Abstract
Purpose:
This study aimed to (i) develop Magnetization-Prepared Golden-angle RAdial Sparse Parallel (MP-GRASP) MRI using a stack-of-stars trajectory for rapid free-breathing T1 mapping and (ii) extend MP-GRASP to multi-echo acquisition (MP-Dixon-GRASP) for fat/water-separated (water-specific) T1 mapping.
Methods:
An adiabatic non-selective 180° inversion-recovery pulse was added to a gradient-echo-based golden-angle stack-of-stars sequence for magnetization-prepared 3D single-echo or 3D multi-echo acquisition. In combination with subspace-based GRASP-Pro reconstruction, the sequence allows for standard T1 mapping (MP-GRASP) or fat/water-separated T1 mapping (MP-Dixon-GRASP), respectively. The accuracy of T1 mapping using MP-GRASP was evaluated in a phantom and volunteers (brain and liver) against clinically accepted reference methods. The repeatability of T1 estimation was also assessed in the phantom and volunteers. The performance of MP-Dixon-GRASP for water-specific T1 mapping was evaluated in a fat/water phantom and volunteers (brain and liver).
Results:
ROI-based mean T1 values are correlated between the references and MP-GRASP in the phantom (R2 = 1.0), brain (R2 = 0.96), and liver (R2 = 0.73). MP-GRASP achieved good repeatability of T1 estimation in the phantom (R2 = 1.0), brain (R2 = 0.99), and liver (R2 = 0.82). Water-specific T1 is different from in-phase and out-of-phase composite T1 (composite T1 when fat and water signal are mixed in phase or out of phase) both in the phantom and volunteers.
Conclusion:
This work demonstrated the initial performance of MP-GRASP and MP-Dixon-GRASP MRI for rapid 3D T1 mapping and 3D fat/water-separated T1 mapping in the brain (without motion) and in the liver (during free breathing). With fat/water-separated T1 estimation, MP-Dixon-GRASP could be potentially useful for imaging patients with fatty-liver diseases.
Keywords: fat/water separation, free-breathing, golden-angle radial, MP-GRASP, MP-Dixon-GRASP, T1 mapping
1 |. INTRODUCTION
Radial sampling in MRI offers various advantages compared to conventional Cartesian sampling, including benign/incoherent undersampling behavior that can be synergistically combined with sparsity-based reconstruction techniques,1–3 inherent robustness to motion for free-breathing body imaging,4–7 and self-navigation capabilities for additional motion management for free-breathing data acquisitions.8–13 Although non-Cartesian imaging has been historically challenging due to its sensitivity to system imperfections (ie, off-resonance and gradient-delay effects),14 radial MR techniques have been extensively optimized in recent years on different vendor platforms, which have facilitated a variety of novel clinical applications. Among different variants of radial sampling, the stack-of-stars trajectory, which combines radial sampling in the kx–ky plane and Cartesian sampling in the kz dimension as a hybrid 3D acquisition scheme,5,7 has attracted particular interest and attention. It combines the advantages of both radial and Cartesian trajectories and offers flexible choices of imaging parameters (ie, field of view and voxel size) in the in-plane and through-plane dimensions.
The performance of stack-of-stars sampling has been previously demonstrated in gradient-echo (GRE) imaging,7,15 fast spin-echo (FSE) imaging,16,17 multi-echo Dixon imaging,9,18,19 and balanced steady-state free precession (bSSFP) imaging.20–22 The stack-of-stars versions of these sequences have been applied for free-breathing dynamic contrast-enhanced MRI (DCE-MRI),3,6,23,24 cardiovascular MRI,9,20–22 fat/water MRI,18,19,25 arterial spin labeling (ASL) MRI26 and others.27 However, to date, the use of stack-of-stars imaging is primarily limited to steady-state acquisitions and its combination with magnetization-prepared data acquisition has been limited to only a few prior studies. Kim et al applied an inversion recovery (IR)-prepared stack-of-stars sequence for motion-insensitive carotid imaging,28 but it is restricted to qualitative imaging for improving image contrast. A similar implementation has also been tested recently for motion-insensitive brain imaging.29 In addition, Maier et al evaluated the use of an IR-prepared stack-of-stars sequence for T1 mapping in the brain,30 and Li et al also demonstrated 3D T1 mapping of the brain and carotid using an IR-prepared stack-of-stars imaging technique.31 Recently, Sharafi et al have extended stack-of-stars sampling with spin-lock preparation for T1rho mapping,32 but the resulting scan time remains long for clinical application. Meanwhile, the use of IR-prepared multi-echo stack-of-stars imaging for fat/water-separated T1 mapping has not been investigated.
During the past years, our group has developed a rapid 3D imaging technique called Golden-angle RAdial Sparse Parallel (GRASP) MRI.3 The performance of GRASP MRI with stack-of-stars sampling has been well-evaluated in various clinical applications,6 and it has been extended to multiple newer versions for improved motion management,11,33 robust removal of undersampling-induced residual streaking artifacts,33,34 higher reconstruction performance35 and increased reconstruction speed.36 In addition, our group has also extended stack-of-stars sampling to multi-echo acquisition, which has been demonstrated for free-breathing fat/water separation.18 The main contributions of this work include three aspects. First, we aimed to implement and demonstrate a magnetization-prepared GRASP framework called MP-GRASP, which combines non-selective IR-prepared stack-of-stars sampling with our latest GRASP components for rapid 3D T1 mapping. Second, we aimed to optimize MP-GRASP to enable motion-corrected image reconstruction for rapid 3D free-breathing T1 mapping of the liver. Third, inspired by several recent works for fat/water-separated T1 mapping,37–44 we aimed to extend MP-GRASP for multi-echo stack-of-stars acquisition (referred to as MP-Dixon-GRASP), which enables fat/water-separated (water-specific) T1 quantification. The performance of MP-GRASP and MP-Dixon-GRASP was evaluated in phantoms and volunteers (brain and liver). Our hypotheses were: (i) MP-GRASP enables accurate and repeatable standard T1 mapping and (ii) MP-Dixon-GRASP allows for water-specific T1 mapping to remove the influence from fat.
2 |. METHODS
2.1 |. IR-prepared stack-of-stars acquisition
An IR-prepared stack-of-stars sequence was developed based on a previously developed stack-of-stars 3D GRE sequence (RAVE: Radial Volumetric Encoding).18 Specifically, an adiabatic non-selective 180° IR pulse was added to the RAVE sequence, which can be periodically played-out to achieve magnetization preparation. The modified sequence allows for both single-echo acquisition and multi-echo acquisition, which can be applied for standard T1 mapping and fat/water-separated T1 mapping to derive water-specific T1 maps. After each IR pulse, a series of radial stacks (referred to as “stack-train”), rotated by a pre-defined rotation scheme, are acquired until the magnetization reaches steady state, as illustrated in Figure 1A. At a given time point, different partitions are acquired in a linear manner with the same rotating angle before moving to the next angle.
FIGURE 1.
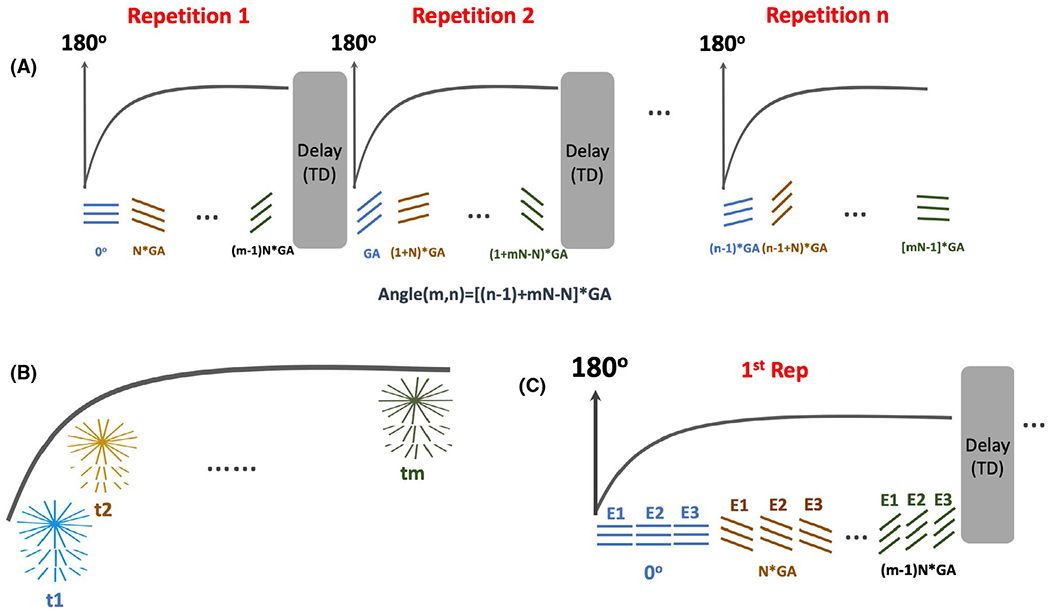
IR-prepared stack-of-stars acquisitions. Imaging sequence was developed based upon a stack-of-stars 3D GRE sequence (RAVE). A, An adiabatic non-selective 180° IR pulse is periodically played-out to achieve magnetization preparation. After each IR pulse, a series of radial stacks rotated by a pre-defined rotation scheme (Equation 1) are acquired until the magnetization reaches steady state. B, After synchronization of all the acquired repetitions, a composite IR-prepared dynamic image series can be generated where k-space at each time point is formed by N consecutive golden-angle rotations to ensure uniform coverage. C, The IR-prepared stack-of-stars sequence can also be performed for multi-echo acquisitions, where rotating angle for different echoes are the same and the number of echoes can be selected by the user
In our study, all acquisitions after each IR pulse are defined as one repetition. Each repetition is ended with an idle period to ensure full (or close-to-full) magnetization recovery before the start of the next repetition (see Figure 1A for details). The length of each stack-train (ie, the number of rotating radial stacks in each repetition) and the number of repetitions can both be pre-selected. The rotating angle of sampling a total of N repetitions is defined as:
| (1) |
Here, m and n denote the mth radial stack in the nth repetition and GA = 111.25° is the golden angle.45 This design ensures that, after synchronization of all acquired repetitions, a composite IR-prepared dynamic image series can be generated where k-space at each time point is formed by N consecutive golden-angle rotations to ensure uniform coverage (Figure 1B). As illustrated in Figure 1C, the IR-prepared stack-of-stars sequence also enables multi-echo acquisition.18 Here, the rotating angle for different echoes are the same and the number of echoes can be pre-selected. To enable motion-corrected reconstruction, the sequence has been modified such that physiological signals from an external device (ie, a respiratory bellow) can be automatically recorded with the raw data. This was implemented to ensure reliable motion detection in the presence of large contrast variation induced by the IR preparations.
2.2 |. Image reconstruction and T1 estimation
2.2.1 |. MP-GRASP reconstruction
Image reconstruction for MP-GRASP is performed utilizing several components that were previously developed for our GRASP framework. It involves the following steps: First, since full sampling is performed along the partition dimension, a kz-dimensional 1D fast Fourier transform (FFT) is performed to disentangle different slices for image reconstruction, which enables slice-by-slice processing. Second, an algorithm called Unstreaking33,34 is applied to remove residual streaking artifacts. Third, all the radial k-space samples are sorted and synchronized to form a composite IR-prepared dynamic image series (see Figure 1B). Self-calibrating GeneRalized autocalibrating partial parallel acquisition Operator Gridding (GROG)46 is then used to shift all the radial k-space data onto a Cartesian grid. This enables fast iterative radial reconstruction entirely on a Cartesian grid without going back to the radial k-space domain.36 Fourth, subspace-based low-rank and sparsity constrained GRASP reconstruction (GRASP-Pro: GRASP MRI with imProved performance35) is performed for dynamic image reconstruction. The reconstruction can be formulated as the following optimization problem:
| (2) |
where y denotes sorted, synchronized and GROG-processed IR-based dynamic k-space, E represents the 2D FFT operation, and S represents coil sensitivities estimated using all acquired radial data with the adaptive combination method.47 UK denotes the first K dominant temporal basis functions for underlying dynamic images and VK represents corresponding coefficients to be reconstructed. The temporal basis UK is estimated from a dictionary generated by solving the Bloch equations, with T1 ranging from 100 to 3000 ms and B1 (to account for flip-angle variation) ranging from 0.8 to 1.2. A spatial total-variation (TV) constraint is enforced directly on the subspace coefficients and is controlled by a parameter λ. For brain image reconstruction, Ω is set as an identity matrix. For liver image reconstruction, Ω represents a motion-weighting matrix generated from the automatically recorded motion signal to perform motion-weighted reconstruction, which controls the contribution of different respiratory phases to the reconstructed images. This ensures that data from the expiratory phases contribute more to the reconstruction, whereas data from the inspiratory phase are attenuated. In this study, motion-weighted reconstruction was performed with four respiratory states spanning from end-expiration to end-inspiration, as we previously implemented in ref.33 After reconstruction, dynamic images (m) can be generated with:
| (3) |
The overall idea of subspace-based reconstruction is to compress dynamic images to a low-dimensional subspace that can be represented by only the K dominant basis components (UK) and associated coefficients (VK).48 This is possible because of extensive spatiotemporal correlations that are typically present in dynamic MR images. The construction of subspace leads to highly reduced degrees of freedom and thus improved reconstruction performance, as shown in prior studies.35,49–54
2.2.2 |. MP-Dixon-GRASP reconstruction
MP-Dixon-GRASP reconstruction involves two steps: The first step is to reconstruct multi-echo dynamic images and the second step is to separate the fat and water signal at different inversion times (TIs). The initial reconstruction step is similar to the MP-GRASP reconstruction (Equation 2). Images from all echoes are jointly reconstructed within a single step. The temporal basis functions are estimated separately for different echoes to account for different echo times (TEs). Once the reconstructed dynamic multi-echo images are available, separated fat/water images are obtained by solving the following cost function:
| (4) |
Here, W and F denote the to-be-separated dynamic water-only and fat-only images, mte represents the dynamic images at echo time te, and Φ denotes the B0 field map estimated from the time-averaged multi-echo images using the B0-NICE algorithm.55 M denotes the forward operator to synthesize multi-echo dynamic images from W, F, and Φ:
| (5) |
where models the fat/water chemical shift according to a multiple peak fat spectrum model with 7 fixed frequencies (Δfp) and relative amplitudes (αp) for a given echo time (tte).19,56
2.2.3 |. T1 map generation
After image reconstruction, pixel-by-pixel fitting based on the following three-parameter model57 is performed to calculate T1 map:
| (6) |
For a given pixel location, S(t) represents the signal at time point t (corresponding TI time), M* and M0 are steady-state and equilibrium magnetization, respectively, and T1* represents the effective relaxation time.57 With the estimated parameters, T1 is then obtained as T1 = M0 · T1*/M*. For MP-Dixon-GRASP, in-phase and out-of-phase composite T1 maps are generated from dynamic images at corresponding echo times. Water-specific T1 maps are generated from the dynamic water-only images.
2.3 |. Imaging experiments
2.3.1 |. Overall experimental design
For evaluation of MP-GRASP, the following six experiments were performed: (1) validating MP-GRASP for T1 mapping against IR-based spin echo (IR-SE) imaging in a T1 mapping phantom; (2) evaluating the repeatability of T1 estimation (for different scan dates and different spatial resolutions) using MP-GRASP in the phantom; (3) comparing MP-GRASP with reference MP2RAGE (Magnetization-Prepared 2 RApid Gradient Echoes imaging)58 for T1 mapping of the brain; (4) evaluating the repeatability of T1 estimation (for both the same spatial resolution and different spatial resolutions) using MP-GRASP in the brain; (5) comparing MP-GRASP with reference BH-MOLLI (Breath-Hold Modified Look-Locker imaging)59 for T1 mapping of the liver; and (6) evaluating the repeatability of T1 estimation in the liver using MP-GRASP.
For evaluation of MP-Dixon-GRASP, the following three experiments were performed: (1) comparing water-specific T1 maps with out-of-phase and in-phase composite T1 maps (estimated from the first-echo and second-echo images, respectively) in a home-made fat/water phantom; (2) comparing water-specific T1 maps with out-of-phase and in-phase composite T1 maps in the brain; and (3) the same comparison in the liver.
A total of 15 volunteers (6 males, 9 females, age = 31.53 ± 6.82y) were recruited for brain imaging. A total of 13 volunteers (5 males, 8 females, age = 31.92 ± 7.10y) were recruited for liver imaging. All human studies are HIPAA-compliant and was approved by the local Institutional Review Board (IRB). Written informed consent was obtained from all subjects before the MR scans.
To avoid region-of-interest (ROI) selection bias, an observer blinded to the main purpose of this study performed all the image analyses in MATLAB (MathWorks, MA) to compare T1 maps generated from different methods.
2.3.2 |. Imaging protocols
Multiple imaging protocols were prepared for brain and liver imaging as listed in Table 1. Phantom imaging used the same imaging protocols. Three MP-GRASP protocols were developed, including two for brain imaging and one for liver imaging. Two MP-Dixon-GRASP protocol were developed for brain imaging and liver imaging, respectively. All imaging studies were performed on a 3T MRI scanner (MAGNETOM Skyra, Siemens Healthcare GmbH, Germany). The idle delay (6 s for the brain protocols and 4.2 s for the liver protocols) was set based on a previous study,60 and 75% partial Fourier was applied along the partition dimension for all radial acquisitions.
TABLE 1.
Relevant imaging parameters of different T1 mapping methods in the brain and the livera.
| Matrix size | Voxel size (mm3) | TR (ms) | TE (ms) | TA (min:s) | IR-Rep | Stack-Train | FA (°) | Bandwidth (Hz/Pixel) | ||
|---|---|---|---|---|---|---|---|---|---|---|
| BRAIN | MP-GRASP Protocol 1 | 224 × 224 × 32 | 1.25 × 1.25 × 3 | 2.83 | 1.32 | 2:32 | 17 | 48 | 5 | 1010 |
| MP-GRASP Protocol 2 | 320 × 320 × 32 | 0.875 × 0.875 × 3 | 3.67 | 1.74 | 2:49 | 17 | 48 | 5 | 710 | |
| MP-Dixon-GRASP (3 echoes) | 224 × 224 × 16 | 1.25 × 1.25 × 3 | 6.12 | 1.41 – 4.61 (ΔTE = 1.6) | 2:37 | 17 | 48 | 5 | 1010 | |
| MP2RAGE | 224 × 224 × 32 | 1.25 × 1.25 × 3 | 4000 | 2.83 | 4:18 | N/A | N/A | 4, 5 | 250 | |
| LIVER | MP-GRASP | 256 × 256 × 32 | 1.37 × 1.37 × 5 | 2.63 | 1.22 | 2:58 | 25 | 48 | 5 | 1150 |
| MP-Dixon-GRASP (4 echoes) | 256 × 256 × 16 | 1.37 × 1.37 × 5 | 6.33 | 1.23 – 4.92 (ΔTE = 1.23) | 3:14 | 25 | 48 | 5 | 1150 | |
| BH-MOLLI | 256 × 256 (2D) | 1.37 × 1.37 × 8 | 2.8 | 1.14 | 0:15 (BH) | N/A | N/A | 35 | 1085 | |
MP-GRASP and MP-Dixon-GRASP were performed during free breathing for liver imaging. MP2RAGE was used as the reference T1 mapping method for brain imaging and 2D breath-hold MOLLI (BH-MOLLI) was used as the T1 mapping reference method for liver imaging. IR-Rep refers to the number of inversion recovery (IR) pulses (see Figure 1) played out during acquisition. Stack-train refers to the number of angles acquired following each IR-Rep.
2.3.3 |. Reconstruction implementation
MP-GRASP and MP-Dixon-GRASP reconstructions were performed offline using the nonlinear conjugate gradient algorithm. The number of temporal basis functions (K in Equation 2) was set as 4 for phantom/brain image reconstruction and 5 for liver image reconstruction. For MP-Dixon-GRASP, fat/water separation was performed on the reconstructed multi-echo images using the limited-memory Broyden–Fletcher–Goldfarb–Shanno (L-BFGS) algorithm. Reconstructed images were then used for fitting T1 maps. All offline reconstruction tasks were performed using MATLAB on a Linux server equipped with 64 CPU cores, 11 GB GPU memory and 256 GB memory. The iterative part of the MP-GRASP reconstruction was implemented to run on a GPU.36 MP-Dixon-GRASP reconstruction was implemented to run on CPUs due to larger data size.
2.3.4 |. MP-GRASP phantom experiment
A vendor-provided T1MES phantom61 was used to evaluate the performance of T1 mapping using MP-GRASP. The phantom contains 9 vials with different T1 values. Two imaging studies were performed with a gap of 12 days to test the repeatability of T1 estimation. In the first study, MP-GRASP brain protocol 1 (see Table 1, referred to as “Day 1 Scan 1”) was applied. IR-SE imaging was then performed to obtain ground-truth T1 values for different phantom vials. IR-SE imaging was performed for a middle slice with the following imaging parameters: field of view (FOV) = 200 x 200 mm2, matrix size = 128 x 128, voxel size = 1.56 x 1.56 mm2, flip angle = 90°, slice thickness = 8 mm, echo time (TE) = 15 ms, repetition time (TR) = 15000 ms, bandwidth = 130 Hz/pixel. 13 images were acquired at inversion delay times (TI) of 30, 50, 100, 200, 300, 400, 500, 700, 1000, 1500, 2000, 3000 and 4500 ms. The total acquisition time was ~7 hours. In the second study, both MP-GRASP brain protocol 1 and MP-GRASP brain protocol 2 were performed (referred to as “Day 2 Scan 1” and “Day 2 Scan 2”, respectively) to assess the repeatability of T1 estimation with the same spatial resolution and a different spatial resolution, respectively.
From the resulting T1 maps, a single circular ROI was manually placed on each phantom vial in a middle slice. Linear regression and Bland-Altman analyses were performed to compare ROI-based mean T1 values (i) between IR-SE and MP-GRASP Day 1 Scan 1 (for assessing accuracy), (ii) between MP-GRASP Day 1 Scan 1 and MP-GRASP Day 2 Scan 1 (for assessing inter-scan repeatability with the same spatial resolution), and (iii) between MP-GRASP Day 1 Scan 1 and MP-GRASP Day 2 Scan 2 (for assessing inter-scan repeatability with different spatial resolutions).
2.3.5 |. MP-GRASP brain experiment
MP-GRASP was performed in all the 15 brain volunteers. For each MRI scan, MP-GRASP was performed first using protocol 1 followed by MP2RAGE. MP-GRASP with protocol 1 was repeated in seven volunteers to assess the repeatability of T1 estimation. In addition, MP-GRASP using protocol 2 with a higher spatial resolution was performed in 14 volunteers to assess the repeatability of T1 estimation cross different spatial resolutions. The protocol 2 was performed for validating that T1 values is independent of spatial resolution. For simplicity, MRI scans using MP-GRASP protocol 1, repeated protocol 1, and protocol 2 are referred to as “Scan 1”, “Scan 2” and “Scan 3,” respectively.
For all brain T1 maps, five ROIs were manually placed, including two ROIs in the white matter (WM), one ROI in the thalamus, one ROI in the putamen and one ROI in the caudate. Linear regression and Bland-Altman analyses were performed to compare ROI-based mean T1 values (i) between MP2RAGE and MP-GRASP Scan 1 (for assessing agreement), (ii) between MP-GRASP Scan 1 and MP-GRASP Scan 2 (for assessing repeatability at the same spatial resolution), and (iii) between MP-GRASP Scan 1 and MP-GRASP Scan 3 (for assessing repeatability at different spatial resolutions).
2.3.6 |. MP-GRASP liver MRI experiment
MP-GRASP was performed in all the 13 liver volunteers. A respiratory bellow was placed on the abdomen of the volunteers to record respiratory motion signals during the scans. The recording was done automatically without any user interaction. For each MRI scan, MP-GRASP was performed first during free breathing (MP-GRASP Scan 1, Table 1). Based on the MP-GRASP images reconstructed online, BH-MOLLI was then performed for three different slices, each of which was performed during one breath hold. For 12 subjects, MP-GRASP with the same protocol was repeated again in the end of each MRI exam (MP-GRASP Scan 2) to assess the repeatability of liver T1 mapping.
For all liver T1 maps, two ROIs was manually placed on each slice of BH-MOLLI T1 map (a total of six ROIs for three slices of BH-MOLLI T1 map) for each dataset. ROIs were selected in the liver parenchyma without hepatic vessel involvement. Corresponding six ROIs were then placed on the T1 maps from MP-GRASP Scan 1 at matching locations. In addition, 10 ROIs were placed on the liver parenchyma in T1 maps from MP-GRASP Scan 1 and MP-GRASP Scan 2 at matching locations. Linear regression and Bland-Altman analyses were performed to compare ROI-based mean T1 values (i) between BH-MOLLI and MP-GRASP Scan 1 (for assessing agreement) and (ii) between MP-GRASP Scan 1 and MP-GRASP Scan 2 (for assessing repeatability).
2.3.7 |. MP-Dixon-GRASP phantom experiment
A home-made fat/water phantom was used to evaluate the performance of fat/water-separated T1 mapping using MP-Dixon-GRASP. The phantom contains 6 vials, including one with pure water, one with pure peanut oil, two with the same gadolinium concentration (0.07936 mol/L MultiHance) but different fat fractions (0% and 20%), and two without gadolinium and with different fat fractions (0% and 20%). The phantom was made following a procedure described in ref.62 Fat and water were manually mixed with agarose. Imaging was performed using the MP-Dixon-GRASP liver protocol with four echoes. From the resulting T1 maps, circular ROIs were placed on each phantom vial. ROI-based mean T1 values from the water-specific T1 map, the out-of-phase composite T1 map (from the first echo, TE = 1.23 ms), and the in-phase composite T1 map (from the second echo, TE = 2.46 ms) were compared. The phantom study was performed to validate the hypothesis that water-specific T1 is independent of fat fractions.
2.3.8 |. MP-Dixon-GRASP brain experiment
MP-Dixon-GRASP was performed in seven brain volunteers using the MP-Dixon-GRASP brain protocol from Table 1. Five brain ROIs, as described above, were manually placed on different T1 maps to compare water-specific T1 with out-of-phase composite T1 and in-phase composite T1 for different tissue types. Bland-Altman analysis and two-tailed paired student t test were performed to assess the differences between composite and water-specific T1, where P < .05 indicated statistical significance.
2.3.9 |. MP-Dixon-GRASP liver experiment
MP-Dixon-GRASP was performed in six volunteers during free breathing using the MP-Dixon-GRASP liver protocol from Table 1. One subject was later confirmed to have liver steatosis by a body radiologist. A total of 10 ROIs in the liver parenchyma were selected on different slices to compare water-specific T1 with out-of-phase composite T1 and in-phase composite T1. Bland-Altman analysis and two-tailed paired student t test were performed to assess the differences between composite and water-specific T1, where P<0.05 indicated statistical significance.
3 |. RESULTS
Figure 2A compares T1 maps obtained with IR-SE and different MP-GRASP scans in the phantom. Visually, both imaging methods generated similar T1 maps. The linear regression and Bland-Altman plots in Figure 2B confirm the visual observation. Mean T1 values from different phantom tubes are highly correlated between IR-SE and MP-GRASP (R2 = 0.998), and they do not show significant bias in T1 estimation according to the Bland-Altman plots. MP-GRASP T1 values obtained at different days and different spatial resolutions are also highly correlated (R2 = 1.0 and R2 = 0.999 for scan/rescan with the same spatial resolution and with different spatial resolutions, respectively). These results suggest good repeatability of MP-GRASP for T1 estimation in the phantom.
FIGURE 2.
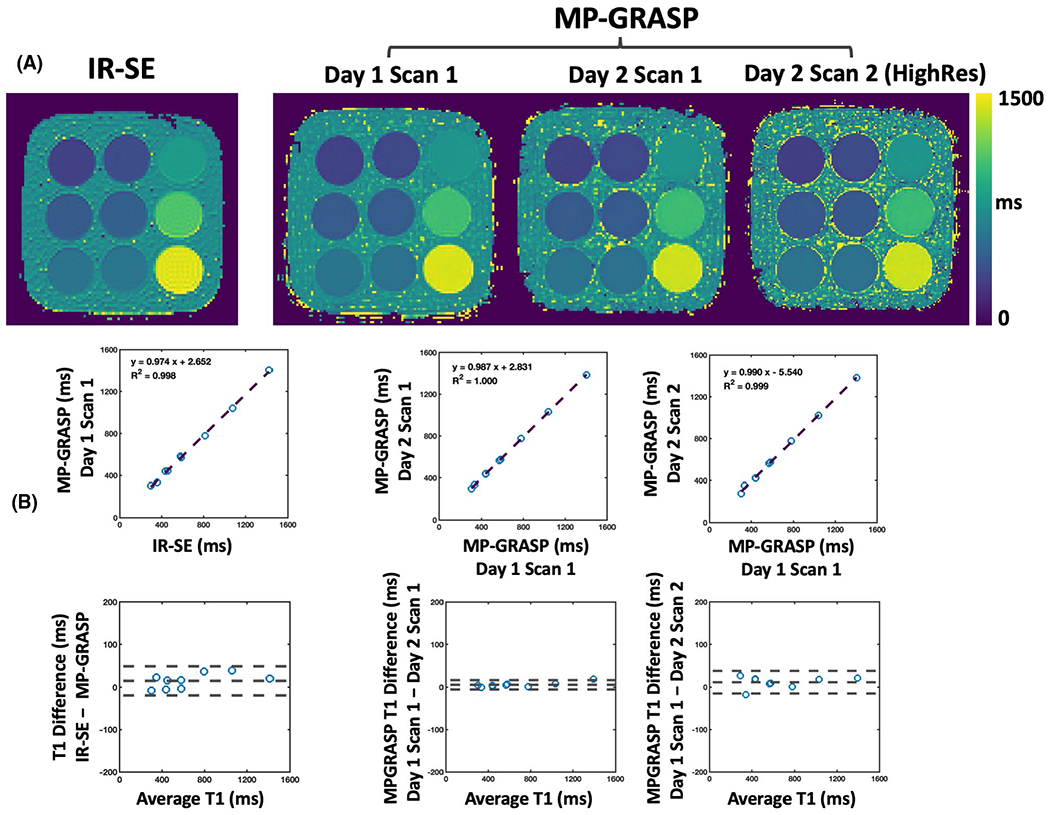
Comparisons of T1 maps obtained from IR-SE and different MP-GRASP scans in phantom imaging. A, All imaging methods show similar T1 maps in different phantom vials. B, Corresponding linear regression and Bland-Altman plots. T1 values from different phantom tubes are highly correlated between IR-SE and MP-GRASP (R2 = 0.998) and do not show significant bias for T1 estimation. MP-GRASP T1 values obtained at different time points and different spatial resolutions are also highly correlated (R2 = 1.0 between MP-GRASP Day 1 Scan 1 and MP-GRASP Day 2 Scan 1 with the same spatial resolution; R2 = 0.999 between MP-GRASP Day 1 Scan 1 and MP-GRASP Day 2 Scan 2 with different spatial resolutions) without significant bias
Figure 3 shows comparisons of brain T1 maps obtained from MP2RAGE and MP-GRASP in one volunteer and linear regression and Bland-Altman plots of mean T1 values in all the selected ROIs across all subjects. Other than the CSF and the skull, the brain T1 maps are visually comparable between the two methods. Although they exhibit good correlation (R2 = 0.955), the Bland-Altman plot suggests that MP2RAGE yielded lower T1 values than MP-GRASP. Supporting Information Figure S1 presents brain T1 maps from 20 consecutive slices in one subject, showing the performance of 3D T1 mapping in the brain.
FIGURE 3.
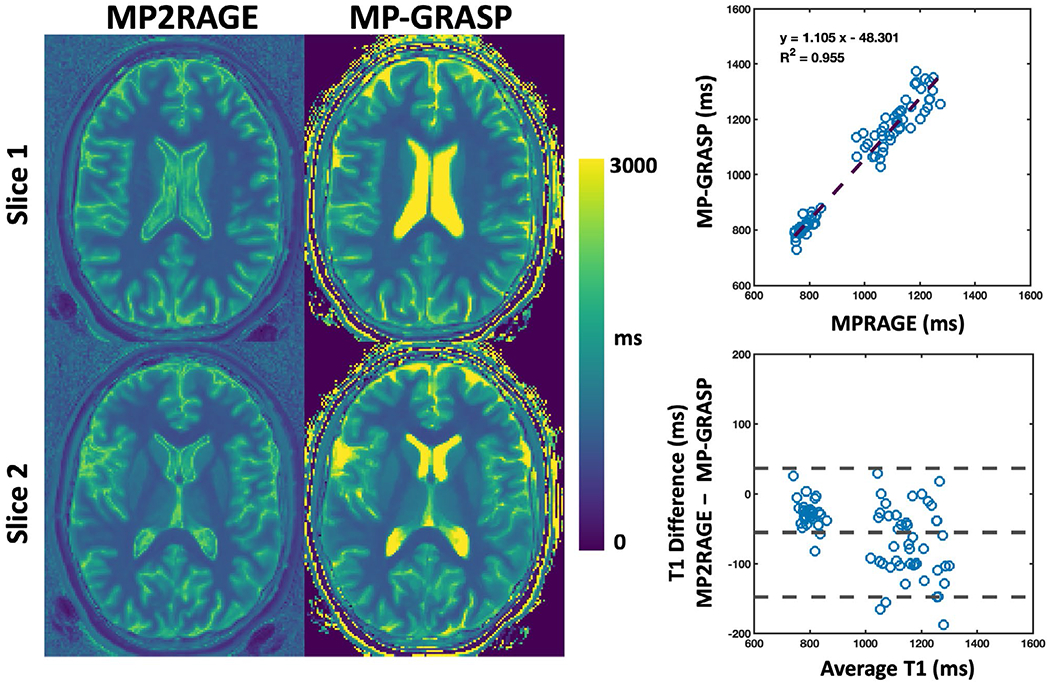
Comparison of brain T1 maps obtained from MP2RAGE and MP-GRASP in one volunteer. The T1 maps are visually comparable except for the CSF and the skull region. The linear regression shows that mean T1 values across all the subjects exhibit a good correlation (R2 = 0.955). The Bland-Altman plot suggests that MP2RAGE yielded lower T1 values compared to those from MP-GRASP
Comparison of brain T1 maps from MP-GRASP Scan 1 (protocol 1), repeated MP-GRASP Scan 2 (protocol 1) and MP-GRASP Scan 3 (protocol 2, higher spatial resolution) is shown in Figure 4. The T1 maps are visually comparable except for the finer structure visible in MP-GRASP Scan 3 due to smaller voxel size. The T1 values from the skull fat region differ between the two imaging protocols (red arrows), which is likely due to the lower bandwidth in MP-GRASP protocol 2 causing chemical-shift blurring in radial acquisition. The statistical analyses indicate that the mean T1 values from the selected ROIs obtained from the scan and rescan are highly correlated (R2 = 0.99) (see Supporting Information Figure S2 for Bland-Altman plots). These results demonstrated good repeatability of T1 estimation with MP-GRASP even at different spatial resolutions. Three slices of MP-GRASP brain images at different TIs are shown in Supporting Information Figure S3.
FIGURE 4.
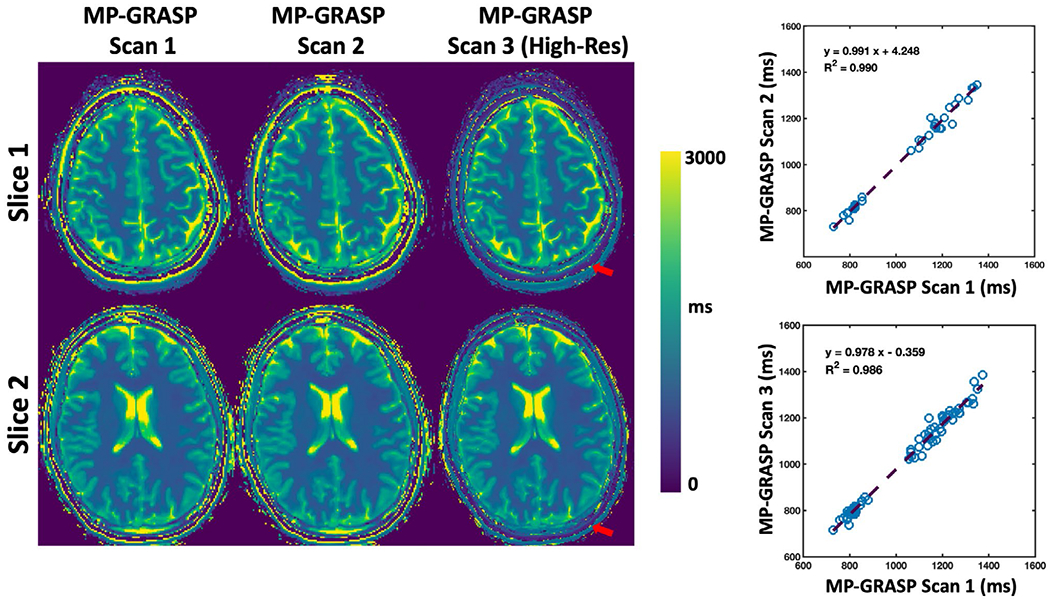
Comparison of brain T1 maps from MP-GRASP Scan 1 (MP-GRASP protocol 1), MP-GRASP Scan 2 (repeated MP-GRASP protocol 1), and MP-GRASP Scan 3 (MP-GRASP protocol 2, higher spatial resolution). The maps are visually comparable except for the finer structure display in MP-GRASP Scan 3 due to increased spatial resolution. The linear regression plots indicate that the mean T1 values from the selected ROIs obtained from the scan and rescan are high-correlated (R2 = 0.99). These results demonstrated good repeatability of T1 estimation using MP-GRASP even at different spatial resolutions
Liver T1 maps from BH-MOLLI and MP-GRASP are compared in Figure 5 for two different slices from one volunteer. The T1 maps are visually comparable and the linear regression indicates moderate correlation (R2 = 0.734). Based on the Bland-Altman plot, T1 values estimated from BH-MOLLI are lower than those from MP-GRASP in all subjects. 20 slices of MP-GRASP liver T1 maps from this subject are shown in Supporting Information Figure S4. Figure 6 shows comparisons of liver T1 maps between MP-GRASP Scan 1 and MP-GRASP Scan 2 in another subject. Despite respiratory motion that may have been different during the two scans, the resulting T1 maps are visually comparable as confirmed by the linear regression (R2 = 0.82) and Bland-Altman plots from all the subjects.
FIGURE 5.
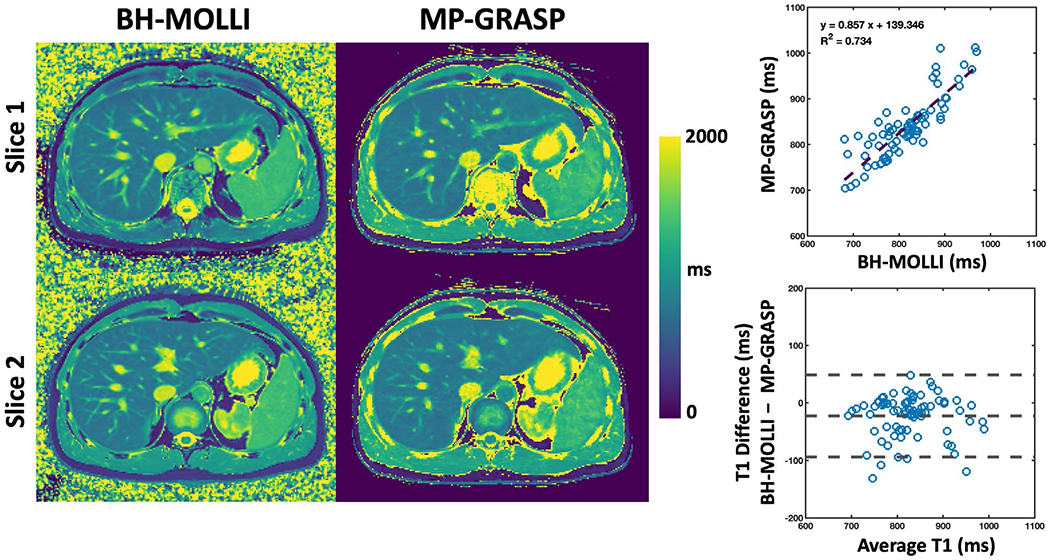
Comparison of liver T1 maps from BH-MOLLI and MP-GRASP for two different slices from one volunteer. The T1 maps are visually comparable and the linear regression plot indicates moderate T1 correlation (R2 = 0.734). T1 values estimated from BH-MOLLI are lower compared to those from MP-GRASP based on the Bland-Altman plot
FIGURE 6.
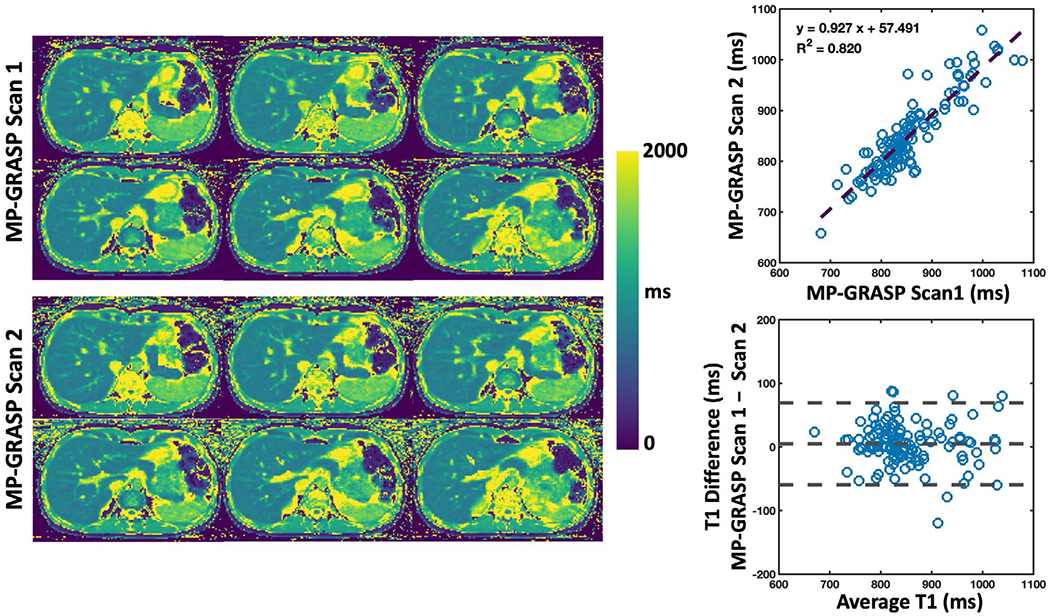
Comparison of liver T1 maps between MP-GRASP Scan 1 and MP-GRASP Scan 2 in one volunteer. Despite respiratory motion that may be different during the two scans, the resulting T1 maps are comparable as confirmed by the linear regression and Bland-Altman plots
Figure 7A shows a picture of the fat/water phantom. Figure 7B compares T1-weighted out-of-phase (TE = 1.23 ms) and in-phase (TE = 2.46 ms) images from the fat/water phantom, together with corresponding separated water and fat images. In the out-of-phase image, vials 1 is brighter than vial 2 and vial 5 is brighter than vial 6. Vial 4 (pure oil) is brighter than vial 3 (pure water) in both out-of-phase and in-phase images. Figure 7C compares the composite T1 maps with the water-specific T1 map from the phantom. When fat and water are not mixed, vials 1, 3, 4, and 5 have similar T1 values across different T1 maps. Note that vial 4 (pure oil) does not contain signal in the water-specific T1 map as confirmed in the water image. When fat and water are mixed (vial 2 and vial 6), larger out-of-phase composite T1 and smaller in-phase composite T1 were obtained compared to corresponding vials without fat (vial 1 and vial 5, respectively). The water-specific T1 map indicates that the influence of fat can be removed, and similar underlying T1 values for [vial 1, vial 2] and [vial 5, vial 6] can be obtained despite different fat fractions. The T1 values from Figure 7C confirm the image contrast presented in corresponding T1-weighted images (Figure 7B).
FIGURE 7.
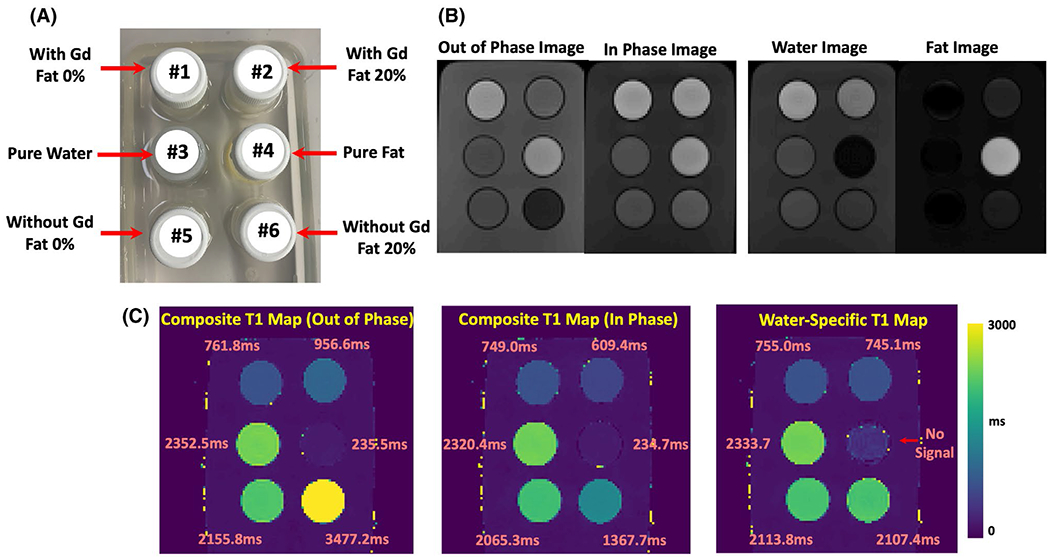
Phantom evaluation of MP-Dixon-GRASP. A, A picture of the fat/water phantom structure. B, In the out-of-phase image, vials 1 is brighter than vial 2 and vial 5 is brighter than vial 6. Vial 4 (pure oil) is brighter than vial 3 (pure water) in both out-of-phase and in-phase images. C, Comparison of the composite T1 maps with the water-specific T1 map from the phantom. When fat and water are not mixed, vials 1, 3, 4, and 5 have similar T1 values across different T1 maps. Vial 4 (pure oil) does not contain signal in the water-specific T1 map as confirmed in the water image. When fat and water are mixed (vial 2 and vial 6), larger out-of-phase composite T1 and smaller in-phase composite T1 were obtained compared to corresponding vials without fat (vial 1 and vial 5, respectively). The water-specific T1 map indicates that the influence of fat can be removed, and similar underlying T1 values for [vial 1, vial 2] and [vial 5, vial 6] can be obtained despite different fat fractions
Figure 8 compares composite T1 maps with water-specific T1 maps in the brain in two volunteers. Bland-Altman analysis of all the subjects is also shown pooling all the ROIs together. The out-of-phase composite T1, in-phase composite T1, and water-specific T1 values are 798.59 ± 29.75 ms, 799.79 ± 27.83 ms, and 800.73 ± 28.71 ms, respectively, for the white matter; 1037.28 ± 29.83 ms, 1031.1 ± 30.68 ms, and 1036.76 ± 30.65 ms, respectively, for the thalamus; 1151.91 ± 58.25 ms, 1144.03 ± 48.73 ms, and 1148.43 ± 54.26 ms, respectively, for the putamen; 1265.83 ± 73.47 ms, 1254.79 ± 76.78 ms, and 1255.32 ± 69.67 ms, respectively, for the caudate. No difference was found between the out-of-phase composite T1 and water-specific T1 (P > .1) except for the Caudate (P < .05). No difference was found between in-phase composite T1 and water-specific T1 in all the tissue types (P > .1).
FIGURE 8.
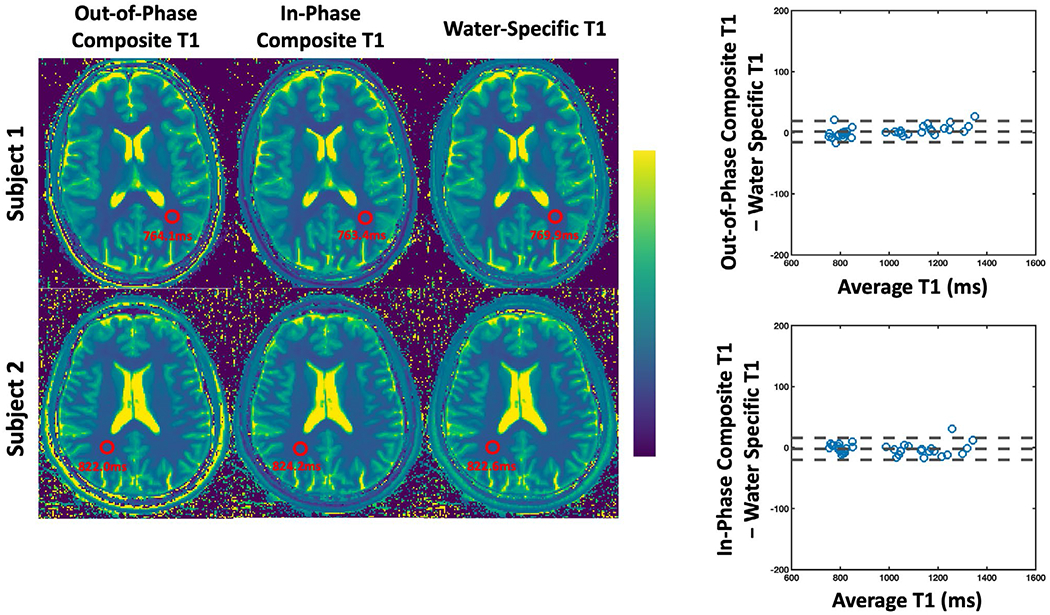
Comparison of out-of-phase and in-phase composite T1 maps with water-specific T1 map generated from MP-Dixon-GRASP in two brain volunteers. No difference was found between out-of-phase composite T1 and water-specific T1 (P > .1) except for the Caudate (P < .05). No difference was found between in-phase composite T1 and water-specific T1 in all the tissue types (P > .1)
Figure 9 compares composite T1 maps with water-specific T1 maps in the liver in two volunteers. The second subject was confirmed to have liver steatosis. The red arrows indicate that the fat signal was removed in the water-specific T1 maps. Bland-Altman plots are shown pooling all the ROIs from all the subjects together. The out-of-phase composite T1, in-phase composite T1, and water-specific T1 values of the liver are 854.31 ± 90.65 ms, 801.94 ± 71.46 ms, and 833.29 ± 67.86 ms, respectively. The water-specific T1 is significantly higher than the in-phase composite T1 (P < .001) and is significantly lower than the out-of-phase composite T1 (P < .05) in the liver. The differences were found to be larger in the subject with liver steatosis compared to others. This finding is consistent with the fat/water phantom results.
FIGURE 9.
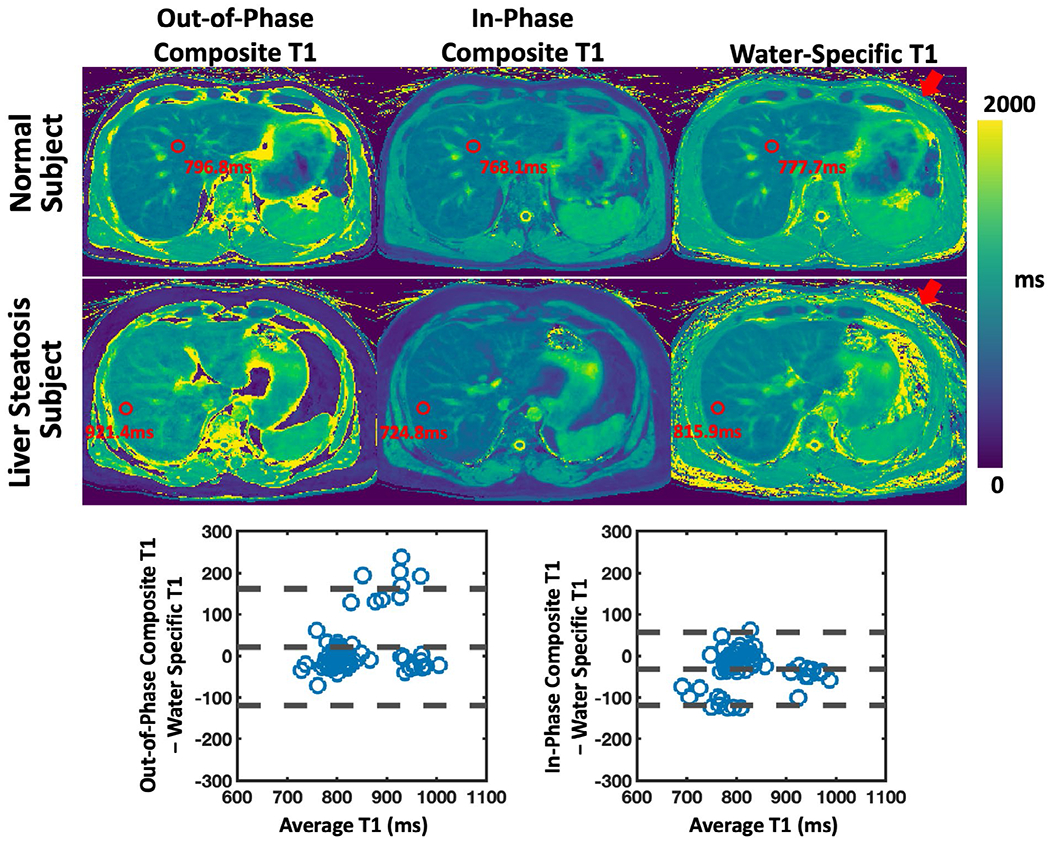
Comparison of out-of-phase and in-phase composite T1 maps with water-specific T1 map generated from MP-Dixon-GRASP in two liver subjects. One subject was confirmed to have liver stenosis. Water-specific T1 is significantly higher than in-phase composite T1 in the liver (P < .001) and is significantly lower than out-of-phase composite T1 in the liver (P < .05). The differences were found to be larger in the subject with liver steatosis compared to others. The red arrows show that the fat signal was removed in the water-specific T1 maps. This finding is consistent with the fat/water phantom results
The scan-rescan test of MP-Dixon-GRASP for water-specific T1 mapping of the brain and the liver in one volunteer is shown in Supporting Information Figure S5. Supporting Information Figure S6 shows fat/water-separated brain and liver images at one TI from one volunteer.
The average image reconstruction time was 21.39 ± 1.59 min for the 3D MP-GRASP brain datasets, 38.18 ± 2.09 min for the 3D MP-GRASP liver datasets, 97.15 ± 6.97 min for the 3D MP-Dixon-GRASP brain datasets, and 166.92 ± 4.74 min for the 3D MP-Dixon-GRASP liver datasets.
4 |. DISCUSSION
In this study, we described two quantitative MRI techniques for 3D T1 mapping (MP-GRASP) and fat/water-separated T1 mapping (MP-Dixon-GRASP). The acquisition is based on IR-prepared golden-angle stack-of-stars sampling, and the image reconstruction is based on subspace low-rank and sparsity constraints.35 The accuracy of MP-GRASP for T1 mapping was evaluated in the T1MES phantom, in the brain and the liver. We also assessed the repeatability of T1 estimation both in phantom and in volunteer scans. Based on MP-GRASP, MP-Dixon-GRASP moves one step further, allowing for fat/water-separated T1 mapping to remove the influence of fat. MP-Dixon-GRASP was evaluated in a fat/water phantom, the brain and liver. We have investigated the difference between water-specific T1 and out-of-phase and in-phase composite T1. In the following subsections, we discuss our experimental findings, differences between our technique with other related techniques, and limitations of our current study.
4.1 |. Experimental findings
For T1 mapping of the brain, T1 values estimated from MP-GRASP were found to be well-correlated with those from MP2RAGE. However, MP2RAGE generated relatively lower T1 values than MP-GRASP (Figure 3). It should be noted, however, that MP2RAGE was used as an exemplary reference. It has been previously reported that MP2RAGE underestimates T1 at different scales.63,64 In addition, as shown in a previous study,58 MP2RAGE cannot accurately estimate T1 > 3 s and T1 < 0.5 s at 3T, which led to substantial T1 underestimation in the CSF (Figure 3).
For T1 mapping of the liver, T1 values estimated from MP-GRASP were found to be moderately correlated with those from BH-MOLLI. Meanwhile, T1 values from BH-MOLLI were found to be lower than those from MP-GRASP. It has been known that BH-MOLLI tends to underestimate T1.65 This can be due to several reasons. First, BH-MOLLI typically implements bSSFP acquisition, which is sensitive to magnetic field inhomogeneity. Second, BH-MOLLI employs 2D acquisition that may require additional correction for excitation profile. Third, BH-MOLLI involves 180° IR pulses that yield large RF energy, which can cause effects such as magnetization transfer (MT) during in-vivo imaging. Despite these issues, BH-MOLLI remains a well-accepted T1 mapping method for both cardiac and liver exams.65,66 Although MP-GRASP uses 3D GRE acquisition that may be less sensitive to field inhomogeneity with better excitation accuracy, it also uses 180° IR pulses. From this perspective, MP-GRASP may also underestimate T1 at certain degrees.
Regarding the repeatability of T1 estimation using MP-GRASP. Good results were obtained in the phantom and volunteers. We performed inter-scan repeatability test for the phantom and intra-scan repeatability test for the brain and the liver. The spatial resolution was changed for the phantom and brain scans to test the repeatability at different spatial resolutions. We noticed certain fat/water blurring in high resolution brain T1 maps (Figure 4, red arrows). This is because no fat suppression was implemented in MP-GRASP, so that a lower bandwidth (restricted by small voxel size) can cause stronger chemical shift effects. Although this is not a major problem in the brain due to limited fat, this could be a potential problem in other organs with more fat content and will require better reconstruction strategies.18
For fat/water-separated T1 mapping, phantom imaging suggested that mixed fat/water leads to decreased T1 in in-phase composite T1. This is expected, since fat is known to have short T1. Thus, a combination of fat and water in phase will lead to a weighted combination of their respective T1. On the other hand, mixed fat/water leads to increased out-of-phase composite T1. This is likely due to the fact that fat and water have phase cancellation when they are out of phase. This finding is consistent with clinical T1-weighted fat/water liver MRI, in which lipid-rich lesions have brighter intensity in fat/water in-phase images (due to decreased composite T1) while low intensity in fat/water out-of-phase images (due to increased composite T1).67 Water-specific T1 mapping is able to remove the influence of fat, generating consistent underlying T1 values despite different fat fractions. The phantom results were confirmed by the in-vivo fat/water-separated T1 mapping results, particularly in the subject with liver steatosis (Figure 9). Compared to the brain, the difference between composite T1 and water-specific T1 is larger in the liver, which may be due to the fact that the liver typically has more fat content than the brain even in volunteers.
4.2 |. Imaging techniques
As an extension of GRASP for magnetization-prepared imaging, MP-GRASP and MP-Dixon-GRASP incorporate several components from our previous GRASP developments. These include approaches to remove residual streaking artifacts (Unstreaking33,34), to increase reconstruction speed (GROG-GRASP36), to improve reconstruction quality based on subspace low-rank and sparsity constraints (GRASP-Pro35), and to reduce motion blurring (motion-weighted GRASP33,68). In addition, we have modified the imaging sequence to automatically record a respiratory bellow signal with the raw data for motion management. Self-navigation (motion detection from acquired k-space) was not used because of the extensive contrast variation caused by the IR preparations (Supporting Information Figure S7), which affect the signal-variation patterns of the z projections and make motion detection less reliable. Motion extraction from a respiratory bellow is not affected by the contrast change. Supporting Information Figure S8 compares MP-GRASP reconstruction for one liver dataset with and without motion-weighted reconstruction. The influence of MP-GRASP reconstruction parameters (eg, the size of subspace and TV regularization parameter) on resulting T1 maps was also investigated (see Supporting Information Figures S9 and S10).
In addition to quantitative T1 mapping, MP-GRASP could also be used for conventional qualitative assessment (Supporting Information Figure S3). In particular, MP-GRASP may server as an improved alternative to clinical MP-RAGE imaging. First, radial sampling enables motion-robust imaging. Second, conventional MP-RAGE acquisition requires selection of a delay time after the IR pulse (known as TD) to generate the optimal image contrast. This, however, results in substantial deadtime that reduces scan efficiency. By employing a continuous acquisition immediately after each IR predation, MP-GRASP can address this issue and can generate contrast-resolved dynamic images, and the most desired contrast can then be selected retrospectively from the reconstructed dynamic image series.
For MP-Dixon-GRASP, a two-step reconstruction scheme has been implemented. IR-prepared multi-echo dynamic images are reconstructed first and the resulting images are used for fat/water separation. These two steps can potentially be integrated into a single step, so that dynamic water and fat images can be directly reconstructed from the dynamic multi-echo k-space data using model-based reconstruction.18,69 Model-based reconstruction may also potentially improve reconstruction performance by employing the fat/water chemical shift model as an intrinsic constraint.
In this work, we only demonstrated the use of MP-GRASP and MP-Dixon-GRASP for T1 mapping. The framework can also be further extended for T2 mapping by employing T2 preparation, or for joint T1 and T2 mapping using multiple preparations, as shown in prior studies.70–73 It can also be further extended to include estimation of R2* and proton density fat fraction (PDFF)25,37,43 and to further remove the influence of iron in T1 estimation. This would lead to a comprehensive free-breathing quantitative imaging framework.
4.3 |. Comparison with related techniques
There are several prior studies that use magnetization-prepared radial sampling for T1 mapping or multiparametric mapping.30,31,43,53,70,71,73–78 However, most of these works use a 2D radial acquisition. A few studies have employed IR-prepared 3D radial sampling based on the Koosh-ball geometry.71,73,78 However, 3D radial sampling typically requires data acquisition with isotropic resolution, which prolongs scan time78 and is less attractive for liver imaging. IR-prepared stack-of-stars sampling was used previously for T1 mapping of the brain and neck,30,31 but there are several key differences between these prior work and our work. After each IR preparation, the study in ref30 aims to acquire all spokes for a given slice followed by the next IR preparation to acquire spokes for a different slice. Such sampling scheme provides reduced motion robustness for free-breathing imaging. The study in ref31 proposed a 3D T1 mapping technique using slab-selective excitation, which may suffer from inhomogeneous excitation profiles. Recently, several works have been presented for fat/water-separated T1 mapping in the liver37,41,43,44 and the heart.38,40,42 These techniques were implemented either for breath-hold 2D imaging or 3D Cartesian imaging. However, stack-of-stars sampling offers improved motion robustness,79 and free-breathing 3D T1 quantification may be more desired for evaluating disease heterogeneity in the whole organ.
Compared to MR fingerprinting (MRF) that aims to encode multiple dimensions of information in one acquisition, our technique is fast and can be more attractive for applications that only require 3D T1 information. For example, MP-GRASP would be ideal when combined with our GRASP MRI technique for free-breathing quantitative DCE-MRI, because the MP-GRASP protocol can be exactly matched with the GRASP DCE-MRI scan. Thus, accurate pixel-wise contrast concentration can be obtained, which would otherwise be challenging with conventional 3D T1 mapping methods.
4.4 |. Limitations of the study
This study has several limitations that require discussion. First, MP-Dixon-GRASP acquires only half of the volumetric coverage compared to MP-GRASP. This is because of the prolonged TR (Table 1) in MP-Dixon-GRASP protocols. To ensure a temporal resolution that is sufficient for reliable T1 quantification, we decided to sacrifice volumetric coverage. A possible solution to overcome this limitation is to accelerated the kz dimension of the stack-of-stars trajectory, as demonstrated in our previous work.80 This can provide a better balance of the hybrid radial-Cartesian k-space coverage in stack-of-stars imaging. Second, the TEs for the MP-Dixon-GRASP brain protocol are not exactly in phase and out of phase. This is due to the smaller voxel size in brain imaging, which restricts the minimum TE. Thus, we had to set the first TE as 1.41. Third, the current version of MP-Dixon-GRASP does not consider the R2* effect along the multi-echo dimension. Although this may not be a big concern for volunteer imaging, it may create bias in patients with iron overload. This can be overcome by further extending MP-Dixon-GRASP for joint R2* mapping and fat/water-separated T1 mapping, which will be explored in future work. Finally, we evaluated our techniques in volunteers only. Further imaging studies in patients, particularly in patients with fatty-liver diseases, will be necessary to fully investigate the performance of the methods. It will also be necessary to further study the differences between composite T1 and water-specific T1 in patients.
5 |. CONCLUSIONS
This work presents MP-GRASP and MP-Dixon-GRASP, two free-breathing techniques for 3D T1 mapping and 3D fat/water-separated T1 mapping. The new methods can be applied for static organs like the brain or for moving organs like the liver. With fat/water-separated T1 estimation, MP-Dixon-GRASP could be potentially useful for patients with fatty-liver diseases.
Supplementary Material
Figure S1 Brain T1 maps from 20 consecutive slices in one volunteer
Figure S2 Bland Altman plots for assessing the repeatability of T1 mapping in the brain using MP-GRASP. Left plot compares MP-GRASP Scan 1 with MP-GRASP Scan 2 (repeatability at the same spatial resolution). Right plot compares MP-GRASP Scan 1 with MP-GRASP Scan 3 (repeatability at different spatial resolutions)
Figure S3 Three slices of MP-GRASP brain images at different TIs (TI times shown on the top of the figure). In addition to quantitative information, MP-GRASP also generates a contrast-resolved image series, from which an image can be selected to emulate the clinical MP-RAGE contrast for qualitative use
Figure S4 Liver T1 maps from 20 consecutive slices in one volunteer
Figure S5 Scan-rescan test for MP-Dixon-GRASP in the brain and in the liver
Figure S6 Representative fat/water-separated brain and liver images obtained from MP-Dixon-GRASP
Figure S7 Respiratory motion signals obtained from the bellow device superimposed on underlying z projection, showing good correlation between them. The z projection profiles were obtained through the centers of stack-of-stars k-space. The bellow signal can be automatically recorded with the raw data
Figure S8 Comparison of liver T1 maps from MP-GRASP reconstruction with and without motion correction in two subjects. Motion-weighted reconstruction improves image sharpness with reduced motion blurring as indicated by the red arrows
Figure S9 The influence of subspace size on T1 maps. K = 2 generated totally wrong T1 maps. T1 quantification seems not sensitive to the size of subspace within a certain range (eg, K = 4 to 6). For the brain dataset (without motion), T1 quantification seems to be insensitive to the selection of K. This can be explained by the fact that T1 fitting can also be seen as a process to remove noise and incoherent artifacts (eg, residual streaking artifacts). For the liver dataset, however, the quality of T1 quantification reduces with a large value of K
Figure S10 The TV regularization parameter was selected empirically by comparing a range of values, as done in most constrained iterative reconstruction studies. TV regularization helps to improve reconstruction quality and resulting T1 maps. The use of TV is not expected to affect temporal fidelity since it is only enforced on spatial dimensions. However, a larger value can lead to spatial blurring
Figure S11 In one dataset, we retrospectively used every 2nd, 4th, 6th, 8th, and 10th time points only for image reconstruction and T1 map generation. In other words, we used [1, 3, 5, 7…], [1, 5, 9, 13…], [1, 7, 13, 19…], [1, 9, 17, 25…], and [1, 11, 21, 31…] TI points only for testing the influence of temporal resolution on image reconstruction and T1 quantification. We found that good T1 maps can be obtained by using up to every 4th time points. When the number of points was further reduced, T1 maps start seeing loss of details. This finding suggests that we may potentially increase the number of partitions quite a bit in our imaging method before seeing significant effects. It also suggests that the short T1 components are more severely affected by reduced temporal resolution, which is expected because short T1 components recover faster than long T1 components
Figure S12 In the same dataset as in Figure S11, we retrospectively used only the first N time points for image reconstruction and T1 fitting, where N equaled 30, 20, 10 and 5. This is similar to acquiring for less time after each IR preparation. We found that good T1 maps can be obtained when using less time points (eg, first 30 time points). However, when the number of points was reduced to ~10, T1 maps start to lose details
ACKNOWLEDGMENTS
The authors thank Renata Pyzik for help with the volunteer recruitments, Kamil Banibaker and Dewey Chu for help with the MRI scans, and Dr. Sara Lewis for reading the liver images.
Footnotes
CONFLICT OF INTEREST
Li Feng and Kai Tobias Block are named co-inventors of a patent (Patent number 9921285) on the GRASP imaging technique. Kai Tobias Block and Thomas Benkert are employees of Siemens Healthcare GmbH, Germany.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section.
REFERENCES
- 1.Block KT, Uecker M, Frahm J. Undersampled radial MRI with multiple coils. Iterative image reconstruction using a total variation constraint. Magn Reson Med. 2007;57:1086–1098. [DOI] [PubMed] [Google Scholar]
- 2.Adluru G, McGann C, Speier P, Kholmovski EG, Shaaban A, Dibella EVR. Acquisition and reconstruction of undersampled radial data for myocardial perfusion magnetic resonance imaging. J Magn Reson Imaging. 2009;29:466–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng L, Grimm R, Block KT, et al. Golden-angle radial sparse parallel MRI: combination of compressed sensing, parallel imaging, and golden-angle radial sampling for fast and flexible dynamic volumetric MRI. Magn Reson Med. 2014;72:707–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glover GH, Pauly JM. Projection reconstruction techniques for reduction of motion effects in MRI. Magn Reson Med. 1992;28:275–289. [DOI] [PubMed] [Google Scholar]
- 5.Block KT, Chandarana H, Milla S, et al. Towards routine clinical use of radial stack-of-stars 3D gradient-echo sequences for reducing motion sensitivity. J Korean Soc Magn Reson Med. 2014;18:87. [Google Scholar]
- 6.Feng L, Benkert T, Block KT, Sodickson DK, Otazo R, Chandarana H. Compressed sensing for body MRI. J Magn Reson Imaging. 2017;45:966–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandarana H, Block KT, Rosenkrantz AB, et al. Free-breathing radial 3D fat-suppressed T1-weighted gradient echo sequence. Invest Radiol. 2011;46:648–653. [DOI] [PubMed] [Google Scholar]
- 8.Larson AC, White RD, Laub G, McVeigh ER, Li D, Simonetti OP. Self-gated cardiac cine MRI. Magn Reson Med. 2004;51:93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J, Spincemaille P, Codella NCF, Nguyen TD, Prince MR, Wang Y. Respiratory and cardiac self-gated free-breathing cardiac CINE imaging with multiecho 3D hybrid radial SSFP acquisition. Magn Reson Med. 2010;63:1230–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piccini D, Littmann A, Nielles-Vallespin S, Zenge MO. Respiratory self-navigation for whole-heart bright-blood coronary MRI: methods for robust isolation and automatic segmentation of the blood pool. Magn Reson Med. 2012;68:571–579. [DOI] [PubMed] [Google Scholar]
- 11.Feng L, Axel L, Chandarana H, Block KT, Sodickson DK, Otazo R. XD-GRASP: golden-angle radial MRI with reconstruction of extra motion-state dimensions using compressed sensing. Magn Reson Med. 2016;75:775–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piccini D, Feng L, Bonanno G, et al. Four-dimensional respiratory motion-resolved whole heart coronary MR angiography. Magn Reson Med. 2017;77:1473–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng L, Coppo S, Piccini D, et al. 5D whole-heart sparse MRI. Magn Reson Med. 2018;79:826–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Block KT. Advanced Methods for Radial Data Sampling in Magnetic Resonance Imaging [PhD Thesis]. Germany: University of Goettingen; 2008. [Google Scholar]
- 15.Chandarana H, Block KT, Winfeld MJ, et al. Free-breathing contrast-enhanced T1-weighted gradient-echo imaging with radial k-space sampling for paediatric abdominopelvic MRI. Eur Radiol. 2014;24:320–326. [DOI] [PubMed] [Google Scholar]
- 16.Benkert T, Mugler JP, Rigie DS, Sodickson DK, Chandarana H, Block KT. Hybrid T2 - and T1 -weighted radial acquisition for free-breathing abdominal examination. Magn Reson Med. 2018;80:1935–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keerthivasan MB, Saranathan M, Johnson K, et al. An efficient 3D stack-of-stars turbo spin echo pulse sequence for simultaneous T2-weighted imaging and T2 mapping. Magn Reson Med. 2019;82:326–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benkert T, Feng L, Sodickson DK, Chandarana H, Block KT. Free-breathing volumetric fat/water separation by combining radial sampling, compressed sensing, and parallel imaging. Magn Reson Med. 2017;78:565–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Armstrong T, Dregely I, Stemmer A, et al. Free-breathing liver fat quantification using a multiecho 3D stack-of-radial technique. Magn Reson Med. 2018;79:370–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, Wieben O, Jung Y, Samsonov AA, Reeder SB, Block WF. Single breathhold cardiac CINE imaging with multi-echo three-dimensional hybrid radial SSFP acquisition. J Magn Reson Imaging. 2010;32:434–440. [DOI] [PubMed] [Google Scholar]
- 21.Haji-Valizadeh H, Collins JD, Aouad PJ, et al. Accelerated, free-breathing, noncontrast, electrocardiograph-triggered, thoracic MR angiography with stack-of-stars k-space sampling and GRASP reconstruction. Magn Reson Med. 2019;81:524–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X, Xie G, Lu NA, et al. 3D self-gated cardiac cine imaging at 3 Tesla using stack-of-stars bSSFP with tiny golden angles and compressed sensing. Magn Reson Med. 2019;81:3234–3244. [DOI] [PubMed] [Google Scholar]
- 23.Chandarana H, Feng L, Block KT, et al. Free-breathing contrast-enhanced multiphase MRI of the liver using a combination of compressed sensing, parallel imaging, and golden-angle radial sampling. Invest Radiol. 2013;48:10–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim SG, Feng L, Grimm R, et al. Influence of temporal regularization and radial undersampling factor on compressed sensing reconstruction in dynamic contrast enhanced MRI of the breast. J Magn Reson Imaging. 2016;43:261–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider M, Benkert T, Solomon E, et al. Free-breathing fat and R2* quantification in the liver using a stack-of-stars multi-echo acquisition with respiratory-resolved model-based reconstruction. Magn Reson Med. 2020;84:2592–2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song HK, Yan L, Smith RX, et al. Noncontrast enhanced four-dimensional dynamic MRA with golden angle radial acquisition and K-space weighted image contrast (KWIC) reconstruction. Magn Reson Med. 2014;72:1541–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang L, Armstrong T, Li X, Wu HH. A variable flip angle golden-angle-ordered 3D stack-of-radial MRI technique for simultaneous proton resonant frequency shift and T1-based thermometry. Magn Reson Med. 2019;82:2062–2076. [DOI] [PubMed] [Google Scholar]
- 28.Kim SE, Roberts JA, Eisenmenger LB, et al. Motion-insensitive carotid intraplaque hemorrhage imaging using 3D inversion recovery preparation stack of stars (IR-prep SOS) technique. J Magn Reson Imaging. 2017;45:410–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Solomon E, Hu HH, Block KT, Sodickson DK, Chandarana HMP-RAVE. IR-Prepared T1-weighted radial stack-of-stars 3D GRE imaging with retrospective motion correction. In ISMRM 2020 Annual Meeting Proceedings, 2020. p. 561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maier O, Schoormans J, Schloegl M, et al. Rapid T1 quantification from high resolution 3D data with model-based reconstruction. Magn Reson Med. 2019;81:2072–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Z, Bilgin A, Martin DR, Altbach MI. Rapid high-resolution 3D T1 mapping using a highly accelerated radial inversion-recovery FLASH technique. In ISMRM 2017 Annual Meeting Proceedings, Honolulu, HI, USA. p. 3720. [Google Scholar]
- 32.Sharafi A, Baboli R, Zibetti M, et al. Volumetric multicomponent T1ρ relaxation mapping of the human liver under free breathing at 3T. Magn Reson Med. 2020;83:2042–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng L, Huang C, Shanbhogue K, Sodickson DK, Chandarana H, Otazo R. RACER-GRASP: Respiratory-weighted, aortic contrast enhancement-guided and coil-unstreaking golden-angle radial sparse MRI. Magn Reson Med. 2018;80:77–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng L, Chandarana H, Sodickson DK, Otazo R. Unstreaking: radial MRI with automatic streaking artifact reduction. In ISMRM 2017 Annual Meeting Proceedings, Honolulu, HI, USA. p. 4001. [Google Scholar]
- 35.Feng L, Wen Q, Huang C, Tong A, Liu F, Chandarana H. GRASP-Pro: imProving GRASP DCE-MRI through self-calibrating subspace-modeling and contrast phase automation. Magn Reson Med. 2020;83:94–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benkert T, Tian Y, Huang C, DiBella EVR, Chandarana H, Feng L. Optimization and validation of accelerated golden-angle radial sparse MRI reconstruction with self-calibrating GRAPPA operator gridding. Magn Reson Med. 2018;80:286–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson RB, Chow K, Mager D, Pagano JJ, Grenier J. Simultaneous proton density fat-fraction and R2* imaging with water-specific T1 mapping (PROFIT1): application in liver. Magn Reson Med. 2021;85:223–238. [DOI] [PubMed] [Google Scholar]
- 38.Liu Y, Hamilton J, Eck B, Griswold M, Seiberlich N. Myocardial T1 and T2 quantification and water-fat separation using cardiac MR fingerprinting with rosette trajectories at 3T and 1.5T. Magn Reson Med. 2021;85:103–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nezafat M, Nakamori S, Basha TA, Fahmy AS, Hauser T, Botnar RM. Imaging sequence for joint myocardial T1 mapping and fat/water separation. Magn Reson Med. 2019;81:486–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jaubert O, Cruz G, Bustin A, et al. Water-fat Dixon cardiac magnetic resonance fingerprinting. Magn Reson Med. 2020;83:2107–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jaubert O, Arrieta C, Cruz G, et al. Multi-parametric liver tissue characterization using MR fingerprinting: Simultaneous T1, T2, T2*, and fat fraction mapping. Magn Reson Med. 2020;84:2625–2635. [DOI] [PubMed] [Google Scholar]
- 42.Milotta G, Bustin A, Jaubert O, Neji R, Prieto C, Botnar RM. 3D whole-heart isotropic-resolution motion-compensated joint T1/T2 mapping and water/fat imaging. Magn Reson Med. 2020;84:3009–3026. [DOI] [PubMed] [Google Scholar]
- 43.Li Z, Pauly J, Vasanawala SS. Rapid multi-slice fat and water separated T1 and composite R2* mapping using a dual-echo radial inversion recovery SPGR pulse sequence. In ISMRM 2020 Annual Meeting Proceedings, 2020. p. 312. [Google Scholar]
- 44.Mozes FE, Tunnicliffe EM, Moolla A, et al. Mapping tissue water T1 in the liver using the MOLLI T1 method in the presence of fat, iron and B0 inhomogeneity. NMR Biomed. 2019;32:e4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winkelmann S, Schaeffter T, Koehler T, Eggers H, Doessel O. An optimal radial profile order based on the golden ratio for time-resolved MRI. IEEE Trans Med Imaging. 2007;26:68–76. [DOI] [PubMed] [Google Scholar]
- 46.Seiberlich N, Breuer F, Blaimer M, Jakob P, Griswold M. Self-calibrating GRAPPA operator gridding for radial and spiral trajectories. Magn Reson Med. 2008;59:930–935. [DOI] [PubMed] [Google Scholar]
- 47.Walsh DO, Gmitro AF, Marcellin MW. Adaptive reconstruction of phased array MR imagery. Magn Reson Med. 2000;43:682–690. [DOI] [PubMed] [Google Scholar]
- 48.Liang ZP. Spatiotemporal imaging with partially separable functions. In: Proc. of 2007 Joint Meet. of the 6th Int. Symp. on Noninvasive Functional Source Imaging of the Brain and Heart and the Int. Conf. on Functional Biomedical Imaging, NFSI and ICFBI 2007; 2007:181–182. [Google Scholar]
- 49.Tamir JI, Uecker M, Chen W, et al. T2 shuffling: sharp, multicontrast, volumetric fast spin-echo imaging. Magn Reson Med. 2017;77:180–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao B, Lu W, Hitchens TK, Lam F, Ho C, Liang ZP. Accelerated MR parameter mapping with low-rank and sparsity constraints. Magn Reson Med. 2015;74:489–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao B, Setsompop K, Adalsteinsson E, et al. Improved magnetic resonance fingerprinting reconstruction with low-rank and subspace modeling. Magn Reson Med. 2018;79:933–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang C, Graff CG, Clarkson EW, Bilgin A, Altbach MI. T 2 mapping from highly undersampled data by reconstruction of principal component coefficient maps using compressed sensing. Magn Reson Med. 2012;67:1355–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Z, Bilgin A, Johnson K, et al. Rapid high-resolution T1 mapping using a highly accelerated radial steady-state free-precession technique. J Magn Reson Imaging. 2019;49:239–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pfister J, Blaimer M, Kullmann WH, Bartsch AJ, Jakob PM, Breuer FA. Simultaneous T1 and T2 measurements using inversion recovery TrueFISP with principle component-based reconstruction, off-resonance correction, and multicomponent analysis. Magn Reson Med. 2019;81:3488–3502. [DOI] [PubMed] [Google Scholar]
- 55.Liu J, Drangova M. Method for B0 off-resonance mapping by non-iterative correction of phase-errors (B0-NICE). Magn Reson Med. 2015;74:1177–1188. [DOI] [PubMed] [Google Scholar]
- 56.Hu HH, Börnert P, Hernando D, et al. ISMRM workshop on fat-water separation: Insights, applications and progress in MRI. Magn Reson Med. 2012;68:378–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deichmann R, Haase A. Quantification of T1 values by SNAPSHOT-FLASH NMR imaging. J Magn Reson. 1992;96:608–612. [Google Scholar]
- 58.Marques JP, Kober T, Krueger G, van der Zwaag W, Van de Moortele PF, Gruetter R. MP2RAGE, a self bias-field corrected sequence for improved segmentation and T1-mapping at high field. NeuroImage. 2010;49:1271–1281. [DOI] [PubMed] [Google Scholar]
- 59.Messroghli DR, Radjenovic A, Kozerke S, Higgins DM, Sivananthan MU, Ridgway JP. Modified Look-Locker Inversion Recovery (MOLLI) for high-resolution T1 mapping of the heart. Magn Reson Med. 2004;52:141–146. [DOI] [PubMed] [Google Scholar]
- 60.Chen Y, Lee GR, Aandal G, et al. Rapid volumetric t1 mapping of the abdomen using three-dimensional through-time spiral GRAPPA. Magn Reson Med. 2016;75:1457–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Captur G, Gatehouse P, Keenan KE, et al. A medical device-grade T1 and ECV phantom for global T1 mapping quality assurance - the T1 Mapping and ECV Standardization in cardiovascular magnetic resonance (T1MES) program. J Cardiovasc Magn Reson. 2016;18:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hines CDG, Yu H, Shimakawa A, McKenzie CA, Brittain JH, Reeder SB. T1 independent, T2* corrected MRI with accurate spectral modeling for quantification of fat: validation in a fat-water-SPIO phantom. J Magn Reson Imaging. 2009;30:1215–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hilbert T, Xia D, Block KT, et al. Magnetization transfer in magnetic resonance fingerprinting. Magn Reson Med. 2020;84:128–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rioux JA, Levesque IR, Rutt BK. Biexponential longitudinal relaxation in white matter: characterization and impact on T1 mapping with IR-FSE and MP2RAGE. Magn Reson Med. 2016;75:2265–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kellman P, Hansen MS. T1-mapping in the heart: accuracy and precision. J Cardiovasc Magn Reson. 2014. January 4;16:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yoon JH, Lee JM, Paek M, Han JK, Choi BI. Quantitative assessment of hepatic function: modified look-locker inversion recovery (MOLLI) sequence for T1 mapping on Gd-EOB-DTPA-enhanced liver MR imaging. Eur Radiol. 2016;26:1775–1782. [DOI] [PubMed] [Google Scholar]
- 67.Outwater EK, Blasbalg R, Siegelman ES, Vala M. Detection of lipid in abdominal tissues with opposed-phase gradient-echo images at 1.5 T: techniques and diagnostic importance. Radiographics. 1998;18:1465–1480. [DOI] [PubMed] [Google Scholar]
- 68.Chen L, Zeng X, Ji B, et al. Improving dynamic contrast-enhanced MRI of the lung using motion-weighted sparse reconstruction: initial experiences in patients. Magn Reson Imaging. 2020;68:36–44. [DOI] [PubMed] [Google Scholar]
- 69.Benkert T, Feng L, Gerges L, et al. Comprehensive T1-weighted dynamic liver MRI during free-breathing using fat/water separation, radial sampling, compressed sensing, parallel imaging, and motion-weighted reconstruction. In ISMRM 2017 Annual Meeting Proceedings, Honolulu, HI, USA. p. 902. [Google Scholar]
- 70.Christodoulou AG, Shaw JL, Nguyen C, et al. Magnetic resonance multitasking for motion-resolved quantitative cardiovascular imaging. Nat Biomed Eng. 2018;2:215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Qi H, Sun J, Qiao H, et al. Simultaneous T1 and T2 mapping of the carotid plaque (SIMPLE) with T2 and inversion recovery prepared 3D radial imaging. Magn Reson Med. 2018;80:2598–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guo R, Chen Z, Herzka DA, Luo J, Ding H. A three-dimensional free-breathing sequence for simultaneous myocardial T1 and T2 mapping. Magn Reson Med. 2019;81:1031–1043. [DOI] [PubMed] [Google Scholar]
- 73.Qi H, Bustin A, Cruz G, et al. Free-running simultaneous myocardial T1/T2 mapping and cine imaging with 3D whole-heart coverage and isotropic spatial resolution. Magn Reson Imaging. 2019;63:159–169. [DOI] [PubMed] [Google Scholar]
- 74.Cloos MA, Knoll F, Zhao T, et al. Multiparametric imaging with heterogeneous radiofrequency fields. Nat Commun. 2016;7:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shaw JL, Yang Q, Zhou Z, et al. Free-breathing, non-ECG, continuous myocardial T1 mapping with cardiovascular magnetic resonance multitasking. Magn Reson Med. 2019;81:2450–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang X, Roeloffs V, Klosowski J, et al. Model-based T1 mapping with sparsity constraints using single-shot inversion-recovery radial FLASH. Magn Reson Med. 2018;79:730–740. [DOI] [PubMed] [Google Scholar]
- 77.Wang X, Kohler F, Unterberg-Buchwald C, Lotz J, Frahm J, Uecker M. Model-based myocardial T1 mapping with sparsity constraints using single-shot inversion-recovery radial FLASH cardiovascular magnetic resonance. J Cardiovasc Magn Reson. 2019. September 19;21:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Qi H, Jaubert O, Bustin A, et al. Free-running 3D whole heart myocardial T1 mapping with isotropic spatial resolution. Magn Reson Med. 2019;82:1331–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Feng L, Chandarana H, Zhao T, Bruno Mary, Sodickson DK, Otazo R Golden-angle sparse liver imaging: radial or cartesian sampling? In ISMRM 2017 Annual Meeting Proceedings, Honolulu, HI, USA. p. 1285. [Google Scholar]
- 80.Feng L, Zhao T, Chandarana H, Sodickson DK, Otazo R. Motion-Resolved golden-angle radial sparse MRI using variable-density stack-of-stars sampling. In ISMRM 2016 Annual Meeting Proceedings, Singapore. p. 1091. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Brain T1 maps from 20 consecutive slices in one volunteer
Figure S2 Bland Altman plots for assessing the repeatability of T1 mapping in the brain using MP-GRASP. Left plot compares MP-GRASP Scan 1 with MP-GRASP Scan 2 (repeatability at the same spatial resolution). Right plot compares MP-GRASP Scan 1 with MP-GRASP Scan 3 (repeatability at different spatial resolutions)
Figure S3 Three slices of MP-GRASP brain images at different TIs (TI times shown on the top of the figure). In addition to quantitative information, MP-GRASP also generates a contrast-resolved image series, from which an image can be selected to emulate the clinical MP-RAGE contrast for qualitative use
Figure S4 Liver T1 maps from 20 consecutive slices in one volunteer
Figure S5 Scan-rescan test for MP-Dixon-GRASP in the brain and in the liver
Figure S6 Representative fat/water-separated brain and liver images obtained from MP-Dixon-GRASP
Figure S7 Respiratory motion signals obtained from the bellow device superimposed on underlying z projection, showing good correlation between them. The z projection profiles were obtained through the centers of stack-of-stars k-space. The bellow signal can be automatically recorded with the raw data
Figure S8 Comparison of liver T1 maps from MP-GRASP reconstruction with and without motion correction in two subjects. Motion-weighted reconstruction improves image sharpness with reduced motion blurring as indicated by the red arrows
Figure S9 The influence of subspace size on T1 maps. K = 2 generated totally wrong T1 maps. T1 quantification seems not sensitive to the size of subspace within a certain range (eg, K = 4 to 6). For the brain dataset (without motion), T1 quantification seems to be insensitive to the selection of K. This can be explained by the fact that T1 fitting can also be seen as a process to remove noise and incoherent artifacts (eg, residual streaking artifacts). For the liver dataset, however, the quality of T1 quantification reduces with a large value of K
Figure S10 The TV regularization parameter was selected empirically by comparing a range of values, as done in most constrained iterative reconstruction studies. TV regularization helps to improve reconstruction quality and resulting T1 maps. The use of TV is not expected to affect temporal fidelity since it is only enforced on spatial dimensions. However, a larger value can lead to spatial blurring
Figure S11 In one dataset, we retrospectively used every 2nd, 4th, 6th, 8th, and 10th time points only for image reconstruction and T1 map generation. In other words, we used [1, 3, 5, 7…], [1, 5, 9, 13…], [1, 7, 13, 19…], [1, 9, 17, 25…], and [1, 11, 21, 31…] TI points only for testing the influence of temporal resolution on image reconstruction and T1 quantification. We found that good T1 maps can be obtained by using up to every 4th time points. When the number of points was further reduced, T1 maps start seeing loss of details. This finding suggests that we may potentially increase the number of partitions quite a bit in our imaging method before seeing significant effects. It also suggests that the short T1 components are more severely affected by reduced temporal resolution, which is expected because short T1 components recover faster than long T1 components
Figure S12 In the same dataset as in Figure S11, we retrospectively used only the first N time points for image reconstruction and T1 fitting, where N equaled 30, 20, 10 and 5. This is similar to acquiring for less time after each IR preparation. We found that good T1 maps can be obtained when using less time points (eg, first 30 time points). However, when the number of points was reduced to ~10, T1 maps start to lose details


