Abstract
The existing CRISPR-mediated diagnostic tests require a two-step procedure (DNA or RNA amplification followed by CRISPR-mediated sequence-specific detection) for nucleic acid detection, which increases complexity and the risk of sample cross-contamination. Here, we report a new CRISPR-mediated test, called CRISPR-top (CRISPR-mediated testing in one-pot), which integrates simultaneous target pre-amplification with CRISPR/cas12b-mediated detection into a one-pot reaction mixture, performed at a constant temperature. The novel CRISPR-top assay was applied to detect severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent for COVID-19 (coronavirus disease 2019). COVID-19 CRISPR-top targets the ORF1ab (opening reading frame 1a/b) and NP (nucleoprotein) genes of SARS-CoV-2, and operates at 59 °C for 40 min with minimal instrument. The COVID-19 CRISPR-top assay can return results within 60-min and is easily interpreted by visual fluorescence or lateral flow readouts. The analytical limit of detection (LoD) for COVID-19 CRISPR-top is 10 copies (for each detection target) per reaction with no cross-reactivity observed from non-SARS-CoV-2 templates. Among clinically collected non-COVID-19 samples, the assay's specificity was 100% (80/80 oropharynx swab samples). Among 52 COVID-19 positive clinical samples collected, the COVID-19 CRISPR-top assay yielded 38 (73.1%) positive results using fluorescence readout and 35 (67.3%) positive results with lateral-flow readout. These diagnostic results were similar to those obtained using RT-PCR (34 positive (65.4%)). These data indicate that COVID-19 CRISPR-top is a simple, rapid, accurate and highly sensitive method for SARS-CoV-2 detection which can be used in the clinic, field laboratories and primary care facilities in resource-challenged settings.
Keywords: CRISPR, Cas12b, COVID-19, SARS-CoV-2, CRISPR-Top, Nucleic acid detection
Graphical abstract
1. Introduction
Coronavirus disease 2019 (COVID-2019), which is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is a highly contagious disease and has become a major global health threat [1]. Epidemiological data indicates that SARS-CoV-2 has strong human-to-human transmissibility and can be rapidly spread through direct contact, droplets yielded by sneezing and coughing, or other possible transmission pathways (e.g., fecal-mouth transmission route) [[2], [3], [4]]. One of the major challenges for controlling the rapid spread of SARS-CoV-2 is the ability to rapidly and accurately identify and isolate COVID-19 cases (especially in asymptomatic cases) preventing transmission of SARS-CoV-2 to close contacts [4]. Thus, effective diagnostic techniques are required to quickly and sensitively determine the spread of SARS-CoV-2 in the population.
Since the start of the COVID-19 pandemic, molecular tests, including nucleic acid detection, antigen and antibody tests, have been devised to diagnose the COVID-19 [5]. Of note, polymerase chain reaction (PCR)-based assays (e.g., RT-PCR, RT-qPCR) are used as the standard, and play a crucial role in the diagnosis of SARS-CoV-2 infection [6]. Despite their strong analytical performance, there are several limitations associated with PCR-based techniques for COVID-19 diagnosis including a relatively long detection procedure (2–4 h), the requirement for expensive equipment (thermal cyclers), limited availability (they are currently restricted to first-line public health laboratories), and a high-proportion of false-negatives results (only about 47–60% of positive COVID-19 cases were detected by PCR-based assays) [7,8]. In particular, the enzymes after COVID-19 RT-PCR or COVID-19 RT-qPCR amplification can not be reused, which would not be financially beneficial [9]. During the SARS-CoV-2 pandemic, there was a shortage of many PCR reagents due to a lack of international transport and/or insufficient production capacity. Together, these limitations hinder the ability of PCR-based diagnostic tests to satisfy the current challenges of testing large numbers of asymptomatic individuals, suspected patients and close contacts. Therefore, alternatives to PCR-based assays for detection of SARS-CoV-2, which can overcome these limitations mentioned above, are needed.
Recently, prokaryotic CRISPR-Cas immune systems (Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated system) have opened the door to new biotechnological applications (e.g., genome editing and transcription regulation) [10]. In particular, the recent discovery of collateral cleavage activity in some Cas effectors (e.g., Cas12a, Cas12b, Cas13a and Cas14) has paved the way for new diagnostic platforms to be developed that offer portable, highly specific and ultra-sensitive tests through nucleic acid sensing [[10], [11], [12]]. Compared to PCR-based tests, CRISPR-based diagnostics offer reduced dependency on specialized instruments and can be conducted at physiological temperature (even room temperature) [13]. Up to now, several promising CRISPR-based detection techniques have been developed and verified [14]. For example, CRISPR-Cas12a-based DETECTR (DNA Endonuclease Targeted CRISPR Tans Reporter) and HOLMES (One-Hour-Low cost Multipurpose highly Efficient System) were used for the detection of HPV (human papillomavirus) and SNPs [11,15]. Similarly, a CRISPR-Cas14-based DETECTR assay was developed for the diagnosis of HPV [16] while a CRISPR-Cas13a-based SHERLOCK (Specific High-Sensitivity Enzymatic Reporter UnLOCKing) assay has been used for DENV (Dengue virus), ZIKV (Zika virus) and HPV detection with single-base mismatch specificity and attomolar sensitivity [17].
Currently, the existing CRISPR-based tests requires two separate reaction steps: (1) pre-amplification of the target nucleic acid using PCR or isothermal amplification assays (e.g., Recombinase polymerase amplification and Loop-mediated isothermal amplification), and (2) detection of the resulting amplification products through CRISPR-based collateral reporter decoding. These two reaction steps (pre-amplification and CRISPR-mediated detection) require multiple manual operation and liquid handling, thus not only complicating the diagnostic procedures but also increasing the risk of cross-contamination and hampering their wider application in various fields. Another limitation of existing CRISPR-based assays is that in order to activate the Cas proteins, the target sequence must contain a protospacer adjacent motif (PAM) site at an appropriate location. This restricts the ability of CRISPR-based assays to detect nucleic acid sequences which do not contain these PAM sites.
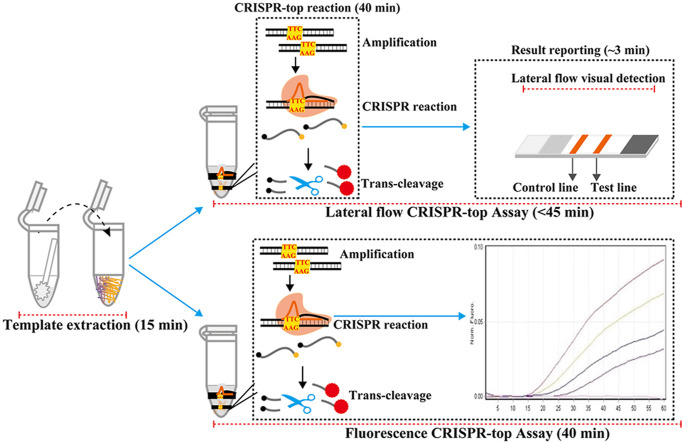
In this report, we devised a novel CRISPR-based detection technique (Fig. 1 and Fig. S1) called CRISPR-top (CRISPR-mediated testing in one-pot), which is an integrated one-step assay, and only requires one fluid-handling step. CRISPR-top is performed at a single temperature and the detection results can be returned using a simple lateral flow readout or real-time fluorescence readout. Additionally, sequences, which lack PAM sites, can also be detected by our CRISPR-top assay using engineered primers. Here, we report a CRISPR-top test developed for SARS-CoV-2 called COVID-19 CRISPR-top (Fig. 2 ).
Fig. 1.
Outline of the CRISPR-top design.
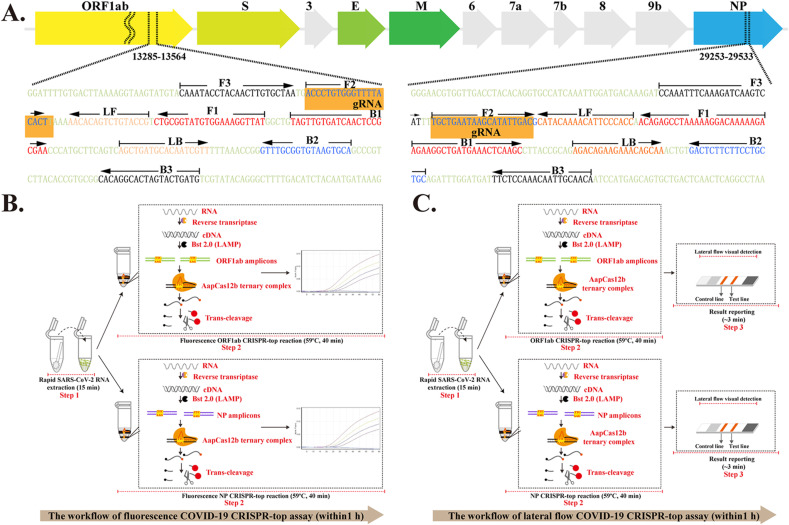
Fig. 2.
The principle of the COVID-19 CRISPR-top test.
2. Materials and methods
2.1. Primer and gRNA design
Two primer sets, which targeted OFR1ab (Opening reading frame 1a/b) and NP (nucleoprotein) genes of SARS-CoV-2 (GenBank MN908947, Wuhan-Hu-1), were designed using Primer explorer (v. 4) (https://primerexplorer.jp/lamp4.0.0/index.html) based on the principles of LAMP and CRISPR-top (Fig. 2, Fig. S2 and Fig. S3) [18]. The specificity of the two primer sets was examined using NCBI BLAST while the secondary structure and primer dimer were examined using OligoAnalyzer (v. 3.1) (Integrated DNA Technologies, Coralville, IA). Two guide RNAs (gRNAs) were designed according to the CRISPR-top principle. Additional details on primer design, locations, sequences and gRNAs are shown in Fig. S2, Fig. S3 and Table S1. LAMP primers were synthesized and purified by TianYi-Huiyan Biotech. Co., Ltd. (Beijing, China) at HPLC purification grade, and gRNAs were synthesized by GeneScript Biotech. Co, Ltd. (Nanjingjing, China) at HPLC purification grade.
2.2. CRISPR-top reaction
The CRISPR-top assay was performed in a 25-μl mixture containing 12.5 μl 2 × isothermal reaction buffer (HuiDeXin, Biotech. Co., Ltd, Tianjing, China), 1 μl Bst 2.0 DNA polymerase (8U), 1.6 μM of FIP and BIP, 0.8 μM of LF and LB, 0.4 μM of F3 and B3, 4.0 μl AapCas12b-gRNA complex, 0.5 μl probe (100 μM of 5′-FAM-TTATTATTAT-Biotin-3′ for lateral flow assay or 100 μM of 5′-FAM-TTATTATTAT-BHQ1-3′ for real-time assay) and template (1 μl for the pure template or 5 μl for clinical samples). The CRISPR-top reactions were performed at 59 °C for 40 min. The Cas12b-gRNA complex mixture was prepared using 100 nM of AapCas12b protein and 150 nM of gRNA in TOLOBuffer (TOLO Biotech. Co., Ltd, Shanghai, China) and incubated at 37 °C for 10 min. The AapCas12b-gRNA complex was used immediately or stored at 4 °C for up to 12 h before use.
2.3. Lateral flow CRISPR-top assay
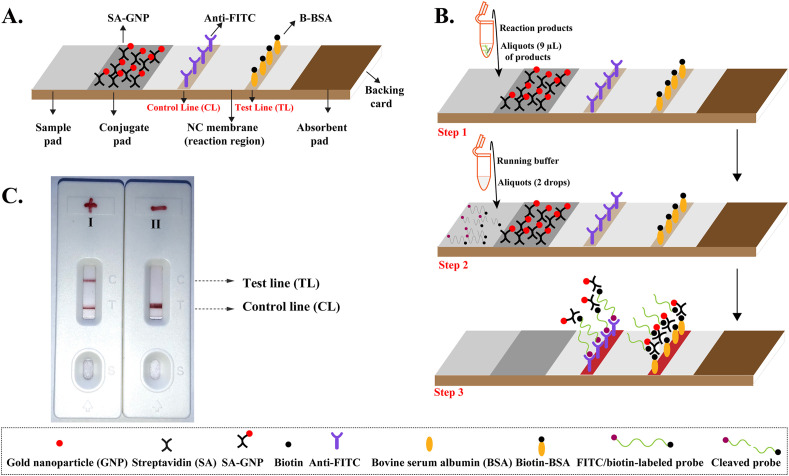
Lateral flow biosensors (LFB, Fig. 3 A) were used to visually readout the CRISPR-top results. The LFB used contained a sample region, a conjugate region, a nitrocellulose membrane and an absorbent pad which were assembled onto a backing card. Streptavidin-immobilized gold nanoparticles (SA-GNPs) were deposited onto the conjugate region of the LFB and used as the indicator reagent. Then, rabbit anti-fluorescein antibodies (anti-FITC) and biotinylated bovine serum albumin (B-BSA) were dispensed onto the nitrocellulose membrane to form the control line (CL, conjugated with anti-FITC)) and test line (TL, conjugated with B-BSA), respectively. The LFB were manufactured by HuiDeXing Biotech. Co., Ltd. (Tianjing, China) according to our design.
Fig. 3.
The principle of biosensor for visualization of CRISPR-top products.
For visual detection using LFB, an aliquot (0.42 μl) of CRISPR-top products were added to the sample pad along with three drops of running buffer (100 mM PBS, PH7.4 with 1% Tween 20). The CRISPR-top results was then visualized within 2 min in the form of red bands appearing on the nitrocellulose membrane area.
2.4. Sensitivity and specificity of the COVID-19 CRISPR-top assay
To examine the sensitivity of the COVID-19 CRISPR-top assay, two plasmid standards (named as ORF1ab-plasmid and NP-plasmid, commercially available from Tianyihuiyuan Biotech. Co., Ltd (Beijing, China)), containing the ORF1ab and NP sequences, respectively, were used. Ten-fold serial dilutions of ORF1ab- and NP-plasmids (from 1 × 106 copies/μl to 1 × 10−1 copies/μl) were prepared and 1 μl aliquots of plasmid templates were used for each CRISPR-top reaction. Real-time CRISPR-top test and lateral flow CRISPR-top test were performed independently to define the analytical LoD (limit of detection) of COVID-19 CRISPR-top. To determine the specificity of the COVID-19 CRISPR-top assay, templates extracted from different viruses, bacteria and fungi (Table S2), were used.
2.5. Validation of the COVID-19 CRISPR-top assay to clinical samples
To validate the COVID-19 CRISPR-top assay, a total of 52 clinical COVID-19 positive samples were tested including pharyngeal, nasal, anal swabs as well as fecal and sputum samples. These samples were collected from asymptomatic, acute and convalescent cases at the Guizhou Province Center for Disease Control and Prevention (GZ-CDC, Table S3). Approval for CRISPR-top COVID-19 test validation on clinical RNA samples was provided by GZ-CDC. RNA templates extracted from clinical samples were collected after rRT-PCR detection in GZ-CDC which was performed using an officially approved clinical RT-PCR kit. Aliquots of 5 μl of RNA templates were used to perform the RT-PCR and COVID-19 CRISPR-top assays. In addition, 80 pharyngeal swabs samples (Table S4) obtained from non-COVID-19 individuals were used to confirm the clinical specificity of the COVID-19 CRISPR-top assays.
3. Result
3.1. CRISPR-top design
The CRISPR-top design and reaction mechanism are shown in Fig. 1 and Fig. S1. The CRISPR-top detection platform, which integrates isothermal amplification (Loop-mediated isothermal amplification; LAMP) with CRISPR/AapCas12b-based detection (AapCas12b trans-cleavage detection), can be performed in a single reaction step and temperature (58-59 °C) for nucleic acid detection. In the CRISPR-top system, we inserted a PAM site (TTC) within the linker region (which is target sequence independent) of the FIP (Forward inner primer) or BIP (Backward inner primer) LAMP primers (Fig. 1 and Fig. S1). By using the PAM site engineered FIP or BIP primers, the LAMP-amplified products will contain a newly acquired TTC PAM site (Fig. 1, Step 1 and 2) which can then be recognized by the corresponding AapCas12b/sgRNA system (Fig. 1, Step 3 and 4). The recognition of the PAM site in the LAMP products activates Cas12b (Fig. 1, Step 5) resulting in trans-cleavage of the reporter molecule (single-strand DNA (ssDNA)) (Fig. 1, Step 6). Importantly, the CRISPR-top assay using modified FIP or BIP primers can detect any target sequence (even sequences that do not contain suitable PAM sites for activation of the CRISPR-AapCas12b/gRNA complex) as long as they meet the design requirements for LAMP.
The CRISPR-top results can then be visualized on a lateral flow biosensor (Fig. 1B, top row) and the whole test, including rapid template preparation (15 min), CRISPR-top reaction (40 min) and LFB result detection (~3 min), can be completed within 1 h. Alternatively, the results for the CRISPR-top assay can also be detected using a real-time fluorescence readout (Fig. 1B, bottom row), thus the real-time fluorescence CRISPR-top assay can also be completed within 1 h (rapid template extraction (15 min) and fluorescence CRISPR-top reaction (up to 40 min)).
3.2. COVID-19 CRISPR-top test
In this study, we report the development and initial validation of an CRISPR-top assay (called COVID-19 CRISPR-top) for the rapid and reliable detection of SARS-CoV-2. Two LAMP primer sets were designed which targeted the ORF1ab and N genes of SARS-CoV-2 (Fig. 2A and Fig. S2 and Fig. S3). The ORF1ab- and NP-FIP LAMP primers were modified with a PAM site (TTC) at the linker region to meet the detection requirement for CRISPR-top (Table S1).
The COVID-19 CRISPR-top assay involves simultaneous reverse transcription, isothermal LAMP amplification and CRISPR-Cas12b-based detection performed within a single-tube and at a constant temperature (59 °C) (Fig. 2B and C). After rapid extraction of SARS-CoV-2 RNA (Fig. 2B and C, Step 1), the RNA templates are first converted to cDNA using reverse transcriptase. The cDNA will then act as templates for isothermal amplification (Fig. 2B and C, Step 2). By using modified ORF1ab-FIP and NP-FIP primers, the ORF1ab- and NP-LAMP products will contain an acquired PAM site (TTC) that is recognized by the AapCas12b/gRNA complex for activation of Cas12b trans-cleavage activity (Fig. 2B and C, Step 2). The ssDNA reporter molecules in the COVID-19 CRISPR-top reaction will then be rapidly sheared by AapCas12b effectors, and the resulting signals can been visualized using a real-time fluorescence instrument (Fig. 2B) or a lateral flow biosensor (Fig. 2C). Therefore, the real-time COVID-19 CRISPR-top test only requires two steps for SARS-CoV-2 detection (rapid RNA extraction (15 min) and real-time CRISPR-top reaction (up to 40 min)) (Fig. 2B) while the lateral flow biosensor requires three (rapid RNA extraction (15 min), CRISPR-top reaction (40) and result reporting (~2 min)) (Fig. 2C). Importantly, both tests can be finished within 1 h.
3.3. The principle of biosensor for visualization of CRISPR-top results
The design details for lateral flow biosensor are shown in Fig. 3A. After CRISPR-top reaction, a volume of 0.42 μl of CRISPR-top product is added to the sample region (Fig. 3B, Step 1) along with three drops of detection buffer (Fig. 3B, Step 2). The detection buffer moves along the biosensor through capillary action and rehydrates the functional nanoparticles (SA-GNPs) in the conjugate region. For negative reactions, anti-FITC antibodies immobilized on the control line (CL) can capture FAM molecules labeled at the end of the reporter molecule while the biotin labeled at other end of the reporter molecule bind SA-GNPs for visualization (Fig. 3B, Step 3). For positive reactions, the reporter molecules are cleaved by the activated AapCas12b effector, thus separating FAM and biotin. The B-BSA immobilized on the test line (TL) can capture biotin/SA-GNPs complex, which indicates a positive result (Fig. 3B, Step 3). The interpretation of the CRISPR-top method using biosensor is depicted in Fig. 3C and Fig. S4.
3.4. Optimal conditions for CRISPR-top assay
We first confirmed the feasibility of the ORF1ab- and NP-LAMP primer sets. As shown in Fig. S5, the ORF1ab- and NP-LAMP primer sets successfully amplified the corresponding templates (ORF1ab-plasmid and NP-plasmid), and no amplification was seen for non-target templates. Thus, the ORF1ab- and NP-LAMP primer sets were suitable candidates for developing the CRISPR-top method for SARS-CoV-2 detection. Next, to determine the optimal amplification temperature for the CRISPR-top assay, real-time ORF1ab-CRISPR-top reaction was tested under 6 different temperatures (from 56 °C to 61 °C, with 1 °C intervals) (Fig. S6). The results indicate that the optimal reaction temperature for the CRISPR-top assay was between 58 °C and 59 °C (Fig. S6, A3 and A4) based on amplification time and fluorescence intensity. In this report, 59 °C was selected as the CRISPR-top reaction temperature for the rest of the study as at 59 °C, a higher fluorescence intensity was observed compared to 60 °C and 61 °C, while a faster amplification time was observed at 59 °C compared to 56 °C, 57 °C and 58 °C (Fig. S6, B and C).
3.5. Sensitivity of the COVID-19 CRISPR-top assay
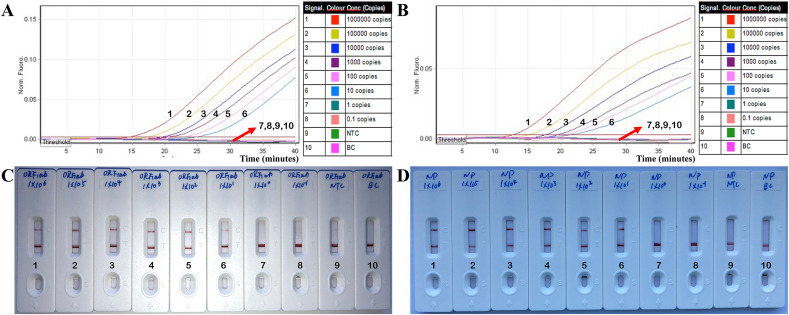
Using eight serial dilutions of the ORF1ab- and NP-plasmid standards, the analytical sensitivity of the COVID-19 CRISPR-top assay was 10 copies per reaction for both ORF1ab and NP (Fig. 4 ). For the real-time fluorescence COVID-19 CRISPR-top assay, amplification and fluorescence was detected from 1 × 106 to 1 × 101 copies of ORF1ab- and NP-plasmid templates (Fig. 4A and B). Similarly, the analytical limit of detection (LoD) using the lateral flow COVID-19 CRISPR-top assay was also 10 copies per reaction (for ORF1ab- and NP-plasmids) (Fig. 4C and D) in agreement with the fluorescence COVID-19 CRISPR-top method. The visual bands for the presence or absence of target templates from the lateral flow biosensors were easy-to-interpret at all detectable levels (Fig. 4C and D).
Fig. 4.
Sensitivity of the COVID-19 CRISPR-top assay.
3.6. Specificity of the COVID-19 CRISPR-top assay
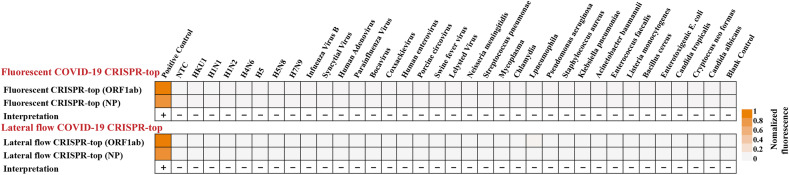
Specificity of the COVID-19 CRISPR-top assay was confirmed using various templates extracted from different viruses, bacteria and fungi. As shown in Fig. 5 , all positive results were obtained from positive controls (ORF1ab- and NP-plasmid templates), whereas all negative results were obtained from non-ORF1ab- and NP-plasmid templates (Fig. S7, S8 and Table S2). The results from the fluorescence COVID-19 CRISPR-top assay (Fig. 5, top row; Fig. S7) were in complete accordance with the lateral flow COVID-19 CRISPR-top method (Fig. 5, bottom row; Fig. S8). Together, these results indicate that our COVID-19 CRISPR-top fluorescence and lateral flow COVID-19 diagnostic test is highly specific for the detection of SARS-CoV-2 with no non-specific cross-reactions observed.
Fig. 5.
Specificity of the COVID-19 CRISPR-top assay. Heat-maps of COVID-19 CRISPR-top assay results on various templates with the test interpretation shown. Top row: fluorescence COVID-19 CRISPR-top assay results. Bottom row: lateral flow COVID-19 CRISPR-top assay results as quantified by ImageJ tools.
3.7. Validation of the COVID-19 CRISPR-top method to clinical samples
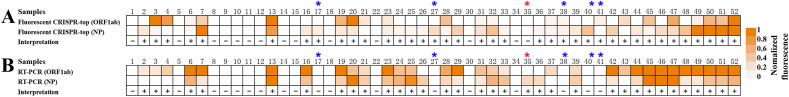
To validate the use of our COVID-19 CRISPR-top method as a SARS-CoV-2 diagnostic tool,we tested RNA extracted from 52 COVID-19 positive clinical samples and 80 non-COVID-19 respiratory swab samples (Table S3 and S4). Using fluorescence COVID-19 CRISPR-top, SARS-CoV-2 RNA was detected in 38/52 (73.1%) clinical samples (Fig. 6 ) while lateral flow COVID-19 CRISPR-top detected 35/52 (67.5%) positive samples (Table S3). The diagnostic results obtained using COVID-19 CRISPR-top assays were similar to those obtained using RT-PCR tests (34/52, 65.4%) (Fig. 6, Table S3). No positive detection was obtained from non-SARS-CoV-2 respiratory samples using COVID-19 CRISPR-top tests (Table S4). These initial results suggest that the COVID-19 CRISPR-top test can be used as a valuable diagnostic tool for SARS-CoV-2 detection.
Fig. 6.
Results of the COVID-19 CRISPR-top assay for COVID-19 positive clinical samples.
4. Discussion
CRISPR -based detection techniques have offered a nascent diagnostic platform for rapid, ultrasensitive and highly specific detection of various nucleic acids [13]. However, the existing CRISPR-based detection techniques usually employ Cas12a or Cas13 to decoding the signals after primer-specific amplification [19], which not only complicate the diagnostic tests, but also increases the risk sample cross-contamination [[15], [16], [17]]. Here, to overcome these shortcomings posed by conventional CRSPIR-based detection, we have successfully devised CRISPR-top technique using AapCas12b, which is a thermophilic RNA-guided endonuclease from the type V–B CRISPR-Cas system of Alicyclobacillu acidophilus, and works at temperatures from 35 °C to 65 °C [20]. Thus, AapCas12b enzyme can be amenable to several isothermal nucleic acid amplification methodologies (such as LAMP and multiple cross displacement amplification) as these methods amplify at 55 °C to 70 °C.
In this report, we integrate LAMP with CRISPR-AapCas12b-mediated diagnostic test to develop our CRISPR-top method, which is a one-step, one-pot reaction mixture test (Fig. 1A). Furthermore, to broaden the application of CRISPR-based diagnostic assays, we modified the LAMP assay by incorporating a PAM site (TTC) into the linker region of the core primer (FIP or BIP). This modification produces LAMP products containing newly acquired PAM sites that can be used for recognition and cutting by the AapCas12b/gRNA complex (Fig. 1A and Fig. S1). Thus, our CRISPR-top technique using the PAM-modified LAMP can detect any target sequence (even sequences that do not contain available PAM sites) as long as they meet the requirements for LAMP design. In addition, the CRISPR-top assay developed is versatile and compatible with both real-time fluorescence (Fig. 1B, bottom row) and lateral-flow detection (Fig. 1B, top row).
Then, we applied our CRISPR-top method to develop a novel SARS-CoV-2 diagnostic test, called COVID-19 CRISPR-top (Fig. 2), which targets the ORF1ab and NP genes of SARS-CoV-2 (Fig. 2A, Fig. S2 and Fig. S3). To satisfy the CRISPR-top design requirements, the core primers of ORF1ab-FIP and NP-FIP were modified with PAM sites (TTC) at the linker region (Table S1). To achieve SARS-CoV-2 RNA detection, the COVID-19 CRISPR-top system simultaneously integrates reverse transcription, LAMP reaction with CRISPR-Cas12b-mediated nucleic acid detection into a one-pot mixture. The results of COVID-19 CRISPR-top can be visualized by fluorescence (Fig. 2B) or lateral flow readouts (Fig. 2C). Compared to existing SARS-CoV-2 CRISPR-mediated tests, our COVID-19 CRISPR-top method only requires a single liquid-handing step, therefore decreasing the risk of cross-contamination between samples. Similar to the standard LAMP technique, our COVID-19 CRISPR-top test operates at a single constant temperature, thus CRISPR-top alleviates the need for sophisticated and expensive equipment (e.g., thermocyclers) making it suitable for point-of-care use and field-based laboratories. Particularly, only a simple heater that is portable, battery-powered, $500 USD device supports 96 CRISPR-top reaction per assay. The CRISPR-top reaction, including LAMP amplification and AapCas12b-based detection, costs ~$3.5 USD. The cost of biosensor devised in our report is estimated to be $1.5 USD per test. Herein, we deem that a CRISPR-top assay would cost ~$ 5 USD per test, and $500 USD for a dedicated instrument.
To determine the optimal temperature conditions, six reaction temperatures (56 °C to 61 °C, with 1 °C intervals) were tested for the fluorescence ORF1ab CRISPR-top assay. As expected, the CRISPR-top assay amplified nucleic acid at all reaction temperatures when the target template (ORF1ab plasmid) was present (Fig. S6A). The CRISPR-top method produced stronger fluorescence signal (>10,000 AU) at lower reaction temperatures (56 °C to 59 °C) (Fig. S6B) while at higher reaction temperatures (59 °C to 61 °C), the amplification speed was faster (<15 min) (Fig. S6C). To achieve a reliable and rapid detection, a reaction temperature of 59 °C was chosen as the operating temperature for our COVID-19 CRISPR-top test (Fig. S6).
The COVID-19 CRISPR-top assay is highly sensitive and can detect down to 10 copies of ORF1ab- and NP-plasmids (Fig. 4). This analytical LoD is similar to other previously reported COVID-19 LAMP-based methods [21]. The high sensitivity of COVID-19 CRISPR-top may be attributed to its use of reverse transcription LAMP (RT-LAMP) for RNA amplification. Previous studies have demonstrated that LAMP-based tests were more sensitive than PCR-based tests as well as other isothermal amplification techniques (e.g., cross-priming amplification and helicase-dependent isothermal amplification) [22]. Positive amplification for COVID-19 CRISPR-top can be obtained in as little as 15 min (Fig. 4A and B, Signal 1) while at analytical LoD levels only 25 min is required for positive results (Fig. 4A and B, Signal 6). To produce a sufficient and discriminative positive result, we recommend a COVID-19 CRISPR-top reaction time of 40 min for diagnosis of clinical samples. Thus, the entire procedure for both real-time fluorescence COVID-19 CRISPR-top and lateral flow CRISPR-top can be completed within 1 h (Fig. 2B and C). The analytical sensitivity of lateral flow COVID-19 CRISPR-top method was also 10 copies of OFR1ab- and NP-plasmids, which was in complete accordance with the fluorescence COVID-19 test (Fig. 4).
As expected, our COVID-19 CRISPR-top assay was highly specific for SARS-CoV-2 detection and achieved 100% inclusivity and 100% exclusivity when it was applied to various templates extracted from viruses, bacteria and fungi (Fig. 5, S7, S8 and Table S2). Amplification was only observed in the assay for ORF1ab- and NP-plasmids but not for non-OFR1ab- and NP-templates. The use of LAMP, which is a validated molecular technique with extremely high specificity, for amplification in the CRISPR-top method ensured the reliability and high specificity of our CRISPR-top assay (Figs. 1 and 2 and Fig. S1). Moreover, the addition of CRISPR-mediated detection in our CRISPR-top assay further increased the reliability, precision and sensitivity of our diagnostic test as CRISPR is a highly specific detection technique (with single nucleotide target specificity) that utilizes target-dependent gRNA and a PAM site for target sequence detection. Therefore, the CRISPR-top assay developed in this study is a powerful and precise nucleic acid diagnostic tool. The analytical specificity results demonstrate that the COVID-19 CRISPR-top assay can be used for the reliable molecular detection of SARS-CoV-2.
A total of 132 samples, including 52 samples collected from COVID-19 patients (pharyngeal, nasal and anal swabs) and 80 samples obtained non-COVID-19 individuals, were tested to demonstrate the feasibility of COVID-19 CRISPR-top for clinical applications (Fig. 6, Table S3 and S4). No positive readouts were seen in non-SARS-CoV-2 patients (Table S4), which verified the analytical specificity of our COVID-19 CRISPR-top tests for clinical application. In addition, of the COVID-19 positive samples, 38 positive results were obtained using the fluorescence COVID-19 CRISPR-top assay and 35 positive results by lateral flow COVID-19 CRISPR-top, whereas only 34 RNA samples were positive by the RT-PCR method used in GZ-CDC (Fig. 6 and Table S3). These data indicate that COVID-19 CRISPR-top assay was more sensitive in diagnosing COVID-19 patients, especially for samples with very low virus loads. The lower detection rate of RT-PCR may be due to the presence of inhibitors which specifically affected RT-PCR or the lower copy number of templates (SARS-CoV-2 RNA) in the clinical samples which was below the analytical LoD for PCR. Furthermore, the CRISPR-top assay, which uses LAMP to amplify target nucleic acids, might share some advantages associated with isothermal amplification techniques. For examples, the CRISPR-top assay may be less sensitive to PCR inhibitors, can tolerate the inhibitory effect of various nucleic acids, or are less affected by the presence of large amounts of salts in the sample buffer [23]. Although this study validated the use of COVID-19 CRISPR-top on clinical samples, the number of clinical samples collected from COVID-19 individuals was small, and that other types of samples (such as urine and blood) were not tested. Hence, further evaluation of COVID-19 CRISPR-top assays using more or different types of clinical samples is warranted.
The real-time fluorescence COVID-19 CRISPR-top showed better performance for clinical sample detection (38/52) compared to lateral flow COVID-19 CRISPR-top (35/52) (Fig. 6 and Table S3). Thus, we recommend the use of real-time fluorescence COVID-19 CRISPR-top method as a rapid screening tool for SARS-CoV-2 detection. However, the advantage of using biosensor for result readout is that they are easy-to-use, disposable, does not require specific equipment and can be made available for large-scale tests. These advantages make lateral flow biosensors suitable for use in field and first-line laboratories. One limitation of the biosensors for COVID-19 CRISPR-top readout is the requirement to open the reaction tube during the detection stage. This may lead to the production of aerosol droplets containing high concentrations of CRISPR-top amplicons thus increasing the risk of carryover contamination. Our data have demonstrated that CRISPR-top is an ultra-sensitive technique (Fig. 4, Fig. 6), which may render it highly susceptible to carryover contamination. If repeated detection of SARS-CoV-2 using lateral flow COVID-19 CRISPR-top method is performed in the same laboratory, carryover contamination may occur through contaminated pipettes, reagents, skin of the personnel performing the test or work space surfaces. Therefore, laboratories using lateral flow CRISPR-top assay should implement careful preventative methods, because once carryover contamination occurs, the procedure to decontaminate is time-consuming and costly.
5. Conclusion
In this report, we devised a novel, one-step CRISPR-based diagnostic test called CRISPR-top, which incorporates simultaneous nucleic acid amplification with CRISPR/AapCas12b-mediated sequence-specific detection into a one-pot reaction. The CRISPR-top technique was successfully applied to detect SARS-CoV-2 and preliminarily verified using plasmid standards and clinical samples. The COVID-19 CRISPR-top method allowed detection of the ORF1ab and NP genes from SARS-CoV-2 and eliminated the use of complex apparatus. The COVID-19 CRISPR-top method is versatile and rapid, returning easily interpretable results by fluorescence or lateral flow readouts within 1 h. The analytical sensitivity and specificity results in this study suggest that the COVID-19 CRISPR-top assay is an ultra-sensitive, highly specific and feasible method for the diagnosis of COVID-19 in a variety of settings including clinical, field, and resource-limited environments. As a proof-of-concept technology, the CRISPR-top method can also be reconfigured to detect various sequences from different organisms or pathogens by redesigning the primers and gRNAs.
Funding
This study was funded by grants from Science and Technology Department of Guizhou Province (Grant No. Qian Ke He Support Plan [2020]4Y182), Qian Ke He Platform talent [2018]-5606, Qian Ke He [2016]-4021 and by Qian Ke He Platform [2018]-5767).
Credit author statement
Yi Wang conceived and designed this study. Shijun Li, Jun Tai and Yi Wang supervised the study. Shijun Li, Junfei Huang, Lijuan Ren, Weijia Jiang, Ming Wang, Li Zhuang, Qinni Zheng, Rui Yang, Yi Zeng and Yi Wang performed the experiments. Shijun Li, Junfei Huang, Lijuan Ren and Yi Wang analyze the data. Shijun Li, Junfei Huang, Lijuan Ren, Weijia Jiang, Ming Wang, Li Zhuang, Qinni Zheng, Rui Yang and Yi Zeng contributed the reagents and analysis tools. Shijun Li, Junfei Huang, Lijuan Ren, Weijia Jiang, Ming Wang, Li Zhuang, Qinni Zheng, Rui Yang and Yi Zeng contributed the materials. Yi Wang conducted the software. Shijung Li and Yi Wang drafted the manuscript. Laurence Don Wai Luu and Yi Wang revised the manuscript.
Data sharing
No additional data available.
Transparency declaration
The lead author and guarantor affirms that the manuscript is an honest, accurate, and transparent account of the study being reported; that no important aspects of the study have been omitted; and that any discrepancies from the study as planned and registered have been explained.
Declaration of competing interest
The authors declare that they have no competing interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.talanta.2021.122591.
A. Schematic illustration of the principle of CRISPR-top detection system. In the CRISPR-top system, the linker region of the FIP primer was modified with a PAM site (TTC). After LAMP amplification, the amplicons contain a newly acquired CRISPR-AapCas12b recognition site, which was derived from the modified FIP primer (Step 1 and 2). The resultant products can be recognized by the corresponding Cas12b/sgRNA system (Step 3 and 4), and activates the AapCas12b effector (Step 5). The activated AapCas12b protein can then digest the reporter molecule (ssDNA) using trans-cleavage activity (Step 6). B. Schematic of the CRISPR-top workflow. The cleaved ssDNA reporters in the CRISPR-top reaction can release signals that can be detected using lateral flow biosensor (Top row) or real-time fluorescence (Bottom row). The lateral flow CRISPR-top assay (Top row) employs three closely linked steps: rapid template preparation, CRISPR-top reaction and biosensor analysis while the real-time fluorescence CRISPR-top assay (Bottom row) requires two closely linked steps (rapid template preparation and real-time CRISPR-top reaction).
A. Primer design for the COVID-19 CRISPR-top assay. Top row: SARS-CoV-2 genome organization (GenBank: MN908947, Wuhan-Hu-1). The length of all genes is not displayed to scale (ORF1ab, Open reading frame 1a/b; S, Spike protein; E, Envelope protein; M, Membrane protein; NP, Nucleoprotein; Accessory proteins, 3, 6, 7a, 7b, and 9b). Bottom row: The details of the COVID-19 CRISPR-top primer design. Part of the nucleotide sequences for ORF1ab (Left) and NP (Right) are listed. The sites for the primer sequences are underlined. Left arrows and right arrows display the complementary and sense sequences that are used. B. The workflow for the real-time COVID-19 CRISPR-top assay. Two steps, including rapid RNA extraction (15 min) and real-time CRISPR-top reaction (up to 40 min), are required for COVID-19 CRISPR-top detection. The whole process can be completed within 60 min. C. The workflow for the lateral flow COVID-19 CRISPR-top assay. Three steps, including rapid RNA extraction (15 min), CRISPR-top reaction (40 min) and result reporting (~3 min), are required for conducting lateral flow COVID-19 CRISPR-top detection, and the whole test can be finished within 60 min.
(A), The schematic of the biosensor design. (B), The principle of biosensor for CRISPR-top products. (C), Interpretation of the CRISPR-top results. I, positive results for CRISPR-top test (CL and TL appear simultaneously on the biosensor); II, negative (only the control line appears on the biosensor).
A and B, Sensitivity results for real-time fluorescence COVID-19 CRISPR-top using serial dilutions of (A) ORF1ab-plasmid and (B) NP-plasmid. C and D, Sensitivity results for lateral flow biosensor COVID-19-CRISPR-top detection (C for ORF1ab-CRISPR-top result, and D for NP-CRISPR-top result). Serial dilutions of plasmid standards from 1 × 106 to 1 × 10−1 copies (labeled as 1–8 with 9 and 10 representing no template control (NTC) and black control (BC), respectively) were used.
Heat-maps of (A) fluorescence COVID-19 CRISPR-top and (B) RT-PCR results on 52 clinical samples collected from COVID-19 patients. CRISPR-top and RT-PCR heat-maps represent normalized mean fluorescence values.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.of the International C.S.G. Nat. Microbiol. 2020;5:536. [Google Scholar]
- 2.Bai Y., Yao L., Wei T., Tian F., Jin D.-Y., Chen L., Wang M. Jama. 2020;323:1406–1407. doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S., Lau E.H., Wong J.Y. New England J. Med. 2020 [Google Scholar]
- 4.Xu M., Wang D., Wang H., Zhang X., Liang T., Dai J., Li M., Zhang J., Zhang K., Xu D. Clin. Transl. Med. 2020;10:e158. doi: 10.1002/ctm2.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plikusiene I., Maciulis V., Ramanaviciene A., Balevicius Z., Buzavaite-Verteliene E., Ciplys E., Slibinskas R., Simanavicius M., Zvirbliene A., Ramanavicius A. J. Colloid and Interface Sci. 2021;594:195–203. doi: 10.1016/j.jcis.2021.02.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li S., Jiang W., Huang J., Liu Y., Ren L., Zhuang L., Zheng Q., Wang M., Yang R., Zeng Y., Wang Y. Eur. Respir. J. 2020:2002060. doi: 10.1183/13993003.02060-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li C., Debruyne D., Spencer J., Kapoor V., Liu L.Y., Zhang B., Lee L., Feigelman R., Burdon G., Liu J., Oliva A., Borcherding A., Xu J., Urban A.E., Liu G., Liu Z. bioRxiv. 2020 2020.2003.2012.988246. [Google Scholar]
- 8.Zhu X., Wang X., Han L., Chen T., Wang L., Li H., Li S., He L., Fu X., Chen S., Xing M., Chen H., Wang Y. Biosens. Bioelectron. 2020;166:112437. doi: 10.1016/j.bios.2020.112437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dronina J., Bubniene U.S., Ramanavicius A. Biosens. Bioelectron. 2021;175:112867. doi: 10.1016/j.bios.2020.112867. [DOI] [PubMed] [Google Scholar]
- 10.Sashital D.G. Genome Med. 2018;10:32. doi: 10.1186/s13073-018-0543-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J.S., Ma E., Harrington L.B., Da Costa M., Tian X., Palefsky J.M., Doudna J.A. Science. 2018;360:436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu X., Wang X., Li S., Luo W., Zhang X., Wang C., Chen Q., Yu S., Tai J., Wang Y. ACS Sens. 2021;6:881–888. doi: 10.1021/acssensors.0c01984. [DOI] [PubMed] [Google Scholar]
- 13.Tsang J., LaManna C.M. CRISPR J. 2020;3:142–145. doi: 10.1089/crispr.2020.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mukama O., Wu J., Li Z., Liang Q., Yi Z., Lu X., Liu Y., Liu Y., Hussain M., Makafe G.G. Biosens. Bioelectron. 2020:112143. doi: 10.1016/j.bios.2020.112143. [DOI] [PubMed] [Google Scholar]
- 15.Li S.-Y., Cheng Q.-X., Wang J.-M., Li X.-Y., Zhang Z.-L., Gao S., Cao R.-B., Zhao G.-P., Wang J. Cell Discov. 2018;4:1–4. doi: 10.1038/s41421-018-0028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrington L.B., Burstein D., Chen J.S., Paez-Espino D., Ma E., Witte I.P., Cofsky J.C., Kyrpides N.C., Banfield J.F., Doudna J.A. Science. 2018;362:839–842. doi: 10.1126/science.aav4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., Verdine V., Donghia N., Daringer N.M., Freije C.A. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Nucleic Acids Res. 2000;28 doi: 10.1093/nar/28.12.e63. e63-e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drobysh M., Ramanaviciene A., Viter R., Ramanavicius A. Micromachines. 2021;12:390. doi: 10.3390/mi12040390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang H., Gao P., Rajashankar K.R., Patel D.J. Cell. 2016;167:1814–1828. doi: 10.1016/j.cell.2016.11.053. e1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subsoontorn P., Lohitnavy M., Kongkaew C. medRxiv. 2020 doi: 10.1038/s41598-020-79237-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Obande G.A., Singh K.K.B. Infect. Drug Resist. 2020;13:455. doi: 10.2147/IDR.S217571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaneko H., Kawana T., Fukushima E., Suzutani T. J. Biochem. Biophys. Methods. 2007;70:499–501. doi: 10.1016/j.jbbm.2006.08.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.