Summary
Salt stress has detrimental effects on crop growth and yield, and the area of salt‐affected land is increasing. Soybean is a major source of vegetable protein, oil and feed, but considered as a salt‐sensitive crop. Cultivated soybean (Glycine max) is domesticated from wild soybean (G. soja) but lost considerable amount of genetic diversity during the artificial selection. Therefore, it is important to exploit the gene pool of wild soybean. In this study, we identified 34 salt‐tolerant accessions from wild soybean germplasm and found that a 7‐bp insertion/deletion (InDel) in the promoter of GsERD15B (early responsive to dehydration 15B) significantly affects the salt tolerance of soybean. GsERD15B encodes a protein with transcriptional activation function and contains a PAM2 domain to mediate its interaction with poly(A)‐binding (PAB) proteins. The 7‐bp deletion in GsERD15B promoter enhanced the salt tolerance of soybean, with increased up‐regulation of GsERD15B, two GmPAB genes, the known stress‐related genes including GmABI1, GmABI2, GmbZIP1, GmP5CS, GmCAT4, GmPIP1:6, GmMYB84 and GmSOS1 in response to salt stress. We propose that natural variation in GsERD15B promoter affects soybean salt tolerance, and overexpression of GsERD15B enhanced salt tolerance probably by increasing the expression levels of genes related to ABA‐signalling, proline content, catalase peroxidase, dehydration response and cation transport.
Keywords: GsERD15B, GWAS, haplotype, InDel, salt stress, soybean
Introduction
Soil salinity causes severe leaf chlorosis, dehydration, necrosis of plants (Xie et al., 2008) and ultimately limits their growth and development (Zhu, 2001). Salinity‐alkalinity stress reduces crop quality and yield (Xiang et al., 2016), which is one of the major environmental factors affecting food security. Saline soils currently account for 19.6% of irrigated land and over 2.1% of non‐irrigated land (FAO, 2018). Meanwhile, the area of salt‐affected agricultural land is predicted to double by year 2050 for irrigated and some semi‐arid areas due to inefficient fertilizer practices, salt‐water intrusion and use of poor quality irrigation water (Rengasamy, 2006).
Soybean [Glycine max (L.) Merr.] is a major source of vegetable protein and oil worldwide (Do et al., 2016), providing about 70% of the world protein meal consumption and 28% of the world vegetable oil consumption in 2019 (www.soystats.com). Soybean is generally classified as a salt‐sensitive crop compared with other major crops such as wheat, rice and cotton (Do et al., 2016; Munns and Tester, 2008). Under salt stress, soybean yield was reduced by 20% at the electrical conductivity (ECe) of 4.0 dS/m and by 54% at ECe of 6.7 dS/m (Katerji et al., 2003). Thus, it is important to identify salt‐tolerant soybean germplasm and genes to breed elite cultivars with greater salt tolerance.
The identification of natural alleles underlying salt tolerance indicates there is a great potential to improve the yield of soybean under saline conditions by genetic improvement (Parker et al., 1983). Previous studies have shown that there is great variation in salt tolerance among soybean germplasm, and a large genetic difference between wild soybean (G. soja) and cultivated soybean (G. max) has been observed (Ha et al., 2013; Qi et al., 2014; Zhou et al., 2015). Several salt‐tolerant soybean accessions (including G. soja) have been identified (Chen et al., 2013; Guan et al., 2014; Qi et al., 2014; Shao et al., 1986). Due to the artificial selection and loss of some important genes in cultivated soybean during domestication, wild soybean population exhibits higher genetic and allelic diversity than the cultivated soybean (Hyten et al., 2006; Wang et al., 2015; Zhou et al., 2015), which is therefore a valuable genetic pool for breeding new salt‐tolerant varieties.
Tolerances to environmental stresses are controlled by a large number of genes scattered throughout the genome (Guan et al., 2014). Quantitative trait locus (QTL) controlling salt tolerance had been identified in soybean (Chen et al., 2008; Hamwieh et al., 2011; Hamwieh and Xu, 2008; Lee et al., 2004). A major salt tolerance QTL was mapped to the region between simple repeat sequence (SSR) markers Sat_091 and Satt237 on chromosome (Chr.) 3 by using 106 F2:5 recombinant inbred lines (RILs) derived from an across between S‐100 and Tokyo (Lee et al., 2004). This QTL region was also identified in other populations (Ha et al., 2013; Hamwieh et al., 2011; Hamwieh and Xu, 2008). Recently, Glyma03g32900 (GmCHX1, encoding a cation H+ exchanger) within this major salt tolerance QTL on Chr.3, was found as a novel ion transporter and associated with salt tolerance (Qi et al., 2014). Another study showed that GmCHX1 is the same gene as GmSALT3, a gene associated with shoot sodium ion accumulation and salt tolerance (Guan et al., 2014). In addition, other salt‐tolerant genes have also been identified from wild soybean. For example, GsCLC‐c2 was identified through genome‐wide association study (GWAS), which improved salt tolerance through the sequestration of excess Cl− into the vacuoles of root cells (Wei et al., 2019). Overexpression of GsMYB15 (Shen et al., 2018), GsRR (Chen et al., 2018) or GsCHX19.3 (Jia et al., 2017) conferred salt and alkali tolerance of Arabidopsis thaliana. Overexpression of GsSRK increased the salt tolerance of transgenic Medicago sativa and A. thaliana (Sun et al., 2018).
Although some salt tolerance QTL and a limited number of salt tolerance genes have been reported in soybean, more diverse natural alleles for salt tolerance from wild soybean need to be discovered. Therefore, in this study, the salt tolerance candidate genes were identified by integrating GWAS on salt tolerance of 182 wild soybean (G. soja) accessions, with differential gene expression and sequence variation analyses between salt‐tolerant and sensitive soybean varieties. Further haplotype analysis revealed the polymorphism of an insertion/deletion (InDel) at the promoter region of one candidate gene, GsERD15B (Glycine soja Early Responsive to Dehydration 15 B), was significantly associated with salt tolerance. The effect of different natural alleles of GsERD15B on salt tolerance was determined by transforming soybean hairy roots with vectors containing different promoters (including two natural alleles) to express GsERD15B gene. The possible molecular mechanism of GsERD15B in response to salt stress was investigated by yeast two‐hybrid (Y2H) experiments, bimolecular fluorescence complementation (BiFC) assay, protein–protein interaction network and expression analysis of the known salt stress‐related genes, as well as promoter luciferase (LUC) assay. A derived Cleaved Amplified Polymorphic Sequences (dCAPS) marker was also developed to distinguish the two natural alleles of GsERD15B. This study identified the natural alleles underlying the salt tolerance in wild soybean (G. soja) and revealed the role of GsERD15B in salt tolerance.
Results
Population structure and variation in salt tolerance of 182 wild soybean accessions
A total of 182 wild soybean (G. soja) accessions originated from 24 regions in China (Table S1) were genotyped by RAD‐seq, and a total of 141,425 SNPs with missing rate < 30% and minor allele frequency (MAF) > 0.01 were retained (the heterozygous SNPs were treated as missing) (He et al., 2017). The missing SNPs were then imputed, and a total of 72,574 genome‐wide SNPs with MAF > 0.05 were used for further analysis. Based on the PCA and population structure analysis, these 182 accessions were clearly grouped into two subpopulations, group 1 and group 2 (Figure S1). From delta K value, the most likely K value was 2 (Figure S1a) and ADMIXTURE analysis (Figure S1b) inferred two ancestral groups, which was supported by the PCA result (Figure S1c). Neighbour‐joining tree (Figure 1a) also classified these 182 soybean accessions into two groups, reflecting their geographical origin (Figure 1b). Based on the soybean Eco‐Region classification in China (Gai and Wang, 2001), the majority (94%) of the accessions within group 1 originated from Eco‐Region I and II, while group 2 mainly (89%) originated from Eco‐Region III and IV.
Figure 1.
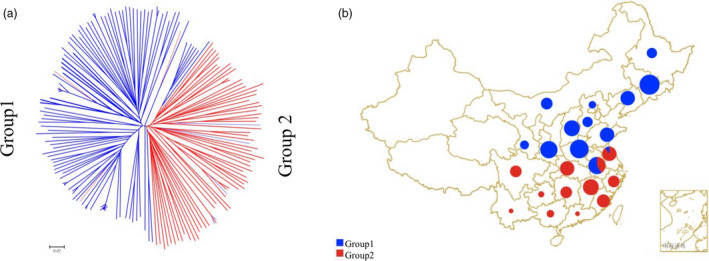
Genetic relationship and geographic origins of the two groups within 182 wild soybean accessions. Two groups, group1 and group2, within the 182 wild soybean (G. soja) accessions identified by Population Structure analysis, are shown in blue and red, respectively. (a) Neighbour‐joining tree of the 182 accessions using Nei’s genetic distance. (b) The geographic origins of the 182 accessions. The size of the pie represents the number of accessions within the location.
The salt tolerance of these 182 soybean accessions was evaluated using STR at seedling stage as described previously (Hamwieh et al., 2011; Shao et al., 1986; Tuyen et al., 2010). The heritability of STR was estimated as 91.0% based on the variance analysis (Table S2), suggesting that the phenotypic variation in salt tolerance of this population was mainly attributed to genetic effects. The correlation coefficients of STR between repeated experiments were quite high (Table S3), indicating high repeatability of STR. There was great variation in salt tolerance among these wild soybean accessions, with STR ranging from 1 to 5, where 1 represents most salt‐tolerant and 5 as most salt‐sensitive (Figure S2a). Due to good repeatability, the BLUP of STR (Figure S2b) is consistent with the average value of three biological replications (Figure S2c, d). The phenotypes of two soybean accessions representing most salt‐tolerant (STR = 1) and salt‐sensitive (STR = 5) under 200 mm NaCl for 16 days showed obvious difference: the salt‐tolerant accession was almost normal with green healthy leaves while the sensitive accession appeared dead with yellow and wilted leaves (Figure S3).
Association analysis and identification of candidate genes for salt tolerance in G. soja
We performed GWAS on salt tolerance (BLUP value of STR) in 182 wild soybean accessions (Figure S4), using mixed model in EMMAX (Zhang et al., 2010), which corrects the confounding effects of population structure and relatedness between individuals. We used the suggestive threshold P‐value < 2.79 × 10−4 (−log10 P‐value > 3.55) to select the potential STR‐associated SNPs, which is non‐conservative but allow us to select the possible candidate genes by integrating GWAS with other experimental evidence without missing too many true candidates. Using this threshold, 19 suggestive SNPs were associated with salt tolerance (Figure S4, Table S4), and 11 of them showed significant association (P < 0.05) with STR in the multi‐locus mixed model (Table S4). To select the candidate genes underlying these 11 STR‐associated SNPs, annotations for the 209 genes within ± 115 kb (average LD decay distance across all 20 chromosomes) of these 11 SNPs were obtained (Table S5), and nine genes had the annotations related to salt tolerance (Table S4).
Next, we investigated the expression patterns of these nine candidate genes in ten salt‐tolerant and ten salt‐sensitive soybean accessions in response to salt stress (Figure S5) by qRT‐PCR. The relative expression levels of four genes, GsPRX, GsJMT, GsCHX and GsERD15B, were significantly higher in response to salt stress in salt‐tolerant soybean accessions than those salt‐sensitive accessions (Figure S5), and the STR was significantly correlated with the relative expressions of these four genes in these 20 soybean accessions (Table S6), indicating their potential roles in salt tolerance of soybean. GsCHX had been identified as the known salt tolerance gene GmCHX1/GmSALT3/Ncl (Glyma03g32900) in soybean (Guan et al., 2014; Qi et al., 2014), while the roles of other three candidate genes in salt tolerance are unknown. Therefore, we sequenced the coding and promoter regions of these three newly identified salt tolerance candidate genes (GsPRX, GsJMT, GsERD15B) from ten salt‐tolerant and ten salt‐sensitive soybean accessions. The results showed that there was sequence variation only in the promoter region of GsERD15B between salt‐tolerant and salt‐sensitive soybean accessions (Figure S6), while there was no variation in the sequences of other genes or the CDS region of GsERD15B, which showed same as the reference genome sequence of Williams 82. Nine out of ten salt‐sensitive accessions had a 7‐bp insertion (ATTTTTT) at −525~−519 bp upstream of the start codon (ATG) of GsERD15B, while eight out of ten salt‐tolerant accessions did not have this insertion (Figure S6). Therefore, we selected GsERD15B as the salt tolerance candidate gene for further analysis.
Haplotype analysis and dCAPS marker development for salt tolerance candidate gene GsERD15B
To confirm the relationship between GsERD15B and salt tolerance, the promoter regions of 48 salt‐tolerant and 54 salt‐sensitive accessions were sequenced and compared (Table S7). Two haplotypes (Hap) were observed in this region among these 102 accessions, including 57 accessions as Hap1 and 45 accessions as Hap2 (Table S7). Hap1 had the 7‐bp insertion at −525~−519 bp of GsERD15B, while Hap2 contained a 7‐bp deletion in this promoter region (Figure 2a). Most (42/48 = 87.5%) salt‐tolerant accessions belong to Hap2 except that six salt‐tolerant accessions belong to Hap1, while most (51/54 = 94.4%) salt‐sensitive accessions belong to Hap1 except that three salt‐sensitive materials belong to Hap2 (Table S7). There was significant (P = 1.47 × 10−12, two‐sided Wilcoxon test) difference in salt tolerance between Hap1 and Hap2: the average STR of Hap1 is 4.20, while the average STR of Hap2 is 1.64 (Figure 2b).
Figure 2.
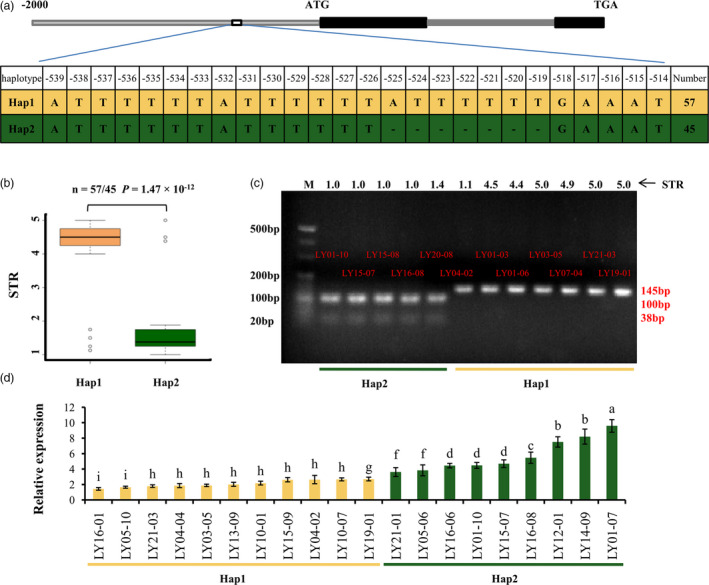
Haplotype and relative expression analyses of GsERD15B. (a) Schematic graph shows the Insertion/Deletion (InDel) variation in the promoter region of GsERD15B gene and the corresponding two haplotypes, Hap1 and Hap2. (b) Boxplot of salt tolerance rating (STR) for two haplotypes. Statistical significance (P = 1.47 × 10−12) of the difference between two haplotypes was determined by two‐sided Wilcoxon test. The centre bold line represents the median; box edges indicate the upper and lower quantiles; whiskers show the 1.5 × interquartile range and points indicate outliers. (c) Electrophoresis shows dCAPS marker polymorphism for GsERD15B. Hap2 type can be cleaved by Ssp I enzyme, which produced DNA fragments of 100‐bp and 38‐bp. Hap1 type cannot be digested by Ssp I enzyme, leading to a single band of 145‐bp. DNA molecular weight marker is 20‐bp DNA Ladder. (d) Relative expression of GsERD15B in soybean response to salt stress. The relative expression of GsERD15B in soybean root tips (0–2 mm) at 12 h after salt stress (180 mm NaCl) was quantified by qRT‐PCR, using GmUKN1 as the reference gene and the corresponding samples under 0 mm NaCl as controls. Data represent the mean ± standard deviation of three replicates. Same letters above bars indicate no significant difference according to Duncan’s multiple range test at α = 0.05.
Since the 7‐bp InDel in GsERD15B promoter was found significantly associated with the salt tolerance in wild soybean, we designed a dCAPS marker for this 7‐bp InDel. After PCR amplification using the specific primers (Table S8), Hap2 type produced 138‐bp amplicons containing ‘TTATAA’ sequence, which can be digested by the restriction enzyme Ssp I, resulting in a 100‐bp band and a 38‐bp band after electrophoresis, while Hap1 type produced 145‐bp amplicons, which cannot be digested by Ssp I and therefore showed a 145‐bp band after electrophoresis (Figure 2c), which was consistent with the sequencing results (Figure S6; Table S7). We used this dCAPS marker to genotype the rest of soybean accessions, and the polymorphisms were consistent with sequencing results (Figure S7; Table S7). Therefore, this dCAPS marker can be used to differentiate the two natural alleles of the 7‐bp InDel in GsERD15B promoter, which would be useful to select other soybean accessions with Hap2 type GsERD15B allele, and also can enhance the efficiency of marker assisted selection (MAS) programme to breed for salt‐tolerant soybean cultivars in future research.
Natural alleles of GsERD15B promoter affect salt tolerance of soybean hairy roots
Based on the observation that the salt‐induced expression levels of GsERD15B were significantly higher in salt‐tolerant soybean accessions than those in salt‐sensitive accessions (Figure S5), and the Hap2 allele of GsERD15B had significantly better salt tolerance than Hap1 allele (Figure 2b), therefore we hypothesized that Hap2 allele might have higher salt‐induced expression levels of GsERD15B than Hap1 allele, then leading to salt tolerance. We used the 20 soybean accessions with extreme STR to test this, and the results showed that, on average, Hap2 allele of GsERD15B conferred significantly better salt tolerance (Figure S8a) and had higher salt‐induced expression levels of GsERD15B than Hap 1 allele (Figure S8b; Figure 2d; Figure S9).
In order to confirm the function of GsERD15B in salt tolerance experimentally by reverse genetics, the coding region of GsERD15B gene was expressed in fusion with GFP, driven by different promoters including the 35S, ProHap1 and ProHap2 (GsERD15B promoter), respectively (Figure 3a), and transformed into soybean cotyledon hairy roots using the salt‐sensitive (STR = 5) soybean accession Tianlong1 which also contains the Hap1 type promoter. The positive transgenic hairy roots were identified through fluorescence signal of GFP (Figure 3b). Under normal condition (0 mM NaCl), all soybean hairy roots grew well, with no obvious difference (Figure 3c, d; Figure S10). Under 100 mM NaCl stress, the average fresh weight of ProHap2:GsERD15B transformed soybean hairy roots was significantly higher than that of ProHap1:GsERD15B or 35S:GsERD15B transformed hairy roots, and much higher than that of the empty vector transformed roots (Figure 3d; Figure S10).
Figure 3.
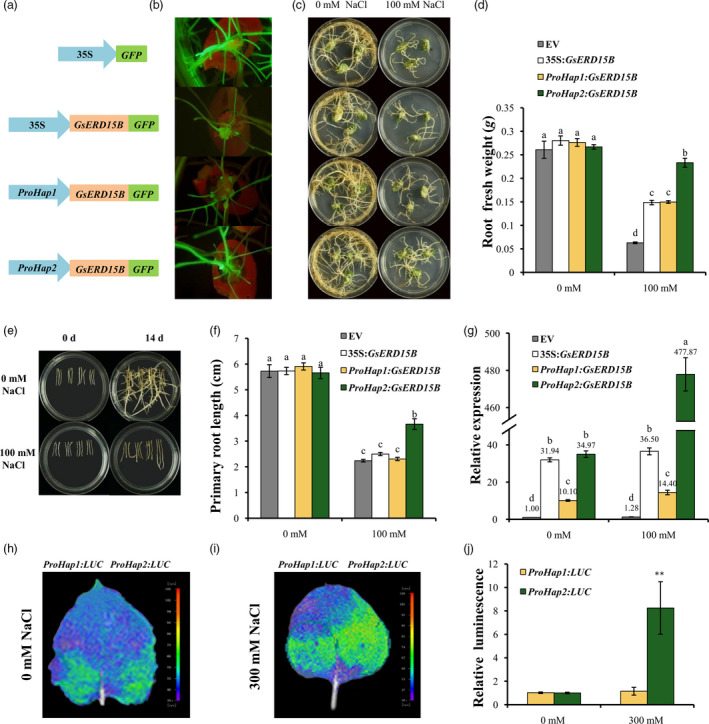
Effect of different GsERD15B promoters on salt tolerance of soybean hairy roots and promoter activities. (a) Schematic diagram of different constructs using pBinGFP4 as the backbone vector, including the empty vector (EV) 35S:GFP, 35S:GsERD15B, ProHap1:GsERD15B and ProHap2:GsERD15B. ProHap1 and ProHap2 represent the promoters from two haplotypes (Hap1 and Hap2) of GsERD15B gene, respectively. The GsERD15B gene was expressed in fusion with green florescence protein (GFP). (b) The fluorescence signal of transgenic soybean hairy roots. (c) Phenotypes of transgenic soybean hairy roots under control (0 mm NaCl) or salt stress (100 mM NaCl). Photographs were taken 14 day after treatment. From top to bottom represent the hairy roots of EV, 35S:GsERD15B, ProHap1:GsERD15B and ProHap2:GsERD15B, respectively. (d) Fresh weights of soybean hairy roots under control (0 mM NaCl) or salt stress (100 mM NaCl) for 14 day. (e) Phenotypes of regenerated transgenic soybean hairy roots under 0 or 100 mM NaCl treatment. Photographs were taken at 0 day and 14 day after treatment, respectively. From left to right in each Petri dish represents empty vector (EV, pBinGFP4), 35S:GsERD15B, ProHap1:GsERD15B and ProHap2:GsERD15B, respectively. (f) Primary root lengths at 14 day after treatment. (g) Relative expression of GsERD15B gene in soybean hairy roots at 3 h after 0 or 100 mM NaCl treatment by qRT‐PCR. For relative expression calculation, the sample from soybean hairy roots with empty vector (EV, pBinGFP4) under 0 mM NaCl was used as control and GmUKN1 was the reference gene. Same letters above bars indicate no significant differences according to Duncan’s multiple range test at 0.05 level. Soybean variety of Tianlong1 was used. Data represent the mean ± standard deviation of three biological replications and each replication has at least four independent roots for each genotype (n ≥ 12). (h‐j) Promoter activity of two GsERD15B haplotypes by transient expression in tobacco leaves after 0 or 300 mM NaCl treatment for 16 h. The LUC reporter gene was driven by each haplotype promoter. The photographs were taken using in vivo plant imaging system, and the luminescence intensity in h and i was shown in j. ** indicates significant difference in the LUC activity between ProHap1:LUC and ProHap2:LUC at 0.01 level by Student’s t‐test (n = 6).
In order to further validate the function of GsERD15B gene and different alleles of its promoter in salt tolerance, we also performed the gain‐of‐function tests using regenerated soybean hairy roots (Figure 3e‐g; Figure S11). Phenotypes of all regenerated hairy roots had no obvious difference under normal growing condition (Figure 3e, f; Figure S11). The primary root length of ProHap2:GsERD15B transformed roots was significantly (P < 0.05) longer than that of the empty vector, 35S:GsERD15B or ProHap1:GsERD15B transformed roots under salt stress (Figure 3f; Figure S11). We confirmed the overexpression level of GsERD15B in ProHap2:GsERD15B or ProHap1:GsERD15B transformed soybean hairy roots, which was over 34 times or 10‐fold (on average) of that in the empty vector transformed hairy roots under 0 mM NaCl, respectively (Figure 3g; Figure S11). The up‐regulation of GsERD15B mRNA expression at 3 h post‐salt stress in ProHap2:GsERD15B or ProHap1:GsERD15B transformed soybean hairy roots was 13.67‐fold (477.87/34.97) or 1.43‐fold (14.40/10.10) on average comparing 100 mM NaCl treatment with 0 mM NaCl condition, respectively (Figure 3g; Figure S11). Furthermore, promoter‐LUC transient expression assays in tobacco leaves revealed that the Hap2 type promoter had a significantly stronger activity than Hap1 type under salt stress but not control condition (Figure 3h‐j), suggesting that Hap2 type promoter is more responsive to salt stress than Hap1 type. Taken together, the Hap2 type promoter can increase the GsERD15B expression rapidly in response to salt stress and therefore enhanced the salt tolerance of soybean hairy roots.
GsERD15B has transcriptional activation function and interacts with poly(A)‐binding (PAB) proteins
Phylogenetic analyses of GsERD15B with 22 ERD15 proteins (Aalto et al., 2012) from plant species showed that GsERD15B was closely related with GmERD15A and MtERD15A (Figure 4a), and these ERD proteins share common domain structures, including the PAM2 and PAE1 motifs in the N‐terminus and a C‐terminal IqQPR motif (Figure 4b). The transient expression of 35S:GsERD15B‐GFP in N. benthamiana leaf cells showed that GsERD15B localized in the nucleus and cytomembrane (Figure 4c).
Figure 4.
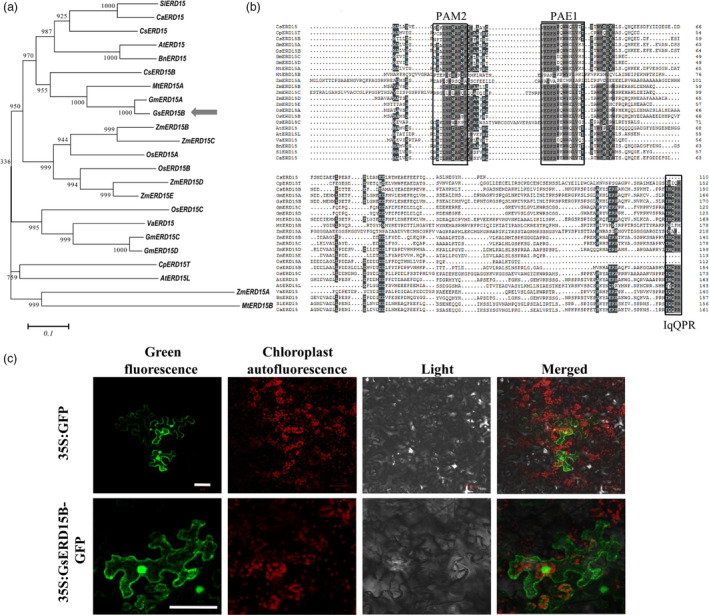
Phylogenetic analysis, sequence alignment and subcellular localization of GsERD15B protein. (a) Phylogenetic tree of ERD15 proteins. The 1000 bootstrap values are shown on the branches. (b) Sequence alignment of ERD15 proteins. The conserved PAM2, PAE1 and IqQPR domains are indicated by boxes. Both figures are generated using the full‐length amino acid sequences of the 22 proteins that are homologous with GsERD15B, including AtERD15 (Arabidopsis thaliana, AT2G41430), AtERD15L (Arabidopsis thaliana, AT4G14270), BnERD15 (Brassica napus, ADP37978.1), CaERD15 (Capsicum annuum, ABB89735.1), CpERD15T (Carica papaya, evm.TU.supercontig_197.11), CsERD15B (Cucumis sativus, Cucsa.335550), GmERD15A (Glycine max, Glyma04g28560), GsERD15B (Glycine soja, XP_028191281.1), GmERD15C (Glycine max, Glyma02g42860), GmERD15D (Glycine max, Glyma14g05980), MtERD15A (Medicago truncatula, Medtr3g023110), MtERD15B (Medicago truncatula, Medtr5g091120), OsERD15A (Oryza sativa, Q7XXS2), OsERD15B (Oryza sativa, Q7EZY8), OsERD15C (Oryza sativa, Q5W6M4), SlERD15 (Solanum lycopersicum, NP_001234461), VaERD15 (Vitis amurensis, JQ687321), ZmERD15A (Zea mays, GRMZM2G093325), ZmERD15B (Zea mays, GRMZM2G181551), ZmERD15C (Zea mays, GRMZM2G045178), ZmERD15D (Zea mays, GRMZM2G327692) and ZmERD15E (Zea mays, GRMZM2G037189). (c) Confocal microscopy images showing the subcellular localization of GsERD15B in leaf cells of Nicotiana benthamiana. Bars, 50 μm.
Since ERD15 proteins contain the conserved PAM2 domain that has the potential to interact with the PABC domain found mainly in PAB proteins (Aalto et al., 2012; Kozlov et al., 2004), Y2H assays were carried out to analyse whether GsERD15B possesses transcriptional activation function and if GsERD15B (the PAM2 domain of GsERD15B) could interact with the known PAB (PAB2, PAB4 and PAB8) proteins (Aalto et al., 2012). The results showed that GsERD15B has transcriptional activation function (Figure 5a) while the GsERD15B‐PAM2 (Figure 5a), PAB2, PAB4 and PAB8 (Figure S12) did not have self‐activation activity. GsERD15B‐PAM2 interacts with PAB2, PAB4 and PAB8 proteins (Figure 5a). The interactions between GsERD15B and PAB2, PAB4, as well as PAB8 were confirmed by BiFC assays (Figure 5b; Figure S13).
Figure 5.
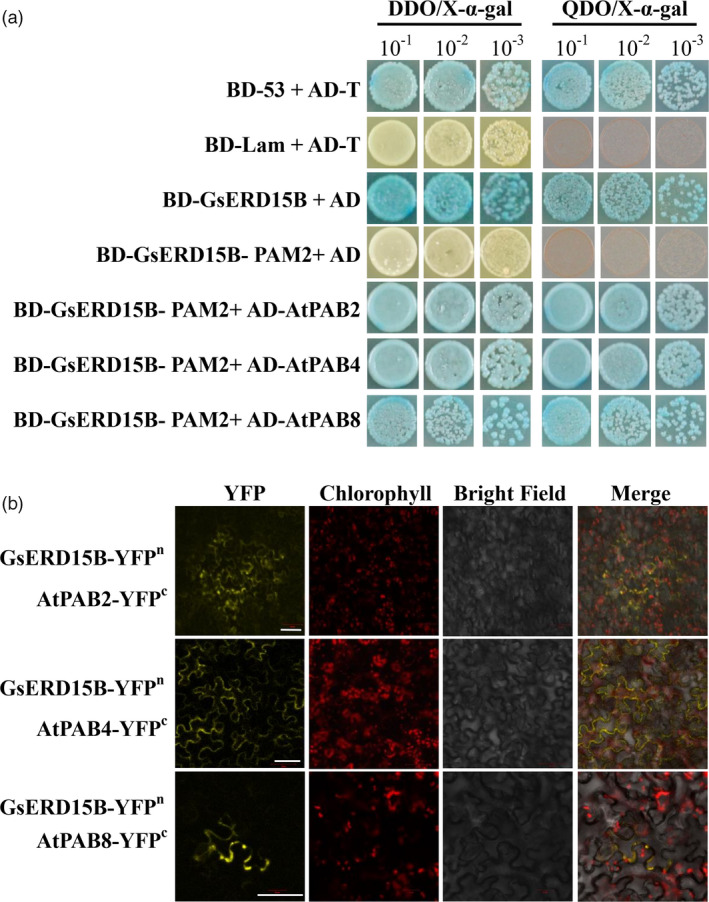
Transactivation and protein interaction analyses of GsERD15B. (a) Yeast two‐hybrid (Y2H) assay showing that GsERD15B protein has self‐activation activity but GsERD15B‐PAM2 domain does not have self‐activation activity, and GsERD15B interacts with AtPAB2, AtPAB4, AtPAB8. DDO/X‐α‐gal and QDO/X‐α‐gal represent double dropout medium (without Leu/Trp) and quadruple dropout medium (without Ade/Leu/Trp/His), respectively. BD: the GAL4 DNA binding domain in pGBKT7 vector; AD: the GAL4 activating domain in pGADT7 vector. The BD‐Lam + AD‐T is the negative control and BD‐53 + AD‐T is the positive control. (b) Bimolecular Fluorescence Complementation (BiFC) assay showing that GsERD15B interacts with AtPAB2, AtPAB4, AtPAB8, in leaf cells of Nicotiana benthamiana. YFP, yellow fluorescent protein. Scale bar, 50 μm.
To further investigate the potential interacting proteins of GsERD15B, the protein–protein interaction network analysis was performed using GmERD15B (having the same sequence as GsERD15B) as a query. Interestingly, five out of the top 10 potential interacting proteins (score > 0.9) of GmERD15B were PAB proteins (Figure 6a). These five GmPAB proteins have the same conserved domains including the PABC domain as PAB2, PAB4, and PAB8 (Figure 6b), and two of the GmPAB genes, GmPAB‐14G and GmPAB‐17G, have the annotations related to response to salt stress. Further, the BiFC experiments demonstrated that GmPAB‐14G and GmPAB‐17G could interact with GsERD15B/GmERD15B (Figure 6c; Figure S13). The coding sequences of GmPAB‐14G and GmPAB‐17G are identical with GsPAB‐14G and GsPAB‐17G, respectively. The relative expression of these two GmPAB genes in the transgenic soybean hairy roots (in the genotype background of Tianlong1) with ProHap2:GsERD15B showed significantly greater increase than that in hairy roots containing ProHap1:GsERD15B, 35S:GsERD15B or the empty vector at 3 h after salt treatment (Figure 6d), suggesting that the transcriptional changes of these two GmPAB genes in response to salt stress are affected by GsERD15B expression.
Figure 6.
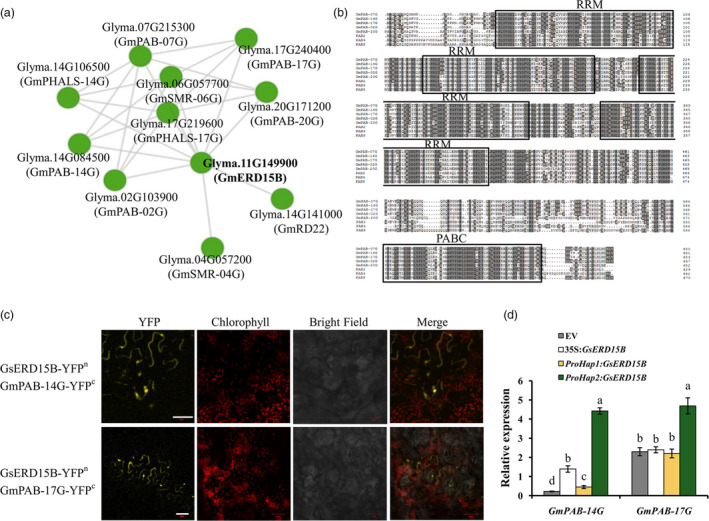
Prediction, sequence alignment, confirmation and relative expression of the GsERD15B‐interacting PAB proteins/genes. (a) The predicted protein–protein interaction network of GmERD15B using STRING database and Cytoscape software. Nodes represent proteins and edges represent interactions. (b) Sequence alignment of PAB proteins, including the five soybean PAB proteins that might interact with GmERD15B (GmPAB‐07G, GmPAB‐14G, GmPAB‐17G, GmPAB‐02G and GmPAB‐20G) and three known PAB proteins (PAB2, PAB4 and PAB8) from Arabidopsis, using full‐length amino acid sequence. The conserved RRM and PABC domains are indicated by boxes. (c) Bimolecular Fluorescence Complementation (BiFC) assay showing that GsERD15B (having the same sequence as GmERD15B) interacts with GmPAB‐14G, GmPAB‐17G in leaf cells of Nicotiana benthamiana. YFP, yellow fluorescent protein. Scale bar, 50 μm. (d) Relative expression levels of the two GmPAB genes at 3 h after 100 mM NaCl treatment in soybean (Tianlong1) hairy roots transformed by empty vector (EV, pBinGFP4), 35S:GsERD15B, ProHap1:GsERD15B, or ProHap2:GsERD15B, respectively. Root samples without salt stress (0 mM NaCl) for each genotype were used as the corresponding controls, and GmUKN1 was the reference gene. Data represent mean ± standard deviation of four biological replications (n ≥ 12). Same letters above bars for each gene indicate no significant difference between genotypes according to Duncan’s multiple range test at 0.05 level.
Overexpression of GsERD15B improved the expression levels of stress‐related genes
In order to investigate the possible mechanisms of GsERD15B‐mediated salt tolerance, the expression patterns of GsERD15B in different soybean tissues (Figure S14), as well as the effect of GsERD15B overexpression on the expression levels of the known stress‐related genes, were investigated (Figure 7a). Under 0 mM NaCl, the expression of GsERD15B was higher in roots and stems than leaves (Figure S14a). Under salt stress, the relative expression of GsERD15B in roots was higher than leaves (Figure S14b), and the relative expression levels in the roots and stems of salt‐tolerant soybean varieties were significantly higher than those in salt‐sensitive soybean varieties (Figure S14b), which suggest that GsERD15B might play important roles in soybean roots under salt stress. Further, the relative expression of ten stress‐related genes in five categories were compared between GsERD15B overexpressing soybean hairy roots and control roots, including three ABA‐related genes, GmABI1, GmABI2 and GmbZIP1 (Gao et al., 2011; Leung et al., 1994; Meyer et al., 1994), one proline synthesis‐related gene GmP5CS (Zegaoui et al., 2017), one gene (GmCAT4) encoding catalase peroxidase (Sun et al., 2016), two water loss related genes, GmMYB84 and GmPIP1:6 (Wang, Zhang et al., 2017; Zhou et al., 2014) and three genes (GmSOS1, GmSALT3, GmNHX1) encoding ion exchangers (Guan et al., 2014; Zhang et al., 2019). The relative expression of GmABI1, GmABI2, GmbZIP1, GmP5CS, GmCAT4, GmMYB84, GmPIP1:6 and GmSOS1 in transgenic soybean hairy roots with ProHap2:GsERD15B showed significantly greater increase than that in hairy roots containing ProHap1:GsERD15B or the empty vector at 3 h after salt treatment (Figure 7a), suggesting that the induced expression of GsERD15B driven by Hap2 type promoter under salt stress can enhance the expression of these stress‐related genes. We then selected the top three genes (GmABI2, GmP5CS and GmbZIP1) with highest relative expression (Figure 7a) for further investigation. We found that the overexpression of GsERD15B enhanced the promoter activities of these three genes in the promoter‐LUC assays (Figure 7b‐g). Thus, we propose that overexpression of GsERD15B enhanced salt tolerance probably by increasing the expression levels of genes related to ABA and dehydration response, proline content, catalase peroxidase and cation transport.
Figure 7.
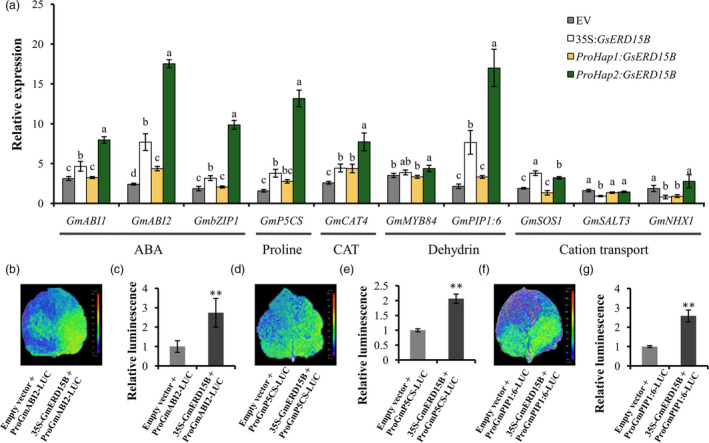
Relative expression patterns of stress‐related genes in transgenic soybean hairy roots and promoter activity assays. (a) Relative expression levels of stress‐related genes at 3 h after 100 mM NaCl treatment in soybean hairy roots transformed by empty vector (EV, pBinGFP4), 35S:GsERD15B, ProHap1:GsERD15B or ProHap2:GsERD15B, respectively. Root samples without salt stress (0 mM NaCl) were used as the controls. Data represent mean ± standard deviation of four biological replications (n ≥ 12). Same letters above bars for each gene indicate no significant difference between genotypes according to Duncan’s multiple range test at 0.05 level. (b, d, f) Promoter luciferase (LUC) assays in tobacco leaves to reveal the promoter activity of GmABI2, GmP5CS and GmPIP1:6. (c, e, g) Quantitative analyses of the LUC activities from tobacco leaves shown in b, d and f. ** represent significant difference at 0.01 level by Student’s t‐test (n = 6).
Discussion
The genetic diversity of soybean germplasm is of great value for soybean improvement and association mapping of important traits (Kan et al., 2015). Due to domestication, the genetic diversity in modern cultivated crops has been gradually reduced (Zhang et al., 2017). Wild soybean has higher phenotypic and genetic diversity than cultivated soybean (Lam et al., 2010; Munoz et al., 2017). Long‐term natural selection and environmental stresses also enable the surviving wild soybean varieties to possess superior characteristics of biotic stress resistance (Qi et al., 2014) and abiotic stress tolerance (Vigueira et al., 2016). In this study, the salt tolerance of 182 wild soybean accessions were evaluated and 34 salt‐tolerant (STR < 1.5) accessions were identified (Table S1), which would be useful for further genetic analysis and breeding for salt tolerance in soybean.
The LD between SNPs was estimated by r2 in our mapping population. As expected, the r 2 value declined as the physical distance between SNPs increased (Figure S15). In addition, the LD decay distance for each chromosome was different, ranging from approximately 32 kb to 230 kb. The average LD decay distance across all 20 chromosomes was estimated at approximately 115 kb, which is less than that in landraces (~500 kb) (Hwang et al., 2014), but more than the previous estimation of 75 kb in wild soybean (Lam et al., 2010). The use of different populations and different sets of molecular markers might cause the difference in estimation of LD decay distance.
Several salt tolerance QTL have been reported in soybean, which are mainly distributed on Chr.3 (Lee et al., 2004), Chr.6 (Cho et al., 2002; Kan et al., 2015), Chr.7 (Kan et al., 2016), Chr.17 (Tuyen et al., 2010) and Chr.19 (Lee et al., 2004). In this study, the single‐locus mixed model of GWAS suggested 19 SNPs associated with salt tolerance, which are distributed on Chr.3, Chr.6, Chr.11 ~ 14, Chr.18 and Chr.20. The loci of four SNPs on Chr.3 and Chr.6 have been reported in previous studies (Cho et al., 2002; Hamwieh et al., 2011; Hamwieh and Xu, 2008; Kan et al., 2015; Lee et al., 2004). The other 15 SNPs on Chr.11, Chr.12, Chr.14, Chr.18 and Chr.20 represent novel salt tolerance loci detected in this study. Different populations might lead to the identification of different QTL (Famoso et al., 2011). Previous reports showed that Chr.3 is the major genomic region controlling salt tolerance in soybean (Ha et al., 2013; Hamwieh et al., 2011; Hamwieh and Xu, 2008; Lee et al., 2004), which includes the salt tolerance gene GmCHX1/GmSALT3 in this region (Guan et al., 2014; Qi et al., 2014). This genomic region on Chr.3 is also detected by GWAS in this study (Figure S4), which is in the LD with the significantly STR‐associated SNPs. The closest SNP to GmCHX1/GmSALT3 in this study is Gm03_40722843 with a P‐value of 8.29 x 10‐5 in the single‐locus mixed model (Table S4), which passed the suggestive threshold (P < 2.79 × 10‐4) but did not reach the stringent threshold of Bonferroni corrected P‐value (false discovery rate, FDR0.05) = 4.38 × 10−7 (0.05/72 574). However, this SNP did show significant (P < 0.05) association with salt tolerance in our multi‐locus mixed model (Table S4). Therefore, we used the suggestive threshold for single‐locus mixed model and P < 0.05 in the multi‐locus mixed model as criteria to choose candidate QTL/SNPs, then chose candidate genes by integrating GWAS results with gene annotation, expression and sequence variation.
A total of 11 SNPs showed significant association with STR in the multi‐locus mixed model (Table S4). In the genomic regions within ± 115kb of these 11 SNPs, nine candidate genes have the functional annotations related to plant response to salt stress (Table S4), and four of them showed higher relative expression in response to salt stress in the ten salt‐tolerant accessions than the ten salt‐sensitive soybean accessions (Figure S5), including one known salt tolerance gene (GmCHX1/GmSALT3) and three novel genes. Sequencing analysis of the coding and promoter regions of the three novel genes showed that only the promoter region of GsERD15B had sequence variation among soybean accessions with extreme salt tolerance (Figure S6). Furthermore, in our mapping population, the SNP (Gm11_17854357) near GsERD15B showed more significant association with salt tolerance than the SNP (Gm03_40722843) near the known salt tolerance gene GsCHX in multi‐locus mixed model (Table S4); therefore, the function of GsERD15B was investigated in more detail.
To further examine the relationship between the variation in GsERD15B promoter and salt tolerance, we sequenced the promoter regions of 48 salt‐tolerant accessions and 54 salt‐sensitive accessions. The results showed that the majority of salt‐tolerant accessions belong to Hap2, while most salt‐sensitive accessions belong to Hap1 (Figure 2b; Table S7). The outliers such as the six salt‐tolerant accessions with Hap1 type promoter suggest there are other loci/genes controlling the salt tolerance, which also reflects the quantitative nature of this trait. In QTL studies, a single locus can only explain part of the phenotypic variation for the complex trait, and the effect of a locus/gene might depend on the genetic background. The SNP near GsERD15B explained 7.79% of the phenotypic variation in the whole population in this study (Table S4). The effects of GsERD15B haplotypes/alleles (two different promoters containing 7‐bp insertion or deletion) on salt tolerance have been compared using transgenic soybean hairy roots (in the genetic background of a salt‐sensitive variety Tianlong1), and the results showed that Hap2 type promoter can increase the GsERD15B expression rapidly in response to salt stress and enhanced the salt tolerance of soybean hairy roots, which confirmed the role of GsERD15B in salt tolerance. To investigate the natural alleles of ERD15B promoter in cultivated soybean, we further sequenced the promoter regions of 10 salt‐tolerant and 10 salt‐sensitive cultivated soybean accessions (Table S9). We found that all 10 salt‐tolerant cultivated soybean accessions belong to Hap2, while six out of 10 salt‐sensitive cultivated soybean accessions belong to Hap1. There was significant (P = 3.93 × 10−3, two‐sided Wilcoxon test) difference in STR between Hap1 and Hap2 among these 20 cultivated soybean accessions: the average STR of Hap1 is 4.61, while the average STR of Hap2 is 2.07 (Figure S16), which is consistent with the observation in wild soybean. Taken together, these findings inferred that the variation in GsERD15B promoter play an important role in soybean adaptation to salt stress.
Cotyledon hairy roots could be used in rapid gain‐of‐function tests of candidate genes (Qi et al., 2014). Soybean hairy roots transformed with GmCHX1 from the salt‐tolerant soybean W05 showed significantly higher root fresh weights than the control under NaCl treatments. It was also found that overexpression of GmCLC1 in soybean hairy roots enhanced the tolerance of soybean composite plants to salt stress (Wei et al., 2016). To confirm the role of two natural alleles of GsERD15B in salt tolerance, we transformed soybean cotyledon hairy roots with the empty vector, 35S:GsERD15B, ProHap1:GsERD15B, or ProHap2:GsERD15B (Figure 3). The ProHap2:GsERD15B transformed soybean hairy roots showed significantly higher root fresh weight (Figure 3c, d) and longer primary root length (Figure 3e, f) than the empty vector, 35S:GsERD15B or ProHap1:GsERD15B transformed roots under salt stress. All these findings inferred that the natural variation (7‐bp InDel) in the promoter region of GsERD15B plays a vital role in soybean salt tolerance. This 7‐bp InDel in the promoter resulted in different transcriptional changes of GsERD15B in response to salt stress: there was more up‐regulation of GsERD15B expression in ProHap2:GsERD15B transformed soybean hairy roots than that in ProHap1:GsERD15B transformed roots (13.67‐fold and 1.43‐fold, respectively) in response to salt stress (Figure 3g; Figure S11). Consistently, promoter‐LUC transient expression assays in tobacco leaves also revealed that the Hap2 type promoter of GsERD15B had a significantly stronger activity than Hap1 type under salt stress (Figure 3i, j), suggesting that Hap2 type promoter is more responsive to salt stress than Hap1 type.
We further analysed the cis‐elements and sequence characteristics of the 2‐kb promoter region (Figure S17) of GsERD15B in two soybean accessions, LY01‐06 (Hap1) and LY01‐10 (Hap2). Although the 7‐bp deletion (−525~−519 bp) in Hap2 disrupted the original cis‐elements of BoxI (light responsive element, with core sequence as TTTGAAA) and ERE (ethylene‐responsive element, with core sequence as TTTGAAAT), it still has the two complete BoxI and ERE cis‐elements (Figure S17). However, the Hap2 type sequence formed an 8‐bp inverted sequence ‘ATTTnnnnAAAT’ due to the 7‐bp deletion. A previous study showed that a 12‐bp palindromic sequence (inverted repeat sequence with no intervening nucleotides between the initial sequence and its downstream reverse complement) in the promoter of NRP‐B is the binding site of ERD15 to activate transcription (Alves et al., 2011). Therefore, the inverted sequence in Hap2 type promoter might be the binding site of a transcription factor in salt signalling pathway to enhance the expression of GsERD15B in response to salt stress.
ERD15 appears ubiquitous in the plant kingdom and may be an essential component in stress responses in plants (Aalto et al., 2012). ERD genes can be induced by various abiotic stresses, such as drought (Kiyosue et al., 1994), low temperature (Kiyosue et al., 1998), abscisic acid (Aalto et al., 2012) and salinity (Jian et al., 2016). ERD15 family has a conserved domain of PAM2 (Aalto et al., 2012), which has been shown to interact with the PABC domain of PAB proteins (Belostotsky and Meagher, 1996). We found that GsERD15B also has a conserved PAM2 domain, and GsERD15B‐PAM2 interacts with PAB2, PAB4, PAB8 (Figure 5) as expected. Taken together, these results revealed that the GsERD15B gene encodes an ERD15 protein with transcriptional activation function, and GsERD15B can interact with the known PAB proteins through its conserved PAM2 domain. Interestingly, the protein–protein interaction network predication and BiFC assay suggested that GsEDR15B also interact with other soybean PAB proteins which have the same highly conserved domains as PAB2, PAB4 and PAB8 (Figure 6). Further, the relative expression of GmABI1, GmABI2, GmbZIP1, GmP5CS, GmCAT4, GmMYB84, GmPIP1:6 and GmSOS1 in transgenic hairy roots with ProHap2:GsERD15B showed significantly greater increase than that in hairy roots containing ProHap1:GsERD15B or the empty vector at 3 h after salt treatment (Figure 7a), and GsERD15B overexpression enhanced the promoter activities of GmABI2, GmP5CS and GmbZIP1 in the tobacco leaf assays (Figure 7b‐g), which suggests that overexpression of GsERD15B enhanced salt tolerance probably by increasing the expression levels of genes related to ABA and dehydration response, proline content, catalase peroxidase and cation transport.
Experimental procedures
Plant materials
A total of 182 wild soybean (G. soja) accessions (Table S1), a well‐known salt‐tolerant soybean variety Lee 68 (Xu et al., 2011), and a transformation recipient soybean variety (salt‐sensitive) Tianlong1 (Li et al., 2017), were obtained from the National Center for Soybean Improvement (Nanjing, China). The 182 wild soybean accessions originated from 24 locations covering four soybean Eco‐Regions in China (Table S1), including Northern Single Cropping Spring Planting Varietal Eco‐Region (Eco‐Region I, n = 47), Huang‐Huai‐Hai Double Cropping Spring and Summer Planting Varietal Eco‐Region (Eco‐Region II, n = 64), Middle and Lower Changjiang Valley Double Cropping Spring and Summer Planting Varietal Eco‐Region (Eco‐Region III, n = 44), and Central South Multiple Cropping Spring Summer and Autumn Planting Varietal Eco‐Region (Eco‐Region IV, n = 27).
Salt tolerance evaluation in greenhouse
Salt tolerance of soybean was evaluated by salt tolerance rating (STR) according to previously published methods with modifications (Guan et al., 2014; Hamwieh et al., 2011; Shao et al., 1986; Tuyen et al., 2010). Details are described in Methods S1.
SNP genotyping
SNP genotyping was performed with RAD‐seq (Restriction‐site Associated DNA sequencing) using Illumina HiSeq 2000 at BGI (Shenzhen, China) (Andolfatto et al., 2011; He et al., 2017). Soybean genomic DNA was extracted using CTAB method (Murray and Thompson, 1980) and digested by Taq I to produce 400–600 bp fragments, which were then ligated with P1 and P2 adapters before pair‐end sequencing (90‐bp in length). The sequencing reads were aligned to the soybean reference genome (Williams 82) sequence (Schmutz et al., 2010) using SOAP2 (Li et al., 2009), and SNPs were called by RealSFS (Yi et al., 2010). FastPHASE (Scheet and Stephens, 2006) was used to impute missing SNPs.
Population structure and linkage disequilibrium (LD) analysis
The principle component analysis (PCA) was conducted using EIGENSTRAT (Price et al., 2006). The software fastStructure (Alexander et al., 2009) was used to analyse the population structure with K = 1–10 and determine the optimal K. MEGA6 was used to build neighbour‐joining tree (Tamura et al., 2013). LD was estimated using Haploview (Barrett et al., 2005). We used 500‐kb sliding window along each chromosome and calculated LD between all pairs of SNPs. Parameter r 2 was used to estimate the degree of LD (Li et al., 2016).
GWAS and candidate gene annotation
GWAS was performed with the efficient mixed model association eXpedited (EMMAX), using the model:
where X stands for the corresponding SNP vector, β is the coefficient vector for SNP effect, Z is the corresponding design matrix, u is the random effect accounting for population structure and relatedness, and ε is the random error (Kang et al., 2010).
The threshold for suggestive association was calculated based on previously published method (King et al., 2013; Zhang et al., 2019), which is P < 2.79 × 10‐4 (the average threshold across 20 soybean chromosomes using 1/n, where n is the number of markers on each chromosome). Multi‐locus mixed model was then used to further screen significant (P < 0.05) association (Wang et al., 2016). Genes within ± 115 kb (the average LD decay distance across all 20 chromosomes) of the significantly STR‐associated SNPs were annotated using Soybase (https://www.soybase.org/), and those with functional annotations related to salt tolerance (hyperosmotic response, calcium ion transport, response to salt stress, osmotic stress, cellular potassium ion homeostasis, sodium ion transmembrane transport, response to oxidative stress, dehydration‐induced protein, water transport) were selected as candidate genes for further analysis.
RNA isolation and quantitative reverse transcription PCR (qRT‐PCR)
Total RNAs were isolated from soybean tissues using RNAprep Pure Plant Kit (Tiangen Biotech, China), and first‐strand cDNA was synthesized by PrimeScript™ 1st Strand cDNA Synthesis Kit (TaKaRa, Japan). The primers (Table S8) were designed using Primer Premier 5 software (http://www.premierbiosoft.com/primerdesign/) (Wang, Jiang et al., 2017). GmUKN1 (Guan et al., 2014) was used as the reference gene for qRT‐PCR. The amplification efficiencies (E) of primer pairs were estimated (Table S8) by qRT‐PCR using 1:10, 1:20, 1:40, 1:80, and 1:160 dilutions of cDNA templates, according to the equation: E = [10−1/slope]−1 (Pfaffl, 2001). The qRT‐PCR was performed using SYBR Premix ExTaq™ II Mix (TaKaRa, Japan) on a Roche 480 real‐time detection system (Roche Diagnostics, Switzerland) according to the manufacturers’ instructions. Each experiment was performed in triplicates. The expression levels of all candidate genes were analysed by the 2−△△CT methods (Livak and Schmittgen, 2001).
Gene cloning and sequence analysis
Nucleotide sequences for selected candidate genes were downloaded from phytozome v10 (https://phytozome.jgi.doe.gov/pz/portal.html). The 2000‐bp upstream of the start codon ATG was considered as the promoter region. Primers (Table S8) were designed and synthesized by Invitrogen (Shanghai, China). The full‐length coding sequences were amplified using soybean cDNA as template and the promoter regions were amplified using soybean genomic DNA as template. The amplicons were sequenced at GenScript (Nanjing, China). Protein domain analysis was performed using SMART (http://smart.embl‐heidelberg.de/).
Development and test of dCAPS marker
The dCAPS marker was developed using dCAPS Finder 2.0 (Neff et al., 2002), which identifies the optimum restriction enzyme to detect sequence polymorphism (Kushanov et al., 2016). Specific primers (Table S8) were designed, and amplification products were purified using TaKaRa MiniBEST DNA Fragment Purification Kit Ver.4.0 (TaKaRa, Japan) after verified by agarose electrophoresis. The purified PCR products were digested by Ssp I (NEB, USA) in a total volume of 10 μL according to the manufacturers’ instructions. The digested products were observed by electrophoresis using 3% agarose gel and stained with ethidium bromide, then photo‐documented using Gel Imaging Documentation System (Bio‐Rad, USA).
Construction of plant expression vectors
Plant expression vectors were constructed using the One Step Cloning Kit (Vazyme, China). Details are described in Methods S1.
Genetic transformation and salt tolerance analysis of soybean hairy roots
ProHap1:GsERD15B, ProHap2:GsERD15B, 35S:GsERD15B and the empty vector pBinGFP4 were separately transformed into Agrobacterium rhizogenes strain K599 (Kereszt et al., 2007), which was further used to infect soybean cotyledons to obtain transgenic hairy roots (Xue et al., 2017). Transgenic soybean hairy roots were grown on Murashige and Skoog (MS) medium containing 500 mg/L carbenicillin and 50 mg/L cefotaxime (Sangon Biotech, China). After 15 days, the positive hairy roots were identified by green fluorescence signal of GFP using a stereoscopic fluorescence microscope (Mshot, China), and the non‐transgenic roots were cut‐off. The positive transgenic soybean hairy roots with similar length and volume were selected and transferred onto white solid medium (pH = 5.8, MDBio, China) containing 0 or 100 mM NaCl, respectively. Hairy roots were collected after two weeks to measure their fresh weight.
To re‐generate transgenic soybean hairy roots, the positive transgenic cotyledon soybean hairy roots were cultured on white solid medium until the length is greater than 3 cm, then the 3‐cm root tips were cut‐off and transferred onto white solid medium containing 0 or 100 mM NaCl for two weeks, respectively, and the photographs were taken, and the primary root lengths were measured using ImageJ (Schindelin et al., 2015). The roots under salt stress or control were collected for relative gene expression analysis.
Promoter‐LUC assays in tobacco leaves
The promoter activities were analysed by promoter‐LUC transient expression assays in tobacco (Nicotiana benthamiana) leaves, according to the procedures described in Methods S1.
Sequence alignment and phylogenetic analysis
The full sequences of GsERD15B and other ERD15 proteins obtained from Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html), were used for multiple sequence alignments by ClustalW2 (https://www.ebi.ac.uk/Tools/msa/clustalw2/) (Larkin et al., 2007). The unrooted phylogenetic tree was then constructed using MEGA 6.0 (Tamura et al., 2013) based on the Maximum Likelihood (ML) algorithm with 1000 bootstraps.
Y2H assays
The MatchmakerTM Gold Yeast Two‐Hybrid System (Clontech, China) was used for Y2H experiments. The yeasts were cultured on double dropout (DDO/X‐α‐gal) medium (SD/‐Leu/‐Trp/X‐α‐gal, TaKaRa, Japan) or quadruple dropout (QDO/X‐α‐gal) medium (SD/‐Ade/‐His/‐Leu/‐Trp/X‐α‐gal, TaKaRa, Japan), respectively.
Subcellular localization and BiFC assays
The constructs of 35S:GsERD15B‐GFP and 35S: GFP (control) in the backbone vector pBinGFP4 were used for subcellular localization. For BiFC assays, the recombinant plasmids were transferred into A. tumefaciens strain EHA105 and then transformed into the N. benthamiana leaves as described previously (Waadt and Kudla, 2008). The signals of GFP and yellow fluorescence protein (YFP) were observed using a confocal laser‐scanning microscope (Zeiss LSM 780, Germany).
Protein‐protein interaction network analysis
The Search Tool for the Retrieval of Interacting Genes (STRING, https://string‐db.org/) was employed to predicate protein‐protein interaction network, using the combined_score > 0.9. Cytoscape (http://www.cytoscape.org/) software (Shannon et al., 2003) was used for visualization of the network. The annotations of the potential GsERD15B interacting proteins were obtained from gene ontology (GO) database (http://www.geneontology.org/).
Statistical analyses
Analysis of variance (ANOVA) of STR across three environments was performed using PROC GLM by SAS 9.1 (SAS Institute, Cary, NC). The heritability was estimated as , where and are the estimated variances of genotype, genotype‐by‐environment and random error, respectively, where s is the number of environments and r is the number of replications (He et al., 2017). The best linear unbiased predictor (BLUP) of STR from three environments was calculated by R software, using the linear model. Differences between groups or genotypes were analysed using two‐sided Wilcoxon test, Duncan’s multiple range test or Student’s t‐test by R software.
Conflict of interest
The authors have patents pending related to this work.
Author contributions
TJ and YL conceived and designed the experiments. TJ, YYS, ZS and NW performed the experiments. TJ and JH analysed the data. TJ generated the pictures. TJ and YL wrote and revised the manuscript. JG and YL contributed reagents/materials and interpretation of the results. All authors read, revised and approved the final manuscript.
Supporting information
Figure S1 The population structure of 182 Chinese wild soybean accessions.
Figure S2 Distribution of salt tolerance rating (STR) in 182 wild soybean accessions.
Figure S3 Phenotype of two wild soybean accessions with extreme salt tolerance rating (STR).
Figure S4 Manhattan plot (left) and quantile‐quantile (Q‐Q) plot (right) of genome‐wide association study on the salt tolerance rating in 182 Chinese wild soybean accessions using mixed linear model by EMMAX.
Figure S5 Relative expression of nine candidate genes in soybean response to salt stress.
Figure S6 Sequence variation in the promoter region of GsERD15B from 20 different wild soybean accessions and Tianlong1.
Figure S7 The dCAPS polymorphism in soybean accessions with extreme salt tolerance by electrophoresis.
Figure S8 Boxplot of STR (a) and relative expression of GsERD15B (b) for two haplotypes in 20 wild soybean accessions with extreme STR.
Figure S9 Relative expression of GsERD15B in soybean varieties carrying Hap1 or Hap2 type of promoter.
Figure S10 Three biological replications for salt tolerance analysis of transgenic soybean cotyledon hairy roots.
Figure S11 Four biological replications for salt tolerance analysis of regenerated transgenic soybean hairy roots.
Figure S12 Transcriptional activation of PAB proteins in yeast.
Figure S13 Negative controls for the Bimolecular Fluorescence Complementation (BiFC) assays.
Figure S14 Expression of GsERD15B in the root, stem and leaf of salt‐tolerant and salt‐sensitive soybean under control (a) and salt stress (b).
Figure S15 Decay of linkage disequilibrium (LD) in wild soybean (G. soja) genome.
Figure S16 Boxplot of salt tolerance rating (STR) for two haplotypes in cultivated soybean accessions.
Figure S17 The cis‐element analysis of the 2‐kb region upstream of start codon ATG of GsERD15B using the Hap1 type sequence.
Table S1 List of 182 wild soybean accessions from China.
Table S2 Analysis of variance in salt tolerance rating among 182 wild soybean accessions.
Table S3 Correlation coefficients of salt tolerance rating (STR) between environments.
Table S4 Significantly salt tolerance rating (STR)‐associated SNPs and predicted candidate genes for salt tolerance in wild soybean (G. soja).
Table S5 Genes within ±115 kb region of 11 SNPs significantly associated with salt tolerance rating (STR) in multi‐locus model.
Table S6 Correlation coefficients between salt tolerance rating (STR) and relative expression of nine genes under salt stress.
Table S7 Haplotypes of 102 wild soybean accessions with extreme salt tolerance.
Table S8 Primers used for dCAPS marker, qRT‐PCR, and gene cloning.
Table S9 Haplotypes of 20 cultivated soybean varieties with extreme salt tolerance.
Methods S1 Supporting experimental procedures.
Acknowledgments
We would like to thank Professor Daolong Dou, Professor Yongmei Bao and Professor Xinyuan Huang (Nanjing Agricultural University) for kindly providing us the vectors of pBinGFP4, pUC‐SPYNE, pUC‐SPYCE and pGreenII0800‐LUC, and thank Professor Peter Gresshoff’s laboratory (University of Queensland) developed A. rhizogenes strain K599 and shared freely around the world. This work was supported by the National Key R & D Program for Crop Breeding (2016YFD0100304), the Fundamental Research Funds for the Central Universities (KYT201801), the Program for Changjiang Scholars and Innovative Research Team in University (IRT_17R55) and the Natural Science Foundation of Jiangsu Province (BK20170713).
Jin, T. , Sun, Y. , Shan, Z. , He, J. , Wang, N. , Gai, J. and Li, Y. (2021) Natural variation in the promoter of GsERD15B affects salt tolerance in soybean. Plant Biotechnol J, 10.1111/pbi.13536
References
- Aalto, M.K. , Helenius, E. , Kariola, T. , Pennanen, V. , Heino, P. , Horak, H. , Puzorjova, I. et al. (2012) ERD15–an attenuator of plant ABA responses and stomatal aperture. Plant Sci. 182, 19–28. [DOI] [PubMed] [Google Scholar]
- Alexander, D.H. , Novembre, J. and Lange, K. (2009) Fast model‐based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves, M.S. , Reis, P.A. , Dadalto, S.P. , Faria, J.A. , Fontes, E.P. and Fietto, L.G. (2011) A novel transcription factor, ERD15 (Early Responsive to Dehydration 15), connects endoplasmic reticulum stress with an osmotic stress‐induced cell death signal. J. Biol. Chem. 286, 20020–20030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto, P. , Davison, D. , Erezyilmaz, D. , Hu, T.T. , Mast, J. , Sunayama‐Morita, T. and Stern, D.L. (2011) Multiplexed shotgun genotyping for rapid and efficient genetic mapping. Genome Res. 21, 610–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett, J.C. , Fry, B. , Maller, J. and Daly, M.J. (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics, 21, 263–265. [DOI] [PubMed] [Google Scholar]
- Belostotsky, D.A. and Meagher, R.B. (1996) A pollen‐, ovule‐, and early embryo‐specific poly(A) binding protein from Arabidopsis complements essential functions in yeast. Plant Cell, 8, 1261–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. , Cui, S. , Fu, S. , Gai, J. and Yu, D. (2008) Identification of quantitative trait loci associated with salt tolerance during seedling growth in soybean (Glycine max L.). Australian J. Agric. Res. 59, 1086–1091. [Google Scholar]
- Chen, C. , Liu, A. , Ren, H. , Yu, Y. , Duanmu, H. , Duan, X. , Sun, X. et al. (2018) Genome‐wide analysis of Glycine soja response regulator GsRR genes under alkali and salt stresses. Front. Plant Sci. 9, 1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, P. , Yan, K. , Shao, H. and Zhao, S. (2013) Physiological mechanisms for high salt tolerance in wild soybean (Glycine soja) from Yellow River Delta, China: photosynthesis, osmotic regulation, ion flux and antioxidant capacity. PLoS One, 8, e83227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, Y. , Njiti, V.N. , Chen, X. , Triwatayakorn, K. , Kassem, M.A. , Meksem, K. , Lightfoot, D.A. et al. (2002) Quantitative trait loci associated with foliar trigonelline accumulation in Glycine Max L. J. Biomed. Biotechnol. 2, 151–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do, T.D. , Chen, H. , Hien, V.T. , Hamwieh, A. , Yamada, T. , Sato, T. , Yan, Y. et al. (2016) Ncl synchronously regulates Na+, K+, and Cl‐ in soybean and greatly increases the grain yield in saline field conditions. Scientific Rep. 6, 19147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famoso, A.N. , Zhao, K. , Clark, R.T. , Tung, C.W. , Wright, M.H. , Bustamante, C. , Kochian, L.V. et al. (2011) Genetic architecture of aluminum tolerance in rice (Oryza sativa) determined through genome‐wide association analysis and QTL mapping. PLoS Genet. 7, 747–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gai, J.Y. and Wang, Y.S. (2001) A study on the varietal Eco‐regions of soybeans in China. Sci. Agric. Sin. 34, 139–145. [Google Scholar]
- Gao, S.Q. , Chen, M. , Xu, Z.S. , Zhao, C.P. , Li, L. , Xu, H.J. , Tang, Y.M. et al. (2011) The soybean GmbZIP1 transcription factor enhances multiple abiotic stress tolerances in transgenic plants. Plant Mol. Biol. 75, 537–553. [DOI] [PubMed] [Google Scholar]
- Guan, R. , Qu, Y. , Guo, Y. , Yu, L. , Liu, Y. , Jiang, J. , Chen, J. et al. (2014) Salinity tolerance in soybean is modulated by natural variation in GmSALT3 . Plant J. 80, 937–950. [DOI] [PubMed] [Google Scholar]
- Ha, B.K. , Vuong, T.D. , Velusamy, V. , Nguyen, H.T. , Shannon, J.G. and Lee, J.D. (2013) Genetic mapping of quantitative trait loci conditioning salt tolerance in wild soybean (Glycine soja) PI 483463. Euphytica, 193, 79–88. [Google Scholar]
- Hamwieh, A. , Tuyen, D.D. , Cong, H. , Benitez, E.R. , Takahashi, R. and Xu, D.H. (2011) Identification and validation of a major QTL for salt tolerance in soybean. Euphytica, 179, 451–459. [Google Scholar]
- Hamwieh, A. and Xu, D.H. (2008) Conserved salt tolerance quantitative trait loci (QTL) in wild and cultivated soybean. Breed. Sci. 58, 355–359. [Google Scholar]
- He, J. , Meng, S. , Zhao, T. , Xing, G. , Yang, S. , Li, Y. , Guan, R. et al. (2017) An innovative procedure of genome‐wide association analysis fits studies on germplasm population and plant breeding. Theoret. Appl. Genet 130, 2327–2343. [DOI] [PubMed] [Google Scholar]
- Hwang, E.Y. , Song, Q. , Jia, G. , Specht, J.E. , Hyten, D.L. , Costa, J. and Cregan, P.B. (2014) A genome‐wide association study of seed protein and oil content in soybean. BMC Genom. 15, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyten, D.L. , Song, Q. , Zhu, Y. , Choi, I.Y. , Nelson, R.L. , Costa, J.M. , Specht, J.E. et al. (2006) Impacts of genetic bottlenecks on soybean genome diversity. Proc. Natl Acad. Sci. USA, 103, 16666–16671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, B. , Sun, M. , DuanMu, H. , Ding, X. , Liu, B. , Zhu, Y. and Sun, X. (2017) GsCHX19.3, a member of cation/H+ exchanger superfamily from wild soybean contributes to high salinity and carbonate alkaline tolerance. Scientific Rep. 7, 9423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian, H. , Wang, J. , Wang, T. , Wei, L. , Li, J. and Liu, L. (2016) Identification of rapeseed microRNAs involved in early stage seed germination under salt and drought stresses. Front. Plant Sci. 7, 658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan, G. , Ning, L. , Li, Y. , Hu, Z. , Zhang, W. , He, X. and Yu, D. (2016) Identification of novel loci for salt stress at the seed germination stage in soybean. Breed. Sci. 66, 530–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan, G. , Zhang, W. , Yang, W. , Ma, D. , Zhang, D. , Hao, D. , Hu, Z. et al. (2015) Association mapping of soybean seed germination under salt stress. Mol. Genet. Genom. 290, 2147–2162. [DOI] [PubMed] [Google Scholar]
- Kang, H.M. , Sul, J.H. , Service, S.K. , Zaitlen, N.A. , Kong, S.Y. , Freimer, N.B. , Sabatti, C. et al. (2010). Variance component model to account for sample structure in genome‐wide association studies. Nat. Genet. 42, 348–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katerji, N. , Hoorn, J.W.V. , Hamdy, A. and Mastrorilli, M. (2003) Salinity effect on crop development and yield, analysis of salt tolerance according to several classification methods. Agric. Water Manage. 62, 37–66. [Google Scholar]
- Kereszt, A. , Li, D. , Indrasumunar, A. , Nguyen, C.D. , Nontachaiyapoom, S. , Kinkema, M. and Gresshoff, P.M. (2007) Agrobacterium rhizogenes‐mediated transformation of soybean to study root biology. Nat. Protocols, 2, 948–952. [DOI] [PubMed] [Google Scholar]
- King, K.E. , Peiffer, G.A. , Reddy, M. , Lauter, N. , Lin, S.F. , Cianzio, S. and Shoemaker, R.C. (2013) Mapping of iron and zinc quantitative trait loci in soybean for association to iron deficiency chlorosis resistance. J. Plant Nutrit. 36, 2132–2153. [Google Scholar]
- Kiyosue, T. , Abe, H. , Yamaguchi‐Shinozaki, K. and Shinozaki, K. (1998) ERD6, a cDNA clone for an early dehydration‐induced gene of Arabidopsis, encodes a putative sugar transporter. Biochem. Biophys. Acta, 1370, 187–191. [DOI] [PubMed] [Google Scholar]
- Kiyosue, T. , Yamaguchi‐Shinozaki, K. and Shinozaki, K. (1994) Cloning of cDNAs for genes that are early‐responsive to dehydration stress (ERDs) in Arabidopsis thaliana L.: identification of three ERDs as HSP cognate genes. Plant Mol. Biol. 25, 791–798. [DOI] [PubMed] [Google Scholar]
- Kozlov, G. , De Crescenzo, G. , Lim, N.S. , Siddiqui, N. , Fantus, D. , Kahvejian, A. , Trempe, J. et al. (2004) Structural basis of ligand recognition by PABC, a highly specific peptide‐binding domain found in poly(A)‐binding protein and a HECT ubiquitin ligase. EMBO J. 23, 272–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushanov, F.N. , Pepper, A.E. , Yu, J.Z. , Buriev, Z.T. , Shermatov, S.E. , Saha, S. , Ulloa, M. et al. (2016) Development, genetic mapping and QTL association of cotton PHYA, PHYB, and HY5‐specific CAPS and dCAPS markers. BMC Genom. 17, 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, H.M. , Xu, X. , Liu, X. , Chen, W. , Yang, G. , Wong, F.L. , Li, M.W. et al. (2010) Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat. Genet. 42, 1053–1059. [DOI] [PubMed] [Google Scholar]
- Larkin, M.A. , Blackshields, G. , Brown, N.P. , Chenne, R. , McGettigan, P.A. , McWilliam, H. , Valentin, F. et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics, 23, 2947–2948. [DOI] [PubMed] [Google Scholar]
- Lee, G.J. , Carter, T.E. Jr , Villagarcia, M.R. , Li, Z. , Zhou, X. , Gibbs, M.O. and Boerma, H.R. (2004) A major QTL conditioning salt tolerance in S‐100 soybean and descendent cultivars. Theoret. Appl. Genet. 109, 1610–1619. [DOI] [PubMed] [Google Scholar]
- Leung, J. , Bouvier‐Durand, M. , Morris, P.C. , Guerrier, D. , Chefdor, F. and Giraudat, J. (1994) Arabidopsis ABA response gene ABI1: features of a calcium‐modulated protein phosphatase. Science, 264, 1448–1452. [DOI] [PubMed] [Google Scholar]
- Li, S. , Cong, Y. , Liu, Y. , Wang, T. , Shuai, Q. , Chen, N. , Gai, J. et al.(2017) Optimization of Agrobacterium‐mediated transformation in soybean. Front. Plant Sci. 8, 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L. , Guo, N. , Niu, J. , Wang, Z. , Cui, X. , Sun, J. , Zhao, T. et al. (2016) Loci and candidate gene identification for resistance to Phytophthora sojae via association analysis in soybean [Glycine max (L.) Merr]. Mol. Genet. Genom. 291, 1095–1103. [DOI] [PubMed] [Google Scholar]
- Li, R. , Yu, C. , Li, Y. , Lam, T.W. , Yiu, S.M. , Kristiansen, K. and Wang, J. (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics, 25, 1966–1967. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2‐△△CT Method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Meyer, K. , Leube, M.P. and Grill, E. (1994) A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana . Science, 264, 1452–1455. [DOI] [PubMed] [Google Scholar]
- Munns, R. and Tester, M. (2008) Mechanisms of salinity tolerance. Annual Rev. Plant Biol. 59, 651–681. [DOI] [PubMed] [Google Scholar]
- Munoz, N. , Liu, A. , Kan, L. , Li, M.W. and Lam, H.M. (2017) Potential uses of wild germplasms of grain legumes for crop improvement. Int. J. Mol. Sci. 18, 382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, M.G. and Thompson, W.F. (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff, M.M. , Turk, E. and Kalishman, M. (2002) Web‐based primer design for single nucleotide polymorphism analysis. Trends Genet. 18, 613–615. [DOI] [PubMed] [Google Scholar]
- Parker, M.B. , Gascho, G.J. and Gaines, T.P. (1983) Chloride toxicity of soybeans grown on Atlantic Coast flatwoods soils1 . Agronomy J. 75, 439–443. [Google Scholar]
- Pfaffl, M.W. (2001) A new mathematical model for relative quantification in real‐time RT‐PCR. Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, A.L. , Patterson, N.J. , Plenge, R.M. , Weinblatt, M.E. , Shadick, N.A. and Reich, D. (2006) Principal components analysis corrects for stratification in genome‐wide association studies. Nat. Genet. 38, 904–909. [DOI] [PubMed] [Google Scholar]
- Qi, X. , Li, M.W. , Xie, M. , Liu, X. , Ni, M. , Shao, G. , Song, C. et al. (2014) Identification of a novel salt tolerance gene in wild soybean by whole‐genome sequencing. Nat. Commun. 5, 4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengasamy, P. (2006) World salinization with emphasis on Australia. J. Exp. Bot. 57, 1017–1023. [DOI] [PubMed] [Google Scholar]
- Scheet, P. and Stephens, M. (2006) A fast and flexible statistical model for large‐scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am. J. Human Genet. 78, 629–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin, J. , Rueden, C.T. , Hiner, M.C. and Eliceiri, K.W. (2015) The ImageJ ecosystem: an open platform for biomedical image analysis. Mol. Reprod. Develop. 82, 518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz, J. , Cannon, S.B. , Schlueter, J. , Ma, J. , Mitros, T. , Nelson, W. , Hyten, D.L. et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature, 463, 178–183. [DOI] [PubMed] [Google Scholar]
- Shannon, P. , Markiel, A. , Ozier, O. , Baliga, N.S. , Wang, J.T. , Ramage, D. , Amin, N. et al. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, G.H. , Song, J.Z. and Liu, H.L. (1986) Preliminary studies on the evaluation of salt tolerance in soybean varieties. Sci. Agric. Sin. 6, 30–35. [Google Scholar]
- Shen, X.J. , Wang, Y.Y. , Zhang, Y.X. , Guo, W. , Jiao, Y.Q. and Zhou, X.A. (2018) Overexpression of the wild soybean R2R3‐MYB transcription factor GsMYB15 enhances resistance to salt stress and Helicoverpa Armigera in transgenic Arabidopsis . Int. J. Mol. Sci. 19, 3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, M. , Qian, X. , Chen, C. , Cheng, S. , Jia, B. , Zhu, Y. and Sun, X. (2018) Ectopic expression of GsSRK in Medicago sativa reveals its involvement in plant architecture and salt stress responses. Front. Plant Sci. 9, 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X. , Sun, M. , Jia, B. , Qin, Z. , Yang, K. , Chen, C. , Yu, Q. et al. (2016) A Glycine soja methionine sulfoxide reductase B5a interacts with the Ca2+/CAM‐binding kinase GsCBRLK and activates ROS signaling under carbonate alkaline stress. Plant J. 86, 514–529. [DOI] [PubMed] [Google Scholar]
- Tamura, K. , Stecher, G. , Peterson, D. , Filipski, A. and Kumar, S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuyen, D.D. , Lal, S.K. and Xu, D.H. (2010) Identification of a major QTL allele from wild soybean (Glycine soja Sieb. & Zucc.) for increasing alkaline salt tolerance in soybean. Theoret. Appl. Genet. 121, 229–236. [DOI] [PubMed] [Google Scholar]
- Vigueira, C.C. , Small, L.L. and Olsen, K.M. (2016) Long‐term balancing selection at the Phosphorus Starvation Tolerance 1 (PSTOL1) locus in wild, domesticated and weedy rice (Oryza). BMC Plant Biol. 16, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt, R. and Kudla, J. (2008) In planta visualization of protein interactions using Bimolecular Fluorescence Complementation (BiFC). Cold Spring Harbor Protocols, 2008, 4995. [DOI] [PubMed] [Google Scholar]
- Wang, S.B. , Feng, J.Y. , Ren, W.L. , Huang, B. , Zhou, L. , Wen, Y.J. , Zhang, J. et al. (2016) Improving power and accuracy of genome‐wide association studies via a multi‐locus mixed linear model methodology. Sci. Rep. 6, 19444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W. , Jiang, W. , Liu, J. , Li, Y. , Gai, J. and Li, Y. (2017) Genome‐wide characterization of the aldehyde dehydrogenase gene superfamily in soybean and its potential role in drought stress response. BMC Genom. 18, 518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y.H. , Zhang, X.J. and Fan, S.J. (2015) Genetic diversity of wild soybean populations in Dongying, China, by simple sequence repeat analysis. Genet. Mol. Res. 14, 11613–11623. [DOI] [PubMed] [Google Scholar]
- Wang, N. , Zhang, W. , Qin, M. , Li, S. , Qiao, M. , Liu, Z. and Xiang, F. (2017) Drought tolerance conferred in soybean (Glycine max. L) by GmMYB84, a novel R2R3‐MYB transcription factor. Plant Cell Physiol. 58, 1764–1776. [DOI] [PubMed] [Google Scholar]
- Wei, P. , Che, B. , Shen, L. , Cui, Y. , Wu, S. , Cheng, C. , Liu, F. et al. (2019) Identification and functional characterization of the chloride channel gene, GsCLC‐c2 from wild soybean. BMC Plant Biol. 19, 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, P. , Wang, L. , Liu, A. , Yu, B. and Lam, H.M. (2016) GmCLC1 confers enhanced salt tolerance through regulating chloride accumulation in soybean. Front. Plant Sci. 7, 1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, L. , Hu, L. , Xu, W. , Zhen, A. , Zhang, L. and Hu, X. (2016) Exogenous γ‐aminobutyric acid improves the structure and function of photosystem II in muskmelon seedlings exposed to salinity‐alkalinity stress. PLoS One, 11, e0164847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z. , Duan, L. , Tian, X. , Wang, B. , Eneji, A.E. and Li, Z. (2008) Coronatine alleviates salinity stress in cotton by improving the antioxidative defense system and radical‐scavenging activity. J. Plant Physiol. 165, 375–384. [DOI] [PubMed] [Google Scholar]
- Xu, X.Y. , Fan, R. , Zheng, R. , Li, C.M. and Yu, D.Y. (2011) Proteomic analysis of seed germination under salt stress in soybeans. J. Zhejiang University Sci. B. 12, 507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, Y.B. , Xiao, B.X. , Zhu, S.N. , Mo, X.H. , Liang, C.Y. , Tian, J. , Liao, H. et al. (2017) GmPHR25, a GmPHR member up‐regulated by phosphate starvation, controls phosphate homeostasis in soybean. J. Exp. Bot. 68, 4951–4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, X. , Liang, Y. , Huerta‐Sanchez, E. , Jin, X. , Cuo, Z.X.P. , Pool, J.E. , Xu, X. et al. (2010) Sequencing of 50 human exomes reveals adaptation to high altitude. Science, 329, 75–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegaoui, Z. , Planchais, S. , Cabassa, C. , Djebbar, R. , Belbachir, O.A. and Carol, P. (2017) Variation in relative water content, proline accumulation and stress gene expression in two cowpea landraces under drought. J. Plant Physiol. 218, 26–34. [DOI] [PubMed] [Google Scholar]
- Zhang, Z. , Ersoz, E. , Lai, C.Q. , Todhunter, R.J. , Tiwari, H.K. , Gore, M.A. , Bradbury, P.J. et al. (2010) Mixed linear model approach adapted for genome‐wide association studies. Nat. Genet. 42, 355–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W. , Liao, X. , Cui, Y. , Ma, W. , Zhang, X. , Du, H. , Ma, Y. et al. (2019) A cation diffusion facilitator, GmCDF1, negatively regulates salt tolerance in soybean. PLoS Genet. 15, e1007798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Song, Q. , Griffin, J.D. and Song, B.H. (2017) Genetic architecture of wild soybean (Glycine soja) response to soybean cyst nematode (Heterodera glycines). Mol. Genet. Genom. 292, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Z. , Jiang, Y. , Wang, Z. , Gou, Z. , Lyu, J. , Li, W. , Yu, Y. et al. (2015) Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat. Biotechnol. 33, 408–414. [DOI] [PubMed] [Google Scholar]
- Zhou, L. , Wang, C. , Liu, R. , Han, Q. , Vandeleur, R.K. , Du, J. , Tyerman, S. et al. (2014) Constitutive overexpression of soybean plasma membrane intrinsic protein GmPIP1;6 confers salt tolerance. BMC Plant Biol. 14, 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J.K. (2001) Plant salt tolerance. Trends Plant Sci. 6, 66–71. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 The population structure of 182 Chinese wild soybean accessions.
Figure S2 Distribution of salt tolerance rating (STR) in 182 wild soybean accessions.
Figure S3 Phenotype of two wild soybean accessions with extreme salt tolerance rating (STR).
Figure S4 Manhattan plot (left) and quantile‐quantile (Q‐Q) plot (right) of genome‐wide association study on the salt tolerance rating in 182 Chinese wild soybean accessions using mixed linear model by EMMAX.
Figure S5 Relative expression of nine candidate genes in soybean response to salt stress.
Figure S6 Sequence variation in the promoter region of GsERD15B from 20 different wild soybean accessions and Tianlong1.
Figure S7 The dCAPS polymorphism in soybean accessions with extreme salt tolerance by electrophoresis.
Figure S8 Boxplot of STR (a) and relative expression of GsERD15B (b) for two haplotypes in 20 wild soybean accessions with extreme STR.
Figure S9 Relative expression of GsERD15B in soybean varieties carrying Hap1 or Hap2 type of promoter.
Figure S10 Three biological replications for salt tolerance analysis of transgenic soybean cotyledon hairy roots.
Figure S11 Four biological replications for salt tolerance analysis of regenerated transgenic soybean hairy roots.
Figure S12 Transcriptional activation of PAB proteins in yeast.
Figure S13 Negative controls for the Bimolecular Fluorescence Complementation (BiFC) assays.
Figure S14 Expression of GsERD15B in the root, stem and leaf of salt‐tolerant and salt‐sensitive soybean under control (a) and salt stress (b).
Figure S15 Decay of linkage disequilibrium (LD) in wild soybean (G. soja) genome.
Figure S16 Boxplot of salt tolerance rating (STR) for two haplotypes in cultivated soybean accessions.
Figure S17 The cis‐element analysis of the 2‐kb region upstream of start codon ATG of GsERD15B using the Hap1 type sequence.
Table S1 List of 182 wild soybean accessions from China.
Table S2 Analysis of variance in salt tolerance rating among 182 wild soybean accessions.
Table S3 Correlation coefficients of salt tolerance rating (STR) between environments.
Table S4 Significantly salt tolerance rating (STR)‐associated SNPs and predicted candidate genes for salt tolerance in wild soybean (G. soja).
Table S5 Genes within ±115 kb region of 11 SNPs significantly associated with salt tolerance rating (STR) in multi‐locus model.
Table S6 Correlation coefficients between salt tolerance rating (STR) and relative expression of nine genes under salt stress.
Table S7 Haplotypes of 102 wild soybean accessions with extreme salt tolerance.
Table S8 Primers used for dCAPS marker, qRT‐PCR, and gene cloning.
Table S9 Haplotypes of 20 cultivated soybean varieties with extreme salt tolerance.
Methods S1 Supporting experimental procedures.


