Abstract
The SARS-CoV-2 helicase Nsp13 is a promising target for developing anti-COVID drugs. In the present study, we have identified potential natural product inhibitors of SARS-CoV-2 Nsp13 targeting the ATP-binding site using molecular docking and molecular dynamics (MD) simulations. MD simulation of the prepared crystal structure of SARS-CoV-2 Nsp13 was performed to generate an ensemble of structures of helicase Nsp13 capturing the conformational diversity of the ATP-binding site. A natural product library of more than 14,000 phytochemicals from Indian medicinal plants was used to perform virtual screening against the ensemble of Nsp13 structures. Subsequently, a two-stage filter, first based on protein–ligand docking binding energy value and second based on protein residues in the ligand-binding site and non-covalent interactions between the protein residues and the ligand in the best-docked pose, was used to identify 368 phytochemicals as potential inhibitors of SARS-CoV-2 helicase Nsp13. MD simulations of the top inhibitors complexed with protein were performed to confirm stable binding, and to compute MM-PBSA based binding energy. From among the 368 potential phytochemical inhibitors, the top identified potential inhibitors of SARS-CoV-2 helicase Nsp13 namely, Picrasidine M, (+)-Epiexcelsin, Isorhoeadine, Euphorbetin and Picrasidine N, can be taken up initially for experimental studies.
Graphic abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s11030-021-10251-1.
Keywords: COVID-19, SARS-CoV-2 helicase Nsp13, Phytochemical inhibitors, Molecular docking, Molecular dynamics simulation, Lead compounds
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is an enveloped positive-sense RNA virus belonging to the family Coronaviridae that is the causative pathogen of the ongoing coronavirus disease 2019 (COVID-19) pandemic [1]. In humans, the clinical features of this new disease ranges from asymptomatic state to acute respiratory distress syndrome [2]. Several small molecules with strong in vitro antiviral activity against SARS-CoV-2 have been identified [3, 4] and are under different stage of clinical trials. Remdesivir, one such small molecule, was granted emergency use authorization by US Food and Drug Administration (FDA) for the treatment of COVID-19 [5]. There is an increasing need to develop anti-COVID drugs, especially, those targeting the key proteins of SARS-CoV-2.
The SARS-CoV-2 genome has been reported to have 14 open reading frames which encode for the 27 viral proteins [6]. The 5’-terminus of the genome has two genes orf1ab and orf1a which code for two polyproteins pp1ab and pp1a, respectively. pp1ab and pp1a are cleaved by two proteases namely, papain‐like protease (PLpro or Nsp3) and 3C‐like cysteine protease (3CLpro or Nsp5), into 15 non-structural proteins, Nsp1 to Nsp10 and Nsp12 to Nsp16 [6]. In contrast, the 3’-terminus of the genome codes for the 4 structural proteins, namely, spike (S), envelope (E), matrix (M) and nucleocapsid (N), and 8 accessory proteins (3a, 3b, p6, 7a, 7b, 8b, 9b and orf14) [6]. The 15 non-structural proteins in SARS-CoV-2 comprise the viral replication and transcription complex which is essential for the coronavirus life cycle [7]. Of the 15 non-structural proteins, papain‐like protease (PLpro or Nsp3), 3C‐like cysteine protease (3CLpro or Nsp5), RNA-dependent RNA polymerase (RdRp or Nsp12) and helicase (Nsp13) are among the highly studied targets for identifying anti-COVID drugs [8].
SARS-CoV-2 helicase Nsp13 has both ATPase and helicase activity, as it unwinds the RNA helices in an ATP-dependent manner [9]. Notably, due to its high sequence conservation across the coronavirus family, Nsp13 is considered an attractive target for the development of antiviral drugs [10, 11]. Also, it was shown that SARS-CoV-2 helicase Nsp13 can hydrolyze all types of NTPs including ATP to unwind the RNA helices [9]. Therefore, the known ATP-binding site of the helicase Nsp13 is a promising target for effective inhibition. In this direction, the recently deposited crystal structure of SARS-CoV-2 helicase Nsp13 (PDB 6ZSL) has made development of anti-COVID drugs via targeting of Nsp13 more viable. Notably, the Nsp13 of SARS-CoV and SARS-CoV-2 share 99.8% sequence identity [11, 12]. Moreover, similar to the SARS-CoV Nsp13 structure [10], the SARS-CoV-2 Nsp13 adopts a triangular pyramid shape with five domains namely, the RecA-like domains 1A and 2A, the 2B domain, the zinc-binding domain (ZBD) and the stalk domain (Fig. 1). Thus, potential drugs against SARS-CoV-2 Nsp13 may have broad activity against β–coronaviruses.
Fig. 1.
Cartoon representation of the prepared crystal structure of SARS-CoV-2 helicase Nsp13 (PDB 6ZSL). The figure shows the five domains in the Nsp13 structure, namely, zinc binding domain (ZBD) colored in red, the stalk domain colored in yellow, the 1B domain colored in green, the RecA-like domains 1A and 2A colored in blue and orange, respectively. The ATP-binding site of the Nsp13 with six key residues involved in ATP hydrolysis is shown in expanded view
Given the urgent need to identify anti-COVID drugs, several computational studies in the past year have undertaken virtual screening of small molecule libraries against key SARS-CoV-2 non-structural proteins especially 3CLpro, RdRp and PLpro [13–15]. Some screening studies [12, 16–19] have also shown that SARS-CoV-2 helicase Nsp13 is a good target for potential inhibitors. In this work, we have identified through computational approaches potential natural product inhibitors of SARS-CoV-2 helicase Nsp13 which can target the ATP-binding site.
Plant-based natural products are a major contributor to the existing approved drug space [20]. Further, medicinal plants are integral to the traditional Indian systems of medicine including Ayurveda, Siddha and Unani, and Indian herbs have been used to treat several human diseases for centuries. Recently, several herbal medicines have been proposed for treating COVID-19 [21, 22]. Previously, we have created IMPPAT [23], the largest database on phytochemicals of the Indian medicinal plants. In the present work, we have used a small molecule library of 14,011 phytochemicals from Indian medicinal plants, mainly compiled from IMPPAT database [23], to perform virtual screening with the goal of identification of potential phytochemical inhibitors of SARS-CoV-2 helicase Nsp13 targeting the ATP-binding site. Picrasidine M, (+)-Epiexcelsin, Isorhoeadine, Euphorbetin and Picrasidine N identified here as the top potential phytochemical inhibitors of SARS-CoV-2 helicase Nsp13, to start with, can be taken up for experimental studies.
Methods
Virtual screening library of phytochemicals
We have earlier built the IMPPAT database [23] (https://cb.imsc.res.in/imppat) which is the largest resource on phytochemicals of Indian medicinal plants or herbs to date. In this work, we have used a ligand library of 14,011 phytochemicals, and this screening library was obtained by combining phytochemicals in IMPPAT [23, 24] with additional data from other literature sources [25–40]. Subsequently, an evaluation of these 14,011 phytochemicals based on the widely-used drug-likeness filter, Lipinski’s rule of five (RO5) [41], led to a subset of 10,510 phytochemicals that passed the R05 filter. Thereafter, we retrieved the three-dimensional (3D) structures of these 10,510 drug-like phytochemicals from PubChem [42] for virtual screening. Lastly, the 3D structures of the 10,510 phytochemicals were energy-minimized using OpenBabel [43] and converted to .pdb format from .sdf format.
Protein structure of SARS-CoV-2 Nsp13
In this investigation, we have used the crystal structure with 1.94 Å resolution (PDB 6ZSL) of the SARS-CoV-2 helicase Nsp13 which was downloaded from Protein Data Bank (RCSB PDB; https://www.rcsb.org/). As of 19 February 2021, there are more than 50 crystal structures of SARS-CoV-2 Nsp13 in PDB. However, the crystal structure 6ZSL selected for this investigation has been validated by Wlodawer et al. [44], and furthermore, is the only SARS-CoV-2 Nsp13 structure not from Pan‐Dataset Density Analysis (PanDDA). In the crystal structure 6ZSL for the SARS-CoV-2 Nsp13, we have gap-filled the structural coordinates for three missing amino acid residues (339–341) by aligning to the crystal structure for the SARS-CoV Nsp13 (PDB 6JYT). Subsequently, the gap-filled structure of SARS-CoV-2 Nsp13 was energy-minimized using the minimize structure utility of UCSF Chimera [45].
Figure 1 shows the cartoon representation of this prepared crystal structure of SARS-CoV-2 Nsp13 wherein six important residues in the ATP-binding site have been highlighted [10, 12]. The SARS-CoV-2 Nsp13 adopts a triangular pyramid shape with five domains similar to the SARS-CoV Nsp13 (Fig. 1). The triangular base of the pyramid is formed by three domains in SARS-CoV-2 Nsp13, the 2B domain and the two RecA-like domains 1A and 2A. The remaining two domains in SARS-CoV-2 Nsp13, the N-terminal zinc binding domain (ZBD) and the stalk domain, are directed toward the apex of the pyramid.
Molecular dynamics simulation
In this work, we have performed molecular dynamics (MD) simulations of SARS-CoV-2 Nsp13 protein structures using GROMACS 5.1.5 [46] with GROMOS96 54a7 force field [47]. Specifically, MD simulations were performed for: (a) the prepared crystal structure of SARS-CoV-2 Nsp13 (uncomplexed protein), and (b) the protein–ligand complexes of SARS-CoV-2 Nsp13 with predicted top five phytochemical inhibitors. The topology of the top five phytochemical inhibitors of SARS-CoV-2 Nsp13 was generated using the automated topology builder (ATB) version 3.0 (https://atb.uq.edu.au/) [48].
To prepare the system for MD simulation, the uncomplexed protein or the protein–ligand complex was placed at the center of a cubic box with periodic boundary conditions, and thereafter, the system was solvated by adding simple point-charge (SPC) water and neutralized by adding thirteen chloride (Cl−) ions. Next, the system was energy-minimized using the steepest descent algorithm. Then, the solvated and neutralized system was subjected to a NVT simulation of 1 ns with 2 fs time step, wherein the number of particles, volume and temperature of the system is kept constant. Next, the pressure of the system was equilibrated to 1 bar during a NPT simulation of 1 ns with 2 fs time step, wherein the number of particles, pressure, and temperature of the system is kept constant. The position of both the protein and ligand were restrained during the above-mentioned NVT and NPT simulations.
Thereafter, a production MD run was performed, after removing the position restraint on protein and ligand, for a period of: (a) 100 ns with 2 fs time step for the SARS-CoV-2 Nsp13 uncomplexed protein, and (b) 50 ns with 2 fs time step for the SARS-CoV-2 Nsp13 protein in complex with each of the top five phytochemical inhibitors. During the production MD run, the structural coordinates of the system were written to the trajectory file after every 2 ps. Further, the system temperature and pressure were maintained at 310 K and 1 bar using the v-rescale temperature [49] and Parrinello-Rahman pressure coupling method [50], respectively. Other parameters for the production MD simulation were fixed as per our recently published study [24].
Finally, using GROMACS scripts and the trajectories obtained from the MD simulations, we have computed the following quantities: (a) radius of gyration (Rg) of the protein, (b) root mean square deviation (RMSD) of the Cα atoms of the protein backbone, (c) root mean square fluctuation (RMSF) of the Cα atoms of the protein backbone, (d) root mean square deviation (RMSD) of the heavy atoms of the ligand, and (e) distance between the center of mass of the ligand and center of mass of the key binding site residues of the protein.
Generation of an ensemble of Nsp13 protein structure conformations
The trajectory from the MD simulation of the prepared crystal structure of SARS-CoV-2 Nsp13 protein without any bound ligands was used to generate an ensemble of protein structures that capture the conformational diversity of the ATP-binding site residues of the helicase. The MD simulation trajectory from 60 to 100 ns was subjected to geometric clustering using the Daura algorithm [51] based on the conformations of the ATP-binding site residues within 8 Å from the two phosphate ions in the crystal structure of SARS-CoV-2 Nsp13. Using a clustering cutoff of 1.2 Å, we were able to identify 23 clusters from the SARS-CoV-2 Nsp13 protein simulation, from which we selected ten representative mid-point structures corresponding to the top ten populated clusters accounting for 97.6% of the sampled conformations of the ATP-binding site residues. Thus, along with the prepared crystal structure of SARS-CoV-2 Nsp13, ten structures based on clustering from the MD simulation trajectory of the protein were selected for virtual screening.
Molecular docking
Molecular docking of the energy-minimized 3D structures of phytochemicals in the ligand library against the structures of SARS-CoV-2 Nsp13 was performed using AutoDock Vina [52]. The structures of SARS-CoV-2 Nsp13 considered for molecular docking were: (a) the prepared crystal structure of Nsp13, and (b) ten selected structures from geometric clustering of the MD simulation trajectory between 60 and 100 ns for the Nsp13 protein.
To perform docking, the structures of small molecule ligands and protein in.pdb file format were converted to .pdbqt format using prepare_ligand4.py and prepare_receptor4.py scripts, respectively, from AutoDockTools [53]. Thereafter, the search space center and dimension for the protein–ligand docking in AutoDock Vina for the above-mentioned eleven structures of SARS-CoV-2 Nsp13 were manually determined by considering the residues in the ATP-binding site. Subsequently, protein–ligand docking was performed using AutoDock Vina by setting the exhaustiveness parameter to 8.
Determination of protein–ligand interactions
Using pdb-tools [54], we have prepared a combined structure of the best-docked conformation of a phytochemical obtained from AutoDock Vina (with the lowest binding energy) and the prepared crystal structure of SARS-CoV-2 Nsp13. Thereafter, the combined structure file of a protein–ligand complex was analyzed for non-covalent interactions between ligand and protein residues using a custom script as described in our earlier work [24]. The non-covalent interactions identified were hydrogen bonds and hydrophobic interactions. The atoms of protein and ligand are reported to have hydrophobic interactions if the distance between a carbon atom of protein (or ligand) and a carbon, halogen or sulfur atom of ligand (or protein) is ≤ 4 Å. Note that the hydrogen, chalcogen or halogen bonds identified between the atoms of protein and ligand are not reported again as hydrophobic interactions [24].
ADMET prediction for phytochemical inhibitors
Based on the computed Absorption, Distribution, Metabolism, Excretion and Toxicity (ADMET) properties using SwissADME [55] and vNN-ADMET [56], we have evaluated the pharmacokinetic properties and potential toxicity of the predicted phytochemical inhibitors for SARS-CoV-2 Nsp13 from this study.
MM-PBSA calculation
In this study, the Molecular Mechanics Poisson-Boltzmann Surface Area (MM-PBSA) method was used to compute the binding energy of only the top five phytochemical inhibitors of SARS-CoV-2 Nsp13. From the 50 ns MD simulation trajectories for each of the protein–ligand complex, trajectories were extracted at an interval of 1 ns between 20 and 50 ns after equilibration. These were used to calculate the binding energy using g_mmpbsa [57, 58] of the phytochemical to SARS-CoV-2 Nsp13.
Results and discussion
The helicase Nsp13 is one of the 15 non-structural proteins coded by the SARS-CoV-2 genome and shares a high sequence conservation across the family Coronaviridae [9, 12]. This protein is a critical component of the replication and transcription complex and it unwinds the RNA helices in a ATP-dependent manner similar to the SARS-CoV helicase Nsp13 [9–11].
Virtual screening
In this work, we have identified, using the largest small molecule library of phytochemicals from Indian herbs, potential phytochemical inhibitors of SARS-CoV-2 helicase Nsp13 that can target the ATP-binding site (Fig. 2). From the small molecule library of 14,011 phytochemicals which was compiled based on information in our IMPPAT database [23, 24], we identified 10,510 phytochemicals as drug-like (Methods).
Fig. 2.
Workflow for the identification of potential phytochemical inhibitors of SARS-CoV-2 helicase Nsp13
Figure 1 shows the prepared crystal structure of SARS-CoV-2 Nsp13 with emphasis on the six key residues in the ATP-binding site. Previously, Jia et al. [10] had identified six residues, namely, K288, S289, D374, E375, Q404 and R567, in SARS-CoV Nsp13 to be crucial for ATP hydrolysis. These six key ATP-binding site residues are also conserved in SARS-CoV-2 Nsp13. SARS-CoV-2 Nsp13 and SARS-CoV Nsp13 share 99.8% sequence identity with only one varying residue which is away from the ATP-binding site and RNA-binding site [11, 12].
In the first stage of filtering of ligands for potential inhibitors, we have considered the binding energy of the phytochemicals with the ensemble structures, as described in the Methods section, of SARS-CoV-2 Nsp13 which represented the conformational landscape of the binding site. To decide on the binding energy cutoff for this filtering, we used the known experimental information on binding from the twelve PanDDA co-crystallized structures of SARS-CoV-2 Nsp13 with ligands bound to the ATP-binding site, namely, PDB 5RM2, 5RM7, 5RLW, 5RL9, 5RLO, 5RLY, 5RLJ, 5RLI, 5RLV, 5RLR, 5RLN and 5RLS. The binding energy of the co-crystallized ligand in the experimentally observed binding pose in the respective co-crystallized structure of SARS-CoV-2 Nsp13 was estimated using score_only option of AutoDock Vina (Supplementary Table S1). From the analysis of the twelve PanDDA co-crystallized structures of SARS-CoV-2 Nsp13, the best binding energy value of − 5.14 kcal/mol obtained for ligand UXG in the structure PDB 5RM2 was used as the cutoff to filter phytochemicals in the protein–ligand docking. Overall, this first stage of filtering of ligands based on known experimental evidence led to a filtered set of 5260 phytochemicals.
In the second stage of filtering of ligands for potential inhibitors, we used the interactions with the residues in the ATP-binding site based on molecular docking performed with the crystal structure conformation. Specifically, we checked that the phytochemical either binds to or forms non-covalent interactions with all six key ATP-binding site residues. At the end of the second stage of filtering, we identified 368 phytochemicals (H1–H368) as potential inhibitors of SARS-CoV-2 Nsp13 targeting the crucial ATP-binding site (Supplementary Table S2).
Interestingly, several of the Indian medicinal plants or herbs that produce the 368 phytochemicals H1 to H368 identified here as potential inhibitors of SARS-CoV-2 Nsp13 are mentioned for potential antiviral use in traditional medicine (Supplementary Table S3). Furthermore, an analysis of the best-docked pose with the prepared crystal structure of SARS-CoV-2 Nsp13 shows the extent of non-covalent interactions between the binding site residues of the protein and the complexed inhibitor (Supplementary Table S4). These are supplemented by the predicted physicochemical and ADMET properties of the 368 potential inhibitors of SARS-CoV-2 Nsp13 identified here (Supplementary Table S5).
Description of the top ten potential phytochemical inhibitor complexes of SARS-CoV-2 Nsp13
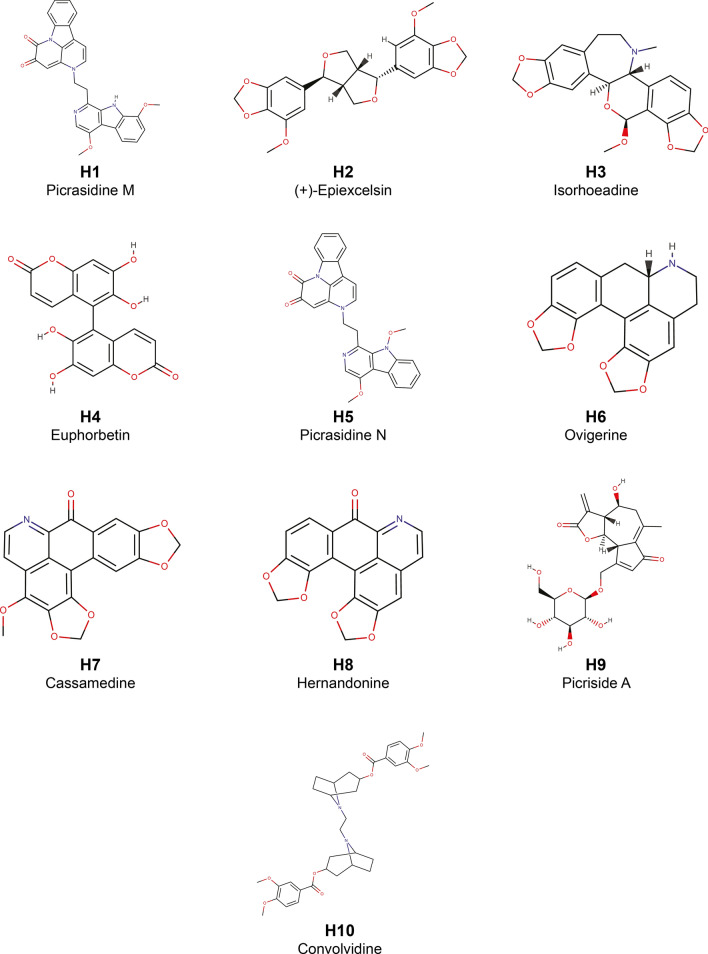
The PubChem chemical identifier, chemical name, IUPAC name and chemical structure in SMILES format have been provided for the 368 potential phytochemical inhibitors (H1–H368) of SARS-CoV-2 Nsp13 identified here (Supplementary Table S2). In this section, a detailed description of the top ten potential phytochemical inhibitors (H1–H10) that have protein–ligand docking binding energies < − 8.5 kcal/mol with the prepared crystal structure of SARS-CoV-2 Nsp13 (Tables 1 and 2) are discussed. Table 1 also provides the list of Indian herbs which can produce these top ten potential phytochemical inhibitors of SARS-CoV-2 Nsp13, whereas Table 2 provides the list of protein residues which form hydrogen bond or hydrophobic interaction with the top potential phytochemical inhibitors H1 to H10. The two-dimensional (2D) chemical structures and the hydrogen bond interactions in the protein–ligand complex of these top ten inhibitors are shown in Figs. 3 and 4, respectively. Of these only the top five, namely, H1 (Picrasidine M), H2 ((+)-Epiexcelsin), H3 (Isorhoeadine), H4 (Euphorbetin) and H5 (Picrasidine N), were examined in more detail through further MD simulations to estimate their binding energies using MM-PBSA (Table 3).
Table 1.
Plant sources of the top ten potential phytochemical inhibitors of SARS-CoV-2 Nsp13
| Phytochemical symbol | Docking binding energy (kcal/mol) | Chemical name | Plant source |
|---|---|---|---|
| H1 | − 10.2 | Picrasidine M | Picrasma quassioides [*] |
| H2 | − 9 | (+)-Epiexcelsin | Litsea verticillata |
| H3 | − 8.9 | Isorhoeadine | Papaver rhoeas [*] |
| H4 | − 8.9 | Euphorbetin | Euphorbia lathyris |
| H5 | − 8.9 | Picrasidine N | Picrasma quassioides [*] |
| H6 | − 8.8 | Ovigerine | Hernandia guianensis,Hernandia nymphaeifolia |
| H7 | − 8.8 | Cassamedine | Cassytha filiformis |
| H8 | − 8.6 | Hernandonine | Hernandia guianensis,Hernandia nymphaeifolia |
| H9 | − 8.6 | Picriside A | Picris hieracioides |
| H10 | − 8.6 | Convolvidine | Convolvulus prostratus [*] |
For each inhibitor, the table gives the phytochemical symbol, docking binding energy in kcal/mol, chemical name and herbal source. Plants for which there is evidence in traditional medicine literature for antiviral use are shown in bold and marked with an [*] sign
Table 2.
The protein residues of SARS-CoV-2 Nsp13 that are involved in hydrogen bond and hydrophobic interaction with the top ten potential phytochemical inhibitors H1 to H10 in the best-docked pose
| Protein–ligand complex | Docking binding energy (kcal/mol) | Number of hydrogen bonds | Hydrogen bond interaction residues | Hydrophobic interaction residues |
|---|---|---|---|---|
| Nsp13-H1 | − 10.2 | 9 | P284, G285, S289, S310, Q404, R443, R567 | G285, G287, K288, S289, A312, A313, E375, M378, G538 |
| Nsp13-H2 | − 9 | 7 | G287, K288, S289, S310, R443, S535 | P284, K288, A312, A313, A316, E375, G538 |
| Nsp13-H3 | − 8.9 | 7 | K288, S289, E375, Q404, R443, R567 | G285, K288 |
| Nsp13-H4 | − 8.9 | 15 | P284, G285, K288, S289, D374, E375, R443, R567 | K288, A313, A316, D374, G538 |
| Nsp13-H5 | − 8.9 | 13 | P284, G285, G287, S289, K320, Q404, R443, G538, R567 | G285, K288, K320, E375, E540 |
| Nsp13-H6 | − 8.8 | 4 | D374, Q404, R443, R567 | G285, K288, A316, E375, G538 |
| Nsp13-H7 | − 8.8 | 7 | S289, A316, Q404, R443, R567 | K288, S289, G538 |
| Nsp13-H8 | − 8.6 | 11 | G285, K288, S289, A316, Q404, R443, G538, R567 | K288, A313, A316, E375 |
| Nsp13-H9 | − 8.6 | 17 | G285, G287, S289, A316, E375, Q404, R443, G538, R567 | G285, G287, K288, E375 |
| Nsp13-H10 | − 8.6 | 10 | S264, G287, S289, D374, E375, Q404, R442 | S264, G285, G287, K288, H290, E375, G400, Q404, L438, R442, G538 |
For each protein–ligand complex the table lists the docking binding energy in kcal/mol, number of hydrogen bonds, and the residues forming hydrogen bond and hydrophobic interactions with the ligand atoms. Note that the hydrophobic interactions listed here are between the carbon atom of the protein residue and the carbon, halogen or sulfur atom of the ligand
Fig. 3.
Chemical name and 2D structure for the top ten potential phytochemical inhibitors of SARS-CoV-2 Nsp13
Fig. 4.
Cartoon representation of the hydrogen bond interactions in the best-docked pose of the top ten potential phytochemical inhibitors of SARS-CoV-2 helicase Nsp13. In this figure, hydrogen bond interactions are shown as yellow colored dotted lines between the residues of Nsp13 and the atoms of a H1, b H2, c H3, d H4, e H5, f H6, g H7, h H8, i H9 and j H10. The carbon atoms of the ligand are colored in green and the carbon atoms of the residues in Nsp13 are colored in cyan. The Nsp13 residues forming hydrogen bond interactions with the ligand are labeled with their one letter amino acid code and their residue number
Table 3.
MM-PBSA based binding energies for the protein–ligand complexes of the top five phytochemical inhibitors of SARS-CoV-2 Nsp13
| Protein–ligand complex | Binding energy (kcal/mol) | Van der waals energy (kcal/mol) | Electrostatic energy (kcal/mol) | Polar solvation energy (kcal/mol) | SASA energy (kcal/mol) |
|---|---|---|---|---|---|
| NSP13-H1 | − 13.211 ± 5.507 | − 52.006 ± 3.019 | − 14.428 ± 2.359 | 58.398 ± 6.429 | − 5.174 ± 0.270 |
| NSP13-H2 | − 21.329 ± 4.067 | − 44.04 ± 2.424 | − 6.027 ± 1.608 | 33.337 ± 5.045 | − 4.599 ± 0.234 |
| NSP13-H3 | − 17.618 ± 3.846 | − 44.531 ± 3.207 | − 5.494 ± 2.005 | 36.548 ± 4.424 | − 4.141 ± 0.320 |
| NSP13-H4 | − 6.564 ± 5.422 | − 23.793 ± 4.342 | − 49.666 ± 7.567 | 70.915 ± 10.753 | − 4.02 ± 0.248 |
| NSP13-H5 | − 11.76 ± 3.253 | − 47.293 ± 2.625 | − 13.453 ± 2.402 | 53.897 ± 4.958 | − 4.912 ± 0.334 |
Phytochemicals H1 (Picrasidine M) and H5 (Picrasidine N) are dimeric β-carboline-type alkaloid produced by the herb Picrasma quassioides. The herb Picrasma quassioides has been reported to have antiviral, antifungal and antiparasitic activities [25, 59, 60]. Additionally, the β-carboline alkaloids from Picrasma quassioides have been experimentally found to inhibit the RNA replication of the plant pathogen Tobacco mosaic virus (TMV) [61]. From Fig. 4a it is seen residue S289 forms 2 hydrogen bonds with H1 (C–H···O type and C–H···N type), and residues Q404 and R567 form 1 N–H···O type hydrogen bond each with H1. In case of H5, residues S289, Q404 and R567, which are among the six key ATP-binding site residues, form 1 C–H···O type, 1 N–H···O type and 2 N–H···O type hydrogen bonds with H5, respectively (Fig. 4e).
Phytochemical H2 ((+)-Epiexcelsin) is a lignan produced by the herb Litsea verticillata [62]. Extract of Litsea verticillata has been experimentally found to have antiviral and anti-HIV activities [63]. From Fig. 4b it is seen, the residues K288 and S289 form 2 hydrogen bonds (C–H···O type and C–H···N type) and 1 hydrogen bond (N–H···O type) with H2, respectively.
Phytochemical H3 (Isorhoeadine) is a rhoeadine alkaloid produced by the herb Papaver rhoeas [59]. H3 forms 3 C–H···O type hydrogen bonds with S289, E375 and Q404, 2 C–H···N type hydrogen bonds with R443 and R567, and 2 N–H···O type hydrogen bonds with K288 and Q404 (Fig. 4c).
Phytochemical H4 (Euphorbetin) is a bicoumarin produced by the herb Euphorbia lathyris [64]. Figure 4d shows the extensive hydrogen bond network between H4 and the protein residues. Residues K288, S289, D374, E375 and R567 form 2 hydrogen bonds (C–H···O type and N–H···O type), 4 hydrogen bonds (2 of N–H···O type, 1 of C–H···O type and 1 of O–H···O type), 1 hydrogen bond (O–H···O type), 1 hydrogen bond (C–H···O type) and 2 hydrogen bonds (N–H···O type) with H4, respectively.
Phytochemicals H6 (Ovigerine) and H8 (Hernandonine) are both produced by the herbs Hernandia guianensis and Hernandia nymphaeifolia. Figure 4f shows the hydrogen bonds H6 forms with residues of Nsp13. In case of H8, residues K288, S289, Q404, and R567 form 1 hydrogen bond (C–H···N type), 4 hydrogen bonds (2 of C–H···O type, 1 of N–H···O type and 1 of C–H···N type), 1 hydrogen bond (N–H···N type) and 1 hydrogen bond (C–H···N type) with H8, respectively (Fig. 4h).
Phytochemical H7 (Cassamedine) is an oxoaporphine alkaloid produced by the herb Cassytha filiformis [25, 59]. From Fig. 4g it is seen, the residues S289, Q404 and R567 form 1 hydrogen bond (O–H···O type), 2 hydrogen bonds (N–H···O type and C–H···N type) and 1 hydrogen bond (C–H···N type) with H7, respectively.
Phytochemical H9 (Picriside A) is a glycoside produced by the herb Picris hieracioides [33]. H9 forms an extensive network of 17 hydrogen bonds with the residues of Nsp13 (Fig. 4i).
Phytochemical H10 (Convolvidine) is a tropane alkaloid produced by the herb Convolvulus prostratus [26]. Residues S289, D374, E375 and Q404 form 2 hydrogen bonds (C–H···O type), 1 hydrogen bond (C–H···O type), 1 hydrogen bond (C–H···O type) and 2 hydrogen bonds (N–H···O type) with H10, respectively (Fig. 4j).
In order to use the knowledge-based information from the PanDDA co-crystallized structures used for our analysis as mentioned earlier, we have examined the ten PanDDA co-crystallized structures of SARS-CoV-2 Nsp13 with ligands bound near the ATP-binding site, namely, PDB 5RM2, 5RM7, 5RLW, 5RL9, 5RLO, 5RLY, 5RLJ, 5RLV, 5RLN and 5RLS, in relation to the top five phytochemicals identified as potential inhibitors of SARS-CoV-2 Nsp13. The ligands from PanDDA co-crystallized structures PDB 5RLI and 5RLR were not considered for comparison as they were found to be structurally similar to the ligand present in PDB 5RLJ. The comparison of the binding modes of the top five phytochemical inhibitors of Nsp13 and the ligands from the PanDDA co-crystallized structures reveals distinct modes of binding for the top inhibitors that are different from the ligands of the PanDDA structures (Supplementary Figure S1a–d). Specifically, the binding mode of the top phytochemical inhibitor H1 in the ATP-binding site of Nsp13 doesn’t overlap with that of the ligand N0E from the PanDDA structure PDB 5RM7, which has the highest 2D structural similarity with H1 (Supplementary Figure S1a, d). The 2D structural similarity was calculated using Tanimoto coefficient along with ECFP4 fingerprint as described in Vivek-Ananth et al. [65]. The above observations suggest the need for further experimental examination through co-crystallized structures and in vitro binding studies for the phytochemical inhibitors uncovered here.
MD analysis of the protein–ligand complexes of the top five phytochemical inhibitors
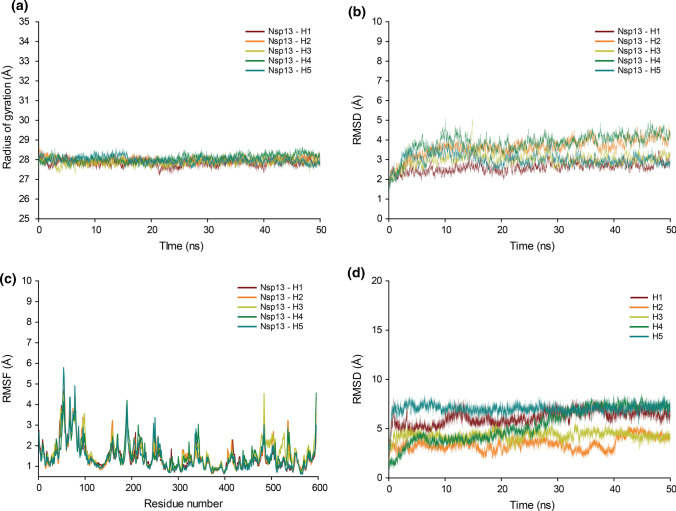
Based on the 50 ns MD simulations of the protein–ligand complexes of the top five phytochemical inhibitors of SARS-CoV-2 Nsp13 identified here, namely, Picrasidine M (H1), (+)-Epiexcelsin (H2), Isorhoeadine (H3), Euphorbetin (H4) and Picrasidine N (H5) (Methods), the structural characteristics of the complexes were evaluated (Fig. 5).
Fig. 5.
Based on the 50 ns MD simulations of the protein–ligand complexes, the figure shows the a Rg, b RMSD and c RMSF of the SARS-CoV-2 Nsp13 in complex with the top five phytochemical inhibitors, namely, Nsp13-H1, Nsp13-H2, Nsp13-H3, Nsp13-H4 and Nsp13-H5, and d RMSD of the top five phytochemical inhibitors H1, H2, H3, H4 and H5
Figure 5a shows the Rg of SARS-CoV-2 Nsp13 in complex with the top five inhibitors. Rg remains largely compact throughout the MD simulation of the five protein–ligand complexes (average Rg values are Nsp13-H1 = 27.836 ± 0.164 Å, Nsp13-H2 = 27.968 ± 0.147 Å, Nsp13-H3 = 27.878 ± 0.154 Å, Nsp13-H4 = 28.071 ± 0.174 Å, and Nsp13-H5 = 27.982 ± 0.167 Å). The RMSD value of the Cα atoms of SARS-CoV-2 Nsp13 in complex with the top five inhibitors stabilizes after 20 ns (Fig. 5b; average RMSD Cα over 20 ns–50 ns are Nsp13-H1 = 2.714 ± 0.222 Å, Nsp13-H2 = 3.823 ± 0.288 Å, Nsp13-H3 = 3.050 ± 0.226 Å, Nsp13-H4 = 4.010 ± 0.295 Å, and Nsp13-H5 = 2.971 ± 0.226 Å). In addition, the RMSF value per residue in the MD simulations of the SARS-CoV-2 Nsp13 in complex with the top five inhibitors closely follows the RMSF value per residue in the MD simulation of the SARS-CoV-2 Nsp13 uncomplexed protein (Fig. 5c; Supplementary Figure S2). The superimposed snapshots at 20 ns, 30 ns, 40 ns and 50 ns from the MD simulations of the protein–ligand complexes Nsp13-H1, Nsp13-H2, Nsp13-H3, Nsp13-H4 and Nsp13-H5 shows the conformational variations upon ligand binding (Supplementary Figure S3a–e). This is also seen by the RMSD of the top inhibitors (H1–H5) (Fig. 5d). The binding of the inhibitors to the protein is good as characterized by the distance between the center of masses of the inhibitors (H1–H5) and the six key ATP-binding site residues namely, K288, S289, D374, E375, Q404 and R567 (Supplementary Figure S4).
Molecular Mechanics Poisson–Boltzmann Surface Area (MM-PBSA) method has been reported to be more accurate in predicting the protein–ligand binding energy in comparison with molecular docking [66]. The MM-PBSA based binding energy of the top five phytochemical inhibitors of SARS-CoV-2 Nsp13 (Table 3) indicates the importance of the relative contributions of the van der Waals energy, the electrostatic energy, the polar solvation energy, and the solvent accessible surface area energy to the binding. The top five phytochemical inhibitors identified here, namely, Picrasidine M (H1), (+)-Epiexcelsin (H2), Isorhoeadine (H3), Euphorbetin (H4) and Picrasidine N (H5), have MM-PBSA based binding energy values of − 13.211 ± 5.507 kcal/mol, − 21.329 ± 4.067 kcal/mol, − 17.618 ± 3.846 kcal/mol, − 6.564 ± 5.422 kcal/mol and − 11.76 ± 3.253 kcal/mol, respectively.
The comparative analysis with the ligands of the PanDDA SARS-CoV-2 Nsp13 co-crystallized structures shows the distinct binding mode of the top inhibitor Picrasidine M (H1) (Supplementary Figure S1a–d). Further, the analysis of the docked pose with SARS-CoV-2 Nsp13 reveals the extent of interactions Picrasidine M makes with the ATP-binding site residues of Nsp13 (Fig. 4a; Table 2; Supplementary Table S4). In the MD simulation, Picrasidine M has exhibited stable interactions with SARS-CoV-2 Nsp13 (Fig. 5; Supplementary Figure S4). Therefore, the phytochemical Picrasidine M is likely to be a good potential inhibitor of SARS-CoV-2 Nsp13 which must be taken forward as a lead compound for experimental and clinical studies.
Conclusions
More than 3.7 million people have died due to COVID-19 which is likely to become endemic past the pandemic phase. Computational approaches such as molecular docking and molecular dynamics simulation can be used to accelerate the identification and development of anti-COVID drugs. SARS-CoV-2 Nsp13 is one of the key components of the viral replication and translation complex, and has been identified as one of the key targets for development of anti-COVID drugs [8, 11]. In this study, we have computationally screened a small molecule library of 14,011 phytochemicals against the ATP-binding site of the SARS-CoV-2 helicase Nsp13.
Natural products have directly or indirectly contributed to many FDA approved drugs [20] and the therapeutic activity space of the diverse natural product space is still largely unmapped and unexplored. This motivated this virtual screening study involving the largest small molecule library of phytochemicals from Indian medicinal plants and the key drug target SARS-CoV-2 helicase Nsp13. We have identified 368 phytochemicals as potential inhibitors of SARS-CoV-2 Nsp13, targeting the ATP-binding site.
Notably, among the top five potential inhibitors, both Picrasidine M (H1) and Picrasidine N (H5) are found to be produced by the herb Picrasma quassioides, which has been used in traditional medicine as antiviral, antifungal and antiparasitic. The herb Litsea verticillata which can produce (+)-Epiexcelsin (H2), has been reported to have antiviral and anti-HIV activities [63]. It should be emphasized that, further experimental studies are required to validate the inhibitory potential of the phytochemical inhibitors of SARS-CoV-2 Nsp13 identified in this study. In conclusion, the potential phytochemical inhibitors of SARS-CoV-2 helicase Nsp13 identified here constitute a potential chemical library for the development of anti-COVID drugs.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors acknowledge the use of the supercomputing machine Nandadevi at The Institute of Mathematical Sciences for the molecular dynamics simulations. The authors also thank the anonymous referees for their constructive suggestions that have helped improve the manuscript.
Funding
AS would like to acknowledge financial support from the Science and Engineering Research Board (SERB) India through the award of a Ramanujan fellowship (SB/S2/RJN-006/2014), and the Max Planck Society Germany for the award of a Max Planck Partner Group. The funders have no role in study design, prediction, analysis or decision to publish this work.
Data availability
The supplementary tables of this manuscript provide all the associated data.
Code availability
Not applicable.
Declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Consent to participate
Not applicable.
Consent for publication
All co-authors have read and approved the manuscript.
Ethical approval
This is a purely computational study and ethics approvals are not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sankaran Krishnaswamy, Email: mkukrishna@imsc.res.in.
Areejit Samal, Email: asamal@imsc.res.in.
References
- 1.Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan JF-W, Yuan S, Kok K-H, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet Lond Engl. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuzikov M, Costanzi E, Reinshagen J, et al. Identification of inhibitors of SARS-CoV-2 3CL-pro enzymatic activity using a small molecule in vitro repurposing screen. ACS Pharmacol Transl Sci. 2021 doi: 10.1021/acsptsci.0c00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braga L, Ali H, Secco I, et al. Drugs that inhibit TMEM16 proteins block SARS-CoV-2 spike-induced syncytia. Nature. 2021 doi: 10.1038/s41586-021-03491-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.US Food and Drug Administration (2020) FDA Issues emergency use authorization for potential COVID-19 treatment. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-issues-emergency-use-authorization-potential-covid-19-treatment
- 6.Wu A, Peng Y, Huang B, et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.V’kovski P, Kratzel A, Steiner S, et al. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li G, De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat Rev Drug Discov. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 9.Shu T, Huang M, Wu D, et al. SARS-coronavirus-2 Nsp13 possesses NTPase and RNA helicase activities that can be inhibited by bismuth salts. Virol Sin. 2020;35:321–329. doi: 10.1007/s12250-020-00242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jia Z, Yan L, Ren Z, et al. Delicate structural coordination of the severe acute respiratory syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res. 2019;47:6538–6550. doi: 10.1093/nar/gkz409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Habtemariam S, Nabavi SF, Banach M, et al. Should we try SARS-CoV-2 helicase inhibitors for COVID-19 therapy? Arch Med Res. 2020;51:733–735. doi: 10.1016/j.arcmed.2020.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.White MA, Lin W, Cheng X. Discovery of COVID-19 inhibitors targeting the SARS-CoV-2 Nsp13 helicase. J Phys Chem Lett. 2020;11:9144–9151. doi: 10.1021/acs.jpclett.0c02421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohamed K, Yazdanpanah N, Saghazadeh A, Rezaei N. Computational drug discovery and repurposing for the treatment of COVID-19: a systematic review. Bioorganic Chem. 2021;106:104490. doi: 10.1016/j.bioorg.2020.104490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sumon TA, Hussain MdA, Hasan MdT, et al. A revisit to the research updates of drugs, vaccines, and bioinformatics approaches in combating COVID-19 pandemic. Front Mol Biosci. 2021;7:585899. doi: 10.3389/fmolb.2020.585899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhowmick S, Saha A, Osman SM, et al. Structure-based identification of SARS-CoV-2 main protease inhibitors from anti-viral specific chemical libraries: an exhaustive computational screening approach. Mol Divers. 2021 doi: 10.1007/s11030-021-10214-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mirza MU, Froeyen M. Structural elucidation of SARS-CoV-2 vital proteins: computational methods reveal potential drug candidates against main protease, Nsp12 polymerase and Nsp13 helicase. J Pharm Anal. 2020;10:320–328. doi: 10.1016/j.jpha.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahmad S, Waheed Y, Ismail S, et al. Structure-based virtual screening identifies multiple stable binding sites at the RecA domains of SARS-CoV-2 helicase enzyme. Molecules. 2021;26:1446. doi: 10.3390/molecules26051446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thurakkal L, Singh S, Roy R, et al. An in-silico study on selected organosulfur compounds as potential drugs for SARS-CoV-2 infection via binding multiple drug targets. Chem Phys Lett. 2021;763:138193. doi: 10.1016/j.cplett.2020.138193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zia M, Muhammad S, Shafiq-urRehman, et al. Exploring the potential of novel phenolic compounds as potential therapeutic candidates against SARS-CoV-2, using quantum chemistry, molecular docking and dynamic studies. Bioorg Med Chem Lett. 2021;43:128079. doi: 10.1016/j.bmcl.2021.128079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newman DJ, Cragg GM. Natural products as sources of new drugs from 1981 to 2014. J Nat Prod. 2016;79:629–661. doi: 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- 21.Ren J, Zhang A-H, Wang X-J. Traditional Chinese medicine for COVID-19 treatment. Pharmacol Res. 2020;155:104743. doi: 10.1016/j.phrs.2020.104743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rastogi S, Pandey DN, Singh RH. COVID-19 pandemic: a pragmatic plan for ayurveda intervention. J Ayurveda Integr Med. 2020 doi: 10.1016/j.jaim.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohanraj K, Karthikeyan BS, Vivek-Ananth RP, et al. IMPPAT: a curated database of Indian medicinal plants. Phytochem Ther Sci Rep. 2018;8:4329. doi: 10.1038/s41598-018-22631-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vivek-Ananth RP, Rana A, Rajan N, et al. In silico identification of potential natural product inhibitors of human proteases key to SARS-CoV-2 infection. Molecules. 2020;25:3822. doi: 10.3390/molecules25173822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.The Wealth of India (2000) The wealth of India: a dictionary of Indian raw materials & industrial products. First supplement series (Raw materials) Vol 1. National Institute of Science Communication, Council of Scientific & Industrial Research, New Delhi
- 26.The Wealth of India (2000) The wealth of India: a dictionary of Indian raw materials & industrial products. First supplement series (Raw materials) Vol 2. National Institute of Science Communication, Council of Scientific & Industrial Research, New Delhi
- 27.The Wealth of India (2000) The wealth of India: a dictionary of Indian raw materials & industrial products. First supplement series (Raw materials) Vol 3. National Institute of Science Communication, Council of Scientific & Industrial Research, New Delhi
- 28.The Wealth of India (2000) The wealth of India: a dictionary of Indian raw materials & industrial products. First supplement series (Raw materials) Vol 4. National Institute of Science Communication, Council of Scientific & Industrial Research, New Delhi
- 29.The Wealth of India (2000) The wealth of India: a dictionary of Indian raw materials & industrial products. First supplement series (Raw materials) Vol 5. National Institute of Science Communication, Council of Scientific & Industrial Research, New Delhi
- 30.Rastogi RP (1990) Compendium of Indian medicinal plants. Vol 1: 1960–1969. Central Drug Research Institute, Lucknow [PubMed]
- 31.Rastogi RP (1991) Compendium of Indian medicinal plants. Vol 2: 1970–1979. Central Drug Research Institute, Lucknow [PubMed]
- 32.Rastogi RP (1993) Compendium of Indian medicinal plants. Vol 3: 1980–1984. Central Drug Research Institute, Lucknow [PubMed]
- 33.Rastogi RP (1995) Compendium of Indian medicinal plants. Vol 4: 1985–1989. Central Drug Research Institute, Lucknow [PubMed]
- 34.Rastogi RP (1998) Compendium of Indian medicinal plants. Vol 5: 1990–1994. Central Drug Research Institute, Lucknow [PubMed]
- 35.Chatterjee A, Pakrashi SC (1991) The treatise on Indian medicinal plants. Vol 1. Publications & Information Directorate, Council of Scientific & Industrial Research (India), New Delhi
- 36.Chatterjee A, Pakrashi SC (1992) The treatise on Indian medicinal plants. Vol 2. Publications & Information Directorate, Council of Scientific & Industrial Research (India), New Delhi
- 37.Chatterjee A, Prakashi SC (1994) The treatise on Indian medicinal plants. Vol 3. Publications & Information Directorate, Council of Scientific & Industrial Research (India), New Delhi
- 38.Chatterjee A, Prakashi SC (1995) The treatise on Indian medicinal plants. Vol 4. Publications & Information Directorate, Council of Scientific & Industrial Research (India), New Delhi
- 39.Chatterjee A, Prakashi SC (1997) The treatise on Indian medicinal plants. Vol 5. Publications & Information Directorate, Council of Scientific & Industrial Research (India), New Delhi
- 40.Chatterjee A, Prakashi SC (2003) The treatise on Indian medicinal plants. Vol 6. Publications & Information Directorate, Council of Scientific & Industrial Research (India), New Delhi
- 41.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 42.Kim S, Chen J, Cheng T, et al. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 2019;47:D1102–D1109. doi: 10.1093/nar/gky1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Boyle NM, Banck M, James CA, et al. Open Babel: an open chemical toolbox. J Cheminform. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wlodawer A, Dauter Z, Shabalin IG, et al. Ligand-centered assessment of SARS-CoV-2 drug target models in the protein data bank. FEBS J. 2020;287:3703–3718. doi: 10.1111/febs.15366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 46.Abraham MJ, Murtola T, Schulz R, et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 47.Schmid N, Eichenberger AP, Choutko A, et al. Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur Biophys J. 2011;40:843. doi: 10.1007/s00249-011-0700-9. [DOI] [PubMed] [Google Scholar]
- 48.Stroet M, Caron B, Visscher KM, et al. Automated topology builder version 3.0: prediction of solvation free enthalpies in water and hexane. J Chem Theory Comput. 2018;14:5834–5845. doi: 10.1021/acs.jctc.8b00768. [DOI] [PubMed] [Google Scholar]
- 49.Berendsen HJC, Postma JPM, van Gunsteren WF, et al. Molecular dynamics with coupling to an external bath. J Chem Phys. 1984;81:3684–3690. doi: 10.1063/1.448118. [DOI] [Google Scholar]
- 50.Parrinello M, Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys. 1981;52:7182–7190. doi: 10.1063/1.328693. [DOI] [Google Scholar]
- 51.Daura X, Gademann K, Jaun B, et al. Peptide folding: when simulation meets experiment. Angew Chem Int Ed. 1999;38:236–240. doi: 10.1002/(SICI)1521-3773(19990115)38:1/2<236::AID-ANIE236>3.0.CO;2-M. [DOI] [Google Scholar]
- 52.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morris GM, Huey R, Lindstrom W, et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rodrigues JPGLM, Teixeira JMC, Trellet M, Bonvin AMJJ. pdb-tools: a swiss army knife for molecular structures. F1000Research. 2018;7:1961. doi: 10.12688/f1000research.17456.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schyman P, Liu R, Desai V, Wallqvist A. vNN web server for ADMET predictions. Front Pharmacol. 2017;8:889. doi: 10.3389/fphar.2017.00889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baker NA, Sept D, Joseph S, et al. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kumari R, Kumar R, Open Source Drug Discovery Consortium, Lynn A (2014) g_mmpbsa—A GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model 54:1951–1962. 10.1021/ci500020m [DOI] [PubMed]
- 59.Khare CP. Indian medicinal plants: an illustrated dictionary. New York: Springer; 2007. [Google Scholar]
- 60.Mohd Jamil MDH, Taher M, Susanti D, et al. Phytochemistry, traditional use and pharmacological activity of Picrasma quassioides: a critical reviews. Nutrients. 2020;12:2584. doi: 10.3390/nu12092584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen J, Yan X-H, Dong J-H, et al. Tobacco mosaic virus (TMV) inhibitors from Picrasma quassioides Benn. J Agric Food Chem. 2009;57:6590–6595. doi: 10.1021/jf901632j. [DOI] [PubMed] [Google Scholar]
- 62.Hoang VD, Tan GT, Zhang H-J, et al. Natural anti-HIV agents—part I: (+)-demethoxyepiexcelsin and verticillatol from Litsea verticillata. Phytochemistry. 2002;59:325–329. doi: 10.1016/S0031-9422(01)00454-X. [DOI] [PubMed] [Google Scholar]
- 63.Guan Y, Wang D, Tan GT, et al. Litsea species as potential antiviral plant sources. Am J Chin Med. 2016;44:275–290. doi: 10.1142/S0192415X16500166. [DOI] [PubMed] [Google Scholar]
- 64.Dutta PK, Banerjee D, Dutta NL. Euphorbetin: a new bicoumarin from Euphorbia lathyris L. Tetrahedron Lett. 1972;13:601–604. doi: 10.1016/S0040-4039(01)84388-4. [DOI] [Google Scholar]
- 65.Vivek-Ananth RP, Sahoo AK, Kumaravel K, et al. MeFSAT: a curated natural product database specific to secondary metabolites of medicinal fungi. RSC Adv. 2021;11:2596–2607. doi: 10.1039/D0RA10322E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Genheden S, Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin Drug Discov. 2015;10:449–461. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The supplementary tables of this manuscript provide all the associated data.
Not applicable.