Abstract
There has long been a crucial tradeoff between spatial and temporal resolution in imaging. Imaging beyond the diffraction limit of light has traditionally been restricted to be used only on fixed samples or live cells outside of tissue labeled with strong fluorescent signal. Current super-resolution live cell imaging techniques require the use of special fluorescence probes, high illumination, multiple image acquisitions with post-acquisition processing, or often a combination of these processes. These prerequisites significantly limit the biological samples and contexts that this technique can be applied to.
Here we describe a method to perform super-resolution (∼140 nm XY-resolution) time-lapse fluorescence live cell imaging in situ. This technique is also compatible with low fluorescent intensity, for example, EGFP or mCherry endogenously tagged at lowly expressed genes. As a proof-of-principle, we have used this method to visualize multiple subcellular structures in the Drosophila testis. During tissue preparation, both the cellular structure and tissue morphology are maintained within the dissected testis. Here, we use this technique to image microtubule dynamics, the interactions between microtubules and the nuclear membrane, as well as the attachment of microtubules to centromeres.
This technique requires special procedures in sample preparation, sample mounting and immobilizing of specimens. Additionally, the specimens must be maintained for several hours after dissection without compromising cellular function and activity. While we have optimized the conditions for live super-resolution imaging specifically in Drosophila male germline stem cells (GSCs) and progenitor germ cells in dissected testis tissue, this technique is broadly applicable to a variety of different cell types. The ability to observe cells under their physiological conditions without sacrificing either spatial or temporal resolution will serve as an invaluable tool to researchers seeking to address crucial questions in cell biology.
Keywords: super-resolution, diffraction limit, time-lapse imaging, live tissue imaging, stem cell, microtubules, centromeres, nuclear membrane, asymmetric cell division, mitosis
SUMMARY:
Presented here is a protocol for super-resolution live-cell imaging in intact tissue. We have standardized the conditions for imaging a highly sensitive adult stem cell population in its native tissue environment. This technique involves balancing temporal and spatial resolution to allow for the direct observation of biological phenomena in live tissue.
INTRODUCTION:
Visualizing subcellular structures and protein dynamics in live cells with resolution beyond the diffraction limit of light is typically very challenging1–3. While multiple super-resolution techniques such as Stochastic-Optical-Reconstruction-Microscopy (STORM), Photo-Activated-Localization-Microscopy (PALM) and Stimulated-Emission-Depletion (STED)4–6 microscopy have been developed, complications in specimen preparation as well as the need to maintain viability and activity ex vivo, limit the use of conventional super-resolution microscopy for imaging live samples. Conventional confocal microscopy cannot reach spatial resolution beyond ∼230 nm XY-resolution and is often insufficient to observe intricate cellular substructures5,6. However, a recent development in confocal microscopy, Airyscan super-resolution imaging, is able to achieve approximately 140 nm (XY-resolution)7,8 and has a relatively simple sample preparation that is compatible with live imaging. Since this imaging detection system requires a long acquisition time, its high spatial resolution does come at the cost of temporal resolution9. Therefore, a method is needed to ensure that live cell imaging is extended with high spatial resolution.
Here, we developed a method for observing live cells in intact tissue at its optimal resolution to decipher subcellular structures with detailed spatial information. This method is designed as such so that samples can be mounted stably for a long period of time (∼ 10 h) without moving or degenerating. The live cell media used in this technique can support cellular function and avoid photobleaching for up to 10 hours under a super-resolution microscope. Finally, this protocol minimizes most stresses caused by the constant illumination of lasers over extended periods of time such as hypoxia, changes in humidity and temperature, as well as nutrient exhaustion.
Using this protocol to image Drosophila male germline stem cells (GSCs), we were able to observe how the asymmetric activity of microtubules allows for preferential interaction with epigenetically distinct sister chromatids10–13. These types of cellular events are highly dynamic and are very difficult to visualize in live cells using other super-resolution imaging methods such as STORM, PALM, or STED. We anticipate that this method will become highly useful for cell biologists as they aim to understand the dynamic subcellular structures of live cells residing in tissues. There are many areas in which this method can be applied to, such as studying the dynamics of proteins; understanding the movement of cells; and lineage tracing and cellular differentiation processes, among other possible applications.
PROTOCOL:
1. Preparation of live cell imaging cocktail (live cell media)
1.1.
Supplement Schneider's Drosophila medium with 15% fetal bovine serum (FBS) and 0.6× penicillin/streptomycin. Adjust the pH to approximately 7.0.
1.2.
Just before using the medium, add insulin to a final concentration of 200 μg/mL.
NOTE: This media is critical for maintaining normal cell division and development of the Drosophila testis during time-lapse imaging10,14.
2. Preparation of glass bottom cell culture dish
2.1.
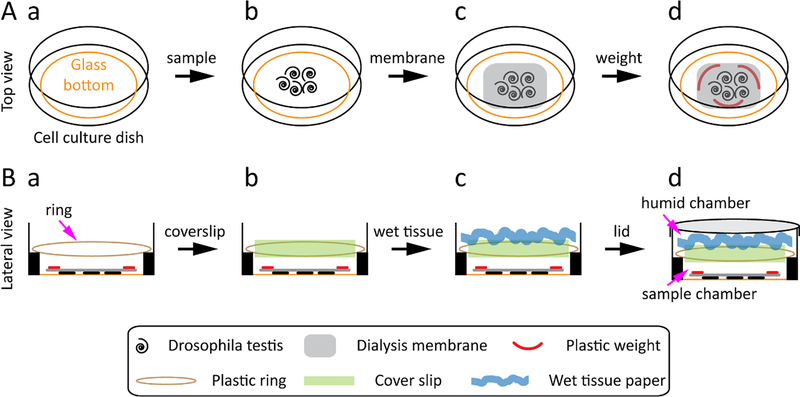
Use a poly L-lysine coated glass-bottom cell culture dish with a diameter of 35 mm for the Drosophila testis. The inner well of the dish has a glass bottom with a diameter of 23 mm and a side with an elevation of 1 mm (Figure 1A–a).
Figure 1: Scheme for the preparation and mounting of Drosophila testis.
(A) Top view: the glass-bottom cell culture dish (A-a), transfer the Drosophila testis toward the center of the dish (A-b), place a dialysis membrane on the top of the testis (A-c), place three plastic weights on the dialysis membrane (A-d). (B) Lateral view: place the plastic ring at the elevated side of the dish (B-a), place a 22 mm × 22 mm coverslip on the plastic ring (B-b), put a swirl and wet tissue paper on the coverslip (B-c), close the dish with the lid (B-d).
NOTE: Choose a glass-bottom dish that allows for a shorter working distance, larger numerical aperture, and higher magnification objective; these features are crucial to obtain a super-resolution image.
2.2.
Use a dialysis membrane (MWCO: 12–14kD) that allows for the gaseous exchange. Cut the membrane into small pieces.
NOTE: The membrane pieces should be smaller than the glass area of the dish but large enough to cover the specimen.
2.3.
Soak the membrane with 100 μL of live cell media for ∼5 min. Do not use a dry membrane on the sample, as it may damage it. Soaking helps prevent hypoxic stress.
2.4.
Prepare 2–3 light-weight glass or plastic pieces to put onto the membrane so that the tissue will not float. To do this, first take the outer ring of the 50 mL centrifuge tube and cut it into small pieces. Then, sterilize the pieces with 70% ethanol before use.
NOTE: This step ensures that the sample remains on the surface during imaging and does not float, which is crucial for obtaining optimal results.
2.5.
Prepare a ring ∼25 mm in diameter and place it at the elevated side of the dish. Put a coverslip on it to generate two chambers. The bottom chamber will contain the sample while the top chamber will serve as a humidity chamber to prevent the sample from drying.
2.5.1.
To make this ring, cut off the outer ring from a 50 mL centrifuge tube, which will allow it to fit securely on the elevated side of the 35 mm dish. A similar ring can be made in other ways, for example, by using a rubber band or plastic twist tie to fit on the elevated side of the dish to properly hold the coverslip without interfering with the sample.
NOTE: This step is critical, especially for samples sensitive to stresses such as hypoxic conditions, and changes in temperature and humidity.
3. Dissection of testes from male flies and mounting
3.1.
Take ∼10 young male flies (2–3 day old) and dissect the testes in the live cell media to obtain ∼10 pairs of Drosophila adult testes.
3.1.1.
Dissect the flies under a dissection microscope in a dissection dish using fine forceps.
NOTE: The Drosophila melanogaster (fruit fly) strain carries UAS-α-Tubulin-GFP transgene driven by an early germ cell driver nanos-Gal4.
3.1.2.
Generate the following knock-in Drosophila melanogaster strains using the CRISPR-Cas9 technology: Lamin-mCherry (C-terminal tag) and CENP-A-Dendra2-CENP-A [tag at the internal site (between 118th - 119th codon)].
NOTE: While only one testis of excellent quality is needed for each experiment, mounting enough number of tissues (15–20) or cells will ensure that there will be at least one healthy sample with excellent fluorescence signals for time-lapse imaging.
3.2.
Wash the testes twice in live cell media in the dissection dish.
3.2.1.
Use a pipette to add media followed by removing the live cell media as a washing step.
3.2.2.
Remove excess tissue using forceps. Any extra tissue will interfere with imaging.
NOTE: Do not use phosphate-buffered saline (PBS) for washing, because it may affect the dynamics of certain cellular components.
3.3.
Add 100–150 μL of live cell media into the dish (prepared in step 2.1) and spread it on the glass surface using a pipette tip. Spreading the media on the surface will allow the tissue to properly stick to the dish.
3.4.
Transfer the testes to the dish using fine forceps and bring them to the center of the dish (Figure 1A–b).
NOTE: Avoid transferring debris because it will interfere with imaging and could reduce the quality of the image.
3.5.
Remove excess live cell media (leave approximately 10 μL of media) to allow for the tissue to flatten and stick properly to the dish. Perform this step quickly to avoid drying the sample.
3.6.
Place the pre-wet membrane (prepared in step 2.2 and 2.3) on top of the testes (Figure 1A–c).
3.7.
Put 2–3 small plastic weights (prepared in Step 2.4) on the membrane so that the sample will not float and quickly add 100–150 μL of live cell media (Figure 1A–d).
NOTE: Adding media quickly and gently ensures that the sample will not dry out or become displaced.
3.8.
Place the plastic ring (prepared in step 2.5) on the elevated side of the dish (Figure 1B–a).
3.9.
Place a 22 mm × 22 mm coverslip on top of the ring (Figure 1B–b). This generates two chambers.
3.10.
Take a piece of tissue paper, dampen it with water, then swirl it and put it on the coverslip, to create a humid chamber (Figure 1B–c). Close the lid of the dish (Figure 1B–d) and begin live cell imaging.
NOTE: If the sample is light sensitive, perform section 3 in the dark.
4. Live cell imaging of Drosophila male germline stem cells (GSC) in situ
4.1.
Place the dish under the super-resolution microscope and secure it with the stage clamps.
4.1.1.
Open the imaging software, turn on the transmitted light, and use 63× objective to focus on the testis tissue with the focus knob.
NOTE: If there is a difficulty in locating a sample with the 63× objective, then first use the 40× or 20× objective before switching to the 63×.
4.1.2.
Turn on the lasers, click Live, and find the GSC with the optimal position and condition. Avoid testes that have a low fluorescence signal or have the hub (niche) away from the surface.
NOTE: In Drosophila testes, the GSCs are attached to the hub.
4.2.
Adjust the focus for the GSCs and identify the right settings for the sample, including laser power, electron-multiplying (EM) gain, averaging, and zoom for the Airyscan mode. Use the following settings as an example for Drosophila testes expressing EGFP tagged α-tubulin in early-stage germ cells.
4.2.1.
Set the frame size between 512 × 512 pixels and 1024 × 1024 pixels by clicking Frame Size.
4.2.2.
Set the frame average to 1 or 2 by clicking Averaging.
NOTE: Increasing the frame size (>1024 × 1024 pixels) and performing more averaging (i.e., more than 2) could result in photobleaching or phototoxicity.
4.2.3.
Set the laser power to 1%-2% by clicking Lasers.
NOTE: Boosting the laser power may also lead to photobleaching or phototoxicity.
4.2.4.
Set the EM gain below the recommended level (< 800) by clicking Master Gain.
NOTE: A higher EM gain may generate artifacts.
4.2.5.
Zoom in to the region or cell of interest to reduce the image acquisition time and reduce photobleaching. This also allows the specimen to survive for a longer time.
4.3.
Start the time-lapse image capture using the Airyscan mode by clicking Start Experiment.
4.3.1.
Before clicking Start Experiment, make sure that the acquisition is optimally configured.
4.3.2.
If it displays a warning with the notice "Airyscan acquisition is not configured optimally", then click Optimal in frame size section, click Optimal in z-stack section, and click Optimal for scan area section (a minimum of 1.3 zoom is required for super-resolution imaging).
4.4.
Optimize the time interval, number of z-slices, and duration of the time-lapse imaging according to the experimental design and the type of specimen, so that the quality of the image is not compromised and the cells will not be arrested at particular stages of the cell cycle.
4.4.1.
As an example, the parameters for the time-lapse imaging of Drosophila testes expressing EGFP-tagged α-tubulin in early germline are shown in the following steps.
4.4.2.
For more than 5 h time-lapse imaging, image at a 10 min interval with 1% laser power and take up to 30 z-slices at each time point.
4.4.3.
For highly dynamic cellular processes, such as microtubule dynamics, set a 2 min interval between time points, perform 1–2 h time-lapse imaging at 1% laser power, and take up to 30 z-slices at each time point.
4.4.4.
For rare cellular events, such as anaphase to early telophase chromatin dynamics (2–3 min events), set live cell imaging with a 1 min interval between time points, perform 30 min time-lapse imaging at 1% laser power, and take up to 30 z-slices at each time point.
NOTE: These parameters may vary for different samples, so alteration will be needed for optimal use for the specific sample.
4.5.
Perform either time-lapse imaging or live snapshot imaging, depending on the sensitivity of the sample and the experimental design.
NOTE: A short movie is 15–30 min, and a long movie is more than 5 h.
4.6.
If the sample is very sensitive and cannot be studied by time-lapse imaging with a super-resolution microscope, as is often the case with Drosophila male GSCs, perform a super-resolution live snapshot (SRLS) of the sample at different cell cycle stages (as shown in Figure 2, Figure 3, Figure 4).
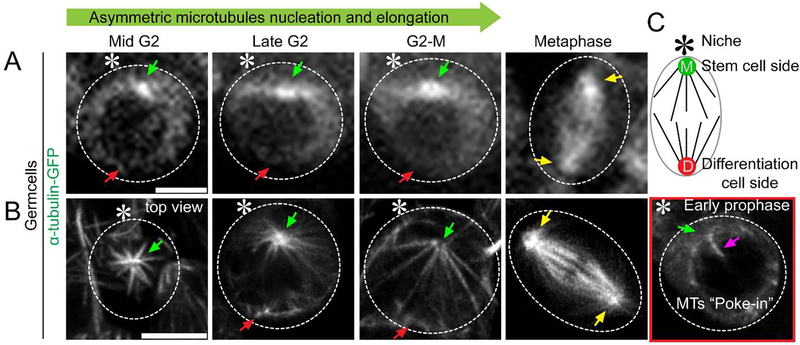
Figure 2: Super-resolution time-lapse imaging of microtubule dynamics in Drosophila tissue (testes) expressing α-tubulin-GFP.
(A) Conventional confocal microscopy live cell image showing microtubule dynamics in mid G2 phase to metaphase. (B) Airyscan microscope live cell image showing asymmetric microtubule emanating from mother versus daughter centrosomes in mid G2 phase to metaphase, with detailed structural information. A cartoon depicts the Drosophila male germline stem cell. Scale bar: 5μm; asterisk: hub (niche); green arrow: mother centrosome (stem cell side [M]); red arrow: daughter centrosome (differentiating daughter cell side [D]); yellow arrow: centrosome in spermatogonia cell (progenitor non-stem germ cell); magenta arrow: microtubule poking in nucleus.
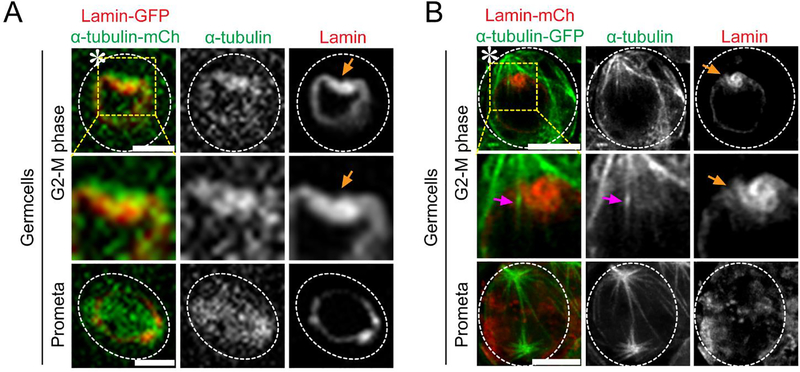
Figure 3: Super-resolution time-lapse imaging of microtubule poking-in activity and asymmetric nuclear envelope breakdown in Drosophila tissue (testes) co-expressing Lamin-mCherry and α-Tubulin-GFP.
(A) Conventional confocal microscopy live cell image showing microtubule dynamics from G2-M phase to mitosis. Image showing higher α-Tubulin-GFP intensity at the stem cell side and its interaction with the nuclear membrane. (B) Airyscan microscope live cell image showing microtubule poking-in and asymmetric NEBD at the stem cell side. Scale bar: 5μm; asterisk: hub (niche); orange arrow: site of microtubule poking in nuclear membrane; magenta arrow: microtubules that poke in at stem cell side.
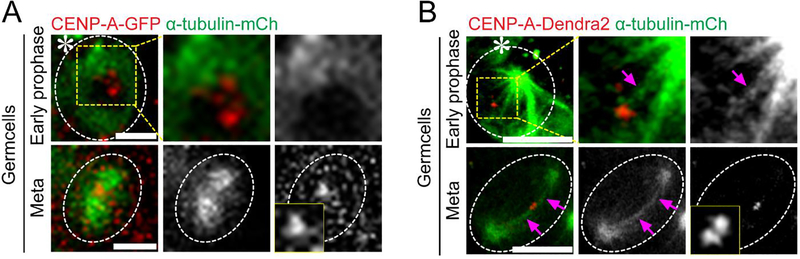
Figure 4: Super-resolution time-lapse imaging of microtubule and centromere attachment in Drosophila tissue (testes) expressing α-Tubulin-mCherry and CENP-A-Dendra2.
(A) Conventional confocal microscopy live cell image showing microtubule and centromere interaction from G2-M phase to metaphase. Image showing higher α-Tubulin-mCherry intensity at stem cell side and CENP-A-GFP signal. (B) Airyscan microscope live cell image showing microtubules emanating from the mother centrosome and attachment to the stronger centromeres in early prophase. At metaphase, centromeres are attached to opposite pole (bi-orientation). Scale bar: 5μm; asterisk: hub (niche); magenta arrow: kinetochore fiber or K-fiber (microtubules attached to the centromere).
4.6.1.
If a rare cell biological event is being captured or the protein of interest has low expression level, then perform SRLS.
4.6.2.
For SRLS, find a cell at the specific cell cycle stage you are interested in. For example, if focusing on microtubules-kinetochore binding, then use a cell at prometaphase or metaphase.
NOTE: This will help ensure that you get a high-quality image of the cell of interest at the right time without risking phototoxicity to the sample or bleaching of the fluorophores, which may occur during a longer movie.
4.6.3.
If using SRLS, image different cell cycle stages of interest across multiple cells and arrange these images in chronological order.
NOTE: This can be used to construct a temporal order of events while maximizing signal and tissue health, to obtain excellent temporal resolution of the event for particularly sensitive samples or samples with low signal.
5. Airyscan processing of the live cell images
5.1.
After image acquisition with the imaging software, select Processing, click Batch, and then select Airyscan Processing.
5.1.1.
Select the images to be processed and then click Run/Process to obtain the super-resolution images.
5.1.2.
Perform the processing using the default setting.
NOTE: Airyscan processing will only work if the imaging settings are properly established for Airyscan imaging.
REPRESENTATIVE RESULTS:
Live cell imaging beyond the diffraction limit in Drosophila tissue, especially for GSCs, provides an opportunity to investigate the dynamics of subcellular events in the context of cell cycle progression. Recently, a study utilizing this protocol has shown that microtubule activities at the mother centrosome versus the daughter centrosome are temporally asymmetric in GSCs10. The mother centrosome emanates microtubules approximately 4 hours prior to the mitotic entry, whereas the daughter centrosome only emanates microtubules at the onset of mitosis. The highly active microtubules from the mother centrosome interact with the nuclear membrane and induce polarized Nuclear Envelope Break Down (NEBD). The local NEBD is proximal to the hub and allows microtubules from the mother centrosome to enter the nucleus and preferentially attach to the stronger sister kinetochore and sister centromere to ensure that they segregate to the future stem cell. These observations suggest that an asymmetric mitotic axis exists in which the microtubules, nuclear membrane, kinetochore, and centromeres interact in a spatiotemporally controlled manner, in order to ensure proper nonrandom chromosome segregation ultimately10–12.
Each of these results was supported using the SRLS approach outlined here. By performing live cell imaging of Drosophila testes expressing α-tubulin-GFP in early-stage germ cells, we were able to identify temporal asymmetry in microtubule activity in GSCs (Figure 2). We were also able to see the asymmetric intensity of GFP signals at the two centrosomes as a brighter signal at the mother centrosome and a relatively weaker signal at the daughter centrosome side using a spinning disk confocal microscope (Figure 2A). The difference in brightness is likely reflected by the temporal asymmetry of microtubule nucleation, but the detailed morphology and quantity of microtubules could not be resolved using spinning disk confocal microscopy (Figure 2A). In contrast, live cell super-resolution imaging allowed us, to visualize the morphology and quantify the number of microtubules (Figure 2B). Its improved resolution revealed patterns of asymmetric microtubule nucleation, elongation, and increased interaction with the nuclear membrane (Figure 2B). As examples, the time-lapse movies of α-tubulin-GFP-expressing GSCs have been published in a primary research paper (Videos S3 and Video S5)10.
Next, we performed live cell imaging on Drosophila testes co-expressing either α-Tubulin-GFP with Lamin-mCherry or α-Tubulin-mCherry with Lamin-GFP in early-stage germ cells. Our results show that asymmetric microtubule activity coincided with asymmetric NEBD in GSCs (Figure 3). Using the spinning-disk confocal microscope, we could visualize the asymmetric nuclear membrane invagination but not the individual microtubules that directly entered the nucleus (Figure 3A). In contrast, this live cell super-resolution technique allowed us to directly observe these events by imaging both the microtubules and the nuclear lamina simultaneously (Figure 3B).
Next, we performed live cell imaging on Drosophila testes expressing α-Tubulin-mCherry and CENP-A-Dendra2 or CENP-A-GFP (Centromere Protein A) in early-stage germ cells. To better visualize the microtubule and centromere attachment, we live imaged them at a lower temperature (∼18 °C) to stabilize their attachment. If needed, the sample can be briefly chilled with ice or ice-cold live cell media for 4–5 minutes to stabilize the microtubule—centromere attachment and depolymerize any unattached microtubule. Our results show that the microtubules emanating from the early active mother centrosome preferentially attach to the stronger sister centromere (Figure 4). Using the spinning-disk confocal microscope, the α-Tubulin-mCherry signals displayed an asymmetric brightness near the two centrosomes. We could also detect both sister centromeres as one signal, as shown in Figure 4A(metaphase), but were not able to visualize the microtubule—centromere attachment (Figure 4A, metaphase). In contrast, the super-resolution live imaging allowed us to visualize the microtubule-centromere attachment (Figure 4B). Our results show that mother centrosome-emanating microtubules attach to the centromere prior to the daughter centrosome-emanating microtubules (Figure 4B, early prophase).
DISCUSSION:
Super-resolution microscopy methods provide spatial resolution as high as 10s of nanometers4–6. The STORM and PALM microscopy methods allow resolution up to 20 to 50 nm (XY-resolution), while STED microscopy offers resolution of 20 to 100 nm (XY-resolution). The spatial resolution of SIM microscopy is limited to 100 to 130 nm15. However, due to its high-photon density and lengthy acquisition time, it is extremely challenging to use these techniques for live cell imaging. Imaging subcellular structures, such as the cytoskeleton, chromosomes, and organelles in live cells within tissues, requires the technique to be both ultrasensitive and noninvasive while still having a high spatial resolution. The Airyscan super-resolution technique presented here combines confocal imaging with a 0.2 Airy Unit pinhole, together with pixel reassignment and deconvolution. This technique can achieve a 1.7× higher resolution than that of conventional confocal microscopy7. The ∼ 220–230 nm resolution of conventional confocal microscopy is restricted by the diffraction limit of light, but this imaging technique is able to achieve approximately 140 nm XY-resolution, which is comparable to other widely used super-resolution techniques, such as SIM (resolution at 110–120 nm)16.
The adaptability and versatility of Airyscan confocal microscopy will allow this protocol to be applicable to various other types of tissues and cells to study a diverse number of cell biology processes7,16. Using this technique, researchers can obtain time-lapse images at a super-resolution level to visualize cellular dynamics with unprecedented spatiotemporal detail.
The critical step in this technique is mounting the sample onto the dish so that it does not move or float during time-lapse imaging. Keeping the specimen close to or stuck to the imaging surface (i.e., the glass bottom of the dish) is critical for acquiring super-resolution images and maintaining the optimal microenvironment for the sample. This ensures that the sample is not stressed due to conditions such as hypoxia and changes in temperature or humidity.
Despite the usefulness of this protocol, there are a few caveats. Time-lapse imaging with the Airyscan mode is a relatively slow process compared to conventional confocal microscope imaging. Therefore, even the slightest physical shift of the sample could result in poor resolution. This situation can be prevented by using an immobilization method to adhere specimens to the imaging surface (e.g., poly L-lysine coating or other coating compatible with the sample). Also, placing a dialysis membrane on top of the tissue and adding a few small weights as described in this protocol helps prevent tissue movement or floating while also allowing for gaseous exchange. Time-lapse imaging cells that are located deep in the tissue with the super-resolution mode requires high illumination and can cause photobleaching. This can be minimized by adjusting the number of z-slices, illumination power, and averaging algorithm to achieve the optimal settings for the sample. If the cell or tissue is sensitive to laser toxicity, hypoxia, or any other stress during imaging, then cell cycle arrest may occur. Also, if a protein has an extremely low expression, short half-life, or low fluorescence signal, then photobleaching may occur which will result in an image with bad quality. This can be avoided by taking a SRLS, making a short movie, or shooting with long intervals between consecutive time points. Additionally, the microscope setting and the time-lapse imaging parameter (as described in this protocol) must be optimized to fit the sample.
Since it is essential that the sample does not move or degenerate during the process, prolonged time-lapse imaging lasting overnight or more than a day without compromising the image resolution may be challenging.
The existing methods mainly focus on optimizing the super-resolution microscopy and time-lapse imaging parameters separately1,9. However, these methods do not provide details regarding sample preparation or mounting parameters, which are critical to ensuring the viability of the sample and the adaptability of the microscope to achieve desired super-resolution during time-lapse imaging. In this method, we have optimized the imaging parameters, sample preparation and mounting parameters so that time-lapse imaging can be performed for an extended period of time to acquire super-resolution images without risking both the degeneration of the sample and the resolution.
Materials.
| Name | Company | Catalog Number | Comments |
|---|---|---|---|
| Adobe Illustrator CS6 (figure making software) | Adobe | N/A | |
| Dialysis membrane | Spectra/Por 1–4 Standard RC Dialysis Membrane | Cat No. 08-67121 | |
| FBS | Thermo Fisher Scientific | Cat. no. 26140079 | 15% (V/V) |
| Fiji (analysis software) | NIH | N/A | Image fluorescence intensity quantification |
| Glass bottom cell culture dishes (FluroDish) | World Precision Instrument, Inc. | FD35PDL-100 | |
| Imaris (image reconstruction software) | Bitplane | N/A | 3D image reconstruction |
| Imerssion oil | Zeiss | Immersol 518F/30 °C | |
| Insulin | Sigma | Cat. No. 15550 | 200 μg/ml |
| Penicillin/streptomycin | Invitrogen | Cat No. - 15140-122 | 0.6x |
| Schneider Drosophila media | Invitrogen | Cat No. - 11720-034 | |
| Spinning disc confocal microscope | Zeiss | N/A | equipped with an evolve camera (Photometrics), using a 63x Zeiss objective (1.4 NA). |
| Tissue paper | Kimwipe | N/A | Wet to form humid chamber |
| LSM 800 confocal microscope with AiryScan super-resolution module | Zeiss | N/A | equipped with highly sensitive GaAsP (Gallium Arsenide Phosphide) detectors using a 63x Zeiss objective (1.4 NA) |
| ZEN (imaging software) | Zeiss | N/A |
ACKNOWLEDGMENTS:
The authors would like to thank Integrated Imaging Core facility at Johns Hopkins University for microscopes and data analysis software. We thank J. Snedeker and Q. E. Yu for proofreading and suggestions, and X.C. lab members for helpful discussions and suggestions. Supported by NIGMS/NIH R35GM127075, the Howard Hughes Medical Institute, the David and Lucile Packard Foundation, and Johns Hopkins University startup funds (X.C.).
Footnotes
DISCLOSURES:
The authors declare no conflict of interest.
REFERENCES:
- 1.Breedijk RMP et al. A live-cell super-resolution technique demonstrated by imaging germinosomes in wild-type bacterial spores. Scientific Reports. 10 (1), 5312 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maddox PS et al. Imaging the mitotic spindle. Methods in Enzymology. 505, 81–103 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Frigault MM, Lacoste J, Swift JL, Brown CM Live-cell microscopy — tips and tools. Journal of Cell Science. 122 (6), 753–767 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Bates M, Jones SA, Zhuang X Stochastic optical reconstruction microscopy (STORM): A method for super resolution fluorescence imaging. Cold Spring Harbor Protocols. 2013 (6), pdb.top075143 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Shroff H, Galbraith CG, Galbraith JA, Betzig E Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nature Methods. 5 (5), 417–423 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vicidomini G, Bianchini P, Diaspro A STED super-resolved microscopy. Nature Methods. 15 (3), 173–182 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Huff J The Airyscan detector from ZEISS: confocal imaging with improved signal-to-noise ratio and super-resolution. Nature Methods. 12 (12), i–ii (2015). [Google Scholar]
- 8.Huff J et al. The new 2D superresolution mode for ZEISS Airyscan. Nature Methods. 14 (12), 1223–1223 (2017). [Google Scholar]
- 9.Korobchevskaya K, Lagerholm B, Colin-York H, Fritzsche M Exploring the potential of Airyscan microscopy for live cell imaging. Photonics. 4 (4), 41 (2017). [Google Scholar]
- 10.Ranjan R, Snedeker J, Chen X Asymmetric centromeres differentially coordinate with mitotic machinery to ensure biased sister chromatid segregation in germline stem cells. Cell Stem Cell. 25 (5), 666–681 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kahney EW, Ranjan R, Gleason RJ, Chen X Symmetry from asymmetry or asymmetry from symmetry? Cold Spring Harbor Symposia on Quantitative Biology. 82, 305–318 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wooten M, Ranjan R, Chen X Asymmetric histone inheritance in asymmetrically dividing stem cells. Trends in Genetics. 36 (1), 30–43 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wooten M et al. Asymmetric histone inheritance via strand-specific incorporation and biased replication fork movement. Nature Structural & Molecular Biology. 26 (8), 732–743 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prasad M, Jang AC-C, Starz-Gaiano M, Melani M, Montell DJ A protocol for culturing Drosophila melanogaster stage 9 egg chambers for live imaging. Nature Protocols. 2 (10), 2467–2473 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Schermelleh L, Heintzmann R, Leonhardt H A guide to super-resolution fluorescence microscopy. Journal of Cell Biology. 190 (2), 165–175 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sivaguru M et al. Comparative performance of airyscan and structured illumination superresolution microscopy in the study of the surface texture and 3D shape of pollen. Microscopy Research and Technique. 81 (2), 101–114 (2018). [DOI] [PubMed] [Google Scholar]