Abstract
Background
Resistance to clarithromycin and levofloxacin in Helicobacter pylori which resulted in treatment failures has become a major challenge for physicians worldwide. The resistance is mainly mediated by mutations in a specific domain of the 23S rRNA, gyrA and gyrB genes for clarithromycin and levofloxacin respectively. Hence in this study, we aimed to investigate the current status of H. pylori resistance in our hospital to these two antibiotics based on the molecular approach.
Materials and Methods
Gastric biopsy samples were obtained from treatment-naïve patients. Bacterial genomic DNA was extracted using a commercial kit and continued with DNA amplification using polymerase chain reaction (PCR) with specific primers. The PCR amplicons were subjected to sequencing on 23S rRNA gene targeting nucleotide positions at 2,146, 2,147, 2,186 and amino acids at gyrA positions 87 and 91 and gyrB positions 436, 438, 481, 484 to investigate the possible mutations or polymorphisms of genes that lead to clarithromycin and levofloxacin resistance respectively.
Results
Sixty-one urease-positive gastric biopsy samples were studied. The findings revealed the primary resistance rates to clarithromycin was 14.8% and to levofloxacin was 3.3% in our current scenario based on detection of reported resistance-related mutations of A2147G and D91N in 23S rRNA and gyrA genes, respectively. Interestingly, we found a high rate of silent mutations of the gyrA codon 87Asn (32.8%, 20/61) and two polymorphisms of the gyrB D481E (16.4%, 10/61) and R484K (21.3%, 13/61). The role of these polymorphisms in gyrB remained to be elucidated whether the levels of levofloxacin resistance are related to the position/amino acid.
Conclusion
The primary resistance rate of H. pylori to clarithromycin has increased compared to the previous report in Malaysia. Therefore, molecular screening could aid and is important for the selection of antibiotics for H. pylori eradication therapies.
Keywords: 23S rRNA, gyrA, gyrB, H. pylori, Malaysia, Primary resistance
Introduction
Helicobacter pylori susceptibility to antibiotics is crucial to the treatment success of H. pylori eradication treatment regimens. Rising H. pylori resistance to clarithromycin has been implicated as the cause of the declining success of the 1-week standard triple therapy which incorporates clarithromycin in the regimen (Buzas & Jozan, 2006; Graham & Fischbach, 2010). Graham and co-workers have persuasively opined that susceptibility testing should be carried out in every case of H. pylori infection (Graham & Shiotani, 2008; Graham & Dore, 2016).
Although culturing of the bacterium for susceptibility testing is widely carried out, the success of culture is low even in specialized laboratories (Mégraud et al., 2015). Molecular methods of detecting resistance to antibiotics, on the other hand, is a more robust technique that does not require stringent transport and laboratory requirements for the primary culture of the bacterium. These techniques have now been established with recognized mutations and provide a practical and viable alternative to the traditional methods of testing susceptibility using the culture of the bacterial strains.
Antimicrobial resistance in H. pylori is primarily due to the acquisition of point mutations. Thus far, point mutations in 23S rRNA, particularly two specific adjacent nucleotide positions, 2,142 (A to G/C) and 2,143 (A to G), are responsible for most clarithromycin resistance cases observed (Raymond et al., 2007). Using ribosome isolation and erythromycin assays as well as transformation experiments, the role of these two mutations is confirmed involved in the CLA resistance (Binh et al., 2014; Occhialini et al., 1997; Wang & Taylor, 1998; Wang et al., 2020).
For levofloxacin, published data showed that resistance is due to point mutations in the quinolones resistance-determining region (QRDR) of the gyrA mainly at codons N87 or D91 (Garcia et al., 2012; Nishizawa & Suzuki, 2014). Using transformation assays, Kim et al. (2005) and Rimbara et al. (2012) demonstrated that resistant transformed H. pylori cells harboured the mutation at positions 87 and 91 showed high-level resistance to fluoroquinolone. Besides, the study from Rimbara and co-workers also reported one mutation located closed to QRDR of the gyrB at codon E463 conferred norfloxacin resistance in H. pylori. In 2006, Miyachi et al. have reported E463 mutation in gyrB were detected in highly levofloxacin resistant strains, but occurred together with gyrA mutations.
Background testing of H. pylori resistance to antibiotics has consistently shown a low prevalence of resistance to clarithromycin and levofloxacin in Malaysia. In the most recent studies, Teh et al. (2014) showed a primary resistance rate 6.8% to both clarithromycin of and levofloxacin. Hanafiah et al. (2019) showed a primary resistance rate of 1.2% and 17.1% to clarithromycin and fluoroquinolones.
In this report, we present our current prevalence of antibiotic resistance to clarithromycin and levofloxacin using molecular-based methods in our local population in Malaysia.
Materials and Methods
Gastric biopsy samples collection
The study was performed according to the International Conference on Harmonisation-Good Clinical Practice (ICHC-GCP) guidelines from 1st June 2019–31st May 2020. The study was approved by the ethics committee (MREC ID No: 2019101-7882) of the University of Malaya Medical Centre. The written informed consents were obtained from all participants before gastric biopsy samples were collected from the antrum and body of treatment-naïve patients who were tested H. pylori-positive based on the rapid urease test at the endoscopy unit of the University of Malaya Medical Centre. The biopsy samples from the antrum and body of the stomach for each patient were put together and stored in Stuart’s transport medium and kept in a refrigerator at 4 °C until they were sent to the molecular laboratory for analysis.
Bacterial DNA extraction
Gastric biopsy samples (mixture of antrum and body) obtained were cut into small pieces and bacterial genomic DNA was extracted using QIAmp DNA mini kit (Qiagen, Germantown, MD, USA) according to the manufacturer’s protocol. The extracted DNA was quantitated by using a nano spectrophotometer (Implen, Germany) and stored at −20 °C.
Molecular detection of known mutations/polymorphisms in 23S rRNA, gyrA and gyrB
According to the reference sequence 26,695 (GenBank accession no. AE000511.1), the primer pairs were designed to detect point mutations/polymorphisms. To detect 23S rRNA gene mutations at positions of 2,146, 2,147 (formerly known as 2,142 and 2,143) and polymorphism at position 2,186 (formerly known as 2,182), forward primer: 5′-ATGAAGCGTTGAATTGAAGC-3′ and reverse primer: 5′-CAACTCTCTTACACTCAGTAAGGC-3′ were employed. To detect mutations associate with levofloxacin resistance, amino acid substitutions at positions 87 (N to L/I/A/K) and 91(D to G/N/A/Y/H) in gyrA were carried out using forward primer: 5′CCGAGTTCCACCCATTAG-3′ and reverse primer: 5′ATTTCACTCATCGCGTCTC-3′. For gyrB, forward primer: 5′CAATTCGTTAAAGACAGCG-3′ and reverse primer: 5′TTGTAAAGAGGGGCTTGAG-3′ were used to detect mutation at position 463 (E to K/G) and polymorphisms 438 (F to S), 481(D to E/N) and 484 (R to K). DNA amplification was performed using polymerase chain reaction (PCR) with a mixture that contained 60 µL of 10 µM of forward primer, 10 µM of reverse primer, DNA template and RNase-free water using DreamTaq polymerase (Thermo Scientific, Waltham, MA, USA). The PCR mixture was denatured at 95 °C for 3 mins, followed by 35 cycles of 95 °C denaturation for 30 s, annealing duration for 30 s (annealing temperature: 23S rRNA—60 °C, gyrA—60 °C, gyrB—58 °C), and 72 °C extension for 1 min and a final extension cycle, 72 °C for 7 min. The amplicons were electrophoresed on a 1% (w/v) agarose gel, purified and sent for sequencing (First Base Laboratories Sdn. Bhd., Malaysia).
Results
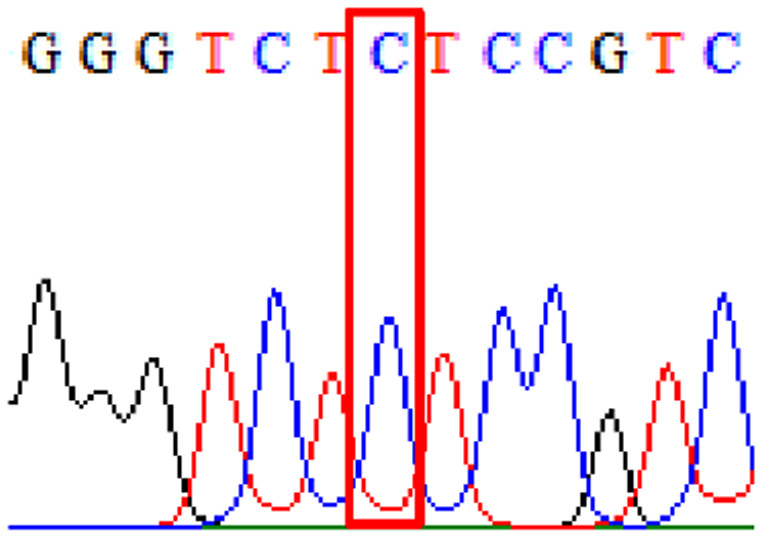
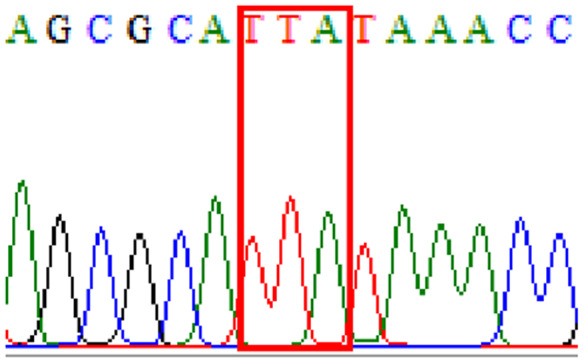
Sixty-one treatment-naïve gastric biopsy samples were studied. Point mutations/polymorphisms detected which are responsible for primary resistance in H. pylori in the present study were listed in Tables 1 and 2. For mutations in the 23S rRNA gene corresponds to the situation of clarithromycin resistance, no A2146G (aka A2142G) mutation was found in this study, but resistance-related mutation A2147G (aka A2143G) and polymorphism T2186C (aka 2,182) were detected with the rate of 14.8% (9/61) and 72.1% (44/61), respectively. Seven samples (11.5%) carried both A2147G and T2186C. For levofloxacin, resistance-related mutation D91N in gyrA was detected at a rate of 3.3% (2/61) while E463(G/K) mutation was not found in our study. The polymorphism rates of D481E (16.4%, 10/61) and R484K (21.3%, 13/61) were found in gyrB. Silent mutations were detected at base triplet 87 of gyrA gene, AAT to AAC (32.8%, 20/61) which do not change the amino acid asparagine. Another similar phenomenon was observed in gyrB gene at base triplet 438, whereby seven samples (11.4%) carried a TTC triplet that code for phenylalanine that was similar to wild type. The presence of resistance-related mutations (A2147G and D91N) to the two antibiotics under study was detected in one sample.
Table 1. Detection of known mutations present in 23S rRNA, gyrA and gyrB genes in treatment-naïve patients infected with Helicobacter pylori.
| Drug | Gene | Mutation point | Forward sequence | Reverse sequence | Frequency (%) |
|---|---|---|---|---|---|
| Clarithromycin | 23S rRNA | A2147G (aka A2143G) |
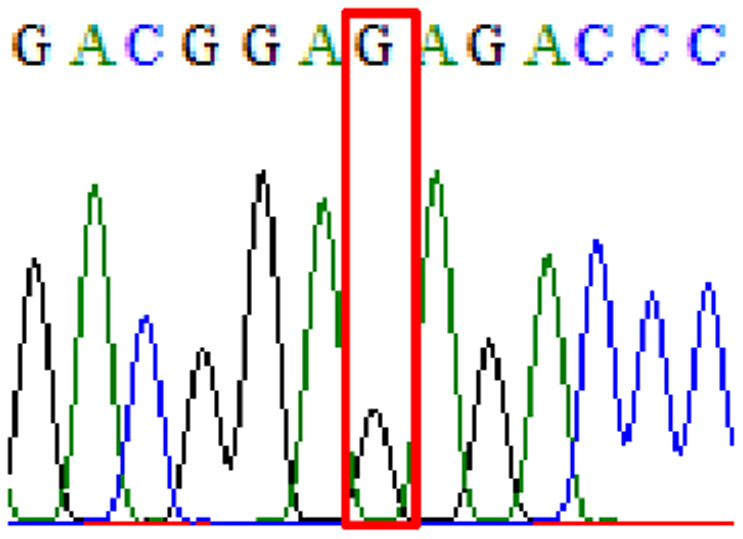 |
 |
14.8% (9/61) |
| Levofloxacin | gyrA | D91N | 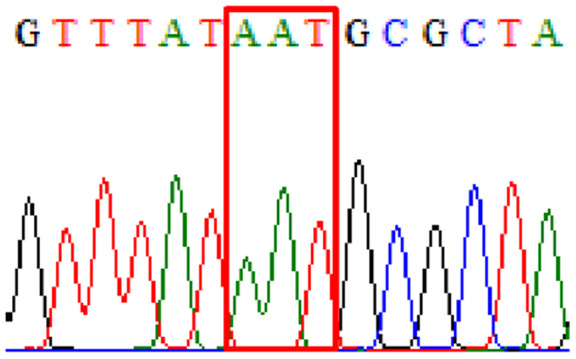 |
 |
3.3% (2/61) |
Table 2. Detection of polymorphisms in antibiotic resistance genes (23S rRNA, gyrA and gyrB) from treatment-naïve patients infected with Helicobacter pylori.
| Drug | Gene | Polymorphisms | Forward sequence | Reverse sequence | Frequency (%) |
|---|---|---|---|---|---|
| Clarithromycin | 23S rRNA | T2186C (aka T2182C) |
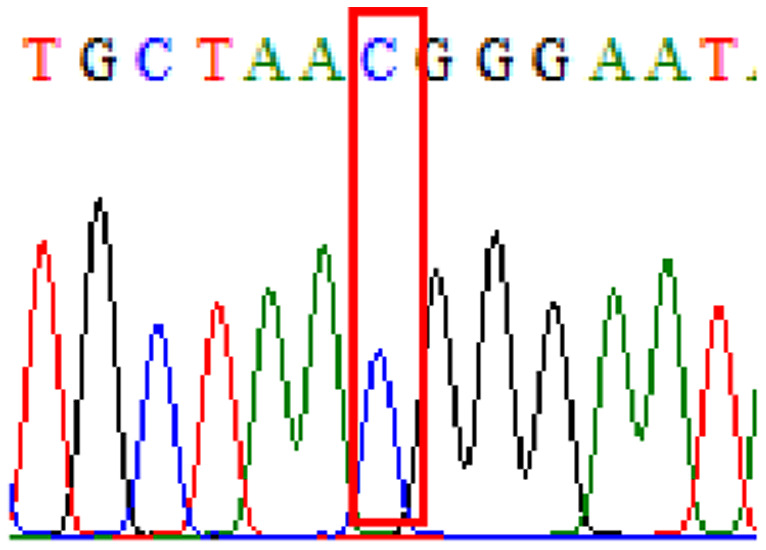 |
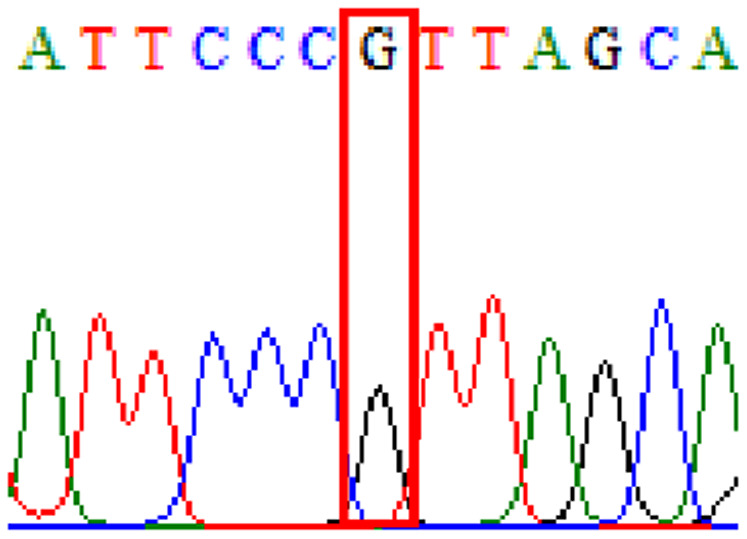 |
72.1% (44/61) |
| Levofloxacin | gyrA | Silent mutation at 87Asn (AAT to AAC at position 261) |
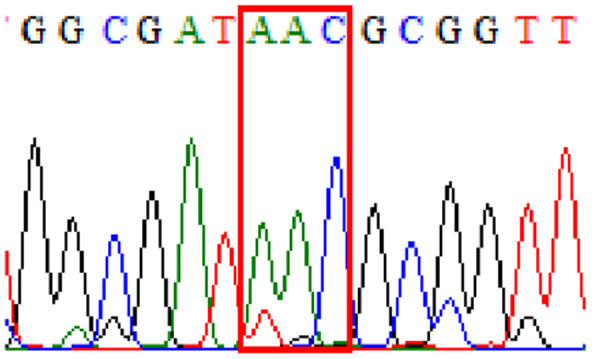 |
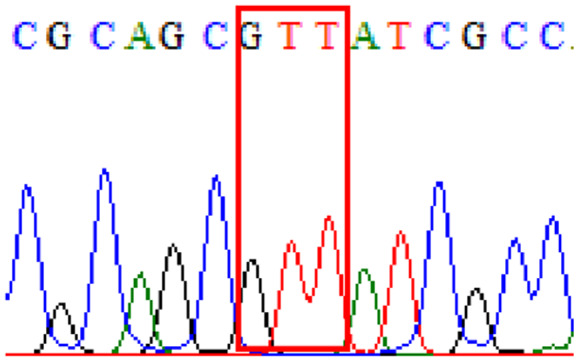 |
32.8% (20/61) |
| gyrB | Silent mutation at 438Phe (TTC to TTT at position 1314) |
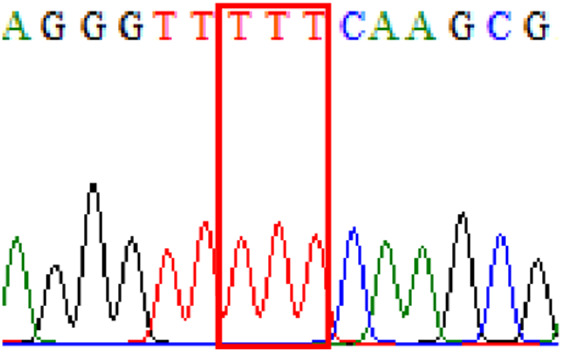 |
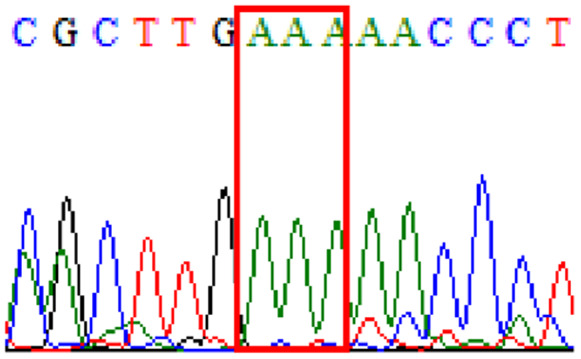 |
11.4% (7/61) |
|
| D481E | 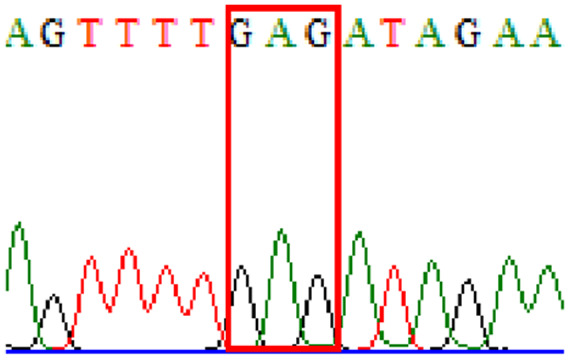 |
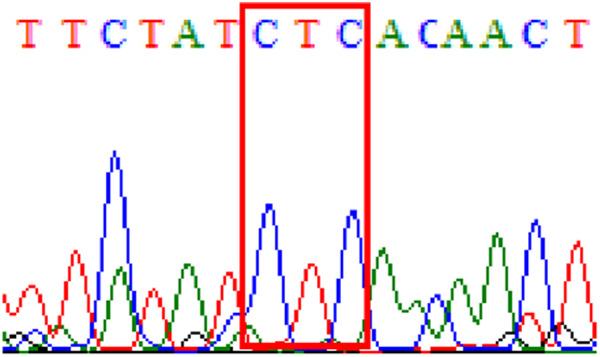 |
16.4% (10/61) |
||
| R484K | 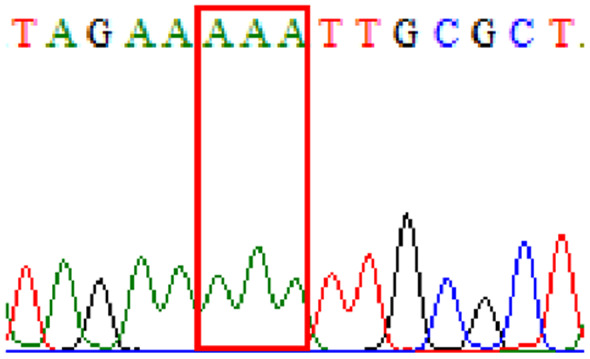 |
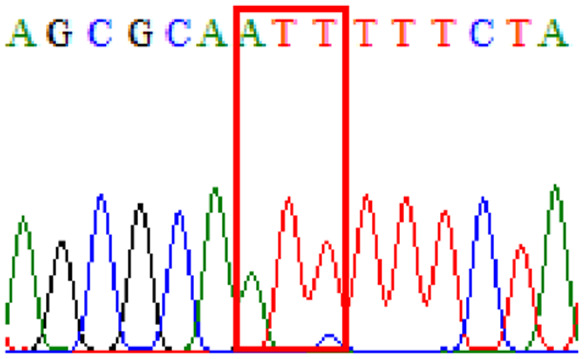 |
21.3% (13/61) |
In this study, based on reported resistance-related mutations in 23S rRNA and gyrA genes, our detected resistance rates to clarithromycin was 14.8% and to levofloxacin 3.3%. Those samples demonstrated D481E and R484K in gyrB polymorphisms concomitantly had mutations at base triplet 87 in gyrA gene—46% (6/13) and 50% (5/10), respectively.
Discussion
Determination of bacterial resistance to antibiotics is crucial to the successful treatment of any bacterial infection. This is particularly so with H. pylori, which affects many people in a population of a country. Monitoring of background susceptibility patterns of H. pylori to antibiotics allows a rational prescription of a treatment regimen for patients. This background pattern of susceptibility to antibiotics must however be continuously monitored and updated over time.
The traditional method of determining bacterial resistance has been to culture the bacterium and test it with several methods such as agar dilution, disc diffusion and E-test. However, this technique is laborious and time-consuming and requires up to a week to yield results on susceptibility to antibiotics (Mégraud & Lehours, 2007). The success of culture with H. pylori is also not high as the bacterium needs to be properly and quickly transported to the bacteriology laboratory (Mégraud & Lehours, 2007). In our practice, we have even resorted to direct plating of the bacteria onto special ox-blood culture media plates to improve the success of the primary culture.
In contrast, molecular methods of detecting resistance to antibiotics are a robust way of testing for resistance to an antibiotic (Mégraud et al., 2015). Collection of samples is easy from the endoscopy room where in addition to routine biopsies for the rapid urease test, biopsies are also taken and stored at 4 °C in a tube that contains a gel transport media. Bacterial DNA is well preserved in this media and remains unaltered even at ambient temperature for long periods. Results can be obtained very quickly within 3 days (from DNA extraction, DNA amplification using PCR and detection using Sanger sequencing). Molecular methods of testing for bacterial resistance are a viable and more efficient alternative to culture and antibiogram. This becomes even more so with growing requests for individual susceptibility testing for all H. pylori infections before treatment.
Clarithromycin and levofloxacin are two antibiotics that are commonly used in H.pylori treatment regimens in Malaysia as in many parts of the world. Mutations in the bacterial 23S rRNA at positions 2,142 and 2,143 are now well established for clarithromycin resistance (Mégraud et al., 2015). Although more difficult to perform, determination of mutations at key sites in topoisomerase II (DNA gyrase) or topoisomerase IV, enzymes has also been established as indicative of resistance to fluoroquinolones. As H. pylori lacks the topoisomerase IV enzyme, thus fluoroquinolone resistance commonly associated to point mutations in the quinolone resistance-determining region (QRDR) of the gyrA and gyrB, encoding the DNA gyrase (Miyachi et al., 2006). Thus far, gyrB gene has yet been well studied as the gyrA gene but previous reports had also described the presence of mutation in the gyrB gene in levofloxacin-resistant strains (Miyachi et al., 2006; Chung et al., 2012; Rimbara et al., 2012).
Previous reports showed mutations at Asn-78 and/or Asp-91 of the gyrA were predominantly in levofloxacin resistance (Miyachi et al., 2006; Chung et al., 2012). Intriguingly, in our study, sequencing of the gyrA gene revealed that 32.8% of samples carried AAC triplet at base 87 which was different from AAT triplet demonstrated by the H. pylori reference strains (accession numbers 26995, U27270 and L29481) available in GenBank database. This nucleotide substitution does not alter amino acid change known as silent mutation or variant at base 87. As for the gyrB subunit, no reported resistance-related mutation at position 463 was detected.
In our local setting in Malaysia, clarithromycin resistance has been tested for many years (Ahmad et al., 2009; Alfizah et al., 2014; Hanafiah et al., 2019; Goh et al., 1997, 2000; Ho et al., 2010; Goh & Navaratnam, 2011; Parasakthi & Goh, 1995; Teh et al., 2014). Results have consistently shown to be zero or very low (Table 3). In one study performed from our centre using molecular methods, Ho et al. (2010) showed a resistance rate of 2.9%. Later studies have shown an increase in resistance rates to 6.8% and 12.2% but they are still relatively low (Teh et al., 2014; Hanafiah et al., 2019). Clarithromycin is a key antibiotic that we continue to recommend as first-line therapy. A recently published paper from our centre has shown a high success rate with 2-week STT incorporating clarithromycin which is reflective of the susceptibility of our local strains to clarithromycin (Hwong-Ruey Leow, Chang & Goh, 2020). However, with the rise in resistance rates as shown in this present study, success rates of this therapy will drop in the future. Reports from other countries in the Asian Pacific region and Europe have already demonstrated a high as well as a rising clarithromycin resistance rate (Horiki et al., 2009; Kuo et al., 2017; Megraud et al., 2013).
Table 3. Primary antibiotic resistance over time in Malaysia.
| Study | Year of study | Sample size (n) | Methodology | % Resistance to clarithromycin | % Resistance to levofloxacin |
|---|---|---|---|---|---|
| Parasakthi & Goh, 1995 | 1994 | 20 | E-test | 0 | - |
| Goh et al., 1997 | 1996 | 63 | E-test | 0 | - |
| Goh et al., 2000 | 1999 | 74 | E-test | 0 | - |
| Ho et al., 2010 | 2002 | 107 | Molecular | 2.9 | - |
| Alfizah et al., 2014 | 2004–2007 | 161 | E-test | 1.2 | 1.9 |
| Ahmad et al., 2009 | 2005–2007 | 187 | E-test | 2.1 | - |
| Goh & Navaratnam, 2011 | 2009 | 90 | E-test | 0 | - |
| Teh et al., 2014 | 2014 | 102 | E-test | 6.8 | 6.8 |
| Hanafiah et al., 2019 | 2014–2015 | 41 | E-test | 12.2 | 17.1 |
| Present study, 2021 | 2019–2020 | 62 | Molecular | 14.8 | 3.3 |
Note:
- Not done.
Levofloxacin has also been increasingly used in clinical practice in Malaysia. We have reported its use as an effective rescue therapy previously (Goh, Manikam & Qua, 2012). Although previously reported to be low, levofloxacin resistance has also increased compared to previous reports, in the present study and this is also alarming to us. Background resistance rates to levofloxacin have been shown to have increased in several countries in the Asian Pacific region (Hu, Zhu & Lu, 2017; Liou et al., 2015; Shu et al., 2018; Lee et al., 2019a, 2019b). Nevertheless, most past studies reported that levofloxacin is mainly affected by gyrA but the gyrB mutation has a steady relationship with gyrA 87–91 mutations (Rimbara et al., 2012; Miyachi et al., 2006; Miftahussurur et al., 2016; Miftahussurur et al., 2019; Wang et al., 2010). Recently, Miftahussur et al. reported the detection of 48.3% (14/29) double polymorphisms (D481E and R484K) simultaneously in the gyrB gene in most of the garenoxacin-resistant H. pylori strains in Nepal and Bangladesh (Miftahussurur et al., 2019). In our study, we observed the presence of both polymorphisms D481E (16.4%) and R484K (21.3%). Hence, the relevance of these two polymorphisms for levofloxacin resistance either individually or in combination warrants further investigation in future study to confirm their clinical significance.
In this study, out of 61 samples, one sample was found to carry reported resistance-related mutations corresponding to clarithromycin (A2147G in 23S rRNA) and levofloxacin (D91N in gyrA) from the same patient before antibiotic treatment. However, one limitation of our study is unable to rule out whether the detection of these two mutations in a single sample containing biopsies taken from two different gastric sites is due to multiple infections or heteroresistance. Susceptible and homoresistant/heteroresistant H. pylori strains may co-exist at the same as well as different gastric regions. Therefore, detection of heteroresistance cases by exploring how the H. pylori loads differ between at least two gastric regions from the same patient or using pure isolates should be taken into account in the assessment of resistance in the future study.
Empirical therapy is widely recommended for the eradication of H. pylori in many countries including Malaysia with a lack of in vitro antimicrobial resistance. Unrestricted use of clarithromycin against H. pylori attributes to suboptimal results and antibiotic resistance. In Malaysia, a low resistance rate for clarithromycin (less than 6.8%) has been reported, therefore clarithromycin (500 mg) is continued to be used as our treatment regimens together with amoxicillin (1 g) and pantoprazole 40 mg (Qua, Manikam & Goh, 2010) or rabeprazole (20 mg) (Goh, Manikam & Qua, 2012; Leow et al., 2018). However, unsatisfactory efficacy was seen with an overall eradication rate of only 71.2% in a comparative, randomized and open-labeled study (Leow et al., 2018). In 2018, the meta-analysis of the global distribution of antibiotic resistance in H. pylori included 178 studies comprising 66,142 isolates from 65 countries including Malaysia showed worrisome levels of resistance rates to clarithromycin, and levofloxacin in all WHO regions (Savoldi et al., 2018). In the South-East Asia region, the pooled prevalence of clarithromycin resistance significantly increased from 13% (95% CI [4%–22%]) in 2006–2008 to 21% (95% CI [1%–42%]) in 2012–2016 (P < 0.001). The pooled prevalence for resistance rates for levofloxacin was 29% (95% CI [16%–42%]) in 2012–2016 (Savoldi et al., 2018).
Compared with empiric chosen treatments, molecular-based tailored therapy is a better alternative for H pylori eradication. In many countries, the selection of antibiotics for H. pylori eradication therapies is made based on the background pattern of resistance to antibiotics. It is therefore important that constant monitoring of background antibiotic resistance to H. pylori is carried out. Molecular methods of detecting mutation in H. pylori strains allows us to do this fairly easily. The low number of samples was the limitation of the present study and using a large sample size is needed for further study to provide comprehensive and up-to-date information on the antibiotic resistance of H. pylori in Malaysia. A further step ahead is to make available individual testing of all H. pylori strains particularly for treatment failures where the choice of antibiotics would be crucial.
Supplemental Information
Mutations on 23S rRNA ( A2147G, T2186C); mutations on gyrA ( 87Asn, D91N) and mutations on gyrB (438Phe, D481E, R484K)
Funding Statement
This work was supported by grants from the Impact-Oriented Interdisciplinary Research Grant Programme (IIRG029A-2019 and IIRG029B-2019), University of Malaya. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare that they have no competing interests.
Author Contributions
Suat Moi Puah performed the experiments, analyzed the data, authored or reviewed drafts of the paper, and approved the final draft.
Khean Lee Goh conceived and designed the experiments, analyzed the data, authored or reviewed drafts of the paper, and approved the final draft.
Heng Kang Ng performed the experiments, analyzed the data, prepared figures and/or tables, and approved the final draft.
Kek Heng Chua conceived and designed the experiments, analyzed the data, authored or reviewed drafts of the paper, and approved the final draft.
Human Ethics
The following information was supplied relating to ethical approvals (i.e., approving body and any reference numbers):
The study was approved by the ethics committee of the University of Malaya Medical Centre (MREC ID No: 2019101-7882).
Data Availability
The following information was supplied regarding data availability:
The nucleotide sequences (n = 105) encompassing the mutations determined in this study are available at GenBank:
23S rRNA, mutation point A2147G: MW341345–MW341353; polymorphism point T2186C are MW341355–MW341398; gyrA, mutation 87Asn: MW375361, MW375363–MW375381; mutation D91N: MW375362, MW375382; gyrB, polymorphisms 438Phe: MW375406–MW375412; D481E: MW375383, MW375385–MW375393; and R484K: MW375384, MW375394–MW375405.
The accessions are also available in the Supplemental File.
References
- Ahmad et al. (2009).Ahmad N, Zakaria WR, Abdullah SA, Mohamed R. Characterization of clarithromycin resistance in Malaysian isolates of Helicobacter pylori. World Journal of Gastroenterology. 2009;15(25):3161–3165. doi: 10.3748/wjg.15.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfizah et al. (2014).Alfizah H, Norazah A, Hamizah R, Ramelah M. Resistotype of Helicobacter pylori isolates: the impact on eradication outcome. Journal of Medical Microbiology. 2014;63(Pt. 5):703–709. doi: 10.1099/jmm.0.069781-0. [DOI] [PubMed] [Google Scholar]
- Binh et al. (2014).Binh TT, Shiota S, Suzuki R, Matsuda M, Trang TT, Kwon DH, Iwatani S, Yamaoka Y. Discovery of novel mutations for clarithromycin resistance in Helicobacter pylori by using next-generation sequencing. Journal of Antimicrobial Chemotherapy. 2014;69(7):1796–1803. doi: 10.1093/jac/dku050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzas & Jozan (2006).Buzas GM, Jozan J. First-line eradication of H. pylori infection in Europe: a meta-analysis based on congress abstracts, 1997–2004. World Journal of Gastroenterology. 2006;12(33):5311–5319. doi: 10.3748/wjg.v12.i33.5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung et al. (2012).Chung J-W, Lee GH, Jeong J-Y, Lee SM, Jung JH, Choi KD, Song HJ, Jung H-Y, Kim J-H. Resistance of Helicobacter pylori strains to antibiotics in Korea with a focus on fluoroquinolone resistance. Journal of Gastroenterology and Hepatology. 2012;27(3):493–497. doi: 10.1111/j.1440-1746.2011.06874.x. [DOI] [PubMed] [Google Scholar]
- Garcia et al. (2012).Garcia M, Raymond J, Garnier M, Cremniter J, Burucoa C. Distribution of spontaneous gyrA mutations in 97 fluoroquinolone-resistant Helicobacter pylori isolates collected in France. Antimicrob Agents Chemother. 2012;56:550–551. doi: 10.1128/AAC.05243-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh, Manikam & Qua (2012).Goh KL, Manikam J, Qua CS. High-dose rabeprazole-amoxicillin dual therapy and rabeprazole triple therapy with amoxicillin and levofloxacin for 2 weeks as first and second line rescue therapies for Helicobacter pylori treatment failures. Alimentary Pharmacology & Therapeutics. 2012;35(9):1097–1102. doi: 10.1111/j.1365-2036.2012.05120.x. [DOI] [PubMed] [Google Scholar]
- Goh & Navaratnam (2011).Goh KL, Navaratnam P. High Helicobacter pylori resistance to metronidazole but zero or low resistance to clarithromycin, levofloxacin, and other antibiotics in Malaysia. Helicobacter. 2011;16(3):241–245. doi: 10.1111/j.1523-5378.2011.00841.x. [DOI] [PubMed] [Google Scholar]
- Goh et al. (2000).Goh K, Parasakthi N, Cheah P, Ranjeev C, Rosmawati M, Tan Y, Chin S. Efficacy of a 1-week pantoprazole triple therapy in eradicating Helicobacter pylori in Asian patients. Journal of Gastroenterology and Hepatology. 2000;15(8):910–914. doi: 10.1046/j.1440-1746.2000.02229.x. [DOI] [PubMed] [Google Scholar]
- Goh et al. (1997).Goh KL, Parasakthi N, Chuah SY, Cheah PL, Lo YL, Chin SC. One-week low dose triple therapy with proton-pump inhibitors: optimal therapy for H. pylori eradication? Alimentary Pharmacology and Therapeutics. 1997;11(6):1115–1118. doi: 10.1046/j.1365-2036.1997.00248.x. [DOI] [PubMed] [Google Scholar]
- Graham & Dore (2016).Graham DY, Dore MP. Helicobacter pylori therapy: a paradigm shift. Expert Review of Anti-infective Therapy. 2016;14(6):577–585. doi: 10.1080/14787210.2016.1178065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham & Fischbach (2010).Graham DY, Fischbach L. Helicobacter pylori treatment in the era of increasing antibiotic resistance. Gut. 2010;59(8):1143–1153. doi: 10.1136/gut.2009.192757. [DOI] [PubMed] [Google Scholar]
- Graham & Shiotani (2008).Graham DY, Shiotani A. New concepts of resistance in the treatment of Helicobacter pylori infections. Nature Clinical Practice Gastroenterology & Hepatology. 2008;5(6):321–331. doi: 10.1038/ncpgasthep1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanafiah et al. (2019).Hanafiah A, Binmaeil H, Raja Ali RA, Mohamed Rose I, Lopes BS. Molecular characterization and prevalence of antibiotic resistance in Helicobacter pylori isolates in Kuala Lumpur, Malaysia. Infection and Drug Resistance. 2019;12:3051–3061. doi: 10.2147/IDR.S219069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho et al. (2010).Ho SL, Tan EL, Sam CK, Goh KL. Clarithromycin resistance and point mutations in the 23S rRNA gene in Helicobacter pylori isolates from Malaysia. Journal of Digestive Diseases. 2010;11(2):101–105. doi: 10.1111/j.1751-2980.2010.00423.x. [DOI] [PubMed] [Google Scholar]
- Horiki et al. (2009).Horiki N, Omata F, Uemura M, Suzuki S, Ishii N, Iizuka Y, Fukuda K, Fujita Y, Katsurahara M, Ito T, Cesar GE, Imoto I, Takei Y. Annual change of primary resistance to clarithromycin among Helicobacter pylori isolates from 1996 through 2008 in Japan. Helicobacter. 2009;14(5):86–90. doi: 10.1111/j.1523-5378.2009.00714.x. [DOI] [PubMed] [Google Scholar]
- Hu, Zhu & Lu (2017).Hu Y, Zhu Y, Lu NH. Primary antibiotic resistance of Helicobacter pylori in China. Digestive Diseases and Sciences. 2017;62(5):1146–1154. doi: 10.1007/s10620-017-4536-8. [DOI] [PubMed] [Google Scholar]
- Hwong-Ruey Leow, Chang & Goh (2020).Hwong-Ruey Leow A, Chang JV, Goh KL. Searching for an optimal therapy for H. pylori eradication: high-dose proton-pump inhibitor dual therapy with amoxicillin vs. standard triple therapy for 14 days. Helicobacter. 2020;25(5):e12723. doi: 10.1111/hel.12723. [DOI] [PubMed] [Google Scholar]
- Kim et al. (2005).Kim JM, Kim JS, Kim N, Jung HC, Song IS. Distribution of fluoroquinolone MICs in Helicobacter pylori strains from Korean patients. Journal of Antimicrobial Chemotherapy. 2005;56(5):965–967. doi: 10.1093/jac/dki334. [DOI] [PubMed] [Google Scholar]
- Kuo et al. (2017).Kuo Y-T, Liou J-M, El-Omar EM, Wu J-Y, Leow AHR, Goh KL, Das R, Lu H, Lin J-T, Tu Y-K, Yamaoka Y, Wu M-S. Primary antibiotic resistance in Helicobacter pylori in the Asia-Pacific region: a systematic review and meta-analysis. Lancet Gastroenterology & Hepatology. 2017;2(10):707–715. doi: 10.1016/S2468-1253(17)30219-4. [DOI] [PubMed] [Google Scholar]
- Lee et al. (2019a).Lee JH, Ahn JY, Choi KD, Jung H-Y, Kim JM, Baik GH, Kim B-W, Park JC, Jung HK, Cho SJ, Shin CM, Choi YJ, Lee SH, Kim JH, Lee WS, Sung JK, Chung J-W, Cheung DY, Lee H, Min YW, Kim JJ, Kim SY, Korean College of Helicobacter; Upper Gastrointestinal Research Nationwide antibiotic resistance mapping of Helicobacter pylori in Korea: a prospective multicenter study. Helicobacter. 2019a;24(4):e12592. doi: 10.1111/hel.12592. [DOI] [PubMed] [Google Scholar]
- Lee et al. (2019b).Lee JY, Kim N, Nam RH, In Choi S, Lee JW, Lee DH. Primary and secondary antibiotic resistance of Helicobacter pylori in Korea from 2003 to 2018. Helicobacter. 2019b;24(6):e12660. doi: 10.1111/hel.12660. [DOI] [PubMed] [Google Scholar]
- Leow et al. (2018).Leow AH, Azmi AN, Loke MF, Vadivelu J, Grahan DY, Goh KL. Optimizing first line 7-day standard triple therapy for Helicobacter pylori eradication: prolonging treatment or adding bismuth: which is better? Journal of Digestive Diseases. 2018;9(11):674–677. doi: 10.1111/1751-2980.12679. [DOI] [PubMed] [Google Scholar]
- Liou et al. (2015).Liou J-M, Chang C-Y, Chen M-J, Chen C-C, Fang Y-J, Lee J-Y, Wu J-Y, Luo J-C, Liou T-C, Chang W-H, Tseng C-H, Wu C-Y, Yang T-H, Chang C-C, Wang H‐Po, Sheu B-S, Lin J-T, Bair M-J, Wu M-S, Taiwan Gastrointestinal Disease and Helicobacter Consortium The primary resistance of Helicobacter pylori in Taiwan after the national policy to restrict antibiotic consumption and its relation to virulence factors-a nationwide study. PLOS ONE. 2015;10(5):e0124199. doi: 10.1371/journal.pone.0124199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraud et al. (2013).Megraud F, Coenen S, Versporten A, Kist M, Lopez-Brea M, Hirschl AM, Andersen LP, Goossens H, Glupczynski Y. Helicobacter pylori resistance to antibiotics in Europe and its relationship to antibiotic consumption. Gut. 2013;62(1):34–42. doi: 10.1136/gutjnl-2012-302254. [DOI] [PubMed] [Google Scholar]
- Miftahussurur et al. (2019).Miftahussurur M, Aftab H, Shrestha PK, Sharma RP, Subsomwong P, Waskito LA, Doohan D, Fauzia KA, Yamaoka Y. Effective therapeutic regimens in two South Asian countries with high resistance to major Helicobacter pylori antibiotics. Antimicrobial Resistance & Infection Control. 2019;8:40. doi: 10.1186/s13756-019-0482-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miftahussurur et al. (2016).Miftahussurur M, Shrestha PK, Subsomwong P, Sharma RP, Yamaoka Y. Emerging Helicobacter pylori levofloxacin resistance and novel genetic mutation in Nepal. BMC Microbiology. 2016;16(1):256. doi: 10.1186/s12866-016-0873-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyachi et al. (2006).Miyachi H, Miki I, Aoyama N, Shirasaka D, Matsumoto Y, Toyoda M, Mitani T, Morita Y, Tamura T, Kinoshita S, Okano Y, Kumagai S, Kasuga M. Primary levofloxacin resistance and gyrA/B mutations among Helicobacter pylori in Japan. Helicobacter. 2006;11(4):243–249. doi: 10.1111/j.1523-5378.2006.00415.x. [DOI] [PubMed] [Google Scholar]
- Mégraud et al. (2015).Mégraud F, Bénéjat L, Ontsira Ngoyi EN, Lehours P. Molecular approaches to identify Helicobacter pylori antimicrobial resistance. Gastroenterology Clinics of North America. 2015;44(3):577–596. doi: 10.1016/j.gtc.2015.05.002. [DOI] [PubMed] [Google Scholar]
- Mégraud & Lehours (2007).Mégraud F, Lehours P. Helicobacter pylori detection and antimicrobial susceptibility testing. Clinical Microbiology Reviews. 2007;20(2):280–322. doi: 10.1128/CMR.00033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa & Suzuki (2014).Nishizawa T, Suzuki H. Mechanisms of Helicobacter pylori antibiotic resistance and molecular testing. Frontiers in Molecular Biosciences. 2014;1:19. doi: 10.3389/fmolb.2014.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Occhialini et al. (1997).Occhialini A, Urdaci M, Doucet-Populaire F, Bébéar CM, Lamouliatte H, Mégraud F. Macrolide resistance in Helicobacter pylori: rapid detection of point mutations and assays of macrolide binding to ribosomes. Antimicrobial Agents and Chemotherapy. 1997;41(12):2724–2728. doi: 10.1128/AAC.41.12.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parasakthi & Goh (1995).Parasakthi N, Goh KL. Primary and acquired resistance to clarithromycin among Helicobacter pylori strains in Malaysia. American Journal of Gastroenterology. 1995;90(3):519. [PubMed] [Google Scholar]
- Qua, Manikam & Goh (2010).Qua CS, Manikam J, Goh KL. Efficacy of 1-week proton pump inhibitor triple therapy as first-line Helicobacter pylori eradication regime in Asian patients: is it still effective 10 years on? Journal of Digestive Diseases. 2010;11:244–248. doi: 10.1111/j.1751-2980.2010.00445.x. [DOI] [PubMed] [Google Scholar]
- Raymond et al. (2007).Raymond J, Butucoa C, Pietrini O, Bergeret M, Decoster A, Wann A, Dupont C, Kalach N. Clarithromycin resistance in Helicobacter pylori strains isolated from French children: prevalence of the different mutations and coexistence of clones harbouring two different mutations in the same biopsy. Helicobacter. 2007;12(2):157–163. doi: 10.1111/j.1523-5378.2007.00486.x. [DOI] [PubMed] [Google Scholar]
- Rimbara et al. (2012).Rimbara E, Noguchi N, Kawai T, Sasatsu M. Fluoroquinolone resistance in Helicobacter pylori: role of mutations at position 87 and 91 of GyrA on the level of resistance and identification of a resistance conferring mutation in GyrB. Helicobacter. 2012;17(1):36–42. doi: 10.1111/j.1523-5378.2011.00912.x. [DOI] [PubMed] [Google Scholar]
- Savoldi et al. (2018).Savoldi A, Carrara E, Graham DY, Conti M, Tacconelli E. Prevalence of antibiotic resistance in Helicobacter pylori: a systematic review and meta-analysis in World Health Organization regions. Gastroenterology. 2018;155(5):1372–1382. doi: 10.1053/j.gastro.2018.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu et al. (2018).Shu X, Yin G, Liu M, Peng K, Zhao H, Jiang M. Antibiotics resistance of Helicobacter pylori in children with upper gastrointestinal symptoms in Hangzhou, China. Helicobacter. 2018;23(3):e12481. doi: 10.1111/hel.12481. [DOI] [PubMed] [Google Scholar]
- Teh et al. (2014).Teh X, Khosravi Y, Lee WC, Leow AH, Loke MF, Vadivelu J, Goh KL. Functional and molecular surveillance of Helicobacter pylori antibiotic resistance in Kuala Lumpur. PLOS ONE. 2014;9(7):e101481. doi: 10.1371/journal.pone.0101481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2010).Wang LH, Cheng H, Hu FL, Li J. Distribution of gyrA mutations in fluoroquinolone-resistant Helicobacter pylori strains. World Journal of Gastroenterology. 2010;16(18):2272–2277. doi: 10.3748/wjg.v16.i18.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang & Taylor (1998).Wang G, Taylor DE. Site-specific mutations in the 23S rRNA gene of Helicobacter pylori confer two types of resistance to macrolide-lincosamide-streptogramin B antibiotics. Antimicrobial Agents and Chemotherapy. 1998;42(8):1952–1958. doi: 10.1128/AAC.42.8.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2020).Wang YH, Wang FF, Gong XL, Yan LL, Zhao QY, Song YP, Zhao RL, He YJ, Zhou L, Liu DS, Xie Y. Genotype profiles of Helicobacter pylori from gastric biopsies and strains with antimicrobial-induced resistance. Therapeutic Advances in Gastroenterology. 2020;13:1756284820952596. doi: 10.1177/1756284820952596. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mutations on 23S rRNA ( A2147G, T2186C); mutations on gyrA ( 87Asn, D91N) and mutations on gyrB (438Phe, D481E, R484K)
Data Availability Statement
The following information was supplied regarding data availability:
The nucleotide sequences (n = 105) encompassing the mutations determined in this study are available at GenBank:
23S rRNA, mutation point A2147G: MW341345–MW341353; polymorphism point T2186C are MW341355–MW341398; gyrA, mutation 87Asn: MW375361, MW375363–MW375381; mutation D91N: MW375362, MW375382; gyrB, polymorphisms 438Phe: MW375406–MW375412; D481E: MW375383, MW375385–MW375393; and R484K: MW375384, MW375394–MW375405.
The accessions are also available in the Supplemental File.


