Abstract
The 2014–15 influenza season saw the emergence of an H3N2 antigenic drift variant that formed the 3C.2a HA clade. Whole viral genomes were sequenced from nasopharyngeal swabs of ninety-four patients with confirmed influenza A virus infection and primary human nasal epithelial cell cultures used to efficiently isolate H3N2 viruses. The isolates were classified by HA clade and the presence of a new set of co-selected mutations in NA (a glycosylation site, NAg+) and PB1-F2 (H75P). The NA and PB1-F2 mutations were present in a subset of clade 3C.2a viruses (NAg+F2P), which dominated during the subsequent influenza seasons. In human nasal epithelial cell cultures, a virus with the novel NAg+F2P genotype replicated less well compared with a virus with the parental genotype. Retrospective analyses of clinical data showed that NAg+F2P genotype viruses were associated with increased cough and shortness of breath in infected patients.
Keywords: antigenic drift, H3N2, influenza, symptoms, human, NA, PB1-F2
1. Introduction
Human influenza epidemics are largely influenced by the emergence and circulation of seasonal viral variants containing mutations that evade pre-existing population immunity (Bedford et al. 2015). This necessitates significant virus surveillance and vaccine efficacy monitoring to guide key public health decision-making and ensure proper preparedness for seasonal epidemics and more infrequent pandemics (Bedford et al. 2015; Flannery et al. 2016; Huzly et al. 2016). The seasonal H3N2 virus variants that became dominant during the 2014–15 influenza season were associated with an increase in outpatient influenza-like illness hospital visits, hospitalizations, and deaths attributable to pneumonia and influenza (Epperson et al. 2014; Appiah et al. 2015). Increased disease severity was also detected in other countries across the northern hemisphere (Broberg et al. 2015; Donadeu et al. 2015). The increased number of cases correlated with a marked diminution of serum hemagglutination inhibition and neutralizing antibodies against seasonal virus isolates (Flannery et al. 2016; Skowronski et al. 2016), which was attributed to discreet changes in HA antigenic sites (Chambers et al. 2015; Stucker et al. 2015) involving a novel glycosylation site in the H3 hemagglutinin (HA) protein (Chambers et al. 2015). Further sequence analysis, protein modeling, and serology indicated that mutations in the HA and neuraminidase (NA) genes could be linked to escape from neutralizing antibodies (Broberg et al. 2015; Chambers et al. 2015; McCauley et al. 2015; Stucker et al. 2015; Flannery et al. 2016; Mostafa et al. 2016; Skowronski et al. 2016; Tian et al. 2019; Powell and Pekosz 2020).
It is critical for research and public health laboratories to have the methods in place for rapid reliable detection, recovery, and characterization of influenza seasonal drift variants. These capabilities inform decision making regarding vaccine composition and the public health response to an ongoing epidemic or pandemic (Pebody et al. 2015; Flannery et al. 2016).
In this report, we use influenza virus whole-genome sequencing (WGS) and phylogenetic analysis to identify genetic variants, their frequencies, and their evolutionary relationships. We show that isolation of virus from nasopharyngeal swab (NPS) and nasopharyngeal wash (NPW) specimens using primary human nasal epithelial cell (hNEC) cultures is more efficient than isolation using a cell line. We found that viruses encoding amino acid (AA) changes in the NA (to introduce a glycosylation site near the sialidase pocket) and PB1-F2 (mutation H75P) were associated with altered proteins correlated with a higher incidence of coughing and shortness of breath. Therefore, our data suggest that NA and PB1-F2 changes contributed to increased reporting of symptoms.
2. Materials and methods
2.1. Sample collection and ethics
The human subject protocol was reviewed and approved by the Johns Hopkins School of Medicine Institutional Review Board (IRB00052743). This protocol allowed for collection of residual NPS from influenza-positive patients from the Johns Hopkins Health System in the Baltimore-DC National Capital Region and collection of related, de-identified patient data from the electronic health records (EHR). The majority of specimens were from pediatric (under 18 years of age) and elderly (over 64) individuals, which represent the groups at increased risk for severe seasonal influenza disease (Luk et al. 2001; Morens et al. 2010; Skowronski et al. 2010). Symptoms were not reported for children under the age of 2. Specimens from patients who had severe disease (as characterized by one of the following: supplemental oxygen use, Intensive Care Unit stay, or death) were included. Specimens from age- and sex-matched individuals, who did not have characteristics of severe disease, were also included. Priority was also given to specimens from patients who had known influenza vaccination status documented in the EHR.
An IRB-exempt protocol allowed for access to residual NPW specimens from de-identified patients of the Wright-Patterson Air Force Base (WPAFB). The WPAFB is the central repository for disease specimens from service men and women from around the country. A set of sixteen samples that were collected before September from the WPAFB and grew to titers above 3 Log TCID50 in hNECs were selected for sequence analysis. Patient-level demographic and clinical data were not available for these specimens.
NPS and NPW specimens were collected from each patient and placed in 3 ml of viral transport medium (MicroTest M4RT; Remel, Lenexa, KS, USA). All the specimens were aliquoted and stored at −70 °C.
2.2. Sample processing for genomic analysis
NPS samples were centrifuged for five minutes at 16,000g to remove debris. Clarified supernatants were purified per manufacturer’s recommendations with the Ambion MagMax Viral RNA purification reagent, with the only protocol modification being omission of carrier RNA to the viral lysis enhancer reagent. RNA was resuspended in 20 μl of supplied RNAse-free elution buffer. Ambion Turbo DNA-free reagent (Life Technologies) was used to deplete genomic DNA. Manufacturer’s instructions for stringent DNAse treatments were followed: 2 U of enzyme was added at the beginning of treatment and after 30 minutes of incubation at 37 °C. Total duration of DNA depletion was one hour, after which DNAse inactivation reagent was added at 0.2 volumes.
Reverse transcription of viral RNA was carried out with Superscript III (Invitrogen) per manufacturer’s instructions for gene-specific priming of low input samples using the Opti1 primer set, consisting of Opti1-F1 (GTTACGCGCCAGCAAAAGCAGG), Opti1-F2 (GTTACGCGCCAGCGAAAGCAGG), and Opti1-R1 (GTTACGCGCCAGTAGAAACAAGG). Samples were prepared using either a traditional multi-segment polymerase chain reaction (PCR) approach (Zhou et al. 2009) or a preamplification-free approach, which we term amplification-limited viral RNA sequencing (vRNAseq). Briefly, primers and dNTPs were added to RNA and annealed by heating to 65 °C for five minutes, and then allowed to return to 4 °C for five minutes. Enzyme (400 U) was then used in a total reaction volume of 40 μl, and reverse transcription was carried out at 50 °C. Upon completion, second-strand synthesis was carried out at 16 °C for 2.5 hours using mRNA Second-Strand Synthesis reagent, which comprises Escherichia coli DNA Pol I, E. coli DNA ligase, RNase H (NEB), in a total volume of 80 µl. Double-stranded viral cDNA was purified using one volume of Ampure XP per manufacturer’s instructions (Beckman Coulter). Once beads were completely dry, samples were eluted in 7.5 µl of nuclease-free water for five minutes prior to collection. One microliter was used for quantification with the High Sensitivity Qubit reagent according to manufacturer instructions (Invitrogen).
Nextera XT DNA library preparation was carried out on all samples in accordance with the manufacturer’s instructions (Illumina). For samples that resulted in less than one nanogram or unquantifiable amounts of DNA, library preparation was carried out with the maximum amount of DNA available in 5 μl. After tagmentation and addition of sequencing adapters by twelve cycles of PCR, samples were purified with one volume of Ampure XP and eluted in 10 µl of nuclease-free water. One microliter of the resulting library was then used for quantification with High Sensitivity Qubit reagents (Invitrogen), and multiplexed libraries were then pooled equi-mass, assuming an even size distribution between samples.
2.3. Sequence data analysis
Raw reads from the Illumina MiSeq sequencer were first filtered using Trim Galore! (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) using an adapter sequence of CTGTCTCTTATACACATCT and a quality score cutoff of 30. QC-trimmed reads were then individually classified using Kraken (https://github.com/DerrickWood/kraken) against a custom influenza-specific kraken database containing all complete sequences in EpiFlu with a modified taxonomy, which for flu types A, B, and C, identifies the likely segment, subtype, host, and year for each read. The most likely subtype for each sample was identified from the Kraken results, and eighty-five H3N2 samples were pulled out for further analysis. QC-trimmed reads for all eighty-five H3N2-identified samples were aligned to the A/Victoria/361/2011 reference sequence, and consensus FASTA sequences were generated for each sample. From these consensus FASTA sequences, sixty-three samples had at least eighty-five percent coverage of the HA segment and were used for subsequent phylogenetic analysis.
Reference sequences for the 2014–15 influenza season were obtained from the GISAID EpiFlu database on 22 October 2016, by downloading all human H3N2 isolate sequences with a collection date between 1 October 2014 and 31 May 2015. In total, there were 5,642 unique EpiFlu ID numbers across 5,338 unique strain names. A subset of EpiFlu sequences was selected in two steps. First strains that contained exactly one of each of the eight genome segments were selected, and then for strain names that contained more than one EpiFlu ID number, only the strain with the lowest ID number was chosen. Sequences for the A/Victoria/361/2011 and A/Texas/50/2012 vaccine strains were obtained from NCBI BioProject PRJNA205638. The ultimate subset contained full genomes for 1,478 H3N2 strains. The same method was applied to select isolates for analysis from the 2013–14 and 2015–16 seasons, which led to 99 and 1,140 isolates, respectively. The number of isolates from the 2013–14 season was an order of magnitude lower than the numbers for the other seasons because sequences for genes other than HA were often not available.
2.4. Phylogenetic analysis
Data from each segment were combined into a segment-specific FASTA file for the 1,478 EpiFlu reference strains, the sixty-three sequenced samples with over eighty-five per cent coverage in each segment, and the A/Texas/50/2012 vaccine strain, for a total of 1,542 isolates. Each segment was then aligned using MAFFT (http://mafft.cbrc.jp/alignment/software/), all deletions (‘—‘) were replaced with ‘N’s, and all sequences were padded with ‘N’s to the length of the vaccine strain sequence. Any insertions with respect to the vaccine sequence were removed (any positions in the vaccine sequence that now contain ’N’s), and all sequences were trimmed to the length of the vaccine sequence. Phylogenetic trees were created for each segment using the nucleotide option of FastTree (http://www.microbesonline.org/fasttree/), and trees were then re-rooted to the vaccine strain using the nw_reroot tool from the Newick utilities package (https://github.com/xflouris/newick-tools). Trees were viewed and colored using FigTree (http://tree.bio.ed.ac.uk/software/figtree/) and HA clades were identified by adding known clade references to these trees and identifying branches for the 3C.2a, 3C.3, 3C.3a, and 3C.3b clades. From these branch identifications, a set of rules was developed (see below) so that a clade label could be added to each strain label. All clade information was added to the name of each strain name before tree generation, and then nodes were colored by searching for a specific clade text and coloring all nodes in that selection.
2.5. Clade and genotype identification
Sequence data from each segment of the 1,542 uniquely named isolates with over eighty-five per cent coverage in each segment were uploaded into a database using R. HA clades were identified using rules that selected sequences by AAs that associated with branches (https://hackmd.io/CwVgHAzAhgnAxgRgLQhiAZk4ATBMkwDsAbIUlFOsGMAEzHEAMEYQA===?view). The defining residues used were: 159Y for 3C.2a; 138S or 159S for 3C.3a; 157S for 3C.3b; 142R for 3C.2; 3L and 128A for 3C.3; and 3I and 128T for 3C.2. H3 numbering was used for all positions. Using these rules, all 1,542 strains were labeled according to the branch they fell on with the following exceptions: A/Brazil/9517/2015 is labeled 3C.3a but falls on the 3C.3b branch; A/Manitoba/RV3623/2014 and A/Nevada/30/2014 are labeled 3C.3b but fall on the 3C.3 branch; 01-13-P-0057 is labeled uncertain but falls on the 3C.2a branch; and 01-90-P-0102 is labeled 3C.3 but falls on the 3C.3b branch.
Consensus sequences of the twelve proteins PB2, PB1, PA, HA, NP, NA, M1, NS1 (primary open reading frames (ORFs)) and PB1-F2, PA-X, M2, and NEP (alternative ORFs) were generated for each isolate that had over eighty-five per cent sequence coverage and all of the reference strains. Sequences of the eight primary ORF proteins were generated by translation from the first ATG in each gene to the first stop codon. Sequences of the four alternative ORF proteins were generated with the following set of rules: translation of PB1-F2 was programmed to start at the fourth ATG of segment 2 and stop at the first in frame stop codon; translation of PA-X started from the first start codon in segment 3, included the first 573 nucleotides, then shifted +1 to nt 575 and carried on to the next stop codon in that frame; M2 was translated from the first start codon of the segment 7 through 51 nts, joined to nt 715 and stopped at the first stop codon in frame 2; NEP was translated from the first start codon in segment 8 through the 30th nt, joined with base 503 and stopped at the in-frame stop codon.
Genetic variation among clade 3C.2a isolates was analyzed further. AA alignments were generated using MAFFT and the frequency of each AA residue at every location was determined using Excel (Microsoft). Instances where the consensus AA fell below 75% frequency were noted. Mutations associated with putative NA glycosylation signatures were identified by the residues at positions 245–247. Isolates were labeled as NAg+ if those residues were NAT or NAS and NAg- for all other sequences. Residues at AA position 75 within PB1-F2 were also identified for each isolate.
2.6. Cell culture and serum-free media
Madin-Darby canine kidney cells (MDCK) were cultured as previously described (Grantham et al. 2010). MDCK-SIAT cells were kindly donated by Scott Hensley and cultured exactly the same as MDCK cells. Cultures of human primary, differentiated nasal epithelial cells (hNEC) were obtained from volunteers and differentiated at an air–liquid interface, as previously described ( Ramanathan et al. 2009; Fischer et al. 2015; Forero et al. 2017; Wohlgemuth et al. 2017).
2.7. Virus isolation
Virus isolation with MDCK cells was performed by incubating 100 µl of NPS on twice-PBS-washed cells in forty-eight-well plates for one hour at 32 °C with five per cent CO2 with agitation every fifteen minutes. After inoculation, NPS media was aspirated and cells were incubated again at 32 °C with five per cent CO2 in infection media (Dulbecco’s Modified minimal media; 0.3% bovine serum albumin) with 5 µg/ml N-acetyl trypsin (NAT). Media was collected and replaced daily.
Virus isolation with hNEC cultures was performed by incubating 100 µl of NPS on the twice-PBS-washed apical surface of hNEC cells in twelve-well transwell plates (Corning) for two hours at 32 °C with five per cent CO2. Inoculum was removed and cells were returned to incubate at 32 °C with five per cent CO2 for up to seven days. The hNEC cultures were maintained with media on the basolateral side, which was changed every two to three days. Apical washes, acquired by adding 300-μl infection media to the apical surface of hNECs and incubating for ten minutes at 32 °C with five per cent CO2, were collected daily. All samples were stored at −70 °C.
2.8. Infectious virus quantification
The infectious virus titers were determined by calculating the fifty per cent tissue culture infectious dose (TCID50) using the Reed and Muench method (Reed and Muench 1938; McCown and Pekosz 2005).
2.9. Virus generation and growth analysis
Infected-hNEC culture supernatants, from 4 dpi of hNECs, were inoculated into MDCK cells with a multiplicity of infection (MOI) of 0.01 or 0.001 TCID50 units per cell and incubated in infection media with 5 µg/ml NAT for five days at 32 °C to make virus stocks. A/Columbia/P0041/2014 (Co14) has the NAg-F2H genotype while A/Bethesda/P0055/2015 (Be15) has the NAg+F2P genotype. Infectious virus was quantified by TCID50. HA, NA, and PB1 genes of amplified virus stocks were Sanger sequenced. The sequences of virus stocks were compared with the WGS consensus sequences and the HA of both Co14 and Be15 were found to have the non-synonymous nucleotide mutation C478A, which encoded the mutation T160K (H3 Numbering). This mutation resulted in the loss of a predicted glycosylation site at position 158 in the HA protein. Co14 had a 160K in the nasal swab sequence. Interestingly, Be15 had the 160T mutation in the nasal swab indicating the 160K mutation was selected for during passage on MDCK cells.
For MDCK and MDCK-SIAT cell growth curves, cells in twenty-four-well plates were washed twice with PBS containing Ca2+ (0.9 mM) and Mg2+ (0.5 mM) (Sigma) and infected with MOI 0.001 virus, diluted in 100 µl of infection media. After one-hour inoculation, the supernatant was removed, cells were washed three times with PBS with calcium and magnesium and incubated in infection media with 2.5 µg/ml NAT at 32 °C with five per cent CO2. Samples were collected at indicated times by removing and replacing the media. Supernatants were stored at −70 °C and titrated by TCID50.
For hNEC growth curves, the apical surface of twenty-four-well hNEC cultures was washed twice with PBS and infected with MOI 0.01 virus in 100 µl in infection media. hNECs were incubated with the inoculum for two hours at 32 °C with five per cent CO2. Inoculum was removed and the apical surface was washed twice with PBS before cells were returned to the incubator. Apical wash samples were collected, at indicated times, by incubating 100 µl of infection media on the apical surface for ten minutes at 32 °C as previously described (Fischer et al. 2015). Supernatants were stored at −70 °C and infectious virus quantified by TCID50.
Each virus was inoculated into four wells of each cell type and growth curves were performed on two separate occasions, for a total of eight wells per virus, per cell type, per time point. The average viral titers from the eight equally treated wells were graphed for virus comparisons.
2.10. Plaque assay
MDCK cells, at approximately 100% confluence, were washed twice with PBS with calcium and magnesium. Virus was serially diluted ten-fold in infection media and 250 µl of each dilution was added to cells. The plates were incubated for one hour at 32 °C in five per cent CO2 with agitation, then inoculum was removed and cells were overlaid with 1× MEM with the addition of 0.3 per cent BSA, 10 mM Pen/Strep, 5 mM Glutamine, 1% agarose (Invitrogen), 1 mM Hepes (Gibco), and 5 µg/ml NAT and incubated at 32 °C in five per cent CO2 for seventy-two hours, after which cells were fixed with four per cent paraformaldehyde (Fisher) and stained with Napthol Blue Black (Sigma). One image per well was collected using an Olympus OM-D E-M5 Mark II digital camera. The scale for all images was determined by photographing a ruler. Images were processed in ImageJ v1.49 Plaque images were cropped to remove area outside of the well, then they were turned into eight-bit images. Next, plaques were circled manually using the selection tool. Finally, plaques were analyzed using the ImageJ analyze particle size command with a size minimum of 200 pixels and a circularity of 0.10–1.00. In total forty to sixty plaques (between 10 and 20 plaques per well) were analyzed per virus.
2.11. Statistical analysis
Differences between the average viral titers of isolates grown on MDCK cells or hNECs were determined by paired, two-tailed, t-test. Virus replication differences in low MOI growth curves were determined with repeated-measures two-way ANOVA followed by a Bonferonni post-test. A multivariate logistic regression analysis was performed in R to determine whether or not any co-morbidities or other demographic factors associated with genotype differences and disease symptoms.
3. Results
3.1. Sequence characterization of virus
Ninety-four influenza-positive NPS specimens with demographic and clinical data were collected from three hospitals in the Johns Hopkins Health System, referred to here as Johns Hopkins Medical Institutes (JHMI). Sixteen influenza-positive NPW specimens that lacked demographic and clinical data were provided by the Wright Patterson Airforce Base (WPAFB). Whole-genome sequencing (WGS) was successfully performed on seventy-seven of the JHMI and all sixteen of the WPAFB specimens using an amplification strategy designed to specifically detect influenza genomic sequences. Illumina sequencing and Kraken analyses were used to generate and categorize consensus sequences for the eight influenza virus gene segments in each specimen.
HA coding region sequence reads were used to determine the influenza virus type in eighty-six per cent (66/77) of the JHMI samples and eighty-eight per cent (14/16) of the WPAFB samples (Table 1). The influenza subtype was determined for eighty-four per cent (65/77), and the HA clade was determined for seventy-seven per cent (59/77), of JHMI isolates. The influenza subtype and clade were determined for eighty-eight per cent (14/16) of the WPAFB samples.
Table 1.
Influenza virus classification by WGS.
| Influenza | WGS JHMI | WGS WPAFB | |
|---|---|---|---|
| Type A | 86% (66/77) | 88% (14/16) | |
| Subtype H3N2 | 84% (65/77) | 88% (14/16) | |
| Clade | (determined) | 77% (59/77) | 88% (14/16) |
| 3C.2 | 0% (0/59) | 0% (0/14) | |
| 3C.2a | 93% (55/59) | 57% (8/14) | |
| 3C.2b | 0% (0/59) | 0% (0/14) | |
| 3C.3 | 0% (0/59) | 7% (1/14) | |
| 3C.3a | 5% (3/59) | 14% (2/14) | |
| 3C.3b | 2% (1/59) | 21% (3/14) |
A total of seventy-seven JHMI and sixteen WPAFB nasal swab samples were sequenced and influenza virus type, subtype, and clade were determined.
3.2. HA clade distribution
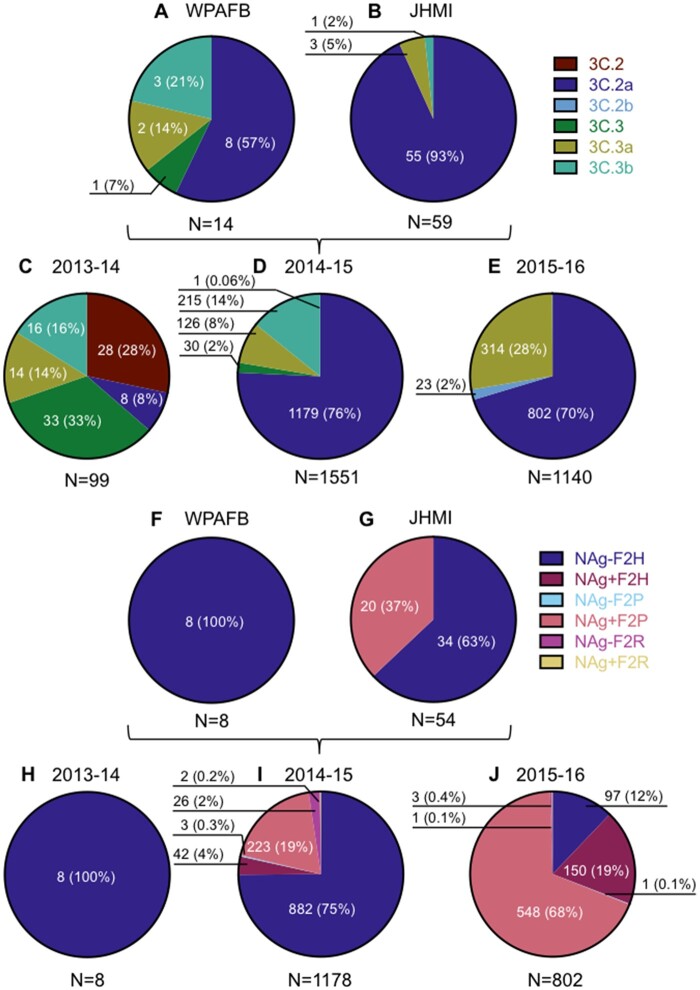
The distribution of HA clades differed between WPAFB and JHMI samples, with fifty-seven per cent of WPAFB and ninety-four per cent of JHMI samples belonging to H3N2 clade 3C.2a (Fig. 1A and B). The remaining isolates were characterized as either 3C.3a or 3C.3b, with one WPAFB isolate characterized as a 3C.3. Clade diversity among the WPAFB viruses was higher compared with the JHMI isolates (Fig. 1A and B). WPAFB samples were collected earlier in the 2014–15 influenza season therefore the timing of collection likely accounts for the differences in clade distribution between these two populations.
Figure 1.
Characterization of human isolates by clade and genotype. The number of virus isolates that belong to clades and genotypes are reported according to sequence only. The prevalence of each clade is represented in pie charts A–E. The prevalence of each 3C.2a genotype, as a percentage of clade 3C.2a isolates, is represented in charts F–J. Pie charts A and F represent isolates from WPAFB, charts B and G represent isolates from JHMI. EpiFlu isolates from the 2013-14 season are represented in C and H and those from the 2015–16 season are represented in E and J. The pie charts for 2014–15 include both EpiFlu sequences and the sequences from this study.
For context, the HA clade distributions of viral isolates with full genomes published on the EpiFlu database were determined for the northern hemisphere winter seasons of 2013–14, 2014–15, and 2015–16. H1N1 dominated during the 2013–14 season, so H3N2 isolates were less frequent and interestingly, their clade distribution was diverse (Fig. 1C). H3N2 viruses dominated during the 2014–15 influenza season and clade 3C.2a viruses were by far most common, as was previously reported (Flannery et al. 2016). JHMI and WPAFB sequences were included in the analysis for the 2014–15 season for a total of 1,551 isolates, of which seventy-six per cent (1,179) belonged to clade 3C.2a (Fig. 1D). While H1N1 viruses dominated the 2015-16 season, 3C.2a clade viruses were the most common H3N2 clade identified (Fig. 1E).
3.3. Identification and frequency of NA glycosylation and PB1-F2 amino acid 75 mutations
Further analysis of the influenza virus sequences revealed that over one-third (37%; 20/54, Fig. 1G) of JHMI isolates had a non-synonymous nucleotide mutation (ntG734A; ORF numbering throughout) in the NA segment. This mutation changed a serine (S) residue to asparagine (N), thereby creating a putative N-linked glycosylation signal in NA at AA residues 245–247 (NAg+). In addition, eighteen of the twenty isolates had another non-synonymous mutation (ntT739A), which changed the Serine residue to Threonine (T) at AA 247. The efficiency of N-linked glycosylation can be affected by the presence of an S or T in the sequon (Kasturi et al. 1995). The 245–247 NAS glycosylation site was previously observed in about a third (27%; 3/11) of 2014–15 influenza virus isolates (Mostafa et al. 2016). A glycan addition at this site reduced enzyme activity, reduced or abolished antibody binding, and reduced antibody-based protection against influenza virus infection in mice (Wan et al. 2019; Powell and Pekosz 2020). Interestingly, the WPAFB samples did not contain the NAg+ mutations (Fig. 1F). Since these samples were collected earlier in the 2014–15 season, the data may indicate that the NAg+ mutation emerged later in the season.
The only other non-synonymous mutation that occurred with greater than twenty-five per cent frequency among the 3C.2a clade JHMI isolates was an ntA318C mutation in the PB1 ORF, which was a synonymous change in the PB1 ORF but resulted in a change from Histidine (H) to Proline (P) at AA 75 (H75P) of the PB1-F2 ORF (F2P). The same twenty JHMI 3C.2a clade viruses that were NAg+ contained the F2P mutation while all other 3C.2a isolates lacked both mutations (NAg-F2H) (Fig. 1G). None of the 3C.2a clade isolates from the WPAFB had the F2P genotype (Fig. 1F). These data indicated that the NAg+ and F2P genotypes were co-selected in clade 3C.2a viruses.
All of the clade 3C.2a influenza winter season isolates from 2013 to 2016 that had WGS were characterized into six genotypes. These were determined by the presence or absence of a glycosylation motif at residues 245–247 in the NA protein and the AA residue at position 75 in the PB1-F2 protein (Fig. 1H–J). Besides Histidine and Proline, the only other AA that was present at position 75 in PB1-F2 was arginine (R). In samples from 2013 to 2014, only the NAg-F2H genotype was detected (Fig. 1H). The NAg-F2H genotype was dominant during the 2014–15 winter season (75%) with the NAg+F2P genotype being the second most common genotype (19%) (Fig. 1I). The frequency of NAg+F2P isolates grew to sixty-eight per cent of all clade 3C.2a isolates in 2015–16 while the NAg-F2H genotype shrank from seventy-five per cent to twelve per cent. The only other genotype to increase in frequency was NAg+F2H, which grew from four per cent to nineteen per cent (Fig. 1I and J). Taken together, the data indicate that the NAg+F2P genotype rapidly became dominant among clade 3C.2a viruses (compare Fig. 1I with J), while the frequency of 3C.2a viruses remained relatively similar (compare Fig. 1D with E) between the 2014–15 and 2015–16 seasons. Viruses with NAg+ accounted for eighty-seven per cent of isolates in the 2015–16 season showing that NAg+ viruses were more frequent than viruses with the PB1-F2P mutation (Fig. 1J).
3.4. Evolution of NA glycosylation and PB1-F275P
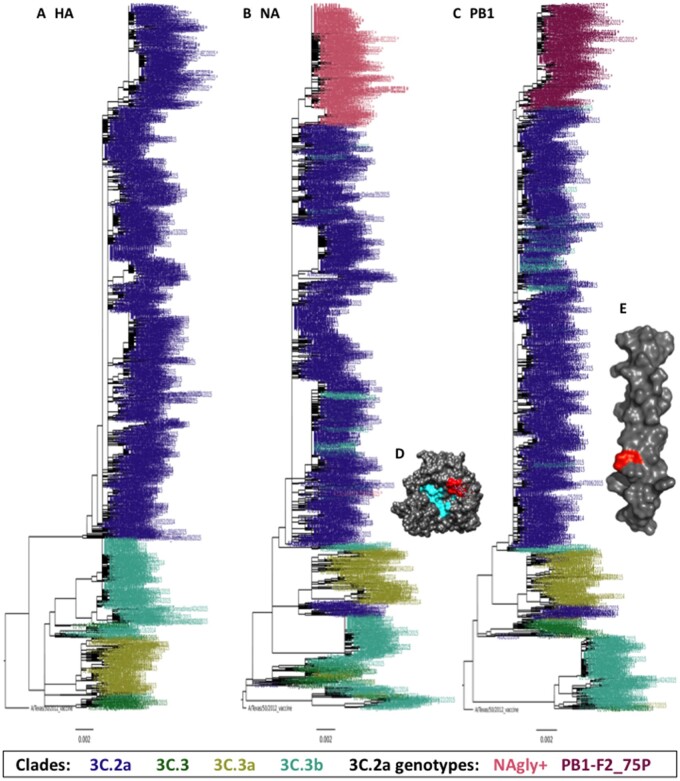
HA, NA, and PB1 phylogenetic trees with the 1487 2014–15 winter season EpiFlu, JHMI, and WPAFB isolate sequences were generated to determine the phylogenetic relationships between genotypes (Fig. 2). HA clades 3C.2a, 3C.3, 3C.3a, and 3C.3b were all represented in the HA phylogenetic tree (Fig. 2A). The branches of the NA and PB1 phylogenetic trees use the HA clade color scheme, with the exception of viruses encoding the NAg+ or F2P mutations, which use the 3C.2a genotype color scheme from Fig. 1 (salmon and burgundy in the NA and PB1 trees, respectively, Fig. 2B and C). In the NA phylogenetic tree, the large set of salmon colored branches at the top indicates that most detected NAg+ isolates arose from a common ancestor. In the PB1 phylogenetic tree, the large set of burgundy colored branches at the top similarly indicates that all detected F2P isolates arose from a common ancestor.
Figure 2.
HA, NA, and PB1 phylogenetic trees and crystal structures. (A, B, C) Each tree was made with sequences from the gene segment indicated. The sequences included 1116 from the EpiFlu database that had full length nucleotide coverage for all ORFs isolated from 10/01/2014 to 5/31/2015 plus 54 JHMI and 8 WPAFB isolates for a total of 1,178 genomes. Phylogenetic trees were made using FastTree and images were made with FigTree. Branches are colored by clade (HA) or clade and genotype (NA and PB1) as indicated in the key. (D) X-ray diffraction crystal structure of NA from A/Tanzania/205/2010 H3N2 (PDB 4GZP). The enzymatic site is colored cyan and the amino acids that encode the putative glycosylation site of NAg+ viruses are colored red. (E) Solution NMR structure of PB1-F2 C-terminal domain AA residues 50–87 from A/Puerto Rico/8/1934 (PDB 2HN8). Residue 75 is colored red. (D, E) Visualized using UCSF Chimera v1.11.1.
Mutations were mapped onto protein structures to determine their location. The putative NA glycosylation site was mapped to the X-ray crystal structure of the NA protein from influenza A/Tanzania/205/2010 (H3N2) (Zhu et al. 2012). It showed that the site was on the surface of the NA protein and directly adjacent to the NA enzymatic site (Fig. 2D). The location of PB1-F2 residue 75 was mapped to the NMR structure of the C-terminal tail of the PB1-F2 protein from influenza A/Puerto Rico/8/1934 (H1N1)(Bruns et al. 2007). It shows that the mutation site is about two-thirds of the way down the C-terminal domain (Fig. 2E).
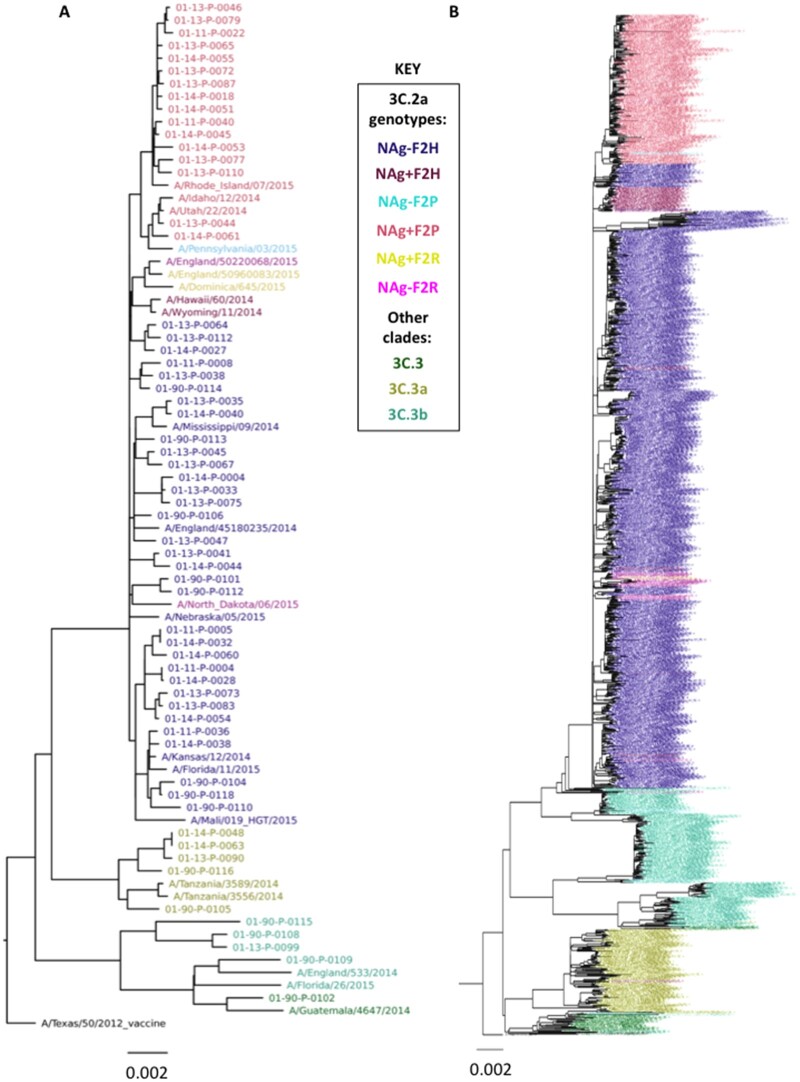
Since the NAg+ and F2P genotypes appear together in most virus strains (Fig. 1I and J), it is likely that one common ancestor led to the dominance of this genotype in 2015–16 clade 3C.2a viruses. To determine the genetic relationships between the JHMI and WPAFB viruses, we performed a phylogenetic analysis on concatenated genome segments from the JHMI and WPAFB samples (Fig. 3A). The NAg+F2P genotype viruses all clustered together from a single branch of the tree (salmon color at top of tree), indicating that a common ancestor acquired both the NAg+ and the F2P mutations. To determine if this result was the same in the general population of viruses, we performed a similar phylogenetic analysis on all H3N2 viruses with full genome sequences available from the 2014–15 and 2015–16 seasons (Fig. 3B). Again, the NAg+F2P viruses clustered together providing further support for the presence of a single common ancestor from which all the NAg+F2P genotype viruses derived.
Figure 3.
Phylogenetic trees of concatemerized influenza virus genomes. (A) Sequences from the 54 JHMI, and 8 WPAFB isolates and some reference strains were concatenated and used to construct a phylogenetic tree. (B) A second phylogenetic tree of concatenated genes was generated using the 1,178 2014–15 isolates used in Fig. 2 plus all 2015–16 isolates from the EpiFlu database. Trees were made with the same methods for Fig. 2. Branches are colored by clade or genotype as indicated in the key.
The locations of the NAg-F2P viruses (cyan branches) within the NAg+F2P viruses (salmon branches) indicate that the NAg-F2P genotype likely arose from mutations that reversed the NAg+ to the NAg- genotype in the NAg+F2P genotype lineage. There was no evidence that the F2P mutation was gained by NAg- viruses. The group of NAg+F2H viruses (plum) that branched from the trunk of the tree before the NAg+F2P group, and the other NAg+F2H virus branch locations, scattered among NAg-F2H branches, suggests that the NAg+ mutation was selected numerous times and that in one case, it was selected slightly before acquisition of the H75P mutation.
NAg+F2R isolates seem to have been selected in the opposite order. The branching of the three NAg+F2R viruses (yellow) among a small group of NAg-F2R viruses (magenta) indicates that a subset of F2R viruses created a background in which NAg+ was selected, but this group was short-lived and did not lead to the emergence of the NAg+F2P genotype. In the PB1 phylogenetic tree, all F2P and all F2R viruses shared a single node (Fig. 3B), indicating that, evolutionarily, these genes were closely related.
The phylogenetic analysis suggests that the two mutations of the NAg+F2P genotype clade 3C.2a viruses descended from a common ancestor. Although the presence of one as a passenger mutation linked with the other cannot be formally excluded, the expansion of NAg+F2P genotype viruses suggests selection according to a fitness advantage of that genotype over other viruses within the 3C.2a clade. Whether both mutations contributed to this fitness advantage remains unclear.
3.5. Virus isolation on hNEC cultures was more successful than on MDCK cells
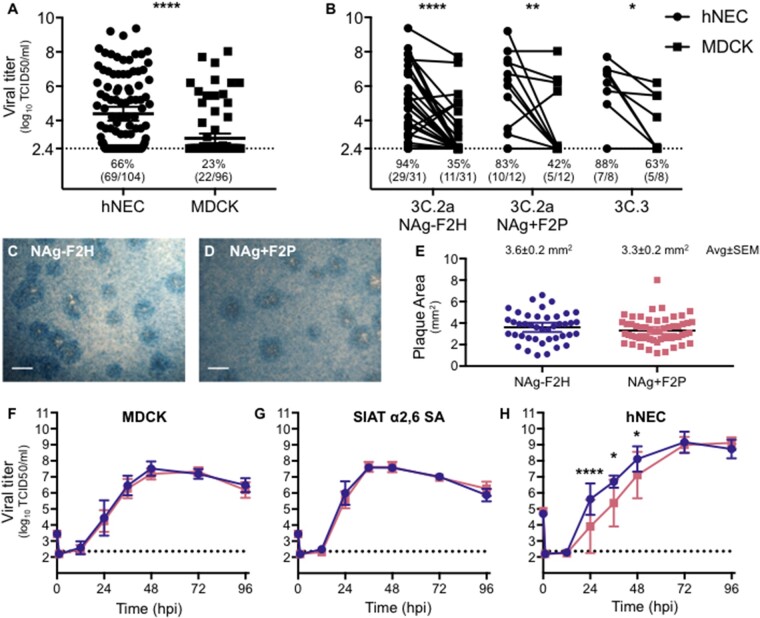
Virus isolation was attempted from 104 of the JHMI and WPAFB NPS specimens. Primary differentiated hNEC cultures were inoculated with all 104 NPS specimens while MDCK cells were inoculated with a subset of ninety-six specimens. Specimen use was dictated by available sample volume. Infectious virus titers were determined by TCID50 assay for supernatants collected from hNEC or MDCK cells at four days post infection (dpi). Inoculation of hNEC cultures led to isolation of viruses from sixty-nine (66%) of the NPS specimens while inoculation of MDCK cells only led to isolation of viruses from twenty-two (23%) of the specimens (Fig. 4A). Isolation using hNEC cultures had a nearly three-fold higher success rate. The median infectious virus titers were 3.6 log10 TCID50/ml from hNEC cultures and 2.4 log10 TCID50/ml, indicating the clinical isolates grew to higher infectious virus titers in hNEC cultures compared to MDCK cells (Fig. 4A). The higher virus isolation frequency and higher viral titers achieved with hNEC cultures were consistently observed, irrespective of the HA clade or genotype of virus isolated (Fig. 4B). These data indicate that primary hNEC cultures, which more accurately represent the cells that influenza viruses encounter in the upper respiratory tract, are more efficient than MDCK cells at isolating virus from clinical specimens.
Figure 4.
H3N2 virus isolation and characterization. (A) Virus isolation was attempted with 104 NPS specimens on hNEC cultures and 96 NPS specimen on MDCK cell cultures. Apical washes and supernatants were collected from hNEC and MDCK cell cultures, respectively, at 4 dpi and infectious virus titers determined by TCID50. Lines represent means and error bars represent 95% CIs. Statistical significance was determined with a Wilcoxon matched pairs signed rank test. (B) Comparison of hNEC and MDCK 4 dpi viral isolation titers was performed for three major genotypes. The percent and ratio of viruses that grew to levels above the limit of detection are noted below each dataset (A, B). Repeated Measures two-way ANOVA indicated that virus had no significant effect on growth and growth in MDCKs was significantly lower than that achieved in hNECs for all three virus genotypes. The limit of detection was 2.36 log10 TCID50/ml. (C–E) Plaque assays were performed by inoculating MDCK cells and incubating for 3 days, under agarose, at 37 °C. Representative images of plaques are shown for each virus (C, D). The area of over 40 plaques for each virus was determined (E) and analyzed using ImageJ. The average of plaque areas between the two viruses was determined by a two-tailed unpaired t-test. (F–H) Viruses isolated on hNECs and expanded once in MDCK cells were used to compare viral growth in MDCK cell (F), MDCK-SIAT cell (G), and hNEC (H) cultures. Cultures were inoculated with MOI 0.001, 0.001, and 0.01, respectively. Supernatants or apical washes were collected at times indicated and infectious virus determined by TCID50. N = 8 wells per virus, per cell type, per time point. Error bars represent SD. Statistical significance was determined with a repeated-measures two-way ANOVA with Bonferroni posttest correction. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
3.6. Virus growth and plaque morphology
Viruses representative of the two 3C.2a genotypes—A/Columbia/P0041/2014 (NAg-F2H) and A/Bethesda/P0055/2015 (NAg+F2P)—were expanded and assessed for differences in replication on three different cell types: MDCK cells, MDCK-SIAT cells, and hNECs. MDCK-SIAT cells overexpress the α2,6-linked sialic acid utilized by human influenza viruses as a cellular receptor (Matrosovich et al. 2003). Plaque assays were performed on MDCK cells and the two genotypes showed similar plaque morphologies (Fig. 4C and D) and areas (Fig. 4E). In low MOI growth curves, the two viruses grew to similar titers on MDCK cells (Fig. 4F) and MDCK-SIAT cells (Fig. 4G). The NAg-F2H virus grew to higher titers on hNEC cultures compared with NAg+ F2P (Fig. 4H) and this difference was statistically significant at 24, 36, and 48 hpi. The replication difference in hNECs was transient and both viruses reached equivalent peak titer at the same day post infection (72 hpi). Taken together, the data indicate that the two genotypes replicate to similar extents in immortalized cell lines but show different replication kinetics in hNEC cultures.
3.7. Clinical characteristics of patients
Clinical and demographic characteristics were retrospectively analyzed for the ninety-four JHMI influenza-positive NPS specimens (Table 2). There were fifty-four per cent males and forty-six per cent females. The racial group distribution was forty-six per cent white, twenty-nine per cent black, and eleven per cent Asian. Vaccination status was known for thirty-six per cent of patients, of whom seventy-three per cent were vaccinated. Sixty-one per cent of patients were treated as outpatients and ten per cent required inpatient care. Thirty per cent of patients required intensive care and most of those (97%) required supplemental oxygen. Five per cent of the patients died. Fifty-seven per cent of patients had one or more co-morbidities. The JHMI samples were supplemented with sixteen nasal wash specimens from the Wright-Patterson Air Force Base (WPAFB) repository. Clinical data for the WPAFB patients were not accessible.
Table 2.
Clinical and demographic characteristics of influenza positive population.
| Total JHH population N = 94 |
Virus |
|||
|---|---|---|---|---|
| Demographics | NAg-F2H N = 34 |
NAg+F2P N = 20 |
P-value | |
| Age, median (range) | 54.5 (0.25–98) | 44 (2–94) | 79 (0.25–98) | 0.124 |
| Gender female, N (%) | 43 (46%) | 16 (47%) | 12 (60%) | 0.4083 |
| Ethnicity, Hispanic, or Latino, N (%) | 4 (4.3%) | 3 (8.8%) | 1 (5%) | >0.99 |
| Race, N (%) | 0.63 | |||
| White | 43 (46%) | 13 (38%) | 11 (55%) | |
| Black or African American | 27 (29%) | 10 (29%) | 6 (30%) | |
| Asian | 10 (11%) | 3 (9%) | 1 (5%) | |
| Other | 14 (15%) | 5 (15%) | 1 (5%) | |
| Influenza vaccination status, N/total known status (%) | ||||
| Vaccinated | 24/33 (73%) | 4/10 (40%) | 6/7 (86%) | 0.057 |
| Co-morbidities, N (%) | 0.1 | |||
| None | 39 (41%) | 17 (50%) | 5 (25%) | |
| One | 23 (24%) | 8 (24%) | 10 (50%) | |
| Multiple | 32 (34%) | 9 (26%) | 5 (25%) | |
| Disease severity, N (%) | ||||
| Pneumonia | 14 (15%) | 3 (9%) | 4 (20%) | 0.4076 |
| Supplemental oxygen | 27 (29%) | 9 (32%) | 7 (44%) | 0.5227 |
| Admitted | 37 (39%) | 32% (11/34) | 50% (10/20) | 0.2527 |
| ICU | 9 (10%) | 3 (9%) | 2 (10%) | >0.99 |
| Hospital length of stay, median (range) | 5 (1–83) | 7 (1–14) | 5 (1–13) | 0.4406 |
| Death | 5 (5%) | 1 (3%) | 2 (10%) | 0.5477 |
Categorical variables assessed by Fisher’s exact or Chi-sq. Continuous variables by Mann–Whitney. Pneumonia as diagnosed by radiological findings.
3.8. Viruses from the 3C.2a NAg+F2P genotype associated with breathing difficulties
The clinical and demographic information from twenty patients infected with the NAg+F2P genotype was compared to the thirty-four patients infected with clade 3C.2a NAg-F2H genotype viruses (Table 3). The only clinical factors that differed between the patient populations was an increased reporting of coughing and shortness of breath in individuals infected with the NAg+F2P genotype. A multivariate logistic regression analysis was performed to determine if any underlying co-morbidities were responsible for the correlations. We found no association of co-morbidities with increased coughing or shortness of breath.
Table 3.
Symptom comparison between individuals infected with NAg-F2H and NAg+F2P genotypes of 3C.2a H3N2 viruses.
| Virus |
|||
|---|---|---|---|
| Symptoms, % (N reported/N total with reported) | NAg-F2H N = 34 |
NAg+F2P N = 20 |
p-value |
| Fever | 91% (31/34) | 71% (12/17) | 0.09 |
| Cough | 78% (25/32) | 100% (19/19) | 0.035* |
| Headache | 43% (9/21) | 40% (4/10) | >0.99 |
| Sore throat | 44% (8/18) | 38% (3/8) | >0.99 |
| Runny nose | 86% (18/21) | 73% (11/15) | 0.42 |
| Body aches | 91% (10/11) | 83% (5/6) | 1 |
| Shortness of breath | 38% (8/21) | 73% (11/15) | 0.049* |
| Wheezing | 35% (7/20) | 67% (8/12) | 0.1437 |
| Chills | 40% (8/20) | 33% (3/9) | >0.99 |
| Fatigue | 60% (12/14) | 33% (3/9) | 0.68 |
| Nausea/vomiting | 9% (2/22) | 21% (3/14) | 0.36 |
Categorical variables assessed by Fisher’s exact or Chi-sq. * indicates p>0.05
4. Discussion
An H3N2 influenza A virus genotype, NAg+F2P, within HA clade 3C.2a with a glycosylation site on the rim of the NA sialidase pocket and the mutation H75P in its PB1-F2 protein was identified. Viruses with the NAg+F2P genotype were first detected in the 2014–15 influenza virus season and they outcompeted the NAg-F2H genotype in the 2015–16 season. This selection occurred despite NAg+F2P viruses showing reduced infectious virus production in hNEC cultures.
The main function of the influenza NA protein is to cleave sialic acid. Cleavage of sialic acid from cellular receptors leads to release of new virions and, alternatively, cleavage from decoy receptors allows virions to breach the mucosal barrier (Matrosovich and Klenk 2003; Blumenkrantz et al. 2013). Glycosylation of NA can assist in protein folding, prevent proteolytic cleavage, mask antigenic regions, and regulate the enzyme’s catalytic activity (Wu et al. 2009; Chen et al. 2012). Antigenic escape epitopes of human seasonal N2 NA proteins have been mapped using monoclonal antibodies and molecular biology and mostly localize around the rim of the sialidase pocket (Lentz et al. 1984; Gulati et al. 2002). NA drift has also been linked to evasion of human polyclonal antibodies (Sandbulte et al. 2011). Recently, the NAg+ mutation was demonstrated to reduce NA enzymatic activity, alter virus replication fitness on hNECs, and contribute to H3N2 NA antigenic drift (Wan et al. 2019; Powell and Pekosz 2020). It follows that the AA mutations S245N and S245T, which encode a glycosylation site near the sialidase rim of the 3C.2a NA, were selected to evade human immunity despite a detrimental effect on virus replication in hNEC cultures. While both the Co14 and Be15 viruses used in this study had lost the HA glycosylation at position 160—which defines the 3C.2a clade—subsequent studies indicated the NAgly+ mutation also attenuates the replication of H3 viruses with a glycan moiety at this position (Powell and Pekosz 2020).
PB1-F2 affects the host innate immune response as well as virus polymerase activity and transmission. The effects of PB1-F2 differ depending on protein sequence, including its length, and the cell type or animal infected. In epithelial cells, PB1-F2 can interact with mitochondrial membrane proteins ANT3 and VDAC1 and induce apoptosis by forming ion pores that release cytochrome c (Zamarin et al. 2005). PB1-F2 can disrupt MAVS signaling by interacting with MAVS, CALCOCO2, TBK1, or IRF3 thus inhibiting transcription of type I IFNs. Interactions with CALCOCO2 promote production of inflammatory cytokines through the TRAF6/NFB pathway. Alternatively, the NFkB pathway can be inhibited when PB1-F2 interacts with IKKß. PB1-F2 can also enhance viral polymerase activity. In macrophages, PB1-F2 interacts with the NLRP3 inflammasome, activates caspases, and leads to the secretion of pro-inflammatory cytokines (reviewed in Kamal et al. 2018). PB1-F2 can affect morbidity and mortality in animal models (Alymova et al. 2011). The full protein or specific residues of PB1-F2 are important for transmission in chickens, turkeys, and ferrets (Deventhiran et al. 2016; James et al. 2016; Zanin et al. 2017). AA at position 75 has undergone changes in human seasonal H3N2 viruses. The 1968 pandemic H3N2 PB1 segment was of avian origin and had R75. In 1987, the PB1-F2 mutation R75H became fixed in human circulating H3N2 influenza viruses (Alymova et al. 2011). The seventy-fifth AA remained H until the 2014–15 season after which, as described here, the H75P mutation became dominant. AA residue 75 is located in the C-terminal portion of the PB1-F2 protein, which plays an important role in mitochondrial localization (Gibbs et al. 2003; Yamada et al. 2004) and is one of four inflammation-associated residues (Alymova et al. 2011). In the influenza virus strain A/Wuhan/359/1995, the PB1-F2 mutation H75R increased pathogenicity in mice. The lungs of mice infected with this virus also showed a trend toward reduced viral titers compared to wild type virus (Alymova et al. 2011).
Co-selection of the NA and PB1-F2 mutations suggests but does not prove that both may contribute to virus fitness during human transmission. It is clear that the NA glycosylation allowed virus to evade antibodies at the cost of enzyme activity and reduced replication efficiency (Wan et al. 2019; Powell and Pekosz 2020). Since the NAg+ mutation was selected before the PB1-F2 mutation, it is worth considering if the PB1-F2 mutation was a passenger of the gene cassette or if it contributed to viral fitness. Experiments with engineered viruses that differ only by the PB1-F2 mutation H75P should be performed to determine its role in increasing transmission.
Primary cells are known to exert more biologically relevant pressures on replicating influenza viruses than the pressures exerted by transformed or immortalized cell lines. Numerous studies used primary cells to show differences in replication of engineered and isolated influenza viruses (Danzy et al. 2014; Elderfield et al. 2014; Fischer et al. 2015; Gerlach et al. 2017; Wohlgemuth et al. 2017; Bui et al. 2019; Monteagudo et al. 2019). Here, we report that hNECs were more permissive for growth of virus isolates than MDCK cell cultures. Indeed, isolation of influenza viruses in hNEC cultures allowed expansion of viruses from two-thirds of positive specimens, almost three times more than the quarter that expanded after MDCK cell culture. This efficient isolation allows more virus genotypes to be studied earlier after their detection than if generating engineered viruses were necessary. We also compared the growth in hNEC cultures of two virus isolates of the NAg-F2H and NAg+F2P genotypes. This comparison showed a difference in virus fitness, but the less fit virus was the one that dominated during natural human transmission in the following season. This suggests that some other advantage was afforded to the viruses containing these mutations, one of which is clearly the escape from preexisting immunity targeting the NA protein. While the NAg+F2P might have reduced in vitro fitness, in the presence of antibodies that inhibit NA activity, this virus could have an advantage over the NAg-F2H virus. In analysis of the NAg+ mutation expressed alone in a recombinant virus, the NAg+ virus was able to evade immunity better than NAg- and replicated in the presence of monoclonal and polyclonal antibodies that recognized N2 protein (Powell and Pekosz 2020).
The 2014–15 Northern hemisphere influenza season had an unusually high number of cases and an increase in disease severity compared to the five preceding years (CDC 2015). Overall vaccine efficacy was nineteen per cent (Zimmerman et al. 2016). This was attributed to antigenic drift of the H3N2 HA protein mediated by a new glycosylation site at AA 160 which partially masked neutralizing antibody epitopes (Chambers et al. 2015; Xie et al. 2015). The NAg-F2H and NAg+F2P genotypes both had the HA mutations associated with antigenic drift, but the eventual dominance of the NAg+F2P genotype suggests a selective advantage over the NAg-F2H genotype.
Associations between different human seasonal influenza A genotypes and altered disease phenotypes have been sought (Galiano et al. 2012), but rarely detected. Within human H1N1 viruses, recent pandemic strains with the HA mutation D225G associated with infection of the lower airways (Iovine et al. 2015), likely due to the increased binding of α2,3-linked sialic acid, and increased disease (van Doremalen et al. 2011). However, these viruses did not outcompete D225 encoding viruses, suggesting that the mutation did not improve transmission. The number of HA clades and frequency of isolates in each clade differ between seasons (Klein et al. 2018). The D225G and NAg+F2P associations with increased pathogenicity or symptoms appeared after a pandemic and after an antigenic drift event that each led to viral population dominance by a single genotype and its descendants. Therefore, it seems that associations between virus genotype and human disease phenotype are easier to detect and pin to specific mutations when genetic variation is small.
Acknowledgements
We thank Samantha Lycett (University of Edinburgh, UK) and Colin Russell (Cambridge University, UK) for sharing their phylogenetic tree branch coloring protocols as well as Monica Galiano (Public Health England) for the discussion about correlations between genotype and phenotype. We are grateful to Anne Hamacher-Brady and Nathan Brady for assistance with microscopy and the members of the Pekosz laboratory, Sabra Klein Laboratory and Kimberley Davis laboratory for helpful discussions. This work was supported by the NIH/NIAID Center of Excellence in Influenza Research and Surveillance contract HHS N272201400007C (Johns Hopkins University), T32 AI007417 and the Richard Eliasberg Family Foundation.
Data Availability Statement
All data is available and present either in the publication or in the indicated databases.
Conflict of interest: None declared.
References
- Alymova I. V. et al. (2011) ‘Immunopathogenic and Antibacterial Effects of H3N2 Influenza a Virus PB1-F2 Map to Amino Acid Residues 62, 75, 79, and 82’, Journal of Virology, 85: 12324–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appiah G. D. et al. Centers for Disease Control and Prevention (CDC) (2015) ‘Influenza Activity - United States, 2014-15 Season and Composition of the 2015-16 Influenza Vaccine’. Morbidity and Mortality Weekly Report. 64, 583–90. [PMC free article] [PubMed] [Google Scholar]
- Bedford T. et al. (2015) ‘Global Circulation Patterns of Seasonal Influenza Viruses Vary with Antigenic Drift’, Nature, 523: 217–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenkrantz D. et al. (2013) ‘The Short Stalk Length of HPAI H5N1 Influenza Neuraminidase Limits Transmission of Pandemic H1N1 Virus in Ferrets’, Journal of Virology, 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broberg E. et al. (2015) ‘Start of the 2014/15 Influenza Season in Europe: Drifted Influenza A(H3N2) Viruses Circulate as Dominant Subtype’, Eurosurveillance, 20: 1–5. [DOI] [PubMed] [Google Scholar]
- Bruns K. et al. (2007) ‘Structural Characterization and Oligomerization of PB1-F2, a Proapoptotic Influenza a Virus Protein’, Journal of Biological Chemistry, 282: 353–63. [DOI] [PubMed] [Google Scholar]
- Bui C. H. T. et al. (2019) ‘Tropism of Influenza B Viruses in Human Respiratory Tract Explants and Airway Organoids’, European Respiratory Journal, 54(2):1900008. doi: 10.1183/13993003.00008 [DOI] [PubMed] [Google Scholar]
- CDC (2015) ‘Summary of the 2014-2015 Infuenza Season [WWW Document]. <https://www.cdc.gov/flu/pastseasons/1415season.htm> accessed 26 Feb 2020).
- Chambers B. S. et al. (2015) ‘Identification of Hemagglutinin Residues Responsible for H3N2 Antigenic Drift during the 2014-2015 Influenza Season’, Cell Reports, 12: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W. et al. (2012) ‘The Evolutionary Pattern of Glycosylation Sites in Influenza Virus (H5N1) Hemagglutinin and Neuraminidase’, PLoS One, 7: e49224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danzy S. et al. (2014) ‘Mutations to PB2 and NP Proteins of an Avian Influenza Virus Combine to Confer Efficient Growth in Primary Human Respiratory Cells’, Journal of Virology, 88: 13436–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deventhiran J. et al. (2016) ‘PB1-F2 Protein Does Not Impact the Virulence of Triple-Reassortant H3N2 Swine Influenza Virus in Pigs but Alters Pathogenicity and Transmission in Turkeys’, Journal of Virology, 90: 222–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donadeu M. et al. (2015) ‘Review of the 2014-2015 Influenza Season in the Northern Hemisphere’, Relevé Épidémiologique Hebdomadaire, 90: 281–96. [PubMed] [Google Scholar]
- Elderfield R. A., MOSAIC Investigators. et al. (2014) ‘Accumulation of Human-Adapting Mutations during Circulation of A(H1N1)pdm09 Influenza Virus in Humans in the United Kingdom’, Journal of Virology, 88: 13269–83., [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epperson S. et al. (2014) ‘Influenza activity - United States, 2013-14 Season and Composition of the 2014-15 Influenza Vaccines’, Morbidity and Mortality Weekly Report, 63: 483–90. [PMC free article] [PubMed] [Google Scholar]
- Fischer W. A. et al. (2015) ‘Restricted Replication of the Live Attenuated Influenza a Virus Vaccine during Infection of Primary Differentiated Human Nasal Epithelial Cells’, Vaccine, 33: 4495–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannery B. et al. (2016) ‘Enhanced Genetic Characterization of Influenza A(H3N2) Viruses and Vaccine Effectiveness by Genetic Group, 2014-2015’, Journal of Infectious Disease, 2015: 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forero A. et al. (2017) ‘Evaluation of the Innate Immune Responses to Influenza and Live-Attenuated Influenza Vaccine Infection in Primary Differentiated Human Nasal Epithelial Cells’, Vaccine, 35: 6112–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galiano M. et al. (2012) ‘Fatal Cases of Influenza A(H3N2) in Children: Insights from Whole Genome Sequence Analysis’, PLoS One, 7: e33166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach T. et al. (2017) ‘pH Optimum of Hemagglutinin-Mediated Membrane Fusion Determines Sensitivity of Influenza a Viruses to the Interferon-Induced Antiviral State and IFITMs’, Journal of Virology, 91: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs J. S. et al. (2003) ‘The Influenza a Virus PB1-F2 Protein Targets the Inner Mitochondrial Membrane via a Predicted Basic Amphipathic Helix That Disrupts Mitochondrial Function’, Journal of Virology, 77: 7214–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham M. L. et al. (2010) ‘Tyrosines in the Influenza a Virus M2 Protein Cytoplasmic Tail Are Critical for Production of Infectious Virus Particles’, Journal of Virology, 84: 8765–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati U. et al. (2002) ‘Antibody Epitopes on the Neuraminidase of a Recent H3N2 Influenza Virus (a/Memphis/31/98)’, Journal of Virology, 76: 12274–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huzly D. et al. (2016) ‘Influenza a Virus Drift Variants Reduced the Detection Sensitivity of a Commercial Multiplex Nucleic Acid Amplification Assay in the Season 2014/15’, Archives of Virology, 161: 2417–23. [DOI] [PubMed] [Google Scholar]
- Iovine N. M. et al. (2015) ‘Severity of Influenza a(H1N1) Illness and Emergence of D225G Variant, 2013–14 Influenza Season, Florida, USA’, Emerging Infectious Diseases, 21: 664–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James J. et al. (2016) ‘Influenza a Virus PB1-F2 Protein Prolongs Viral Shedding in Chickens Lengthening the Transmission Window’, Journal of General Virology, 97: 2516–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamal R. P., Alymova I. V., York I. A. (2018) ‘Evolution and Virulence of Influenza a Virus Protein PB1-F2’, International Journal of Molecular Medical Science, 19: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasturi L. et al. (1995) ‘The Hydroxy Amino Acid in an Asn-X-Ser/Thr Sequon Can Influence N-Linked Core Glycosylation Efficiency and the Level of Expression of a Cell Surface Glycoprotein’, Journal of Biological Chemistry, 270: 14756–61. [DOI] [PubMed] [Google Scholar]
- Klein E. Y. et al. (2018) ‘Stability of the Influenza Virus Hemagglutinin Protein Correlates with Evolutionary Dynamics’, mSphere, 3: e00554–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentz M. R. et al. (1984) ‘Sequence of the Neuraminidase Gene of Influenza Virus a/Tokyo/3/67 and Previously Uncharacterized Monoclonal Variants’, Virology, 135: 257–65. [DOI] [PubMed] [Google Scholar]
- Luk J., Gross P., Thompson W. W. (2001) ‘Observations on Mortality during the 1918 Influenza Pandemic’, Clinical Infectious Diseases, 33: 1375–8. [DOI] [PubMed] [Google Scholar]
- Matrosovich M., Klenk H.-D. (2003) ‘Natural and Synthetic Sialic Acid-Containing Inhibitors of Influenza Virus Receptor Binding’, Reviews in Medical Virology, 13: 85–97. [DOI] [PubMed] [Google Scholar]
- Matrosovich M. et al. (2003) ‘Overexpression of the Alpha-2,6-Sialyltransferase in MDCK Cells Increases Influenza Virus Sensitivity to Neuraminidase Inhibitors’, Journal of Virology, 77: 8418–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley J. W. et al. (2015) Report prepared for the WHO annual consultation on the composition of influenza vaccine for the Northern Hemisphere 2015/16. WHO Influ. Cent. London.
- McCown M. F., Pekosz A. (2005) ‘The Influenza a Virus M2 Cytoplasmic Tail is Required for Infectious Virus Production and Efficient Genome Packaging’, Journal of Virology, 79: 3595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteagudo P. L. et al. (2019) ‘Differential Modulation of Innate Immune Responses in Human Primary Cells by Influenza a Viruses Carrying Human or Avian Nonstructural Protein 1’, Journal of Virology, 94: 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morens D. M. et al. (2010) ‘The 1918 Influenza Pandemic: Lessons for 2009 and the Future’, Critical Care Medicine, 38: e10–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafa A. et al. (2016) ‘Phylogenetic Analysis of Human Influenza a/H3N2 Viruses Isolated in 2015 in Germany Indicates Significant Genetic Divergence from Vaccine Strains’, Archives of Virology, 161: 1505–15. [DOI] [PubMed] [Google Scholar]
- Pebody R. G. et al. (2015) ‘Low Effectiveness of Seasonal Influenza Vaccine in Preventing Laboratory-Confirmed Influenza in Primary Care in the United Kingdom : 2014/15 Mid – Season Results’, Eurosurveillance, 20: 1–7. [PubMed] [Google Scholar]
- Powell H., Pekosz A. (2020) ‘Neuraminidase Antigenic Drift of H3N2 Clade 3c.2a Viruses Alters Virus Replication, Enzymatic Activity and Inhibitory Antibody Binding’, PLoS Pathogens, 16: e1008411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan M. et al. (2007) ‘Sinonasal Epithelial Cell Expression of Toll-like Receptor 9 is Decreased in Chronic Rhinosinusitis with Polyps’, American Journal of Rhinology, 21: 110–6. [DOI] [PubMed] [Google Scholar]
- Reed L., Muench H. (1938) ‘A Simple Method of Estimating Fifty per Cent Endpoints’, American Journal of Epidemiology, 27: 493–7. [Google Scholar]
- Sandbulte M. R. et al. (2011) ‘Discordant Antigenic Drift of Neuraminidase and Hemagglutinin in H1N1 and H3N2 Influenza Viruses’, Proceedings of the National Academy of Sciences, 108: 20748–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowronski D. M. et al. (2016) ‘A Perfect Storm: Impact of Genomic Variation and Serial Vaccination on Low Influenza Vaccine Effectiveness during the 2014-2015 Season’, Clinical Infectious Diseases, 63: 21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowronski D. M. et al. (2010) ‘Prevalence of Seroprotection against the Pandemic (H1N1) Virus after the 2009 Pandemic’, CMAJ, 182: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucker K. M. et al. (2015) ‘Haemagglutinin Mutations and Glycosylation Changes Shaped the 2012/13 Influenza A(H3N2) Epidemic, Houston, Texas’, Eurosurveillance, 20: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T. et al. (2019) ‘Clustering Single-Cell RNA-Seq Data with a Model-Based Deep Learning Approach’, Nature Machine Intelligence, 1: 191–8. [Google Scholar]
- van Doremalen N. et al. (2011) ‘A Single Amino Acid in the HA of pH1N1 2009 Influenza Virus Affects Cell Tropism in Human Airway Epithelium, but Not Transmission in Ferrets’, PLoS One, 6: e25755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H. et al. (2019) ‘The Neuraminidase of A(H3N2) Influenza Viruses Circulating since 2016 is Antigenically Distinct from the a/Hong Kong/4801/2014 Vaccine Strain’, Nature Microbiology, 4: 2216–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlgemuth N. et al. (2017) ‘The M2 Protein of Live, Attenuated Influenza Vaccine Encodes a Mutation That Reduces Replication in Human Nasal Epithelial Cells’, Vaccine, 35: 6691–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z. L. et al. (2009) ‘Active 1918 Pandemic Flu Viral Neuraminidase Has Distinct N-Glycan Profile and is Resistant to Trypsin Digestion’, Biochemical and Biophysical Research Communications, 379: 749–53. [DOI] [PubMed] [Google Scholar]
- Xie H. et al. (2015) ‘H3N2 Mismatch of 2014-15 Northern Hemisphere Influenza Vaccines and Head-to-Head Comparison between Human and Ferret Antisera Derived Antigenic Maps’ Science Report, 5: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H. et al. (2004) ‘Mitochondrial Targeting Sequence of the Influenza a Virus PB1-F2 Protein and Its Function in Mitochondria’, FEBS Letters, 578: 331–6. [DOI] [PubMed] [Google Scholar]
- Yamamoto H. et al. (2011) ‘Dual Role of the Receptor Tom20 in Specificity and Efficiency of Protein Import into Mitochondria’, Proceedings of the National Academy of Sciences, 108: 91–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamarin D. et al. (2005) ‘Influenza Virus PB1-F2 Protein Induces Cell Death through Mitochondrial ANT3 and VDAC1’, PLoS Pathogens, 1: e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanin M. et al. (2017) ‘Molecular Basis of Mammalian Transmissibility of Avian H1N1 Influenza Viruses and Their Pandemic Potential’, Proceedings of the National Academy of Sciences, 114: 11217–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B. et al. (2009) ‘Single-Reaction Genomic Amplification Accelerates Sequencing and Vaccine Production for Classical and Swine Origin Human Influenza a Viruses’, Journal of Virology, 83: 10309–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X. et al. (2012) ‘Influenza Virus Neuraminidases with Reduced Enzymatic Activity That Avidly Bind Sialic Acid Receptors’, Journal of Virology, 86: 13371–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman R.K. et al. (2016). ‘2014-2015 Influenza Vaccine Effectiveness in the United States by Vaccine Type’. Clinical Infectious Diseases. 63, 1564–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]