Abstract
Background
Point-of-care tests (POCT) are promising tools to detect SARS-CoV-2 in specific settings. Initial reports suggest the ID NOW™ COVID-19 assay (Abbott Diagnostics Inc, USA) is less sensitive than standard real-time reverse transcription polymerase chain reaction (rRT-PCR) assays. This has raised concern over false negatives in SARS-CoV-2 POCT.
Objectives
We compared the performance of the ID NOW™ COVID-19 assay to our in-house rRT-PCR assay to assess whether dry swabs used in ID NOW™ testing could be stored in transport media and be re-tested by rRT-PCR for redundancy and to provide material for further investigation.
Methods
Paired respiratory swabs collected from patients at three acute care hospitals were used. One swab in transport media (McMaster Molecular Media (MMM)) was tested for SARS-CoV-2 by a laboratory-developed two-target rRT-PCR assay. The second was stored dry in a sterile container and tested by the ID NOW™ COVID-19 assay. Following ID NOW™ testing, dry swabs were stored in MMM for up to 48 h and re-tested by rRT-PCR. Serially diluted SARS-CoV-2 particles were used to assess the impact of heat inactivation and storage time.
Results
Respiratory swabs (n = 343) from 179 individuals were included. Using rRT-PCR results as the comparator, the ID NOW™ COVID-19 assay had positive (PPA) and negative (NPA) percent agreements of 87.0% (95% CI:0.74–0.94) and 99.7% (95% CI:0.98–0.99). Re-tested swabs placed in MMM following ID NOW testing had PPA and NPA of 88.8% (95% CI:0.76–0.95) and 99.7% (95% CI:0.98–0.99), respectively.
Conclusions
Storing spent dry swabs in transport media for redundancy rRT-PCR testing is a potential approach to address possible false negatives with the ID NOW™ COVID-19 assay.
Keywords: Point-of-care testing, SARS-CoV-2, Abbott ID NOW™ COVID-19, COVID-19
1. Introduction
The emergence and spread of a novel human coronavirus, SARS-CoV-2, has resulted in over 110,000,000 cases and 2440,000 deaths due to coronavirus-19 disease (COVID-19) as of February 20th, 2021 [1]. Laboratory confirmation of SARS-CoV-2 infection is made using real- time reverse transcription polymerase chain reaction (rRT-PCR) assays that amplify and detect specific regions of the SARS-CoV-2 genome [2]. While these assays are the gold standard for SARS-CoV-2 diagnostic testing, they are complex, requiring hours to complete and sufficient technological expertise and equipment that is often only available in accredited diagnostic laboratories.
Limiting SARS-CoV-2 spread is dependent on the rapid identification of individuals who are communicable. Diagnostic methods with short turnaround times are desirable in certain settings, such as in outbreak investigations or in remote areas where rRT-PCR testing is not readily available [3]. The ID NOW™ COVID-19 assay (Abbott Diagnostics Inc, USA), is a rapid (<15 min) molecular in vitro diagnostic test that uses isothermal nucleic acid amplification technology to qualitatively detect SARS-CoV-2 viral RNA directly from nasal, throat or nasopharyngeal (NP) swabs [4]. To date, the assay has demonstrated variable performance, with reported sensitivities ranging from 45% to 94% [[5], [6], [7], [8], [9], [10]]. A recent systematic review noted an average sensitivity of 76.8% (95% CI: 72.9–80.3%) and a specificity of 99.6% (95% CI: 98.4–99.9%) across 5 independent evaluations [9]. For optimal performance of the ID NOW™ COVID-19 assay, the manufacturer recommends that respiratory swabs be tested as soon as possible after collection and not be stored in viral transport media (VTM). It is acceptable for swabs to be held for up to one hour at room temperature prior to testing. Restrictive transport and storage requirements and low sensitivity may limit the use of SARS-CoV-2 POCT on a wider scale. As this assay is designed for use at the point-of-care, testing may be performed by healthcare providers that are not experienced working with live virus. Prior to rRT-PCR assays, SARS-CoV-2 is typically inactivated using heat (65°C for 30 min) or lysis solutions to avoid the biosafety risks associated with aerosol generation [11–13]. This combination of variables presents logistical challenges for specimens requiring transport between sites and could lead to COVID-19 cases evading laboratory detection, requiring a second swab that adds to patient discomfort and laboratory testing volumes.
In this study we (i) compared the performance of the ID NOW™ COVID-19 assay to rRT-PCR via parallel testing of two swabs collected simultaneously from symptomatic and asymptomatic individuals, (ii) assessed whether spent swabs from the ID NOW™ assay could be stored in a viral lysis medium and be re-tested by rRT-PCR, and (iii) examined the effects of heat inactivation, prolonged storage time, and the use of viral lysis media on ID NOW™ COVID-19 assay results.
2. Materials and methods
2.1. Specimen collection
Two NP, mid-turbinate or anterior nasal swabs were collected simultaneously from individuals who presented to a COVID-19 assessment center or were hospitalized for COVID-19 disease at three acute care hospitals in Toronto, Canada between December 9th 2020 and January 8th, 2021. This study met the criteria of a quality improvement project in accordance with institutional guidelines. One swab was stored in McMaster Molecular Medium (MMM) (Bay Area Health Trustee Corp, Canada), a guanidine thiocyanate-based viral inactivation medium for rRT-PCR testing [12], and the second was kept dry in a sterile 50 mL conical tube (Corning Inc., USA) for ID NOW™ testing. Individuals collecting specimens were advised to store the dry swabs for 1 hour at room temperature or 2–8 °C for up to 24 h if delays in transport were anticipated.
2.2. Molecular detection of SARS-CoV-2 by rRT-PCR, ID NOW™ COVID-19 assay and secondary rRT-PCR on used dry swabs
All SARS-CoV-2 testing was performed at the Shared Hospital Laboratory (Toronto, Canada). Real-time RT-PCR testing to detect the E gene and UTR of SARS-CoV-2 was performed as previously described [11,12]. Testing with the ID NOW™ COVID-19 assay was performed as per manufacturer's recommendations (Abbott Diagnostics Inc, USA), with dry swabs being inoculated into the elution buffer within the Sample Receiver cartridge. For secondary rRT-PCR testing, dry swabs that were mixed in the ID NOW™ Sample Receiver buffer were subsequently stored in MMM lysis media. Results from ID NOW™ COVID-19 and subsequent rRT-PCR tests were for research use only and were performed blinded to the initial rRT-PCR result. Sample collection time and time of testing were recorded.
2.3. Effect of heat inactivation, storage time and transport medium on SARS-CoV-2 detection by ID NOW™
All in vitro culture of SARS-CoV-2 was performed in a Biosafety Level 3 facility using techniques described previously [14]. Serial dilutions of cultured SARS-CoV-2 (Vero E6 cells) were used to create contrived positive control specimens to assess whether heat inactivation and storage time can impact the ID NOW™ COVID-19 assay result. For heat-inactivation experiments, purified SARS-CoV-2 particles (50 µL of a 102 TCID50 stock of heat-inactivated SARS-CoV-2) were inoculated onto 11 replicate swabs, held at room temperature for 30 min in a 50 mL conical tube (Sigma-Aldrich, Ontario, Canada) and then incubated at 65 °C for 30 min. Ten of the swabs (HK-1 to HK-10) were then tested by the ID NOW™ COVID-19 assay and placed in MMM for later rRT-PCR testing, while the eleventh (HK-REF) was placed directly into MMM only for rRT-PCR testing. For storage time experiments, purified SARS-CoV-2 particles (50 µL of a 103 TCID50 stock of heat-inactivated SARS-CoV-2) were directly inoculated onto 10 replicate swabs as described above and held at room temperature for 2.5 to 4.2 h. Nine of the swabs (RT-1 to RT-9) were tested by the ID NOW™ COVID-19 assay and placed in MMM for later rRT-PCR testing, while the tenth (RT-10) was placed directly into MMM for rRT-PCR testing only.
To determine compatibility of universal transport media (UTM) with the ID NOW™ COVID-19 assay, an additional set of heat-inactivated patient respiratory swabs in UTM were tested on the ID NOW™ instrument according to manufacturer instructions. Specimens were refrigerated (4–8 °C) for 24 to 72 h prior to testing.
2.4. SARS-CoV-2 inactivation
To verify that the ID NOW™ elution buffer sufficiently inactivates SARS-CoV-2, a 1:5 dilution of 9.28 × 107 TCID50/mL of SARS-CoV-2 particles was made in elution buffer and held for 30 s. For comparison, an equivalent amount of SARS-CoV-2 particles were added to the same amount of Dulbecco's Modified Eagle Medium (DMEM) (Gibco, Mississauga, Canada) while 100μL of DMEM media was added to 400μL of elution buffer to create positive and negative controls, respectively. All three solutions were serially diluted with DMEM media and added to Vero E6 cells, starting with the 1:1000 dilution, in a viral outgrowth assay. Plates were evaluated for cytopathic effect after 14 days.
2.5. Statistical analysis
All statistical analyses were performed using GraphPad Prism v 9.0.1 (Graphpad Software LLC, USA). Percent positive and negative agreement, and Cohen's Kappa were calculated using the laboratory developed rRT-PCR as the reference standard [11,15]. For experiments comparing pre-treatments, differences in threshold cycles (Ct) between treatments were tabulated by comparing Ct values. A significant change in Ct identified as those with a 1 log difference (approximately 3.3 cycles).
3. Results
3.1. Performance of the ID NOW™ COVID-19 assay
A total of 343 respiratory swabs were collected from 179 individuals (n = 255 swabs from symptomatic and n = 88 from asymptomatic) during the study period and produced valid results. Swab types included NP (n = 92), midturbinate (n = 223) or anterior nasal (n = 28). The overall positive (PPA) and negative percent agreements (NPA) between the ID NOW™ COVID-19 and rRT-PCR assays were 87.0% (95% CI:0.74–0.94) and 99.7% (95% CI:0.98–0.99), respectively (Table 1 ). High inter-method agreement was noted by a kappa coefficient of 0.91 (95% CI: 0.84–0.97). Performance was highest among anterior nasal and midturbinate swabs (Table 1). Fifteen (4.1%) swabs tested by ID NOW™ were invalid and not included in the analysis. The median time between sample collection and ID NOW™ testing was 2.4 h (interquartile range: 3.5 to 5.9 h). The limit of detection for the ID NOW™ COVID-19 assay was determined as 1 TCID50/mL based on serially diluted cultured virus [14]. There was a positive predictive value of 97.6% (95% CI: 0.87–0.99) and negative predictive value of 98.0% (95% CI: 0.96–0.99). We noted a >6-log decrease in the recovery of SARS-CoV-2 virions after 30 s of contact time in ID NOW™ COVID-19 elution buffer (Data not shown).
Table 1.
Performance comparison of the ID NOW COVID-19 assay and a laboratory developed rRT-PCR assay to detect SARS-CoV-2 from paired or re-used ID NOW swabs.
| Swab type | rRT-PCR | PPA (95% CI) | NPA (95% CI) | |
|---|---|---|---|---|
| Positive | Negative | |||
| ID NOW (Paired ID NOW and MMM swabs) | ||||
| Anterior nasal swab (n = 28) | ||||
| Positive | 6 | 0 | 100.0% | 100.0% |
| Negative | 0 | 22 | (0.61–1.0) | (0.85–1.0) |
| Midturbinate swab (n = 223) | ||||
| Positive | 18 | 1 | 94.7% | 99.5% |
| Negative | 1 | 203 | (0.75–0.99) | (0.97–0.99) |
| Nasopharyngeal swab (n = 92) | ||||
| Positive | 16 | 0 | 76.2% | 100.0% |
| Negative | 5 | 71 | (0.54–0.89) | (0.95–1.0) |
| Overall (n = 343) | ||||
| Positive | 40 | 1 | 86.9% | 99.7% |
| Negative | 6 | 296 | (0.74–0.94) | (0.98–0.99) |
| ID NOW (Re-used ID NOW swab in MMM) | ||||
| Anterior nasal swab (n = 28) | ||||
| Positive | 6 | 0 | 100.0% | 100.0% |
| Negative | 0 | 22 | (0.61–1.0) | (0.85–1.0) |
| Midturbinate swab (n = 223) | ||||
| Positive | 18 | 1 | 90.0% | 99.5% |
| Negative | 2 | 202 | (0.69–0.98) | (0.97–0.99) |
| Nasopharyngeal swab (n = 92) | ||||
| Positive | 16 | 0 | 84.2% | 100.0% |
| Negative | 3 | 73 | (0.68–0.94) | (0.95–1.0) |
| Overall (n = 343) | ||||
| Positive | 40 | 1 | 88.8% | 99.7% |
| Negative | 5 | 297 | (0.76–0.95) | (0.98–0.99) |
3.2. Effect of heat inactivation and storage time on SARS-CoV-2 detection
Heat inactivation of the dry swab did not impact SARS-CoV-2 detection by either the ID NOW™ COVID-19 assay or downstream rRT-PCR (Table 2 ). We similarly noted that dry swabs remained positive after being held at room temperature for up to 4.2 h. There was no significant difference in Ct values for secondary rRT-PCR testing between the shortest (t = 2.5 h) and longest (t = 4.2 h) room temperature storage times (ΔCt E gene = 0.26 cycles, ΔCt UTR= 0.10 cycles), however, Ct values were highly variable within this time frame despite the same starting viral inoculum (Table 2).
Table 2.
Effect of heat inactivation and storage temperature on SARS-CoV-2 detection.
| Parameter | Heat-inactivationa | Duration of storage at room temperatureb |
|---|---|---|
| No. of replicates or conditions tested | n = 10 | 7 different storage lengths, n = 1 (2.5, 3, 3.5, 3.8, 4.2) or n = 2 (2.7, 3.2 h)b |
| Average Δ Ct E (SD) | 1.56 ± 0.82 | 4.01 ± 1.32 |
| Average Δ Ct 5′ UTR (SD) | 0.99 ± 0.86 | 3.49 ± 1.37 |
| Agreement with ID NOW™ (%) | 100.0 | 100.0 |
dry swabs were heat inactivated prior to ID NOW™ COVID-19 and RT-PCR testing.
dry swabs were stored at room temperature for up to 4.2 h prior to ID NOW™ COVID-19 and RT-PCR testing, the time intervals for RT-1 to RT-10 were 2.5, 2.7, 3, 3.2, 3.5 3.8, 4.2 h.
3.3. Secondary rRT-PCR testing on used ID NOW™ swabs
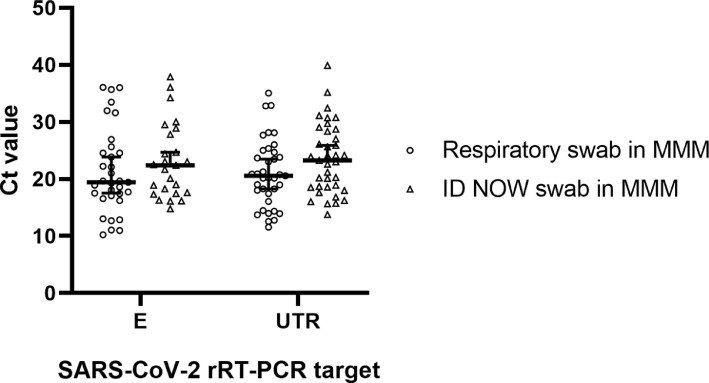
No significant differences in Ct values were noted between dry respiratory swabs tested by ID NOW™ and then stored in MMM for rRT-PCR testing (median Ct E = 22.4, median Ct UTR=23.2) and parallel respiratory swabs collected directly in MMM (median Ct E = 19.4, median Ct UTR=20.6) when tested by RT-PCR (E gene P = 0.33, UTR P = 0.07, Welch's t-test) (Fig. 1 ). The PPA was 88.8% (95% CI:0.76–0.95) and NPA was 99.6% (95% CI:0.98–0.99). The kappa coefficient was 0.91 (95% CI: 0.84 to 0.97). Three dry swabs tested negative by both ID NOW™ and secondary rRT-PCR, but were positive on the initial rRT-PCR with only one of the PCR targets being amplified (two samples with high Ct E genes near the assay cutoff (Ct >35), one sample with low Ct UTR (Ct <15) (Table 3 ). Similarly, three dry swabs that tested negative by ID NOW™ were found to test positive by rRT-PCR and redundancy rRT-PCR testing, with at least one of the two targets typically having a Ct value above 32 (Table 3).
Fig. 1.
rRT-PCR Ct comparison between respiratory swabs stored in MMM and dry swabs tested on ID NOW™ and subsequently stored in MMM. Data are shown with median Ct for each group (horizontal line) with 95% confidence interval.
Table 3.
Discordance in SARS-CoV-2 detection among ID NOW, rRT-PCR and secondary rRT-PCR testing from used swabs.
| Initial rRT-PCR testinga | ID NOW COVID-19 Result | Secondary rRT-PCR testinga | ||
|---|---|---|---|---|
| E gene Ct | UTR Ct | E gene Ct | UTR Ct | |
| 36.0 | nd | Negative | nd | nd |
| 35.7 | nd | Negative | nd | nd |
| n/d | 12.8 | Negative | nd | nd |
| 31.6 | 32.8 | Negative | nd | 28.4 |
| 25.7 | 27.7 | Negative | nd | 32.4 |
| 36.0 | nd | Negative | 36.0 | nd |
nd indicates the target was not detected by rRT-PCR.
3.4. Additional ID NOW™ testing among specimens stored in universal transport media
To confirm whether NP swabs collected in standard UTM (COPAN, Italy) were also compatible with the ID NOW™ COVID-19 assay, we tested 87 patient specimens previously assessed by rRT-PCR (n = 68 SARS-CoV-2 positive, n = 19 SARS-CoV-2 negative), Of these, 72% (n = 63) had been heat inactivated and stored at refrigeration temperature for 24 h while the remaining 28% (n = 24) had been stored for 48 h under the same conditions. We noted a PPA of 89.7% (95% CI:0.80–0.95) and NPA of 94.7% (95% CI:0.75–0.99) between methods. Seven swabs in UTM (8.0%) were discordant, the majority of which had high Ct values in rRT-PCR testing (median Ct E = 34.3, range: 23.6 to 35.7; median Ct UTR=35.8, range: 24.8 to 37.4).
4. Discussion
Rapid identification of individuals with COVID-19 is critical to prevent transmission and ensure patients receive optimal care. In agreement with others, we report that the ID NOW™ COVID-19 assay has high specificity but low sensitivity for detecting SARS-CoV-2 in dry swabs [9,10,16,17]. We noted a percent positivity of 13.4% (46/343) which reflected COVID-19 infection dynamics occurring at the time the study was conducted, which was during the peak of the second pandemic wave in Ontario, Canada. It is difficult to compare agreement of the ID NOW™ assay across studies as they often vary in specimen collection processes and specimen type (e.g. clinical specimens in VTM vs. purified RNA). We noted that heat inactivation of the SARS-CoV-2 virus prior to testing did not significantly impact the results of either the ID NOW™ COVID-19 assay or secondary rRT-PCR. In contrast, extended storage of dry swabs at room temperature introduced variability in Ct values that may have clinical significance.
As the ID NOW™ COVID-19 assay requires respiratory swabs to be processed in the absence of transport media, it presents challenges for the transport and storage of specimens. In this study specimens were transported to a centralized testing laboratory rather than testing at the point of care, in order to ensure assay reliability. A previous examination of swabs collected in VTM prior to ID NOW™ testing despite manufacturer's recommendations found a sensitivity of 71.7% and a specificity of 100% [6]. In our study, we noted that clinical specimens collected in a guanidine thiocyanate-based medium and tested by the ID NOW™ COVID-19 assay showed high agreement with our in-house rRT-PCR method. Based on the results of our secondary rRT-PCR testing, we propose that a single dry swab collected and stored in an empty sterile tube could be saved in transport media for supplemental testing by rRT-PCR after ID NOW™ testing. Considering this assay's performance, supplemental testing could be reserved for situations where ID NOW™ COVID-19 assay results are negative and clinical or epidemiological suspicion for COVID-19 remains high.
Our study has several limitations. Firstly, we were unable to ensure that individuals were tested within the symptomatic window recommended by the ID NOW™ manufacturer (e.g. within 7 days after onset). It is likely that the ID NOW™ test would be used in settings where this information may not be available at the time of collection, and therefore, our assessment of the performance of the ID NOW™ may not be more widely applicable. Secondly, pre-analytical factors such as the timing of specimen collection and specimen quality may have contributed to some of the false negatives identified from the ID NOW™ COVID-19 assay. It is important to note that with the emergence and spread of SARS-CoV-2 variants of concern, there is a need for increased genomic surveillance. Many laboratories are currently performing whole genome sequencing for epidemiologic and other purposes using amplicon-based sequencing approaches which require high quality input materials [18]. There may be some RNA degradation on the dry swab prior to being stored in transport media which may affect the success of some specimens in these downstream applications. However, the approach we have described in this study may facilitate conducting screens for variants of concern and other surveillance activities that would not otherwise be possible if the ID NOW™ swab was not salvaged for additional testing.
5. Conclusions
Our findings suggest that spent dry swabs from the ID NOW™ COVID-19 assay can be stored in guanidine thiocyanate-based media for secondary rRT-PCR testing to overcome the potential for false negatives with the ID NOW™ COVID-19 assay and provide material for additional investigations.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank the laboratory personnel at Shared Hospital Laboratory (Toronto, ON, Canada) for performing the ID NOW™ COVID-19 and conventional RT-PCR testing, as well as the healthcare professionals at Sunnybrook Health Sciences center (Toronto, ON, Canada), North York General Hospital (Toronto, ON, Canada) and Michael Garron Hospital (Toronto, ON, Canada) who participated in the specimen collection. Additionally, we thank Dr. William Stokes and Dr. Graham Tipples (Alberta Health Services, AB, Canada) for sharing their experiences with the ID NOW™ COVID-19 assay.
References
- 1.World Health Organization. 2021. WHO Coronavirus Disease (COVID-19) Dashboard. https://covid19.who.int/. Last accessed February 16, 2021.
- 2.D’Cruz R.J., Currier A.W., Sampson V.B. Laboratory testing methods for novel severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) Front. Cell Dev. Biol. 2020;8:468. doi: 10.3389/fcell.2020.00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hengel B., Causer L., Matthews S., Smith K., Andrewartha K., Badman S., Spaeth B., Tangey A., Cunningham P., Phillips E., Ward J., Watts C., King J., Applegate T., Shephard M., Guy R. A decentralised point-of-care testing model to address inequities in the COVID-19 response. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30859-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li C., Ren L. Recent progress on the diagnosis of 2019 Novel Coronavirus. Transbound. Emerg. Dis. 2020;67:1485–1491. doi: 10.1111/tbed.13620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basu A., Zinger T., Inglima K., Woo K.M., Atie O., Yurasits L., See B., Aguero-Rosenfeld M.E. Performance of Abbott ID Now COVID-19 Rapid nucleic acid amplification test using nasopharyngeal swabs transported in viral transport media and dry nasal swabs in a New York City academic institution. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.01136-20. e01136-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitchell S.L., George K.S. Evaluation of the COVID19 ID NOW EUA assay. J. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhen W., Smith E., Manji R., Schron D., Berry G.J. Clinical evaluation of three sample-to-answer platforms for detection of SARS-CoV-2. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00783-20. e00783-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rhoads D.D., Cherian S.S., Roman K., Stempak L.M., Schmotzer C.L., Sadri N. Comparison of Abbott ID Now, DiaSorin Simplexa, and CDC FDA emergency use authorization methods for the detection of SARS-CoV-2 from nasopharyngeal and nasal swabs from individuals diagnosed with COVID-19. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00760-20. e00760-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dinnes J., Deeks J.J., Adriano A., Berhane S., Davenport C., Dittrich S., Emperador D., Takwoingi Y., Cunningham J., Beese S., Dretzke J., Ferrante di Ruffano L., Harris I.M., Price M.J., Taylor-Phillips S., Hooft L., Leeflang M.M., Spijker R., Van den Bruel A., Cochrane C-DTAG Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst. Rev. 2020;8 doi: 10.1002/14651858.CD013705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Subsoontorn P., Lohitnavy M., Kongkaew C. The diagnostic accuracy of isothermal nucleic acid point-of-care tests for human coronaviruses: a systematic review and meta-analysis. Sci. Rep. 2020;10:22349. doi: 10.1038/s41598-020-79237-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kandel C., Zheng J., McCready J., Serbanescu M.A., Racher H., Desaulnier M., Powis J.E., Vojdani K., Finlay L., Sheldrake E., Vermeiren C., Katz K., McGeer A., Kozak R., Goneau L.W. Detection of SARS-CoV-2 from saliva as compared to nasopharyngeal swabs in outpatients. Viruses. 2020;12:1314. doi: 10.3390/v12111314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kandel C.E., Young M., Serbanescu M.A., Powis J.E., Bulir D., Callahan J., Katz K., McCready J., Racher H., Sheldrake E., Quon D., Vojdani O.K., McGeer A., Goneau L.W., Vermeiren C. Detection of SARS-CoV-2 in outpatients: a multi-centre comparison of self-collected saline gargle, oral swab and combined oral-anterior nasal swab to a provider collected nasopharyngeal swab. Infect. Control Hosp. Epidemiol. 2021 doi: 10.1017/ice.2021.2:1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chin A.W.H., Chu J.T.S., Perera M.R.A., Hui K.P.Y., Yen H.L., Chan M.C.W., Peiris M., Poon L.L.M. Stability of SARS-CoV-2 in different environmental conditions. Lancet Microbe. 2020;1:e10. doi: 10.1016/S2666-5247(20)30093-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banerjee A., Nasir J.A., Budylowski P., Yip L., Aftanas P., Christie N., Ghalami A., Baid K., Raphenya A.R., Hirota J.A., Miller M.S., McGeer A.J., Ostrowski M., Kozak R.A., McArthur A.G., Mossman K., Mubareka S. Isolation, sequence, infectivity, and replication kinetics of severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. 2020;26:2054–2063. doi: 10.3201/eid2609.201495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clinical and Laboratory Standards Institute . 2nd ed. Clinical Laboratory Standards Institute; Wayne PA: 2008. User Protocol for Evaluation of Qualitative Test Performance; Approved Guideline. CLSI Document EP12-A2. [Google Scholar]
- 16.Harrington A., Cox B., Snowdon J., Bakst J., Ley E., Grajales P., Maggiore J., Kahn S. Comparison of Abbott ID Now and Abbott m2000 methods for the detection of SARS-CoV-2 from nasopharyngeal and nasal swabs from symptomatic patients. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00798-20. e00798-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thwe P.M., Ren P. How many are we missing with ID NOW COVID-19 assay using direct nasopharyngeal swabs? Findings from a mid-sized academic hospital clinical microbiology laboratory. Diagn. Microbiol. Infect. Dis. 2020;98 doi: 10.1016/j.diagmicrobio.2020.115123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nasir J.A., Kozak R.A., Aftanas P., Raphenya A.R., Smith K.M., Maguire F., Maan H., Alruwaili M., Banerjee A., Mbareche H., Alcock B.P., Knox N.C., Mossman K., Wang B., Hiscox J.A., McArthur A.G., Mubareka S. A comparison of whole genome sequencing of SARS-CoV-2 using amplicon-based sequencing, random hexamers, and bait capture. Viruses. 2020;12:895. doi: 10.3390/v12080895. [DOI] [PMC free article] [PubMed] [Google Scholar]