Abstract
Bcl-x pre-mRNA splicing serves as a typical example to study the impact of alternative splicing in the modulation of cell death. Dysregulation of Bcl-x apoptotic isoforms caused by precarious equilibrium splicing is implicated in genesis and development of multiple human diseases, especially cancers. Exploring the mechanism of Bcl-x splicing and regulation has provided insight into the development of drugs that could contribute to sensitivity of cancer cells to death. On this basis, we review the multiple splicing patterns and structural characteristics of Bcl-x. Additionally, we outline the cis-regulatory elements, trans-acting factors as well as epigenetic modifications involved in the splicing regulation of Bcl-x. Furthermore, this review highlights aberrant splicing of Bcl-x involved in apoptosis evade, autophagy, metastasis, and therapy resistance of various cancer cells. Last, emphasis is given to the clinical role of targeting Bcl-x splicing correction in human cancer based on the splice-switching oligonucleotides, small molecular modulators and BH3 mimetics. Thus, it is highlighting significance of aberrant splicing isoforms of Bcl-x as targets for cancer therapy.
Keywords: Bcl-x, Alternative splicing, Cell apoptosis, Splicing correction, Splice-switching oligonucleotides, Small molecular modulators
Background
Apoptosis regulator Bcl-extra (Bcl-x), also named BCL2L or BCL2L1, is a typical example of apoptotic response gene impacted by splicing. It is an essential member of B-cell lymphoma 2 (Bcl-2) apoptosis family that regulates cell fate [1, 2]. Bcl-x nascent transcripts are alternatively spliced and mainly encode two antagonistic isoforms. The long isoform Bcl-xL blocks apoptosis by inhibiting pro-apoptotic counterparts of Bcl-2 family, whereas the short isoform Bcl-xS can promote apoptosis [2]. An increasing body of data suggests that dysregulated expression of Bcl-x apoptotic isoforms contributes to multiple hallmarks of human cancers. For example, Bcl-xL level was strongly enhanced in cancer cells at the invasive forefront of human breast carcinomas and simultaneously acquired resistance to apoptotic stimuli [3, 4]. However, Bcl-xS conferred the therapeutic sensitivity by decreasing the apoptosis threshold [5]. The ratio of pro-apoptotic Bcl-xS and anti-apoptotic Bcl-xL proteins plays a vital role in regulating the switch between cell life and death. Hence, in this review, we summarize the patterns and the splicing regulatory network of Bcl-x pre-mRNA splicing. In addition, we describe how this aberrant splicing impacted apoptosis, autophagy, invasion and metastasis, immune response, as well as clinical therapy resistance in cancer. Furthermore, we outline the emerging strategies that modulate the cancerous Bcl-x splicing and restore the balance of Bcl-xL/Bcl-xS ratio. Targeting the oncogenic splicing of Bcl-x is believed to result in sensitized cell death by simultaneously blocking Bcl-xL and enhancing Bcl-xS splicing [6].
Splicing isoforms of Bcl-x
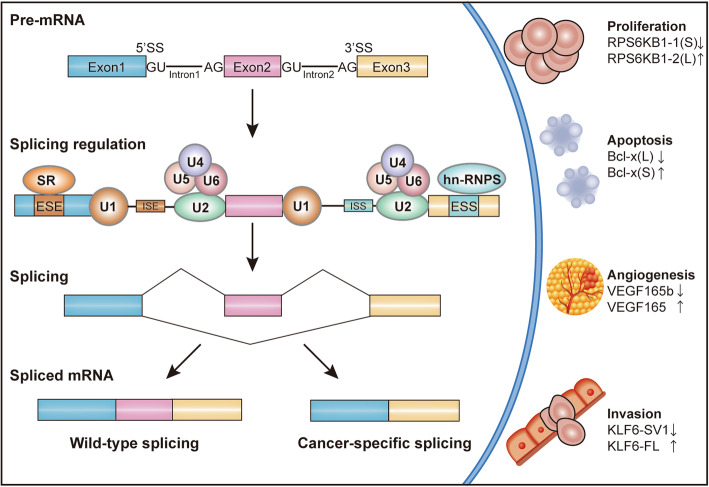
Alternative splicing expands the coding capacity of genomes of eukaryotes significantly through splice site selection (Fig. 1) [7]. Nearly >95% of human multi-exonic genes could be alternatively spliced into mRNAs isoforms [8]. Splicing reaction is orchestrated by a highly dynamic ribonucleoprotein complex known as spliceosome and hundreds of related proteins [9]. The spliceosome recognizes and assembles reversibly on pre-mRNA to catalytic splicing in a stepwise manner. This process is further modulated by a number of cis-acting elements and trans-acting factors (splice factors) bound to them [9]. Indeed, mutations in cis-regulatory sequences and spliceosomal associated proteins are enriched in cancer. These mutations always affect the splicing of cancer-related genes [10, 11]. A growing body of evidence has revealed aberrant splicing events as contributors of hallmarks of tumorigenesis, such as proliferation, angiogenesis, invasion and apoptosis (Fig. 1) [11, 12].
Fig. 1.
Alternative splicing and the effect of aberrant alternative splicing on cancer progression. The spliceosome, consists of five small nuclear ribonucleoproteins particles (U1, U2, U4, U5 and U6) and hundreds of additional proteins, recognizes the consensus sequence of each intron and assembles reversibly on splice sites to catalytic pre-mRNA splicing. SR proteins and hnRNPs bound to exonic or intronic regulatory elements to promote or prevent the use of splice sites thus affecting alternative splicing decisions. The figure displays some examples of cancer-specific splicing events that contribute to distinct hallmarks of cancer. Arrows up and down indicate the corresponding isoforms contributing or suppressing the hallmark respectively
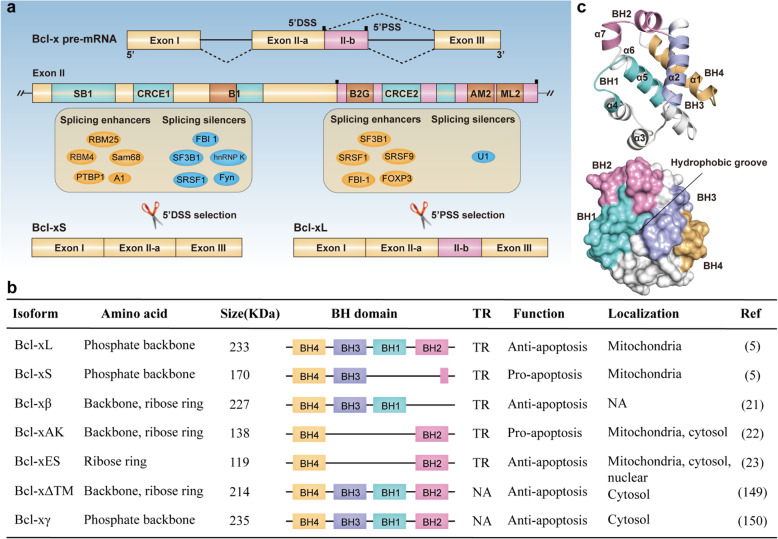
Bcl-x, a critical apoptotic gene of the Bcl-2 family, is located in chromosome 20 (20q11.1). It was first discovered by using Bcl-2 fluorescent probe hybridization in chickens [2]. Subsequently, two antagonistic isoforms of Bcl-x pre-mRNA in human body were isolated, which then were identified as Bcl-xL and Bcl-xS, respectively. Splicing selection closer to the proximal 5' splice site (5'PSS) of exon 2 resulted in the long anti-apoptosis isoform Bcl-xL (Fig. 2a), which contained four exons and was composed of 780 bp. The primary structure of Bcl-xL is composed of 233 aa, which contains C-terminal hydrophobic transmembrane (TM) domain responsible for the anchoring to membranes and all four BH domains (BH1-4) (Fig. 2b,c). When the splicing occurred near the cryptic distal 5' splice site (5' DSS) of exon 2, the short isoform pro-apoptotic Bcl-xS with 591 bp was produced. Stable Bcl-xS expression played an important role in regulating the ability of pro-survival genes to inhibit apoptotic cell death [2]. Bcl-xS protein is encoded by 170 aa containing both BH3, BH4, and TM domains but lacking BH1 and BH2 domains, which might lead to alternation of hydrophobicity. In addition, other splice isoforms encoded by Bcl-x also had been identified in different cell types and performed diverse functions (Fig. 2b) [13–17]. Despite multiple isoforms that Bcl-x could splice, the pro-apoptotic Bcl-xS and anti-apoptotic Bcl-xL are still the predominant isoforms acting in cell fate.
Fig. 2.
Bcl-X pre-mRNA splicing and structures of splicing isoforms. a. Alternative splicing mode and splicing regulation. Splicing occurred closer to the 5'PSS of exon 2 produces the long isoform Bcl-xL. Alternative splicing occurred near the 5' DSS of exon 2 produces the short isoform Bcl-xS. In addition, distinct cis-elements and splice factors bind to cis-elements to influence the alternative 5' splice site selection of Bcl-x pre-mRNA. b. General characteristics of isoforms spliced from Bcl-x pre-mRNA. c. The protein structures of Bcl-xL. The secondary structure of Bcl-xL and the position in the space of BH domains (up). Tertiary structure of Bcl-xL and the BH domain and hydrophobic groove are showed (down)
Regulation of Bcl-x pre-mRNA splicing
Cis-regulatory elements
Cis-regulatory elements are short nucleotide motifs within pre-mRNA transcripts that providing binding sites for specific trans-acting factors. It can be categorized into exonic splicing enhancers (ESEs), exonic splicing silencers (ESSs), intronic splicing enhancers (ISEs) and intronic splicing silencers (ISSs) depending on their position and impact on the use of splice site [18] Bcl-x pre-mRNA contains several cis-elements acting as splicing activators or repressors by interacting with related splicing factors (Fig. 2a). For example, SB1 (361bp), located in the first half of exon 2, was defined as an ESE because splicing to Bcl-xS was even stronger in the absence of SB1. Similarly, DNA damage-induced Bcl-xS splicing only increased in the presence of SB1 [19]. In addition, B1 was a composite element located upstream of 5' DSS of Bcl-x pre-mRNA. The 5' portion of B1 displayed ESE activity, whereas the 3' portion was occupied with ESS element. hnRNP K bound to the silencer portion of B1 to repress the production of Bcl-xS [20]. Moreover, B2G module was a 30-nucleotides G-rich element located immediately downstream of the 5' DSS. Combination of hnRNP F/H to B2G enhanced Bcl-xS splicing. B3 located up-stream of 5'PSS to favor the production of Bcl-xL. The two elements ML2 and AM2 within B3 were identified to enhance Bcl-xL splicing through interacting with SRp30c [21]. Deleting B2G and B3 regions completely abrogated production of Bcl-xS and Bcl-xL respectively [22]. Another two cis-elements CRCE1 and CRCE2 within exon 2 were essential for ceramide-responsive Bcl-x expression. Mutation of CRCE1 or CRCE2 induced a decreased ratio of Bcl-xL/Bcl-xS [23]. In addition to exonic elements, the intron region downstream from Bcl-xL 5'PSS also had been identified to mediate signals from extracellular factors such as interleukin-6 and granulocyte-macrophage colony-stimulating factor (GM-CSF) to repress Bcl-xL splicing [24].
Trans-acting factors
A variety of trans-acting factors are involved in the formation of splicing regulatory network, including SR proteins, hnRNPs as well as some transcription factors (Fig. 2a) [25]. Posttranslational modifications of splicing factors would also change their binding state to cis-elements. For example, Sam68 bound to Bcl-x pre-mRNA specifically and recruited hnRNP A1 to a particular region, which caused the selection of Bcl-xS in a dose-dependent manner. This favor could be inverted by tyrosine phosphorylation of Sam68 [26]. In addition, SRSF1 was known to compete with hnRNPA1 to promote Bcl-xL splicing. However, the activity of SRSF1 itself was antagonized by splicing factors RBM4 and PTBP1 [27, 28]. Moreover, a multicomponent regulatory hub consisting of SRSF10, hnRNP A1/A2, and Sam68 was reported to activate the 5'DSS of Bcl-x in response to DNA damage [29]. Because alternative splicing is coupled to transcription, a large number of transcription factors were found to influence splicing selection. For instance, E2EF1 was suggested to increase Bcl-xS isoform through upregulating SRSF2 [30]. FBI -1 could interact with the full length or C-terminal domain of Sam68 to counteract Sam68-mediated apoptosis [31]. Moreover, TCERG1 [32], FOXP3 [33], ETS [34] as well as cellular signal pathways and other regulatory modes such as exon junction complex, G-rich sequences were all involved in Bcl-x splicing and had been summarized in Table 1 (Table 1).
Table 1.
Trans-acting factors involved in Bcl-x splicing regulation
| Regulation methods | Mechanism | Selection | Ref | |
|---|---|---|---|---|
| RNA binding proteins | Sam68 | Recruit hnRNP A1 to certain regions. | Bcl-xS | [26] |
| SRSF1 (ASF/SF2) | Compete with hnRNP A1. | Bcl-xL | [26, 35] | |
| SRSF10 | Collaborate with hnRNP A1/A2 and Sam68. | Bcl-xS | [29] | |
| SRSF2 (SC35) | As a direct transcriptional target of E2F1. | Bcl-xS | [30] | |
| SRSF3 | Favor the selection of the 5' DSS. | Bcl-xS | [28] | |
| SRSF7 | Favor the selection of the 5' DSS. | Bcl-xL | [28] | |
| SRSF9 (SRp30c) | Bind to ML2 and AM2. | Bcl-xL | [21] | |
| TRA2β | Favored selection of the 5' DSS. | Bcl-xS | [28] | |
| hnRNP F/H | Bind to B2G region. | Bcl-xS | [22, 36] | |
| PTBP1 (hnRNP I) | Bind to polypyrimidine to promote 5' DSS selection | Bcl-xS | [28] | |
| hnRNP A1 | Interact with Sam68. | Bcl-xS | [25, 26] | |
| hnRNP A2/B1 | Regulated by Fyn activity. | Bcl-xL | [37] | |
| hnRNP K | Bind to silencer element of the 5' DSS. | Bcl-xL | [20] | |
| RBM4 | Antagonize oncogenic SRSF1. | Bcl-xS | [38] | |
| RBM10 (S1-1) | Block the GGGUAAG of exon 2. | Bcl-xS | [39] | |
| RBM11 | Antagonize SRSF1. | Bcl-xS | [40] | |
| RBM25 | Bind to CGGGCA sequence within exon 2. | Bcl-xS | [41] | |
| Transcription factors | E2F1 | Upregulate SC35 protein expression. | Bcl-xS | [30] |
| FBI-1 | Interact with Sam68 and affects its binding. | Bcl-xL | [31] | |
| TCERG1 | Increase the elongation rate of RNAPII. | Bcl-xS | [32] | |
| FOXP3 | Repress hnRNPF binding to 5'DSS. | Bcl-xL | [33] | |
| SAP155 (SF3B1) | Bind to CRCE 1 region. | Bcl-xL | [42] | |
| Signal pathway | PKC signal | Through SB1 to repress the 5'DSS splicing. | Bcl-xL | [43] |
| PI3K/PKCι signal | Regulate SAP155-CRCE1 complex formation. | Bcl-xL | [44] | |
| LPS/PRMT2 or TNF-α pathway | Interact with Sam68 and regulate its subcellular localization via its SH3 domain. | Bcl-xL | [45] | |
| G4s and G4s ligands | G-quadruplexes (G4s) | Close to the two alternative 5'SS to compete with other RNA structures or proteins | Bcl-xS or Bcl-xL | [46, 47] |
| G-quadruplex ligands (GQC05) | Stabilize G-quadruplexes. | Bcl-xS | [46] | |
| EJC | Exon junction complex ( EJC ) | RNPS1 and core EJC proteins control Bcl-x splicing through cis-acting elements SB1. | Bcl-xL | [48] |
Epigenetic modifications
Epigenetic modifications had been suggested to interplay intricately with alternative splicing [49]. It had been reported that DNA methylation at exons and splicing sites were involved in over 20% splicing modulation by regulating the elongation rate of RNA polymerase II primarily [50]. In other studies, the dynamic histone acetylation mark of H3K4me3 nucleosome played a critical role in Mcl-1 pre-mRNA splicing [51]. Moreover, N6-methyladenosine modification was indicated to regulate splicing by co-localization with splice sites and reshaping the structure of pre-mRNA [52]. However, there were poor reports about whether and how the epigenetic modifications mentioned above affected the splicing decision of Bcl-x pre-mRNA. To date, ncRNA was the most common epigenetic regulation identified to influence Bcl-x splicing that functions as 'interactors' or 'hijackers' of splicing factors [49]. LncRNA BC200, LINC00162 as well as LncRNA-HEIH had been shown to modulate Bcl-x pre-mRNA splicing effectively [53–55]. To investigate the splicing mechanism regulated by ncRNA in-depth is necessary.
The function of aberrant Bcl-x splicing in cancer
Apoptosis
Apoptosis, characterized by a series of morphological alternations including cell shrinkage, pyknosis and karyorrhexis, plasma membrane blebbing and apoptotic body formation, is a mechanism for all multicellular organisms to modulate cell life development [56]. Abnormalities in apoptosis play a crucial role in the progression of various human disease like cancer [57]. Therefore, targeting apoptotic pathways has been a mainstay for the cancer drug discovery and development. There are two commonly established apoptotic pathways in mammals: the extrinsic pathway of apoptosis mediated by the death receptor and the intrinsic pathway of apoptosis mediated by mitochondria [58]. The extrinsic apoptotic signal begins when extracellular death-inducing factors bind to its receptors (TNFR, TRAIL, FasL), recruiting adapter proteins (TRADD, FADD, caspase 8 and/or caspase 10) to form the death inducing signaling complex [59]. The intrinsic apoptotic pathway is closely regulated by the Bcl-2 family proteins. Bcl-2 proteins mainly were divided into three subgroups up to their BH domain: BH3 only proteins to initiate apoptosis (Bim, Bad, Bid, Noxa, Bmf, Hrk, Bik, Puma), pro-apoptotic proteins act as apoptotic executioner (Bax, Bak, Bok) and anti-apoptotic subfamily (Bcl-2, Bcl-xL, Bcl-W, A1, Bcl-B, Mcl-1) [60]. Initiated by internal stimuli such as DNA damage, hypoxia and oxidative stress, activated-BH3 only proteins inhibit the anti-apoptotic Bcl-2 proteins. Subsequently, activated and oligomerized Bax/Bak located in the mitochondrial outer membrane, promoting mitochondrial outer membrane permeability (MOMP), the release of cytochrome C and caspase activation [61]. Disruption in the balance of pro-apoptotic and anti-apoptotic members of the Bcl-2 family proteins promotes carcinogenesis and cancer cell survival [62]. More importantly, anti-apoptotic Bcl-2 proteins are widely over-expressed in cancers and have been established to contribute to therapy resistance, recurrence and poor prognosis [63, 64].
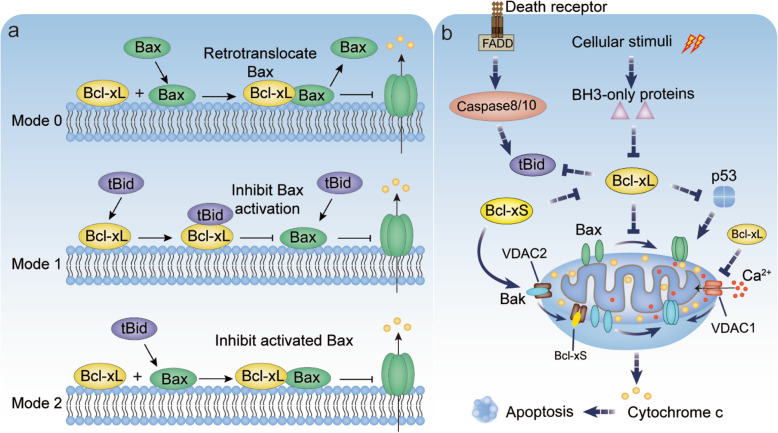
Alternative splicing of Bcl-x pre-mRNA is one of the earliest oncogenic splicing events critical for apoptotic responses of cancer cells. The elevated level of Bcl-xL caused by aberrant splicing has been revealed in a multitude of human cancers and is considered to be a powerful driving force for cell apoptotic resistance (Table 2) [1, 78]. Instead, cells with highly expressed Bcl-xS were more sensitive to apoptosis stimuli [25]. Structural features enabled Bcl-xL to bind to its natural ligands, such as pro-apoptotic Bcl-2 family members that respond to a variety of cellular stimuli [64]. This process had been well documented by the tertiary structure of Bcl-xL/BH3 peptides, that pro-apoptotic BH3 peptide bound to the hydrophobic groove of Bcl-xL via hydrophobic and electrostatic interactions [88]. In general, Bcl-xL distributed on the intracellular membrane appeared to regulate apoptosis mainly by three modes. In mode 0, Bcl-xL could prevent the binding of apoptotic effectors Bax to mitochondrial outer membrane through retrotranslocating Bax from the mitochondria into cytosol constantly (Fig. 2a) [89]. In mode 1, Bcl-xL could sequester BH3-only activators (For example Bid truncated in the death receptor-mediated pathways (Fig. 3b)) to prevent them from binding to and activating Bax (Fig. 3a). In mode 2, Bcl-xL was suggested to directly bind to activated Bax to prevent its oligomerization and pore formation, which prevented the release of caspase activator from mitochondrial outer membrane (Fig. 3a) [90]. However, Bcl-xL sequestration could be derepressed by sensitizer BH3 only proteins (For example Bad), which then induced activation of Bax and MOMP (Fig. 3b) [90]. Bogner et al. suggested that the allosteric regulation by Bcl-xL complexes might play an important role in this process [91]. Moreover, Bcl-xL was suggested to inhibit a weak Bax activation and apoptotic signal via directly sequestrating active cytosolic p53 induced by damage stimuli (Fig. 3b) [92]. The above demonstrated that apoptosis decision of cells was dependent on the relative abundance of Bcl-xL and its pro-apoptotic counterparts [64, 90]. In addition to the already established roles in mitochondrial apoptotic pathways, Bcl-xL was proved to function as an inhibitor of VDAC1 to prevent apoptosis induced by excessive Ca2+ transferred from endoplasmic reticulum to mitochondria (Fig. 3b) [93]. Thus it is imperative to investigate the in-depth mechanism that Bcl-xL used to coordinate apoptotic signals from multiple pathways and ultimately, form an integrated perspective. Compared to Bcl-xL, the short isoform Bcl-xS was reported as a negative regulator of survival because it could inhibit the function of Bcl-xL by forming heterodimers with Bcl-xL through the BH3 domain, or disrupting the VDAC2-Bak complex to cause the release of Bak and activation of MOMP (Fig. 3b) [94, 95]. Furthermore, Bcl-xS induced activation of Bak and promoted apoptosis through apoptosome-dependent and independent pathways [96]. Therefore, the antagonistic roles played by Bcl-x isoforms were critical for cell fate decision. Alternative splicing regulation of Bcl-x to promote Bcl-xS but inhibit Bcl-xL splicing could act as a tumour suppression strategy. For instance, Src family kinase Fyn was found to decrease the phosphorylation of Sam68 but regulate hnRNPA2/B1 expression, which synergistically promoted the splicing of Bcl-xL and inhibited apoptosis of pancreatic cancer cells. Fyn inhibition down-regulated hnRNPA2/B1 expression and increased Bcl-xS splicing [37]. In addition, study on human liver fibrosis suggested that Bcl-xL was preferentially spliced in human hepatic stellate cells and consistent with apoptosis resistance of HSCs. Antisense oligonucleotides inhibiting Bcl-xL splicing induced HSC cell apoptosis [97]. Interestingly, the IE86 gene of human cytomegalovirus was found to inhibit apoptosis and promote proliferation of glioma cells by enhancing the favor splicing of Bcl-xL mediated by hnRNPA2/B1 [98]. Thus, the favored use of 5’PSS splice site in Bcl-x pre-mRNA contributes to the escape of cancer cells from intrinsic programmed apoptosis.
Table 2.
Aberrant Bcl-x splicing in cancers and its clinical application
| Cancer type | Bcl-xL/S | Function | Ref | |
|---|---|---|---|---|
| Apoptosis | Hepatocellular Carcinomas | Bcl-xL↑ | Inhibit apoptosis initiated by cellular stimuli | [65] |
| Colorectal cancer (CRC) | Bcl-xL↑ | Drive tumourigenesis and progression. | [66] | |
| Breast cancer | Bcl-xL↑ | Suppress BETi-induced apoptosis. | [67] | |
| Meningioma | Bcl-xL↑ | Contribute to apoptosis induced by Dovitinib. | [68] | |
| Malignancy | Gastric cancer | Bcl-xL↑ | Associated with high Beclin1 expression. | [69] |
| Tongue Carcinoma | Bcl-xL↑ | Related to the degree of differentiation. | [70] | |
| Hodgkin lymphoma | Bcl-xL↑ | Consistent with the severity of patients. | [71] | |
| Myeloproliferative neoplasms | Bcl-xL↑ | Progressively over-expressed. | [72] | |
| Lymphomas | Bcl-XS/L↓ | Expressed by malignant cells. | [73] | |
| Wilms' tumours | Bcl-XS/L↓ | Negatively correlated with tumour stage. | [74] | |
| Endometrial carcinoma | Bcl-XS/L↓ | Correlated with pathological grading. | [75] | |
| Metastasis | Pancreatic cancer | Bcl-xL↑ | Promote metastasis | [76] |
| Glioblastoma | Bcl-xL↑ | Promote cell migration, invasion, angiogenesis and stemness. | [77] | |
| Melanoma | Bcl-xL↑ | [77] | ||
| Drug-resistance | Chondrosarcoma | Bcl-xL↑ | Confer resistance to chemotherapy. | [78] |
| Ewing sarcoma | Bcl-xL↑ | Resistant to olaparib. | [79] | |
| Ovarian carcinoma (OC) | Bcl-xL↑ | Confer resistance to chemotherapy. | [80] | |
| Hepatocellular carcinoma | Bcl-xL↑ | Chemoresistance and poor prognosis. | [81] | |
| Urothelial Carcinoma | Bcl-xL↑ | Effectively inhibited cisplatin-resistant UCs. | [82] | |
| Radiation | laryngeal cancer | Bcl-xL↑ | Associated with radioresistant. | [83] |
| Non-small cell lung cancer | Bcl-xL↑ | Enhance irradiation resistance. | [84] | |
| Prostate cancer | Bcl-xL↑ | Enhance survival to cells exposured to IR. | [85] | |
| Osteosarcoma | Bcl-xL↑ | Enhance irradiation resistance. | [86] | |
| Malignant pleural mesothelioma | Bcl-xL↑ | Negatively associated with radiosensitivity. | [87] |
Fig. 3.
Cell apoptosis regulated by Bcl-x isoforms. a. Three modes that had been proposed to explain how Bcl-xL regulate MOMP. Mode 0: Bcl-xL prevented the binding of apoptotic effectors Bax to mitochondrial outer membrane through retrotranslocating Bax from the mitochondria into cytosol constantly. Mode 1: Bcl-xL sequestered BH3-only activators (tBid) to prevent them from binding to and activating Bax. Mode 2: Bcl-xL directly bound to activated Bax to prevent its oligomerization and MOMP. b. Cell apoptosis pathways regulated by Bcl-xL and Bcl-xS
Autophagy
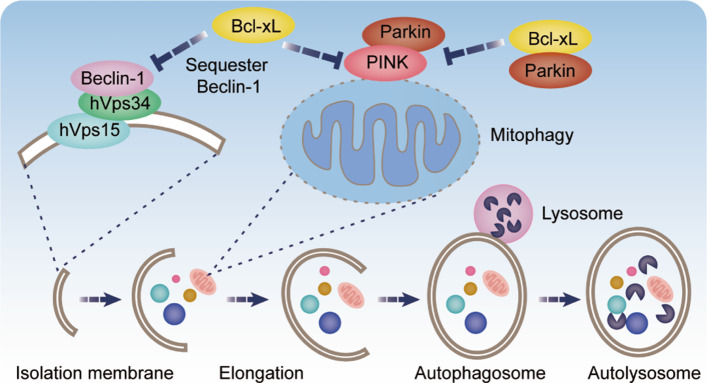
In addition to apoptosis, the long isoform Bcl-xL also had been suggested to be involved in autophagy, which was an evolutionarily conserved pathway and played a double-edged role in tumour progression [84]. A recent study explored that Bcl-xL could inhibit PINK1/Parkin-dependent mitophagy through directly interacting with Parkin and PINK1 to inhibit the translocation of Parkin from cytoplasm into mitochondria (Fig. 4) [99]. More importantly, Bcl-xL was identified to hinder macro-autophagy mediated by class III PI3K pathway through direct interactions with Beclin-1, a new BH-3 only protein that is essential to regulate the initial of autophagy [100, 101]. Low expressed Beclin-1 but highly expressed Bcl-xL is consistent with malignant phenotype and poor prognosis of cancer [69, 102]. Bcl-xL physically interacted with BH3 domain of Beclin-1 and disrupted hVps34–Beclin-1 complex which stimulated autophagosomes formation (Fig. 4) [103, 104]. BH3 mimetics and overexpressed BH3-only proteins could displace Beclin-1 from Bcl-xL and stimulate autophagy. Intriguingly, Beclin-1 was reported to induce apoptosis of glioblastoma cells through binding to Bcl-xL [105], whereas another research suggested that heterooligomers formed by Bcl-xL and Beclin-1 could maintain full anti-apoptotic function in HeLa cells induced by staurosporine [106]. These results support the model that direct interactions between Bcl-xL and Beclin-1. However, Lindqvist LM et al. suggested that Bcl-xL had no measurable effect on autophagy in the absence of Bax/Bak [107]. When Bax/Bak were present, inhibiting the pro-survival Bcl-2 family members stimulated autophagy and correlated with increased cell death, suggesting that inhibition of Bcl-xL on autophagy was an indirect effect generated from apoptosis inhibition by a yet unknow mechanism. In summary, the possible relevance between apoptosis and autophagy in the process of cell death mediated by Bcl-xL includes:(1) Bcl-xL physically interacts with Beclin-1 to regulate apoptosis and autophagy synergistically or antagonisticly; (2) Bcl-xL does not bind to Beclin 1 but instead regulate autophagy by inhibiting Bax/Bak mediated apoptosis [3]. Thus, further research is required to determin the crosstalk between apoptosis and autophagy mediated by Bcl-xL and other Bcl-2 family proteins, which is of great significance for maintaining the overall cell fates.
Fig. 4.
Cell autophagy mediated by Bcl-xL. Bcl-xL inhibited initial steps of autophagy by interacting with the core regulators of autophagy Beclin-1, which disrupted the hVps34–Beclin-1 complex and limited its ability to stimulate autophagosome formation. Bcl-xL also could inhibit PINK1/Parkin-dependent mitophagy through directly interacting with PINK1 and Parkin to inhibit the translocation of Parkin from cytoplasm into mitochondria
Invasion and metastasis
Bcl-xL had been suggested to contribute to invasion and metastasis in multiple cancer types. After knocking Bcl-xL, the invasive and metastatic phenotype of CRC cells were reduced but did not cause spontaneous cell death [108, 109]. Studies of oral tongue cancer and breast cancer found that Bcl-xL expression was significantly high in metastasis tissue [4, 70] . In transformed human mammary epithelial cells, Bcl-xL directly interacted with RAS to modulate RAS signaling through BH4 domain, which was critical for RAS induced stemness and cancer initiating cell phenotype [110]. In addition, overexpressed Bcl-xL in human melanoma was found to promote vasculogenic structures through CXCL8/CXCR2 pathway. Meanwhile, the increased cancer stem cell markers associated with stemness and aggression of tumour cells ( for example HIF-1α, NANOG, OCT4, BMI1, and SOX2) were also observed [111]. Notably, a recent study of Bcl-xL with defection in apoptosis suggested that anti-apoptotic function of Bcl-xL might be separated from its roles in the motility of cancer cells. For example, Choi et al. [76] showed that Bcl-xL defected in anti-apoptotic function promoted epithelial-mesenchymal transition (EMT), migration, invasion, as well as stemness of panNET and breast cancer cells. Additionally, Bessou et al. [112] suggested that the complex formed by BH4 domain of Bcl-xL and VDAC1 acted on MOMP to increase ROS in mitochondrial electron transport chain and inhibit the absorption of Ca2+, thereby promoting migration and metastasis of breast cancer cells independently of apoptotic activity. How Bcl-xL regulates the invasion and metastasis of cancer cells independently of apoptosis still needs further exploration.
Anti-tumor immunity
Anti-apoptotic Bcl-xL has been demonstrated to play a key role in the survival of immune cells and immune responses. Grillot et al. has reported that Bcl-xL is highly expressed in CD4+ CD8+ thymocytes which exhibited increased viability in vitro [113]. Enforced Bcl-xL expression could also restore the development of splenic B Lymphocyte [114]. In addition, Bcl-xL was proved to promote survival of effector T cells and antigen-bearing dendritic cells [115, 116]. Regulatory T cells showed enhanced suppressive capacity through increasing Bcl-xL expression, which provide a new strategy for treatment of tumours through remodelling regulatory T cells [117]. Surprisingly, Bcl-xL was demonstrated to protect tumour cells from Natural Killer cells-mediated suppression and therefore exerted tumour-progressive activity [118, 119]. However, Andersen et al. suggested highly expressed Bcl-xL of cancer cells were the common target recognized by specific T cells [120]. They also speculated that immune responses against apoptosis inhibitors like Bcl-xL might represent a general feature in cancer [120]. Taken together, there is a complex effect of Bcl-xL expression on anti-tumour immune response. It is of great significance to identify the role of overexpressed Bcl-xL in immune escape of tumor cells.
Clinical impaction of Bcl-x splicing in cancer
Chemotherapy
Tolerability generated during chemotherapy such as apoptosis escape and Epithelial mesenchymal transformation (EMT), results in poor prognosis and is currently impeding the success of targeted therapies in cancer treatment [121, 122]. Plenty of evidence suggested that Bcl-xL-dependent apoptotic inhibition was the main reason that promoted chemotherapy resistance in tumours in vitro and vivo (Table 2) [123, 124]. A study on breast cancer showed that cells passed through EMT obtained therapeutic resistance by upregulating Bcl-xL transcripts. However, apparent apoptotic resistance was removed after deleting Bcl-xL [4]. Bcl-xL was also found to mediate doxorubicin resistance of breast cancer through the Ring finger protein 6/Estrogen receptor α/Bcl-xL pathway [125]. Inhibiting Bcl-xL expression in breast cancer cells enhanced the cytotoxicity and apoptosis induced by T-DM1 [126]. Additionally, increased CXCR4 expression in ovarian cancer induced cisplatin resistance through promoting Bcl-xL/S [123]. Upregulated Bcl-xL expression was also found to be involved in resistance to therapy targeting Bcl-2 in mantle-cell lymphoma and Acute Myelocytic Leukemia [57, 127]. Regarding melanoma, it has been demonstrated that forced expression of ectopic Bcl-xL converted drug-sensitive cell lines into drug-resistant ones [128]. However, vivo-Morpholino (vMO) antisense oligomers that used to upregulate Bcl-xS expression but decrease Bcl-xL in chronic myeloid leukemia (CML) increased growth inhibition and apoptotic sensitivity of imatinib mesylate-resistant CML cells [5]. Similarly, overexpressed Bcl-xS in human breast carcinoma cells induced a remarkable increase in sensitivity to chemotherapy agents, but did not affect cell viability by itself [129].
Radiotherapy
The splicing favor of Bcl-xL contributed to long-term radiotherapy resistance (Table 2). Clinical data showed that Bcl-xL was expressed by about 91% of laryngeal cancer patients resistant to radiotherapy, suggesting a critical function of Bcl-xL in radiotherapy [83]. Streffer et al. [130, 131] found that glioma cell lines with high Bcl-xL expression had higher ED50 (2.9 ± 0.8Gy) than cell lines with lower Bcl-xL. However, no association with radiosensitivity was observed for the expression levels of Bcl-xS. Highly expressed Bcl-xL was also found to cause radiation resistance of osteosarcoma cells with both low and high metastasis level, and Bcl-xL downregulation could significantly enhance radiation cytotoxicity of osteosarcoma cells [86]. Moreover, inhibiting the expression level of Bcl-xL were suggested to reverse radio-resistance and regulate radiation-induced apoptosis of mesothelioma, breast cancer, prostate cancer, colorectal cancer as well as non-small cell lung cancer [84, 87, 132, 133]. In addition to therapeutic effects, irradiation was well known to induce increased invasiveness and metastasis of cancer cells. Ho et al. demonstrated that the expression of Bcl-x was elevated after irradiation, which promoted the malignant actions of lung cancer cells [134]. A recent study also suggested that upregulated Bcl-xL induced invasion of cancer cells that underwent sublethal doses of irradiation by stimulating respiratory complex I and increasing additional ROS production, which might be involved in the local recurrence or distal metastasis of somne patients after radiotherapy [135]. Interestingly, the expression of Bcl-xL could enhance energy metabolism and prevent oxidative stress, which might be involved in the alleviation of mitochondrial oxidative stress induced by radiation [136]. In addition, inhibition targeting Bcl-xL/2 had been found to reverse the pulmonary fibrosis induced by ionizing radiation [137]. These results provided a wealth of evidence that inhibition the endogenous expression of Bcl-xL might promote both radiation sensitization and radiation protection. However, combination of γ-irradiation and genetic loss but not pharmacological inhibition of Bcl-xL was found to cause fatal kidney damage and secondary anemia in adult mice, and the underlying mechanism remained unclear [138]. This finding demonstrated the protective role of Bcl-xL in adult kidney, which also represents challenges for the radio-sensitization targeting Bcl-xL.
Strategies modulating Bcl-x splicing in cancer
Splice-switching oligonucleotides
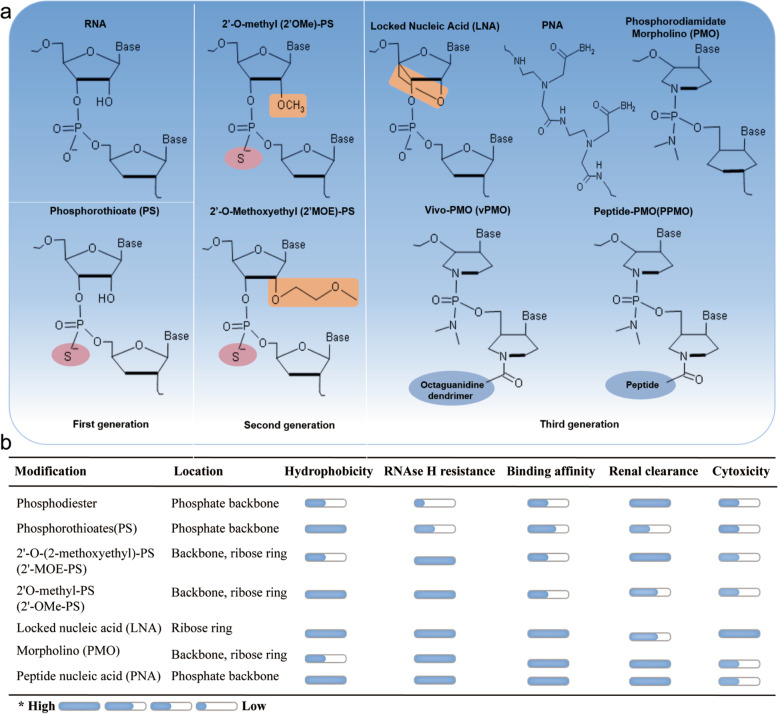
SSOs, typically 15-30 nucleotides, is a kind of synthetic, modified, steric block antisense oligonucleotides which have been widely used to disrupt the splicing mode of pre-mRNA through Watson-Crick base pairing. The generated steric hindrance but not degradation of targeted transcripts affected accessibility of splicing factors and visibility of spliceosome, which led to splicing isoforms switching ultimately [139]. Notably, natural oligonucleotides had been proved to be quite ineffective due to their defects such as easy to be degraded, lower affinity, and higher off-target effect. Therefore, various chemical modifications on phosphate backbone or ribose rings of SSOs had been developed to allow for improved stability and binding affinity, meanwhile, reduced cytotoxicity and immunogenicity [139]. Common types of oligonucleotides chemistry have been shown in Fig. 5. Notably, the third generation of antisense oligonucleotides was featured by furanose ring modifications of nucleotides including phosphorodiamidate morpholinos (PMOs), locked nucleic acid and peptide nucleic acid. PMOs was a type of neutrally charged nucleic acid, in which the furanose ring was substituted by a morpholine ring while each unit was bridged with a phosphorodiamidate linkage. PMOs usually needed to be conjugated to cell-penetrating peptides or covalently linked to an octaguanidine dendrimer for efficient delivery due to their rapid renal clearance. To date, PMOs modified SSOs drugs eteplirsen and golodirsen had been approved by the FDA for clinical therapy of Duchenne muscular dystrophy and spinal muscular atrophy, respectively [140, 141]. In addition, for effective SSOs of target genes, the optimized length, sequence constitution, secondary structures, accessibility, as well as thermodynamic properties were all critical factors [92]. Generally, SSOs base-paired to the alternative splice siteof Bcl-x pre-mRNA could block arrival of spliceosome and binding of splicing factors to their target motif, which led to the redirection of splicing favor (Fig. 6a). Bcl-xSSOs could promote apoptosis and drug sensitivity of cancer cells by correcting Bcl-xL splicing to Bcl-xS efficiently [142]. Some Bcl-xSSOs sequences used in preclinical had been summarized in Table 3. Mercatante et al. proved that endogenous highly expressed Bcl-xL was positively correlated with cellular response to Bcl-xSSOs induced splice shift [147], which indicated that normal cells with low expressed Bcl-xL might be more resistant to Bcl-xSSOs therapy. 2'-OMe-PS modified Bcl-xSSOs caused a splice shift from Bcl-xL to Bcl-xS and increased apoptosis of prostate cancer, breast cancer, and hepatic stellate cells [97, 147]. In addition, splice redirection of Bcl-x pre-mRNA induced by 2'-MOE modified Bcl-xSSOs in glioma cells and melanoma xenograft models showed pro-apoptotic effect and reduced tumour load respectively [142, 143]. Moreover, vMO modified Bcl-xSSOs was found to correct aberrant splicing of Bcl-x in CML cells and improve therapeutic sensitivity to imatinib mesylate significantly [5, 148]. Therefore, highly expressed Bcl-xL could be reversed by modified Bcl-xSSOs, which allowed the redirection of aberrant splicing and rebalanced the ratio of Bcl-xL/Bcl-xS [6].
Fig. 5.
Chemical modifications of splice switching oligonucleotides. a. Chemical modifications on phosphate backbone and ribose ring of SSOs. Unmodified RNA is shown for reference. PS, one of the phosphate backbone oxygen atom is replaced by a sulphur atom; 2′-MOE and 2′-OMe, PS-SSOs are often combined with ribose modifications including 2′-O-(2-methoxyethyl) or 2′O-methyl; PMO, charge-neutral nucleic acid, in which the six-membered morpholine ring replaces the five-membered ribose heterocycle; PPMO, positively charged peptides in PPMO dramatically improve intracellular uptake of PMO. VPMO, covalently linking MO to an octaguanidine dendrimer to improve delivery efficacy. LNA, the second and fourth of ribose form a rigid structure by shrinkage. PNA, a pseudo peptide polymer backbone substitutes for the phosphate backbone of RNA. b. Properties comparison of the common chemistries of antisense oligonucleotides
Fig. 6.
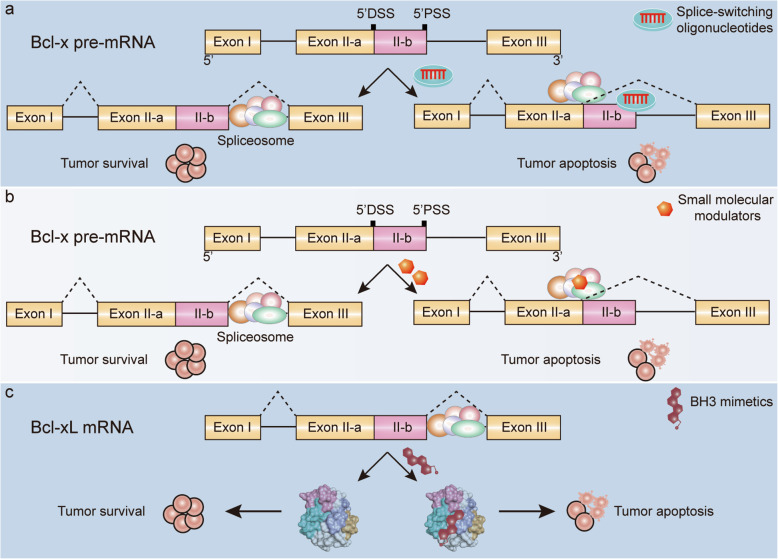
Strategies modulating Bcl-x splicing in cancer. a. An SSO that binds to the proximal 5' splice site (5'PSS) prevents binding of spliceosome, leading to a splicing shift to the short isoform Bcl-xS. b. a. The small molecular modulators that bind to spliceosomal components affect splice-site accessibility, leading to an inhibition of Bcl-xL splicing. c. At the protein level, BH3-mimetics could occupy the hydrophobic pockets of Bcl-Xl, thus blocking their anti-apoptotic activity and resulting in the ignition of apoptosis
Table 3.
Splice switching oligonucleotids used to modulate Bcl-x pre-mRNA splicing
| Cells types | Sequence | Length | Chemistry | Ref |
|---|---|---|---|---|
| K562 | 5'-GCTTGGTTCTTACCCAGCCGCCGTT-3' | 25 mer | vMO | [5] |
| Primary HSCs | 5'-TGGTTCTTACCCAGCCGCCG-3' | 20 mer | 2'-OMe-PS | [97] |
| U87, U251 | 5'-TGGTTCTTACCCAGCCGCCG-3' | 20 mer | 2'-MOE-PS | [142] |
| B16F10 | 5'-TGGTTCTTACCCAGCCGCCG-3' | 20 mer | 2'-MOE-PS | [143] |
| Human RPE | 5'-TGGTTCTTACCCAGCCGCCG-3' | 20 mer | 2'-MOE | [144] |
| A549 | 5'-CTGGATCCAAGGCTCTAGGT-3' | 20 mer | 2'-MOE | [145] |
| PC-3 | 5'-ACCCAGCCGCCGUUCUCC-3' | 18 mer | 2'-OMe-PS | [146] |
| MCF-7 | 5'-ACCCAGCCGCCGUUCUCC-3' | 18 mer | 2'-OMe-PS | [146] |
| Hela | 5'-ACCCAGCCGCCGUUCUCC-3' | 18 mer | 2'-OMe-PS | [146] |
Small molecular modulators redirect Bcl-x splicing
A class of natural or synthetic small molecular modulators had been identified to perform anti-tumour activity through inhibiting the activity of targeted splicing factors (Fig. 6b) [149, 150]. These compounds modulated RNA splicing in two ways generally. One way was to directly target pre-mRNA splicing factors. For example, the crude extract of the South African Medicinal Plant (Cotyledon orbiculata) could induce a splicing shift from Bcl-xL to Bcl-xS and apoptosis of cancer cells [151]. However, it was interesting that Bcl-x exhibited resistance to splicing perturbation induced by SF3B1 inhibitors E7107. Splicing modulation of E7107 was sensitized after Bcl-xL knockdown, which suggested that Bcl-xL could function as a sensitizing gene or as a biomarker for splicing modulator treatment [152]. The other way was to target kinases that were involved in post-translation modulation of splicing factors. For example, potent protein synthesis inhibitor emetine had been proved to enhance Bcl-xS splicing in a time and dose-dependent manner in multiple cancers, however, this effect could be hindered by protein phosphatase 1 (PP1 )[153]. Alkaloid Homoharringtonine approved for CML treatment by the FDA as well as antihypertensive agent amiloride and its derivatives BS008 were all proved to normalize oncogenic splicing pattern of Bcl-x in cancer cells depending on PP1 activation [154–156]. Intriguingly, Moore et al. found that inhibitors of cell cycle factors aurora kinase A (for example ZM447439 and VX-680) could induce endogenous Bcl-xS splicing significantly, revealing a complex alternative splicing network coordinating cell cycle and apoptosis [157]. In general terms, studying the mechanism of splice switch induced by small molecular modulators is essential for splicing therapies and antitumour agent discovery based on splicing correction. Notably, although the effectiveness that small molecules showed in splicing modulation, they usually lacked specificity and caused off-targeted or on-targeted cytotoxicity.
BH3 mimetics inhibit Bcl-xL isoform
Selective or multi-targeted BH3 mimetics had been developed to antagonize anti-apoptotic proteins of Bcl-2 family through competitively occupying the hydrophobic pockets and thus blocking their anti-apoptotic activity (Fig. 6c) [158]. Table 4 listed BH3-mimetics targeting anti-apoptotic Bcl-2 family proteins selectively. A-1331852 and WEHI-539 selectively targeted to Bcl-xL were all proved to enhance death signals of cancer cells synergistically with radiation or chemotherapy agents [158, 187]. In addition, compounds DT2216 and XZ424 converted from BH3 mimetics by proteolysis-targeting chimera showed improved anticancer potency and reduced cytotoxicity based on E3 ligase mediated degradation of Bcl-xL [162, 163]. However, use of BH3-mimetics in chronic lymphocytic leukaemia and other solid tumours exhibited on-target and off-target effects of Bcl-xL dependent cells and pathways [158, 188–190]. In addition, efficacy of BH3 mimetics was intensely dependent on the membrane localization of Bcl-xL and the nature of interactions between Bcl-xL and pro-apoptotic proteins, which might contribute to a chemotherapeutic resistance of BH3 mimetics [191]. These are still the obstacles for clinical application of BH3 mimetics. Consequently, optimizing the pharmacological effect and concurrent targets of BH3 mimetics to make them promising therapeutic regimens of cancer has been challenging.
Table 4.
Clinical application of BH3-mimetics targeting anti-apoptotic Bcl-2 family proteins. (clinicaltrials.gov)
| Multiple targets | Compounds | Origin | Stage | Ref |
|---|---|---|---|---|
| Bcl-xL | A-1155463 | Structure-based design. | Preclinical | [159] |
| A-1331852 | Structure-based design. | Preclinical | [160] | |
| WEHI-539 | Structure- based design | Preclinical | [161] | |
| DT2216 | Proteolysis targeting chimera | Preclinical | [162] | |
| XZ424 | Proteolysis targeting chimera | Preclinical | [163] | |
| ABBV-155 | Structure-based design | Phase I | NCT03595059 | |
|
Bcl-xL Bcl-2 |
AZD4320 | Structure-based design | Preclinical | [164] |
| BM-957 | Structure-based design | Preclinical | [165] | |
| BM-1197 | Structure-based design | Preclinical | [166] | |
| S44563 | Structure-based design | Preclinical | [167] | |
| APG-1252 | Structure-based design | Phase I/II | [168] | |
|
Bcl-xL Bcl-2 Bcl-w |
Ch282-5 | Gossypol derivative | Preclinical | [169] |
| ABT-737 | Synthetic, acylsulfonamide-based | Phase I/II | [170] | |
| ABT-263 (Navitoclax) | Derivant of ABT-737 | Phase I/II/III | [171] | |
| Bcl-xL, Bcl-2, Mcl-1 | BH3-M6 | Synthetic terphenyl scaffold | Preclinical | [172] |
|
Bcl-xL, Bcl-2, Bcl-w, Mcl-1 |
TW-37 | Benzenesulfonyl derivative of gossypol | Preclinical | [173] |
| BI-97C1 (Sabutoclax) | Diastereoisomer of Apogossypol | Preclinical | [174] | |
| BIM-SAHB | Stapled Bim peptide | Preclinical | [175] | |
| GX15-070 (Obatoclax) | Synthetic indolyl-dipyrromethene | Phase I/II/III | [176] | |
| AT-101 | (−)-gossypol enantiomer | Phase I/II/III | [177] | |
| Bcl-2 | S55746 | Structure-based design | Phase I | [178] |
| ABT-199 (Venetoclax) | Derivant of ABT-263 | Phase I/II/III | [127] | |
| Mcl-1 | A-1210477 | Structure-based design | Preclinical | [179] |
| UMI-77 | Structure-based design | Preclinical | [180] | |
| VU661013 | Fragment-based lead generation | Preclinical | [181] | |
| S63845 | Structure-based design | Preclinical | [182] | |
| AMG176, | Structure-based design | Phase I | [183] | |
| AZD5991 | Structure-based design | Phase I | [184] | |
| S64315 | Fragment-based lead generation | Phase I/II | [182] | |
| Bcl-2, Mcl-1 | S1-6 | Structure-based design | Preclinical | [185] |
| Nap-1 | Derivant of S1-6 | Preclinical | [186] |
Conclusions
The inactivation or dysfunction of essential genes caused by defective splicing is emerging as a potential driver of cancer development. Therefore, controlling splicing is of great therapeutic benefit. Dysregulated Bcl-x splicing plays a key role in promoting malignant phenotypes of cancers and weakening the toxicity of treatment. Bcl-xL contributed to the invasion, metastasis, and angiogenesis of cancers. On the contrary, Bcl-xS overexpression was suggested to sensitize apoptosis induced by drugs [129]. Bcl-x splicing correction by SSOs and small molecular modulators showed efficiency in apoptosis regulation of cancer cells. However, the on-targeted toxicity to Bcl-xL-dependent cell types posed challenges to the exploitation and delivery of splicing modulation drugs, which was expected to be addressed by the breakthrough of drug chemistry and carrier system [6]. In addition, the inhibitors of specific splicing factors for Bcl-x splicing correction are needed to be identified. Generally, induction of the balanced ratio of Bcl-xL/Bcl-xS has been shown anti-tumour activity by targeting multiple hallmarks of tumour, but it is still imperative that we understood this biomolecule. It is still unknown what is the intracellular mechanism that induced the preferred splicing of long isoform Bcl-xL. In addition, to discover the interplay of apoptosis and autophagy regulated by Bcl-xL means great significance to Bcl-xL targeted therapy. Moreover, whether the diverse domains of Bcl-xL execute biological functions independently and how does membrane localization affect its biological function in vivo remains unknown. Little is known about the biological function of Bcl-xS beyond its canonical function of lowering apoptosis threshold. In addition to Bcl-x, the anti-apoptotic family members including Bcl-2, Mcl-1 and Bcl-w also have a variety of splice isoforms, however, the elaborate coordination of biological roles played by multiple splice isoforms of Bcl-2 family members is unclear. Thus, much more remains to be researched about this gene in the future.
Acknowledgements
Not applicable
Abbreviations
- Bcl-x
Bcl-extra
- Bcl-2
B-cell lymphoma 2
- SSOs
Splicing-switch oligonucleotide
- 5' PSS
Proximal 5' splice site
- 5' DSS
Distal 5' splice site
- TM
Transmembrane
- IDR
Intrinsically disordered region
- MOMP
Mitochondrial outer membrane permeabilization
- ROS
Reactive oxygen species
- PKC
Protein kinase C inhibitor
- GM-CSF
Granulocyte-macrophage colony-stimulating factor
- SR
Serine/arginine-rich
- hnRNPs
Heterogeneous nuclear ribonucleoproteins
- EMT
Epithelial-mesenchymal transition
- CML
Chronic myeloid leukaemia
- vMO
Vivo-Morpholino
- PS
Phosphorothiate
- 2'-OMe
2'-O-methyl
- 2'-MOE
2'-O-methoxyethyl
- ESS
Exonic splicing silencer
- ESE
Exonic splicing enhancer
- PP1
Protein phosphatase 1
Authors’ contributions
Study concept and design: Cuixia Di and Hong Zhang. Drafting of the manuscript: Zhihui Dou. Critical revision of the manuscript for important intellectual content: Dapeng Zhao, Xiaohua Chen, Caipeng Xu, Xiaodong Jin, Xuetian Zhang, Yupei Wang, Xiaodong Xie, and Qiang Li. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the National Key R&D project of the Chinese Ministry of Science and Technology [2018YFE0205100]; the Key Program of the National Natural Science Foundation of China [U1632270]; the National Natural Science Foundation of China [11675234].
Availability of data and materials
Not applicable
Declarations
Consent for publication
All authors have agreed on the contents of the manuscript.
Competing interests
The authors have declared that no competing interest exists.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Cuixia Di, Email: dicx@impcas.ac.cn.
Hong Zhang, Email: zhang.h@impcas.ac.cn.
References
- 1.Pistritto G, Trisciuoglio D, Ceci C, Garufi A, D'orazi G. Apoptosis as anticancer mechanism: function and dysfunction of its modulators and targeted therapeutic strategies. Aging (Albany NY) 2016;8:603–619. doi: 10.18632/aging.100934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boise LH, González-García M, Postema CE, Ding L, Lindsten T, Turka LA, Mao X, Nuñez G, Thompson CB. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-N. [DOI] [PubMed] [Google Scholar]
- 3.Li M, Wang D, He J, Chen L, Li H. Bcl-X(L): A multifunctional anti-apoptotic protein. Pharmacol Res. 2020;151:104547. doi: 10.1016/j.phrs.2019.104547. [DOI] [PubMed] [Google Scholar]
- 4.Keitel U, Scheel A, Thomale J, Halpape R, Kaulfuß S, Scheel C, Dobbelstein M. Bcl-xL mediates therapeutic resistance of a mesenchymal breast cancer cell subpopulation. Oncotarget. 2014;5:11778–11791. doi: 10.18632/oncotarget.2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang J, Wang Y, Li SQ, Fang L, Wang XZ, Li J, Zhang HB, Huang B, Xu YM, Yang WM, et al. Correction of Bcl-x splicing improves responses to imatinib in chronic myeloid leukaemia cells and mouse models. Br J Haematol. 2020;189:1141–1150. doi: 10.1111/bjh.16472. [DOI] [PubMed] [Google Scholar]
- 6.Roberts TC, Langer R, Wood MJA. Advances in oligonucleotide drug delivery. Nat Rev Drug Discov. 2020:1–22. [DOI] [PMC free article] [PubMed]
- 7.Irimia M, Blencowe BJ. Alternative splicing: decoding an expansive regulatory layer. Curr Opin Cell Biol. 2012;24:323–332. doi: 10.1016/j.ceb.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 8.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 9.Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 10.Lee SC, Abdel-Wahab O. Therapeutic targeting of splicing in cancer. Nat Med. 2016;22:976–986. doi: 10.1038/nm.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonnal SC, López-Oreja I, Valcárcel J. Roles and mechanisms of alternative splicing in cancer - implications for care. Nat Rev Clin Oncol. 2020;17:457–474. doi: 10.1038/s41571-020-0350-x. [DOI] [PubMed] [Google Scholar]
- 12.Paschalis A, Sharp A, Welti JC, Neeb A, Raj GV, Luo J, Plymate SR, de Bono JS. Alternative splicing in prostate cancer. Nat Rev Clin Oncol. 2018;15:663–675. doi: 10.1038/s41571-018-0085-0. [DOI] [PubMed] [Google Scholar]
- 13.Yang XF, Weber GF, Cantor H. A novel Bcl-x isoform connected to the T cell receptor regulates apoptosis in T cells. Immunity. 1997;7:629–639. doi: 10.1016/S1074-7613(00)80384-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang W, Rivard JJ, Mueller DL, Behrens TW. Cloning and molecular characterization of mouse bcl-x in B and T lymphocytes. J Immunol. 1994;153:4388–4398. [PubMed] [Google Scholar]
- 15.Hossini AM, Geilen CC, Fecker LF, Daniel PT, Eberle J. A novel Bcl-x splice product, Bcl-xAK, triggers apoptosis in human melanoma cells without BH3 domain. Oncogene. 2006;25:2160–2169. doi: 10.1038/sj.onc.1209253. [DOI] [PubMed] [Google Scholar]
- 16.Schmitt E, Paquet C, Beauchemin M, Bertrand R. Bcl-xES, a BH4- and BH2-containing antiapoptotic protein, delays Bax oligomer formation and binds Apaf-1, blocking procaspase-9 activation. Oncogene. 2004;23:3915–3931. doi: 10.1038/sj.onc.1207554. [DOI] [PubMed] [Google Scholar]
- 17.Ban J, Eckhart L, Weninger W, Mildner M, Tschachler E. Identification of a human cDNA encoding a novel Bcl-x isoform. Biochem Biophys Res Commun. 1998;248:147–152. doi: 10.1006/bbrc.1998.8907. [DOI] [PubMed] [Google Scholar]
- 18.Naftelberg S, Schor IE, Ast G, Kornblihtt AR. Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu Rev Biochem. 2015;84:165–198. doi: 10.1146/annurev-biochem-060614-034242. [DOI] [PubMed] [Google Scholar]
- 19.Shkreta L, Michelle L, Toutant J, Tremblay ML, Chabot B. The DNA damage response pathway regulates the alternative splicing of the apoptotic mediator Bcl-x. J Biol Chem. 2011;286:331–340. doi: 10.1074/jbc.M110.162644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Revil T, Pelletier J, Toutant J, Cloutier A, Chabot B. Heterogeneous nuclear ribonucleoprotein K represses the production of pro-apoptotic Bcl-xS splice isoform. J Biol Chem. 2009;284:21458–21467. doi: 10.1074/jbc.M109.019711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cloutier P, Toutant J, Shkreta L, Goekjian S, Revil T, Chabot B. Antagonistic effects of the SRp30c protein and cryptic 5' splice sites on the alternative splicing of the apoptotic regulator Bcl-x. J Biol Chem. 2008;283:21315–21324. doi: 10.1074/jbc.M800353200. [DOI] [PubMed] [Google Scholar]
- 22.Garneau D, Revil T, Fisette JF, Chabot B. Heterogeneous nuclear ribonucleoprotein F/H proteins modulate the alternative splicing of the apoptotic mediator Bcl-x. J Biol Chem. 2005;280:22641–22650. doi: 10.1074/jbc.M501070200. [DOI] [PubMed] [Google Scholar]
- 23.Massiello A, Salas A, Pinkerman RL, Roddy P, Roesser JR, Chalfant CE. Identification of two RNA cis-elements that function to regulate the 5' splice site selection of Bcl-x pre-mRNA in response to ceramide. J Biol Chem. 2004;279:15799–15804. doi: 10.1074/jbc.M313950200. [DOI] [PubMed] [Google Scholar]
- 24.Li CY, Chu JY, Yu JK, Huang XQ, Liu XJ, Shi L, Che YC, Xie JY. Regulation of alternative splicing of Bcl-x by IL-6, GM-CSF and TPA. Cell Res. 2004;14:473–479. doi: 10.1038/sj.cr.7290250. [DOI] [PubMed] [Google Scholar]
- 25.Stevens M, Oltean S. Modulation of the apoptosis gene Bcl-x function through alternative splicing. Front Genet. 2019;10:804. doi: 10.3389/fgene.2019.00804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paronetto MP, Achsel T, Massiello A, Chalfant CE, Sette C. The RNA-binding protein Sam68 modulates the alternative splicing of Bcl-x. J Cell Biol. 2007;176:929–939. doi: 10.1083/jcb.200701005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kędzierska H, Piekiełko-Witkowska A. Splicing factors of SR and hnRNP families as regulators of apoptosis in cancer. Cancer Lett. 2017;396:53–65. doi: 10.1016/j.canlet.2017.03.013. [DOI] [PubMed] [Google Scholar]
- 28.Bielli P, Bordi M, Di Biasio V, Sette C. Regulation of BCL-X splicing reveals a role for the polypyrimidine tract binding protein (PTBP1/hnRNP I) in alternative 5' splice site selection. Nucleic Acids Res. 2014;42:12070–12081. doi: 10.1093/nar/gku922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cloutier A, Shkreta L, Toutant J, Durand M, Thibault P, Chabot B. hnRNP A1/A2 and Sam68 collaborate with SRSF10 to control the alternative splicing response to oxaliplatin-mediated DNA damage. Sci Rep. 2018;8:2206. doi: 10.1038/s41598-018-20360-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merdzhanova G, Edmond V, De Seranno S, Van den Broeck A, Corcos L, Brambilla C, Brambilla E, Gazzeri S, Eymin B. E2F1 controls alternative splicing pattern of genes involved in apoptosis through upregulation of the splicing factor SC35. Cell Death Differ. 2008;15:1815–1823. doi: 10.1038/cdd.2008.135. [DOI] [PubMed] [Google Scholar]
- 31.Bielli P, Busà R, Di Stasi SM, Munoz MJ, Botti F, Kornblihtt AR, Sette C. The transcription factor FBI-1 inhibits SAM68-mediated BCL-X alternative splicing and apoptosis. EMBO Rep. 2014;15:419–427. doi: 10.1002/embr.201338241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Montes M, Cloutier A, Sánchez-Hernández N, Michelle L, Lemieux B, Blanchette M, Hernández-Munain C, Chabot B, Suñé C. TCERG1 regulates alternative splicing of the Bcl-x gene by modulating the rate of RNA polymerase II transcription. Mol Cell Biol. 2012;32:751–762. doi: 10.1128/MCB.06255-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du J, Wang Q, Ziegler SF, Zhou B. FOXP3 interacts with hnRNPF to modulate pre-mRNA alternative splicing. J Biol Chem. 2018;293:10235–10244. doi: 10.1074/jbc.RA117.001349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao X, Littlejohn J, Rodarte C, Zhang L, Martino B, Rascoe P, Hamid K, Jupiter D, Smythe WR. Up-regulation of Bcl-xl by hepatocyte growth factor in human mesothelioma cells involves ETS transcription factors. Am J Pathol. 2009;175:2207–2216. doi: 10.2353/ajpath.2009.090070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naro C, Barbagallo F, Chieffi P, Bourgeois CF, Paronetto MP, Sette C. The centrosomal kinase NEK2 is a novel splicing factor kinase involved in cell survival. Nucleic Acids Res. 2014;42:3218–3227. doi: 10.1093/nar/gkt1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shkreta L, Toutant J, Durand M, Manley JL, Chabot B. SRSF10 connects DNA damage to the alternative splicing of transcripts encoding apoptosis, cell-cycle control, and DNA repair factors. Cell Rep. 2016;17:1990–2003. doi: 10.1016/j.celrep.2016.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen ZY, Cai L, Zhu J, Chen M, Chen J, Li ZH, Liu XD, Wang SG, Bie P, Jiang P, et al. Fyn requires HnRNPA2B1 and Sam68 to synergistically regulate apoptosis in pancreatic cancer. Carcinogenesis. 2011;32:1419–1426. doi: 10.1093/carcin/bgr088. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y, Chen D, Qian H, Tsai YS, Shao S, Liu Q, Dominguez D, Wang Z. The splicing factor RBM4 controls apoptosis, proliferation, and migration to suppress tumor progression. Cancer Cell. 2014;26:374–389. doi: 10.1016/j.ccr.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Inoue A, Yamamoto N, Kimura M, Nishio K, Yamane H, Nakajima K. RBM10 regulates alternative splicing. FEBS Lett. 2014;588:942–947. doi: 10.1016/j.febslet.2014.01.052. [DOI] [PubMed] [Google Scholar]
- 40.Pedrotti S, Busà R, Compagnucci C, Sette C. The RNA recognition motif protein RBM11 is a novel tissue-specific splicing regulator. Nucleic Acids Res. 2012;40:1021–1032. doi: 10.1093/nar/gkr819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou A, Ou AC, Cho A, Benz EJ, Jr, Huang SC. Novel splicing factor RBM25 modulates Bcl-x pre-mRNA 5' splice site selection. Mol Cell Biol. 2008;28:5924–5936. doi: 10.1128/MCB.00560-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Massiello A, Roesser JR, Chalfant CE. SAP155 Binds to ceramide-responsive RNA cis-element 1 and regulates the alternative 5' splice site selection of Bcl-x pre-mRNA. Faseb j. 2006;20:1680–1682. doi: 10.1096/fj.05-5021fje. [DOI] [PubMed] [Google Scholar]
- 43.Revil T, Toutant J, Shkreta L, Garneau D, Cloutier P, Chabot B. Protein kinase C-dependent control of Bcl-x alternative splicing. Mol Cell Biol. 2007;27:8431–8441. doi: 10.1128/MCB.00565-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shultz JC, Vu N, Shultz MD, Mba MU, Shapiro BA, Chalfant CE. The Proto-oncogene PKCι regulates the alternative splicing of Bcl-x pre-mRNA. Mol Cancer Res. 2012;10:660–669. doi: 10.1158/1541-7786.MCR-11-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vhuiyan MI, Pak ML, Park MA, Thomas D, Lakowski TM, Chalfant CE, Frankel A. PRMT2 interacts with splicing factors and regulates the alternative splicing of BCL-X. J Biochem. 2017;162:17–25. doi: 10.1093/jb/mvw102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weldon C, Dacanay JG, Gokhale V, Boddupally PVL, Behm-Ansmant I, Burley GA, Branlant C, Hurley LH, Dominguez C, Eperon IC. Specific G-quadruplex ligands modulate the alternative splicing of Bcl-X. Nucleic Acids Res. 2018;46:886–896. doi: 10.1093/nar/gkx1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weldon C, Behm-Ansmant I, Hurley LH, Burley GA, Branlant C, Eperon IC, Dominguez C. Identification of G-quadruplexes in long functional RNAs using 7-deazaguanine RNA. Nat Chem Biol. 2017;13:18–20. doi: 10.1038/nchembio.2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Michelle L, Cloutier A, Toutant J, Shkreta L, Thibault P, Durand M, Garneau D, Gendron D, Lapointe E, Couture S, et al. Proteins associated with the exon junction complex also control the alternative splicing of apoptotic regulators. Mol Cell Biol. 2012;32(5):954–67. [DOI] [PMC free article] [PubMed]
- 49.Zhu LY, Zhu YR, Dai DJ, Wang X, Jin HC. Epigenetic regulation of alternative splicing. Am J Cancer Res. 2018;8:2346–2358. [PMC free article] [PubMed] [Google Scholar]
- 50.Lev Maor G, Yearim A, Ast G. The alternative role of DNA methylation in splicing regulation. Trends Genet. 2015;31:274–280. doi: 10.1016/j.tig.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 51.Khan DH, Gonzalez C, Tailor N, Hamedani MK, Leygue E, Davie JR. Dynamic histone acetylation of H3K4me3 nucleosome regulates MCL1 Pre-mRNA splicing. J Cell Physiol. 2016;231:2196–2204. doi: 10.1002/jcp.25337. [DOI] [PubMed] [Google Scholar]
- 52.Chen XY, Zhang J, Zhu JS. The role of m (6) A RNA methylation in human cancer. Mol Cancer. 2019;18:103. doi: 10.1186/s12943-019-1033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singh R, Gupta SC, Peng WX, Zhou N, Pochampally R, Atfi A, Watabe K, Lu Z, Mo YY. Regulation of alternative splicing of Bcl-x by BC200 contributes to breast cancer pathogenesis. Cell Death Dis. 2016;7:e2262. doi: 10.1038/cddis.2016.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zong L, Hattori N, Yasukawa Y, Kimura K, Mori A, Seto Y, Ushijima T. LINC00162 confers sensitivity to 5-Aza-2'-deoxycytidine via modulation of an RNA splicing protein, HNRNPH1. Oncogene. 2019;38:5281–5293. doi: 10.1038/s41388-019-0792-8. [DOI] [PubMed] [Google Scholar]
- 55.Cui C, Zhai D, Cai L, Duan Q, Xie L, Yu J. Long Noncoding RNA HEIH Promotes Colorectal Cancer Tumorigenesis via Counteracting miR-939–Mediated Transcriptional Repression of Bcl-xL. Cancer Res Treat. 2018;50:992–1008. doi: 10.4143/crt.2017.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carneiro BA, El-Deiry WS. Targeting apoptosis in cancer therapy. Nat Rev Clin Oncol. 2020;17:395–417. doi: 10.1038/s41571-020-0341-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goldar S, Khaniani MS, Derakhshan SM, Baradaran B. Molecular mechanisms of apoptosis and roles in cancer development and treatment. Asian Pac J Cancer Prev. 2015;16:2129–2144. doi: 10.7314/APJCP.2015.16.6.2129. [DOI] [PubMed] [Google Scholar]
- 59.Schneider P, Tschopp J. Apoptosis induced by death receptors. Pharm Acta Helv. 2000;74:281–286. doi: 10.1016/S0031-6865(99)00038-2. [DOI] [PubMed] [Google Scholar]
- 60.O'Neill KL, Huang K, Zhang J, Chen Y, Luo X. Inactivation of prosurvival Bcl-2 proteins activates Bax/Bak through the outer mitochondrial membrane. Genes Dev. 2016;30:973–988. doi: 10.1101/gad.276725.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ashkenazi A, Fairbrother WJ, Leverson JD, Souers AJ. From basic apoptosis discoveries to advanced selective BCL-2 family inhibitors. Nat Rev Drug Discov. 2017;16:273–284. doi: 10.1038/nrd.2016.253. [DOI] [PubMed] [Google Scholar]
- 62.Kang MH, Reynolds CP. Bcl-2 inhibitors: targeting mitochondrial apoptotic pathways in cancer therapy. Clin Cancer Res. 2009;15:1126–1132. doi: 10.1158/1078-0432.CCR-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Amundson SA, Myers TG, Scudiero D, Kitada S, Reed JC, Fornace AJ., Jr An informatics approach identifying markers of chemosensitivity in human cancer cell lines. Cancer Res. 2000;60:6101–6110. [PubMed] [Google Scholar]
- 64.Delbridge AR, Grabow S, Strasser A, Vaux DL. Thirty years of BCL-2: translating cell death discoveries into novel cancer therapies. Nat Rev Cancer. 2016;16:99–109. doi: 10.1038/nrc.2015.17. [DOI] [PubMed] [Google Scholar]
- 65.Takehara T, Liu X, Fujimoto J, Friedman SL, Takahashi H. Expression and role of Bcl-xL in human hepatocellular carcinomas. Hepatology. 2001;34:55–61. doi: 10.1053/jhep.2001.25387. [DOI] [PubMed] [Google Scholar]
- 66.Scherr AL, Gdynia G, Salou M, Radhakrishnan P, Duglova K, Heller A, Keim S, Kautz N, Jassowicz A, Elssner C, et al. Bcl-xL is an oncogenic driver in colorectal cancer. Cell Death Dis. 2016;7:e2342. doi: 10.1038/cddis.2016.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gayle SS, Sahni JM, Webb BM, Weber-Bonk KL, Shively MS, Spina R, Bar EE, Summers MK, Keri RA. Targeting BCL-xL improves the efficacy of bromodomain and extra-terminal protein inhibitors in triple-negative breast cancer by eliciting the death of senescent cells. J Biol Chem. 2019;294:875–886. doi: 10.1074/jbc.RA118.004712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Das A, Martinez Santos JL, Alshareef M, Porto GBF, Infinger LK, Vandergrift WA, 3rd, Lindhorst SM, Varma AK, Patel SJ, Cachia D. In vitro effect of dovitinib (TKI258), a multi-target angiokinase inhibitor on aggressive meningioma cells. Cancer Invest. 2020;38:349–355. doi: 10.1080/07357907.2020.1773844. [DOI] [PubMed] [Google Scholar]
- 69.Zhou WH, Tang F, Xu J, Wu X, Yang SB, Feng ZY, Ding YG, Wan XB, Guan Z, Li HG, et al. Low expression of Beclin 1, associated with high Bcl-xL, predicts a malignant phenotype and poor prognosis of gastric cancer. Autophagy. 2012;8:389–400. doi: 10.4161/auto.18641. [DOI] [PubMed] [Google Scholar]
- 70.Zhang K, Jiao K, Xing Z, Zhang L, Yang J, Xie X, Yang L. Bcl-xL overexpression and its association with the progress of tongue carcinoma. Int J Clin Exp Pathol. 2014;7:7360–7377. [PMC free article] [PubMed] [Google Scholar]
- 71.Adams CM, Mitra R, Vogel AN, Liu J, Gong JZ, Eischen CM. Targeting BCL-W and BCL-XL as a therapeutic strategy for Hodgkin lymphoma. Leukemia. 2020;34:947–952. doi: 10.1038/s41375-019-0611-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Petiti J, Lo Iacono M, Rosso V, Andreani G, Jovanovski A, Podestà M, Lame D, Gobbi M, Fava C, Saglio G, et al. Bcl-xL represents a therapeutic target in Philadelphia negative myeloproliferative neoplasms. J Cell Mol Med. 2020;24:10978–10986. doi: 10.1111/jcmm.15730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xerri L, Parc P, Brousset P, Schlaifer D, Hassoun J, Reed JC, Krajewski S, Birnbaum D. Predominant expression of the long isoform of Bcl-x (Bcl-xL) in human lymphomas. Br J Haematol. 1996;92:900–906. doi: 10.1046/j.1365-2141.1996.423958.x. [DOI] [PubMed] [Google Scholar]
- 74.Basta-Jovanović G, Radonjic V, Stolic I, Nenadovic M, Brasanac D, Jovanovic D, Radojevic-Skodric S. Significance of proto-oncogene Bcl-X(S/L) expression in Wilms tumor. Ren Fail. 2005;27:13–18. doi: 10.1081/JDI-42856. [DOI] [PubMed] [Google Scholar]
- 75.Ma X, Zhao Y, Li Y, Lu H, He Y. Relevance of Bcl-x expression in different types of endometrial tissues. J Exp Clin Cancer Res. 2010;29:14. doi: 10.1186/1756-9966-29-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Choi S, Chen Z, Tang LH, Fang Y, Shin SJ, Panarelli NC, Chen YT, Li Y, Jiang X, Du YN. Bcl-xL promotes metastasis independent of its anti-apoptotic activity. Nat Commun. 2016;7:10384. doi: 10.1038/ncomms10384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Trisciuoglio D, Tupone MG, Desideri M, Di Martile M, Gabellini C, Buglioni S, Pallocca M, Alessandrini G, D'Aguanno S, Del Bufalo D. BCL-X(L) overexpression promotes tumor progression-associated properties. Cell Death Dis. 2017;8:3216. doi: 10.1038/s41419-017-0055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.de Jong Y, Monderer D, Brandinelli E, Monchanin M, van den Akker BE, van Oosterwijk JG, Blay JY, Dutour A, Bovée J. Bcl-xl as the most promising Bcl-2 family member in targeted treatment of chondrosarcoma. Oncogenesis. 2018;7:74. doi: 10.1038/s41389-018-0084-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Heisey DAR, Lochmann TL, Floros KV, Coon CM, Powell KM, Jacob S, Calbert ML, Ghotra MS, Stein GT, Maves YK, et al. The Ewing Family of tumors relies on BCL-2 and BCL-X(L) to escape PARP inhibitor toxicity. Clin Cancer Res. 2019;25:1664–1675. doi: 10.1158/1078-0432.CCR-18-0277. [DOI] [PubMed] [Google Scholar]
- 80.Simonin K, N'Diaye M, Lheureux S, Loussouarn C, Dutoit S, Briand M, Giffard F, Brotin E, Blanc-Fournier C, Poulain L. Platinum compounds sensitize ovarian carcinoma cells to ABT-737 by modulation of the Mcl-1/Noxa axis. Apoptosis. 2013;18:492–508. doi: 10.1007/s10495-012-0799-x. [DOI] [PubMed] [Google Scholar]
- 81.Cheng J, Qian D, Ding X, Song T, Cai M, Dan X, Wang Y, Zhao J, Liu Z, Wu Z, et al. High PGAM5 expression induces chemoresistance by enhancing Bcl-xL-mediated anti-apoptotic signaling and predicts poor prognosis in hepatocellular carcinoma patients. Cell Death Dis. 2018;9:991. doi: 10.1038/s41419-018-1017-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kuo KL, Liu SH, Lin WC, Hsu FS, Chow PM, Chang YW, et al. Trifluoperazine, an antipsychotic drug, effectively reduces drug resistance in cisplatin-resistant urothelial carcinoma cells via suppressing Bcl-xL: an in vitro and in vivo study. Int J Mol Sci. 2019;20. [DOI] [PMC free article] [PubMed]
- 83.Nix P, Cawkwell L, Patmore H, Greenman J, Stafford N. Bcl-2 expression predicts radiotherapy failure in laryngeal cancer. Br J Cancer. 2005;92:2185–2189. doi: 10.1038/sj.bjc.6602647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang Z, Jin F, Lian X, Li M, Wang G, Lan B, He H, Liu GD, Wu Y, Sun G, et al. Genistein promotes ionizing radiation-induced cell death by reducing cytoplasmic Bcl-xL levels in non-small cell lung cancer. Sci Rep. 2018;8:328. doi: 10.1038/s41598-017-18755-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Su ZZ, Lebedeva IV, Sarkar D, Emdad L, Gupta P, Kitada S, Dent P, Reed JC, Fisher PB. Ionizing radiation enhances therapeutic activity of mda-7/IL-24: overcoming radiation- and mda-7/IL-24-resistance in prostate cancer cells overexpressing the antiapoptotic proteins bcl-xL or bcl-2. Oncogene. 2006;25:2339–2348. doi: 10.1038/sj.onc.1209271. [DOI] [PubMed] [Google Scholar]
- 86.Wang ZX, Yang JS, Pan X, Wang JR, Li J, Yin YM, De W. Functional and biological analysis of Bcl-xL expression in human osteosarcoma. Bone. 2010;47:445–454. doi: 10.1016/j.bone.2010.05.027. [DOI] [PubMed] [Google Scholar]
- 87.Jackson MR, Ashton M, Koessinger AL, Dick C, Verheij M, Chalmers AJ. Mesothelioma cells depend on the antiapoptotic protein Bcl-xL for survival and are sensitized to ionizing radiation by BH3-Mimetics. Int J Radiat Oncol Biol Phys. 2020;106:867–877. doi: 10.1016/j.ijrobp.2019.11.029. [DOI] [PubMed] [Google Scholar]
- 88.Lee EF, Fairlie WD. The Structural Biology of Bcl-x(L). Int J Mol Sci. 2019;20. [DOI] [PMC free article] [PubMed]
- 89.Edlich F, Banerjee S, Suzuki M, Cleland MM, Arnoult D, Wang C, Neutzner A, Tjandra N, Youle RJ. Bcl-x(L) retrotranslocates Bax from the mitochondria into the cytosol. Cell. 2011;145:104–116. doi: 10.1016/j.cell.2011.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Llambi F, Moldoveanu T, Tait SW, Bouchier-Hayes L, Temirov J, McCormick LL, Dillon CP, Green DR. A unified model of mammalian BCL-2 protein family interactions at the mitochondria. Mol Cell. 2011;44:517–531. doi: 10.1016/j.molcel.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bogner C, Kale J, Pogmore J, Chi X, Shamas-Din A, Fradin C, Leber B, Andrews DW. Allosteric regulation of BH3 proteins in Bcl-x(L) complexes enables switch-like activation of Bax. Mol Cell. 2020;77:901–912.e909. doi: 10.1016/j.molcel.2019.12.025. [DOI] [PubMed] [Google Scholar]
- 92.Aartsma-Rus A, van Vliet L, Hirschi M, Janson AA, Heemskerk H, de Winter CL, de Kimpe S, van Deutekom JC, t Hoen PA, van Ommen GJ. Guidelines for antisense oligonucleotide design and insight into splice-modulating mechanisms. Mol Ther. 2009;17:548–553. doi: 10.1038/mt.2008.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Monaco G, Decrock E, Arbel N, van Vliet AR, La Rovere RM, De Smedt H, Parys JB, Agostinis P, Leybaert L, Shoshan-Barmatz V, Bultynck G. The BH4 domain of anti-apoptotic Bcl-XL, but not that of the related Bcl-2, limits the voltage-dependent anion channel 1 (VDAC1)-mediated transfer of pro-apoptotic Ca2+ signals to mitochondria. J Biol Chem. 2015;290:9150–9161. doi: 10.1074/jbc.M114.622514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chang BS, Kelekar A, Harris MH, Harlan JE, Fesik SW, Thompson CB. The BH3 domain of Bcl-x(S) is required for inhibition of the antiapoptotic function of Bcl-x(L) Mol Cell Biol. 1999;19:6673–6681. doi: 10.1128/MCB.19.10.6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Plötz M, Gillissen B, Hossini AM, Daniel PT, Eberle J. Disruption of the VDAC2-Bak interaction by Bcl-x(S) mediates efficient induction of apoptosis in melanoma cells. Cell Death Differ. 2012;19:1928–1938. doi: 10.1038/cdd.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Peña-Blanco A, García-Sáez AJ. Bax, Bak and beyond - mitochondrial performance in apoptosis. FEBS J. 2018;285:416–431. doi: 10.1111/febs.14186. [DOI] [PubMed] [Google Scholar]
- 97.Wu L, Mao C, Ming X. Modulation of Bcl-x Alternative Splicing Induces Apoptosis of Human Hepatic Stellate Cells. Biomed Res Int. 2016;2016:7478650. doi: 10.1155/2016/7478650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhao R, Hu M, Liang S, Wang B, Yu B, Yang G, Qian D. IE86 Inhibits the apoptosis and promotes the cell proliferation of glioma cells via the hnRNP A2/B1-mediated alternative splicing of Bcl-x. Int J Clin Exp Pathol. 2019;12:2775–2785. [PMC free article] [PubMed] [Google Scholar]
- 99.Yu S, Du M, Yin A, Mai Z, Wang Y, Zhao M, Wang X, Chen T. Bcl-xL inhibits PINK1/Parkin-dependent mitophagy by preventing mitochondrial Parkin accumulation. Int J Biochem Cell Biol. 2020;122:105720. doi: 10.1016/j.biocel.2020.105720. [DOI] [PubMed] [Google Scholar]
- 100.Zhou F, Yang Y, Xing D. Bcl-2 and Bcl-xL play important roles in the crosstalk between autophagy and apoptosis. FEBS J. 2011;278:403–413. doi: 10.1111/j.1742-4658.2010.07965.x. [DOI] [PubMed] [Google Scholar]
- 101.Li X, He S, Ma B. Autophagy and autophagy-related proteins in cancer. Mol Cancer. 2020;19:12. doi: 10.1186/s12943-020-1138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lin HX, Qiu HJ, Zeng F, Rao HL, Yang GF, Kung HF, Zhu XF, Zeng YX, Cai MY, Xie D. Decreased expression of Beclin 1 correlates closely with Bcl-xL expression and poor prognosis of ovarian carcinoma. PLoS ONE. 2013;8:e60516. doi: 10.1371/journal.pone.0060516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 105.Huang X, Qi Q, Hua X, Li X, Zhang W, Sun H, Li S, Wang X, Li B. Beclin 1, an autophagy-related gene, augments apoptosis in U87 glioblastoma cells. Oncol Rep. 2014;31:1761–1767. doi: 10.3892/or.2014.3015. [DOI] [PubMed] [Google Scholar]
- 106.Wen J, Mai Z, Zhao M, Wang X, Chen T. Full anti-apoptotic function of Bcl-XL complexed with Beclin-1 verified by live-cell FRET assays. Biochem Biophys Res Commun. 2019;511:700–704. doi: 10.1016/j.bbrc.2019.02.107. [DOI] [PubMed] [Google Scholar]
- 107.Lindqvist LM, Heinlein M, Huang DC, Vaux DL. Prosurvival Bcl-2 family members affect autophagy only indirectly, by inhibiting Bax and Bak. Proc Natl Acad Sci U S A. 2014;111:8512–8517. doi: 10.1073/pnas.1406425111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Koehler BC, Scherr AL, Lorenz S, Urbanik T, Kautz N, Elssner C, Welte S, Bermejo JL, Jäger D, Schulze-Bergkamen H. Beyond cell death - antiapoptotic Bcl-2 proteins regulate migration and invasion of colorectal cancer cells in vitro. PLoS ONE. 2013;8:e76446. doi: 10.1371/journal.pone.0076446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yang J, Sun M, Zhang A, Lv C, De W, Wang Z. Adenovirus-mediated siRNA targeting Bcl-xL inhibits proliferation, reduces invasion and enhances radiosensitivity of human colorectal cancer cells. World J Surg Oncol. 2011;9:117. doi: 10.1186/1477-7819-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Carné Trécesson S, Souazé F, Basseville A, Bernard AC, Pécot J, Lopez J, Bessou M, Sarosiek KA, Letai A, Barillé-Nion S, et al. BCL-X(L) directly modulates RAS signalling to favour cancer cell stemness. Nat Commun. 2017;8:1123. doi: 10.1038/s41467-017-01079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gabellini C, Gómez-Abenza E, Ibáñez-Molero S, Tupone MG, Pérez-Oliva AB, de Oliveira S, Del Bufalo D, Mulero V. Interleukin 8 mediates bcl-xL-induced enhancement of human melanoma cell dissemination and angiogenesis in a zebrafish xenograft model. Int J Cancer. 2018;142:584–596. doi: 10.1002/ijc.31075. [DOI] [PubMed] [Google Scholar]
- 112.Bessou M, Lopez J, Gadet R, Deygas M, Popgeorgiev N, Poncet D, Nougarède A, Billard P, Mikaelian I, Gonzalo P, et al. The apoptosis inhibitor Bcl-xL controls breast cancer cell migration through mitochondria-dependent reactive oxygen species production. Oncogene. 2020;39:3056–3074. doi: 10.1038/s41388-020-1212-9. [DOI] [PubMed] [Google Scholar]
- 113.Grillot DA, Merino R, Núñez G. Bcl-XL displays restricted distribution during T cell development and inhibits multiple forms of apoptosis but not clonal deletion in transgenic mice. J Exp Med. 1995;182:1973–1983. doi: 10.1084/jem.182.6.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Amanna IJ, Dingwall JP, Hayes CE. Enforced bcl-xL gene expression restored splenic B lymphocyte development in BAFF-R mutant mice. J Immunol. 2003;170:4593–4600. doi: 10.4049/jimmunol.170.9.4593. [DOI] [PubMed] [Google Scholar]
- 115.Kohlhapp FJ, Haribhai D, Mathew R, Duggan R, Ellis PA, Wang R, et al. Venetoclax increases intra-tumoral effector T cells and anti-tumor efficacy in combination with immune checkpoint blockade. Cancer Discov. 2020. [DOI] [PubMed]
- 116.Chen S, Li X, Zhang W, Zi M, Xu Y. Inflammatory compound lipopolysaccharide promotes the survival of GM-CSF cultured dendritic cell via PI3 kinase-dependent upregulation of Bcl-x. Immunol Cell Biol. 2018;96:912–921. doi: 10.1111/imcb.12051. [DOI] [PubMed] [Google Scholar]
- 117.Issa F, Milward K, Goto R, Betts G, Wood KJ, Hester J. Transiently activated human regulatory T cells upregulate BCL-XL expression and acquire a functional advantage in vivo. Front Immunol. 2019;10:889. doi: 10.3389/fimmu.2019.00889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wallin RP, Sundquist VS, Bråkenhielm E, Cao Y, Ljunggren HG, Grandien A. Angiostatic effects of NK cell-derived IFN-γ counteracted by tumour cell Bcl-xL expression. Scand J Immunol. 2014;79:90–97. doi: 10.1111/sji.12134. [DOI] [PubMed] [Google Scholar]
- 119.Carrington EM, Tarlinton DM, Gray DH, Huntington ND, Zhan Y, Lew AM. The life and death of immune cell types: the role of BCL-2 anti-apoptotic molecules. Immunol Cell Biol. 2017;95:870–877. doi: 10.1038/icb.2017.72. [DOI] [PubMed] [Google Scholar]
- 120.Andersen MH, Reker S, Kvistborg P, Becker JC, thor Straten P. Spontaneous immunity against Bcl-xL in cancer patients. J Immunol. 2005;175:2709–2714. doi: 10.4049/jimmunol.175.4.2709. [DOI] [PubMed] [Google Scholar]
- 121.Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741–4751. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mohammad RM, Muqbil I, Lowe L, Yedjou C, Hsu HY, Lin LT, Siegelin MD, Fimognari C, Kumar NB, Dou QP, et al. Broad targeting of resistance to apoptosis in cancer. Semin Cancer Biol. 2015;35(Suppl):S78–s103. doi: 10.1016/j.semcancer.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhang J, Quan LN, Meng Q, Wang HY, Wang J, Yu P, Fu JT, Li YJ, Chen J, Cheng H, et al. miR-548e sponged by ZFAS1 regulates metastasis and cisplatin resistance of OC by targeting CXCR4 and let-7a/BCL-XL/S signaling axis. Mol Ther Nucleic Acids. 2020;20:621–638. doi: 10.1016/j.omtn.2020.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Liu R, Page C, Beidler DR, Wicha MS, Núñez G. Overexpression of Bcl-x(L) promotes chemotherapy resistance of mammary tumors in a syngeneic mouse model. Am J Pathol. 1999;155:1861–1867. doi: 10.1016/S0002-9440(10)65505-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zeng Y, Xu X, Wang S, Zhang Z, Liu Y, Han K, Cao B, Mao X. Ring finger protein 6 promotes breast cancer cell proliferation by stabilizing estrogen receptor alpha. Oncotarget. 2017;8:20103–20112. doi: 10.18632/oncotarget.15384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zoeller JJ, Vagodny A, Taneja K, Tan BY, O'Brien N, Slamon DJ, Sampath D, Leverson JD, Bronson RT, Dillon DA, Brugge JS. Neutralization of BCL-2/X(L) enhances the cytotoxicity of T-DM1 In Vivo. Mol Cancer Ther. 2019;18:1115–1126. doi: 10.1158/1535-7163.MCT-18-0743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pan R, Hogdal LJ, Benito JM, Bucci D, Han L, Borthakur G, Cortes J, DeAngelo DJ, Debose L, Mu H, et al. Selective BCL-2 inhibition by ABT-199 causes on-target cell death in acute myeloid leukemia. Cancer Discov. 2014;4:362–375. doi: 10.1158/2159-8290.CD-13-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wolter KG, Verhaegen M, Fernández Y, Nikolovska-Coleska Z, Riblett M, de la Vega CM, Wang S, Soengas MS. Therapeutic window for melanoma treatment provided by selective effects of the proteasome on Bcl-2 proteins. Cell Death Differ. 2007;14:1605–1616. doi: 10.1038/sj.cdd.4402163. [DOI] [PubMed] [Google Scholar]
- 129.Sumantran VN, Ealovega MW, Nuñez G, Clarke MF, Wicha MS. Overexpression of Bcl-XS sensitizes MCF-7 cells to chemotherapy-induced apoptosis. Cancer Res. 1995;55:2507–2510. [PubMed] [Google Scholar]
- 130.Streffer JR, Rimner A, Rieger J, Naumann U, Rodemann HP, Weller M. BCL-2 family proteins modulate radiosensitivity in human malignant glioma cells. J Neurooncol. 2002;56:43–49. doi: 10.1023/A:1014448721327. [DOI] [PubMed] [Google Scholar]
- 131.Strik H, Deininger M, Streffer J, Grote E, Wickboldt J, Dichgans J, Weller M, Meyermann R. BCL-2 family protein expression in initial and recurrent glioblastomas: modulation by radiochemotherapy. J Neurol Neurosurg Psychiatry. 1999;67:763–768. doi: 10.1136/jnnp.67.6.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Li JY, Li YY, Jin W, Yang Q, Shao ZM, Tian XS. ABT-737 reverses the acquired radioresistance of breast cancer cells by targeting Bcl-2 and Bcl-xL. J Exp Clin Cancer Res. 2012;31:102. doi: 10.1186/1756-9966-31-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhu L, Zhu B, Yang L, Zhao X, Jiang H, Ma F. RelB regulates Bcl-xl expression and the irradiation-induced apoptosis of murine prostate cancer cells. Biomed Rep. 2014;2:354–358. doi: 10.3892/br.2014.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ho JN, Kang GY, Lee SS, Kim J, Bae IH, Hwang SG, Um HD. Bcl-XL and STAT3 mediate malignant actions of gamma-irradiation in lung cancer cells. Cancer Sci. 2010;101:1417–1423. doi: 10.1111/j.1349-7006.2010.01552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jung CH, Kim EM, Song JY, Park JK, Um HD. Mitochondrial superoxide dismutase 2 mediates γ-irradiation-induced cancer cell invasion. Exp Mol Med. 2019;51:1–10. doi: 10.1038/s12276-019-0207-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Borrás C, Mas-Bargues C, Román-Domínguez A, Sanz-Ros J, Gimeno-Mallench L, Inglés M, et al. BCL-xL, a Mitochondrial Protein Involved in Successful Aging: From C. elegans to Human Centenarians. Int J Mol Sci. 2020:21. [DOI] [PMC free article] [PubMed]
- 137.Pan J, Li D, Xu Y, Zhang J, Wang Y, Chen M, Lin S, Huang L, Chung EJ, Citrin DE, et al. Inhibition of Bcl-2/xl with ABT-263 selectively kills senescent Type II pneumocytes and reverses persistent pulmonary fibrosis induced by ionizing radiation in mice. Int J Radiat Oncol Biol Phys. 2017;99:353–361. doi: 10.1016/j.ijrobp.2017.02.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Brinkmann K, Waring P, Glaser SP, Wimmer V, Cottle DL, Tham MS, et al. BCL-XL exerts a protective role against anemia caused by radiation-induced kidney damage. EMBO J. 2020:e105561. [DOI] [PMC free article] [PubMed]
- 139.Havens MA, Hastings ML. Splice-switching antisense oligonucleotides as therapeutic drugs. Nucleic Acids Res. 2016;44:6549–6563. doi: 10.1093/nar/gkw533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Charleston JS, Schnell FJ, Dworzak J, Donoghue C, Lewis S, Chen L, Young GD, Milici AJ, Voss J, DeAlwis U, et al. Eteplirsen treatment for Duchenne muscular dystrophy: Exon skipping and dystrophin production. Neurology. 2018;90:e2146–e2154. doi: 10.1212/WNL.0000000000005680. [DOI] [PubMed] [Google Scholar]
- 141.Michelson D, Ciafaloni E, Ashwal S, Lewis E, Narayanaswami P, Oskoui M, Armstrong MJ. Evidence in focus: Nusinersen use in spinal muscular atrophy: Report of the Guideline Development, Dissemination, and Implementation Subcommittee of the American Academy of Neurology. Neurology. 2018;91:923–933. doi: 10.1212/WNL.0000000000006502. [DOI] [PubMed] [Google Scholar]
- 142.Li Z, Li Q, Han L, Tian N, Liang Q, Li Y, Zhao X, Du C, Tian Y. Pro-apoptotic effects of splice-switching oligonucleotides targeting Bcl-x pre-mRNA in human glioma cell lines. Oncol Rep. 2016;35:1013–1019. doi: 10.3892/or.2015.4465. [DOI] [PubMed] [Google Scholar]
- 143.Bauman JA, Li SD, Yang A, Huang L, Kole R. Anti-tumor activity of splice-switching oligonucleotides. Nucleic Acids Res. 2010;38:8348–8356. doi: 10.1093/nar/gkq731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhang N, Peairs JJ, Yang P, Tyrrell J, Roberts J, Kole R, Jaffe GJ. The importance of Bcl-xL in the survival of human RPE cells. Invest Ophthalmol Vis Sci. 2007;48:3846–3853. doi: 10.1167/iovs.06-1145. [DOI] [PubMed] [Google Scholar]
- 145.Taylor JK, Zhang QQ, Wyatt JR, Dean NM. Induction of endogenous Bcl-xS through the control of Bcl-x pre-mRNA splicing by antisense oligonucleotides. Nat Biotechnol. 1999;17:1097–1100. doi: 10.1038/15079. [DOI] [PubMed] [Google Scholar]
- 146.Mercatante DR, Bortner CD, Cidlowski JA, Kole R. Modification of alternative splicing of Bcl-x pre-mRNA in prostate and breast cancer cells. analysis of apoptosis and cell death. J Biol Chem. 2001;276:16411–16417. doi: 10.1074/jbc.M009256200. [DOI] [PubMed] [Google Scholar]
- 147.Mercatante DR, Mohler JL, Kole R. Cellular response to an antisense-mediated shift of Bcl-x pre-mRNA splicing and antineoplastic agents. J Biol Chem. 2002;277:49374–49382. doi: 10.1074/jbc.M209236200. [DOI] [PubMed] [Google Scholar]
- 148.Subbotina E, Koganti SR, Hodgson-Zingman DM, Zingman LV. Morpholino-driven gene editing: A new horizon for disease treatment and prevention. Clin Pharmacol Ther. 2016;99:21–25. doi: 10.1002/cpt.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Corrionero A, Miñana B, Valcárcel J. Reduced fidelity of branch point recognition and alternative splicing induced by the anti-tumor drug spliceostatin A. Genes Dev. 2011;25:445–459. doi: 10.1101/gad.2014311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Han T, Goralski M, Gaskill N, Capota E, Kim J, Ting TC, et al. Anticancer sulfonamides target splicing by inducing RBM39 degradation via recruitment to DCAF15. Science. 2017;356. [DOI] [PubMed]
- 151.Makhafola TJ, Mbele M, Yacqub-Usman K, Hendren A, Haigh DB, Blackley Z, Meyer M, Mongan NP, Bates DO, Dlamini Z. Apoptosis in cancer cells is induced by alternative splicing of hnRNPA2/B1 through splicing of Bcl-x, a mechanism that can be stimulated by an extract of the South African Medicinal Plant, Cotyledon orbiculata. Front Oncol. 2020;10:547392. doi: 10.3389/fonc.2020.547392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Aird D, Teng T, Huang CL, Pazolli E, Banka D, Cheung-Ong K, Eifert C, Furman C, Wu ZJ, Seiler M, et al. Sensitivity to splicing modulation of BCL2 family genes defines cancer therapeutic strategies for splicing modulators. Nat Commun. 2019;10:137. doi: 10.1038/s41467-018-08150-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sun Q, Yogosawa S, Iizumi Y, Sakai T, Sowa Y. The alkaloid emetine sensitizes ovarian carcinoma cells to cisplatin through downregulation of bcl-xL. Int J Oncol. 2015;46:389–394. doi: 10.3892/ijo.2014.2703. [DOI] [PubMed] [Google Scholar]
- 154.Chang JG, Yang DM, Chang WH, Chow LP, Chan WL, Lin HH, Huang HD, Chang YS, Hung CH, Yang WK. Small molecule amiloride modulates oncogenic RNA alternative splicing to devitalize human cancer cells. PLoS One. 2011;6:e18643. doi: 10.1371/journal.pone.0018643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lee CC, Chang WH, Chang YS, Yang JM, Chang CS, Hsu KC, Chen YT, Liu TY, Chen YC, Lin SY, et al. Alternative splicing in human cancer cells is modulated by the amiloride derivative 3,5-diamino-6-chloro-N-(N-(2,6-dichlorobenzoyl)carbamimidoyl)pyrazine-2-carboxide. Mol Oncol. 2019;13:1744–1762. doi: 10.1002/1878-0261.12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sun Q, Li S, Li J, Fu Q, Wang Z, Li B, Liu SS, Su Z, Song J, Lu D. Homoharringtonine regulates the alternative splicing of Bcl-x and caspase 9 through a protein phosphatase 1-dependent mechanism. BMC Complement Altern Med. 2018;18:164. doi: 10.1186/s12906-018-2233-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Moore MJ, Wang Q, Kennedy CJ, Silver PA. An alternative splicing network links cell-cycle control to apoptosis. Cell. 2010;142:625–636. doi: 10.1016/j.cell.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Opydo-Chanek M, Gonzalo O, Marzo I. Multifaceted anticancer activity of BH3 mimetics: current evidence and future prospects. Biochem Pharmacol. 2017;136:12–23. doi: 10.1016/j.bcp.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 159.Tao ZF, Hasvold L, Wang L, Wang X, Petros AM, Park CH, Boghaert ER, Catron ND, Chen J, Colman PM, et al. Discovery of a potent and selective BCL-XL inhibitor with in vivo activity. ACS Med Chem Lett. 2014;5:1088–1093. doi: 10.1021/ml5001867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wang L, Doherty GA, Judd AS, Tao ZF, Hansen TM, Frey RR, Song X, Bruncko M, Kunzer AR, Wang X, et al. Discovery of A-1331852, a first-in-class, potent, and orally-bioavailable BCL-X(L) inhibitor. ACS Med Chem Lett. 2020;11:1829–1836. doi: 10.1021/acsmedchemlett.9b00568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lessene G, Czabotar PE, Sleebs BE, Zobel K, Lowes KN, Adams JM, Baell JB, Colman PM, Deshayes K, Fairbrother WJ, et al. Structure-guided design of a selective BCL-X(L) inhibitor. Nat Chem Biol. 2013;9:390–397. doi: 10.1038/nchembio.1246. [DOI] [PubMed] [Google Scholar]
- 162.Khan S, Zhang X, Lv D, Zhang Q, He Y, Zhang P, Liu X, Thummuri D, Yuan Y, Wiegand JS, et al. A selective BCL-X(L) PROTAC degrader achieves safe and potent antitumor activity. Nat Med. 2019;25:1938–1947. doi: 10.1038/s41591-019-0668-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zhang X, Thummuri D, He Y, Liu X, Zhang P, Zhou D, Zheng G. Utilizing PROTAC technology to address the on-target platelet toxicity associated with inhibition of BCL-X(L) Chem Commun (Camb) 2019;55:14765–14768. doi: 10.1039/C9CC07217A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Balachander SB, Criscione SW, Byth KF, Cidado J, Adam A, Lewis P, Macintyre T, Wen S, Lawson D, Burke K, et al. AZD4320, a dual inhibitor of Bcl-2 and Bcl-x(L), induces tumor regression in hematologic cancer models without dose-limiting thrombocytopenia. Clin Cancer Res. 2020;26:6535–6549. doi: 10.1158/1078-0432.CCR-20-0863. [DOI] [PubMed] [Google Scholar]
- 165.Chen J, Zhou H, Aguilar A, Liu L, Bai L, McEachern D, Yang CY, Meagher JL, Stuckey JA, Wang S. Structure-based discovery of BM-957 as a potent small-molecule inhibitor of Bcl-2 and Bcl-xL capable of achieving complete tumor regression. J Med Chem. 2012;55:8502–8514. doi: 10.1021/jm3010306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bai L, Chen J, McEachern D, Liu L, Zhou H, Aguilar A, Wang S. BM-1197: a novel and specific Bcl-2/Bcl-xL inhibitor inducing complete and long-lasting tumor regression in vivo. PLoS One. 2014;9:e99404. doi: 10.1371/journal.pone.0099404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Loriot Y, Mordant P, Dugue D, Geneste O, Gombos A, Opolon P, Guegan J, Perfettini JL, Pierre A, Berthier LK, et al. Radiosensitization by a novel Bcl-2 and Bcl-XL inhibitor S44563 in small-cell lung cancer. Cell Death Dis. 2014;5:e1423. doi: 10.1038/cddis.2014.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Yi H, Qiu MZ, Yuan L, Luo Q, Pan W, Zhou S, Zhang L, Yan X, Yang DJ. Bcl-2/Bcl-xl inhibitor APG-1252-M1 is a promising therapeutic strategy for gastric carcinoma. Cancer Med. 2020;9:4197–4206. doi: 10.1002/cam4.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wang X, Zhang C, Yan X, Lan B, Wang J, Wei C, Cao X, Wang R, Yao J, Zhou T, et al. A novel bioavailable BH3 mimetic efficiently inhibits colon cancer via cascade effects of mitochondria. Clin Cancer Res. 2016;22:1445–1458. doi: 10.1158/1078-0432.CCR-15-0732. [DOI] [PubMed] [Google Scholar]
- 170.Pan R, Ruvolo VR, Wei J, Konopleva M, Reed JC, Pellecchia M, Andreeff M, Ruvolo PP. Inhibition of Mcl-1 with the pan-Bcl-2 family inhibitor (-)BI97D6 overcomes ABT-737 resistance in acute myeloid leukemia. Blood. 2015;126:363–372. doi: 10.1182/blood-2014-10-604975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kivioja JL, Thanasopoulou A, Kumar A, Kontro M, Yadav B, Majumder MM, Javarappa KK, Eldfors S, Schwaller J, Porkka K, Heckman CA. Dasatinib and navitoclax act synergistically to target NUP98-NSD1(+)/FLT3-ITD(+) acute myeloid leukemia. Leukemia. 2019;33:1360–1372. doi: 10.1038/s41375-018-0327-2. [DOI] [PubMed] [Google Scholar]
- 172.Kazi A, Sun J, Doi K, Sung SS, Takahashi Y, Yin H, Rodriguez JM, Becerril J, Berndt N, Hamilton AD, et al. The BH3 alpha-helical mimic BH3-M6 disrupts Bcl-X(L), Bcl-2, and MCL-1 protein-protein interactions with Bax, Bak, Bad, or Bim and induces apoptosis in a Bax- and Bim-dependent manner. J Biol Chem. 2011;286:9382–9392. doi: 10.1074/jbc.M110.203638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ahn CH, Lee WW, Jung YC, Shin JA, Hong KO, Choi S, Swarup N, Kim J, Ahn MH, Jung M, et al. Antitumor effect of TW-37, a BH3 mimetic in human oral cancer. Lab Anim Res. 2019;35:27. doi: 10.1186/s42826-019-0028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wei J, Stebbins JL, Kitada S, Dash R, Placzek W, Rega MF, Wu B, Cellitti J, Zhai D, Yang L, et al. BI-97C1, an optically pure Apogossypol derivative as pan-active inhibitor of antiapoptotic B-cell lymphoma/leukemia-2 (Bcl-2) family proteins. J Med Chem. 2010;53:4166–4176. doi: 10.1021/jm1001265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Gavathiotis E, Suzuki M, Davis ML, Pitter K, Bird GH, Katz SG, Tu HC, Kim H, Cheng EH, Tjandra N, Walensky LD. BAX activation is initiated at a novel interaction site. Nature. 2008;455:1076–1081. doi: 10.1038/nature07396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Cournoyer S, Addioui A, Belounis A, Beaunoyer M, Nyalendo C, Le Gall R, Teira P, Haddad E, Vassal G, Sartelet H. GX15-070 (Obatoclax), a Bcl-2 family proteins inhibitor engenders apoptosis and pro-survival autophagy and increases Chemosensitivity in neuroblastoma. BMC Cancer. 2019;19:1018. doi: 10.1186/s12885-019-6195-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhang L, Zhou Y, Chen K, Shi P, Li Y, Deng M, Jiang Z, Wang X, Li P, Xu B. The pan-Bcl2 Inhibitor AT101 Activates the Intrinsic Apoptotic Pathway and Causes DNA Damage in Acute Myeloid Leukemia Stem-Like Cells. Target Oncol. 2017;12:677–687. doi: 10.1007/s11523-017-0509-2. [DOI] [PubMed] [Google Scholar]
- 178.Casara P, Davidson J, Claperon A, Le Toumelin-Braizat G, Vogler M, Bruno A, Chanrion M, Lysiak-Auvity G, Le Diguarher T, Starck JB, et al. S55746 is a novel orally active BCL-2 selective and potent inhibitor that impairs hematological tumor growth. Oncotarget. 2018;9:20075–20088. doi: 10.18632/oncotarget.24744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Bruncko M, Wang L, Sheppard GS, Phillips DC, Tahir SK, Xue J, Erickson S, Fidanze S, Fry E, Hasvold L, et al. Structure-guided design of a series of MCL-1 inhibitors with high affinity and selectivity. J Med Chem. 2015;58:2180–2194. doi: 10.1021/jm501258m. [DOI] [PubMed] [Google Scholar]
- 180.Abulwerdi F, Liao C, Liu M, Azmi AS, Aboukameel A, Mady AS, Gulappa T, Cierpicki T, Owens S, Zhang T, et al. A novel small-molecule inhibitor of mcl-1 blocks pancreatic cancer growth in vitro and in vivo. Mol Cancer Ther. 2014;13:565–575. doi: 10.1158/1535-7163.MCT-12-0767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ramsey HE, Fischer MA, Lee T, Gorska AE, Arrate MP, Fuller L, Boyd KL, Strickland SA, Sensintaffar J, Hogdal LJ, et al. A novel MCL1 inhibitor combined with venetoclax rescues venetoclax-resistant acute myelogenous leukemia. Cancer Discov. 2018;8:1566–1581. doi: 10.1158/2159-8290.CD-18-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Szlávik Z, Ondi L, Csékei M, Paczal A, Szabó ZB, Radics G, Murray J, Davidson J, Chen I, Davis B, et al. Structure-guided discovery of a selective Mcl-1 inhibitor with cellular activity. J Med Chem. 2019;62:6913–6924. doi: 10.1021/acs.jmedchem.9b00134. [DOI] [PubMed] [Google Scholar]
- 183.Yi X, Sarkar A, Kismali G, Aslan B, Ayres M, Iles LR, Keating MJ, Wierda WG, Long JP, Bertilaccio MTS, Gandhi V. AMG-176, an Mcl-1 antagonist, shows preclinical efficacy in chronic lymphocytic leukemia. Clin Cancer Res. 2020;26:3856–3867. doi: 10.1158/1078-0432.CCR-19-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Tron AE, Belmonte MA, Adam A, Aquila BM, Boise LH, Chiarparin E, Cidado J, Embrey KJ, Gangl E, Gibbons FD, et al. Discovery of Mcl-1-specific inhibitor AZD5991 and preclinical activity in multiple myeloma and acute myeloid leukemia. Nat Commun. 2018;9:5341. doi: 10.1038/s41467-018-07551-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zhang Z, Wu G, Xie F, Song T, Chang X. 3-Thiomorpholin-8-oxo-8H-acenaphtho [1,2-b]pyrrole-9-carbonitrile (S1) based molecules as potent, dual inhibitors of B-cell lymphoma 2 (Bcl-2) and myeloid cell leukemia sequence 1 (Mcl-1): structure-based design and structure-activity relationship studies. J Med Chem. 2011;54:1101–1105. doi: 10.1021/jm101181u. [DOI] [PubMed] [Google Scholar]
- 186.Wang Z, Guo Z, Song T, Zhang X, He N, Liu P, Wang P, Zhang Z. Proteome-wide identification of on- and off-targets of Bcl-2 inhibitors in native biological systems by using affinity-based probes (AfBPs) Chembiochem. 2018;19:2312–2320. doi: 10.1002/cbic.201800380. [DOI] [PubMed] [Google Scholar]
- 187.Shen HP, Wu WJ, Ko JL, Wu TF, Yang SF, Wu CH, Yeh CM, Wang PH. Effects of ABT-737 combined with irradiation treatment on uterine cervical cancer cells. Oncol Lett. 2019;18:4328–4336. doi: 10.3892/ol.2019.10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Tahir SK, Smith ML, Hessler P, Rapp LR, Idler KB, Park CH, Leverson JD, Lam LT. Potential mechanisms of resistance to venetoclax and strategies to circumvent it. BMC Cancer. 2017;17:399. doi: 10.1186/s12885-017-3383-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Mérino D, Khaw SL, Glaser SP, Anderson DJ, Belmont LD, Wong C, Yue P, Robati M, Phipson B, Fairlie WD, et al. Bcl-2, Bcl-x(L), and Bcl-w are not equivalent targets of ABT-737 and navitoclax (ABT-263) in lymphoid and leukemic cells. Blood. 2012;119:5807–5816. doi: 10.1182/blood-2011-12-400929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Gandhi L, Camidge DR, Ribeiro de Oliveira M, Bonomi P, Gandara D, Khaira D, Hann CL, EM MK, Litvinovich E, Hemken PM, et al. Phase I study of Navitoclax (ABT-263), a novel Bcl-2 family inhibitor, in patients with small-cell lung cancer and other solid tumors. J Clin Oncol. 2011;29:909–916. doi: 10.1200/JCO.2010.31.6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Pécot J, Maillet L, Le Pen J, Vuillier C, Trécesson SC, Fétiveau A, Sarosiek KA, Bock FJ, Braun F, Letai A, et al. Tight sequestration of BH3 Proteins by BCL-xL at subcellular membranes contributes to apoptotic resistance. Cell Rep. 2016;17:3347–3358. doi: 10.1016/j.celrep.2016.11.064. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable