Abstract
This research work aims to identify COVID-19 through deep learning models using lung CT-SCAN images. In order to enhance lung CT scan efficiency, a super-residual dense neural network was applied. The experimentation has been carried out using benchmark datasets like SARS-COV-2 CT-Scan and Covid-CT Scan. To mark COVID-19 as positive or negative for the improved CT scan, existing pre-trained models such as XceptionNet, MobileNet, InceptionV3, DenseNet, ResNet50, and VGG (Visual Geometry Group)16 have been used. Taking CT scans with super resolution using a residual dense neural network in the pre-processing step resulted in improving the accuracy, F1 score, precision, and recall of the proposed model. On the dataset Covid-CT Scan and SARS-COV-2 CT-Scan, the MobileNet model provided a precision of 94.12% and 100% respectively.
Keywords: COVID-19, CT Scan, MobileNet, Transfer learning, Pandemic, Deep learning
Graphical abstract
1. Introduction
The World Health Organization (WHO) got the first update related to Corona virus disease 2019 (COVID-19) on December 31, 2019. On January 30, 2020, WHO announced the COVID-19 spread as a global health emergency. Corona virus is a zoonotic virus, which means it began in animals before spreading to humans. The transmission of the virus occurred in humans after coming into contact with animals. Corona virus can transmit from one person to another through respiratory droplets when a person exhales, coughs, sneezes, or chats with others [1]. It is also believed that the virus may have transferred from bats to other species, such as snakes or pangolins, and then to humans. Multiple COVID-19 complications leading to liver problems, pneumonia, respiratory failure, cardiovascular diseases, septic shock, etc. have been prompted by a condition called cytokine release syndrome or a cytokine storm. This occurs when an infection activates the immune system to leak inflammatory proteins known as cytokines into the bloodstream which can kill tissues and organs in human beings.
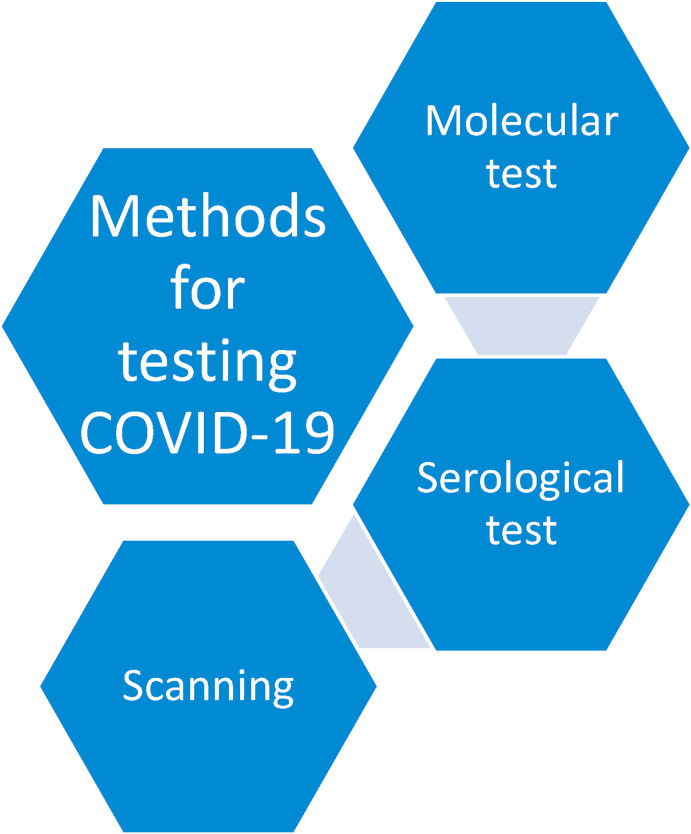
Fig. 1 highlights the various methods used for testing corona virus. These include molecular, serological, and scanning. Molecular tests look for signs of an infection which are still active. Swabbing the back of the throat for a sample is normally done using a cotton swab. A polymerase chain reaction (PCR) test is performed on the sample. This test provides the signs of the virus's genetic material. A PCR test after detecting two unique SARS-CoV-2 genes confirms a COVID-19 diagnosis. Only existing cases of COVID-19 can be detected through molecular tests; and it cannot be said whether anyone has recovered from this infection. Serological testing can identify antibodies generated by the body to fight the virus. Normally, a blood sample is needed for a serological test; and such a test is particularly helpful in identifying infections that have mild to no symptoms [2]. Anyone who has recovered from COVID-19 possesses these antibodies which can be found in blood and tissues all over the body. Apart from swab and serological tests, organs and structures of the chest can be imaged using X-rays (radiography) or computed tomography (CT) scans [3]. A chest CT scan, a common imaging approach for detecting pneumonia, is a rapid and painless procedure. It has appeared through new research that sensitivity of CT is 98% for COVID-19 infection which is greater as compared to 71% for Reverse Transcription Polymerase Chain Reaction (RT-PCR) testing. The COVID-19 classification based on the Thorax CT necessitates the involvement of a radiology specialist which incurs a lot of time. Thus, an automated processing of Thorax CT images is desirable in order to help the medical specialists to save their precious time. This automation may also assist in the avoidance of medicinal delays.
Fig. 1.
Methods used to test the subject for COVID-19 as positive or negative.
2. Literature review
Panwar et al. [4] conducted a binary image classification study to detect COVID-19 patients. A fine-tuned VGG model was used to classify the input photos. The researchers used the Grad-CAM technique to apply a color visualization approach to make the proposed deep learning model more explainable and interpretable. Ni et al. [5] developed a lesion detection, segmentation, and position deep learning algorithm. With chest CT images, the proposed approach was validated on 14,435 participants. During the outbreak, the algorithm was checked on a non-overlapping dataset of 96 reported COVID-19 patients in three hospitals across China. Xiao et al. [6] built and validated a deep learning-based model (ResNet34) using residual convolutional neural networks and multiple instance learning.
Deep CNN-based image classification strategies were evaluated by Mishra et al. [7] in order to distinguish COVID-19 cases taking chest CT scan images. Ko et al. [8] created the fast-track COVID-19 classification network (FCONet) through a 2-D Deep learning system to diagnose COVID-19 taking image of chest CT. As a backbone, FCONet was built using transfer learning which included pre-trained deep learning models, viz. VGG16, Inception-v3, Xception or ResNet-50. Song et al. [9] developed a large-scale bi-directional generative adversarial network (BigBiGAN) architecture to construct an end-to-end representation learning system. The researchers extracted semantic features from CT images used in a linear classifier. ResNet-50, a CNN-based model for using chest CT to distinguish COVID-19 and Non-COVID-19, was proposed by E. Matsuyama [10]. Without clipping any parts of the image, the CNN model fed the wavelet coefficients of the whole picture as input. To identify the patients having COVID-19, Jaiswal et al. [11] provided the DenseNet201-based deep transfer learning (DTL) model. The proposed model extracted features from the ImageNet dataset using its own trained weights and a convolutional neural structure. Loey et al. [12] chose five distinct deep convolutional neural network-based models viz. AlexNet, VGGNet19, VGGNet16, ResNet50, and GoogleNet to identify the Coronavirus-infected person using chest CT radiograph digital images for their analysis. The authors used CGAN and traditional data augmentations to create additional photos to help in the identification of COVID-19. The COVID-19 patients were classified using CNN by Singh et al. [13]. The multi-objective differential evolution (MODE) was used to tune the initial parameters of CNN. Using visual features extracted through volumetric chest CT scans, Li et al. [14] developed a deep learning model called COVNet to detect COVID-19. To assess the model's robustness, CT scans of community-acquired pneumonia (CAP) as well as other non-pneumonia anomalies were considered. The regions under the receiver operating characteristic (ROC) curve, specificity and sensitivity were applied to test the proposed model's diagnostic efficiency. Cuong Do et al. [15] investigated VGG-16, Inception-V3, VGG19, Xception, InceptionResNet-V2, DenseNet-169, DenseNet121, and DenseNet201 pre-trained models. Overfitting was avoided by dropping and augmenting instances during training. Wang et al. [16] employed a two-step transfer learning methodology to identify CT as positive or negative. Where, in the first phase, the researchers gathered CT scans of 4106 lung cancer patients who had a CT scan and had their epidermal growth factor receptor (EGFR) gene sequenced. The DL system picked up features that could represent the relationship between a chest CT image and a micro-level functional abnormality in the lungs by using this large CT-EGFR training data collection. The scientists then employed a large, geographically diversified COVID-19 dataset (n = 1266) to train and validate the DL system's diagnostic and prognostic results.
He et al. [17] suggested the Self-Trans method for classifying COVID-19 instances which incorporated transfer learning and contrastive self-supervised learning to learn efficient as well as unbiased feature representations while reducing the possibility of overfitting. To model architecture, validate, practice, and test, Sakagianni et al. [18] used Google AutoML Cloud Vision. The underlying framework that enabled this platform to find the best way to train deep learning models has been called Neural Architecture Search (NAS). To achieve the best performance, Alshazly et al. [19] used advanced deep network architecture and suggested a transfer learning strategy that utilized custom-sized input optimized for deep architecture. Silva et al. [20] proposed Efficient CovidNet, a model for detecting COVID-19 patterns in CT images that involves a voting-based approach and cross-dataset analysis. On a small COVID-19 CT scan dataset, Ewen et al. [21] calculated the usefulness of self-supervision classification. Ragab et al. [22] introduced a novel CAD method called FUSI-CAD centered on the fusion of many distinct CNN architectures taking features like grey level co-occurrence matrix (GLCM) and discrete wavelet transform (DWT). The dataset(s) utilized by the researchers earlier, as well as the results obtained for various assessment criteria, are summarized in Table 1 .
Table 1.
Datasets used in various studies showing their Sensitivity (Se)/Specificity (Sp)/Accuracy (Acc).
| S.No. | Author(s) | Dataset | Dataset Source | Se/Sp/Acc |
|---|---|---|---|---|
| 1. | Panwar et al. [4] | SARS-COV-2 CT | https://www.kaggle.com/plameneduardo/sarscov2-ctscan-dataset/notebooks | 94.04/95.84/95.00 |
| 2. | Jaiswal et al. [11] | 96.29/96.21/96.25 | ||
| 3. | Singh et al. [13] | 90.5/90.5/93 | ||
| 4. | Alshazly et al. [19] | 99.10/99.60/99.40 | ||
| 5. | Silva et al. [20] | 98.80/NA/98.99 | ||
| 6. | Dina A. Ragab and Omneya Attallah [22] | 99/NA/99 | ||
| 7. | Mishra et al. [7] | COVID-CT | https://github.com/UCSD-AI4H/COVID-CT | 88.00/90.00/88.00 |
| 8. | E. Matsuyama [10] | 90.40/93.30/92.20 | ||
| 9. | Loey et al. [12] | 77.60/87.62/82.91 | ||
| 10. | Cuong Do and Lan Vu [15] | NA/NA/85.00 | ||
| 11. | He et al. [17] | NA/NA/86.00 | ||
| 12. | Sakagianni et al. [18] | 90.20/86.10/88.30 | ||
| 13. | Ewen et al. [21] | NA/NA/86.21 | ||
| 14. | Wang et al. [16] | Not available/disclosed | Not available/disclosed | 79.35/71.43/85.00 |
| 15. | Li et al. [14] | Data collected from 6 different hospitals | 90.00/95.00/NA | |
| 16. | Ni et al. [5] | Data collected from three Chinese hospitals | 100/NA/NA | |
| 17. | Xiao et al. [6] | Data collected from patients of the People's Hospital of Honghu | NA/NA/81.90 | |
| 18. | Ko et al. [8] | COVID-19 Database | https://www.sirm.org/en/category/articles/covid-19-database/ | 99.50/100/99.80 |
| 19. | Song et al. [9] | Images of COVID-19 positive and negative pneumonia patients | https://data.mendeley.com/datasets/kk6y7nnbfs/1 | 80.00/75.00/NA |
The approach proposed here in this research paper has achieved an accuracy score of 94.12% on COVID-CT- Dataset, and an accuracy of 100% on SARS-COV-2 CT Scan Dataset with MobileNet model. Although the accuracy scores claimed by Panwar et al. [4], Jaiswal et al. [11], Alshazly et al. [19], and Ko et al. [8] are 95.00, 96.25, 99.40, and 99.80% respectively, but the findings produced by the researchers [4,11,19], and [8] still have some gaps. The work against reference [1] suffers from the flaw that it stops learning after a few learning epochs, limiting the resulting neural network to the class of semi-linear models and preventing learning from developing complex non-linear structures in the neural network.
In their research work, the authors [11] missed the realistic scenario, where poorly contrasted CT scan images could lead to the chances of misclassification; and the researchers did not pre-processed the image w.r.t. its quality enhancement. Their models were validated using a segment of the testing dataset [8]. As a result, the training and research datasets were derived from the same sources. This is likely to raise the concern about generalizability and overfitting. The authors, in their work [19], have not provided any proper reasoning for taking the specific values of parameters like number of epochs, and customized size of the input image.
However, in the work proposed here, CT images were enhanced during the pre-processing phase before feeding them to the deep learning models for classification. By converting the normal CT image into its corresponding super-resolution image, a major concern over compromising the image quality has been settled. Overfitting has been avoided through augmentation. This investigation also ceased model implementation at the time when loss of validation began to rise. It helps to resolve the problem of early stopping.
3. Proposed methodology
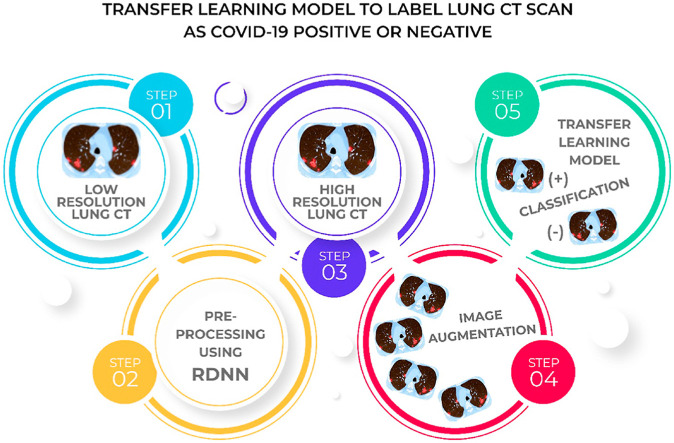
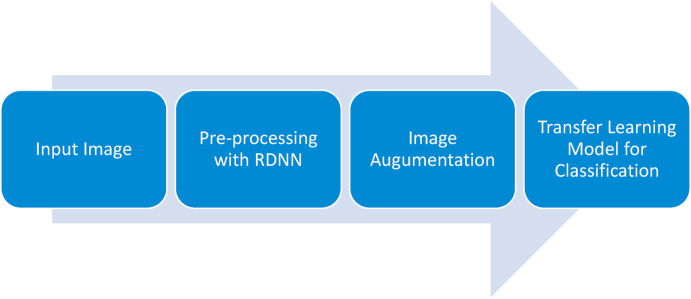
The processes, in this section, are described to assess if a COVID-19 lung CT scan is positive or negative. Fig. 2 presents a general block diagram of the research.
Fig. 2.
Steps followed for classifying lung CT into normal and abnormal for COVID-19.
Under most of the circumstances, the spatial resolution in CT pictures is not perfect because of restrictions in CT machine hardware configurations. Pre-processing through the use of RDNN allows the spatial resolution of CT images to be increased, resulting in lower system costs and complexity. Variation in CT picture resolution can also be caused by changes in radiation dosage and slice thickness which might make radiologists' diagnostic abilities questionable. As a result, boosting clarity and crispness in low-quality CT scans is highly desired. The use of linear and non-linear functions in classical SR approaches results in jagged edges and blurring in pictures which generates unrequired noise in the data. Deep learning algorithms have been successful in mitigating these anomalies by extracting high-dimensional and non-linear information from CT scans, resulting in improved SR CT pictures. Due to the use of hierarchical features via dense feature fusion, the RDN can recover sharper and cleaner edges when compared to other deep learning models [[23], [24], [25]].
3.1. Image dataset
SARS-COV-2 CT [26] and COVID-CT [27] have been used as the benchmark datasets for the transfer learning models in this research work. The datasets were divided into two sections; taking 80% of the total dataset for training, and 20% for testing. The COVID-CT-Dataset contained 349 COVID-19 CT images from 216 patients, and 463 non-COVID-19 CTs. The SARS-COV-2 CT included 2482 CT scan images from 1252 SARSCoV-2 infected patients. In SARS-COV-2 CT, of the total 2482 images, the non-COVID-19 subjects accounted for 1230 CT scans.
3.2. Pre-processing through RDNN
Medical imaging modalities include both anatomical and functional details about the human body structure. Resolution limits, on the other hand, often reduce the diagnostic value of medical images. The primary medical imaging modalities, including computerized tomography (CT), magnetic resonance imaging (MRI), functional MRI (fMRI), and positron emission tomography (PET) can be enhanced with Super Resolution (SR). The purpose of SR is to improve the resolution of medical images while preserving true isotropic 3-D images.
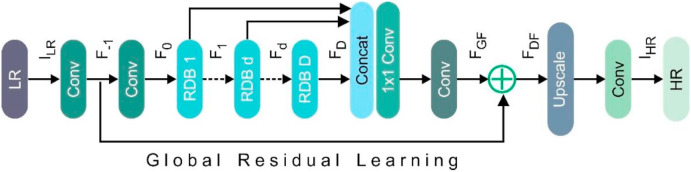
For SR, an extremely deep convolutional neural network (CNN) has been used, but the reported ones have not fully exploited the hierarchical features of the source low-resolution (LR) images, resulting in poor performance. In this study, the residual dense network (RDN) was employed to address the LR problem. RDN uses residual dense block (RDB) where densely connected convolutional layers extract a large number of local features. The RDB's local feature fusion is then utilized to learn more efficient features from the previous and existing local features in an adaptive manner. Global feature fusion has been used to learn global hierarchical features holistically after dense local features.
Fig. 3 depicts the RDN's internals, which have been divided into four sections: Residual dense blocks (RDBs), shallow feature extraction net (SFENet), up-sampling net (UPNet) and dense feature fusion (DFF). denotes the low-resolution lung CT, while represents the high-resolution lung CT obtained by RDN. To remove the shallow features, two convolutional layers were used. The first convolutional layer takes the LR input and extracts features [Equation (1)].
| (1) |
where, denotes convolution operation. The shallow function was extracted, and global residual learning was performed using . Thus, Equation (2) appeared as below:
| (2) |
denotes the second shallow feature extraction layer's convolution process which has been used as an input to residual dense blocks. In the case of D residual dense blocks, the output of the RDB can be obtained by Equations (3), (4)
| (3) |
| (4) |
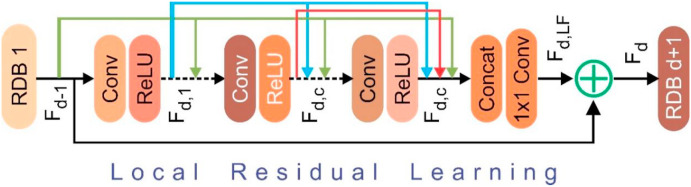
where, denotes the operations of the RDB. is an amalgamation of convolution and rectified linear unit (ReLU). can be taken as a local feature, as it has been produced by the RDB by fully utilizing each convolutional layer within the block. Dense feature fusion (DFF) involving global residual learning (GRL) and global feature fusion (GFF) were used to extract hierarchical features using a collection of RDBs. Fig. 4 displays the architecture of RDB, where, DFF makes a full use of features from all the preceding layers and can be represented as Equation (5):
| (5) |
Where, denotes the DFF output feature-maps obtained through a composite function .
Fig. 3.
Architecture of the residual dense network (RDN).
Fig. 4.
Internals of the residual dense block (RDB).
In the HR space, up-sampling net (UPNet) was stacked after extracting global and local features in the LR space. Equation (6) shows the output obtained from the RDN:
| (6) |
The prime variables used here in the RNN approach are as follows:
– count of RDB; – total count of inner layers (convolutional) for an RDB; – feature maps count for the convolutional layer (inside RDB); and – feature maps count for the convolutional layer (outside RDB).
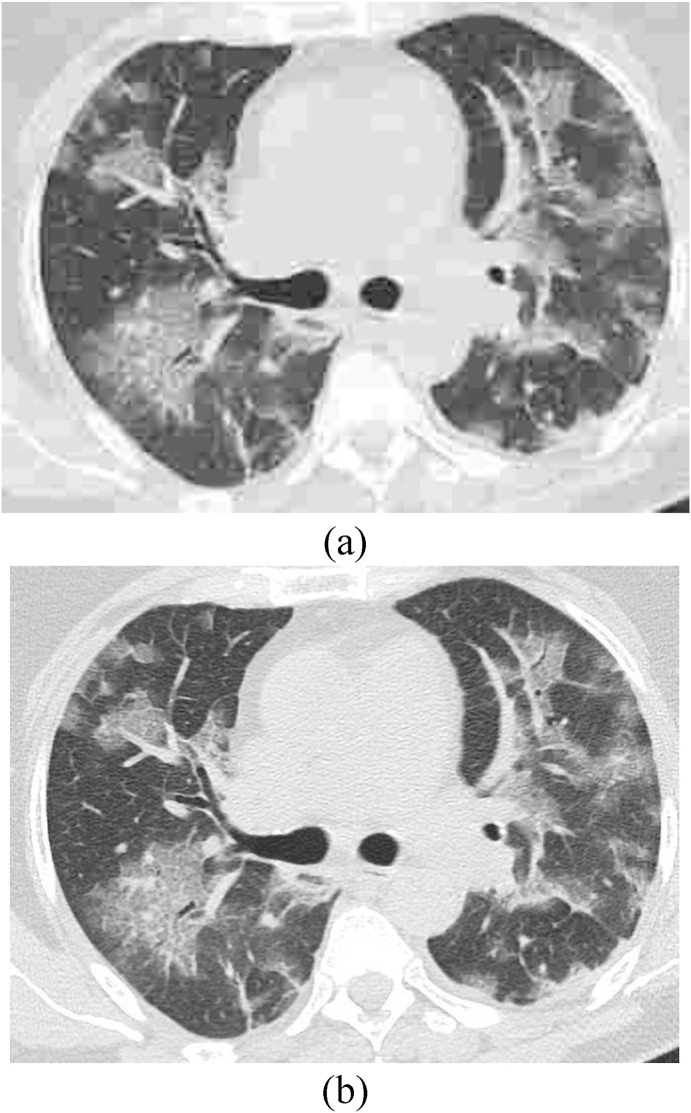
The model was trained on the DIV2K [28] dataset taking the following values for the various parameters , , , and . From a low resolution CT image, an augmented patch of has been used taking 86 epochs and batch size as 1000. Fig. 5 shows the sample lung CT images taken randomly from the benchmark datasets under consideration and as obtained after employing image super resolution module on the same images.
Fig. 5.
(a) Lung CT image without super resolution, and (b) CT obtained after super resolution.
3.3. Image augmentation
A large dataset is a technique used to categorize deep learning effectively and successfully. But it is not always possible to have a vast dataset. In the domains of machine learning and deep learning, the data size can be raised to enhance classification accuracy. Data enhancement approaches increase the algorithm's learning and network capability significantly. Because of few shortcomings, texture, color, and geometric-based data augmentation techniques are not equally common. While this strategy provides data diversity, it has limitations such as more memory, training time, and expenses associated with conversion measurement. To supplement the lung CT images in the current investigation, only geometric alterations were performed. The count of images in dataset becomes tentatively three times larger after executing data augmentation. A randomly generated number has been used to rotate the lung CT images counter clockwise from to .
3.4. Transfer learning models for classification
Transfer learning is the process of moving the variables of a neural network trained on one dataset, and task to a new data repository and task. Several deep neural networks trained on natural images share a peculiar function: on the first layer, model learns features that seem to be universal, in the sense that these can be applied to a wide range of datasets and tasks. The network's final layers must gradually transition features from general to particular. Transfer learning can be an imposing method for enabling training without overfitting when the target dataset is a fraction of the size of the base dataset [29,30]. Here, in this work, DenseNet121, MobileNet, VGG16, ResNet50, InceptionV3, and XceptionNet have been used as the base models, pre-trained on the ImageNet dataset for object detection. ImageNet is a 1.28 million natural image dataset that is open to the public; and it is divided into 1,000 categories. Python 3.6, Scikit-Learn 0.20.4, Keras 2.3.1, and TensorFlow 1.15.2 have been used here to deploy the proposed methods. All the tests were run on a computer with an Intel Core i7 8th generation processor running at 1.9 GHz with 8 GB of RAM, Intel UHD Graphics 620, and Windows 10 installed.
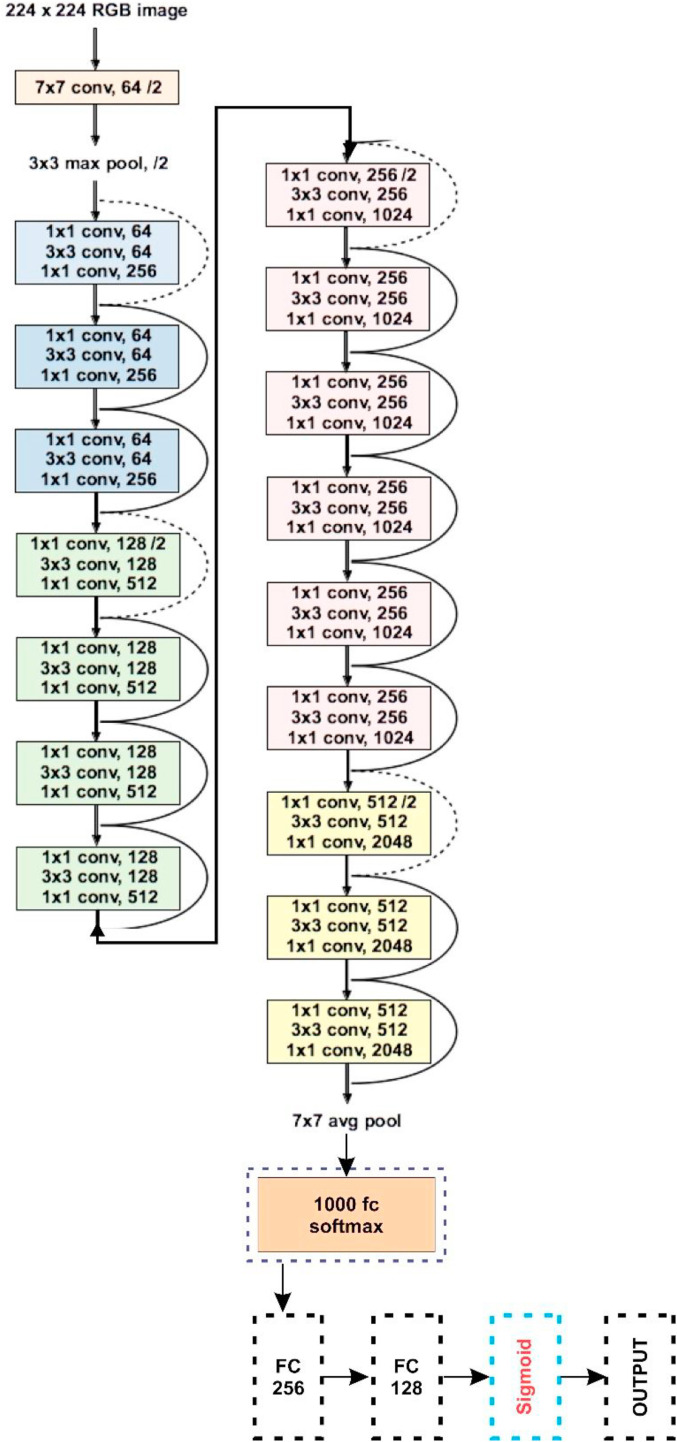
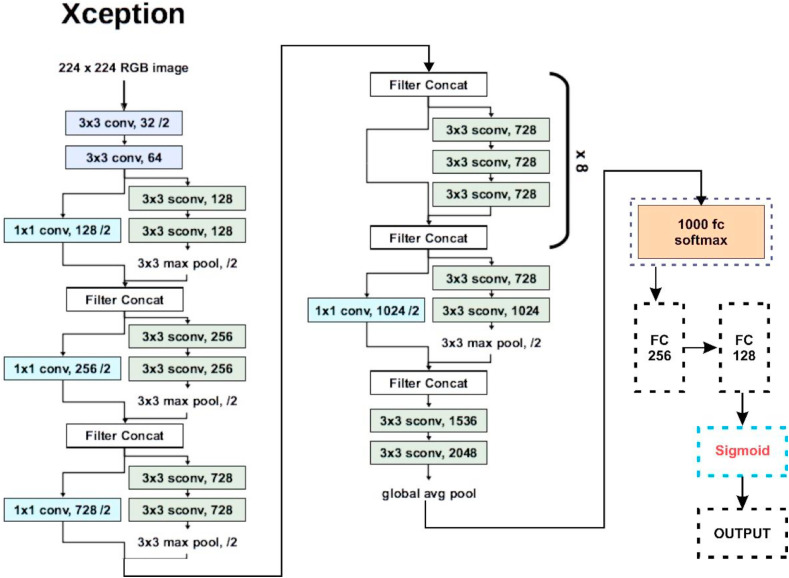
Fig. 6, Fig. 7, Fig. 8, Fig. 9, Fig. 10 highlight the basic architecture of the transfer learning models and their customization that has been finally deployed to obtain the classification results here in this work.
Fig. 6.
Basic architecture and customization in ResNet50.
Fig. 7.
Basic architecture and customization in Xception.
Fig. 8.
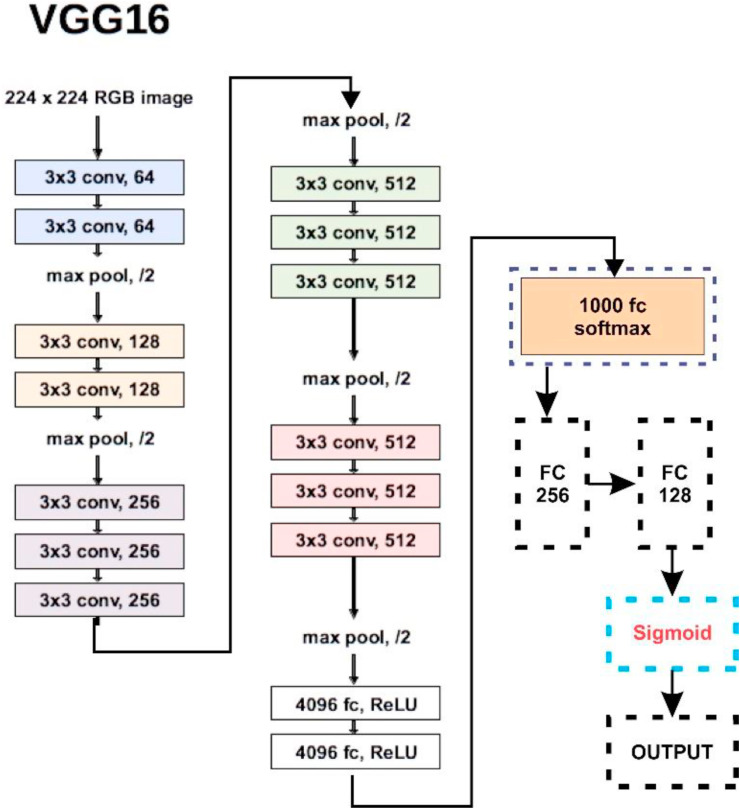
Basic architecture and customization in VGG16.
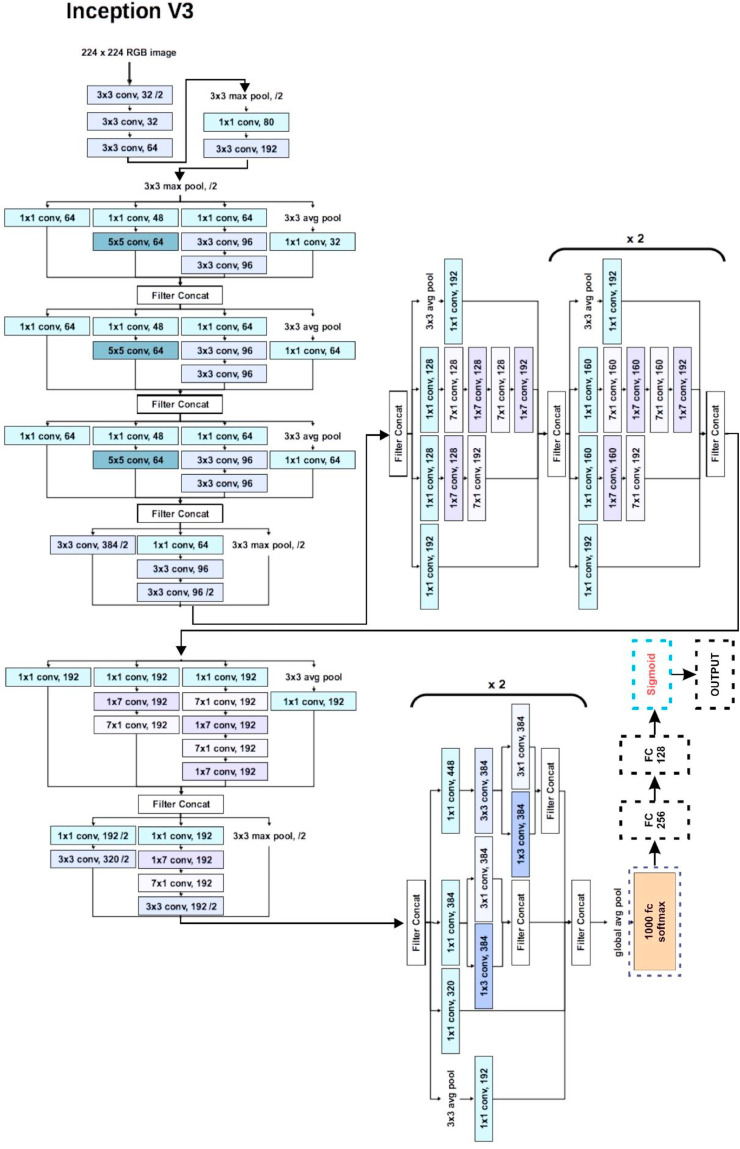
Fig. 9.
Basic architecture and customization in InceptionV3.
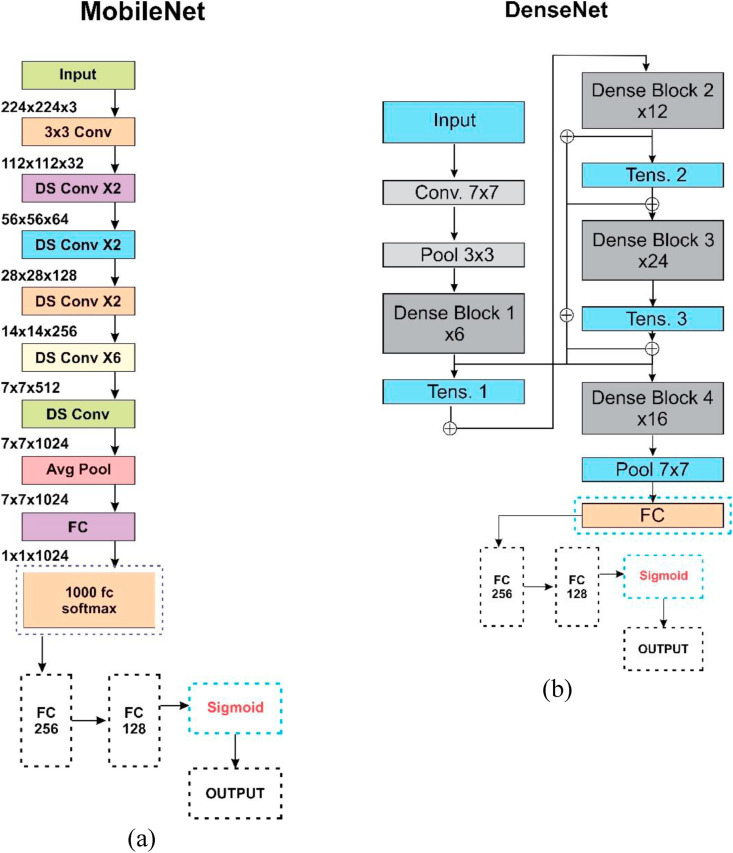
Fig. 10.
Basic architecture and customization in (a) MobileNet, and (b) DenseNet.
4. Results and experiments
Here, in this research work, COVID-19 positive patients who have been correctly categorized as COVID-19 positive are denoted by true positive (TP), while false positive (FP) subjects are normal, but have been incorrectly labelled as corona positive. True Negative (TN) denotes the COVID-19 negative subjects who have been recognized correctly as negative. Finally, COVID-19 positive patients, misclassified as negative, have been denoted with False Negative (FN). To ensure the reliability of the proposed F1 score, Accuracy, Precision, and Recall have been taken as evaluation matrices.
Accuracy represents total number of correctly identified cases, and can be formulated as per Equation (7):
| (7) |
When dataset is imbalanced, the precision-recall serves as a useful matric for predicting the performance. Precision is a measure of outcome relevancy in information retrieval, while recall is a measure of how often genuinely valid items are retrieved. Equations (8), (9) represent Precision and Recall respectively:
| (8) |
| (9) |
Also, the F1 score is considered to be good as compared to the Accuracy score, especially when the dataset is not balanced one. It is sometimes referred as the harmonic mean of Precision and Recall that can be evaluated as given in Equation (10):
| (10) |
To prove the role of super resolution in the proposed methodology, all the transfer learning models were first executed on the CT images taken from COVID-CT-Dataset [27] and SARS-COV-2 CT [26] without doing any image super resolution. The results obtained for Accuracy, Precision, Recall and F1 Score have been exhibited in Table 2 . Table 3 presents the results taking the same evaluation matrices, but after performing image super resolution during the pre-processing phase. The values for the attributes obtained after deploying super resolution were better than the scenario when no super resolution was employed.
Table 2.
Accuracy, Recall, Precision and F1 Score obtained by executing MobileNet, DenseNet121, ResNet50, VGG16, InceptionV3 and XceptionNet taking COVID-CT-Dataset without employing any image super resolution operation.
| Model Name | Accuracy | Recall | Precision | F1 Score |
|---|---|---|---|---|
| MobileNet | 86.60 | 95.70 | 85.04 | 90.02 |
| DenseNet121 | 93.00 | 93.00 | 94.10 | 92.50 |
| ResNet50 | 82.60 | 91.90 | 71.40 | 79.70 |
| VGG16 | 86.60 | 93.80 | 86.70 | 90.09 |
| InceptionV3 | 89.30 | 92.80 | 89.00 | 90.09 |
| XceptionNet | 85.30 | 90.20 | 85.90 | 88.04 |
Table 3.
Accuracy, Recall, Precision and F1 Score obtained by executing MobileNet, DenseNet121, ResNet50, VGG16, InceptionV3 and XceptionNet taking COVID-CT-Dataset and employing super resolution operation.
| Model Name | Accuracy | Recall | Precision | F1 Score |
|---|---|---|---|---|
| MobileNet | 94.12 | 96.11 | 96.11 | 96.11 |
| DenseNet121 | 88.24 | 92.36 | 84.72 | 88.36 |
| ResNet50 | 73.53 | 75.49 | 70.11 | 72.60 |
| VGG16 | 85.29 | 83.38 | 93.37 | 87.54 |
| InceptionV3 | 94.10 | 96.53 | 92.82 | 94.57 |
| XceptionNet | 85.29 | 87.28 | 85.79 | 86.52 |
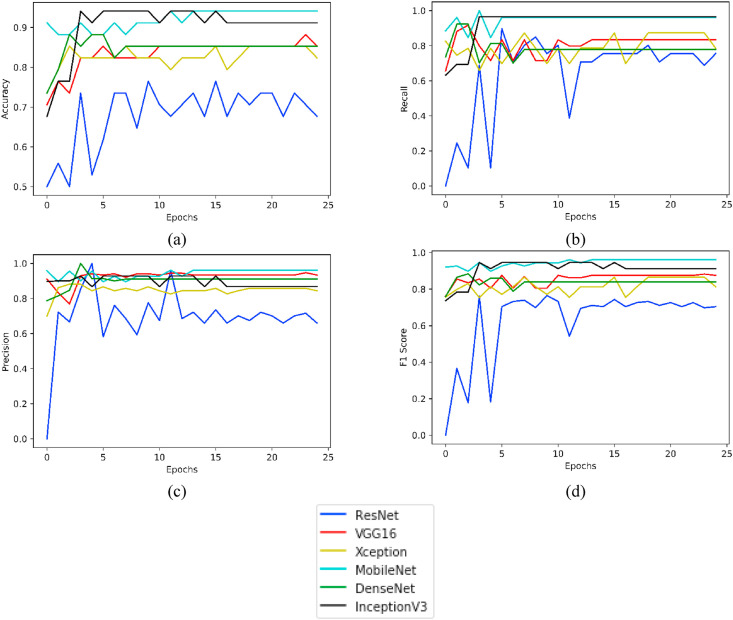
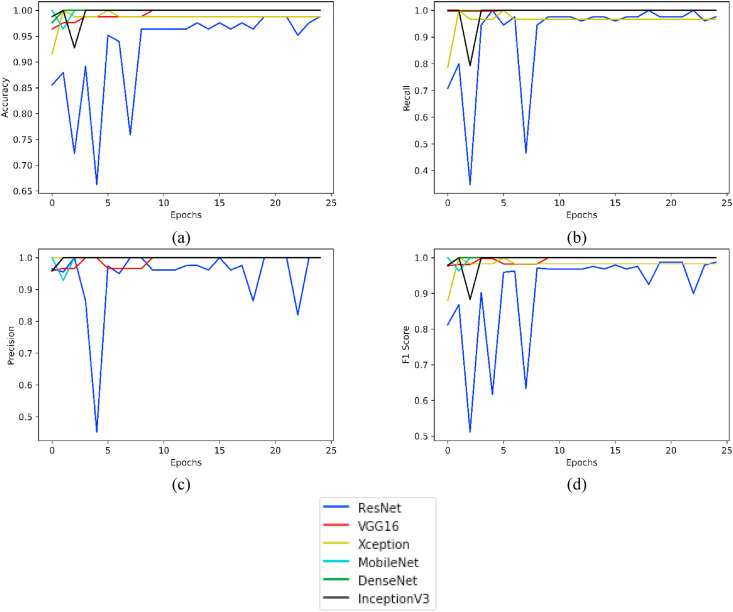
For the COVID-CT dataset (taken after having super resolution), Fig. 11 highlights plotting for the parameters Accuracy, Recall, Precision and F1 Score taken for 25 epochs of the various models under consideration.
Fig. 11.
Plots of various evaluation parameters, viz. (a) Accuracy, (b) Recall, (c) Precision and (d) F1 Score as obtained from ResNet, VGG16, Xception, MobileNet, DenseNet and InceptionV3 using COVID-CT- Dataset (with super resolution).
Similarly, for the dataset SARS-COV-2 CT, the results obtained for all the selected parameters are presented in Table 4 . Table 5 provides the results evaluated on the same parameters, but after performing the image super resolution. The values for the attributes obtained after deploying super resolution were better than the scenario when no super resolution was employed.
Table 4.
Accuracy, Recall, Precision and F1 Score obtained by executing MobileNet, DenseNet121, ResNet50, VGG16, InceptionV3 and XceptionNet taking SARS-COV-2 CT dataset and without employing any image super resolution operation.
| Model Name | Accuracy | Recall | Precision | F1 Score |
|---|---|---|---|---|
| MobileNet | 98.00 | 98.40 | 98.00 | 98.20 |
| DenseNet121 | 98.00 | 98.50 | 94.90 | 96.60 |
| ResNet50 | 93.50 | 93.60 | 95.30 | 93.50 |
| VGG16 | 98.00 | 98.00 | 99.40 | 99.20 |
| InceptionV3 | 95.10 | 97.80 | 93.80 | 95.70 |
| XceptionNet | 95.10 | 95.20 | 96.60 | 95.90 |
Table 5.
Accuracy, Recall, Precision and F1 Score obtained by executing MobileNet, DenseNet121, ResNet50, VGG16, InceptionV3 and XceptionNet taking SARS-COV-2 CT dataset and employing super resolution operation.
| Model Name | Accuracy | Recall | Precision | F1 Score |
|---|---|---|---|---|
| MobileNet | 100.00 | 100.00 | 100.00 | 100.00 |
| DenseNet121 | 100.00 | 100.00 | 100.00 | 100.00 |
| ResNet50 | 97.59 | 97.57 | 97.57 | 97.50 |
| VGG16 | 100.00 | 100.00 | 100.00 | 100.00 |
| InceptionV3 | 100.00 | 100.00 | 100.00 | 100.00 |
| XceptionNet | 98.80 | 96.68 | 100.00 | 98.30 |
For the SARS-COV-2 CT Dataset (taken after having super resolution), Fig. 12 displays plotting for the parameters Accuracy, Recall, Precision and F1 Score taken for 25 epochs of the various models under consideration.
Fig. 12.
Plots of various evaluation parameters, viz. (a) Accuracy, (b) Recall, (c) Precision and (d) F1 Score as obtained from ResNet, VGG16, Xception, MobileNet, DenseNet, and InceptionV3 using SARS-COV-2 CT Dataset (with super resolution).
5. Conclusion and future scope
CT scans were taken from two benchmark datasets for this research study; and all the images were enhanced in the pre-processing phase using super resolution deep neural networks. Transfer learning models were employed to label the images as positive and negative for COVID-19. The MobileNet model produced better results as compared to its peer models. On the benchmark datasets, viz. Covid-CT Scan and SARS-COV-2 CT-Scan, for the MobileNet model, the sensitivity scores were found to be 96.11% and 100% respectively; precision scores were 96.11% and 100% respectively; F-1 scores were recorded as 96.11% and 100% respectively; and accuracy was to the tune of 94.12% and 100% respectively. The proposed work can be customized further by stacking hybrid pre-trained algorithms.
Author contributions
Conceptualization, Vinay Arora and Eddie Yin-Kwee Ng; Data curation, Rohan Leekha and Medhavi Darshan; Formal analysis, Eddie Yin-Kwee Ng; Investigation, Vinay Arora; Methodology, Rohan Singh Leekha and Medhavi Darshan; Project administration, Eddie Yin-Kwee Ng and Vinay Arora; Validation, Eddie Yin-Kwee Ng, Medhavi Darshan, Arshdeep Singh; Visualization, Vinay Arora, Rohan Leekha and Eddie Yin-Kwee Ng; Writing – original draft, Vinay Arora, Medhavi Darshan; Writing – review & editing, Vinay Arora and Rohan Singh Leekha.
Funding statement
No funding has been received.
Ethical compliance
Research experiments conducted in this article with animals or humans were approved by the Ethical Committee and responsible authorities of our research organization(s) following all guidelines, regulations, legal, and ethical standards as required for humans or animals. (Yes/No/Not applicable).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Tomar A., Gupta N. Prediction for the spread of COVID-19 in India and effectiveness of preventive measures. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kushwaha S., Bahl S., Bagha A.K., Parmar K.S., Javaid M., Haleem A., Singh R.P. Significant applications of machine learning for COVID-19 pandemic. J. Industrial Integr. Manag. 2020;5 doi: 10.1142/S2424862220500268. [DOI] [Google Scholar]
- 3.Jiang Y., Guo D., Li C., Chen T., Li R. High-resolution CT features of the COVID-19 infection in Nanchong City: initial and follow-up changes among different clinical types. Radiol. Infect. Dis. 2020;7:71–77. doi: 10.1016/j.jrid.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Panwar H., Gupta P., Siddiqui M.K., Morales-Menendez R., Bhardwaj P., Singh V. A deep learning and grad-CAM based color visualization approach for fast detection of COVID-19 cases using chest X-ray and CT-Scan images. Chaos, Solit. Fractals. 2020;140 doi: 10.1016/j.chaos.2020.110190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ni Q., Sun Z.Y., Qi L., Chen W., Yang Y., Wang L., Zhang X., Yang L., Fang Y., Xing Z., Zhou Z., Yu Y., Lu G.M., Zhang L.J. A deep learning approach to characterize 2019 coronavirus disease (COVID-19) pneumonia in chest CT images. Eur. Radiol. 2020;30:6517–6527. doi: 10.1007/s00330-020-07044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiao L.S., Li P., Sun F., Zhang Y., Xu C., Zhu H., Cai F.Q., He Y.L., Zhang W.F., Ma S.-C., Hu C., Gong M., Liu L., Shi W., Zhu H. Development and validation of a deep learning-based model using computed tomography imaging for predicting disease severity of Coronavirus disease 2019. Front. Bioeng. Biotechnol. 2020;8:898. doi: 10.3389/fbioe.2020.00898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mishra A.K., Das S.K., Roy P., Bandyopadhyay S. Identifying COVID19 from chest CT images: a deep convolutional neural networks based approach. J. Healthcare Eng. 2020:2020. doi: 10.1155/2020/8843664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ko H., Chung H., Kang W.S., Kim K.W., Shin Y., Kang S.J., Lee J.H., Kim Y.J., Kim N.Y., Jung H., Lee J. COVID-19 pneumonia diagnosis using a simple 2D deep learning framework with a single chest CT image: model development and validation. J. Med. Internet Res. 2020;22 doi: 10.2196/19569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song J., Wang H., Liu Y., Wu W., Dai G., Wu Z., Zhu P., Zhang W., Yeom K.W., Deng K. End-to-end automatic differentiation of the coronavirus disease 2019 (COVID-19) from viral pneumonia based on chest CT. Eur. J. Nucl. Med. Mol. Imag. 2020;47:2516–2524. doi: 10.1007/s00259-021-05267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuyama E. A deep learning interpretable model for novel coronavirus disease (COVID-19) screening with chest CT images. J. Biomed. Sci. Eng. 2020;13:140. doi: 10.4236/jbise.2020.137014. [DOI] [Google Scholar]
- 11.Jaiswal A., Gianchandani N., Singh D., Kumar V., Kaur M. Classification of the COVID-19 infected patients using DenseNet201 based deep transfer learning. J. Biomol. Struct. Dyn. 2020:1–8. doi: 10.1080/07391102.2020.1788642. [DOI] [PubMed] [Google Scholar]
- 12.Loey M., Manogaran G., Khalifa N.E.M. A deep transfer learning model with classical data augmentation and cgan to detect covid-19 from chest ct radiography digital images. Neural Comput. Appl. 2020:1–13. doi: 10.1007/s00521-020-05437-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh D., Kumar V., Vaishali, Kaur M. Classification of COVID-19 patients from chest CT images using multi-objective differential evolution–based convolutional neural networks. Eur. J. Clin. Microbiol. Infect. Dis. 2020;39:1379–1389. doi: 10.1007/s10096-020-03901-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B., Bai J., Lu Y., Fang Z., Song Q., Cao K., Liu D., Wang G., Xu Q., Fang X., Zhang S., Xia J., Xia J. Using artificial intelligence to detect COVID-19 and community-acquired pneumonia based on pulmonary CT: evaluation of the diagnostic accuracy. Radiology. 2020;296:E65–E71. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Do C., Vu L. International Society for Optics and Photonics; 2020. An Approach for Recognizing COVID-19 Cases Using Convolutional Neural Networks Applied to CT Scan Images, Applications of Digital Image Processing XLIII. 1151034. [DOI] [Google Scholar]
- 16.Wang S., Zha Y., Li W., Wu Q., Li X., Niu M., Wang M., Qiu X., Li H., Yu H., Gong W., Bai Y., Li L., Zhu Y., Wang L., Tian J. A fully automatic deep learning system for COVID-19 diagnostic and prognostic analysis. Eur. Respir. J. 2020;56 doi: 10.1183/13993003.00775-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He X., Yang X., Zhang S., Zhao J., Zhang Y., Xing E., Xie P. 2020. Sample-efficient Deep Learning for Covid-19 Diagnosis Based on Ct Scans. MedRxiv. [DOI] [Google Scholar]
- 18.Sakagianni A., Feretzakis G., kalles D., Koufopoulou C. Setting up an easy-to-use machine learning pipeline for medical decision support: a case study for COVID-19 diagnosis based on deep learning with CT scans. Importance Health Informat. Publ. Health during a Pandemic. 2020;272:13. doi: 10.3233/shti200481. [DOI] [PubMed] [Google Scholar]
- 19.Alshazly H., Linse C., Barth E., Martinetz T. Explainable COVID-19 detection using chest CT scans and deep learning. Sensors. 2021;21:455. doi: 10.3390/s21020455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silva P., Luz E., Silva G., Moreira G., Silva R., Lucio D., Menotti D. COVID-19 detection in CT images with deep learning: a voting-based scheme and cross-datasets analysis. Inform. Med. Unlocked. 2020;20:100427. doi: 10.1016/j.imu.2020.100427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ewen N., Khan N. 2020. Targeted Self Supervision for Classification on a Small COVID-19 CT Scan Dataset. arXiv preprint arXiv:2011.10188. [Google Scholar]
- 22.Ragab D.A., Attallah O., FUSI-CAD Coronavirus (COVID-19) diagnosis based on the fusion of CNNs and handcrafted features. PeerJ Comput. Sci. 2020;6 doi: 10.7717/peerj-cs.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Z., Yu S., Qin W., Liang X., Xie Y., Cao G. 2020. CT Super Resolution via Zero Shot Learning. arXiv preprint arXiv:2012.08943. [Google Scholar]
- 24.Zhang Y., Tian Y., Kong Y., Zhong B., Fu Y. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2018. Residual dense network for image super-resolution; pp. 2472–2481. Salt Lake City, Utah, June 18-22. [Google Scholar]
- 25.Li M., Shen S., Gao W., Hsu W., Cong J. In: Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. DLMIA 2018, ML-CDS. Stoyanov D., et al., editors. vol. 11045. Springer; Cham: 2018. Computed tomography image enhancement using 3D convolutional neural network. Lecture Notes in Computer Science. [DOI] [Google Scholar]
- 26.Dataset SARS-COV-2 CT https://www.kaggle.com/plameneduardo/sarscov2-ctscan-dataset as accessed on November 2020.
- 27.Dataset COVID-CT https://github.com/UCSD-AI4H/COVID-CT as accessed on 15th December 2020.
- 28.DIV2K Dataset https://data.vision.ee.ethz.ch/cvl/DIV2K/ as accessed on 25th December 2020.
- 29.Sufian A., Ghosh A., Sadiq A.S., Smarandache F. A survey on deep transfer learning to edge computing for mitigating the COVID-19 pandemic. J. Syst. Architect. 2020;108 doi: 10.1016/j.sysarc.2020.101830. [DOI] [Google Scholar]
- 30.Zhou T., Lu H., Yang Z., Qiu S., Huo B., Dong Y. The ensemble deep learning model for novel COVID-19 on CT images. Appl. Soft Comput. 2021;98 doi: 10.1016/j.asoc.2020.106885. [DOI] [PMC free article] [PubMed] [Google Scholar]