Abstract
Dubowitz syndrome (DubS) is considered a recognizable syndrome characterized by a distinctive facial appearance and deficits in growth and development. There have been over 200 individuals reported with Dubowitz or a “Dubowitz-like” condition, although no single gene has been implicated as responsible for its cause. We have performed exome (ES) or genome sequencing (GS) for 31 individuals clinically diagnosed with DubS. After genome-wide sequencing, rare variant filtering and computational and Mendelian genomic analyses, a presumptive molecular diagnosis was made in 13/27 (48%) families. The molecular diagnoses included biallelic variants in SKIV2L, SLC35C1, BRCA1, NSUN2; de novo variants in ARID1B, ARID1A, CREBBP, POGZ, TAF1, HDAC8 and copy-number variation at1p36.11(ARID1A), 8q22.2(VPS13B), Xp22 and Xq13(HDAC8). Variants of unknown significance in known disease genes, and also in genes of uncertain significance, were observed in 7/27 (26%) additional families. Only one gene, HDAC8, could explain the phenotype in more than one family (N=2). All but two of the genomic diagnoses were for genes discovered, or for conditions recognized, since the introduction of next-generation sequencing. Overall, the DubS-like clinical phenotype is associated with extensive locus heterogeneity and the molecular diagnoses made are for emerging clinical conditions sharing characteristic features that overlap the DubS phenotype.
Keywords: Dubowitz syndrome, exome sequencing, genome sequencing, microarray, genetic heterogeneity
1. INTRODUCTION
Dubowitz syndrome (DubS) was first clinically described in 1965 (Dubowitz, 1965) as a disorder characterized by mild short stature, microcephaly, eczema, as well as mild delays in development and cognition (Grosse, Gorlin, & Opitz, 1971; Opitz, Pfeiffer, Hermann, & Kushnick, 1973). A susceptibility to malignancy and hematological disorders was also observed in early cases (Sauer & Spelger, 1977; Walters & Desposito, 1985). A recognizable pattern of facial dysmorphology includes a sloping forehead, ptosis, telecanthus, blepharophimosis, facial asymmetry, and micrognathia (Dubowitz, 1965; Grosse et al., 1971). Since its original description, a significant degree of phenotypic variability has been recognized to be part of DubS (Innes, McInnes, & Dyment, 2018; Stewart et al., 2014; Tsukahara & Opitz, 1996). In the absence of a laboratory diagnostic biomarker or pathognomonic feature, the phenotypic expansion associated with the syndrome has led to discussion regarding its existence as a single unifying diagnosis (Dyment et al., 2018; O’Donnell-Luria et al., 2018). Nevertheless, with over 200 individuals reported in the literature with this clinical diagnosis, DubS is an important disease phenotype under consideration for many clinicians as they formulate their differential diagnoses. A better understanding of the underlying biology, and potential disease gene(s) and variant alleles contributing to DubS, may provide further insights for the individuals, families and caring physicians.
The observation of affected siblings in some families has suggested a potential autosomal recessive disease trait; however, no single gene, group of related genes, or common pathway has been identified to explain DubS (Innes et al., 2018). Biallelic variants in LIG4 have been identified in two families with a clinical diagnosis of DubS (Gruhn et al., 2007; Stewart et al., 2014). These children experienced mild cognitive delays and an increased risk of malignancy in adulthood. Other genes implicated in single individuals clinically diagnosed with DubS include NSUN2, PCNT, RNU4ATAC, ACTB, and STAT3 (Beitzke et al., 2011; Dieks, Baumer, Wilichowski, Rauch, & Sigler, 2014; Johnston et al., 2013; Kariminejad et al., 2017; Krøigård et al., 2016; Martinez et al., 2012). In each case, these genes are linked to known multisystem genetic disorders that present with some clinical phenotypic similarity to DubS. The routine use of chromosomal microarray analysis has also identified pathogenic copy number variants (CNV) in individuals diagnosed with DubS-like phenotypes; in particular, deletions at chromosome 13q31, 14q32, 17q24.2-q24.3, and 19q13 show phenotypic overlap with DubS, though only the deletion on chromosome 17 has been reported in more than one family (Hancarova et al., 2018).
By leveraging the genomics resources, exome and genome sequence data (ES/GS), and clinical phenotypic information from two international gene-discovery programs, we sought to identify the underlying genetic cause(s) for DubS. We used a family-based genomics approach and studied 31 individuals clinically diagnosed with DubS from 27 families and, when possible, their unaffected parents and/or affected relatives.
2. MATERIAL AND METHODS
The Centers for Mendelian Genomics (CMG), and the FORGE/Care4Rare Canada Consortium (FORGE/C4R) are collaborative research projects with the goal of identifying pathogenic variants responsible for rare childhood diseases. Local institutional research ethics board approval for Care4Rare was obtained prior to enrollment of any participant. Cases recruited through the CMGs were enrolled through individual research studies with local institutional research ethics board approval followed by research ethics approval for sequencing and analysis of de-identified samples at a CMG.
Recruitment
FORGE/Care4Rare:
The Finding of Rare Disease Genes Canada Consortium (FORGE; 2010–2014), which subsequently became the Care4Rare Canada Consortium (Care4Rare; 2014–2022), is a national consortium funded by Genome Canada, the Canadian Institutes of Health Research, and other funders, to rapidly identify pathogenic variants responsible for a wide spectrum of rare, pediatric and adult-onset diseases using ES and emerging technologies. The consortium comprises over 170 members (clinical geneticists, clinical subspecialists, bioinformaticians and molecular biologists) from 21 genetics centers and 3 science and technology innovation centers from across Canada (Beaulieu et al., 2014). A call to the members of the Canadian consortium was made for any individuals with a clinical diagnosis of DubS or with a diagnosis of DubS strongly considered as part of the differential diagnosis by an experienced clinical geneticist or pediatrician. DubS was one of the initial recognizable malformation syndromes selected for investigation as part of the original FORGE gene discovery project in 2010. The initial call took place in 2010 and has subsequently repeated every 1–2 years. While most of the families recruited were from Canadian centers, families from outside of Canada with a DubS diagnosis considered by a geneticist or pediatrician (two probands in this series) were also included. In total, 11 individuals from 11 families were recruited into the FORGE/Care4Rare arm of the study.
Centers for Mendelian Genomics:
The CMGs are a National Institutes of Health-funded initiative in the United States established in 2012 and renewed in 2016, formed to identify novel genes underlying Mendelian phenotypes using exome- and genome-level sequencing (Bamshad et al., 2012; Posey et al., 2019). The current iteration of the CMGs includes centers based at Baylor College of Medicine/Johns Hopkins University (BH-CMG), the Broad Institute (B-CMG), the University of Washington (UW-CMG), and Yale University (Y-CMG). Independent investigators may apply for research sequencing and data analysis support through site-specific web portals. In 2017, the CMG Data Analysis Working Group put out a coordinated call for any previously sequenced CMG cases with a clinical diagnosis of DubS. No strict phenotypic criteria were applied aside from a physician having made, or strongly considered, the diagnosis of DubS. In total, 20 individuals from 16 families were identified for this cohort study.
Free and informed consent was provided by all probands with DubS and family members participating in the FORGE/Care4Rare or CMG associated studies.
Sequencing and analysis:
Exome capture and high-throughput sequencing of genomic DNA was performed for the proband and parents/siblings/relatives of each kindred when available. For the families recruited by FORGE/Care4Rare, targeted exon capture was performed using the Agilent SureSelect All Exon 50 MB (V5) exome enrichment kit and sequenced on an Illumina Hi-Seq 2000 using 2×100bp chemistry. Read alignment, variant calling, and annotation were done as outlined for previous FORGE and Care4Rare projects (Beaulieu et al., 2014; Srour et al., 2012) with a pipeline based on Burrows-Wheeler Aligner (BWA) (Li & Durbin, 2009), Picard (http://picard.sourceforge.net/), ANNOVAR (Wang, Li, & Hakonarson, 2010), and custom annotation scripts. Analyses were performed under X-linked, recessive, and dominant modes of inheritance. The variants were prioritized by allele frequency (less than 1% in our local Care4Rare database). One individual had their sequencing performed by a commercial company (GeneDx) and BAM files were re-analyzed with the Care4Rare annotation pipeline.
Each of the four CMGs contributed sequence data which were collected over the course of several years. Multiple exome capture platforms were used, including Roche/Nimblegen SeqCap EZ v2.0 2×75bp (UW-CMG, Y-CMG), Nimblegen core design (BH-CMG) and Nextera DNA Exome 2×75bp (B-CMG, BH-CMG) and IDT xGen 2×101bp (Y-CMG). Similarly, sequencing was performed on different sequencers including the HiSeq2500 (BH-CMG, UW-CMG), HiSeq2000 (BH-CMG) and HiSeq4000 (B-CMG, Y-CMG). Genomes were sequenced on a HiSeqX (B-CMG). Joint variant calling was performed using Haplotype Caller from GATK (V3.2), with a detailed description available online (http://uwcmg.org/#/instruction), and shared with each CMG. Each CMG analyzed the data in parallel through their standard pipelines and results were compared and discussed for consensus. Here, we briefly summarize the UW-CMG approach as a representative example, noting that each CMG applied similar strategies with different software such as seqr (B-CMG; https://github.com/macarthur-lab/seqr). Sample quality control, including ancestry and pedigree checks, were performed with peddy (Pedersen & Quinlan, 2017). Sequence variants were annotated by Variant Effect Predictor (VEP; v83) (McLaren et al., 2016), then loaded into a GEMINI (v0.19.1) database (Paila, Chapman, Kirchner, & Quinlan, 2013). Analyses were performed under recessive, de novo dominant, X-linked, and dominant models depending on pedigree information, and variants were required to meet the following criteria: minimum depth ≥6, minimum genotype quality ≥ 20, GATK FILTER value of “PASS” or “SBFilter”, maximum alternate allele frequency (AAF) ≤ 0.005 across the subpopulations represented in the 1000 genomes (Auton et al., 2015), Exome Sequencing Project (Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP), Seattle, WA (URL: http://evs.gs.washington.edu/EVS/) [January, 2018]), UK10K (Walter et al., 2015), and the Exome Aggregation Consortium and Genome Aggregation Database (Karczewski et al., 2020; Lek et al., 2016) reference sets, UW-CMG AAF ≤ 0.05, and VEP impact severity of “MED” or “HIGH”. Strong candidate variants passing these filtering criteria were evaluated with the Integrative Genomics Viewer (Thorvaldsdóttir, Robinson, & Mesirov, 2013) for evidence of genotyping error. Genes containing variants passing the filtering criteria were evaluated for evidence of related phenotypes in humans or model organisms using the Monarch Initiative (Mungall et al., 2017), ClinVar (Landrum et al., 2018), and DECIPHER databases (Firth et al., 2009). Novel candidate disease genes were submitted to the Matchmaker Exchange through GeneMatcher, matchbox, or MyGene2 nodes (Arachchi et al., 2018; Chong et al., 2016; Philippakis et al., 2015; N. Sobreira, Schiettecatte, Valle, & Hamosh, 2015). Copy-number variant (CNV) analysis on exome data was performed with GATK-gCNV best practices. Briefly, read coverage was first calculated for each exome using GATK CollectReadCounts and samples batched based on a principal components analysis of sequencing read counts and then a model trained for the batch. All raw CNVs were aggregated across all batches and post-processed using quality- and frequency-based filtering to produce a final CNV callset that was annotated with known disease genes. Any identified CNVs were confirmed by chromosomal microarray analysis using the Illumina Infinium CoreExome-24 (v1.2) platform in GenomeStudio. The mean LogRRatio of markers within the CNV position was consistent with heterozygous parents, homozygous affected siblings and wild type unaffected sibling in a family with known consanguinity.
Pathway and Network analyses
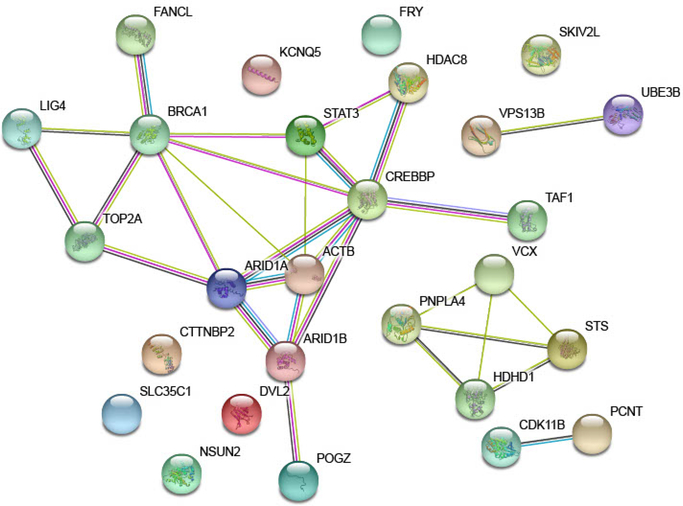
We hypothesized that the genes implicated by our analyses share biological pathways with each other and the genes underlying single gene disorders previously implicated in individuals diagnosed with DubS). The novel candidate genes, the genes driving alternative diagnoses among our cases and the genes implicated in the literature for individuals clinically diagnosed with DubS (Innes et al., 2018) were provided to STRING-db (v11.0) (Szklarczyk et al., 2019) to assess for common pathways or interactions. This was performed by using human reference data, only the queried proteins, protein-protein interaction (PPI) data from known interactions (curated databases, experimentally determined), predicted interactions based on gene fusions and gene co-occurrence, co-expression, protein homology and text mining; medium confidence (0.400) was the minimum required for interaction scores. The gene list was tested for protein-protein interaction enrichment, and functional enrichment in Gene Ontology (GO) biological processes.
3. RESULTS
It was apparent that the presentation of the 31 individuals showed a significant degree of clinical variability, and that these individuals likely represented several clinical entities despite sharing core features of DubS (Table 1 and Supplemental Table 1). This was in contrast to several other recognizable malformation syndrome cohorts studied in parallel by both CMG and FORGE consortia that showed marked homogeneity, such as Nager syndrome (Bernier et al., 2012), Kabuki syndrome (Ng et al., 2010) and Floating Harbor syndrome (Hood et al., 2012). Nevertheless, elements of the common facial gestalt (a sloping forehead, hypertelorism/ptosis/blepharophimosis and micrognathia) were shared among all individuals included for study (Table 1). Other features of the DubS phenotype were also common: 11/25 (44%) were observed to be less than 2 standard deviations for height, 20/25 (80%) were reported to be microcephalic and 20/21 (95%) presented mild to severe deficits in cognitive abilities (Table 1 and Supplemental Table 1). There were four instances of a familial recurrence, including three concordant sibling pairs and a concordant aunt-niece pair (Supplemental Table 1). Eczema was observed in 17/27 (63%) and a high-pitched voice was also reported in several individuals, 9/20 (45%; Table 1). No individuals in the cohort presented with all of the commonly observed “core” elements previously reported (IUGR, short stature, microcephaly, cognitive deficits, hyperactivity, a high-pitched voice, eczema, and the facial gestalt) (Grosse et al., 1971).
Table 1:
Features reported in individuals diagnosed with Dubowitz syndrome
| Feature | Total Frequency (%)(n=31*) | Those with a molecular diagnosis (n=14*) | Those without a molecular diagnosis† (n=8*) |
|---|---|---|---|
| Intrauterine growth restriction | 16/26 (62%) | 9/14 | 2/5 |
| Short stature (less than -2SD) | 11/25** (44%) | 7/13 | 2/6** |
| Microcephaly (less than -2SD) | 20/25 (80%) | 11/13 | 4/5 |
| Intellectual disability (includes mild, moderate, severe and profound when specified) | 20/21 (95%) | 13/13 | 4/5 |
| Behavioral concerns (eg ADHD) | 15/22 (68%) | 8/12 | 4/4 |
| Sloping forehead | 7/23 (30%) | 4/11 | 3/5 |
| Telecanthus/hypertelorism/ptosis/ blepharphimosis | 26/27 (96%) | 14/14 | 4/5 |
| High-pitched voice | 9/20 (45%) | 5/11 | 2/3 |
| Micrognathia | 17/23 (74%) | 8/12 | 1/3 |
| History of eczema | 17/27 (68%) | 7/13 | 6/6 |
| Malignancy | 1/23 (4%) | 1/13 | 0/4 |
The denominator varies for each feature and is dependent on the available clinical information for the 31 reported individuals with the Dubowitz phenotype.
Two individuals were described as “short” with no measurements provided, 13/27 (48%).
This column is comprised of 8 individuals without (1) a formal diagnosis or without a compelling VUS in a known gene or without a compelling variant in a gene of interest.
Genomic diagnoses:
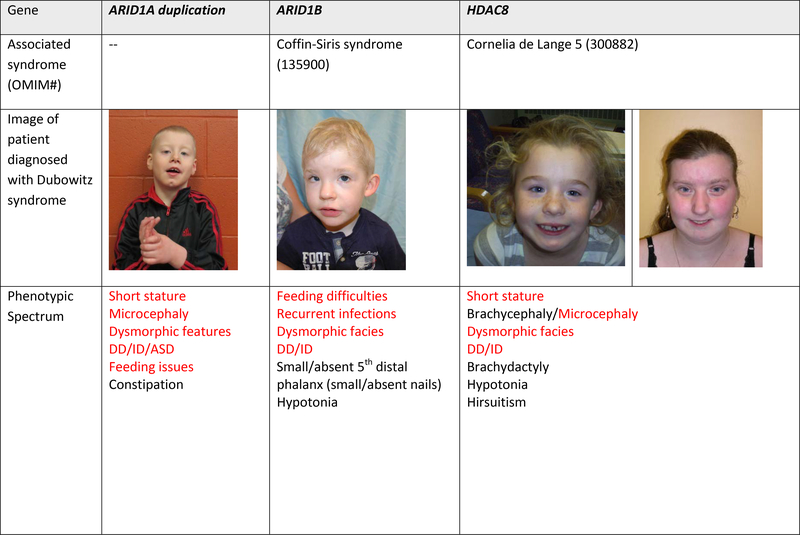
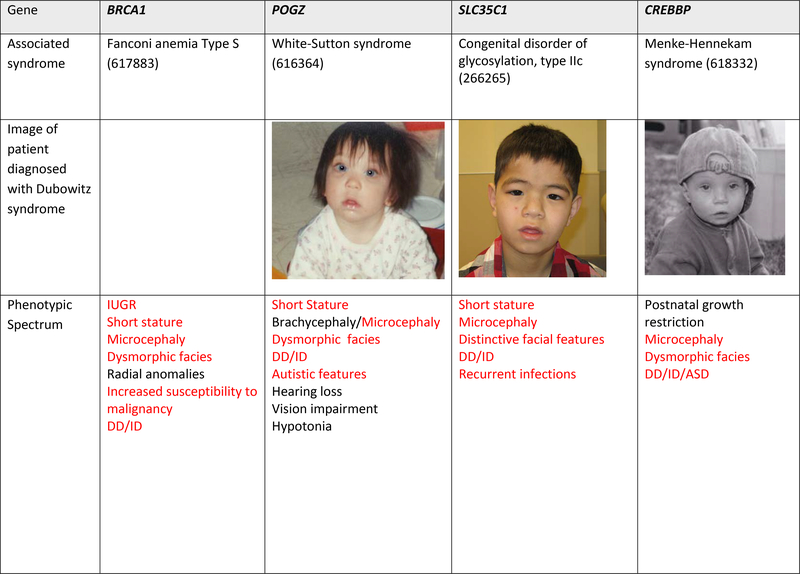
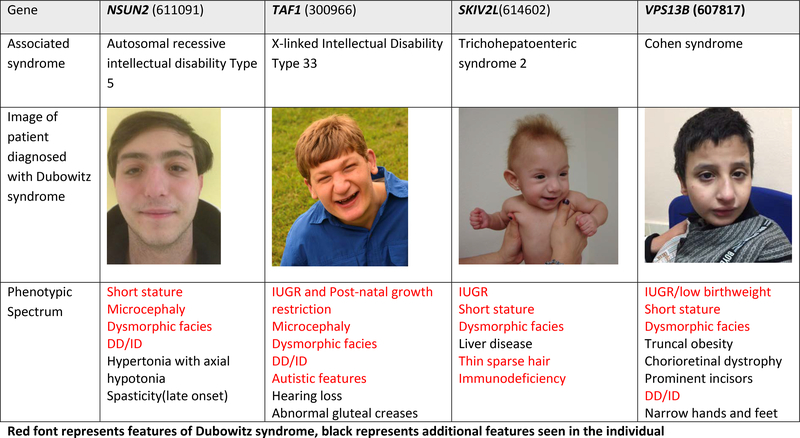
Fourteen of the 31 (45%) individuals from 27 families were diagnosed with other disorders by the genomic studies of microarray or ES/GS data (Table 2). Three individuals were diagnosed based on re-analysis of previous microarray findings that were not initially appreciated to be the explanation for the clinical presentation, including a duplication of ARID1A, a deletion of HDAC8 and a deletion of several contiguous X-linked genes (STS, VCX, PUDP, PNPLA4). An additional 11 diagnoses were made based on the ES/GS analyses (Table 2). The molecular diagnoses included pathogenic biallelic variants (VPS13B, SKIV2L, SLC35C1, BRCA1, and NSUN2), de novo and inherited dominant variants (ARID1B, CREBBP and POGZ) and de novo X-linked variants (HDAC8 and TAF1) (Table 2). Only variation at HDAC8 was seen in more than one family in this series.
Table 2.
Alternative diagnoses in those individuals with a clinical diagnosis of Dubowitz syndrome.
| Case ID | Gene | Variants | Alternative Diagnosis (OMIM#) | Inheritance | Year of first reference of gene to phenotype |
|---|---|---|---|---|---|
| DubS21 | BRCA1 | ENST00000357654.9 c.594_597delTGTG;5095C>T, ENSP00000418960 (p.Ser198ArgfsX35;Arg1699Trp) | Fanconi Anemia S (617883) | Autosomal recessive (compound heterozygous) | 2013 |
| DubS22 | HDAC8 | Arr chrXq13.1–13.2 (71632632–72449647*) | Cornelia de Lange 5 (33082) | X-linked de novo | 2012 |
| DubS24 | CREBBP | ENST00000262367.10,c.5612–5614del, ENSP00000262367, p.1871_1872del | Menke-Hennekam Syndrome 1 (618332) | De novo dominant | 2016 |
| DubS26 | SLC35C1 | ENST00000314134.4, c.887A>G, ENSP00000313318, p.His296Arg | Congenital disorder of glycosylation, type IIc (266265) | Autosomal recessive (homozygous) | 2001 |
| DubS28 | ARID1B | ENST00000350026.10, :c.5737C>T, ENSP00000344546, p.Arg1913Ter | Coffin-Siris 1 (135900) | De novo dominant | 2011 |
| DubS29 | ARID1A | Arr 1p36 (27,001,498–27,110,331)x3 | ARID1A duplication associated intellectual disability syndrome | De novo dominant | 2017 |
| DubS30 | SKIV2L | ENST00000375394.7, c.235C>T, ENSP00000364543, p.Arg79Ter | Trichohepatoenteric syndrome 2 (614602) | Autosomal recessive (homozygous) | 2012 |
| DubS11 | POGZ | ENST00000271715.2:c.1679–3C>G (splice region variant) | White-Sutton syndrome (616364) | De novo dominant | 2016 |
| DubS12 | TAF1 | ENST00000276072.3:c.61A>T; ENSP00000276072.3:p.Met21Leu | X-linked syndromic intellectual disability Type 33 (300966) | X-linked de novo | 2015 |
| DubS1 | HDAC8 | ENST00000373568.2:c.638–2A>G (splice acceptor variant) | Cornelia de Lange 5 (30082) | X-linked de novo | 2012 |
| DubS18 | VCX-PUDP-STS-PNPLA4 | Arr Xp22.31 (6454182–8115193)x1 | X-linked deletion syndrome | X-linked | 2013 |
| DubS6 and DubS7 | VPS13B | Arr 8q22.2 (99096530–99142877)x4 | Cohen syndrome (216550) | Autosomal recessive | 2003 |
| DubS2 | NSUN2 | ENST00000264670.6:c.1903A>G/c.529C>T ENSP00000264670.6: p.Asn635Asp/p.His177Tyr | Autosomal recessive intellectual disability type 5 (611091) | Autosomal recessive (compound heterozygote) | 2012 |
Coordinates have been converted to GRCh37/Hg19 with liftover (available at https://genome.ucsc.edu/util.html)
Variants of unknown clinical significance in known disease genes:
Compelling variants of unknown significance (VUS) were observed in 4 individuals from 3 families, implicating FRY, KCNQ5,and FANCL(Supplemental Table 2). Proband DubS27 had biallelic variants in FRY, ENST00000380250.3:c.2647C>G, p.Arg883Gly and c.4338G>C, p.Glu1446Asp. Recessive variants in FRY have been reported to be responsible for a rare intellectual disability syndrome in 3 families to date (Paulraj et al., 2019; Riazuddin et al., 2017). The variant seen in DubS27 has not been reported before. Probands DubS19 and 20 carry a rare missense variant in the channel gene, KCNQ5 (ENST00000342056.2:c.1829C>T; p.Thr610Ile) and have been reported previously to have Dubowitz syndrome (Grosse et al., 1971; Jones, 1997; Jones, 2006; Swartz et al., 2003). The parent of the niece (and sister of the aunt) is unaffected and incomplete penetrance would be necessary to explain the inheritance pattern. KCNQ5 has been associated with a neurodevelopmental disorder in 4 individuals who reportedly did not show evidence for growth restriction, microcephaly or dysmorphic features (Lehman et al., 2017) as observed in DubS19 and 20. There are no images of individuals with variants in either FRY or KCNQ5 in the literature for comparison with the individuals studied in this series. Lastly, a homozygous VUS within the 3’ UTR of FANCL (NM_001114636.1:c.*83dup) was also observed in proband DubS23. Biallelic pathogenic variation in FANCL results in Fanconi Anemia (Vetro et al., 2015). Chromosomal breakage studies were planned to aid in variant interpretation; however, the individual was subsequently lost to follow-up.
Genes of uncertain significance:
Candidates for novel disease genes, or a novel mechanism in a known disease-associated gene, were observed in 5/31 (16%) individuals (Supplemental Table 2), although no candidate gene was implicated in multiple families. These genes included biallelic variants in CDK11B and CTTNBP2, and monoallelic variants in DVL2 and TOP2A. Biallelic variants observed in CDK11B segregated in two sisters, DubS8 and DubS9, that had previously been reported as having brachymorphism-onychodysplasia-dysphalangism syndrome (Ounap, Justus, & Lipping-Sitska, 1998). Murine knock-out models for CDK11B show embryonic growth arrest and abnormal skin, while Drosophila models are small and have neuroanatomy phenotypes (Mungall et al., 2017). Proband DubS25 had biallelic missense variants in CTTNBP2, a gene in which de novo dominant variants have been previously associated with autism (Guo et al., 2018). Proband DubS4 carried a de novo missense variant in TOP2A, a gene in which animal models share features with DubS: zebrafish null mutants have abnormal retinas, body shape, head/brain morphology, and a central nervous system phenotype, while Drosophila mutants have neuroanatomy defects and small body size (Mungall et al., 2017). Proband DubS2 had a de novo frameshift variant in DVL2. This heterozygous frameshift was present in the last exon and is predicted to escape nonsense mediated decay, similar to the loss-of-function variants in the last exon of DVL1 and DVL3 causing Robinow syndrome, an autosomal dominant skeletal dysplasia characterized by distinctive facial features with some phenotypic overlap with DubS (hypertelorism, long philtrum, small chin) (J. White et al., 2015; J. J. White et al., 2016). Similar variants in DVL2 cause a Robinow-like phenotype in dogs (Mansour et al., 2018), while knockout/knockdown models cause microcephaly in zebrafish (Carvalho et al., 2014) and neural tube defects in mice (Hamblet et al., 2002; Mungall et al., 2017). Furthermore, a homozygous nonsense variant in DVL2 has been reported in a child described as having short stature, ptosis, developmental delay, facial dysmorphism and a cardiomyopathy (Monies et al., 2017).
Each of these candidate genes require additional evidence to demonstrate, definitively, that they contributed to their respective clinical presentations of the DubS phenotype. Sharing candidate genes through the Matchmaker Exchange (N. L. M. Sobreira et al., 2017) has not yet provided additional cases with sufficiently similar phenotypes, although this work is ongoing.
Network and Pathway analyses
The combined set of 6 novel candidate genes (Supplemental Table 2), 16 genes offering an alternative diagnosis (Table 2), and 8 DubS genes from the literature (Innes et al., 2018) exhibited significant protein-protein interaction (PPI) enrichment: 29 observed edges, 12 expected, PPI enrichment p-value = 4.5e-05. The network, shown in Figure 2, captures four clusters: the two smallest clusters connecting our novel candidate genes to genes previously implicated in DubS under a recessive model: CDK11B to PCNT and VPS13B to UBE3B (Dieks et al., 2014; Innes et al., 2018); a second cluster driven by the genes within the chromosome X deletion observed in one case (note HDHD1 is another name for PUDP); and a large cluster capturing many genes underlying syndromes similar to DubS and one of our novel candidate genes, TOP2A. This set of 27 genes is significantly enriched in 37 GO:Biological Processes (Supplemental Table 3). The top 10 GO:Biological Processes are listed in Table 3. We observe particular enrichment in GO:Biological Processes related to cell cycle, cellular and chromosomal organization, and gene regulation. These results suggest that the genes underlying the phenotype of DubS and related disorders share dysregulation of basic biological processes.
Figure 2.
Protein networks shared by candidate genes and genes offering alternative diagnoses among individuals sharing Dubowitz Syndrome diagnoses.
Table 3.
The top 10 GO:Biological Processes with significant evidence of enrichment among candidate genes and genes offering an alternative diagnosis. Significance: False Discovery Rate (FDR) value < 0.05. Complete table of 37 enriched terms is available in Supplemental Table 3.
| GO:term | Description | Count | Matching proteins found in network | FDR |
|---|---|---|---|---|
| GO:0051276 | chromosome organization | 9 of 999 | ARID1A, ARID1B, CREBBP, HDAC8, LIG4, POGZ, TAF1, TOP2A, VCX | 0.0067 |
| GO:0006996 | organelle organization | 14 of 3131 | ACTB, ARID1A, ARID1B, BRCA1, CREBBP, FRY, HDAC8, LIG4, PCNT, POGZ, STAT3, TAF1, TOP2A, VCX | 0.0089 |
| GO:0007049 | cell cycle | 9 of 1263 | BRCA1, CDK11B, HDAC8, LIG4, NSUN2, PCNT, POGZ, TAF1, TOP2A | 0.0089 |
| GO:0040029 | regulation of gene expression, epigenetic | 5 of 251 | ACTB, ARID1A, ARID1B, BRCA1, STAT3 | 0.0089 |
| GO:0051726 | regulation of cell cycle | 9 of 1129 | ACTB, BRCA1, CDK11B, HDAC8, NSUN2, PCNT, STAT3, TAF1, TOP2A | 0.0089 |
| GO:0071417 | cellular response to organonitrogen compound | 6 of 485 | ACTB, ARID1B, BRCA1, HDAC8, STAT3, TAF1 | 0.0102 |
| GO:0045815 | positive regulation of gene expression, epigenetic | 3 of 51 | ACTB, ARID1A, ARID1B | 0.0107 |
| GO:0071407 | cellular response to organic cyclic compound | 6 of 505 | ACTB, ARID1A, BRCA1, HDAC8, STAT3, TAF1 | 0.0107 |
| GO:0048096 | chromatin-mediated maintenance of transcription | 2 of 9 | ARID1A, ARID1B | 0.0150 |
| GO:2000615 | regulation of histone H3-K9 acetylation | 2 of 11 | BRCA1, HDAC8 | 0.0174 |
4. DISCUSSION
No single gene was identified as responsible for the majority, or even a significant minority, of the individuals clinically diagnosed with DubS. Potential explanations for the lack of a common cause are that we have sequenced a clinically heterogeneous cohort, DubS is genetically heterogeneous, or DubS as a clinical entity is a nonspecific collection of relatively common clinical features. Historically, the key features of DubS are mild intellectual disability, short stature, microcephaly, sloping forehead, ptosis, telecanthus, eczema and a high-pitched voice (Table 1) (Grosse et al., 1971). However, there are no formal phenotypic criteria to diagnose this syndrome. We recognize that the lack of strict phenotypic criteria for diagnosis and the reliance on the clinical impression of the referring physician is a limitation to any study involving DubS. Nonetheless it is a well-studied syndrome and present in several editions of Smith’s Recognizable Patterns of Human Malformation and thus should be familiar to most clinical geneticists.
We also recognize that none of the historical clinical features of DubS would be considered an overly specific, or ‘hard-handle’, and hence the clinical presentation may show overlap with atypical or mild forms of different syndromes associated with intellectual disability (Innes et al., 2018). Furthermore, over the decades since its original description, an extremely broad phenotypic expansion has occurred such that the published clinical features of DubS now range from normal stature to dwarfism, normal intellect to severe intellectual disability, microcephaly to macrocephaly, the absence of ptosis to the presence of bilateral ptosis (Tsukahara & Opitz, 1996). Upon inspection of the clinical features seen in this cohort (Figure 1; Table 1), we observe most cases report elements of the DubS facial gestalt, microcephaly, and developmental delay, but the short stature, eczema and high-pitched voice features of DubS were relatively less likely to be shared.
Figure 1. Images and alternative diagnoses for individuals with a clinical diagnosis of Dubowitz syndrome.
Red font represents features of Dubowitz syndrome, black represents additional features seen in the individual
The molecular diagnoses made and candidate genes nominated in this study offer insight in shared disease mechanisms and pathways between DubS cases and other syndromes. We see the DubS phenotype may be explained by the alternative diagnoses of Fanconi anemia, Coffin-Siris syndrome, or Cornelia de Lange syndrome. Individual DubS cases in our cohort may be explained by other syndromes sharing core biological processes, including DNA repair and chromatin remodeling. Three families in our cohort may be explained by genetic variants influencing DNA repair. One and possibly two additional individuals were diagnosed with rare forms of Fanconi anemia (Types S and L). The first child carried biallelic pathogenic variants in BRCA1 and has been published previously (Sawyer et al., 2015). The biallelic variants result in an intellectual disability syndrome with dysmorphic features, early onset of breast cancer and functional studies consistent with defective DNA repair (Sawyer et al., 2015). A second case may represent FANCL due to homozygous variant in the 3’UTR; however, the child was lost to follow-up despite multiple attempts to re-contact and chromosomal breakage studies were not performed. In the literature, LIG4 has been highlighted as a DubS gene (O’Driscoll et al., 2001). While it is not a gene in the Fanconi Anemia DNA repair pathway, it does function to repair double strand breaks in DNA by non-homologous end joining and biallelic variants were observed in one of the original families with DubS (Gruhn et al., 2007; Stewart et al., 2014). These individuals with biallelic LIG4 variants are also prone to malignancy in adulthood. Given the phenotypic overlap between DubS and Fanconi Anemia types S and L, we suspect that DNA breakage and repair mechanisms may represent a shared pathway to these shared phenotypic features in a subset of individuals.
Pathogenic variation was seen in several genes associated with chromatin remodeling within our DubS cohort. In fact, the only pathogenic variants seen in a single gene in more than one individual/family of the cohort was HDAC8 that encodes histone deacetylase 8. These children, one of whom was previously published, have a de novo X-linked dominant form of Cornelia de Lange syndrome (Deardorff et al., 2012). Clinical features (Figure 1) show overlap with DubS and, as might be expected in the context of X-inactivation, the intellectual disability can be mild to absent in female individuals. ARID1A and ARID1B encode subunits of the SWI/SNF complex involved in chromatin remodeling and Coffin-Siris syndrome, and two diagnoses in the cohort were associated with these genes. The child with the pathogenic ARID1B variant did not have the characteristic fifth digit hypoplasia of classical Coffin-Siris syndrome, although certainly since the delineation of the molecular basis of this syndrome, it is recognized that this is not a mandatory feature and in particular is frequently absent in ARID1B-related Coffin-Siris syndrome (van der Sluijs et al., 2019). The child with the ARID1A duplication was only recently determined to have this newly described syndrome as a result of the de novo duplication (Bidart et al., 2017). The functional impact of the duplication on chromatin remodeling has yet to be fully elucidated though downstream dysregulation of several canonical pathways have been observed (Bidart et al., 2017). CREBBP is another gene associated with transcription co-activation following chromatin remodeling. This individual has a variant in one of the final two exons of CREBBP, consistent with the emerging Menke-Hennekam syndrome, a disorder clinically distinct from Rubinstein-Taybi syndrome (Angius et al., 2019; Banka et al., 2019; Menke et al., 2016). Lastly, POGZ was observed in one individual who showed overlap with the previously described White-Sutton syndrome, but with relatively mild cognitive involvement and lacking the characteristic behavioral and gastrointestinal manifestations (Stessman et al., 2016) POGZ is a heterochromatin protein 1 α-binding protein and it functions as a transcriptional regulator in neurons by modifying chromatin structure(Stessman et al., 2016).
NSUN2 has been presented, albeit cautiously, as a gene responsible for a DubS-like syndrome in sibling pairs (Martinez et al., 2012). The clinical diagnosis was based on the clinical overlap with the historical cases (mild microcephaly and ID, blepharophimosis and hypertelorism, broad nasal bridge); however, the siblings did not have all features (for example, voice differences, triangular face, or a round nose tip and prominent ears (Dubowitz, 1965; Grosse et al., 1971)). Furthermore, pathogenic variants in NSUN2 have been reported as an explanation for syndromic intellectual disability (Abbasi-Moheb et al., 2012; Khan et al., 2012; Yavarna et al., 2015) and even as a gene responsible for a Noonan-like syndrome (Fahiminiya et al., 2014). Given the extent of the phenotypic overlap, it is not surprising that at least one individual in the cohort who presented with moderate intellectual disability, short stature and microcephaly carried pathogenic NSUN2 variants (Supplemental Table 1).
We studied one aunt-niece pair that has been previously reported in the literature (Grosse et al., 1971; Swartz et al., 2003). Indeed these two individuals represent historically ‘typical’ examples of DubS as the aunt was published in one of the earliest publications and both the aunt and the niece have been included as representative individuals with DubS in Smith’s Recognizable Patterns of Human Malformation. These 2 individuals were found to share a rare missense variant of unknown clinical significance in the gene KCNQ5, associated with MRD46 (OMIM 617601). To date only 4 individuals with sequence variants in this gene have been published in the literature (Lehman et al., 2017) and while these individuals are described as non-dysmorphic, there are no published photographs. Affected individuals have mild to profound developmental impairment with OFC measurements between −1 and −2 SD below the mean (Lehman et al., 2017).
The majority of the solved individuals had their initial diagnostic investigations performed and subsequent enrollment into these two gene discovery projects before the molecular causes had been well-established or recognized for their respective syndromes. As such, their diagnoses only became evident with time as additional cases with similar variants were reported. This is intuitive given that many of these cases were identified as suitable candidates for research-based gene discovery in the very early days of ES implementation, and it has been demonstrated that a significant proportion of all Mendelian gene discoveries (and increasingly a proportion of novel phenotype reports) have occurred since 2010 (Bamshad, Nickerson, & Chong, 2019). It follows that many of these novel gene discoveries from the “next-generation sequencing era” are, in contrast to earlier ‘phenotype-first’ syndrome delineations, in individuals with rather non-specific phenotypes that have been studied in ‘reverse’ following the gene discovery. Consistent with the rather non-specific and increasingly broad phenotypic spectrum of DubS in the published literature over the last three decades, it is understandable that pathogenic variants in any of a large and growing number of non-specific syndromic ID genes could be associated with such individuals. This speaks to the rapid increase in knowledge of underlying variants associated with the increasing use of next-generation sequencing.
Seventeen individuals did not receive a molecular or cytogenetic diagnosis. Subjectively, some of these individuals did show overlap with the historical cases though there were still no shared candidate variants to suggest a novel DubS gene. One could presume that by diagnosing 14 individuals we have reduced the heterogeneity of the entire series and perhaps further inspection of these individuals may further homogenize the cohort to a greater extent. However, this was not apparent when we reviewed the frequency of the component features of DubS though numbers were small (Table 1; Supplemental Table 1). Nevertheless it may be a useful strategy to subjectively identify the most representative cases mirroring historical DubS and pursue additional studies, such as GS or transcriptome sequencing to elucidate an underlying cause or pathway in these select cases.
The results of this work do show that if there is a clinical suspicion of DubS, then a chromosomal microarray analysis and ES should be considered as first-line investigations. These results suggest the diagnostic rate can be 48% or as high as 70–80% should the variant or gene of unknown significance be shown to be the explanations for the respective presentations. This diagnostic rate exceeds the published yield of clinical ES in diverse cohorts of individuals of 20–30% (Bowling et al., 2017; Lee et al., 2014; Retterer et al., 2016; Yang et al., 2014). Other investigations that should be considered in those with DubS would include chromosomal breakage studies in those with microcephaly, malignancy, familial recurrence or consanguinity suggesting an autosomal recessive condition. Immunoglobulin or other immune-related studies may also be pursued if recurrent infections are present.
Another key message of this project is that the individuals we considered as having a clinical diagnosis of DubS were often found to have alternative diagnoses despite being diagnosed by experienced clinicians. In this study, these diagnoses tended to fall in broad categories that include genes of (1) chromatin remodeling and transcription, typically de novo dominant as well as (2) DNA repair genes, typically autosomal recessive. Should DubS be a consideration, a careful clinical assessment of these respective syndromes should also be undertaken. By providing alternative diagnoses by molecular means, we have ruled out the diagnosis of DubS in a large proportion of individuals. Furthermore, by not observing compelling variants in shared genes in the remaining individuals, we conclude that the majority of individuals diagnosed with DubS do not in fact have a shared syndrome.
Supplementary Material
Supplemental Table 1. Dubowitz cohort detailed phenotype data
Supplemental Table 2. Variants of unknown clinical significance (VUS) and variants in genes of unknown significance (GUS) seen in individuals with a clinical diagnosis of Dubowitz syndrome
Supplemental Table 3. The complete list of 37 GO:Biological Processes with significant evidence for enrichment among candidate genes, genes offering alternative diagnoses, and DubS genes from the literature.
ACKNOWLEDGEMENT
We would like to acknowledge the families and individuals that have provided samples.
The Care4Rare Canada Consortium work was funded by Genome Canada and the Ontario Genomics Institute (OGI-147), the Canadian Institutes of Health Research, Ontario Research Fund, Genome Alberta, Genome British Columbia, Genome Quebec, and Children’s Hospital of Eastern Ontario Foundation. Funding was also provided by the National Organization of Rare Disorders (NORD).
The Baylor Hopkins Center for Mendelian Genomics, Broad Institute Harvard Center for Mendelian Genomics, University of Washington Center for Mendelian Genomics, and Yale Center for Mendelian Genomics were funded by the National Human Genome Research Institute (NHGRI)/ National Heart Lung and Blood Institute (NHLBI) awards UM1 HG006542, UM1 HG008900, UM1 HG006493, and UM1 HG006504, respectively. Analysis was additionally supported by National Human Genome Research Institute grant R01 HG009141. Funds were also provided under the National Heart, Lung, and Blood Institute (NHLBI) under the Trans-Omics for Precision Medicine Program (TOPMed), and the National Eye Institute (NEI). The GSP Coordinating Center (U24 HG008956) contributed to cross-program scientific initiatives and provided logistical and general study coordination. A.H.O.-L. was supported by National Institute of Child Health and Human Development (NICHD) K12 HD052896 and a Boston Children’s Hospital OFD Career Development Award. KÕ and SP are supported by Estonian Research Council grants PRG471 and PUTJD827. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This work was financially supported by grants from the Dutch Organization for Health Research and Development (ZON-MW grants 917-86-319 and 912-12-109 to B.B.A.d.V.).
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
DATA AVAILABILITY
Data that support the findings of this study are available on request from the corresponding authors (EB/AMI). The data are not publicly available due to privacy or ethical restrictions of the research participants. A subset of participants has provided consent for sharing in a controlled access repository (dbGaP, AnVIL). This information will be shared within this controlled setting and with appropriate permission from respective co-author(s).
References:
- Abbasi-Moheb L, Mertel S, Gonsior M, Nouri-Vahid L, Kahrizi K, Cirak S, … Kuss AW (2012). Mutations in NSUN2 cause autosomal-recessive intellectual disability. Am J Hum Genet, 90(5), 847–855. doi: 10.1016/j.ajhg.2012.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angius A, Uva P, Oppo M, Persico I, Onano S, Olla S, … Crisponi L (2019). Confirmation of a new phenotype in an individual with a variant in the last part of exon 30 of CREBBP. Am J Med Genet A, 179(4), 634–638. doi: 10.1002/ajmg.a.61052 [DOI] [PubMed] [Google Scholar]
- Arachchi H, Wojcik MH, Weisburd B, Jacobsen JOB, Valkanas E, Baxter S, … Rehm HL (2018). matchbox: An open-source tool for patient matching via the Matchmaker Exchange. Hum Mutat, 39(12), 1827–1834. doi: 10.1002/humu.23655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, … Consortium, G. P. (2015). A global reference for human genetic variation. Nature, 526(7571), 68–74. doi: 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad MJ, Nickerson DA, & Chong JX (2019). Mendelian Gene Discovery: Fast and Furious with No End in Sight. Am J Hum Genet, 105(3), 448–455. doi: 10.1016/j.ajhg.2019.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad MJ, Shendure JA, Valle D, Hamosh A, Lupski JR, Gibbs RA, … Genomics, C. f. M. (2012). The Centers for Mendelian Genomics: a new large-scale initiative to identify the genes underlying rare Mendelian conditions. Am J Med Genet A, 158A(7), 1523–1525. doi: 10.1002/ajmg.a.35470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banka S, Sayer R, Breen C, Barton S, Pavaine J, Sheppard SE, … Clayton-Smith J (2019). Genotype-phenotype specificity in Menke-Hennekam syndrome caused by missense variants in exon 30 or 31 of CREBBP. Am J Med Genet A, 179(6), 1058–1062. doi: 10.1002/ajmg.a.61131 [DOI] [PubMed] [Google Scholar]
- Beaulieu CL, Majewski J, Schwartzentruber J, Samuels M, Fernandez B, Bernier F, … Boycott KM (2014). FORGE Canada Consortium: Outcomes of a 2-Year National Rare Disease Gene Discovery Project. American Journal of Human Genetics(Submitted). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beitzke M, Enzinger C, Windpassinger C, Pfeifer D, Fazekas F, Woellner C, … Kroisel PM (2011). Community acquired Staphylococcus aureus meningitis and cerebral abscesses in a patient with a hyper-IgE and a Dubowitz-like syndrome. J Neurol Sci, 309(1–2), 12–15. doi: 10.1016/j.jns.2011.07.045 [DOI] [PubMed] [Google Scholar]
- Bernier FP, Caluseriu O, Ng S, Schwartzentruber J, Buckingham KJ, Innes AM, … Consortium, F. C. (2012). Haploinsufficiency of SF3B4, a component of the pre-mRNA spliceosomal complex, causes Nager syndrome. Am J Hum Genet, 90(5), 925–933. doi: 10.1016/j.ajhg.2012.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidart M, El Atifi M, Miladi S, Rendu J, Satre V, Ray PF, … Coutton C (2017). Microduplication of the ARID1A gene causes intellectual disability with recognizable syndromic features. Genet Med, 19(6), 701–710. doi: 10.1038/gim.2016.180 [DOI] [PubMed] [Google Scholar]
- Bowling KM, Thompson ML, Amaral MD, Finnila CR, Hiatt SM, Engel KL, … Cooper GM (2017). Genomic diagnosis for children with intellectual disability and/or developmental delay. Genome Med, 9(1), 43. doi: 10.1186/s13073-017-0433-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM, Vasanth S, Shinawi M, Russell C, Ramocki MB, Brown CW, … Lupski JR (2014). Dosage changes of a segment at 17p13.1 lead to intellectual disability and microcephaly as a result of complex genetic interaction of multiple genes. Am J Hum Genet, 95(5), 565–578. doi: 10.1016/j.ajhg.2014.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong JX, Yu JH, Lorentzen P, Park KM, Jamal SM, Tabor HK, … Bamshad MJ (2016). Gene discovery for Mendelian conditions via social networking: de novo variants in KDM1A cause developmental delay and distinctive facial features. Genet Med, 18(8), 788–795. doi: 10.1038/gim.2015.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deardorff MA, Bando M, Nakato R, Watrin E, Itoh T, Minamino M, … Shirahige K (2012). HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature, 489(7415), 313–317. doi: 10.1038/nature11316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieks JK, Baumer A, Wilichowski E, Rauch A, & Sigler M (2014). Microcephalic osteodysplastic primordial dwarfism type II (MOPD II) with multiple vascular complications misdiagnosed as Dubowitz syndrome. Eur J Pediatr, 173(9), 1253–1256. doi: 10.1007/s00431-014-2368-5 [DOI] [PubMed] [Google Scholar]
- Dubowitz V (1965). Familial low birthweight dwarfism with an unusual facies and a skin eruption. J Med Genet, 2(1), 12–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyment D, McInnes B, Hartley T, Kernohan K, Lauzon J, Lowry B, … Innes M (2018). Alternative diagnoses for individuals referred for exome sequencing with a clinical diagnosis of Dubowitz syndrome. In (4 ed., Vol. 179, pp. 693). American Journal of Medical Genetics: Wiley. [Google Scholar]
- Fahiminiya S, Almuriekhi M, Nawaz Z, Staffa A, Lepage P, Ali R, … Ben-Omran T (2014). Whole exome sequencing unravels disease-causing genes in consanguineous families in Qatar. Clin Genet, 86(2), 134–141. doi: 10.1111/cge.12280 [DOI] [PubMed] [Google Scholar]
- Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D, … Carter NP (2009). DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am J Hum Genet, 84(4), 524–533. doi: 10.1016/j.ajhg.2009.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco B, Meroni G, Parenti G, Levilliers J, Bernard L, Gebbia M, … Ballabio A (1995). A cluster of sulfatase genes on Xp22.3: mutations in chondrodysplasia punctata (CDPX) and implications for warfarin embryopathy. Cell, 81(1), 15–25. doi: 10.1016/0092-8674(95)90367-4 [DOI] [PubMed] [Google Scholar]
- Grosse R, Gorlin J, & Opitz JM (1971). The Dubowitz syndrome. Z Kinderheilkd, 110(3), 175–187. [DOI] [PubMed] [Google Scholar]
- Gruhn B, Seidel J, Zintl F, Varon R, Tönnies H, Neitzel H, … Schindler D (2007). Successful bone marrow transplantation in a patient with DNA ligase IV deficiency and bone marrow failure. Orphanet J Rare Dis, 2, 5. doi: 10.1186/1750-1172-2-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Wang T, Wu H, Long M, Coe BP, Li H, … Xia K (2018). Inherited and multiple de novo mutations in autism/developmental delay risk genes suggest a multifactorial model. Mol Autism, 9, 64. doi: 10.1186/s13229-018-0247-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamblet NS, Lijam N, Ruiz-Lozano P, Wang J, Yang Y, Luo Z, … Wynshaw-Boris A (2002). Dishevelled 2 is essential for cardiac outflow tract development, somite segmentation and neural tube closure. Development, 129(24), 5827–5838. doi: 10.1242/dev.00164 [DOI] [PubMed] [Google Scholar]
- Hancarova M, Malikova M, Kotrova M, Drabova J, Trkova M, & Sedlacek Z (2018). Association of 17q24.2-q24.3 deletions with recognizable phenotype and short telomeres. Am J Med Genet A, 176(6), 1438–1442. doi: 10.1002/ajmg.a.38711 [DOI] [PubMed] [Google Scholar]
- Hood RL, Lines MA, Nikkel SM, Schwartzentruber J, Beaulieu C, Nowaczyk MJ, … Consortium, F. C. (2012). Mutations in SRCAP, encoding SNF2-related CREBBP activator protein, cause Floating-Harbor syndrome. Am J Hum Genet, 90(2), 308–313. doi: 10.1016/j.ajhg.2011.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innes AM, McInnes BL, & Dyment DA (2018). Clinical and genetic heterogeneity in Dubowitz syndrome: Implications for diagnosis, management and further research. Am J Med Genet C Semin Med Genet, 178(4), 387–397. doi: 10.1002/ajmg.c.31661 [DOI] [PubMed] [Google Scholar]
- Johnston JJ, Wen KK, Keppler-Noreuil K, McKane M, Maiers JL, Greiner A, … Center, N. I. S. (2013). Functional Analysis of a De Novo ACTB Mutation in a Patient with Atypical Baraitser-Winter Syndrome. Hum Mutat, 34(9), 1242–1249. doi: 10.1002/humu.22350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KL (1997). Smith’s Recognizable Patterns of Human Malformation (Fifth Edition ed.). Philadelphia, Pennsylvania: Elsevier Saunders. [Google Scholar]
- Jones KL (2006). Smith’s Recognizable Patterns of Human Malformation (Sixth Edition ed.). Philadelphia, Pennsylvania: Elsevier Saunders [Google Scholar]
- Karczewski K, Francioli L, Tiao G, Cummings B, Alföldi J, Wang Q, … MacArthur D.(2020. ). The mutational constraint spectrum quantified from variation in 141,456 humans In (pp. p. 531210). bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariminejad A, Ajeawung NF, Bozorgmehr B, Dionne-Laporte A, Molidperee S, Najafi K, … Campeau PM (2017). Kaufman oculo-cerebro-facial syndrome in a child with small and absent terminal phalanges and absent nails. J Hum Genet, 62(4), 465–471. doi: 10.1038/jhg.2016.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MA, Rafiq MA, Noor A, Hussain S, Flores JV, Rupp V, … Vincent JB (2012). Mutation in NSUN2, which encodes an RNA methyltransferase, causes autosomal-recessive intellectual disability. Am J Hum Genet, 90(5), 856–863. doi: 10.1016/j.ajhg.2012.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krøigård AB, Jackson AP, Bicknell LS, Baple E, Brusgaard K, Hansen LK, & Ousager LB (2016). Two novel mutations in RNU4ATAC in two siblings with an atypical mild phenotype of microcephalic osteodysplastic primordial dwarfism type 1. Clin Dysmorphol, 25(2), 68–72. doi: 10.1097/MCD.0000000000000110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, … Maglott DR (2018). ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res, 46(D1), D1062–D1067. doi: 10.1093/nar/gkx1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Deignan JL, Dorrani N, Strom SP, Kantarci S, Quintero-Rivera F, … Nelson SF (2014). Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA, 312(18), 1880–1887. doi: 10.1001/jama.2014.14604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman A, Thouta S, Mancini GMS, Naidu S, van Slegtenhorst M, McWalter K, … Study, E. (2017). Loss-of-Function and Gain-of-Function Mutations in KCNQ5 Cause Intellectual Disability or Epileptic Encephalopathy. Am J Hum Genet, 101(1), 65–74. doi: 10.1016/j.ajhg.2017.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, … Consortium, E. A. (2016). Analysis of protein-coding genetic variation in 60,706 humans. Nature, 536(7616), 285–291. doi: 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, & Durbin R (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 25(14), 1754–1760. doi: 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour TA, Lucot K, Konopelski SE, Dickinson PJ, Sturges BK, Vernau KL, … Bannasch DL (2018). Whole genome variant association across 100 dogs identifies a frame shift mutation in DISHEVELLED 2 which contributes to Robinow-like syndrome in Bulldogs and related screw tail dog breeds. PLoS Genet, 14(12), e1007850. doi: 10.1371/journal.pgen.1007850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez FJ, Lee JH, Lee JE, Blanco S, Nickerson E, Gabriel S, … Gleeson JG (2012). Whole exome sequencing identifies a splicing mutation in NSUN2 as a cause of a Dubowitz-like syndrome. J Med Genet, 49(6), 380–385. doi: 10.1136/jmedgenet-2011-100686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, … Cunningham F (2016). The Ensembl Variant Effect Predictor. Genome Biol, 17(1), 122. doi: 10.1186/s13059-016-0974-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke LA, van Belzen MJ, Alders M, Cristofoli F, Ehmke N, Fergelot P, … Study, D. (2016). CREBBP mutations in individuals without Rubinstein-Taybi syndrome phenotype. Am J Med Genet A, 170(10), 2681–2693. doi: 10.1002/ajmg.a.37800 [DOI] [PubMed] [Google Scholar]
- Monies D, Abouelhoda M, AlSayed M, Alhassnan Z, Alotaibi M, Kayyali H, … Alkuraya FS (2017). The landscape of genetic diseases in Saudi Arabia based on the first 1000 diagnostic panels and exomes. Hum Genet, 136(8), 921–939. doi: 10.1007/s00439-017-1821-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungall CJ, McMurry JA, Köhler S, Balhoff JP, Borromeo C, Brush M, … Haendel MA (2017). The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species. Nucleic Acids Res, 45(D1), D712–D722. doi: 10.1093/nar/gkw1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, McMillin MJ, Gildersleeve HI, … Shendure J (2010). Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet, 42(9), 790–793. doi: 10.1038/ng.646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell-Luria A, Blue E, England E, Lopez-Giraldez F, Sobreira N, Akdemir Z, … Nickerson D (2018). Cohort analysis reveals diverse genetic etiology underlying Dubowitz syndrome. In (4 ed., Vol. 179, pp. 693–694). American Journal of Medical Genetics A Wiley. [Google Scholar]
- O’Driscoll M, Cerosaletti KM, Girard PM, Dai Y, Stumm M, Kysela B, … Concannon P (2001). DNA ligase IV mutations identified in patients exhibiting developmental delay and immunodeficiency. Mol Cell, 8(6), 1175–1185. [DOI] [PubMed] [Google Scholar]
- Opitz JM, Pfeiffer RA, Hermann JP, & Kushnick T (1973). Studies of malformation syndromes of man XXIV B: the Dubowitz syndrome. Further observations. Z Kinderheilkd, 116(1), 1–12. [DOI] [PubMed] [Google Scholar]
- Ounap K, Justus I, & Lipping-Sitska M (1998). Two sisters with growth failure, microcephaly, peculiar facies and apical dystrophy: the presentation of brachymorphism-onychodysplasia-dysphalangism syndrome? Clin Dysmorphol, 7(1), 45–50. [PubMed] [Google Scholar]
- Paila U, Chapman BA, Kirchner R, & Quinlan AR (2013). GEMINI: integrative exploration of genetic variation and genome annotations. PLoS Comput Biol, 9(7), e1003153. doi: 10.1371/journal.pcbi.1003153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulraj P, Bosworth M, Longhurst M, Hornbuckle C, Gotway G, Lamb AN, & Andersen EF (2019). A Novel Homozygous Deletion within the FRY Gene Associated with Nonsyndromic Developmental Delay. Cytogenet Genome Res, 159(1), 19–25. doi: 10.1159/000502598 [DOI] [PubMed] [Google Scholar]
- Pedersen BS, & Quinlan AR (2017). Who’s Who? Detecting and Resolving Sample Anomalies in Human DNA Sequencing Studies with Peddy. Am J Hum Genet, 100(3), 406–413. doi: 10.1016/j.ajhg.2017.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippakis AA, Azzariti DR, Beltran S, Brookes AJ, Brownstein CA, Brudno M, … Rehm HL (2015). The Matchmaker Exchange: a platform for rare disease gene discovery. Hum Mutat, 36(10), 915–921. doi: 10.1002/humu.22858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posey JE, O’Donnell-Luria AH, Chong JX, Harel T, Jhangiani SN, Coban Akdemir ZH, … Genomics, C. f. M. (2019). Insights into genetics, human biology and disease gleaned from family based genomic studies. Genet Med, 21(4), 798–812. doi: 10.1038/s41436-018-0408-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retterer K, Juusola J, Cho MT, Vitazka P, Millan F, Gibellini F, … Bale S (2016). Clinical application of whole-exome sequencing across clinical indications. Genet Med, 18(7), 696–704. doi: 10.1038/gim.2015.148 [DOI] [PubMed] [Google Scholar]
- Riazuddin S, Hussain M, Razzaq A, Iqbal Z, Shahzad M, Polla DL, … UK10K. (2017). Exome sequencing of Pakistani consanguineous families identifies 30 novel candidate genes for recessive intellectual disability. Mol Psychiatry, 22(11), 1604–1614. doi: 10.1038/mp.2016.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer O, & Spelger G (1977). [Dubowitz syndrome with immunodeficiency and solid malignant tumor in two siblings (author’s transl)]. Monatsschr Kinderheilkd, 125(10), 885–887. [PubMed] [Google Scholar]
- Sawyer SL, Tian L, Kähkönen M, Schwartzentruber J, Kircher M, Majewski J, … Consortium, F. C. (2015). Biallelic mutations in BRCA1 cause a new Fanconi anemia subtype. Cancer Discov, 5(2), 135–142. doi: 10.1158/2159-8290.CD-14-1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobreira N, Schiettecatte F, Valle D, & Hamosh A (2015). GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat, 36(10), 928–930. doi: 10.1002/humu.22844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobreira NLM, Arachchi H, Buske OJ, Chong JX, Hutton B, Foreman J, … Consortium, M. E. (2017). Matchmaker Exchange. Curr Protoc Hum Genet, 95, 9.31.31–39.31.15. doi: 10.1002/cphg.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srour M, Schwartzentruber J, Hamdan FF, Ospina LH, Patry L, Labuda D, … Consortium, F. C. (2012). Mutations in C5ORF42 cause Joubert syndrome in the French Canadian population. Am J Hum Genet, 90(4), 693–700. doi: 10.1016/j.ajhg.2012.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stessman HAF, Willemsen MH, Fenckova M, Penn O, Hoischen A, Xiong B, … Kleefstra T (2016). Disruption of POGZ Is Associated with Intellectual Disability and Autism Spectrum Disorders. Am J Hum Genet, 98(3), 541–552. doi: 10.1016/j.ajhg.2016.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart DR, Pemov A, Johnston JJ, Sapp JC, Yeager M, He J, … Savage SA (2014). Dubowitz syndrome is a complex comprised of multiple, genetically distinct and phenotypically overlapping disorders. PLoS One, 9(6), e98686. doi: 10.1371/journal.pone.0098686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz KR, Resnick D, Iskandar BJ, Wargowski D, Brockmeyer D, & Opitz J (2003). Craniocervical anomalies in Dubowitz syndrome. Three cases and a literature review. Pediatr Neurosurg, 38(5), 238–243. doi:69822 [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, … Mering CV (2019). STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res, 47(D1), D607–D613. doi: 10.1093/nar/gky1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, & Mesirov JP (2013). Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform, 14(2), 178–192. doi: 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukahara M, & Opitz JM (1996). Dubowitz syndrome: review of 141 cases including 36 previously unreported patients. Am J Med Genet, 63(1), 277–289. doi: [DOI] [PubMed] [Google Scholar]
- van der Sluijs PJ, Jansen S, Vergano SA, Adachi-Fukuda M, Alanay Y, AlKindy A, … Santen GWE (2019). The ARID1B spectrum in 143 patients: from nonsyndromic intellectual disability to Coffin-Siris syndrome. Genet Med, 21(6), 1295–1307. doi: 10.1038/s41436-018-0330-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetro A, Iascone M, Limongelli I, Ameziane N, Gana S, Della Mina E, … Zuffardi O (2015). Loss-of-Function FANCL Mutations Associate with Severe Fanconi Anemia Overlapping the VACTERL Association. Hum Mutat, 36(5), 562–568. doi: 10.1002/humu.22784 [DOI] [PubMed] [Google Scholar]
- Walter K, Min JL, Huang J, Crooks L, Memari Y, McCarthy S, … Consortium, U. K. (2015). The UK10K project identifies rare variants in health and disease. Nature, 526(7571), 82–90. doi: 10.1038/nature14962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters TR, & Desposito F (1985). Aplastic anemia in Dubowitz syndrome. J Pediatr, 106(4), 622–623. [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, & Hakonarson H (2010). ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res, 38(16), e164. doi: 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J, Mazzeu JF, Hoischen A, Jhangiani SN, Gambin T, Alcino MC, … Genomics, B.-H. C. f. M. (2015). DVL1 frameshift mutations clustering in the penultimate exon cause autosomal-dominant Robinow syndrome. Am J Hum Genet, 96(4), 612–622. doi: 10.1016/j.ajhg.2015.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JJ, Mazzeu JF, Hoischen A, Bayram Y, Withers M, Gezdirici A, … Genomics, B.-H. C. f. M. (2016). DVL3 Alleles Resulting in a −1 Frameshift of the Last Exon Mediate Autosomal-Dominant Robinow Syndrome. Am J Hum Genet, 98(3), 553–561. doi: 10.1016/j.ajhg.2016.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Muzny DM, Xia F, Niu Z, Person R, Ding Y, … Eng CM (2014). Molecular findings among patients referred for clinical whole-exome sequencing. JAMA, 312(18), 1870–1879. doi: 10.1001/jama.2014.14601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yavarna T, Al-Dewik N, Al-Mureikhi M, Ali R, Al-Mesaifri F, Mahmoud L, … Ben-Omran T (2015). High diagnostic yield of clinical exome sequencing in Middle Eastern patients with Mendelian disorders. Hum Genet. doi: 10.1007/s00439-015-1575-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. Dubowitz cohort detailed phenotype data
Supplemental Table 2. Variants of unknown clinical significance (VUS) and variants in genes of unknown significance (GUS) seen in individuals with a clinical diagnosis of Dubowitz syndrome
Supplemental Table 3. The complete list of 37 GO:Biological Processes with significant evidence for enrichment among candidate genes, genes offering alternative diagnoses, and DubS genes from the literature.
Data Availability Statement
Data that support the findings of this study are available on request from the corresponding authors (EB/AMI). The data are not publicly available due to privacy or ethical restrictions of the research participants. A subset of participants has provided consent for sharing in a controlled access repository (dbGaP, AnVIL). This information will be shared within this controlled setting and with appropriate permission from respective co-author(s).