed an improvement to 99.4% from 98.7% for COVID-19 category. This shows an improvement in the detection of COVID-19 category with the synthetic images for the above models. Table 5 shows various performance metrics estimated from the confusion matrix.
Table 5.
Performance metrics for the five models trained with synthetic COVID-19 image.
| Models | Class | TPR | TNR | FPR | FNR | PPV | NPV | PL | NL | DOR |
|---|---|---|---|---|---|---|---|---|---|---|
| VGG16 | COVID-19 Pneumonia Normal |
99.2% 98.8% 96.9% |
99.7% 98.4% 99.5% |
0.3% 1.6% 0.5% |
0.8% 1.2% 3.1% |
99.4% 98.4% 97.7% |
0.6% 1.6% 2.3% |
330.73 63.75 206.21 |
0.0078 0.0119 0.0309 |
42401.3 5357.1 6673.5 |
| MobileNetV2 | COVID-19 Pneumonia Normal |
97.3% 86.9% 72.3% |
90.4% 90.5% 98.9% |
9.6% 9.5% 1.1% |
2.7% 13.1% 27.7% |
83.7% 89.9% 93% |
16.3% 10.1% 7% |
10.15 9.14 65.72 |
0.0298 0.1446 0.2800 |
340.6 63.2 234.7 |
| Xception | COVID-19 Pneumonia Normal |
99.4% 98.4% 94.6% |
99.8% 97.8% 99.2% |
0.2% 2.2% 0.8% |
0.6% 1.6% 5.4% |
99.6% 97.8% 96% |
0.4% 2.2% 4% |
497.05 44.94 121.29 |
0.0058 0.0118 0.0544 |
85698.3 3808.5 2229.6 |
| NASNetmobile | COVID-19 Pneumonia Normal |
90.4% 98.3% 81.3% |
100% 87.6% 98.9% |
0% 12.4% 1.1% |
9.6% 1.7% 18.7% |
100% 88.8% 93.4% |
0% 11.2% 6.6% |
∞ 7.90 73.97 |
0.096 0.0195 0.1883 |
∞ 405.1 392.8 |
| InceptionResNet V2 |
COVID-19 Pneumonia Normal |
99.2% 96.3% 95% |
99.2% 97.9% 98.3% |
0.8% 2.1% 1.7% |
0.8% 3.7% 5% |
98.5% 97.9% 92.2% |
1.5% 2.1% 7.8% |
125.59 46.76 55.23 |
0.0078 0.0374 0.0508 |
16101.3 1250.3 1087.2 |
As VGG16, Xception and InceptionResNetV2 revealed an improved detection of COVID-19 category, the DOR values for these models have been compared. It can be observed that, the highest DOR value has been obtained for COVID-19 using Xception when compared to the other three models. The DOR values for VGG16 has been highest for normal and COVID-19 categories. Although the DOR value for NASNetMobile is found to be infinity in both the cases, the lower TPR value suggests higher FNR for the COVID-19 category.
From the above analysis two best models each from the studies using augmented and synthetic COVID-19 images have been identified. In the case of augmented COVID-19 images, MobileNetV2 and Xception have been selected whereas with synthetic COVID-19 images VGG16 and Xception have been selected based on the performance metrics for further deployment in the smartphone.
3.3. Deployment in a smartphone and performance assessment
As the health workers are severely stressed, there are probabilities of false diagnosis of this disease. With wide availability of smartphones, diagnosis of COVID-19 using X-ray images can become easier. But, the deployment of the trained deep learning model directly in a cost-effective smartphone is a challenge as it exploits the computing resources. To counteract this problem, the trained TensorFlow model has been converted to TesnorFlow Lite model using the TensorFlow Lite converter which is an open source tool. The converted model has been then integrated with the Android application by modifying the sample code obtained from the source (https://github.com/tensorflow/examples/blob/master/lite/examples/image_classification/android/EXPLORE_THE_CODE.md.). The process has to be repeated for each model and the input dimensions of the X-ray images have to be modified so as to be implemented in this code. Trials using the smartphone have been performed with the images in the test dataset which consists of 271, 441 and 48 images pertaining to normal, pneumonia and COVID-19 categories respectively. As it is a preliminary study and due to the difficulty in obtaining the actual X-ray images for COVID-19 category, the digitized images on the computer screen have been scanned by the smartphone camera for diagnosis. In addition, the number of available COVID-19 images were limited and hence it has been augmented to produce 48 images for the test dataset.
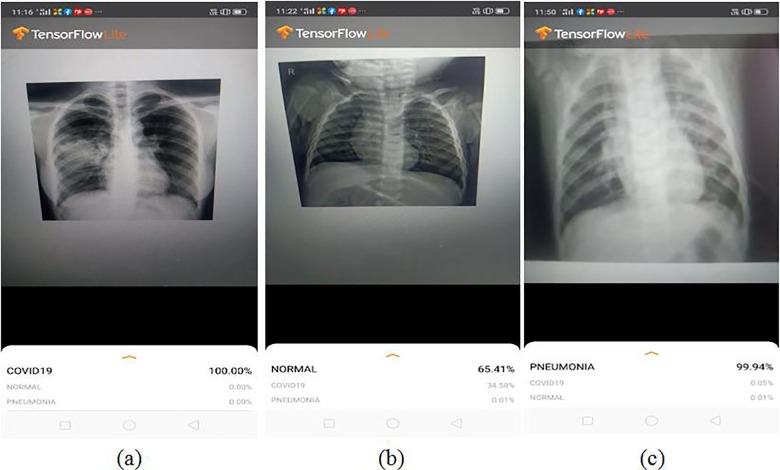
The smartphone used for the study was Realme (Model: RMX 1831) which has 3 GB of RAM with a camera resolution of 13MP. A sample output of the predicted results obtained using the smartphone application is shown in Fig. 8 .
Fig. 8.
A sample screenshot of the diagnosis using X-ray images with the smartphone application integrated with the deep learning model (a) COVID-19; (b) Normal and (c) Pneumonia.
Initially the trials using MobileNetV2 and Xception trained with augmented COVID-19 images detected COVID-19 with an accuracy of 100% for both the models. Similarly, pneumonia has been detected with an accuracy of 97% and 99.3% using MobileNetV2 and Xception respectively. But the performance of the models for the prediction of normal category came out lower which resulted in many false positives to the pneumonia category and a few to the COVID-19 category. In the second case, VGG16 and Xception trained with synthetic COVID-19 images have been evaluated using the smartphone application. Xception and VGG16 resulted in an accuracy of 100% and 97.3% respectively for the detection of COVID-19. Surprisingly, VGG16 resulted in the better performance for detection of the pneumonia category with an accuracy of 100% while Xception produced only 97.7%. VGG16 also depicted an improved discrimination ability for the detection of normal category with decreased false positives to COVID-19. The detection of false positives to COVID-19 will result in patient stress and causes a strain on the already compromised healthcare system. But despite the improved performance produced by VGG16, it consumes more computing resources and higher prediction time as shown in Table 6 .
Table 6.
Comparison of the best performing models integrated with smartphone application.
| Models | Memory space | Average Prediction time | Diagnosis test for COVID-19 |
||
|---|---|---|---|---|---|
| PL | NL | DOR | |||
| MobileNetV2 | 9.84 MB | 169.2 ms | 9.7 | 0 | ∞ |
| Xception | 95.3 MB | 2281.7ms | 27.3 | 0 | ∞ |
| Xception with synthetic image | 10.6 | 0 | ∞ | ||
| VGG16 | 106 MB | 3627.2ms | 97.9 | 0.021 | 4617.9 |
Although the DOR values for all the three models except VGG16 tends to infinity, lower TNR has affected the PL values for the diagnosis of COVID-19. Due to the acceptable performance metrics of VGG16 (trained with synthetic COVID-19 X-ray images) compared to other models, it turned out to be one of the best suitable models for the diagnosis of COVID-19 with the smartphone-based application.
Currently, in its existing form the smartphone-based application is still not suitable for deployment in diagnosis of the COVID-19. There exist some practical issues that may result in false diagnosis due to variations in distance between the smartphone and X-ray images and also due to the effect of various distortions during image acquisition. Camera resolution also contributes significantly to the false diagnosis. Further, although the dataset has been increased artificially, lack of sufficient number of original chest X-ray images pertaining to COVID-19 and other categories have affected the overall performance of the model. Hence a large dataset is required to further improve performance and confidence in its predictions. RT-PCR and antigen tests are the prime methods for diagnosis of COVID-19 in which, the RT-PCR test is considered to be the gold standard. If the performance of the smartphone-based application can be improved by incorporating a large dataset, then it can act as an effective support system for COVID-19 diagnosis. Not many literature discussed in Section 1 have reported the deployment of AI based models on smartphones for scanning chest X-ray images to diagnose COVID-19. Hence this study is a small step in the journey towards development of efficient and reliable smartphone based applications for swift and accurate diagnosis of COVID-19.
4. Conclusion
Higher wait time and low detection rate are the major disadvantages of RT-PCR test. This test may take a couple of days to a week to produce a result and to confirm whether a person is infected with COVID-19 or not, the test needs to be repeatedly conducted. Fast diagnosis will help in moving the infected person to quarantine and provide medical attention in a swift manner. SARS-CoV-2 causes abnormalities in the chest X-rays of the infected individual and hence these X-ray images can be a tool that can be used in conjunction with RT-PCR test for improved diagnosis of COVID-19. But there is a need of qualified experts to understand and analyze these X-rays images. It has been proved by several researchers that, entailing various AI techniques with chest X-rays images can encourage the experts to accurately analyze these images in little time. Moreover, implementing such techniques in widely available platforms like smartphones or tablets can aid in the effective diagnosis of this disease. Hence in this study, a smartphone based system has been developed for scanning chest X-ray images so as to predict whether they belong to normal, pneumonia or COVID-19 categories. Publicly available X-ray image dataset pertaining to normal, pneumonia and COVID-19 categories have been used for the study. But the number of images in COVID-19 category is very less compared to the other two categories. Hence using traditional augmentation strategy and GAN, the number of images in COVID-19 dataset has been increased. The performance using the images generated employing these two strategies has been compared using five pre-trained CNN models namely NASNetMobile, InceptionResNetV2, VGG16, Xception and MobileNetV2. In the case of training with augmented COVID-19 images, MobileNetV2 and Xception provided an overall accuracy of 97.9% and 98% respectively. Xception and VGG16 emerged as models with the highest validation accuracy of 98.1% and 98.6% respectively in the case of models trained with synthetic images generated using GAN. Considering the overall accuracy and other performance metrics, these models have been selected for further deployment in the smartphone. VGG16 trained using the synthetic images, with some compromise in computing resources and prediction time, turned out to be the best suitable model for the diagnosis of COVID-19 using the smartphone-based application. Any technological intervention, big or small, to support the front-end warriors will come handy in this war against COVID-19. This study and its outcome are preliminary steps to assist and encourage qualified experts in providing their service without error and in a timely manner in these times of distress.
CRediT authorship contribution statement
Aravind Krishnaswamy Rangarajan: Conceptualization, Methodology, Software, Formal analysis, Investigation, Writing - original draft. Hari Krishnan Ramachandran: Conceptualization, Validation, Investigation, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
Nil.
Funding details
Nil.
References
- Abdelhafiz D., Yang C., Ammar R., Nabavi S. Deep convolutional neural networks for mammography: Advances, challenges and applications. BMC Bioinformatics. 2019;20(11):75–94. doi: 10.1186/s12859-019-2823-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- P. Afshar S. Heidarian F. Naderkhani A. Oikonomou K.N. Plataniotis A. Mohammadi COVID-CAPS: A Capsule Network-based Framework for Identification of COVID-19 cases from X-ray Images Retrieved from 2020 http://arxiv.org/abs/2004.02696. [DOI] [PMC free article] [PubMed]
- Agarwal R., Díaz O., Yap M.H., Lladó X., Martí R. Deep learning for mass detection in Full Field Digital Mammograms. Computers in Biology and Medicine. 2020;121 doi: 10.1016/j.compbiomed.2020.103774. [DOI] [PubMed] [Google Scholar]
- Al Mubarok A.F., Dominique J.A.M., Thias A.H. Pneumonia detection with deep convolutional architecture. Proceeding - 2019 International Conference of Artificial Intelligence and Information Technology. ICAIIT. 2019;2019:486–489. doi: 10.1109/ICAIIT.2019.8834476. [DOI] [Google Scholar]
- Anderson C., Henner T., Burkey J. Tablet computers in support of rural and frontier clinical practice. International Journal of Medical Informatics. 2013;82(11):1046–1058. doi: 10.1016/j.ijmedinf.2013.08.006. [DOI] [PubMed] [Google Scholar]
- Apostolopoulos I.D., Mpesiana T.A. Covid-19: Automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Physical and Engineering Sciences in Medicine. 2020;43(2):635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvidsson, I., Overgaard, N. C., Astrom, K., & Heyden, A. (2019). Comparison of different augmentation techniques for improved generalization performance for gleason grading. Proceedings - International Symposium on Biomedical Imaging, 2019-April, 923–927. https://doi.org/10.1109/ISBI.2019.8759264.
- Brunese L., Mercaldo F., Reginelli A., Santone A. Explainable deep learning for pulmonary disease and coronavirus COVID-19 Detection from X-rays. Computer Methods and Programs in Biomedicine. 2020;196 doi: 10.1016/j.cmpb.2020.105608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Z. Cao Z. Yang X. Liu et al. Deep Learning Based Lesion Detection For Mammograms Proceedings- IEEE International Conference on Healthcare Informatics (ICHI) 2019 1 3 https://doi.ieeecomputersociety.org/10.1109/ICHI.2019.8904695.
- Chen X., Li J., Zhang Y., Lu Y., Liu S. Automatic feature extraction in X-ray image based on deep learning approach for determination of bone age. Future Generation Computer Systems. 2020;110:795–801. doi: 10.1016/j.future.2019.10.032. [DOI] [Google Scholar]
- F. Chollet Xception: Deep learning with depthwise separable convolutions Proceedings - 30th IEEE Conference on Computer Vision and Pattern Recognition 2017 10.1109/CVPR.2017.195.
- J.P. Cohen P. Morrison L. Dao COVID-19 Image Data Collection Retrieved from 2020 http://arxiv.org/abs/2003.11597.
- Ezzat D., Hassanien A.E., Ella H.A. An optimized deep learning architecture for the diagnosis of COVID-19 disease based on gravitational search optimization. Applied Soft Computing. 2020;106742 doi: 10.1016/j.asoc.2020.106742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- M. Farooq A. Hafeez COVID-ResNet: A Deep Learning Framework for Screening of COVID19 from Radiographs Retrieved from 2020 http://arxiv.org/abs/2003.14395.
- FDA. Coronavirus Testing Basics. (2020). Retrieved July 26, 2020, from https://www.fda.gov/consumers/consumer-updates/coronavirus-testing-basics.
- Glas A.S., Lijmer J.G., Prins M.H., Bonsel G.J., Bossuyt P.M.M. The diagnostic odds ratio: A single indicator of test performance. Journal of Clinical Epidemiology. 2003;56(11):1129–1135. doi: 10.1016/S0895-4356(03)00177-X. [DOI] [PubMed] [Google Scholar]
- I.J. Goodfellow J. Pouget-Abadie M. Mirza B. Xu D. Warde-Farley S. Ozair et al. Generative adversarial nets Advances in Neural Information Processing Systems 3 January 2014 2672 2680 Retrieved from https://arxiv.org/pdf/1406.2661.pdf.
- Guan B., Zhang G., Yao J., Wang X., Wang M. Arm fracture detection in X-rays based on improved deep convolutional neural network. Computers and Electrical Engineering. 2020;81 doi: 10.1016/j.compeleceng.2019.106530. [DOI] [Google Scholar]
- Hartmann M., Hashmi U.S., Imran A. Edge computing in smart health care systems: Review, challenges, and research directions. Trans Emerging Tel Technology. 2019;3710 doi: 10.1002/ett.3710. [DOI] [Google Scholar]
- E.-E.-D. Hemdan M.A. Shouman M.E. Karar COVIDX-Net: A Framework of Deep Learning Classifiers to Diagnose COVID-19 in X-Ray Images Retrieved from 2020 http://arxiv.org/abs/2003.11055.
- Islam, S. R., Maity, S. P., Ray, A. K., & Mandal, M. (2019). Automatic Detection of Pneumonia on Compressed Sensing Images using Deep Learning. 2019 IEEE Canadian Conference of Electrical and Computer Engineering, CCECE 2019. https://doi.org/10.1109/CCECE.2019.8861969.
- Ismael A.M., Şengür A. Deep Learning Approaches for COVID-19 Detection Based on Chest X-ray Images. Expert Systems with Applications. 2021;164 doi: 10.1016/j.eswa.2020.114054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain G., Mittal D., Thakur D., Mittal M.K. A deep learning approach to detect Covid-19 coronavirus with X-Ray images. Biocybernetics and Biomedical Engineering. 2020;40(4):1391–1405. doi: 10.1016/j.bbe.2020.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthik R., Menaka R., Hariharan M. Learning distinctive filters for COVID-19 detection from chest X-ray using shuffled residual CNN. Applied Soft Computing Journal. 2020;106744 doi: 10.1016/j.asoc.2020.106744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermany D.S., Goldbaum M., Cai W., Valentim C.C.S., Liang H., Baxter S.L., et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172(5):1122–1131.e9. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- Liang G., Zheng L. A transfer learning method with deep residual network for pediatric pneumonia diagnosis. Computer Methods and Programs in Biomedicine. 2020;187 doi: 10.1016/j.cmpb.2019.06.023. [DOI] [PubMed] [Google Scholar]
- Loey M., Smarandache F., Khalifa N.E.M. Within the lack of chest COVID-19 X-ray dataset: A novel detection model based on GAN and deep transfer learning. Symmetry. 2020;12(4):651. doi: 10.3390/SYM12040651. [DOI] [Google Scholar]
- E. Luz P.L. Silva R. Silva L. Silva G. Moreira D. Menotti Towards an Effective and Efficient Deep Learning Model for COVID-19 Patterns Detection in X-ray Images Retrieved from 2020 http://arxiv.org/abs/2004.05717.
- Mahmud T., Rahman M.A., Fattah S.A. CovXNet: A multi-dilation convolutional neural network for automatic COVID-19 and other pneumonia detection from chest X-ray images with transferable multi-receptive feature optimization. Computers in Biology and Medicine. 2020;122 doi: 10.1016/j.compbiomed.2020.103869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minaee S., Kafieh R., Sonka M., et al. Deep-COVID: Predicting COVID-19 from chest X-ray images using deep transfer learning. Medical Image Analysis. 2020;65 doi: 10.1016/j.media.2020.101794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- P. Mooney Chest X-Ray Images (Pneumonia) | Kaggle Retrieved August 3, 2020, from https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia/data 2018.
- A. Narin C. Kaya Z. Pamuk Automatic Detection of Coronavirus Disease (COVID-19) Using X-ray Images and Deep Convolutional Neural Networks Retrieved from 2020 http://arxiv.org/abs/2003.10849. [DOI] [PMC free article] [PubMed]
- Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Rajendra Acharya U. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Computers in Biology and Medicine. 2020;121 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panth M., Acharya A.S. The unprecedented role of computers in improvement and transformation of public health: An emerging priority. Indian Journal of Community Medicine. 2015;40(1):8–13. doi: 10.4103/0970-0218.149262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- P. Rajpurkar J. Irvin K. Zhu B. Yang H. Mehta T. Duan et al. CheXNet: Radiologist-Level Pneumonia Detection on Chest X-Rays with Deep Learning Retrieved from 2017 http://arxiv.org/abs/1711.05225.
- Ribili D., Horváth A., Unger Z., Pollner P., Csabai I. Detecting and classifying lesions in mammograms with Deep Learning. Scientific Reports. 2018;8:4165. doi: 10.1038/s41598-018-22437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler M., Howard A., Zhu M., Zhmoginov A., Chen L.C. Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition. 2018. MobileNetV2: Inverted Residuals and Linear Bottlenecks; pp. 4510–4520. [DOI] [Google Scholar]
- Sethy P.K., Behera S.K., Ratha P.K., Biswas P. Detection of Coronavirus Disease (COVID-19) Based on Deep Features. International Journal of Mathematical, Engineering and Management Sciences. 2020;5(4):643–651. doi: 10.20944/preprints202003.0300.v1. [DOI] [Google Scholar]
- Shen D., Wu G., Suk H. Deep learning in medical image analysis. Annual Review of Biomedical Engineering. 2017;19:221–248. doi: 10.1146/annurev-bioeng-071516-044442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonyan K., Zisserman A. 3rd International Conference on Learning Representations, ICLR 2015 - Conference Track Proceedings. Retrieved from. 2015. Very deep convolutional networks for large-scale image recognition. [Google Scholar]
- A.-M. Šimundić Measures of Diagnostic Accuracy: Basic Definitions EJIFCC 19 4 2009 203 211 Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/27683318. [PMC free article] [PubMed]
- Szegedy C., Ioffe S., Vanhoucke V., Alemi A.A. 31st AAAI Conference on Artificial Intelligence. 2017. Inception-v4, inception-ResNet and the impact of residual connections on learning. [Google Scholar]
- Tharwat A. Classification assessment methods. Applied Computing and Informatics. 2018 doi: 10.1016/j.aci.2018.08.003. [DOI] [Google Scholar]
- Toğaçar M., Ergen B., Cömert Z. COVID-19 detection using deep learning models to exploit Social Mimic Optimization and structured chest X-ray images using fuzzy color and stacking approaches. Computers in Biology and Medicine. 2020;121 doi: 10.1016/j.compbiomed.2020.103805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsochatzidis L., Costaridou L., Pratikakis I. Deep Learning for Breast Cancer Diagnosis from Mammograms—A Comparative Study. Journal of Imaging. 2019;5(37):1–11. doi: 10.3390/jimaging5030037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ucar F., Korkmaz D. COVIDiagnosis-Net: Deep Bayes-SqueezeNet based diagnosis of the coronavirus disease 2019 (COVID-19) from X-ray images. Medical Hypotheses. 2020;140 doi: 10.1016/j.mehy.2020.109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waheed A., Goyal M., Gupta D., Khanna A., Al-Turjman F., Pinheiro P.R. CovidGAN: Data Augmentation Using Auxiliary Classifier GAN for Improved Covid-19 Detection. IEEE Access. 2020;8:91916–91923. doi: 10.1109/ACCESS.2020.2994762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L. Wang A. Wong COVID-Net: A Tailored Deep Convolutional Neural Network Design for Detection of COVID-19 Cases from Chest X-Ray Images Retrieved from 2020 http://arxiv.org/abs/2003.09871. [DOI] [PMC free article] [PubMed]
- Wang X., Peng Y., Lu L., Lu Z., Bagheri M., Summers R.M. Proceedings - 30th IEEE Conference on Computer Vision and Pattern Recognition. 2017. ChestX-ray8: Hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. [DOI] [Google Scholar]
- WHO. WHO Coronavirus Disease (COVID-19) Dashboard. (2020). Retrieved January 17, 2021, from https://covid19.who.int/.
- Zhang B., Yu K., Ning Z., et al. Deep learning of lumbar spine X-ray for osteopenia and osteoporosis screening: A multicenter retrospective cohort study. Bone. 2020;140 doi: 10.1016/j.bone.2020.115561. [DOI] [PubMed] [Google Scholar]
- Zoph B., Vasudevan V., Shlens J., Le Q.V. Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition. 2018. Learning Transferable Architectures for Scalable Image Recognition; pp. 8697–8710. [DOI] [Google Scholar]