Abstract
Intrinsically disordered proteins and proteins containing intrinsically disordered regions are highly abundant in the proteome of eukaryotes and are extensively involved in essential biological functions. More recently, their role in the organization of biomolecular condensates has become evident and along with their misregulation in several neurologic disorders. Currently, most studies involving these proteins are carried out in vitro and using purified proteins. Given that in cells, condensate‐forming proteins are exposed to high, millimolar concentrations of cellular metabolites, we aimed to reveal the interactions of cellular metabolites and a representative condensate‐forming protein. Here, using the arginine–glycine/arginine–glycine–glycine (RG/RGG)‐rich cold inducible RNA binding protein (CIRBP) as paradigm, we studied binding of the cellular metabolome to CIRBP. We found that most of the highly abundant cellular metabolites, except nucleotides, do not directly bind to CIRBP. ATP, ADP, and AMP as well as NAD+, NADH, NADP+, and NADPH directly interact with CIRBP, involving both the folded RNA‐recognition motif and the disordered RG/RGG region. ATP binding inhibited RNA‐driven phase separation of CIRBP. Thus, it might be beneficial to include cellular metabolites in in vitro liquid–liquid phase separation studies of RG/RGG and other condensate‐forming proteins in order to better mimic the cellular environment in the future.
Keywords: ATP, CIRBP, disordered protein, liquid–liquid phase separation, RG/RGG region, RNA‐binding protein
Abbreviations
- CIRBP
cold inducible RNA binding protein
- CSP
chemical shift perturbation
- DIC
differential interference contrast
- FUS
fused in sarcoma
- HSQC
heteronuclear single quantum coherence
- IDP
intrinsically disordered protein
- IDR
intrinsically disordered region
- LLPS
liquid–liquid phase separation
- NMR
nuclear magnetic resonance
- NOE
nuclear Overhauser enhancement
- RBPs
RNA‐binding protein
- RG/RGG
arginine–glycine/arginine–glycine–glycine
- RNP
ribonucleoprotein
- RRM
RNA‐recognition‐motif
- RSY
arginine–serine–tyrosine
- SG
stress granule
1. INTRODUCTION
Intrinsically disordered proteins (IDPs) and proteins harboring intrinsically disordered regions (IDRs) constitute 33% of the eukaryotic and 44% of the human proteome. 1 , 2 , 3 Their functional implications include essential biological processes like cell signaling, 4 , 5 gene transcription, 6 RNA splicing, 7 , 8 and cell cycle regulation. 9 , 10 Over the last decades, a number of studies have demonstrated the essential role of IDPs/IDRs in the formation of biomolecular condensates also referred as membraneless organelles. 4 , 11 , 12 Ribonucleoprotein (RNP) granules, such as stress granules (SGs), P‐bodies, Cajal bodies, or paraspeckles constitute a large class of these condensates and are composed of a dynamic assembly of RNA and RNA‐binding proteins (RBPs). 13 , 14 , 15 IDRs in RBPs can undergo liquid–liquid phase separation (LLPS) in vitro which constitute a physical mechanism allowing the formation of these biological assemblies with dynamic, liquid‐like behavior in vivo. 14 , 16 , 17 Mutations in these regions are often associated with aberrant localization of RBPs and the formation of pathological aggregates, 14 , 18 , 19 and associate with a large variety of diseases including cancers and neurodegenerative diseases. 18 , 20 Therefore, these RBPs are considered as potential therapeutic targets and, thus, there is a need for a better understanding of the cellular mechanisms involved in the regulation of the formation of RNP granules in vivo as well as RBPs LLPS in vitro. Cellular metabolites are one of the most abundant molecules in living cells with a total intracellular concentration of about 200 mM in mammalian iBMK cells. 21 Therefore, in this study we addressed the question whether some of these highly abundant metabolites could regulate LLPS of RBPs in vitro.
Arginine–glycine/arginine–glycine–glycine (RG/RGG) rich regions constitute a large fraction of IDRs in RBPs and have been shown to contribute to LLPS in vitro and associate with several biomolecular condensates in vivo, 22 , 23 respectively. Furthermore, recent studies demonstrated that adenine triphosphate (ATP), a highly abundant cellular metabolite, can directly bind to the RG/RGG region, as well as the RNA‐recognition‐motif (RRM) of the RBP fused in sarcoma (FUS), thereby affecting both FUS LLPS and fibrillirization. 24 , 25 Whether ATP regulates phase separation of other RBPs containing RG/RGG regions and whether this is a general mechanism controlling their association with RNP granules is still unclear. Furthermore, whether other metabolites bind RBPs and share similar LLPS‐modulatory functions as ATP remains unknown.
We recently showed that the RG/RGG region of the cold inducible RNA binding protein (CIRBP) is essential to mediate both CIRBP phase separation in vitro and stress‐granule association in cells. 26 We therefore used CIRBP as model system to study the influence of human cell metabolites on CIRBP LLPS. CIRBP consists of a conserved folded RNA recognition motif (RRM, 1–90), a disordered RG/RGG region (90–137) and a disordered arginine–serine–tyrosine (RSY) rich region (138–172) (Figure 1a). CIRBP belongs to the family of cold shock proteins. 27 , 28 In response to cellular stresses like ultraviolet irradiation, mild cold shock and hypoxia, the expression level of CIRBP is upregulated 29 , 30 , 31 and CIRBP is translocated from the nucleus to the cytoplasm where it associates with SGs. 32 Cytoplasmic CIRBP stabilizes and protects mRNA from stress conditions. 33 , 34 In addition, it is involved in cancer development by acting both as tumor promotor and tumor suppressor in different biological contexts. 35
FIGURE 1.
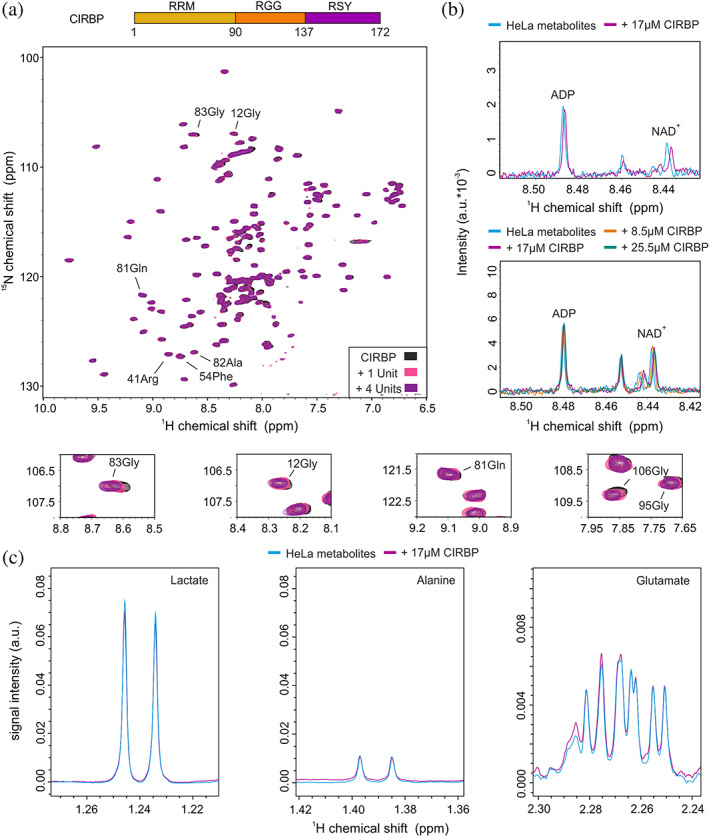
HeLa Cell metabolites contain binding partners of CIRBP. (a) 1H‐15N HSQC spectrum of 15N‐labeled CIRBP at 17 μM without (black) or with addition of 1 or 4 units of HeLa metabolites extract (pink and magenta, respectively). Overlay of 1D 1H‐CPMG spectra of HeLa metabolites extract in absence (cyan) or presence of increasing concentrations of CIRBP ranging from 8.5 to 25.5 μM (orange, magenta and green) and zoomed in a region corresponding to ADP and NAD+ in (b) and lactate, alanine and glutamate in (c)
Using nuclear magnetic resonance (NMR) spectroscopy we showed that among the most abundant cellular metabolites, only ATP, ADP, and AMP (nucleotides) and NAD+, NADH, NADP+, and NADPH (dinucleotides) directly bind to CIRBP and that this involved both the RRM as well as the RG/RGG rich region. Using turbidity assays and differential interference contrast (DIC) microscopy we showed that ATP binding prevents CIRBP LLPS by competing with RNA‐driven RG/RGG phase separation. Summarizing, our study provides new mechanisms involved in the regulation of phase separation of RBPs as well as a better comprehension of the role of cellular metabolites involved in such processes.
2. RESULTS
2.1. HeLa cell metabolites contain binding partners of CIRBP
In order to validate our hypothesis that CIRBP can interact with cellular metabolites, we tested binding of extracted HeLa cell metabolites to a solution of recombinant and isotope labeled CIRBP using NMR spectroscopy. Stepwise addition of HeLa metabolites extract resulted in progressive chemical shift perturbations (CSPs) of several CIRBP 1H‐15N HSQC cross‐peaks (Figure 1a), indicating the direct interaction between CIRBP and some yet undefined HeLa metabolites. To determine which metabolites are specifically involved in the interaction, we further performed NMR analysis of HeLa metabolites extract upon addition of CIRBP. The analysis revealed minor CSPs of 1H metabolite peaks upon stepwise addition of CIRBP. The affected peaks were assigned to the 1H peaks corresponding to ADP as well as NAD+ (Figure 1b). Most of the other metabolite 1H peaks remained unaffected upon addition of CIRBP, including those of the most abundant metabolites lactate, alanine and glutamate (Figure 1c). This shows that the majority of Hela metabolites do not contribute to CIRBP binding aside from a specific subset including the nucleotides ADP and NAD+.
2.2. CIRBP interacts directly with (di)nucleotides, but none of the other highly abundant cellular metabolites
In order to validate our results, we screened the most abundant metabolites in mammalian cells for CIRBP binding. It has been previously shown that among them and across organisms, amino‐acids followed by central carbon metabolites and nucleotides are by far the most abundant with concentrations up to 64 mM for glutamate. 21 Although lactate concentrations were not included in these studies, it is among the most abundant metabolites in cells and included in our study. Consistent with our results obtained for the HeLa metabolite extract, ADP and NAD+ showed binding to CIRBP (Figure 2a,c). Moreover, the other tested nucleotides (ATP, AMP) as well as dinucleotides (NADH, NADP+ and NADPH) showed significant binding to CIRBP, as demonstrated by CSPs of several CIRBP 1H‐15N cross‐peaks upon addition of the respective nucleotides (Figure 2a,c,e). It is worth noting, that the same set of signals is shifted upon (di)nucleotide addition, with ATP and NADPH showing the strongest shifts. This indicates that the binding mode is similar between these (di)nucleotides and that ATP and NADPH have the highest affinity for CIRBP (Figure 2b,d,f). ATP binding to CIRBP is in the fast exchange regime with an associated KD of 2.9 ± 1.4 mM (Figure 3a,b). The highest CSPs are associated with the CIRBP residues 10Val, 12Gly, 51Phe, 81Gln, and 83Gly and most of the affected and assigned CIRBP 1H‐15N cross‐peaks correspond to amino‐acids clustering within a positively charged surface comprising β‐strands β1, β3, and β4 of the CIRBP RRM (Figure 3c). Interestingly, 83Gly is the most affected CIRBP amino acid both with isolated (di)nucleotides and HeLa metabolite extract (Figures 1a and 2a,c,e), confirming that the CSPs observed upon addition of HeLa metabolites extract are mainly due to the (di)nucleotide components. In agreement, addition of the other highly abundant cellular metabolites, including alanine, aspartate, glutamine, glutamate, lactate, and glucose did not change the 1H‐15N HSQC spectrum of CIRBP, indicating that none of them interact with CIRBP with a comparable or higher affinity (Figure 3d).
FIGURE 2.
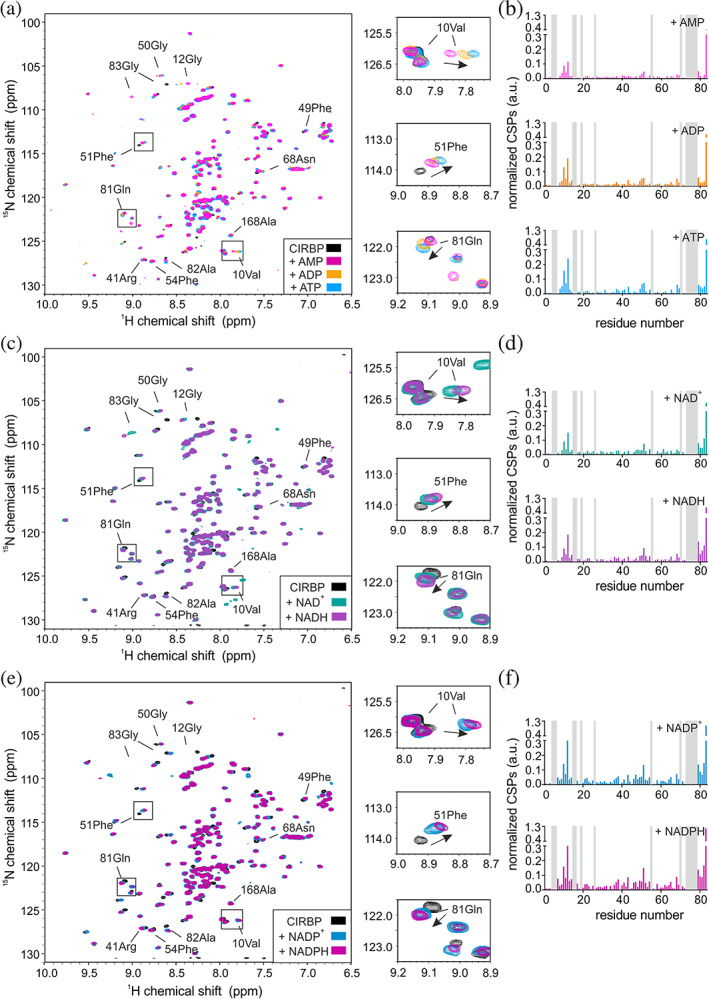
CIRBP directly interacts with (di)nucleotides. (a) 1H‐15N HSQC spectrum of 15N‐labeled CIRBP at 17 μM without (black) or with addition of 10 mM of either AMP, ADP, or ATP (pink, orange, and cyan, respectively). Chemical shift perturbations (CSPs) of the 1H‐15N HSQC CIRBP cross‐peaks between the CIRBP free and either AMP, ADP or ATP bound forms are shown in a bar‐plot in (b). Unassigned residues are indicated in grey. The CIRBP region between amino‐acids 91–172 is not shown as most of the corresponding 1H‐15N HSQC CIRBP cross‐peaks are unassigned. (c) 1H‐15N HSQC spectrum of 15N‐labeled CIRBP at 17 μM without (black) or with addition of 10 mM of either NAD+ or NADH (green and violet, respectively). CSPs of 1H‐15N HSQC CIRBP cross‐peaks between free CIRBP and either NAD+‐ or NADH‐bound forms are shown in bar‐plots in (d). Unassigned residues are indicated in grey. The CIRBP region between amino‐acids 91–172 is not shown as most of the corresponding 1H‐15N HSQC CIRBP cross‐peaks are unassigned. (e) 1H‐15N HSQC spectrum of 15N‐labeled CIRBP at 17 μM without (black) or upon addition of 10 mM of either NADP+ or NADPH (blue and magenta, respectively). CSPs of the 1H‐15N HSQC CIRBP cross‐peaks between free and either NADP+ or NADPH bound forms are shown in a bar‐plot in (f). Unassigned residues are indicated in grey. The CIRBP region between amino‐acids 91–172 is not shown as most of the corresponding 1H‐15N HSQC CIRBP cross‐peaks are unassigned. CIRBP, cold inducible RNA binding protein
FIGURE 3.
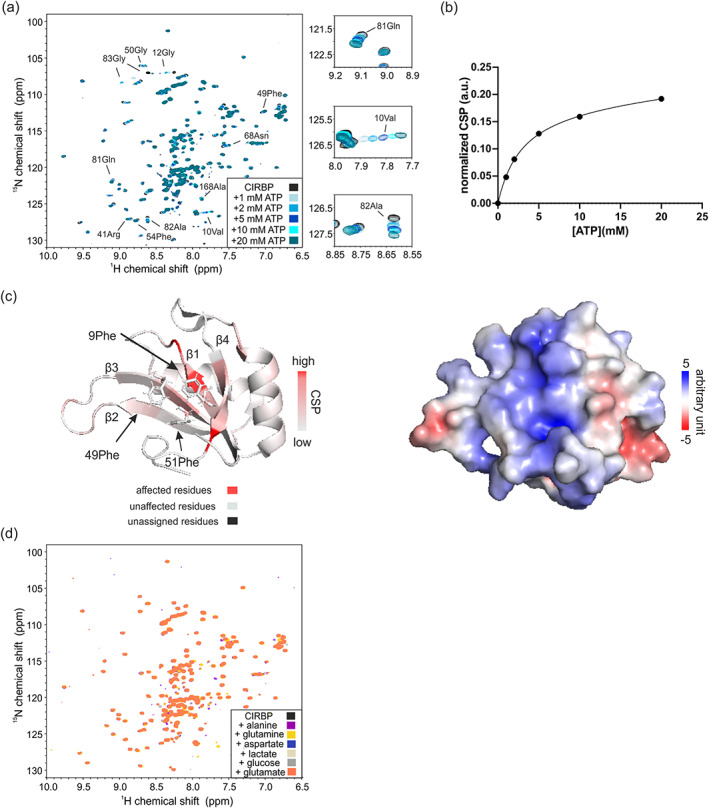
ATP binding to CIRBP overlaps with the putative RNA‐binding interface. (a) 1H, 15N HSQC spectrum of 15N‐labeled CIRBP at 17 μM without (black) or with increasing ATP concentration, ranging from 1 to 20 mM (blue gradient). (b) CSP plot of CIRBP 1H‐15N cross‐peak corresponding to valine 10 as function of ATP concentration. Using GraphPad Prism 8.4 we estimated the corresponding affinity with an associated KD of 2.9 ± 1.4 mM using a nonlinear one binding site fitting function. (c) Mapping of the CSPs of CIRBP1H‐15N cross‐peaks (residue 1–90) upon binding to 10 mM ATP on the published structure of the RRM of CIRBP (PDB code: 5TBX). The electrostatic potential surface representation of the CIRBPRRM is shown. (d) 1H‐15N HSQC spectrum of 15N‐labeled CIRBP at 17 μM without (black) or with addition of 3.3 mM of aspartate or 10 mM of either alanine, glutamine, lactate, glucose or glutamate (blue, magenta, yellow, light grey, grey, and orange, respectively). CIRBP, cold inducible RNA binding protein
Summarizing we showed that among the most concentrated cellular metabolites, only nucleotides binds directly to CIRBP whereas amino acids and central carbon metabolites do not show any binding.
2.3. The RRM and RG/RGG region of CIRBP are involved in (di)nucleotide binding
To explore how independent regions of CIRBP are involved in (di)nucleotide binding, we purified isolated CIRBP regions, CIRBPRRM (1–90), CIRBPRGG (90–137) and CIRBPRSY (138–172) and tested their binding to (di)nucleotides using NMR. Addition of ATP and NAD+ to a solution of isotope labeled CIRBPRSY did not affect the 1H‐15N HSQC spectrum, indicating that the CIRBP RSY region is not involved in ATP and NAD+ binding (Figure 4a). In contrast, upon addition of (di)nucleotides, we observed CSPs of several CIRBPRGG as well as CIRBPRRM 1H‐15N cross‐peaks (Figures 4b,c, 5a–d, and 6a–f), indicating direct interaction between both the RRM and the RG/RGG region of CIRBP with these (di)nucleotides. In case of AMP, NAD+ and NADH, CSPs of CIRBPRGG 1H‐15N cross‐peaks are weak and distributed along the entire RG/RGG region. This suggests unspecific binding of AMP, NAD+, and NADH to the CIRBP RG/RGG region. As for CIRBP full‐length, the same set of signals of the RRM were shifted upon (di)nucleotide addition, with ATP and NADPH showing the strongest effect (Figure 6b,d,e). This indicates that the binding mode is similar between these nucleotides for both CIRBP full length and the isolated RRM, with ATP and NADPH harboring the strongest affinity. Furthermore, using 15N{1H} heteronuclear NOE experiments of isotope labeled CIRBPRGG in presence or absence of ATP, we observed higher heteronuclear NOE values of a set of CIRBPRGG 1H‐15N cross‐peaks upon ATP addition indicating increased rigidity of the RG/RGG region in the ATP bound state (Figure 4d).
FIGURE 4.
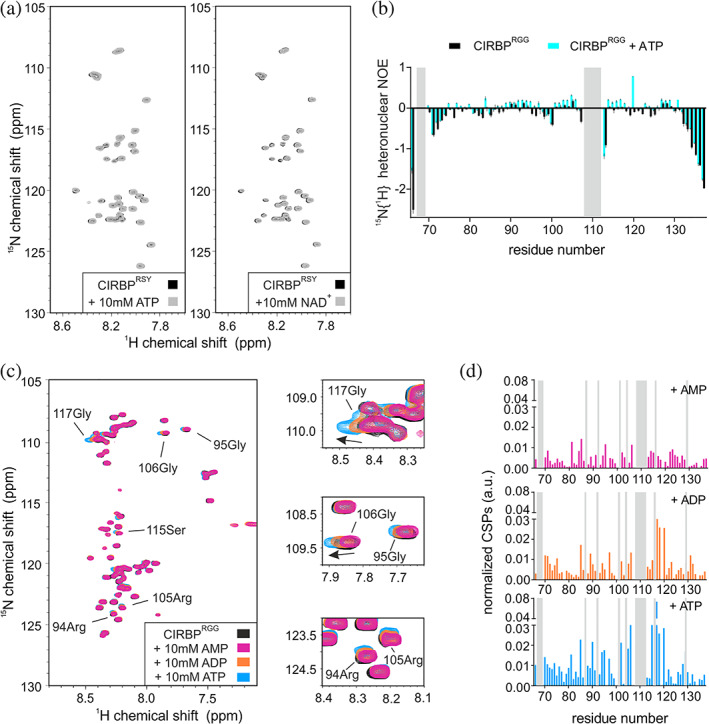
The RG/RGG region of CIRBP are involved in nucleotide binding. (a) 1H‐15N HSQC spectrum of 15N‐labeled CIRBPRSY at 30 μM without (black) or with addition of 10 mM of ATP or NAD+ (grey, left and right panel, respectively). (b) 1H‐15N HSQC spectrum of 15N‐labeled CIRBPRGG at 30 μM without (black) or with addition of 10 mM of either AMP, ADP, or ATP (magenta, orange, and blue, respectively). CSPs of the 1H‐15N HSQC CIRBPRGG cross‐peaks between the CIRBP free and either AMP, ADP or ATP bound forms are shown in a bar‐plot in (c). Unassigned residues are indicated in grey. (d) 15N{1H} heteronuclear NOE values are plotted versus CIRBPRGG residue numbers in presence (cyan) or absence (black) of ATP. Error bars for heteronuclear NOE values were derived from error propagation calculation using standard deviation of 10 arbitrarily chosen noise peaks in saturated and unsaturated spectra. CIRBP, cold inducible RNA binding protein
FIGURE 5.
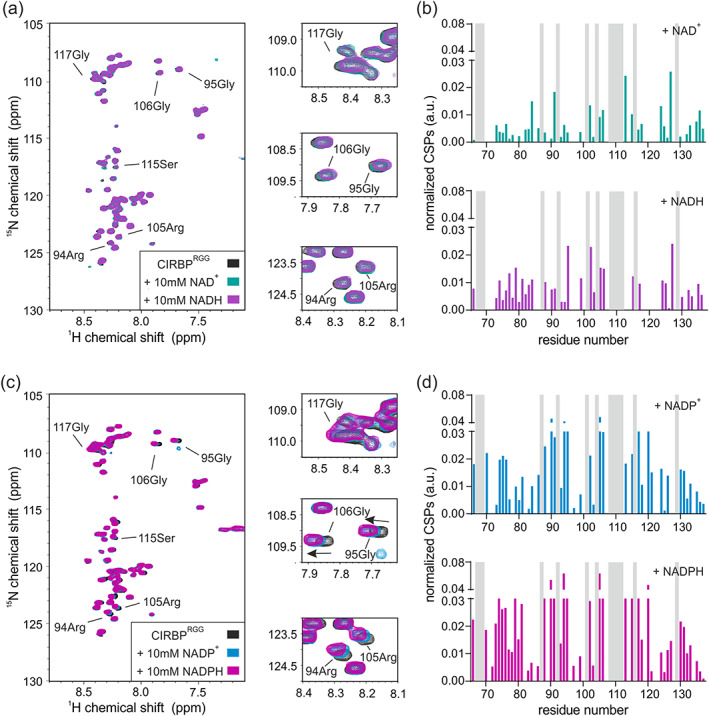
The RG/RGG region of CIRBP is involved in dinucleotide binding. (a) 1H‐15N HSQC spectrum of 15N‐labeled CIRBPRGG at 30 μM without (black) or upon addition of 10 mM of either NAD+ or NADH (green and violet, respectively). CSPs of 1H‐15N HSQC CIRBPRGG cross‐peaks between free and either NAD+ or NADH bound forms are shown in a bar‐plot in (b). The unassigned residues are indicated in grey. (c) 1H‐15N HSQC spectrum of 15N‐labeled CIRBPRGG at 30 μM without (black) or upon addition of 10 mM of either NADP+ or NADPH (blue and magenta, respectively). CSPs of 1H‐15N HSQC CIRBPRGG cross‐peaks between free and either NADP+ or NADPH bound forms are shown in a bar‐plot in (d). unassigned residues are indicated in grey
FIGURE 6.
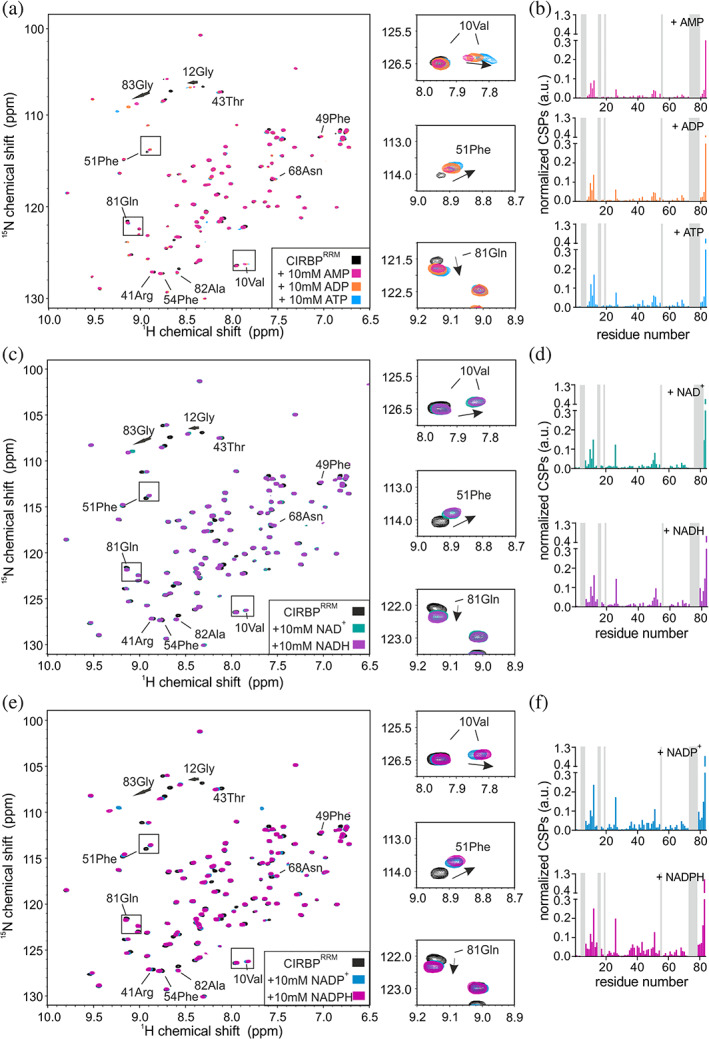
The RRM of CIRBP is involved in (di)nucleotide binding. (a) 1H‐15N HSQC spectrum of 15N‐labeled CIRBPRRM at 30 μM without (black) or upon addition of 10 mM of either AMP, ADP or ATP (magenta, orange, and blue, respectively). CSPs of 1H‐15N HSQC CIRBPRRM cross‐peaks between free and either AMP, ADP or ATP bound forms are shown in a bar‐plot in (b). Unassigned residues are indicated in grey. (c) 1H‐15N HSQC spectrum of 15N‐labeled CIRBPRRM at 30 μM without (black) or upon addition of 10 mM of either NAD+ or NADH (green and violet, respectively). CSPs of 1H‐15N HSQC CIRBPRRM cross‐peaks between free and either NAD+ or NADH bound forms are shown in a bar‐plot in (d). Unassigned residues are indicated in grey. (e) 1H‐15N HSQC spectrum of 15N‐labeled CIRBPRRM at 30 μM without (black) or upon addition of 10 mM of either NADP+ or NADPH (blue and magenta, respectively). CSPs of 1H‐15N HSQC CIRBPRRM cross‐peaks between free and either NADP+ or NADPH bound forms are shown in a bar‐plot in (f). Unassigned residues are indicated in grey. CIRBP, cold inducible RNA binding protein
Summarizing, we showed that both the CIRBP RRM as well as the RG/RGG region can independently interact with (di)nucleotides.
2.4. ATP binding to the CIRBP RG/RGG region attenuates RNA binding and is regulated by arginine methylation
We previously showed that the RG/RGG region of CIRBP can directly interact with RNA. 26 Given that both ATP and RNA are negatively charged, we hypothesized that they could share similar binding modes and compete together for CIRBP RG/RGG binding. We therefore performed an NMR‐based competition assay to access whether ATP competes with RNA while binding to CIRBPRGG. Line broadening of CIRBPRGG 1H‐15N cross‐peaks was observed upon addition of unlabeled (UG)12 RNA to a solution of isotope labeled CIRBPRGG, (Figure 7a, left panel) which indicates direct interaction between the CIRBP RG/RGG region and (UG)12 RNA, as previously shown. 26 In contrast, presence of an excess of ATP leads to reappearance of the CIRBPRGG 1H‐15N cross‐peaks, indicating that ATP can compete with (UG)12 RNA for binding to the RG/RGG region of CIRBP in the experimental conditions tested (Figure 7a, right panel, Figure 7b).
FIGURE 7.
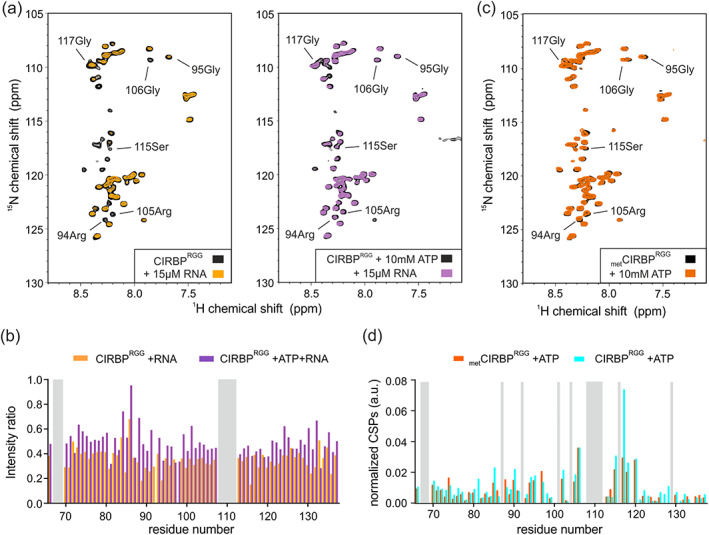
ATP binding to the CIRBP RG/RGG region attenuates RNA binding and is regulated by arginine methylation. (a) 1H‐15N HSQC spectrum of 15N‐labeled CIRBPRGG at 30 μM in presence or absence of 10 mM ATP (right and left panel, respectively) and without (black) or with addition of 15 μM (UG)12 RNA (orange, violet). The intensity ratio of the 1H‐15N HSQC CIRBPRGG cross‐peaks between the ATP (orange) or ATP/RNA (violet) free and bound forms are shown in a bar‐plot in (b). The unassigned residues are indicated in grey. (c) 1H‐15N HSQC spectrum of 15N‐labeled metCIRBPRGG at 30 μM in presence (orange) or absence of 10 mM ATP (black). CSPs of the 1H‐15N HSQC methylated metCIRBPRGG (orange) or unmethylated CIRBPRGG (blue) cross‐peaks between the ATP free and bound forms are shown in a bar‐plot in (d). The unassigned residues are indicated in grey. CIRBP, cold inducible RNA binding protein
As we and others, previously demonstrated that PRMT1‐mediated arginine methylation of the RG/RGG region regulates its binding to RNA, 36 we next aimed to assess whether it can also influence ATP binding. We therefore in vitro methylated CIRBP RG/RGG using recombinant PRMT1, purified the methylated protein (metCIRBPRGG) and tested its binding to ATP by NMR spectroscopy. Addition of ATP to a solution of isotope labeled metCIRBPRGG showed similar effects on the 1H‐15N HSQC spectrum of metCIRBPRGG as compared to the same experiment performed with unmethylated CIRBPRGG, suggesting that methylation of the RG/RGG region of CIRBP has minor impact on its interaction with ATP molecules (Figure 7c,d).
2.5. ATP modulates LLPS of CIRBP
We previously showed that CIRBP can phase separate in vitro in the presence of RNA. 26 As we have shown that ATP can efficiently compete with RNA for CIRBP binding, we hypothesized that ATP can also regulate its propensity to phase separate.
To this end, we monitored phase separation of purified CIRBP in a turbidity assay, which uses the optical density of a protein solution as a measure of phase separation. In line with a previous study, 26 titration of (UG)12 RNA to a sample with a fixed CIRBP concentration (17 μM) led to a progressive turbidity increase at low RNA concentrations, whereas higher amounts of RNA had a suppressive effect on CIRBP LLPS (Figure 8a). Addition of increasing amounts of ATP to RNA‐bound and phase separated CIRBP led to a concentration‐dependent decrease in turbidity, demonstrating that ATP inhibits CIRBP LLPS (Figure 8b). In order to further validate these findings, we monitored CIRBP phase separation by DIC microscopy. In the presence of substoichiometric amounts of RNA, CIRBP immediately formed small condensates that increased in size over time, most likely due to fusion of condensates, and indicating their “liquid‐like” behavior (Figure 8c). In line with the turbidity assay, addition of ATP was able to suppress CIRBP condensate formation (Figure 8c).
FIGURE 8.
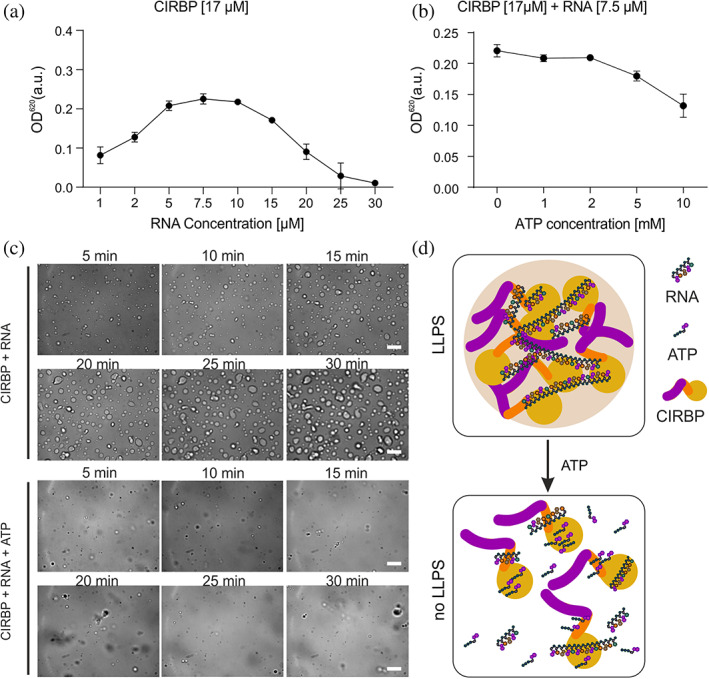
ATP modulates LLPS of CIRBP. Turbidity assay to quantify phase separation of full‐length CIRBP with fixed CIRBP concentration (17 μM) and increasing RNA concentration in (a), with fixed CIRBP and RNA concentration (17 and 7.5 μM, respectively) and increasing ATP concentration in (b). Values represent means ±SEM (n = 3). (c) DIC microscopy images of CIRBP at 17 μM in presence of 7.5 μM RNA and/or 5 mM ATP. Scale bar, 10 μm. (d) Model of ATP/RNA‐mediated regulation of CIRBP LLPS. CIRBP, cold inducible RNA binding protein; DIC, differential interference contrast; LLPS, liquid–liquid phase separation
Summarizing, our data clearly demonstrate that ATP interferes with LLPS of CIRBP in vitro and likely with SGs recruitment in cells. This is probably due to interference between ATP and RNA‐mediated enhancement of CIRBP LLPS.
3. DISCUSSION
Here we showed using NMR spectroscopy that CIRBP, one of the constituent of SGs 26 , 32 directly interact with several (di)nucleotides present in HeLa cell metabolite extracts (Figure 1b). In contrast, other highly abundant metabolites which do not contain an aromatic moiety did not show any detectable binding to CIRBP despite their negatively charged nature. Hence, the carboxyl groups from lactate and other negatively charged amino acids are insufficient to mediate interaction with CIRBP, and possibly other RG/RGG‐containing proteins (Figure 1c). In addition, none of the other highly abundant metabolites alanine, glutamine, or glucose was found to bind to CIRBP. Therefore, our data suggest that both the aromatic and the phosphate moieties of (di)nucleotides contribute to efficient CIRBP interaction allowing recognition of this class of metabolites. To our knowledge, this is the first study demonstrating that the large majority of cellular metabolites do not bind RBP.
Here, we showed that both the RRM and the RG/RGG region of CIRBP can independently interact with (di)nucleotides (Figures 4b,c, 5, and 6). Binding of (di)nucleotides to the C‐terminal disordered RG/RGG region and RRM of CIRBP might involve (a) cation‐π interactions between adenosine aromatic cycles and arginines of the RG/RGG rich region or arginines/lysines of the RNA‐binding interface of the RRM, 23 , 25 , 37 (b) electrostatic contacts between the phosphate groups and/or ribose sugars of the nucleotides and arginines/lysines, 23 , 38 (c) π − π stacking interactions between adenosine aromatic cycles and aromatic amino‐acids highly populated in the IDR regions of RBPs 37 , 39 , 40 and a combination thereof. These interactions make (di)nucleotides probably divalent or multivalent ligands. 41 Similar interactions have been shown for the RBP FUS, where ATP can also efficiently interact with both the RRM and the C‐terminal disordered RG/RGG region. 24 , 25 Therefore, it is tempting to speculate that (di)nucleotide binding to such RNA‐binding domains 25 can be generalized to the large class of RBPs. The affinity of CIRBP for the tested (di)nucleotides can be estimated to be in the millimolar range. Among them, and to our knowledge, only ATP has been reported to reach millimolar intra‐cellular concentrations. 21 Nevertheless, further studies are required to define the exact (sub)cellular, and context‐specific concentrations of these metabolites. Therefore, we cannot infer whether the observed binding for other tested metabolites would be functionally relevant in vivo.
Besides, we observed an increase in rigidity of the CIRBP RG/RGG region upon ATP binding (Figure 4d) implying that RG/RGG regions undergo local ordering in presence of nucleotides. Disordered proteins and nucleotides might originate earlier than folded proteins during evolution, 42 nucleotides might thereupon act as folding chaperones, templates or inducers to enable disordered proteins reaching a suitable conformation for further interactions, 43 as well as increasing solubility of unstructured proteins, thus counteracting their propensity to form pathological aggregates. 44 , 45 , 46
In this regard, and besides its function as co‐factor for enzymes, 47 ATP has more recently been shown to play a function as biological hydrotrope. 44 Here, we showed, using turbidity assays and DIC microscopy, that ATP can efficiently dissolve CIRBP liquid droplets, thus interfering with CIRBP LLPS (Figure 8b,c). Such “chaperone” function of ATP has been shown for others RBPs, including FUS, TAF‐15 and hnRNPA3. 44 These proteins share common features such as RG/RGG regions and RRMs, but the underlying ATP‐driven “chaperone” mechanisms remain unclear. CIRBP RRM possesses the features of a canonical RRM with a typical β1–α1–β2–β3–α2–β4 topology and three conserved aromatic side chains (9Phe, 49Phe, and 51Phe) located on β‐strand 1 and 3 which are essential for RNA binding (Figure 3c). 48 The CIRBP RRM surface has been proposed, based on homology modeling, to mediate CIRBP binding to RNA. 49 This is in agreement with the electrostatic potential surface of the CIRBP RRM showing a stretch of positively charged residues between the β‐strands β3 and β4 (Figure 3c). The same surface is responsible for ATP binding (Figure 3c). Therefore, it is tempting to speculate that ATP binding to the RRM will impair RNA binding.
Our data indicates that ATP‐mediated regulation of LLPS involves the CIRBP RG/RGG, but how this is linked to the in vivo situation remains unclear. We and others previously showed that RNA regulates LLPS of RG/RGG regions through the formation of intermolecular contacts. 23 , 26 Therefore, our data provide new mechanistic evidences suggesting that the intra‐cellular ATP/RNA ratio can influence RNP granule biogenesis, shifting the equilibrium toward granule formation or dissociation (Figure 8d). Concentrations of RNA and ATP in cells change upon cellular stress 50 or aging. 24 During oxidative stress, ATP is depleted from the cell and this correlates with CIRBP recruitment into SGs, a specific class of RNP granules. 32 , 51 Therefore, our findings provide first clues allowing a better understanding of the regulation of biomolecular condensates. They suggest that cellular concentration of nucleotides and derivatives govern intra‐cellular LLPS propensity of multiple RBPs containing RG/RGG regions via interfering with RNA binding. This might be of great biological interest as the mechanisms allowing RNP granule formation and dissociation are mis‐regulated in a plethora of diseases.
4. MATERIALS AND METHODS
4.1. Plasmids
Expression constructs for the fragments of wild type human CIRBP (1–172), CIRBPRRM (1–90), CIRBPRGG (68–137) and CIRBPRSY (138–172) were generated by synthesis of the corresponding optimized CIRBP DNA constructs (Genscript) and insertion of these into pETM11‐His6‐protein A vector containing a Tobacco Etch Virus (TEV) protease cleavage site after protein A. The expression construct for the fragment of rat PRMT1 (11–353) in pET28b‐His6 vector was previously described. 52
4.2. Protein expression and purification
For expression of recombinant unlabeled or 15N‐13C labeled His6‐protein A tagged constructs, the different bacterial expression vectors were transformed into Escherichia coli (E.coli)BL21‐DE3 Star strain and 1 L expression cultures were grown in lysogeny broth (LB) medium (for unlabeled proteins) or in minimal medium supplemented with 2 g of 13C6H12O6 (Cambridge Isotope Laboratories) and 1 g of 15NH4Cl (Sigma) (15N‐13C labeled proteins) at 37°C until OD (600)nm of 0.8 and further induced with 0.5 mM isopropyl β‐D‐1‐thiogalactopyranoside (IPTG) followed by protein expression for 16 hr at 20°C (CIRBPRRM, CIRBPRGG, CIRBPRSY and PRMT1) or 1.5 hr at 37°C (CIRBP). Cell pellets were harvested and sonicated either in denaturating lysis buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 20 mM Imidazole, 6 M urea) for disordered protein fragments (CIRBPRGG and CIRBPRSY) or in non‐denaturating lysis buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 20 mM imidazole, 2 mM tris(2‐carboxyethyl)phosphine [TCEP]) for folded protein fragments (CIRBP, CIRBPRRM and PRMT1). For CIRBPRRM and full‐length CIRBP, cell lysates were incubated for 30 min at room temperature with RNase A (Life Technologies) before centrifugation. Supernatants of His6‐protein A CIRBP, CIRBPRRM, CIRBPRGG, CIRBPRSY were applied on a nickel‐nitrilotriacetic (Ni‐NTA) agarose resin (Qiagen) compacted in gravity columns and eluted with 50 mM Tris–HCl pH 7.5, 1 M NaCl, 500 mM imidazole, 2 mM TCEP, 0.04% NaN3 containing buffer. Eluted His6‐protein A tagged CIRBP and individual regions were cleaved with TEV protease overnight at 4°C. Untagged CIRBP was diluted 10 times in 50 mM Tris, pH 7.5, 6 M Urea containing buffer, isolated using a HiTrap Heparin HP affinity column (Cytiva) and eluted with a buffer containing 50 mM Tris, pH 7.5, 1 M NaCl, 6 M Urea. CIRBP was buffer exchanged in a 10X‐salt buffer containing 50 mM NaH2PO4/Na2HPO4, pH 6.5, 1,500 mM NaCl and further diluted 10 times in the same buffer without NaCl prior usage. Untagged CIRBPRSY, CIRBPRGG and CIRBPRRM were isolated using a second affinity purification using Ni‐NTA beads. A final size exclusion chromatography purification step was performed in the buffer of interest on a gel filtration column (Superdex 75 GE Healthcare for CIRBPRRM and CIRBPRGG and Superdex peptide, GE Healthcare for CIRBPRSY).
The supernatant of His6‐PRMT1 after cell lysis was applied on a 5 ml HisTrap HP column (GE Healthcare) at 4°C and eluted with 50 mM Tris–HCl, pH 7.5, 1 M NaCl, 500 mM imidazole, 2 mM TCEP, 0.04% NaN3 containing buffer. A final size exclusion chromatography purification step was performed in the buffer of interest on a gel filtration column (Superdex 200 GE Healthcare).
Protein concentrations were estimated from their absorbance at 280 nm, assuming that the ε at 280 nm was equal to the theoretical ε value.
4.3. Cell culture
HeLa cells (ATCC, Guernsey, UK) were seeded on 1.5H high precision glass cover slips (Marienfeld‐Superior) and cultured in DMEM supplemented with 2 mM glutamine, 1% PS (100 U/ml penicillin, 100 μg/ml streptomycin) and 10% fetal bovine serum (FBS). Cells were maintained in a humidified incubator at 37°C with 5% CO2.
4.4. Cell metabolite extraction
HeLa cells pellets were suspended in 2:1, ice‐cold methanol:H2O mixture, and transferred to a tube containing Precellys beads (1.4 mm zirconium oxide beads, Bertin Technologies, Villeurbanne, France) for homogenization by Precellys 24 tissue homogenizer (Bertin Technologies, Montigny‐le‐Bretonneux, France). After centrifugation at 13,000 rpm for 45 min (4°C), the supernatant was transferred to a fresh tube, and the samples were lyophilized at <1 Torr, 850 rpm, 25°C for 10 hr in a vacuum‐drying chamber (Savant Speedvac SPD210 vacuum concentrator), with an attached cooling trap (Savant RVT450 refrigerated vapor trap) and vacuum pump (VLP120) (Thermo Scientific, Waltham, MA). The lyophilized metabolites were redissolved into a buffer containing 50 mM NaH2PO4/Na2HPO4 at pH 6.5. The concentration of 10 μl redissolved metabolites extract diluted in 500 μl NMR sample was defined as one arbitrary concentration unit, which was measured to contain 126 μM lactate in three replicates.
4.5. Turbidity assay
CIRBP and RNA (12 × UG repeats) samples were prepared in 50 mM NaH2PO4/Na2HPO4, pH 6.5, 150 mM NaCl. Turbidity measurements were conducted at 620 nm in 96‐well plates with 90 μl samples using a BioTek Power Wave HT plate reader (BioTek). Each experiment was performed in three replicates.
4.6. Differential interference contrast microscopy
CIRBP and RNA (12 × UG repeats) samples were prepared in 50 mM NaH2PO4/Na2HPO4, pH 6.5, 150 mM NaCl. Thirty microliter of sample was plated on a 30 mm No. 1 round glass coverslip and mounted on an Observer D1 microscope with 100×/1.45 oil immersion objective (Zeiss). Protein droplets were viewed using HAL 100 halogen lamp, and images were captured with an OrcaD2 camera (Hamamatsu) using VisiView 4.0.0.13 software (Visitron Systems GmbH). Droplet formation was induced by addition of RNA.
4.7. In vitro arginine methylation
CIRBPRGG and His6‐PRMT1 were equilibrated in methylation buffer containing 50 mM Na2HPO4/NaH2PO4, pH 8.0, 150 mM NaCl, 2 mM DTT. Fifty micromolar CIRBPRGG was incubated with 25 μM His6‐PRMT1 in presence of 2 mM S‐Adenosylmethionine (New England biolabs) for 16 hr at room temperature. Untagged methylated CIRBPRGG (metCIRBPRGG) was then isolated from His6‐PRMT1 performing a second affinity purification using Ni‐NTA beads and further equilibrated in the buffer of interest. metCIRBPRGG was analyzed using NMR 1H‐13C HSQC spectrum to confirm the CIRBP methylation status.
4.8. NMR spectroscopy
All protein samples were equilibrated in buffer containing 50 mM NaH2PO4/Na2HPO4, 150 mM NaCl at pH 6.5. All spectral acquisitions were performed at 298 K on a 600 MHz Bruker Avance Neo NMR spectrometer equipped with a TXI 600S3 probehead. Spectra were processed using Topspin 4.1.
The Carr–Purcell–Meiboom–Gill (CPMG) pulse sequence was used to acquire 1H 1D NMR spectra with pre‐saturation for water suppression (cpmgpr1d, 512 scans, 73,728 points in F1, 11,904.762 Hz spectral width, 1,024 transients, recycle delay 4 s) for all HeLa metabolites binding analysis.
2D 1H‐15N HSQC pulse program using water flip‐back and preservation of equivalent pathways (PEP) (hsqcetfpf3gpsi, 64 scans for CIRBP and 32 scans for CIRBP fragments, 128 points in F1, 1,024 points in F2, 1949.318 Hz spectral width in F1, 9,615.385 Hz spectral width in F2, 1,024 transients) was used to analyze all protein binding and titrations studies.
Phase sensitive ge‐2D 1H,15N HSQC (hsqcnoef3gpsi, 16 scans, 128 points in F1, 1,024 points in F2, 1,278.772 Hz spectral width in F1, 9,615.385 Hz spectral width in F2, 1,024 transients) was used to acquire heteronuclear 15N{1H} NOE of CIRBPRGG. The 15N{1H} heteronuclear NOE experiments were recorded with a saturation period/total interscan delay of 3.0 s.
Normalized CSPs of CIRBP 1H‐15N cross‐peaks upon binding to (di)nucleotides were calculated according to the following equation:
where δ H is the 1H chemical shift difference between CIRBP 1H‐15N cross‐peaks in the bound and free states and δ N is the 15N chemical shift difference between CIRBP 1H‐15N cross‐peaks in the bound and free states.
CONFLICT OF INTEREST
The authors declare no competing interests.
AUTHOR CONTRIBUTIONS
Qishun Zhou: Data curation; formal analysis; investigation; methodology; resources; visualization; writing‐original draft. Sinem Usluer: Data curation; investigation; resources; validation. Fangrong Zhang: Formal analysis; resources; writing‐review & editing. Aneta Lenard: Methodology; resources; writing‐review & editing. Benjamin Bourgeois: Conceptualization; writing‐review & editing. Tobias Madl: Funding acquisition; supervision; writing‐review & editing.
ACKNOWLEDGEMENTS
The work was supported by Austrian Science Fund (FWF) grants P28854, I3792, and DK‐MCD W1226 to T.M. and FWF grant P30162 to U.S.; T.M. was supported by the Austrian Research Promotion Agency (FFG) Grants 864690 and 870454; the Integrative Metabolism Research Center Graz; Austrian Infrastructure Program 2016/2017, the Styrian Government (Zukunftsfonds), and BioTechMed‐Graz (Flagship project DYNIMO). A.L., Q.Z., and S.U. were trained within the frame of the PhD program Metabolic and Cardiovascular Disease (DK‐MCD), and F.Z. was trained within the frame of the PhD program in Molecular Medicine. We thank the Center for Medical Research, Medical University of Graz, Graz, Austria for laboratory access.
Zhou Q, Usluer S, Zhang F, Lenard AJ, Bourgeois BMR, Madl T. ATP regulates RNA‐driven cold inducible RNA binding protein phase separation. Protein Science. 2021;30:1438–1453. 10.1002/pro.4123
Funding information Austrian Science Fund, Grant/Award Numbers: DK‐MCD W1226, I3792, P28854; Österreichische Forschungsförderungsgesellschaft, Grant/Award Numbers: 864690, 870454
REFERENCES
- 1. Deiana A, Forcelloni S, Porrello A, Giansanti A. Intrinsically disordered proteins and structured proteins with intrinsically disordered regions have different functional roles in the cell. PLoS One. 2019;14:e0217889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ward JJ, Sodhi JS, Mcguffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–645. [DOI] [PubMed] [Google Scholar]
- 3. Van Der Lee R, Buljan M, Lang B, et al. Classification of intrinsically disordered regions and proteins. Chem Rev. 2014;114:6589–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 2015;16:18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bürgi J, Xue B, Uversky VN, Van Der Goot FG. Intrinsic disorder in transmembrane proteins: Roles in signaling and topology prediction. PLoS One. 2016;11:e0158594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lakoucheva LM, Brown CJ, Lawson JD, Obradović Z, Dunker AK. Intrinsic disorder in cell‐signaling and cancer‐associated proteins. J Mol Biol. 2002;323:573–584. [DOI] [PubMed] [Google Scholar]
- 7. Hamann F, Schmitt A, Favretto F, et al. Structural analysis of the intrinsically disordered splicing factor Spp2 and its binding to the DEAH‐box ATPase Prp2. Proc Natl Acad Sci U S A. 2020;117:2948–2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Korneta I, Bujnicki JM. Intrinsic disorder in the human spliceosomal proteome. PLoS Comput Biol. 2012;8:e1002641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ou L, Waddell MB, Kriwacki RW. Mechanism of cell cycle entry mediated by the intrinsically disordered protein p27Kip1. ACS Chem Biol. 2012;7:678–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fahmi M, Ito M. Evolutionary approach of intrinsically disordered CIP/KIP proteins. Sci Rep. 2019;9:1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee C‐YS, Putnam A, Lu T, He S, Ouyang JPT, Seydoux G. Recruitment of mRNAs to P granules by condensation with intrinsically‐disordered proteins. Elife. 2020;9:e52896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Buchan JR. mRNP granules. Assembly, function, and connections with disease. RNA Biol. 2014;11:1019–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Protter DSW, Parker R. Principles and properties of stress granules. Trends Cell Biol. 2016;26:668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lin Y, David M, Parker R. Formation and maturation of phase‐separated liquid droplets by RNA‐binding proteins. Mol Cell. 2015;60:208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Souquere S, Mollet S, Kress M, Dautry F, Pierron G, Weil D. Unravelling the ultrastructure of stress granules and associated P‐bodies in human cells. J Cell Sci. 2009;122:3619–3626. [DOI] [PubMed] [Google Scholar]
- 16. Lin Y, Currie SL, Rosen MK. Intrinsically disordered sequences enable modulation of protein phase separation through distributed tyrosine motifs. J Biol Chem. 2017;292:19110–19120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Uversky VN. Intrinsically disordered proteins in overcrowded milieu: Membrane‐less organelles, phase separation, and intrinsic disorder. Curr Opin Struct Biol. 2017;44:18–30. [DOI] [PubMed] [Google Scholar]
- 18. Martinelli A, Lopes F, John E, Carlini C, Ligabue‐Braun R. Modulation of disordered proteins with a focus on neurodegenerative diseases and other pathologies. Int J Mol Sci. 2019;20:1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vance C, Scotter EL, Nishimura AL, et al. ALS mutant FUS disrupts nuclear localization and sequesters wild‐type FUS within cytoplasmic stress granules. Hum Mol Genet. 2013;22:2676–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu X, Chen J. Modulation of p53 transactivation domain conformations by ligand binding and cancer‐associated mutations. Pac Symp Biocomput. 2020;25:195–206. [PMC free article] [PubMed] [Google Scholar]
- 21. Park JO, Rubin SA, Xu Y‐F, et al. Metabolite concentrations, fluxes and free energies imply efficient enzyme usage. Nat Chem Biol. 2016;12:482–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Thandapani P, O'Connor TR, Bailey TL, Richard S. Defining the RGG/RG motif. Mol Cell. 2013;50:613–623. [DOI] [PubMed] [Google Scholar]
- 23. Chong PA, Vernon RM, Forman‐Kay JD. RGG/RG motif regions in RNA binding and phase separation. J Mol Biol. 2018;430:4650–4665. [DOI] [PubMed] [Google Scholar]
- 24. Kang J, Lim L, Song J. ATP enhances at low concentrations but dissolves at high concentrations liquid‐liquid phase separation (LLPS) of ALS/FTD‐causing FUS. Biochem Biophys Res Commun. 2018;504:545–551. [DOI] [PubMed] [Google Scholar]
- 25. Kang J, Lim L, Song J. ATP binds and inhibits the neurodegeneration‐associated fibrillization of the FUS RRM domain. Commun Biol. 2019;2:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bourgeois B, Hutten S, Gottschalk B, et al. Nonclassical nuclear localization signals mediate nuclear import of CIRBP. Proc Natl Acad Sci U S A. 2020;117:8503–8514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nishiyama H, Itoh K, Kaneko Y, Kishishita M, Yoshida O, Fujita J. A glycine‐rich RNA‐binding protein mediating cold‐inducible suppression of mammalian cell growth. J Cell Biol. 1997;137:899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhong P, Huang H. Recent progress in the research of cold‐inducible RNA‐binding protein. Future Sci OA. 2017;3:FSO246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sheikh MS, Carrier F, Papathanasiou MA, et al. Identification of several human homologs of hamster DNA damage‐inducible transcripts. J Biol Chem. 1997;272:26720–26726. [DOI] [PubMed] [Google Scholar]
- 30. Wellmann S, Bührer C, Moderegger E, et al. Oxygen‐regulated expression of the RNA‐binding proteins RBM3 and CIRP by a HIF‐1‐independent mechanism. J Cell Sci. 2004;117:1785–1794. [DOI] [PubMed] [Google Scholar]
- 31. Wang X, Che H, Zhang W, et al. Effects of mild chronic intermittent cold exposure on rat organs. Int J Biol Sci. 2015;11:1171–1180.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. De Leeuw F, Zhang T, Wauquier C, Huez G, Kruys V, Gueydan C. The cold‐inducible RNA‐binding protein migrates from the nucleus to cytoplasmic stress granules by a methylation‐dependent mechanism and acts as a translational repressor. Exp Cell Res. 2007;313:4130–4144. [DOI] [PubMed] [Google Scholar]
- 33. Pan F, Zarate J, Choudhury A, Rupprecht R, Bradley TM. Osmotic stress of salmon stimulates upregulation of a cold inducible RNA binding protein (CIRP) similar to that of mammals and amphibians. Biochimie. 2004;86:451–461. [DOI] [PubMed] [Google Scholar]
- 34. Zhou M, Yang W‐L, Ji Y, Qiang X, Wang P. Cold‐inducible RNA‐binding protein mediates neuroinflammation in cerebral ischemia. Biochim Biophys Acta. 1840;2014:2253–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lujan DA, Ochoa JL, Hartley RS. Cold‐inducible RNA binding protein in cancer and inflammation. Wiley Interdiscip Rev RNA. 2018;9:e1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hofweber M, Hutten S, Bourgeois B, et al. Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell. 2018;173:706–719. [DOI] [PubMed] [Google Scholar]
- 37. Myers JC, Shamoo Y. Human UP1 as a model for understanding purine recognition in the family of proteins containing the RNA recognition motif (RRM). J Mol Biol. 2004;342:743–756. [DOI] [PubMed] [Google Scholar]
- 38. Alayyoubi M, Leser GP, Kors CA, Lamb RA. Structure of the paramyxovirus parainfluenza virus 5 nucleoprotein–RNA complex. Proc Natl Acad Sci U S A. 2015;112:E1792–E1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Baker CM, Grant GH. Role of aromatic amino acids in protein‐nucleic acid recognition. Biopolymers. 2007;85:456–470. [DOI] [PubMed] [Google Scholar]
- 40. Wilson KA, Holland DJ, Wetmore SD. Topology of RNA–protein nucleobase–amino acid π–π interactions and comparison to analogous DNA–protein π–π contacts. RNA. 2016;22:696–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ruff KM, Dar F, Pappu RV. Ligand effects on phase separation of multivalent macromolecules. Proc Natl Acad Sci U S A. 2021;118:e2017184118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kulkarni P, Uversky V. Intrinsically disordered proteins and the Janus challenge. Biomolecules. 2018;8:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cafferty BJ, Fialho DM, Khanam J, Krishnamurthy R, Hud NV. Spontaneous formation and base pairing of plausible prebiotic nucleotides in water. Nat Commun. 2016;7:11328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Patel A, Malinovska L, Saha S, et al. ATP as a biological hydrotrope. Science. 2017;356:753–756. [DOI] [PubMed] [Google Scholar]
- 45. Rice AM, Rosen MK. ATP controls the crowd. Science. 2017;356:701–702. [DOI] [PubMed] [Google Scholar]
- 46. Alberti S, Hyman AA. Are aberrant phase transitions a driver of cellular aging? Bioessays. 2016;38:959–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nagana Gowda GA, Abell L, Lee CF, Tian R, Raftery D. Simultaneous analysis of major coenzymes of cellular redox reactions and energy using ex vivo 1H NMR spectroscopy. Anal Chem. 2016;88:4817–4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Daubner GM, Cléry A, Allain FH‐T. RRM–RNA recognition: NMR or crystallography…and new findings. Curr Opin Struct Biol. 2013;23:100–108. [DOI] [PubMed] [Google Scholar]
- 49. Coburn K, Melville Z, Aligholizadeh E, et al. Crystal structure of the human heterogeneous ribonucleoprotein A18 RNA‐recognition motif. Acta Cryst F. 2017;73:209–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Guillén‐Boixet J, Kopach A, Holehouse AS, et al. RNA‐induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell. 2020;181:346–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang X, Wu XQ, Lu S, Guo YL, Ma X. Deficit of mitochondria‐derived ATP during oxidative stress impairs mouse MII oocyte spindles. Cell Res. 2006;16:841–850. [DOI] [PubMed] [Google Scholar]
- 52. Zhang X, Cheng X. Structure of the predominant protein arginine methyltransferase PRMT1 and analysis of its binding to substrate peptides. Structure. 2003;11:509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]


