Abstract
In this review, we chart the major milestones in the research progress on the DyP-type peroxidase family over the past decade. Though mainly distributed among bacteria and fungi, this family actually exhibits more widespread diversity. Advanced tertiary structural analyses have revealed common and different features among members of this family. Notably, the catalytic cycle for the peroxidase activity of DyP-type peroxidases appears to be different from that of other ubiquitous heme peroxidases. DyP-type peroxidases have also been reported to possess activities in addition to peroxidase function, including hydrolase or oxidase activity. They also show various cellular distributions, functioning not only inside cells but also outside of cells. Some are also cargo proteins of encapsulin. Unique, noteworthy functions include a key role in life-cycle switching in Streptomyces and the operation of an iron transport system in Staphylococcus aureus, Bacillus subtilis and Escherichia coli. We also present several probable physiological roles of DyP-type peroxidases that reflect the widespread distribution and function of these enzymes. Lignin degradation is the most common function attributed to DyP-type peroxidases, but their activity is not high compared with that of standard lignin-degrading enzymes. From an environmental standpoint, degradation of natural antifungal anthraquinone compounds is a specific focus of DyP-type peroxidase research. Considered in its totality, the DyP-type peroxidase family offers a rich source of diverse and attractive materials for research scientists.
Keywords: DyP, DyP-type peroxidase, structure-based sequence alignments, antifungal anthraquinone compounds, lignin degradation, iron uptake, life cycle, hydrolase, oxidase, encapsulin, nano compartment, cargo protein
1. Introduction
General peroxidase research has seemed to wane in recent years, at least in part because peroxidases are ubiquitous and their generally accepted physiological role in plants—removing hydrogen peroxide (H2O2) generated inside of cells and constructing a part of the cell wall such as lignin—has been long and thoroughly studied. In contrast, the research history of DyP-type peroxidases is in its early stages, with the first identification of the enzyme occurring only two decades ago. The first DyP-type peroxidase, dye decolorizing peroxidase (DyP) from Geotrichum candidum, was reported in 1999 [1,2]. The origin of this enzyme was later revised to Bjerkandera adusta [3]. In 2007, DyP-type peroxidases were recognized as being distinguishable from other known peroxidase and were defined as a novel peroxidase family—the DyP-type peroxidase family [4]. To date, DyP-type peroxidases have been found in a variety of organisms (Figure 1). These enzymes also exhibit a varied distribution, functioning not only inside but also outside of cells. In prokaryotes, in particular, the cellular distribution ranges widely from intracellular to extracellular, including periplasmic space. Some secreted DyP-type peroxidases have a Tat (twin arginine translocation) signal, suggesting that they are Tat substrates [5,6,7,8,9,10]. Notably, all secreted DyP-type peroxidases with Tat signal belong to class I, as described in a later section. Moreover, EfeB and YwbN, which are representative DyP-type peroxidases as Tat substrates, reside in the periplasmic space and outside of the cell, respectively. They are homologs of each other and are inferred to play similar physiological roles [8,11]. Among eukaryotes, basidiomycetes have been a source of interesting findings on DyP-type peroxidases. Surprisingly, the DyP-type peroxidases in these organisms are all extracellular, similar to some other peroxidases from basidiomycete. If the main role of DyP-type peroxidases is to scavenge the H2O2 generated inside of the cell or construct a part of cell tissues, it would be unnecessary to secrete the protein into the extracellular milieu. Therefore, secreted DyP-type peroxidases likely serve a function independent of removal of intracellular H2O2 or construct the cell tissues. Interestingly, it appears that some DyP-type peroxidases exist as a cargo protein in encapsulin [12,13]. In particular, there is speculation that MtDyP (class P) provides defense against attacks of host cells. Several DyP-type peroxidases are summarized in outline form in Table 1.
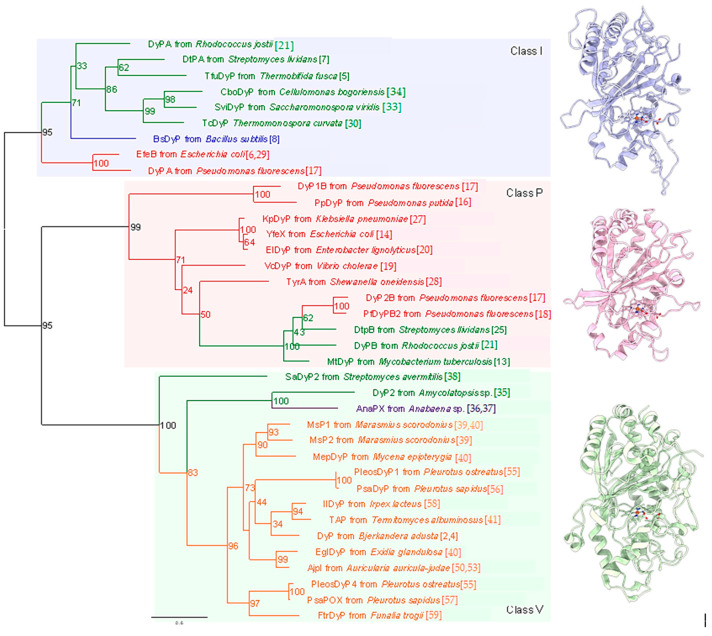
Figure 1.
DyP phylogenetic tree. The tree was constructed with the maximum-likelihood method using RAxML-NG [61] with multiple sequence alignments of DyP amino acid sequences generated by the MAFFT [62] program. Percentages of bootstrap values obtained from 1000 bootstrap replicates are shown at the nodes. The best-fit model of evolution of the alignment was determined using ModelTest-NG [63]. Branches and labels of different phyla are shown in different colors: green, actinobacteria; blue, firmicutes; red, proteobacteria; purple, cyanobacteria; orange, basidiomycota. Reference numbers for each DyP are shown in parentheses. Structures of representative DyPs of the three classes are shown on the right: Class I, DtPA (PDB ID: 6gzw); Class P, DtPB (PDB ID: 6yrj); Class V: DyP (PDB ID: 3afv).
Table 1.
Characteristics of representative DyP-type peroxidase from classes P, I, and V.
| Class | Former Class | Name | Length (aa) |
Reaction with Lignin a | Peroxidase Kinetic Parameters for Anthraquinone Compound | Comp II c | Deduced Radical Sites | Remarkable Comments | Reference | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Km (µM) |
kcat (s−1) |
kcat/Km (s−1M−1) |
Substrate b | |||||||||
| P | B | YfeX | 299 | [14] | ||||||||
| DyPPa | 299 | 107 | 0.024 | 2.2 × 102 | rb5 | [15] | ||||||
| PpDyP | 287 | 40 | 8.0 | 2.0 × 105 | rb5 | manganese oxidizing activity | [16] | |||||
| DyP1B | 295 | ✓ | 120 | 1.0 | 9.0 × 103 | rb4 | manganese oxidizing activity | [17] | ||||
| DyP2B | 324 | ✓ | Not Detected | rb4 | manganese oxidizing activity | [17] | ||||||
| PfDyP B2 | 316 | ✓ | 10 | 1.5 | 1.5 × 105 | rb4 | [18] | |||||
| VcDyP | 302 | 50 | 1.3 | 2.6 × 104 | rb19 | ✓ | [19] | |||||
| ElDyP | 299 | manganese oxidizing activity | [20] | |||||||||
| DyPB | 350 | ✓ | 350 | 0.05 | 1.4 × 102 | rb4 | ✓ | manganese oxidizing activity (improved in N246A), | [12,21,22,23,24] | |||
| encapsulin carrier | ||||||||||||
| DtpB | 316 | [25] | ||||||||||
| Mt-DyP | 335 | encapsulin carrier | [13,26] | |||||||||
| KpDyP | 299 | ✓ | [27] | |||||||||
| TyrA | 311 | 84 | 5.9 | 7.0 × 104 | rb5 | [28] | ||||||
| I | A | EfeB (YcdB) | 423 | Tat signal, EfeUOB operon, deferrochelatase? | [6,29] | |||||||
| DyPA | 436 | ✓ | 210 | 1.9 | 9.0 × 103 | rb4 | [17] | |||||
| TcDyP | 403 | 5 | 41 | 7.8 × 106 | rb19 | ✓ | ✓ | [30,31] | ||||
| TfuDyP | 430 | 29 | 10 | 3.5 × 105 | rb19 | Tat signal | [5] | |||||
| ✓ | 179 | 1.9 | 1.0 × 104 | rb4 | [32] | |||||||
| SviDyP | 404 | ✓ | rb19 | [33] | ||||||||
| DyPA | 428 | 1,000 | 13 | 1.3 × 104 | rb4 | ✓ | [21,24] | |||||
| DtpA | 445 | ✓ | Tat signal, sco, ecuc, Cu-transporter | [7] | ||||||||
| YwbN | 416 | Tat signal | [8,9] | |||||||||
| FepB | 409 | FepABC operon (A,EfeO; B,EfeB; C,EfeU), | [10] | |||||||||
| deferrochelatase? | ||||||||||||
| CboDyP | 387 | 17 | 0.22 | 1.3 × 104 | rb19 | [34] | ||||||
| V | C | DyP2 | 473 | ✓ | 48 | 34 | 7.1 × 105 | rb5 | Mn binding site in crystal structure, | [35] | ||
| Mn-dependent oxidase activity | ||||||||||||
| AnaPX | 469 | 3.6 | 384 | 1.2 × 107 | rb5 | [36,37] | ||||||
| SaDyP2 | 456 | 61 | 0.78 | 1.2 × 104 | ab324 | [38] | ||||||
| D | MsP1 (MscDyP) | 513 | ✓ | β-carotene is a substrate | [39,40] | |||||||
| MsP2 | 510 | β-carotene is substrate | [39] | |||||||||
| TAP | 504 | [41] | ||||||||||
| DyP | 498 | 80 | 980 | 1.2 × 107 | The first found DyP-type peroxidase | [1,2,3,4,42,43,44,45,46,47,48] | ||||||
| AjPI (AauDyP1) | 509 | ✓ | 23 | 114 | 5.0 × 106 | rb5 | ✓ | [40,49,50,51,52] | ||||
| [53,54] | ||||||||||||
| EglDyP | 501 | ✓ | [40] | |||||||||
| MepDyP | 526 | ✓ | [40] | |||||||||
| Pleos-DyP1 | 516 | 45 | 5 | 1.1 × 105 | rb19 | manganese oxidizing activity | [55] | |||||
| Pleos-DyP4 | 504 | 82 | 152 | 1.9 × 106 | rb19 | ✓ | manganese oxidizing activity/ | [55] | ||||
| Mn binding site is the same as DMP binding site from [43]. | ||||||||||||
| PsaDyP | 516 | 24 | 18 | 7.5 × 105 | rb5 | oxidase activity/ | [56] | |||||
| β-carotene, annatto are also substrates | ||||||||||||
| PsaPOX | 504 | Not Detected | rb19 | manganese oxidizing activity/alkene cleavage/ | [57] | |||||||
| β-carotene, annatto are also substrates | ||||||||||||
| Il-DyP4 | 502 | ✓ | 133 | 5,345 | 4.0 × 107 | rb19 | manganese oxidizing activity | [58] | ||||
| FtrDyP | 484 | 187 | 2.6 | 1.4 × 104 | manganese oxidizing activity | [59] | ||||||
| AncDyPD-b1 | 511 | 42 | 22 | 5.3 × 105 | rb19 | ancestral fungal dye-decolorizing peroxidase | [60] | |||||
| AjPII (AauDyP2) | unknown | ✓ | 15 | 256 | 1.7 × 107 | rb5 | [40,50] | |||||
a include R-478 and model compounds of lignin b rb and ab mean reactive blue and acid blue, respectively. c comp II means compound II.
In any case, research performed over the past decade has clarified the unique characteristics of DyP-type peroxidases, converging on the view that they do not exist solely to remove H2O2 but instead have more important specific roles.
2. Importance of Tertiary Structure and Catalytic Mechanism
2.1. Characteristics Based on Tertiary Structure
In early-stage research, DyP-type peroxidases were categorized into classes A, B, C and D according to primary structural homologies (RedOxiBase). Although this orthodox classification is useful, it has limited value for studying catalytic mechanisms and active sites because the primary structure provides only amino acid sequence information. In contrast, tertiary structures define tangible entities that allow for detailed assessments of the relationship between structure and function among members of the DyP-type peroxidase family. In 2015, a new classification scheme was proposed for this family based on structure-based sequence alignments obtained using the multiple three-dimensional alignment tool, MATRAS [42]. Such tertiary structural analyses are apparently effective for further analysis because they revealed genuine structural homologies among different DyP-type peroxidases. Interestingly, this new scheme clearly reclassified the family into three new classes, combining classes C and D into a new class V (adVanced) and renaming classes A and B as classes I (Intermediate) and P (Primitive), respectively [42]. Throughout this review, we apply this new class I, P and V classification scheme. Typical structures of each of the three classes are shown on the right side of Figure 1.
The basic skeletal structure is common among the three classes and consists of a dimeric α + β barrel structure [42]. The structures of classes I and V are constructed based on class P, with some additional regions. Therefore, class P members have the smallest molecular size among the three classes, with members of a class V having the largest size. Notably, all important amino acid residues that define DyP-type peroxidases are included in the skeletal structure (i.e., class P), suggesting that basic functions are determined by this skeletal structure, with extra regions mediating additional functions. For instance, catalytic efficiencies (kcat/Km) toward anthraquinone compounds are different among the three classes. As shown in Table 1, the catalytic efficiencies of most class V members (104 to 107) are the largest among the three classes. By comparison, catalytic efficiencies range from 103 to 106 for class I and from 102 to 105 for class P.
2.2. Catalytic Mechanism
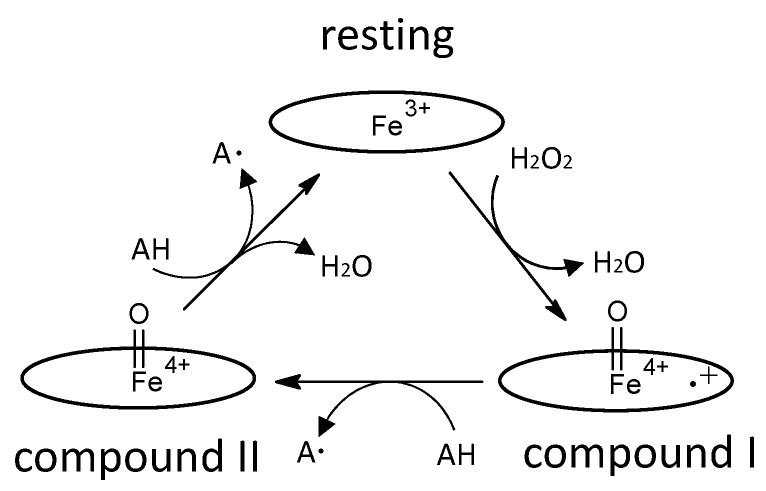
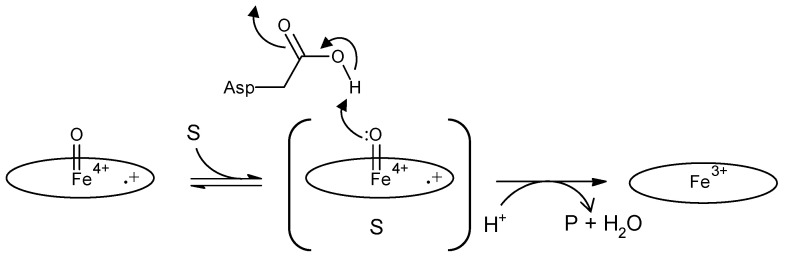
The enzyme commission number for DyP is EC 1.11.1.19, which corresponds to a peroxidase group. Actually, in most cases, DyP-type peroxidases show typical peroxidase activity, such as that shown by horseradish peroxidase (HRP). Therefore, the catalytic cycle appears to be nearly the same as that of ubiquitous peroxidases [64]. As shown in Figure 2, general heme peroxidases cycle through resting, compound I and compound II states. The compound I formation process of DyP-type peroxidase is essentially inconsistent with that of classical heme peroxidases such as lignin peroxidase [4]. In the case of DyP-type peroxidase, resting and compound I states have been confirmed, but compound II is controversial. DyPA (class I) from Rhodococcus jostii shows a spectrum corresponding to compound II at pH 7.5 [21]. TcDyP (class I) from Thermomonospora curvata shows compound II at pH 7.8 with both H2O2 and hydroquinone, but not with H2O2 only [30]. Furthermore, compound II was not observed at pH 3, which is the optimum pH for dye decolorizing [30]. In support of this, Shrestha et al. reported that compound II was not observed at low pH in ElDyP (class P) from Enterobacter lignolyticus, suggesting that the catalytic cycle does not follow a typical two-step process [20]. The authors of this latter study suggested that the enzyme adopts a two-electron reduction process for compound I, as shown in Figure 3. Moreover, no exact compound II has yet been reported at low pH for DyP-type peroxidases belonging to classes P or V. Therefore, further study at low pH—the active pH for dye decolorization—is necessary regardless of whether the two-step catalytic cycle with sequential one-electron reduction of the enzyme occurs or not. On the other hand, excess H2O2 deactivates DyP-type peroxidsaes, a property also characteristic of ubiquitous peroxidases.
Figure 2.
Catalytic cycle of a typical heme peroxidase. Oval denotes the heme plane in the enzyme. AH is a substrate.
Figure 3.
Schematic diagram of two-electron oxidation of substrates by a DyP-type peroxidase, proposed in [20]. Left, compound I; center, ES complex; right, resting state. Oval denotes the heme plane in the enzyme. Asp is the catalytic residue. S and P denote substrate and product, respectively. In a general peroxidase, compound I changes to compound II with one electron reduction, as shown in Figure 2, but in this scheme, compound I of DyP-type peroxidases changes to the resting state directly through two-electron reduction.
2.3. Active or Binding Sites
DyP-type peroxidases show a broad range of substrate specificities, and several active sites have been proposed [31,43,49,65]. It is likely that the binding site(s) of most substrates are different from the H2O2 binding site. We will first focus on the H2O2 binding site, which is located at the distal site of the heme. As described previously, aspartic acid and arginine are absolutely conserved in the H2O2-binding site of most DyP-type peroxidases, whereas general peroxidases have histidine and arginine at this site [44]. In particular, aspartic acid (D) in the GXXDG motif, which is a well-known conserved region among DyP-type peroxidases, replaces the histidine in general peroxidases, accounting for the lower optimum pH of DyP-type peroxidases compared with general peroxidases [4,44]. A D171N point mutation in DyP (class V) from B. adusta results in the disappearance of compound I formation. This causes a drastic decrease in peroxidase activity [4], highlighting the critical importance of aspartic acid in GXXDG. In contrast, Singh et al. reported that the arginine in DyPB (class P) from R. jostii is essential for peroxidase activity [22]. It has similarly been found that the arginine in DtpB (class P) from Streptomyces lividans is also a key residue for peroxidase activity [25]. In this latter study, a serial femtosecond X-ray crystallography approach was used to determine how the distal heme site in DyP-type peroxidase can be tuned to choose either aspartic acid or arginine. In the case of YfeX (class P), the activity toward guaiacol and catechol is retained if the arginine is replaced with other amino acids, suggesting that this arginine may play a limited role [14]. Additionally, it has been reported that the distal aspartic acid of ElDyP (class P) is catalytically more important than the distal arginine and plays a key role in determining the acidic pH optimum of DyPs [20]. Similar effect depending on pH has been reported in a resurrect class V DyP [60].
As noted above, it is clear that DyP forms compound I, indicating two-electron oxidization using hydrogen peroxide. Actually, compound I of DtpB has been directly determined for a peroxidase carrying a porphyrin π cation radical [25]. Although compound I oxidizes various substrates, no reports provide direct evidence to show how and where substrates bind to it. This is one of the least clarified and essential characteristics of DyP-type peroxidases. The active sites of substrates other than H2O2 are probably located apart from the distal heme region, which would otherwise limit access to bulky molecules, such as synthetic dyes. A likely hypothesis is that this site is located at the molecular surface of the enzyme, and that an electron is transferred from the distal area of the heme to the substrate via a long-range electron-transfer route [31,49,66]. A similar speculation has been raised for versatile peroxidases [67]. Several studies have shown that aromatic residues such as tryptophan and tyrosine accumulate and retain radicals, thereby serving as a point of substrate oxidation [19,31,49,66]. This hypothesis seems reasonable, although unfortunately there are no reports of a genuine enzyme–substrate (ES) complex between bulky substrates and aromatic residues in DyP-type peroxidases. In contrast, DMP (2,6-dimethoxyphenol), which is a general substrate for peroxidases, has been reported to associate with asparagine 313 at the molecular surface of DyP from B. adusta (class V), and a hydrogen-bonding network has been reported from there to the propionate of heme [43]. This network is similar to that of ascorbate peroxidase [68]. To date, this is the sole report of an ES complex for a DyP-type peroxidase. Although a Mn2+ binding pocket was observed in an artificial N246A mutant of DypB [23], there was no report of an ES complex in the case of native DypB. Despite these advances, the location of the binding site for bulky substrates such as synthetic dyes, the representative substrates of DyP, has remained elusive.
3. Functions Besides Peroxidase
3.1. Hydrolase
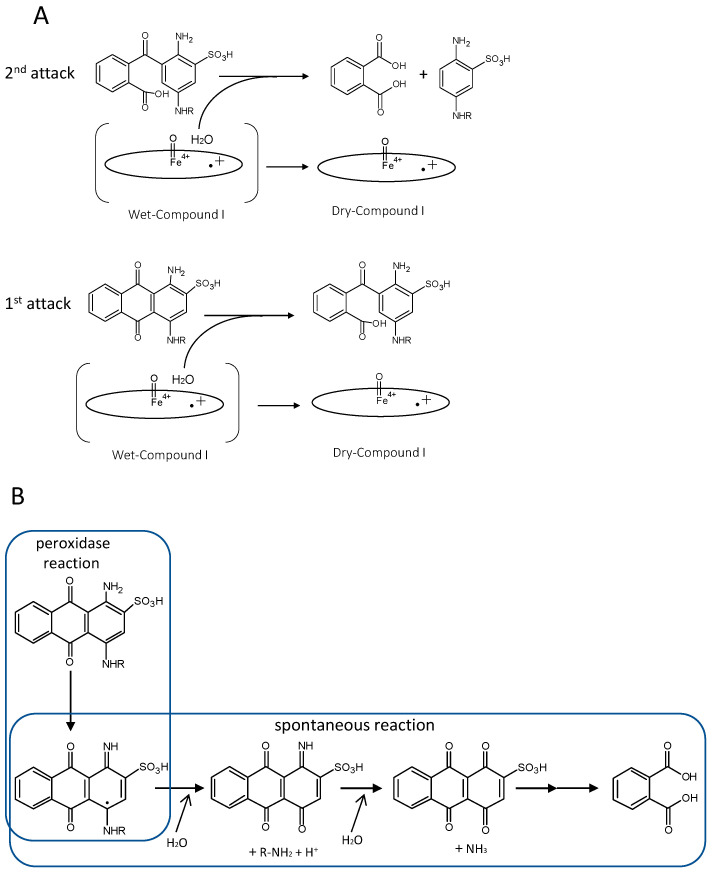
Some DyP-type peroxidases have been reported to have other functions besides peroxidase activity. One of the reaction products confirmed to arise from catalysis of Reactive blue 5—a representative anthraquinone dye—by DyP from Bjerkandera adusta is phthalic acid [44,45]. This indicates that the anthraquinone frame is degraded by hydrolysis. If this is the case, it is reasonable that DyP acts as a hydrolase because this hydrolysis would never proceed in the absence of H2O2, suggesting the necessity of forming H2O from H2O2. The importance of H2O produced from H2O2 is also reported for classical heme peroxidases and catalases. Jones reported that wet (H2O-containing) and dry (non-H2O-containing) forms of compound I apparently play independent roles. He proposed a redox pathway switching mechanism, such that the states for the two electron-equivalent reduction of compound I are accessible in the dry form, but in the wet form only one-electron-equivalent processes are possible, unless the release of water can be stimulated [69]. The importance of a water molecule in the heme distal area was also reported for DtpB from S. lividans (class I) [25]. These reports suggested that H2O from H2O2 might play a specific role in the catalytic mechanism. Together, these observations suggest that DyP from B. adusta is a bifunctional enzyme that exerts hydrolase activity using the H2O released by the peroxidase function. In contrast, Linde et al. have reported that spontaneous hydrolysis would occur if an anthracenetetrone was formed as an intermediate from an anthraquinone, given that the ubiquitous heme peroxidase HRP also decolorizes Reactive blue 5 [66]. Actually, HRP decolorizes Reactive blue 5, but there is no evidence that it degrades the anthraquinone frame because phthalic acid has thus far not been detected. It is probably the case that decolorization with HRP depends on degradation, not of the anthraquinone frame, but of an auxochrome group. Moreover, there are no reports that anthracenetetrone is generated as an intermediate for a DyP-type peroxidase. These two ideas are summarized in Figure 4.
Figure 4.
Comparison of proposed anthraquinone degradation mechanisms. A, H2O from Wet compound I attacks the anthraquinone frame, causing enzymatic hydrolysis [44]. Oval denotes the heme plane in the enzyme. B, Spontaneous hydrolysis generates phthalic acid, but predicted intermediates have not been detected [66].
3.2. Deferrochelatase or Iron Uptake
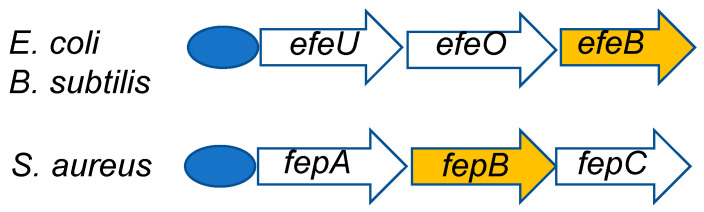
In 2009, Létoffé et al. reported that EfeB (class I), a DyP-type peroxidase from Escherichia coli, functions as a deferrochelatase [70]. They observed that EfeB captures only iron without degradation of heme, which retains an intact tetrapyrrole skeleton. This phenomenon is quite different from that of heme oxygenase, which degrades the heme skeleton and then “picks out” the iron. This study created a profound impression on many researchers of DyP-type peroxidases. However, the corresponding function of FepB (class I), an EfeB homolog from Staphylococcus aureus, is obscure [11]. Moreover, YwbN (class I) from Bacillus subtilis, also a homolog of EfeB, shows no deferrochelatase activity, but does exhibit peroxidase activity [8]. Overall, whether DyP-type peroxidases have deferrochelatase activity remains a matter of controversy. Viewed from another standpoint, EfeB is a part of the EfeUOB operon, which encodes components of an iron transport system [29]. Interestingly, a similar operon has been found in both B. subtilis and S. aureus [8,11], and it has been proposed that both EfeB homologs are typical peroxidases that result in oxidation of Fe2+. Moreover, the efeB gene of B. subtilis is under additional control of several σ factors that are associated with the cell envelope stress response, suggesting a key role in the life-cycle of the organism [8]. Collectively, these studies suggest that the relationship between iron uptake and the role of DyP-type peroxidases must be critical to bacterial viability because iron uptake is essential for life. Further study of this relationship can be expected. A schematic diagram of the EfeUOB operon from the above three organisms is shown in Figure 5.
Figure 5.
Schematic depiction of efeUOB and fepABC operons. Both operons are regulated by the Fur repressor (blue oval). efeB and fepB are orthologs of each other and encode DyP-type peroxidases.
3.3. Oxidase
Peroxidase reactions absolutely depend on peroxides such as H2O2. Surprisingly, MscDyP from Marasmius scorodonius, a typical class V DyP, appears to oxidize β-carotene without H2O2 [39]. If this proves to be the case, it means that MscDyP not only shows peroxidase activity but also possesses oxidase activity, suggesting a bifunctional enzyme. Additionally, catalases, which are similar to heme peroxidases, are also suggested to show phenol oxidase activity in the absence of H2O2 [71]. However, it has been reported that PsaPOX (class V) from Pleurotus sapidus oxidizes β-carotene in the presence of H2O2, suggesting peroxidase activity [57]. Strikingly, β-carotene was found to be transformed by recombinant PsaDyP in both the presence and absence of H2O2, although enzymatic activity was increased by the addition of H2O2 [56]. This suggests that rPsaDyP has an oxidase function in addition to peroxidase activity. PsaPOX was also shown to oxidize several alkenes, such as anethol to anisaldehyde, but the details of the reaction mechanism are still obscure [57].
4. Physiological Role of DyP-Type Peroxidases
One of the accepted physiological role of a plant peroxidase is to form lignin of cell wall consuming H2O2 generated inside of cells. In contrast, it is noteworthy that many DyP-type peroxidases function in the extracellular environment. This suggests that the main physiological role of these peroxidases is not removal of intracellular H2O2, leading to speculation about other cryptic roles. An additional noteworthy characteristic of DyP-type peroxidases is that they are distributed across a broad range of organisms, suggesting possible divergent physiological roles. Probable physiological roles are considered below.
4.1. Anthraquinone Degradation
The DyP (class V) from Bjerkandera adusta Dec 1 is a classic member of this family. This strain was isolated from soil in Japan and exhibits broad decolorizing effects on various synthetic dyes [72]. Notably, DyP showed stronger decolorizing actions against anthraquinone dyes than azo dyes, a unique characteristic compared with other decolorizing enzymes known to date, which have been reported to be effective toward azo dyes [73,74,75,76]. One of the most distinctive characteristics of DyP from B. adusta is its unique ability to degrade the anthraquinone skeleton. Reactive blue 5 is actually completely decolorized through concerted reactions with versatile peroxidases and DyP [46], making these enzymes suitable for treating wastewater containing synthetic dyes. However, such synthetic dyes are not true substrates because they are never generated in nature, suggesting that natural anthraquinone compounds must be true substrates. For example, plants, including trees, synthesize multiple anthraquinone compounds that serve antifungal functions. In this context, DyP from B. adusta degrades alizarin, which is a natural antifungal anthraquinone compound produced by plants [47]. B. adusta is a white rot fungus that parasitizes living trees, which in response generate phytoalexin to protect against infection. Therefore, one probable physiological role is to degrade antifungal anthraquinone compounds and accelerate tree parasitism. This is the only study of its kind to date, but future work on this issue is expected.
4.2. Lignin Degradation
A relationship between lignin degradation and some DyP-type peroxidases has been reported. In particular, there are many reports of this function for class V DyPs, as shown in Table 1. To date, lignin peroxidase (LiP), versatile peroxidase (VP), manganese peroxidase (MnP), and laccase (Lac) from basidiomycetes and ascomycetes have been well studied, as have lignin-degrading enzymes from basidiomycetes, such as white rot fungi [67,76,77,78]. In recent years, several DyP-type peroxidases have been found to degrade lignin or its model compounds [30,79,80,81]. Interestingly, lignin degradation by DyP-type peroxidases has been reported for bacteria as well as basidiomycetes, whereas general lignin-degrading enzymes are mainly isolated from basidiomycetes. DyPA from Pseudomonas fluorescens pf-5 [17], TfuDyP from Thermobifida fusca [5], and SviDyP from Saccharomonospora viridis DSM43017 [33] are class I, whereas DyP1B from Pseudomonas fluorescens pf-5 [17], DyP2B, and DyPB from Rhodococcus jostii are class P. DypB from Rhodococcus jostii, which is an actinomycete, has been reported to degrade lignin in the presence of Mn2+ and H2O2, but its activity is low compared with that of fungal lignin-degrading enzymes [24,35,66]. DypB might be a manganese peroxidase whose primary role is to oxidize Mn2+ to Mn3+ [24]. However, the kcat/Km ratio of DypB for Mn2+ was reported to be 1/31,250th of that of a typical MnP, indicating that Mn2+ is unlikely an essential substrate for DypB [82]. In contrast, DypA (class I) from the same strain shows no lignin-degrading activity, suggesting a difference in physiological roles, despite the fact that both are DyP-type peroxidases [24]. More puzzlingly, TfuDyP (class I) from T. fusca, which is also an actinomycete, was shown to degrade kraft lignin and oxidize a β-aryl ether lignin model compound [32]. Specifically, this latter study demonstrated that Streptomyces sp. S6 grew in medium containing kraft lignin as a sole carbon source and showed apparent LiP activity. However, in the case of DyP, S6 showed low activity—at most 1% compared with LiP [81]. SviDyP from Saccharomonospora viridis showed optimum pH and temperatures of pH 7.0 and 70 °C, respectively. These characteristics seem to offer advantages compared with other DyP-type peroxidases for practical biobleaching of kraft pulp [33]. Furthermore, two lignin-degrading bacteria, Ochrobacterium sp. and Paenibacillus sp., which contain no class P DyP-type peroxidases that have been implicated in lignin degradation in other bacteria, alternatively possess a multi-copper oxidase gene, which shows oxidation activity for β-aryl ether and biphenyl lignin dimer model compounds [83]. Thus, the importance of DyP-type peroxidases in lignin degradation, at least in bacteria, is open to interpretation.
The situation is different in basidiomycetes, for which considerable research supports lignin degradation. White rot fungi, such as Phanerochaete chrysosporium and Pleurotus ostreatus, in particular have demonstrated prominent lignin degradation activity. Their main enzymes are LiP, MnP and VP. Notably, P. chrysosporium does not have a DyP-type peroxidase. Since DyP-type peroxidases were first identified, their potential for lignin degradation has been a focus of research attention [39,40,49,50,51,52,84]. Such studies have confirmed the widespread transcript-level expression of DyP-type peroxidases in almost all samples of fungi from forest floor habitats [85]. The class V DyP-type peroxidases, AjP I, AjP II, EglDyP and MepDyP, from basidiomycetes are secreted outside the cell and degrade non-phenolic lignin model compounds through their peroxidase activity [40,50]. However, the lignin-degrading activity of DyP-type peroxidases from basidiomycetes is at most 4% that of LiP from P. chrysosporium [40,66]. IlDyP (class V) from Irpex lacteus has been characterized and suggested to hydrolyze wheat straw [86]. Because wheat straw contains lignocellulose, IlDyP might function in lignin degradation. Consistent with this, IlDyP-1 and IlDyP-2 were reported to degrade the lignin model compound, DMP [87], although this study reported no data on degradation of lignin itself by IlDyP.
Taken together, these observations indicate that most lignin-degrading organisms that possess DyP also have general lignin-degrading enzymes, such as LiP, MnP, VP and multi-copper oxidase; thus, whether lignin-degrading activity is an essential function of DyP-type peroxidases remains obscure.
4.3. Life Cycle of an Actinomycete
DtpA (class I) from the actinomycete S. lividans appears to have a specific role in the life cycle of the organism. In S. lividans, copper is important for stimulating the developmental switch between vegetative mycelium and aerial hyphae. DtpA uses H2O2 to oxidize Cu+ to Cu2+, which causes maturation of GlxA; thus, DtPA is required for maturation of GlxA. GlxA, in turn, generates H2O2 from O2, thereby providing the H2O2 necessary for DtPA-mediated oxidation of Cu+ to Cu2+. GlxA is a key protein in the shift from vegetative mycelium to aerial hyphae [88]. Mature GlxA acts in concert with the cellulose synthase-like protein, CslA, to form an extracellular glycan specific for aerial hyphae. This role for DtpA in oxidizing metal ions would thus be similar to the function of EfeB, which oxidizes Fe2+ to Fe3+ [8]. Notable in this context, S. lividans also appears to have an efeB homologue, which is located in a gene cluster harboring an iron transporter. Streptomyces avermitilis also has an efeB homologue (SAV_5925), strongly suggesting its specific role.
4.4. Cargo Protein of Encapsulin
An additional unique characteristic of several DyPs is their function as a cargo protein for encapsulin, a bacterial nanocompartment protein. One of the best-studied of these DyPs is MtDyP (class P) from Mycobacterium tuberculosis [13,26], a bacterium that slips through defenses generated by the host immune response and induces tuberculosis. MtDyP, which is shielded from oxidative stress in the nanocompartment through association with encapsulin, appears to play an important role in defending against the immune assault by virtue of its peroxidase activity. DypB (class P) from R. jostii was shown to assemble with encapsulin in vitro [12], suggesting the potential of class P DyPs to act as cargo proteins of encapsulin. A recent high-resolution cryogenic electron microscopy study revealed that a DyP-type peroxidase from Mycobacterium smegmatis is a primary cargo protein of mycobacterial encapsulins [89]. Notably, this latter study noted that the encapsulin shell plays a role in stabilizing DyP in a dodecameric form, which is larger than any previously reported DyP oligomer. Considering this packaging of M. smegmatis encapsulin as a model system for M. tuberculosis would suggest a similar role for DyP from M. smegmatis in protecting cells against oxidative stress. By extension, if DyPs from pathogens indeed serve to defend against immune assault from the host, VC2145 VcDyP (class P) from Vibrio cholerae [67] might be speculated to have a similar role. One more noteworthy characteristic is that encapsulated DyP has a specified target peptide toward encapsulin. If this is the case, screening for the presence of the target peptide is effective to find unknown encapsulated DyP-type peroxidase [90].
5. Perspectives
In this review, we have highlighted the prominent themes in the progression of DyP-type peroxidase research over the past decade. The feature of DyP-type peroxidase that has received the greatest emphasis is the diversity of this family, which displays a widespread expansion of three different classes: P, I and V. Tertiary structural analyses have clearly identified both common and distinct features among the three classes. Substrate specificities and catalytic efficiencies have also been shown to vary among the three classes. Therefore, researchers should continue to apply the current structure–function approach to clarify the catalytic mechanism, which remains unclear. In the end, the catalytic mechanism will likely turn out to be more complicated than that of ubiquitous heme peroxidases, reflecting additional functions such as hydrolase or oxidase activity. One issue in particular that will require further careful study as part of efforts to elucidate the multifunctional feature of DyP-type peroxidase is the wet and dry states of compound I.
Surprisingly, the true physiological roles of DyP-type peroxidases remain unclear; thus, establishing these roles is an important area for future research. As noted above, DyP-type peroxidases have been clearly shown to function in various environments, including extracellular, periplasmic as well as intracellular environments, including within encapsulin. These observations suggest that the physiological role of DyP-type peroxidases is not limited to removal of H2O2 inside of cells, hinting at other critical physiological functions. In particular, MtDyP and DyP from B. adusta seem to have a similar physiological role: defending against counterattacks from the host. Moreover, EfeB and DtpA play important roles in oxidizing Fe2+ and Cu+, respectively, taking part in the life cycle of their organisms. Compared with other peroxidases reported to date, DyP-type peroxidase have unique features that put them in a class of their own, arguably one that is more advanced than that of ubiquitous peroxidases. With future studies, the universe of DyP-type peroxidase will be unveiled, revealing itself to be widespread and deep like the cosmos.
Author Contributions
Conceptualization and organization, writing—original draft preparation, review and editing, supervision Y.S.; data curation and validation T.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kim S.J., Shoda M. Purification and Characterization of a Novel Peroxidase from Geotrichum candisum Dec 1 Involved in Decolorization of Dyes. Appl. Environ. Microbiol. 1999;65:1029–1035. doi: 10.1128/AEM.65.3.1029-1035.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sugano Y., Sasaki K., Shoda M. cDNA Cloning and Genetic Analysis of a Novel Decolorizing Enzyme, Peroxidase Gene dyp from Geotrichum candidum. J. Biosci. Bioeng. 1999;87:411–417. doi: 10.1016/S1389-1723(99)80087-5. [DOI] [PubMed] [Google Scholar]
- 3.Gomi N., Yoshida S., Matsumoto K., Okudomi M., Konno H., Hisabori T., Sugano Y. Degradation of the Synthetic Dye Amaranth by the Fungus Bjerkandera adusta Dec 1: Inference of the Degradation Pathway from an Analysis of Decolorized Products. Biodegradation. 2011;22:1239–1245. doi: 10.1007/s10532-011-9478-9. [DOI] [PubMed] [Google Scholar]
- 4.Sugano Y., Muramatsu R., Ichiyanagi A., Sato T., Shoda M. DyP, a Unique Dye-decolorizing Peroxidase, Represents a Novel Heme Peroxidase Family: Asp171 Replaces the Distal Histidine of Classical Peroxidases. J. Biol. Chem. 2007;282:36652–36658. doi: 10.1074/jbc.M706996200. [DOI] [PubMed] [Google Scholar]
- 5.Van Bloois E., Torres Pazmino D.E., Winter R.T., Fraaije M.W. A Robust and Extracelluar Heme-containing Peroxidase from Thermobifida fusca as Prototype of a Bacterial Peroxidase Superfamily. Appl. Microbiol. Biotechnol. 2010;86:1419–1430. doi: 10.1007/s00253-009-2369-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sturm A., Schierhorn A., Lindenstrauss U., Lilie H., Brüser T. YcdB from Escherichia coli Reveals a Novel Class of Tat-dependently Translocated Hemoproteins. J. Biol. Chem. 2006;281:13972–13978. doi: 10.1074/jbc.M511891200. [DOI] [PubMed] [Google Scholar]
- 7.Petrus M.L., Vijgenboom E., Chaplin A.K., Worrall J.A., van Wezel G.P., Claessen D. The DyP-type Peroxidase DtpA is a Tat-substrate Required for GlxA Maturation and Morphogenesis in Streptomyces. Open Biol. 2016;6:150149. doi: 10.1098/rsob.150149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miethke M., Monteferrante C.G., Marahiel M.A., van Dijl J.M. The Bacillus subtilis EfeUOB Transporter is Essential for High-affinity Acquisition of Ferrous and Ferric Iron. Biochim. Biophys. Acta. 2013;1833:2267–2278. doi: 10.1016/j.bbamcr.2013.05.027. [DOI] [PubMed] [Google Scholar]
- 9.Van der Ploeg R., Mäder U., Homuth G., Schaffer M., Denham E.L., Monteferrante C.G., Miethke M., Marahiel M.A., Harwood C.R., Winter T., et al. Environmental Salinity Determines the Specificity and Need for Tat-dependent Secretion of the YwbN Protein in Bacillus subtilis. PLoS ONE. 2011;6:18140. doi: 10.1371/journal.pone.0018140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biswas L., Biswas R., Nerz C., Ohlsen K., Schlag M., Schäfer T., Lamkemeyer T., Ziebandt A.K., Hantke K., Rosenstein R., et al. Role of the Twin-arginine Translocation Pathway in Staphylococcus. J. Bacteriol. 2009;191:5921–5929. doi: 10.1128/JB.00642-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turlin E., Débarbouillé M., Augustyniak K., Gilles A.-M., Wandersman C. Staphylococus aureus FepA and FepB Proteins Drive Heme Iron Utilization in Escherichia coli. PLoS ONE. 2013;8:e56529. doi: 10.1371/journal.pone.0056529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rahmanpour R., Bugg T.D. Assembly in vitro of Rhodococcus jostii RHA1 Encapsulin and Peroxidase DypB to Form a Nanocompartment. FEBS J. 2013;280:2097–2104. doi: 10.1111/febs.12234. [DOI] [PubMed] [Google Scholar]
- 13.Contreras H., Joens M.S., McMath L.M., Le V.P., Tullius M.V., Kimmey J.M., Bionghi N., Horwitz M.A., Fitzpatrick J.A.J., Goulding C.W. Characterization of a Mycobacterium tuberculosis Nanocompartment and Its Potential Cargo Proteins. J. Biol. Chem. 2014;289:18279–18289. doi: 10.1074/jbc.M114.570119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X., Yuan Z., Wang J., Cui Y., Liu S., Ma Y., Gu L., Xu S. Crystal Structure and Biochemical Features of Dye-decolorizing Peroxidase YfeX from Escherichia coli O157 Asp(143) and Arg(232) Play Divergent Roles toward Different Substrates. Biochem. Biophys. Res. Commun. 2017;484:40–44. doi: 10.1016/j.bbrc.2017.01.081. [DOI] [PubMed] [Google Scholar]
- 15.Li J., Liu C., Li B., Yuan H., Yang J., Zheng B. Identification and Molecular Characterization of a Novel DyP-type Peroxidase from Pseudomonas aeruginosa PKE117. Appl. Biochem. Biotechnol. 2012;166:774–785. doi: 10.1007/s12010-011-9466-x. [DOI] [PubMed] [Google Scholar]
- 16.Mendes S., Brissos V., Gabriel A., Catarino T., Turner D.L., Todorovic S., Martins L.O. An Integrated View of Redox and Cartalytic Properties of B-type PpDyP from Pseudomonas putida MET94 and Its Distal Variants. Arch. Biochem. Biophys. 2015;574:99–107. doi: 10.1016/j.abb.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 17.Rahmanpour R., Bugg T.D. Characterization of Dyp-type Peroxidases from Pseudomonas fluorescens Pf-5: Oxidation of Mn(II) and Polymeric Lignin by Dyp1B. Arch. Biochem. Biophys. 2015;574:93–98. doi: 10.1016/j.abb.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 18.Loncar N., Draskovic N., Bozic N., Romeo E., Simic S., Opsenica I., Vujcic Z., Fraaije M.W. Expression and Characterization of a Dye-decolorizing Peroxidase from Pseudomonas fluorescens Pf0-1. Catalyis. 2019;9:463. doi: 10.3390/catal9050463. [DOI] [Google Scholar]
- 19.Uchida T., Sasaki M., Tanaka Y., Ishimori K. A Dye-decolorizing Peroxidase from Vibrio cholerae. Biochemistry. 2015;54:6610–6621. doi: 10.1021/acs.biochem.5b00952. [DOI] [PubMed] [Google Scholar]
- 20.Shrestha R., Huang G., Meekins D.A., Geisbrecht B.V., Li P. Mechanistic Insights into Dye-decolorizing Peroxidase Revealed by Solvent Isotope and Viscosity Effects. ACS Catal. 2017;7:6352–6364. doi: 10.1021/acscatal.7b01861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts J.N., Singh R., Grigg J.C., Murphy M.E., Bugg T.D., Eltis L.D. Characterization of Dye-decolorizing Peroxidases from Rhodococcus jostii RHA1. Biochemistry. 2011;50:5108–5119. doi: 10.1021/bi200427h. [DOI] [PubMed] [Google Scholar]
- 22.Singh R., Grigg J.C., Armstrong Z., Murphy M.E.P., Eltis L.D. Distal Heme Pocket Residues of B-type Dye-decolorizing Peroxidase: Arginine but Not Aspartate is Essential for Peroxidase Activity. J. Biol. Chem. 2012;287:10623–10630. doi: 10.1074/jbc.M111.332171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh R., Grigg J.C., Qin W., Kadla J.F., Murphy M.E., Eltis L.D. Improved Manganese-oxidizing Activity of DypB, a Peroxidase from a Lignolytic Bacterium. ACS Chem. Biol. 2013;8:700–706. doi: 10.1021/cb300608x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmad M., Roberts J.N., Hardiman E.M., Singh R., Eltis L.D., Bugg T.D.H. Identification of DypB from Rhodococcus jostii RHA1 as a Lignin Peroxidase. Biochemistry. 2011;50:5096–5107. doi: 10.1021/bi101892z. [DOI] [PubMed] [Google Scholar]
- 25.Lučić M., Svistunenko D.A., Wilson M.T., Chaplin A.K., Davy B., Ebrahim A., Axford D., Tosha T., Sugimoto H., Owada S., et al. Serial Femtosecond Zero Dose Crystallography Captures a Water-free Distal Heme Site in a Dye-decolorizing Peroxidase to Reveal a Catalytic Role for an Arginine in Fe(IV) = O Formation. Angew. Chem. Int. 2020;59:21656–21662. doi: 10.1002/anie.202008622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirykowicz A.M., Woodward J.D. Shotgun EM of Mycobacterial Protein Complexes during Phase Stress. Curr. Res. Struct. Biol. 2020;2:204–212. doi: 10.1016/j.crstbi.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfanzagl V., Nys K., Bellei M., Michlits H., Mlynek G., Battistuzzi G., Djinovic-Carugo K., Van Doorslaer S., Furtmuller P.G., Hofbauer S., et al. Roles of Distal Aspartate and Arginine of B-class Dye-decolorizing Peroxidase in Heterolytic Hydrogen Peroxide Cleavage. J. Biol. Chem. 2018;293:14823–14838. doi: 10.1074/jbc.RA118.004773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zubieta C., Joseph R., Krishna S.S., McMullan D., Kapoor M., Axelrod H.L., Miller M.D., Abdubek P., Acosta C., Astakhova T., et al. Idenitification and Structural Characterization of Heme Binding in a Novel Dye-decolorizing Peroxidase, TyrA. Proteins. 2007;69:234–243. doi: 10.1002/prot.21673. [DOI] [PubMed] [Google Scholar]
- 29.Cao J., Woodhall M.R., Alvarez J., Cartron M.L., Andrews S.C. EfeUOB(YcdNOB) is a Tripartite, Acid-induced and CpxAR-regulated. Mol. Microbiol. 2007;65:857–875. doi: 10.1111/j.1365-2958.2007.05802.x. [DOI] [PubMed] [Google Scholar]
- 30.Chen C., Shrestha R., Jia K., Gao P.F., Geisbrecht B.V., Bossmann S.H., Shi J., Li P. Characterization of Dye-decolorizing Peroxidase (DyP) from Thermomonospora curvata Reveals Unique Catalytic Properties of A-type DyPs. J. Biol. Chem. 2015;290:23447–23463. doi: 10.1074/jbc.M115.658807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shrestha R., Chen X., Ramyar K.X., Hayati Z., Carlson E.A., Bossmann S.H., Song L., Geisbrecht B.V., Li P. Identification of Surface-exposed Protein Radicals and a Substrate Oxidation Site in A-class Dye-decolorizing Peroxidase from Thermomonospora curvata. ACS Catal. 2016;6:8036–8047. doi: 10.1021/acscatal.6b01952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rahmanpour R., Rea D., Jamshidi S., Fülöp V., Bugg T.D. Structure of Thermobifida fusca DyP-type Peroxidase and Activity towards Kraft Lignin and Lignin Model Compounds. Arch Biochem Biophys. 2016;594:54–60. doi: 10.1016/j.abb.2016.02.019. [DOI] [PubMed] [Google Scholar]
- 33.Yu W., Liu W., Huang H., Zheng F., Wang X., Wu Y., Li K., Xie X., Jin Y. Application of a Novel Alkali-tolerant Thermostable DyP-type Peroxidase from Saccharomonospora viridis DSM 43017 in Biobleaching of Eucalyptus Kraft Pulp. PLoS ONE. 2014;9:e110319. doi: 10.1371/journal.pone.0110319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Habib M.H., Rozeboom H.J., Fraaije M.W. Characterization of a New DyP-peroxidase from the Alkaliphilic Cellulomonad, Cellulomonas bogoriensis. Molecules. 2019;24:1208. doi: 10.3390/molecules24071208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown M.E., Barros T., Chang M.C.Y. Identification and Characterization of a Multifunctional Dye Peroxidase from a Lignin Reactive Bacterium. ACS Chem. Biol. 2012;7:2074–2081. doi: 10.1021/cb300383y. [DOI] [PubMed] [Google Scholar]
- 36.Ogola H.J., Kamiike T., Hashimoto N., Ashida H., Ishikawa T., Shibata H., Sawa Y. Molecular Characterization of a Novel Peroxidase from the Cyanobacterium Anabaena sp. strain PCC 7120. Appl. Environ. Microbiol. 2009;75:7509–7518. doi: 10.1128/AEM.01121-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshida T., Ogola H.J., Amano Y., Hisabori T., Ashida H., Sawa Y., Tsuge H., Sugano Y. Anabaena sp. DyP-type Peroxidase is a Tetramer Consisting of Two Asymmetric Dimers. Proteins. 2016;84:31–42. doi: 10.1002/prot.24952. [DOI] [PubMed] [Google Scholar]
- 38.Sugawara K., Nishihashi Y., Narioka T., Yoshida T., Morita M., Sugano Y. Characterization of a Novel DyP-type Peroxidase from Streptomyces avermitilis. J. Biosci. Bioeng. 2017;123:425–430. doi: 10.1016/j.jbiosc.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 39.Scheibner M., Hulsdau B., Zelena K., Nimtz M., de Boer L., Berger R.G., Zorn H. Novel Peroxidases of Marasmius scorodonius Degrade Beta-carotene. Appl. Microbiol. Biotechnol. 2008;77:1241–1250. doi: 10.1007/s00253-007-1261-9. [DOI] [PubMed] [Google Scholar]
- 40.Liers C., Pecyna M.J., Kellner H., Worrich A., Zorn H., Steffen K.T., Hofrichter M., Ullrich R. Substrate Oxidation by Dye-decolorizing Peroxidases (DyPs) from Wood- and Litter-degrading Agaricomycetes Compared to Other Fungal and Plant Heme-peroxidases. Appl. Microbiol. Biotechnol. 2013;97:5839–5849. doi: 10.1007/s00253-012-4521-2. [DOI] [PubMed] [Google Scholar]
- 41.Johjima T., Ohkuma M., Kudo T. Isolation and cDNA Cloning of Novel Hydrogen Peroxide-dependent Phenol Oxidase from the Basidimomycete Termitomyces albuminosus. Appl. Microbiol. Biotechnol. 2003;61:220–225. doi: 10.1007/s00253-003-1236-4. [DOI] [PubMed] [Google Scholar]
- 42.Yoshida T., Sugano Y. A Structural and Functional Perspective of DyP-type Peroxidase Family. Arch. Biochem. Biophys. 2015;574:49–55. doi: 10.1016/j.abb.2015.01.022. [DOI] [PubMed] [Google Scholar]
- 43.Yoshida T., Tsuge H., Hisabori T., Sugano Y. Crystal Structures of Dye-decolorizing Peroxidase with Ascorbic Acid and 2,6-dimethoxyphenol. FEBS Lett. 2012;586:4351–4356. doi: 10.1016/j.febslet.2012.10.049. [DOI] [PubMed] [Google Scholar]
- 44.Sugano Y. DyP-type Peroxidases Comprise a Novel Heme Peroxidase Family. Cell. Mol. Life Sci. 2009;66:1387–1403. doi: 10.1007/s00018-008-8651-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sugano Y., Matsushima Y., Tsuchiya K., Aoki H., Hirai M., Shoda M. Degradation Pathway of an Anthraquinone Dye Catalyzed by a Unique Peroxidase DyP from Thanatephorus cucumeris Dec 1. Biodegradation. 2009;20:433–440. doi: 10.1007/s10532-008-9234-y. [DOI] [PubMed] [Google Scholar]
- 46.Sugano Y., Matsushima Y., Shoda M. Complete Decolorization of the Anthraquinone Dye Reactive Blue 5 by the Concerted Action of Two Peroxidases from Thanatephorus cucumeris Dec 1. Appl. Microbiol. Biotechnol. 2006;73:862–871. doi: 10.1007/s00253-006-0545-9. [DOI] [PubMed] [Google Scholar]
- 47.Sugawara K., Igeta E., Amano Y., Hyuga M., Sugano Y. Degradation of Antifungal Anthraquinone Compounds is a Probable Physiological Role of DyP Secreted by Bjerkandera adusta. AMB Express. 2019;9:56. doi: 10.1186/s13568-019-0779-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoshida T., Tsuge H., Konno H., Hisabori T., Sugano Y. The Catalytic Mechanism of Dye-decolorizing peroxidase DyP May Require the Swinging Movement of an Aspartic Acid Residue. FEBS J. 2011;278:2387–2394. doi: 10.1111/j.1742-4658.2011.08161.x. [DOI] [PubMed] [Google Scholar]
- 49.Linde D., Pogni R., Cañellas M., Lucas F., Guallar V., Baratto M.C., Sinicropi A., Sáez-Jiménez V., Coscolín C., et al. Catalytic Surface Radical in Dye-decolorizing Peroxidase: A Computational, Spectroscopic and Site-directed Mutagenesis Study. Biochem. J. 2015;466:253–262. doi: 10.1042/BJ20141211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liers C., Bobeth C., Pecyna M., Ullrich R., Hofrichter M. DyP-like Peroxidases of the Jelly Fungus Auricularia auricula-judae Oxidize Nonphenolic Lignin Model Compounds and High-redox Potential dyes. Appl. Microbiol. Biotechnol. 2010;85:1869–1879. doi: 10.1007/s00253-009-2173-7. [DOI] [PubMed] [Google Scholar]
- 51.Strittmatter E., Wachter S., Liers C., Ullrich R., Hofrichter M., Plattner D.A., Piontek K. Radical Formation on a Conserved Tyrosine Residue is Crucial of DyP Activity. Arch. Biochem. Biophys. 2013;537:161–167. doi: 10.1016/j.abb.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 52.Büttner E., Ullrich R., Strittmatter E., Piontek K., Plattner D.A., Hofrichter M., Liers C. Oxidation and Nitration of Mononitrophenols by a DyP-type Peroxidase. Arch. Biochem. Biophys. 2015;574:86–92. doi: 10.1016/j.abb.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 53.Linde D., Coscolín C., Liers C., Hofrichter M., Martínez A.T., Ruiz-Dueñas F.J. Heterologous Expression and Physicochemical Characterization of a Fungal Dye-decolorizing Peroxidase from Auricularia auricula-judae. Protein Expr. Purif. 2014;103:28–37. doi: 10.1016/j.pep.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 54.Linde D., Ayuso-Fernández I., Ruiz-Dueñas F.J., Martínez A.T. Differential Fungal Peroxidases Oxidize Nitrophenols at a Surface Catalytic Tryptophan. Arch. Biochem. Biophys. 2019;668:23–28. doi: 10.1016/j.abb.2019.05.010. [DOI] [PubMed] [Google Scholar]
- 55.Fernández-Fueyo E., Linde D., Almendral D., López-Lucendo M.F., Ruiz-Dueñas F.J., Martínez A.T. Description of the First Fungal Dye-decolorizing Peroxidase Oxidizing Manganese (II) Appl. Microbiol. Biotechnol. 2015;99:8927–8942. doi: 10.1007/s00253-015-6665-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lauber C., Schwarz T., Nguyen Q.K., Lorenz P., Lochnit G., Zorn H. Identification, Heterologous Expression and Characterization of a Dye-decolorizing Peroxidase of Pleurotus sapidus. AMB Express. 2017;7:164. doi: 10.1186/s13568-017-0463-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krahe N.K., Berger R.G., Ersoy F. A DyP-type Peroxidase of Pleurotus sapidus with Alkene Cleaving Activity. Molecules. 2020;25:1536. doi: 10.3390/molecules25071536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Duan Z., Shen R., Liu B., Yao M., Jia R. Comprehensive Investigation of a Dye-decolorizing Peroxidase and a Manganese Peroxidase from Irpex lacteus F17, a Lignin-degrading Basidiomycete. AMB Express. 2018;8:119. doi: 10.1186/s13568-018-0648-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kolwek J., Behrens C., Linke D., Krings U., Berger R.G. Cell-free One-pot Conversion of (+)-valencene to (+)-nootkatone by a Unique Dye-decolorizing Peroxidase Combined with a Laccase from Funalia trogii. J. Indust. Microbiol. Biotechnol. 2018;45:89–101. doi: 10.1007/s10295-017-1998-9. [DOI] [PubMed] [Google Scholar]
- 60.Zitare U.A., Habib M.H., Rozeboom H., Mascotti M., Todorovic S., Fraaije M.W. Mutational and Structural Analysis of an Ancestral Fungal Dye-decolorizing Peroxidase. FEBS J. 2020:15687. doi: 10.1111/febs.15687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kozlov A.M., Darriba D., Flouri T., Morel B., Stamatakis A. RAxML-NG: A Fast, Scalable and User-friendly Tool for Maximum Likelihood Phylogenetic Inference. Bioinformatics. 2019;35:4453–4455. doi: 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Katoh K., Standley D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Darriba D., Posada D., Kozlov A.M., Stamatakis A., Morel B., Flouri T. ModelTest-NG: A New and Scalable Tool for the Selection of DNA and Protein Evolutionary Models. Mol. Biol. Evol. 2020;37:291–294. doi: 10.1093/molbev/msz189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dunford H.B. Heme Peroxidses. John Wiley and Sons; New York, NY, USA: 1999. [Google Scholar]
- 65.Strittmatter E., Serrer K., Liers C., Ullrich R., Hofrichter M., Piontek K., Schleicher E., Plattner D.A. The Toolbox of Auricularia auricula-judae Dye-decolorizing Peroxidase: Identification of Three New Potential Substrate-interaction Sites. Arch. Biochem. Biophys. 2015;574:75–85. doi: 10.1016/j.abb.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 66.Linde D., Ruiz-Dueñas F.J., Fernández-Fueyo E., Guallar V., Hammel K.E., Pogni R., Martínez A.T. Basidiomycete DyPs: Genomic Diversity, Structural-functional Aspects, Reaction Mechanism and Environmental Significance. Arch. Biochem. Biophys. 2015;574:66–74. doi: 10.1016/j.abb.2015.01.018. [DOI] [PubMed] [Google Scholar]
- 67.Camarero S., Sarkar S., Ruiz-Dueñas F.J., Martínez M.J., Martínez A.T. Description of Versatile Peroxidase Involved in the Natural Degradation of Lignin That Has Both Manganese Peroxidase and Lignin Peroxidase Substrate Intereaction Sites. J. Biol. Chem. 1999;274:10324–10330. doi: 10.1074/jbc.274.15.10324. [DOI] [PubMed] [Google Scholar]
- 68.Macdonald I.K., Badyal S.K., Ghamsari L., Moody P.C.E., Raven E.L. Interaction of Ascorbate Peroxidase with Substrates: A Mechanistic and Structural Analysis. Biochemistry. 2006;45:7808–7817. doi: 10.1021/bi0606849. [DOI] [PubMed] [Google Scholar]
- 69.Jones P. Roles of Water in Heme Peroxidase and Catalase Mechanisms. J. Biol. Chem. 2001;276:13791–13796. doi: 10.1074/jbc.M011413200. [DOI] [PubMed] [Google Scholar]
- 70.Létoffé S., Heuck G., Delepelaire P., Lange N., Wandersman C. Bacteria Capture Iron from Heme by Keeping Tetrapyrol Skeleton Intact. Proc. Natl. Acad. Sci USA. 2009;106:11719–11724. doi: 10.1073/pnas.0903842106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sutay Kocabas D., bakir U., Phillips S.E., McPherson M.J., Ogel Z.B. Purification, Characterization, and Identification of a Novel Bifunctional Catalase-phenol Oxidase from Scytalidium thermophilum. Appl. Microbiol. Biotechnol. 2008;79:407–415. doi: 10.1007/s00253-008-1437-y. [DOI] [PubMed] [Google Scholar]
- 72.Kim S.J., Ishikawa K., Hirai M., Shoda M. Characteristics of a Newly Isolated Fungus, Geotrichum candidum Dec 1, Which Decolorizes Various Dyes. J. Ferment. Bioeng. 1995;79:601–607. doi: 10.1016/0922-338X(95)94755-G. [DOI] [Google Scholar]
- 73.Goszczynski S., Paszczynski A., Pasti-Grigsby M.B., Crawford R.L., Crawford D.L. New Pathway for Degradation of Sulfonated Azo Dyes by Microbial Peroxidases of Phanerochaete chrysosporium and Streptomyces chromofuscus. J. Bacteriol. 1994;176:1339–1347. doi: 10.1128/JB.176.5.1339-1347.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Spadaro J.T., Gold M.H., Renganathan V. Degradatioon of Azo Dyes by the Lignin-degrading Fungus Phanerochaete chrysosporium. Appl. Environ. Microbiol. 1992;58:2397–2401. doi: 10.1128/AEM.58.8.2397-2401.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Paszczynski A., Pasti-Grigsby M.B., Goszczynski S., Crawford R.L., Crawford D.L. Mineralization of Sulfonated Azo Dyes and Sulfanic Acid by Phanerochaete chrysosporium and Streptomyces chromofuscus. Appl. Environ. Microbiol. 1992;58:3598–3604. doi: 10.1128/AEM.58.11.3598-3604.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chivukula M., Renganathan V. Phenolic Azo Dye Oxidation by Laccase from Pyricularia oryzae. Appl. Environ. Microbiol. 1995;61:4374–4377. doi: 10.1128/AEM.61.12.4374-4377.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gold M.H., Kuwahara M., Chiu A.A., Glenn J.K. Purification and Characterization of an Extracellular H2O2-requiring Diarylpropane Oxygenase from the White Rot Basidiomycete, Phanerochaete chrysosporium. Arch. Biochem. Biophys. 1984;234:353–362. doi: 10.1016/0003-9861(84)90280-7. [DOI] [PubMed] [Google Scholar]
- 78.Glenn J.K., Gold M.H. Purification and Characterization of an Extracellular Mn(II)-dependennt Peroxidase from the Lignin-degradaing Basidiomycete, Phanerochaete chrysosporium. Arch. Biochem. Biophys. 1985;242:329–341. doi: 10.1016/0003-9861(85)90217-6. [DOI] [PubMed] [Google Scholar]
- 79.Granja-Travez R.S., Persinoti G.F., Squina F.M., Bugg T.D.H. Functional Genomic Analysis of Bacterial Lignin Degraders: Diversity in Mechanisms of Lignin Oxidation and Metabolism. Appl. Microbiol. Biotechnol. 2020;104:3305–3320. doi: 10.1007/s00253-019-10318-y. [DOI] [PubMed] [Google Scholar]
- 80.Sahinkaya M., Colak D.N., Ozer A., Canakci S., Deniz I., Belduz A.O. Cloning, Characterization and Paper Pulp Applications of a Newly Isolated DyP Type Peroxidase from Rhodococcus sp. T1. Mol. Biol. Rep. 2019;46:569–580. doi: 10.1007/s11033-018-4509-9. [DOI] [PubMed] [Google Scholar]
- 81.Riyadi F.A., Tahir A.A., Yusof N., Sabri N.S.A., Noor M.J.M.M., Akhir F.N.M.D., Othman N., Zakaria Z., Hara H. Enzymatic and Genetic Characterization of Lignin Depolymerization by Streptomyces sp. S6 Isolated from a Tropical Environment. Sci. Rep. 2020;10:7813. doi: 10.1038/s41598-020-64817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Youngs H.L., Sollewijn Gelpke M.D., Li D., Sundaramoorthy M., Gold M.H. The Role of Glu39 in MnII Binding and Oxidation by Manganese Peroxidase from Phanerochaete chrysosporium. Biochemistry. 2001;2001. 40:2243–2250. doi: 10.1021/bi002104s. [DOI] [PubMed] [Google Scholar]
- 83.Granja-Travez R.S., Wilkinson R.C., Persinoti G.F., Squina F.M., Fülöp V., Bugg T.D.H. Structural and Functional Characterization of Multi-copper Oxidase CueO from Lignin-degrading Bacterium Ochrobacterium sp. Reveal Its Activity towards Lignin Model Compounds and Lignosulfonate. FEBS J. 2018;285:1684–1700. doi: 10.1111/febs.14437. [DOI] [PubMed] [Google Scholar]
- 84.Kumar M., Verma S., Gazara R.K., Kumar M., Pandey A., Verma P.K., Thakur I.S. Genomic and Proteomic Analysis of Lignin Degrading and Polyhydroxyalkanoater Accumulating β-proteobacterium Pandoraea sp. ISTKB. Biotechnol. Biofuel. 2018;11:154. doi: 10.1186/s13068-018-1148-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kellner H., Luis P., Pecyna M.J., Barbi F., Kapturska D., Kruger D., Zak D.R., Marmeisse R., Vandenbol M., Hofrichter M. Widespread Occurrence of Expressed Fungal Secretory Peroxidases in Forest Soils. PLoS ONE. 2014;9:e95557. doi: 10.1371/journal.pone.0095557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Salvachúa D., Prieto A., Martínez Á.T., Martínez M.J. Characterization of a Novel Dye-decolorizing Peroxidase (DyP)-type Enzyme from Irpex lacteus and Its Application in Enzymatic Hydrolysis of Wheat Straw. Appl. Environ. Microbiol. 2013;79:4316–4324. doi: 10.1128/AEM.00699-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Qin X., Luo H., Zhang X., Yao B., Ma F., Su X. Dye-decolorizing Peroxidases in Irpex lacteus Combining the Catalytic Properties of Heme Peroxidases and Laccase Play Important Roles in Ligninolytic System. Biotechnol. Biofuels. 2018;11:302. doi: 10.1186/s13068-018-1303-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chaplin A.K., Petrus M.L., Mangiameli G., Hough M.A., Svistunenko D.A., Nicholls P., Claessen D., Vijgenboom E., Worrall J.A. GlxA is a New Structural Member of the Radical Copper Oxidase Family and is Required for Glycan Deposition at Hyphal Tips and Morphogenesis of Streptomyces lividans. Biochem. J. 2015;469:433–444. doi: 10.1042/BJ20150190. [DOI] [PubMed] [Google Scholar]
- 89.Tang Y., Mu A., Zhang Y., Zhou S., Wang W., Lai Y., Zhou X., Liu F., Yang X., Gong H., et al. Cryo-EM Structure of Mycobacterium smegmatis DyP-loaded Encapsulin. Proc. Natl. Acad. Sci. USA. 2021;118:e2025658118. doi: 10.1073/pnas.2025658118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Loncar N., Rozeboom H.J., Franken L.E., Stuart M.C.A., Fraaije M.W. Structure of a Robust Bacterial Protein Cage and Application as a Versatile Biocatalytic Platform through Enzyme Encapsulation. Biochem. Biophys. Res. Commun. 2020;529:548–553. doi: 10.1016/j.bbrc.2020.06.059. [DOI] [PubMed] [Google Scholar]