Abstract
Mutations on γ-secretase subunits are associated with neurologic diseases. Whereas the role of γ-secretase in neurogenesis has been intensively studied, little is known about its role in astrogliogenesis. Recent evidence has demonstrated that astrocytes can be generated from oligodendrocyte precursor cells (OPCs). However, it is not well understood what mechanism may control OPCs to differentiate into astrocytes. To address the above questions, we generated two independent lines of oligodendrocyte lineage-specific presenilin enhancer 2 (Pen-2) conditional KO mice. Both male and female mice were used. Here we demonstrate that conditional inactivation of Pen-2 mediated by Olig1-Cre or NG2-CreERT2 causes enhanced generation of astrocytes. Lineage-tracing experiments indicate that abnormally generated astrocytes are derived from Cre-expressing OPCs in the CNS in Pen-2 conditional KO mice. Mechanistic analysis reveals that deletion of Pen-2 inhibits the Notch signaling to upregulate signal transducer and activator of transcription 3, which triggers activation of GFAP to promote astrocyte differentiation. Together, these novel findings indicate that Pen-2 regulates the specification of astrocytes from OPCs through the signal transducer and activator of transcription 3 signaling.
SIGNIFICANCE STATEMENT Astrocytes and oligodendrocyte (OLs) play critical roles in the brain. Recent evidence has demonstrated that astrocytes can be generated from OL precursor cells (OPCs). However, it remains poorly understood what mechanism governs the differentiation of OPCs into astrocytes. In this study, we took advantage of OL lineage cells specific presenilin enhancer 2 (Pen-2) conditional KO mice. We show that deletion of Pen-2 leads to dramatically enhanced astrocyte differentiation from OPCs in the CNS. Mechanistic analysis reveals that deletion of Pen-2 inhibits Hes1 and activates signal transducer and activator of transcription 3 to trigger GFAP activation which promotes astrocyte differentiation. Overall, this study identifies a novel function of Pen-2 in astrogliogenesis from OPCs.
Keywords: astrocytes, astrogliogenesis, oligodendrocyte differentiation, oligodendrocyte precursor cells, Presenilin enhancer 2
Introduction
As major constituents in the CNS, oligodendrocytes (OLs) and astrocytes are essential for a variety of brain functions (Rowitch and Kriegstein, 2010; McKenzie et al., 2014; Li and Richardson, 2016; Elbaz and Popko, 2019). Whereas OLs are myelin-producing cells (Nave and Trapp, 2008; Bechler et al., 2015; Figlia et al., 2018; Liu et al., 2018), astrocytes play multiple roles in various biological processes, including the formation of the brain–blood barrier and inflammatory responses (Freeman, 2010; Molofsky et al., 2012). Although astrocytes are believed to be generated from neural progenitor cells (NPCs) (Freeman, 2010; Namihira and Nakashima, 2013), the following evidence has shown that astrocytes can also be generated from OPCs. First, glial progenitor cells give rise to both OLs and astrocytes (Raff et al., 1983; Kondo and Raff, 2000). Second, OPCs are found to generate OLs and astrocytes in the CNS (Belachew et al., 2003; Cai et al., 2007; Zhu et al., 2012; Huang et al., 2014; Zuo et al., 2018; Sun et al., 2019). However, little is known about the molecular mechanism that controls OPCs to differentiate into astrocytes during postnatal CNS development.
γ-Secretase is a key protease for Notch receptors and amyloid precursor protein (APP) (De Strooper, 2003). It is composed of four subunits, including presenilin, presenilin enhancer 2 (Pen-2), nicastrin, and anterior pharynx defective 1 (De Strooper, 2003; Kimberly et al., 2003). Recent cryo-electron microscopy studies uncover structural basis for the recognition of γ-secretase to Notch or to APP (Yang et al., 2019; Zhou et al., 2019). The role of γ-secretase in developing and adult brains has been intensively studied (Saura et al., 2004; Shen and Kelleher, 2007; Kim and Shen, 2008; Tabuchi et al., 2009; Dries et al., 2016; Hou et al., 2016; Acx et al., 2017; Liu et al., 2017; Cheng et al., 2019). However, it remains unknown whether γ-secretase in OPCs is important for astrogliogenesis.
To address the above questions, we generated two lines of OL lineage-specific Pen-2 conditional KO (cKO) mice, Pen-2f/f;Olig1-Cre and Pen-2f/f;NG2-CreERT2. We find that conditional inactivation of Pen-2 leads to increased number of astrocytes but unchanged number of neurons in the CNS. Lineage-tracing analyses reveal that GFAP+ cells are derived from Cre-expressing OPCs in Pen-2 cKO mice. We demonstrate that deletion of Pen-2 causes increased levels of signal transducer and activator of transcription 3 (Stat3) in OPCs, and that elevated Stat3 promotes astrocyte differentiation via GFAP activation. Overall, the above observations highlight an essential role of γ-secretase in astrogliogenesis from OPCs during postnatal CNS development.
Materials and Methods
Animals
The generation of Pen-2f/f mice was reported previously (Cheng et al., 2019; Bi et al., 2021). To generate Pen-2 cKO mice (Pen-2f/f;Olig1-Cre), we bred Olig1-Cre mutant (Xin et al., 2005; Wang et al., 2021) with Pen-2f/f to obtain Pen-2f/+;Olig1-Cre. The latter were then crossed to Pen-2f/f to get Pen-2 cKO. Pen-2f/+;Olig1-Cre served as littermate controls. We generated a reporter line expressing LoxP-Stop-LoxP-tdTomato, which was referred to as LSL-tdTomato hereafter and was used to produce Pen-2f/+;Olig1-Cre;LSL-tdTomato, Pen-2f/f;Olig1-Cre;LSL-tdTomato, and Olig1-Cre;LSL-tdTomato mice. The mTmG line (Muzumdar et al., 2007) was used to generate Pen-2f/+;Olig1-Cre;mTmG and Pen-2f/f;Olig1-Cre;mTmG mice. The hGFAP-GFP was used to generate Pen-2f/f;Olig1-Cre;hGFAP-GFP mice. We also generated inducible Pen-2 cKO (Pen-2 icKO) mice by crossing NG2-CreERT2 (Huang et al., 2014) to Pen-2f/f;LSL-tdTomato. Pen-2f/f;NG2-CreERT2;LSL-tdTomato mice served as Pen-2 icKO after receiving tamoxifen from their lactating mothers (Huang et al., 2014).
Both male and female mice were used in this study. The genetic background of the mice was C57BL/6. The animals were group-housed (4 or 5 per cage) throughout the experimental period and had ad libitum access to food and water. The mice were maintained in an SPF-leveled animal room in the core facility of the Model Animal Research Center (MARC) at Nanjing University. The light-cycle of the animal room was automatically controlled. The animal room was maintained under constant humidity and temperature (25 ± 1°C). Mouse breeding was conducted under Institutional Animal Care and Use Committee approved protocols at the MARC. All the experiments were performed in accordance with the Guide for the Care and Use of Laboratory Animals of the MARC at Nanjing University.
Nissl staining
The mice were killed with CO2, perfused with PBS, fixed in 4% PFA overnight at 4°C. Sections were deparaffinized, ethanol rehydrated, and treated with 0.1% cresyl violet for 1 min followed by rinsing with water. Sections were sealed using neutral resin.
Cell counting
Counting on GFAP+, Pdgfrα+, Olig2+, CC1+, Aldh1l1+, S100β+, NeuN+, or Iba1+ cells was conducted using three brain sections per mouse spaced 400 µm apart. For each section, 2 microscopic fields (20× objective lens of an Olympus BX53 microscope) were randomly selected for the cortex or the thalamus. Images for each field (438.6 µm × 330.2 µm) were captured and the total number of cells was counted using ImageJ. The serial numbers were averaged across sections, and the mean was presented as the averaged cell number in each microscopic field for each animal.
Measurement on fluorescence intensity of Olig2 and Sox10
To compare the expression of Olig2 or Sox10 in GFAP+ and GFAP– cells, we analyzed fluorescence intensity of Olig2 or Sox10 using a method described recently (Kelenis et al., 2018). Briefly, images for Olig2+/GFAP+ or Olig2+/Sox10+/GFAP+ cells were captured under the 40× objective lens with a Carl Zeiss LSM880 confocal microscope. Olig2+/GFAP+ or Olig2+/Sox10+/GFAP+ cells were randomly chosen for analysis, and adjacent Olig2+/GFAP– or Olig2+/Sox10+/GFAP– cells were used for comparison. A total of 24 cells for each type from 3 Pen-2 cKO mice at P12 were used for quantification. ImageJ was used to draw an outline around the Olig2 signal in each cell. The integrated density of the Olig2+ or Sox10+ the area was analyzed, and that of the no-signal region was used as the background. Fluorescence intensity of Olig2 or Sox10 for each cell was calculated as the integrated density of the Olig2+ or Sox10+ area minus the background.
Brain lysate preparation
Mice were killed at P14 and P30. Cortical samples were prepared using a method described previously (Peng et al., 2010). Samples were stored at −80˚C until use.
Western blotting
We used a method described previously (Liu et al., 2017). Briefly, normalized volumes for cortical protein samples were loaded onto 8%-15% polyacrylamide gels and separated by electrophoresis for ∼2 h. The gels were transferred into nitrocellulose membrane at 25 V for 2.5 h. After blocking with 5% (w/v) nonfat dry milk for 1 h, membranes were probed with primary antibodies overnight and detected using infrared dye-coupled secondary antibodies (goat anti-rabbit IRdye800, goat anti-rabbit IRdye680, goat anti-mouse IRdye800, and goat anti-mouse IRdye680). Information for primary antibodies was provided in Table 1.
Table 1.
Antibody list
| Antibodies | Source | Identifier |
|---|---|---|
| Mouse anti-Olig2 | Millipore | Catalog #MABN50, RRID:AB_10807410 |
| Rabbit anti-Pdgfrα | Cell Signaling Technology | Catalog #3174, RRID:AB_2162345 |
| Rat anti-BrdU | Abcam | Catalog #ab6326, RRID:AB_305426 |
| Rabbit anti-GFAP | Abcam | Catalog #ab7260, RRID:AB_305808 |
| Mouse anti-Aldh1l1 | Abcam | Catalog #ab 56777, RRID:AB_940204 |
| Rabbit anti-S100β | Abcam | Catalog #ab52642, RRID:AB_882426 |
| Rabbit anti-NeuN | Millipore | Catalog #ABN78, RRID:AB_10807945 |
| Rabbit anti-APP | Sigma-Aldrich | Catalog #A8717, RRID:AB_258409 |
| Rabbit anti-Iba1 | Wako | Catalog #019-19741, RRID:AB_839504 |
| Mouse anti-Stat3 | Cell Signaling Technology | Catalog #9139, RRID:AB_331757 |
| Rabbit anti-Pen-2 | Abclonal | Catalog #A15172, RRID:AB_2762062 |
| Mouse anti-Hdac3 | Cell Signaling Technology | Catalog #3949, RRID:AB_2118371 |
| Rat anti-Mbp | Millipore | Catalog #MAB386, RRID:AB_94975 |
| Rat anti-Plp1 | Millipore | Catalog #MAB388, RRID:AB_177623 |
| Mouse anti-CC1 | Calbiochem | Catalog #OP80, RRID:AB_2057371 |
| Rabbit anti-cleaved caspase 3 | Cell Signaling Technology | Catalog #9661, RRID:AB_2341188 |
| Rabbit anti-phosphorylated- Stat3 (Tyr705) |
Cell Signaling Technology | Catalog #9145, RRID:AB_2491009 |
| Mouse anti-GFAP | Santa Cruz Biotechnology | Catalog #sc-6534, RRID:AB_783553 |
| Goat anti-GFAP | Abcam | Catalog #ab53554, RRID:AB_880202 |
| Rabbit anti-Sox10 | Abcam | Catalog #ab155279, RRID:AB_2650603 |
| Mouse anti-GAPDH | Abcam | Catalog #ab8245, RRID:AB_2107448 |
| Mouse anti-β-Actin | Sigma-Aldrich | Catalog #A1978, RRID:AB_476692 |
Immunohistochemistry (IHC)
Brain sections were deparaffinized, ethanol rehydrated, and then boiled in 0.01 M pH 6.0 sodium citrate buffer solution. The sections were blocked using hydrogen peroxide (30% H2O2 diluted in methanol at 10:1) and then incubated with BSA (5% BSA in PBS). They were incubated overnight at 4°C with primary antibodies. Antibody information was provided in Table 1. Sections were incubated with biotin-labeled secondary antibodies (diluted in PBS), and were treated using the avidin-biotin peroxidase complex (ABC kit, Vector PK-6100), developed by DAB (Vector SK-4100). For fluorescence immunostaining, brain sections were incubated with either AlexaFluor-488 goat anti-mouse/anti-rabbit or AlexaFluor-594 goat anti-mouse/anti-rabbit secondary antibodies (Invitrogen). Images were captured and then analyzed using a Carl Zeiss LSM880 confocal microscope.
Cell culture and transfection
HEK293T cells were maintained in DMEM (Invitrogen) containing 10% FBS (Lonsera). The cells were transfected using polyethyleneimine. The medium was replaced 6 h after transfection, and the cells were harvested at 48 h after transfection.
Purification of Pen-2 KO cells by FACS
The cortex was dissected from control (Pen-2f/+;Olig1-Cre;mTmG) and Pen2 cKO (Pen-2f/f;Olig1-Cre;mTmG) mice. Cortical samples were digested by trypsin and became single-cell suspension containing Cre+ cells expressing green fluorescence protein and Cre– cells expressing red fluorescence protein. Cell suspensions were used by FACS (BD FACS AriaIII) to purify GFP+ cells. The sorting efficiency was larger than 90% for each sample. The resultant cell suspensions were centrifuged (3000 rpm) to discard the supernatants. Cells were treated using TRIzol reagent (Invitrogen) for RNA purification.
OPC and OL cultures
We used a method described previously (Parras et al., 2004). Briefly, cells from cortices from P8 mice were plated in neurosphere medium (DMEM/F12 containing 2 mm L-glutamine, 1× B27, 1× N2, 5 μm HEPES, 0.01% heparin, 100 µg/ml penicillin, 0.1 mg/ml streptomycin) supplemented with 20 ng/ml EGF and 20 ng/ml FGFb. Primary neurospheres were cultured for 3-5 d. Next, the culture medium was changed to DMEM/F12 supplemented with 20 ng/ml PDGF-AA and 10 ng/m FGFb to induce OPC differentiation. The medium was changed once every 4 d. Twelve days after the OPC culture, 10% FBS and 15 nm T3 were added into the culture medium to induce OL differentiation. OLs were further cultured for 4-8 d.
RNA isolation
Total RNA was isolated from control and Pen-2 cKO cortices using TRIzol reagent (Invitrogen).
TUNEL staining
Brain sections were blocked with 5% goat serum for 30 min and then treated with the TUNEL BrightGreen Apoptosis Detection Kit (Vazyme) at 37°C for 1 h.
BrdU pulse-labeling
BrdU (B5002, Sigma-Aldrich) was administered at the concentration of 100 mg/kg. To label proliferating OPCs, BrdU was intraperitoneally injected into mice. Brains were collected 1 h after the injection.
Tamoxifen administration
Tamoxifen (T5648, Sigma-Aldrich) was dissolved in corn oil (C116025, Aladdin), and the concentration was 10 mg/ml. The lactating mothers for Pen-2f/+;NG2-CreERT2;LSL-tdTomato and Pen-2f/f;NG2-CreERT2;LSL-tdTomato mice were intraperitoneally injected with tamoxifen once a day, at the dosage of 100 mg/kg body weight (Huang et al., 2014). The injection lasted 4 d from P1 to P4.
qRT-PCR
Total RNA (1 µg) was reverse-transcribed using PrimeScript RT reagent Kit (Takara). Real-time PCR was performed using the ABI StepOne Plus system. Primer information was provided in Table 2.
Table 2.
List for primers
| 1. Primers for qPCR | ||
|---|---|---|
| Gene name | Species | Sequence 5′-3′ |
| Pen-2 | Mouse gDNA F | GAACTTGGAGCGGGTATCCA |
| Pen-2 | Mouse gDNA R | AGAGGTAGTCCCCAAGAGCG |
| Hes1 | Mouse gDNA F | CAACACGACACCGGACAAAC |
| Hes1 | Mouse gDNA R | CGGAGGTGCTTCACAGTCAT |
| GFAP | Mouse gDNA F | GCAAGAGACAGAGGAGTGGT |
| GFAP | Mouse gDNA R | CTCTTCCTGTTCGCGCATTT |
| Stat3 | Mouse gDNA F | CAGCTGGACACACGCTACCT |
| Stat3 | Mouse gDNA R | GGACTCTTGCAGGAATCGGCT |
| Nfia | Mouse gDNA F | TCTTGTGAAACGAAACGCCC |
| Nfia | Mouse gDNA R | GCCCACAGTGATTCTCAACC |
| Hdac3 | Mouse gDNA F | ATCGCCTGGCATTGACTCAT |
| Hdac3 | Mouse gDNA R | TGTAACGGGAGCAGAACTCG |
| 2. Primers for recombinant DNA | ||
| Gene name | amino acids | Sequence 5′-3′ |
| mNotch1 ICD-F | 1744-2531 | GTGCTGCTGTCCCGCAAGCGC |
| mNotch1 ICD-R | TTTAAATGCCTCTGGAATGTG | |
| mErbB4 ICD-F | 636-1292 | TACCCATGGACGGGCCATTC |
| mErbB4 ICD-R | TCACACCACAGTATTCCGGT | |
| mAPP ICD-F | 722-770 | ATGTTGAAGAAGAAACAGTA |
| mAPP ICD-R | TTAGTTCTGCATTTGCTCAA | |
| mTrkB ICD-F | 451-821 | CTCCTGCTCAAGTTGGCGAG |
| mTrkB ICD-R | CTAGCCTAGGATATCCAGGT | |
| mP75 ICD-F | 272-427 | TATATTGCTTTCAAGAGATG |
| mP75 ICD-R | TCACACAGGGGACGTGGCAG | |
| mLrp1 ICD-F | 4444-4545 | TGGTATAAGCGGCGAGTCCG |
| mLrp1 ICD-R | CTATGCCAAGGGATCTCCTA | |
| mDag ICD-F | 771-893 | ATCTGCTATCGCAAGAAGAG |
| mDag ICD-R | TTAAGGGGGAACATACGGAG | |
| mEphA4 ICD-F | 567-986 | TTTGTGATCAGCCGAAGACG |
| mEphA4 ICD-R | TCAGACAGGAACCATCCTGC | |
| mNG2 ICD-F | 2250-2327 | TACCTCCGCAAACGCAACAA |
| mNG2 ICD-R | TCACACCCAGTACTGGCCAT | |
| mCdh2 ICD-F | 744-906 | GTATGGATGAAACGGCGGGA |
| mCdh2 ICD-R | TCAGTCGTCACCACCGCCGT | |
| mHes1-F | 1-282 | ATGCCAGCTGATATAATGGA |
| mHes1-R | TCAGTTCCGCCACGGTCTCC | |
| mStat3-F | 1-722 | ATGGCTCAGTGGAACCAGCT |
| mStat3-R | TTATTTCCAAACTGCATCAA | |
| 3. Primers for luciferase assay | ||
| Promoter | Region | Sequence 5′-3′ |
| mStat3 promoter-F | −1839∼TSS∼261 | CGTAGTTAAGAGCACTGCTC |
| mStat3 promoter-R | TCCCGAGGGCCGCCTGGCCT | |
| mStat3 promoter fragment 2-F | −1338∼TSS∼261 | GCAGAGTTCATTCTCTATGA |
| mStat3 promoter fragment 2-R | TCCCGAGGGCCGCCTGGCCT | |
| mStat3 promoter fragment 3-F | −834∼TSS∼261 | AACTTGAACTTGAATTTTTA |
| mStat3 promoter fragment 3-R | TCCCGAGGGCCGCCTGGCCT | |
| mStat3 promoter fragment 4-F | −213∼TSS∼261 | TTTCAGCAGGACATTCCGCT |
| mStat3 promoter fragment 4-R | TCCCGAGGGCCGCCTGGCCT | |
| hGFAP promoter-F | −1702∼TSS∼−5 | CATATCCTGGTGTGGAGTAG |
| hGFAP promoter-R | GGGTGCCCCTGGCAACACCC | |
Plasmids
Mouse Hes1, ErbB4 ICD, APP ICD, TrkB ICD, P75 ICD, Lrp1 ICD, Dag ICD, EphA4 ICD, NG2 ICD, Cdh2 ICD, and Stat3 were amplified by PCR from cDNA libraries prepared from mouse brain. Notch1 ICD was purchased from Addgene (#26891). These cDNAs were subcloned into pCDNA5-HA plasmids. WT human Stat3 and inactive human Stat3 constructs were gifts from Prof. Xinyuan Fu. Primer information was included in Table 2.
Luciferase assay
Luciferase assay was performed using a method described recently (Sehgal et al., 2009; Ning et al., 2019). The mouse Stat3 promoter (from −1839 to 261 bp) was cloned from mouse NPCs. The human GFAP promoter (from −1702 to −5 bp) was cloned from DNAs of the hGFAP-Cre mouse. These constructs were inserted into the pGL3-luciferase vector (Promega). Hes1 was cotransfected with Stat3-Luc or hGFAP-Luc into HEK293T cells. Lipofectamine 2000 and 3000 (Invitrogen) was used for transfection. The cells were cultured for 48 h, and cell extracts were assayed for luciferase activity by the dual-luciferase reporter assay system (Promega). Primer information was included in Table 2.
Experimental design and statistical analysis
Data were presented as the mean ± SEM. Two-tailed Student's t test was performed to examine the difference between control and mutant mice. p < 0.05 was considered statistically significant.
Results
Generation and characterization of OL lineage-specific Pen-2 cKO mice
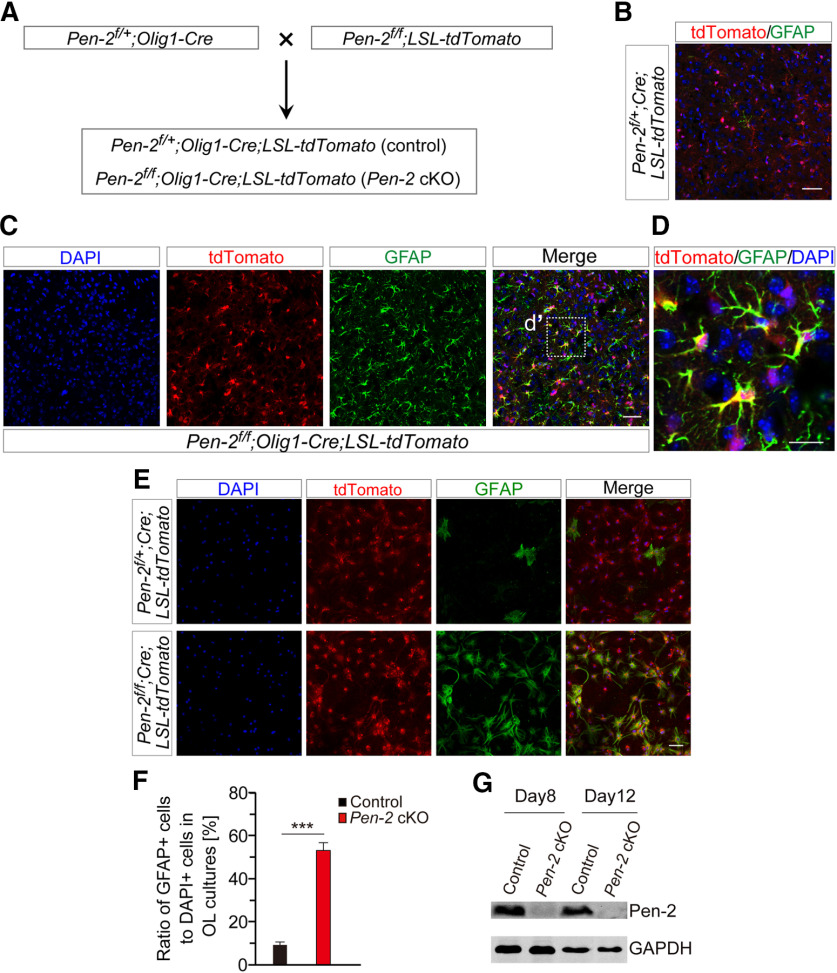
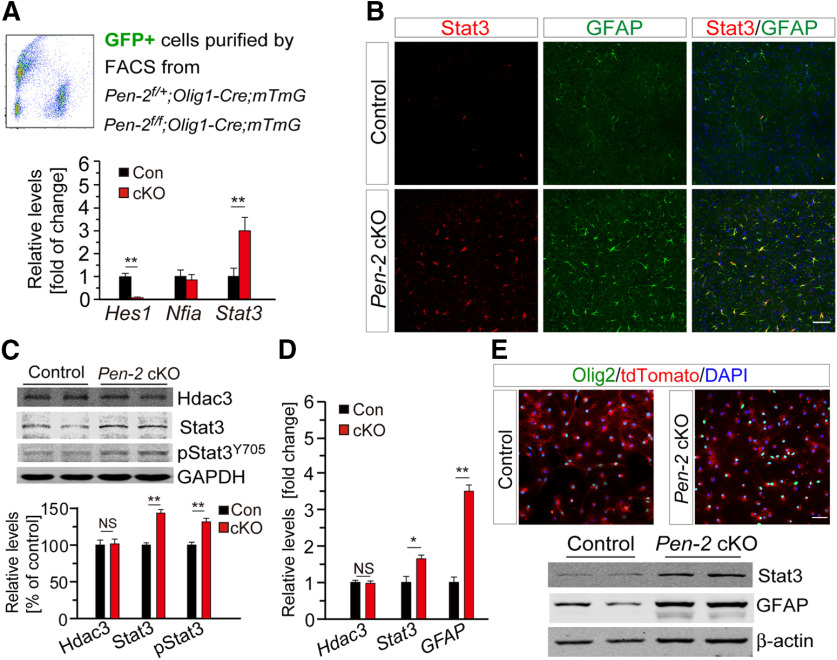
We generated OL lineage cells specific Pen-2 cKO mice through breeding Pen-2f/f with Pen-2f/+;Olig1-Cre (Fig. 1A). Pen-2 cKO mice were born in expected Mendelian ratios and survived to adulthood. Nissl staining showed comparable brain morphology between control and Pen-2 cKO mice at postnatal day 30 (P30) (Fig. 1B). To examine whether Pen-2 was inactivated efficiently, we obtained Pen-2f/+;Olig1-Cre;mTmG and Pen-2f/f;Olig1-Cre;mTmG mice by crossing the mTmG (Muzumdar et al., 2007) to Pen-2f/+;Olig1-Cre (Fig. 1C). RNA samples were prepared using GFP-expressing cells purified from cortical tissues in the above control and cKO mice by the FACS technique (Fig. 1D). qRT-PCR result did show extremely low levels of Pen-2 mRNA in Pen-2f/f;Olig1-Cre;mTmG compared with the control (Fig. 1E).
Figure 1.
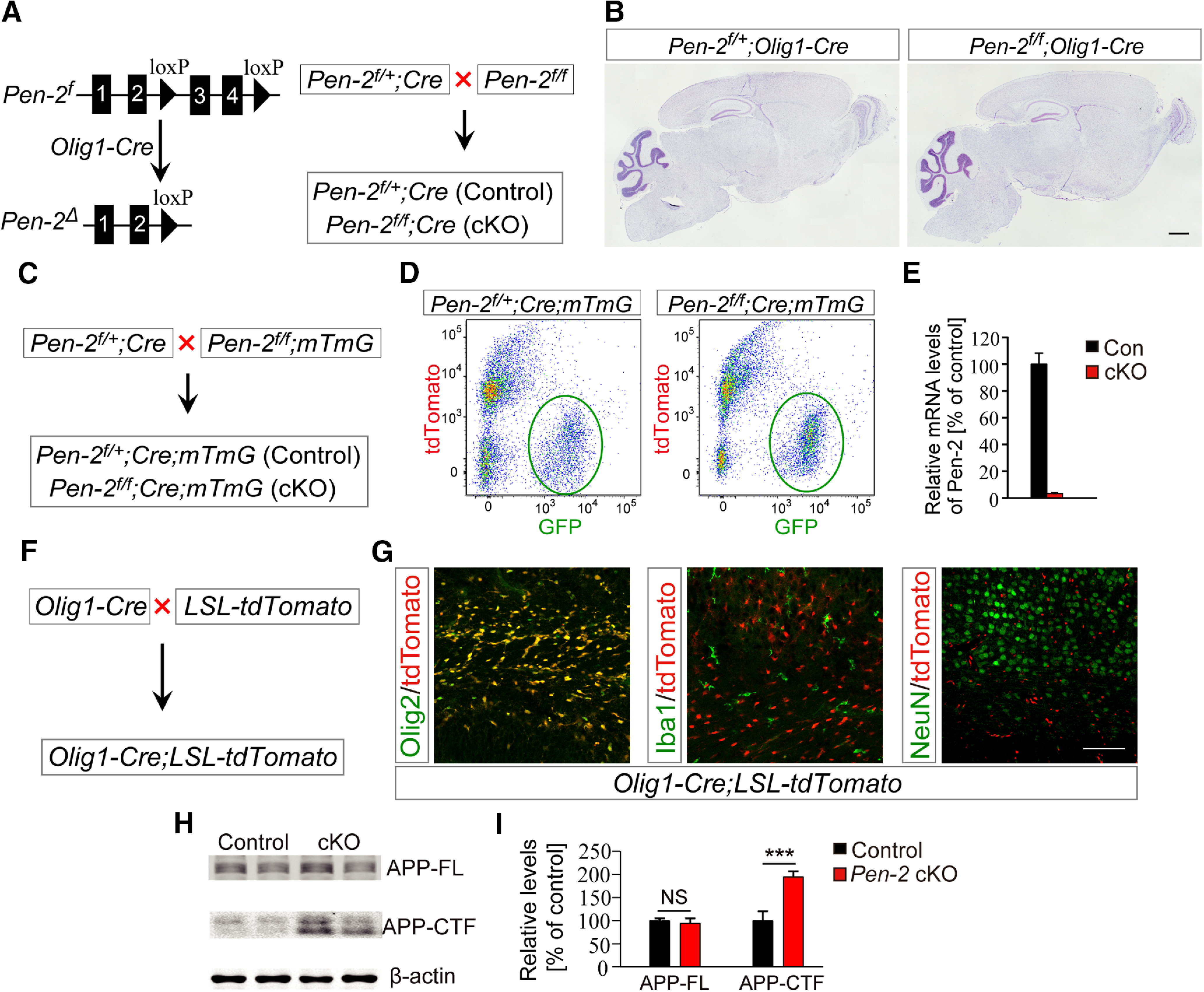
Generation and molecular characterization of OL lineage-specific Pen-2 cKO mice. A, Strategy for the generation of Pen-2 cKO mice. B, Nissl staining. There was no detectable change on brain morphology in Pen-2 cKO mice at P30 compared with controls. C, Breeding strategy for the generation of Pen-2 cKO mice expressing mTmG. D, Purification of GFP+ cells from control and Pen-2 cKO mice by FACS. Cortices from control (Pen-2f/+;Olig1-Cre;mTmG) and Pen-2 cKO (Pen-2f/f;Olig1-Cre;mTmG) mice at P11 were used. E, qRT-PCR analysis on Pen-2 mRNA in GFP+ cells collected from the above mice. There was a highly significant difference between two groups (Control: n = 3; Pen-2 cKO: n = 4). p < 0.001. F, G, Examination for Cre expression pattern using Olig1-Cre;LSL-tdTomato mice (F). TdTomato was costained with Olig2, but not Iba1 or NeuN, in the mutant at P2. H, I, Western analysis for APP-FL and APP-CTF. These two proteins were detected by Western blotting (H). There were significantly increased levels of APP-CTF but not APP-FL in Pen-2 cKO cortical samples compared with controls. ***p < 0.005. β-actin was used as the loading control (Control: n = 3; Pen-2 cKO: n = 4). Scale bars: B, 1 mm; G, 100 µm.
To visualize the expression pattern of Cre, we generated a reporter line expressing LoxP-Stop-LoxP-tdTomato, namely, Olig1-Cre;LSL-tdTomato (Fig. 1F). IHC results showed that tdTomato was costained with Olig2 but not Iba1 or NeuN in Cre-expressing cells in the corpus callosum in Olig1-Cre;LSL-tdTomato mice at P2 (Fig. 1G). Western analysis revealed that levels of full-length APP (APP-FL) were not changed but those for C-terminal fragment of APP (APP-CTF) were increased in Pen-2 cKO cortices compared with controls (Fig. 1H,I). Thus, conditional deletion of Pen-2 caused significantly decreased γ-secretase activity in Pen-2 cKO mice.
Excessive generation of astrocytes in the CNS in Pen-2 cKO mice
To examine astrocytes in Pen-2 cKO mice, we performed IHC on GFAP. We observed increased immunoreactivity of GFAP in the cortex and the thalamus of Pen-2 cKO mice at P14 and P30 compared with controls (Fig. 2A). Cell counting results confirmed significantly increased number of GFAP+ cells in the brain of Pen-2 cKO mice (Fig. 2B). Western blotting also showed significantly increased levels of GFAP in Pen-2 cKO cortices compared with controls (Fig. 2C). Next, we crossed the hGFAP-GFP mouse (Zhuo et al., 1997) to Pen-2 cKO to obtain Pen-2f/f;Olig1-Cre; hGFAP-GFP (Fig. 2D), in which abundant GFP+ cells were detected in the cortex and the thalamus compared with control mice (Pen-2f/+;Olig1-Cre;hGFAP-GFP) (Fig. 2E).
Figure 2.
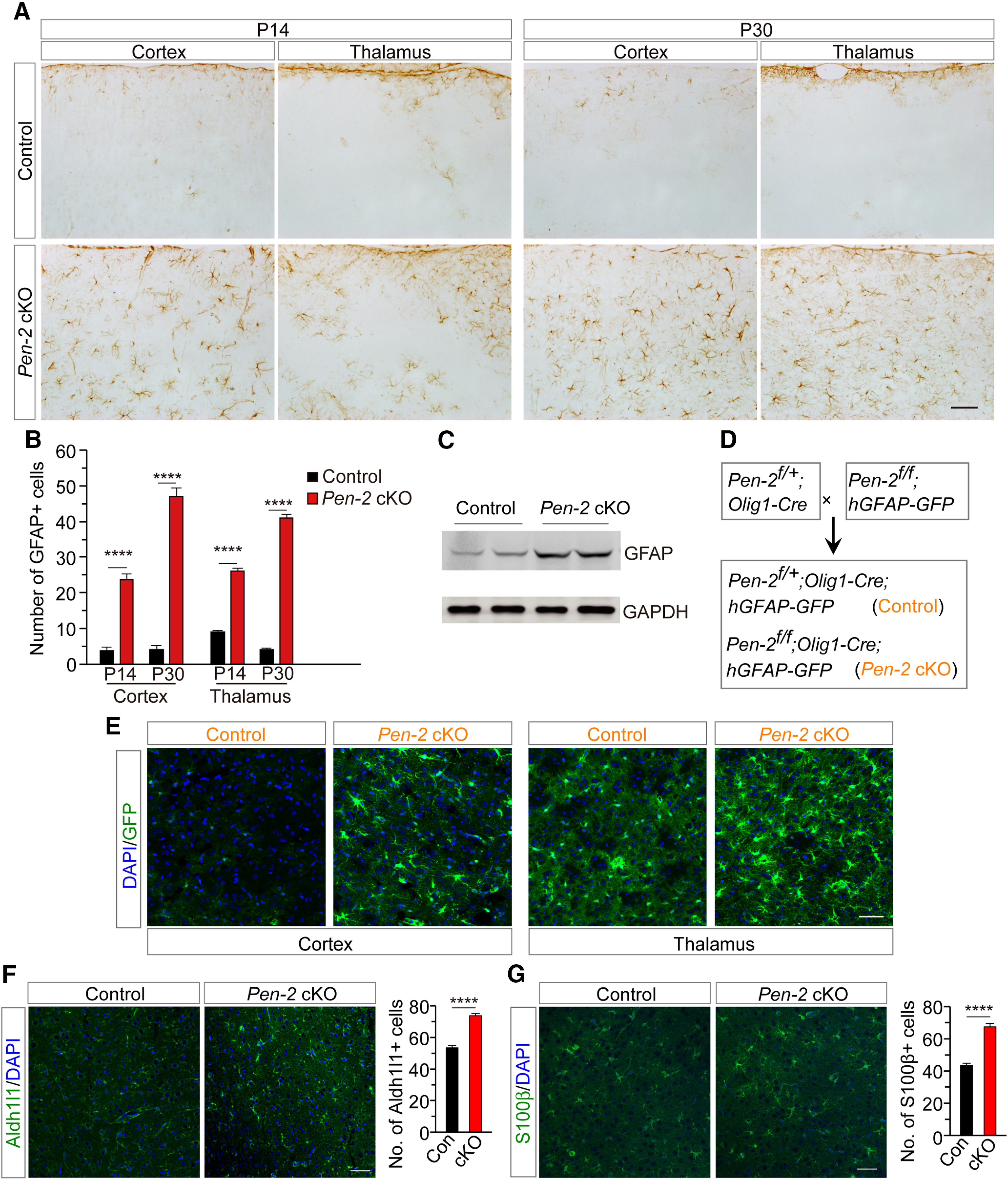
Enhanced generation of astrocytes in Pen-2 cKO mice. A, IHC on GFAP. There was increased immunoreactivity of GFAP in the cortex and the thalamus in Pen-2 cKO mice at P14 and P30 compared with age-matched controls. B, Averaged number of GFAP+ cells. There was a highly significant difference on the averaged number of GFAP+ cells between control and Pen-2 cKO mice at P14 and P30 (n = 6 mice per group). ****p < 0.001. C, Western blotting on GFAP. There was increased GFAP protein in the cortex of Pen-2 cKO mice compared with controls at P14 (Control: n = 3; Pen-2 cKO: n = 4). GAPDH served as the loading control. D, Generation of Pen-2 cKO mice expressing GFP in astrocytes. Pen-2f/+;Olig1-Cre; hGFAP-GFP (control) and Pen-2f/f;Olig1-Cre;hGFAP-GFP (Pen-2 cKO) mice were obtained. E, Abundant GFP+ cells were seen in the cortex and the thalamus in Pen-2 cKO mice at P14 compared with controls. F, Representative images for fluorescence IHC on Aldh1l1. There was a significant difference on the averaged number of Aldh1l1+ cells between control and Pen-2 cKO mice at P30 (n = 4 mice per group). **** p < 0.001. G, Representative images for fluorescence IHC on S100β. There was a significant difference on the averaged number of S100β+ cells between control and Pen-2 cKO mice at P30 (n = 4 mice per group). ****p < 0.001. Scale bars: A, 100 µm; E-G, 50 µm.
Moreover, we used Aldh1l1 and S100β as two additional markers of astrocyte to conduct IHC experiments. First, cell counting results revealed significantly increased number of Aldh1l1+ cells in Pen-2 cKO mice compared with controls at P30 (Fig. 2F). Second, the number of S100β+ cells was also significantly increased in Pen-2 cKO mice (Fig. 2G). The above observations were consistent with those for GFAP+ cells (Fig. 2A,B), confirming excessive generation of astrocytes in Pen-2 cKO mice.
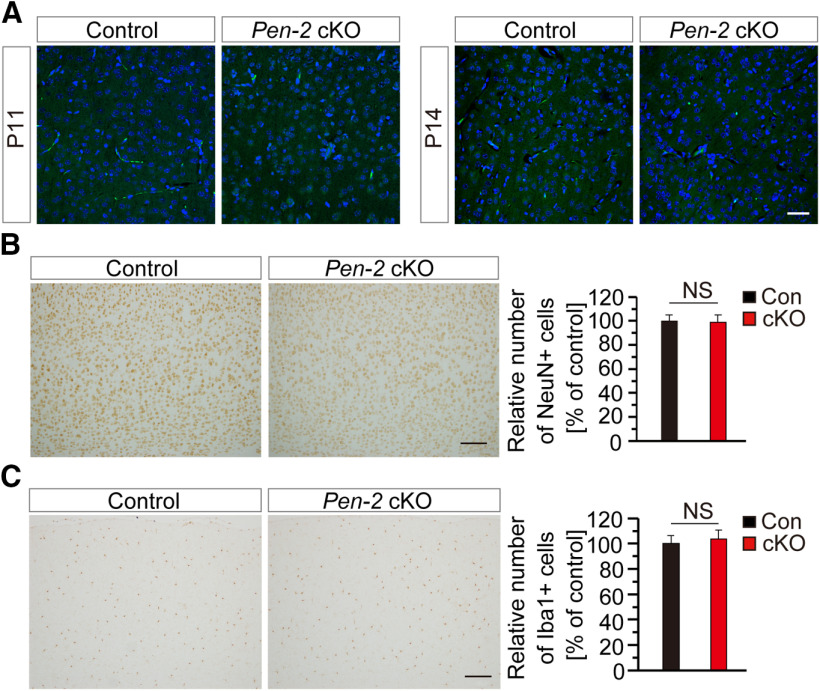
To study whether there was apoptosis in Pen-2 cKO mice, we performed TUNEL staining and IHC on cleaved caspase 3 (CC3) using brain sections at various ages. First, TUNEL+ cells were not observed in the brain of control and Pen-2 cKO mice at P11 (Fig. 3A), P14 (Fig. 3A), or P30 (data not shown). Second, CC3+ cells were not detected in control and Pen-2 cKO mice at any age tested above (data not shown). These findings suggest that deletion of Pen-2 in OPCs does not cause abnormal cell death.
Figure 3.
Unchanged apoptosis and neuroinflammatory responses in Pen-2 cKO mice. A, TUNEL staining. No TUNEL+ cells were observed in the cortex or the thalamus in control and Pen-2 cKO mice at P11 and P14. B, C, IHC on NeuN and Iba1. There was comparable immunoreactivity of NeuN (B) or Iba1 (C) between control and Pen-2 cKO mice. There was no significant difference on averaged number of NeuN+ (B) or Iba1+ (C) cells (in 1 mm2 area) in the cortex of control and Pen-2 cKO mice at P14 (n = 4 per group). Scale bars: A, 50 µm; B, C, 100 µm. NS, not significant.
We then examined whether neurons and microglia were affected in Pen-2 cKO mice. First, NeuN immunoreactivity was comparable between control and Pen-2 cKO mice at P30 (Fig. 3B). Cell counting confirmed no significant difference on the number of NeuN+ cells in the cortex between control and Pen-2 cKO mice (Fig. 3B), suggesting no detectable neuron loss in Pen-2 cKO mice. Western analysis revealed comparable levels for postsynaptic density protein 95 and synaptophysin between control and Pen-2 cKO mice (data not shown). Second, IHC on Iba1 was conducted. However, we did not observe significant change on the number of total Iba1+ cells in Pen-2 cKO cortices compared with controls at P14 (Fig. 3C). Thus, deletion of Pen-2 in OL lineages did not cause abnormal cell death or inflammatory responses.
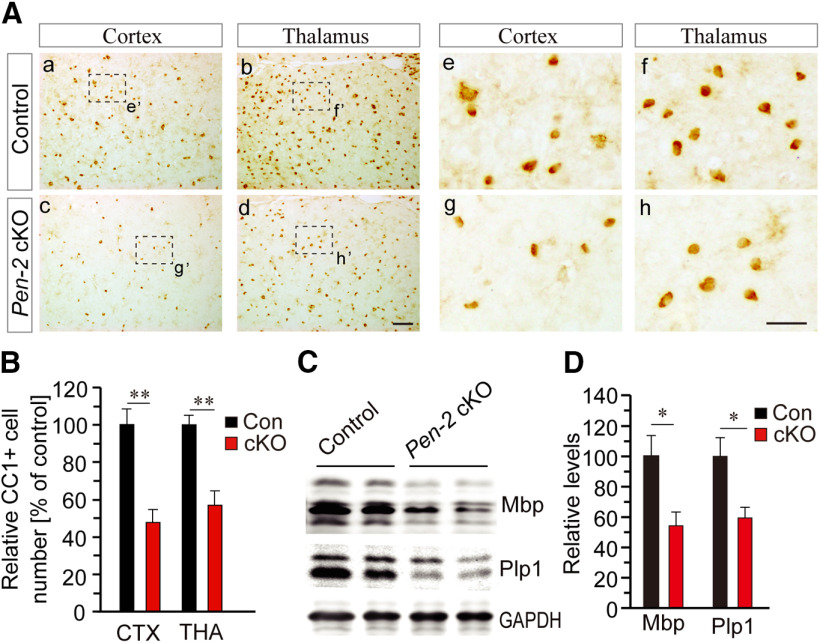
Reduced number of mature OLs and increased number of OPCs in the cortex of Pen-2 cKO mice
To examine whether loss of Pen-2 affected mature OLs, IHC on CC1 was performed (Fig. 4A). We found that the averaged number of CC1+ cells in the cortex and the thalamus was significantly decreased in Pen-2 cKO mice at P14 (Fig. 4B) and P30 (data not shown) compared with controls. Moreover, Western blotting on Mbp and Plp1 was conducted (Fig. 4C). We found that protein levels for these two molecules were significantly decreased in Pen-2 cKO cortices at P14 compared with controls (Fig. 4D). Thus, deletion of Pen-2 caused significant reduction on mature OLs.
Figure 4.
Decreased number of mature OLs in Pen-2 cKO mice. A, Representative images for CC1 IHC in the cortex and the thalamus at P30. The boxed area of e′, f′, g′, or h′ was enlarged as Ae, Af, Ag, or Ah. Scale bars: Aa-Ad, 50 µm; Ae-Ah, 10 µm. B, Relative number of CC1+ cells in 1 mm2 area. There were significant differences on the averaged number of CC1+ cells in the cortex and the thalamus between control and Pen-2 cKO mice at P14 (Control: n = 4; Pen-2 cKO: n = 5). **p < 0.01. C, Western blotting for Mbp and Plp1 at P14. D, Relative protein levels. There were significant differences on protein levels of Mbp and Plp1 between control and Pen-2 cKO mice at P14 (Control: n = 3 mice; Pen-2 cKO: n = 3 mice). *p < 0.05.
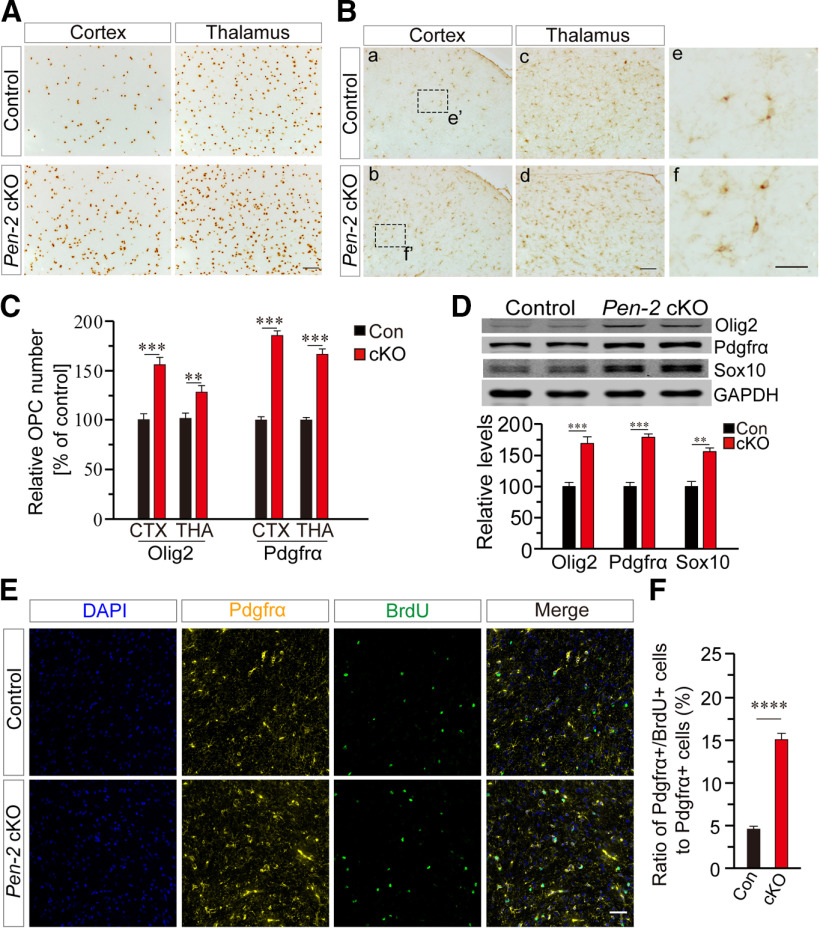
To examine whether OPCs were affected, IHC on Olig2 (Fig. 5A) and Pdgfrα (Fig. 5B) was conducted. We found that the number of Olig2+ cells was increased in the cortex and the thalamus of Pen-2 cKO mice at P14 compared with controls (Fig. 5C). The number of Pdgfrα+ cells was also significantly increased in the cortex and the thalamus in Pen-2 cKO mice at P14 (Fig. 5B,C). Western analysis showed elevated levels of Olig2, Pdgfrα, and Sox10 in Pen-2 cKO cortices at P14 compared with controls (Fig. 5D).
Figure 5.
Increased number of OPCs in Pen-2 cKO mice. A, IHC on Olig2. Images for the cortex and the thalamus at P14 are shown. B, IHC on Pdgfrα. Boxed areas in Ba and Bb were enlarged as Be and Bf, respectively. Scale bars: Ba-Bd, 50 µm; Be, Bf, 10 µm. C, Relative number of Olig2+ and Pdgfrα+ cells in 1 mm2 area in the cortex and the thalamus. There were significant differences on relative numbers for Olig2+ and Pdgfrα+ cells between control and Pen-2 cKO mice at P14 (n = 4 mice per group). **p < 0.01. ***p < 0.005. D, Western blotting for Olig2, Pdgfrα, and Sox10. There were significant differences on relative levels for Olig2, Pdgfrα, and Sox10 between control and Pen-2 cKO mice at P14 (Control: n = 3; Pen-2 cKO: n = 4). **p < 0.01. ***p < 0.005. GAPDH served as the loading control. E, Representative images for double immunostaining for Pdgfrα and BrdU. BrdU was injected into mice at P7. Images were taken from the cortex. Scale bar, 50 µm. F, Percentage of Pdgfrα+/BrdU+ cells to Pdgfrα+ cells in the cortex. There was a highly significant difference between control and Pen-2 cKO mice (Control: 100 ± 2.9%, n = 4 mice; Pen-2 cKO: 168.4 ± 4.4%, n = 4 mice). ****p < 0.001.
To search for cellular mechanism responsible for the change in OPCs in Pen-2 cKO mice, we analyzed OPC proliferation by conducting BrdU pulse-labeling experiments. BrdU was injected intraperitoneally into mice at P7. Double-staining of Pdgfrα/BrdU (Fig. 5E) or Olig2/BrdU was performed. First, we found that the ratio of Pdgfrα+/BrdU+ cells to Pdgfrα+ cells in Pen-2 cKO mice was significantly increased compared with controls (Fig. 5F). Second, similar results were obtained for Olig2+/BrdU+ cells (data not shown). These findings suggest that deletion of Pen-2 causes enhanced proliferation of OPCs, which could explain why Pen-2 cKO mice have larger number of OPCs. The latter may directly result in increased levels for Pdgfrα, Olig2, and Sox10 observed (Fig. 5D).
Generation of GFAP-positive astrocytes from cre-expressing OPCs in vivo and in vitro
To find out how excessive astrocytes were generated, we performed lineage-tracing experiments. The LSL-tdTomato mouse was used to generate Pen-2 cKO-expressing tdTomato (Fig. 6A). Scarce GFAP+ cells were observed in the cortex in Pen-2f/+;Olig1-Cre;LSL-tdTomato (control) at P14, and they were not positive for tdTomato (Fig. 6B). In contrast, GFAP+/tdTomato+ cells were abundantly detected in the cortex in Pen-2f/f;Olig1-Cre;LSL-tdTomato (Pen-2 cKO) mice (Fig. 6C,D). The same results were obtained for the thalamus (data not shown). The above observations suggest that GFAP+ astrocytes are generated from Cre-expressing cells in Pen-2 cKO mice.
Figure 6.
Derivation of astrocytes from OPCs in vivo and in vitro. A, Breeding strategy for the generation of Pen-2 cKO mice expressing tdTomato. Pen-2f/+;Olig1-Cre;LSL-tdTomato and Pen-2f/f;Olig1-Cre;LSL-tdTomato were used. B, C, Double-staining of GFAP/tdTomato for control (B) and Pen-2 cKO (C) mice. There was costaining for GFAP and tdTomato in the cortex of Pen-2 cKO but not control mice. D, Enlarged image for the boxed area in C. E, Double-staining for GFAP and tdTomato in OL cultures. OPCs/OLs were cultured from Pen-2f/+;Olig1-Cre;LSL-tdTomato and Pen-2f/f;Olig1-Cre;LSL-tdTomato cortices. Cultures at DIV8 were used for immunostaining. F, Ratio of GFAP+ cells to DAPI+ cells. There was a significant difference between Pen-2f/+;Olig1-Cre;LSL-tdTomato and Pen-2f/f;Olig1-Cre;LSL-tdTomato cell cultures at DIV8. ***p < 0.005. G, Western blotting on Pen-2. Lysates were prepared from cultured OPCs at the culture day 8 and 12. Pen-2 was detected in control but not Pen-2 cKO OPC cultures. Scale bars: B, C, E, 50 µm; D, 10 µm.
To verify the above in vivo findings, we cultured OPCs using cortices from Pen-2f/+;Olig1-Cre;LSL-tdTomato and Pen-2f/f;Olig1-Cre;LSL-tdTomato mice at P8. OPCs were cultured in OPC medium for 3 weeks, followed by OL medium for 8 d (DIV8). Few cells from control cultures but abundant cells from Pen-2 cKOs were doubly positive for tdTomato and GFAP at DIV8 (Fig. 6E). Cell counting results showed significantly increased number of GFAP+ cells in Pen-2 cKO cultures compared with controls (Fig. 6F). In addition, Western blotting confirmed undetectable expression of Pen-2 in Pen-2 cKO OPCs collected at the culture day 8 or 12 (Fig. 6G). Overall, these in vitro results confirmed accelerated differentiation of OPCs into astrocytes in Pen-2 cKO cultures compared with controls.
Downregulation of Olig2 and Sox10 in OPCs undergoing differentiation into astrocytes in the cortex of Pen-2 cKO mice
We sought to identify a subgroup of OPCs, which underwent the differentiation process to astrocytes in Pen-2 cKO mice. To this end, we performed double-staining for Pdgfrα and GFAP using brain sections at P12. Indeed, we observed numerous Pdgfrα+/GFAP+ cells in the cortex (Fig. 7A) and the thalamus (data not shown) of Pen-2 cKO mice. Moreover, apparent processes were observed in Pdgfrα+/GFAP+ cells, which morphologically looked closer to astrocytes than to OPCs (Fig. 7B). Pdgfrα was mainly expressed in the cytoplasm in Pdgfrα+/GFAP+ and Pdgfrα+/GFAP– cells (Fig. 7A,B). Quantification data showed that the ratio of Pdgfrα+/GFAP+ cells to Pdgfrα+ cells was significantly higher in Pen-2 cKO cortices than in controls (Fig. 7C). Overall, the discovery of abundant Pdgfrα+/GFAP+ cells provided direct evidence supporting the conclusion that deletion of Pen-2 causes enhanced transition of OPCs to astrocytes in the cortex.
Figure 7.

Decreased expression of Olig2 and Sox10 in OPCs undergoing differentiating into astrocytes in Pen-2 cKO mice. A, Representative images for double-staining of Pdgfrα/GFAP in the cortex of Pen-2 cKO mice at P12. Yellow arrowheads indicated Pdgfrα+/GFAP+ cells. B, Enlarged images for the boxed area in A. Yellow arrowheads indicate two Pdgfrα+/GFAP+ cells. Pdgfrα was colocalized with GFAP in the cytoplasm. C, Percentage of Pdgfrα+/GFAP+ cells to Pdgfrα+ cells in P12 cortices. There was a highly significant difference between control and Pen-2 cKO mice. ****p < 0.001. D, Representative images for double-staining of GFAP/Olig2. GFAP+/Olig2+ cells were abundantly detected in the cortex of Pen-2 cKO but not control mice at P12. E, Enlarged images for the boxed area in D. White arrowhead indicates an Olig2+/GFAP+ cell. Yellow arrowheads indicate two adjacent Olig2+/GFAP– cells. F, Percentage of fluorescence intensity of Olig2 in Olig2+/GFAP+ cells to that in Olig2+/GFAP– cells. There was a highly significant difference on relative fluorescence intensity of Olig2 between Olig2+/GFAP+ and Olig2+/GFAP– cells in the cortex of Pen-2 cKO mice at P12 (n = 24 cells per group from 3 animals). ****p < 0.001. G, Representative images for costaining of GFAP/Olig2/Sox10 in the cortex of Pen-2 cKO mice at P12. White arrowhead indicates an Olig2+/Sox10+/GFAP+ cell. Yellow arrowheads indicate two Olig2+/Sox10+/GFAP– cells. H, Percentage of fluorescence intensity of Sox10 in Olig2+/Sox10+/GFAP+ cells to that in Olig2+/Sox10+/GFAP– cells. There was a highly significant difference on relative fluorescence intensity of Sox10 between Olig2+/Sox10+/GFAP+ and Olig2+/Sox10+/GFAP– cells in the cortex of Pen-2 cKO mice (n = 24 cells per group from 3 animals). ****p < 0.001. Scale bars: A, D, 50 µm; B, E, G, 10 µm.
Olig2 is known to play a critical role in regulating the OPC-to-astrocyte switch (Zuo et al., 2018). Since increased levels for Olig2 and Sox10 (Fig. 5D) are mixed effects from all types of cells, we reason that these changes may be because of larger population of OPCs in Pen-2 cKO cortices than in controls. To study whether Olig2 and Sox10 also exhibited upregulation in OPCs, which were differentiating to astrocytes, we compared expression levels of these two molecules in Olig2+ cells, which were either positive or negative (–) for GFAP in Pen-2 cKO mice. First, we conducted double-staining for Olig2 and GFAP using brain sections at P12. There were abundant Olig2+/GFAP+ cells in the cortex (Fig. 7D) and the thalamus (data not shown) in Pen-2 cKO mice but not controls. There results were consistent with those for Pdgfrα+/GFAP+ cells (Fig. 7A). Fluorescence intensity of Olig2 was measured (Fig. 7E), and it was significantly decreased in Olig2+/GFAP+ cells compared with neighboring Olig2+/GFAP– cells in Pen-2 cKO cortices (Fig. 7F). Second, we conducted triple staining for Olig2, Sox10, and GFAP (Fig. 7G). Fluorescence intensity of Sox10 was then quantified. It was significantly reduced in Olig2+/Sox10+/GFAP+ cells compared with Olig2+/Sox10+/GFAP– cells in the same image (Fig. 7H). Overall, these results are in agreement with the notion that Olig2 and Sox10 are downregulated in order that OPCs are able to exit the OL lineage program (Zhang et al., 2016; Zuo et al., 2018).
Enhanced OPC-to-astrocyte differentiation in inducible Pen-2 cKO mice
To confirm the finding that Pen-2 is important for the OPC-to-astrocyte differentiation, we crossed the NG2-CreERT2 mouse to Pen-2f/f to generate OPC-specific Pen-2 icKO mice using a protocol shown in Figure 8A. We then used a method described recently (Huang et al., 2014) to induce Cre expression (Fig. 8B). To check the expression pattern of Cre, we performed costaining of tdTomato/Olig2 using brain sections prepared from NG2-CreERT2;LSL-tdTomato mice at P21 (Fig. 8C). We found that the ratio of tdTomato+/Olig2+ cells to Olig2+ cells was >60% (Fig. 8D). Thus, the above induction method was quite efficient.
Figure 8.
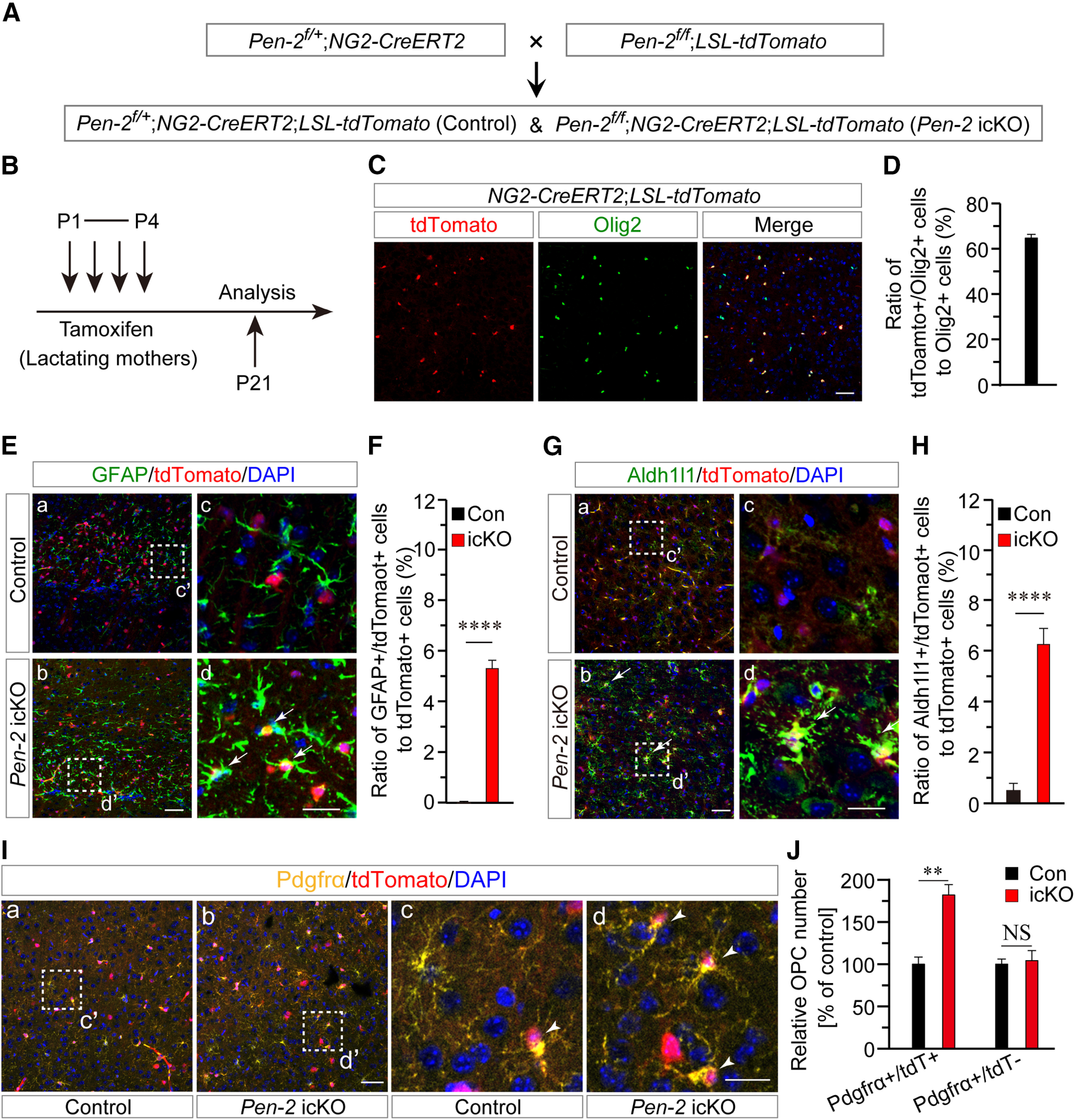
Increased differentiation of OPCs to astrocytes in Pen-2 icKO mice. A, Breeding strategy for Pen-2 icKO mice. Pen-2f/+;NG2-CreERT2;LSL-tdTomato and Pen-2f/f;NG2-CreERT2;LSL-tdTomato were used as control and Pen-2 icKO, respectively. B, Pen-2f/+;NG2-CreERT2;LSL-tdTomato and Pen-2f/f;NG2-CreERT2;LSL-tdTomato mice received tamoxifen from P1 to P4 from their lactating mothers by intraperitoneal injection. C, Costaining of Olig2/tdTomato in the cortex of NG2-CreERT2;LSL-tdTomato mice at P21. Scale bar, 50 µm. D, Percentage of Olig2+/tdTomato+ cells to Olig2+ cells. The ratio was ∼64% in NG2-CreERT2;LSL-tdTomato mice at P21 (n = 4 mice). E, Representative images for costaining of GFAP/tdTomato in cortices of control and Pen-2 icKO mice. Boxed areas in Ea and Eb were enlarged as Ec and Ed, respectively. Scale bars: Ea, Eb, 50 µm; Ec, Ed, 10 µm. F, Percentage of GFAP+/tdTomato+ cells to tdTomato+ cells. There was a highly significant difference between control and Pen-2 icKO mice at P21 (n = 4 mice per group). ****p < 0.001. G, Representative images for costaining of Aldh1l1/tdTomato in cortices of control and Pen-2 icKO mice. Boxed areas in Ga and Gb were enlarged as Gc and Gd, respectively. Scale bars: Ga, Gb, 50 µm; Gc, Gd, 10 µm. H, Percentage of Aldh1l1+/tdTomato+ cells to tdTomato+ cells. There was a highly significant difference between control and Pen-2 icKO mice at P21 (n = 4 mice per group). ****p < 0.001. I, Representative images for costaining of Pdgfrα/tdTomato. Boxed areas in Ia and Ib were enlarged as Ic and Id, respectively. Scale bars: Ia, Ib, 50 µm; Ic, Id, 10 µm. J, Relative number of Pdgfrα+/tdTomato+ cells in Pen-2 icKO mice to controls. There was a significant difference on Pdgfrα+/tdTomato+, but not Pdgfrα+/tdTomato– cells, between control and Pen-2 icKO mice (n = 4 mice per group). **p < 0.01, NS, not significant.
To find out whether NG2-CreERT2-mediated deletion of Pen-2 affected astrogliogenesis, we first conducted costaining of GFAP/tdTomato using brain sections at P21 (Fig. 8E). We found that Pen-2 icKO cortices exhibited significantly increased ratio of GFAP+/tdTomato+ cells to tdTomato+ cells compared with controls (Fig. 8F). Second, we conducted costaining of Aldh1l1/tdTomato (Fig. 8G). There was significantly increased ratio of Aldh1l1+/tdTomato+ cells to tdTomato+ cells in Pen-2 icKO cortices (Fig. 8H). Since GFAP+/tdTomato+ and Aldh1l1+/tdTomato+ cells represented astrocytes that were derived from Cre-expressing OPCs in Pen-2 icKO mice, the above results suggest that NG2-CreERT2-mediated deletion of Pen-2 causes increased differentiation of OPCs into astrocytes.
To study whether there were changes in OPCs in Pen-2 icKO mice, we conducted costaining of Pdgfrα/tdTomato using P21 brain sections (Fig. 8I). Quantification data revealed that relative number of Pdgfrα+/tdTomato+ cells was significantly increased in Pen-2 icKO cortices compared with controls (Fig. 8J). As expected, relative number of Pdgfrα+/tdTomato– cells in the cortex was not different between control and Pen-2 icKO mice (Fig. 8J), indicating that OPCs without Cre expression were not significantly affected.
Enhanced expression of Stat3 in Pen-2 cKO mice
To identify the underlying molecular mechanisms, we focused on Nfia and Stat3, since these two transcriptional factors are critical for NPCs to differentiate into astrocytes (Fan et al., 2005; Tiwari et al., 2018). We prepared RNA samples from GFP+ cells collected from cortices of Pen-2f/+;Olig1-Cre;mTmG and Pen-2f/f;Olig1-Cre;mTmG mice at P11 by the FACS (Fig. 9A). First, qRT-PCR analysis revealed significantly reduced levels of Hes1 in Pen-2 cKO GFP+ cells compared with controls (Fig. 9A), suggesting that inactivation of Pen-2 inhibits the Notch signaling. Second, we observed highly increased Stat3, but not Nfia, in Pen-2 cKO GFP+ cells (Fig. 9A).
Figure 9.
Increased expression of Stat3 in Pen-2 cKO mice. A, qRT-PCR analyses on Hes1, Nfia, and Stat3. GFP+ cells were sorted from Pen-2f/+;Olig1-Cre;mTmG (control) and Pen-2f/f;Olig1-Cre;mTmG (Pen-2 cKO) cortices. There were highly significant differences on Hes1 and Stat3 but not Nfia between two genotype groups (n = 4 per group). **p < 0.01. B, Double-staining for Stat3 and GFAP. There was increased immunoreactivity of Stat3 in the cortex of Pen-2 cKO mice at P14. GFAP-expressing cells were also positive for Stat3 in Pen-2 cKO mice. C, Western analyses on Hdac3, Stat3, and pStat3Y705 in the cortex. There were significant differences on levels of Stat3 and pStat3Y705 but not Hdac3 between control and Pen-2 cKO mice at P14 (Control: n = 3; Pen-2 cKO: n = 4). **p < 0.01 for Stat3 and pStat3Y705. GAPDH served as the loading control. D, qRT-PCR analyses on Hdac3, Stat3, and GFAP mRNAs in the cortex. There were significant differences on levels of Stat3 and GFAP but not Hdac3 between control and Pen-2 cKO mice at P14 (n = 4 mice per group). *p < 0.05 for Stat3. **p < 0.01 for GFAP. E, Costaining for Olig2 and tdTomato in cultured OLs at DIV8. Cortices from Pen-2f/+;Olig1-Cre;LSL-tdTomato and Pen-2f/f;Olig1-Cre;LSL-tdTomato were used for OPC cultures. Western blotting was conducted for Stat3 and GFAP. There were increased levels of Stat3 and GFAP in Pen-2 cKO cultures at DIV8 compared with controls (n = 4 per group). Scale bars: B, E, 50 µm. NS, not significant.
To visualize the expression pattern of Stat3 in Pen-2 cKO mice, we performed IHC on Stat3. There was increased Stat3 immunoreactivity in the cortex (Fig. 9B) and the thalamus (data not shown) in Pen-2 cKO mice at P14 compared with controls. In addition, numerous Stat3+/GFAP+ cells were detected in the brain of Pen-2 cKO but not control mice (Fig. 9B). Western blotting showed increased levels of Stat3 and pStat3Y705 but not Hdac3 in the cortex of Pen-2 cKO mice compared with controls (Fig. 9C). qRT-PCR analysis confirmed significantly increased levels of Stat3 and GFAP but not Hdac3 in cortical RNA samples in Pen-2 cKO mice at P14 (Fig. 9D). Furthermore, we cultured OPCs from cortices of control and Pen-2 cKO mice (Fig. 9E). Western blotting revealed increased levels of Stat3 and GFAP in Pen-2 cKO cultures compared with controls (Fig. 9E).
Regulation on Stat3 by Pen-2 via Hes1
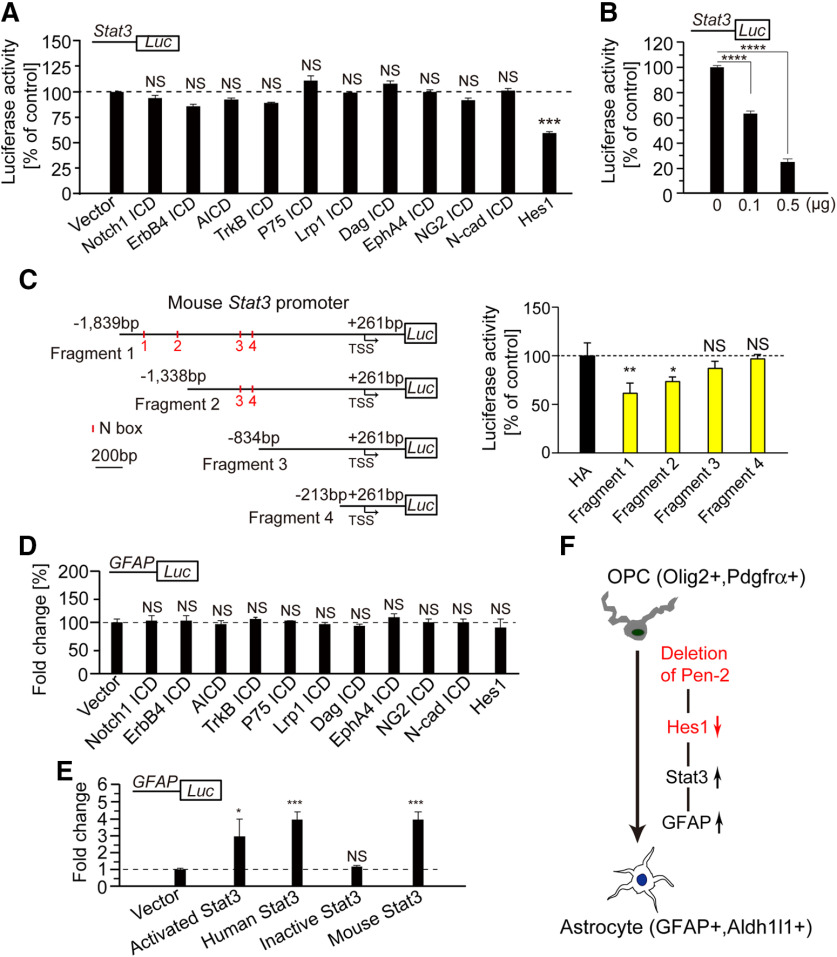
Notch, APP, ErbB2 receptor tyrosine kinase 4 (ErbB4), neurotrophic tyrosine kinase receptor Type 2 (TrkB), p75 neurotrophin receptor (P75), low-density lipoprotein receptor-related protein 1 (Lrp1), lymphocyte antigen 6 complex (Dag), Eph receptor A4 (EphA4), and chondroitin sulfate proteoglycan 4 (NG2) are well-known substrates to be cleaved by γ-secretase to produce different types of intracellular domains (ICDs) (Lee et al., 2002; De Strooper, 2003; Mei and Nave, 2014). To examine whether Stat3 could be regulated by any of the above γ-secretase cleavage products, we conducted a number of luciferase experiments. Plasmid expressing Notch1 ICD, AICD, ErbB4 ICD, TrkB ICD, P75 ICD, Lrp1 ICD, Dag ICD, EphA4 ICD, NG2 ICD, N-cadherin ICD (N-cad ICD) or Hes1 was cotransfected with the luciferase reporter under the Stat3 promoter in HEK293T cells. We found that expression of Hes1, but none of the above ICDs repressed the Stat3 promoter activity (Fig. 10A). Moreover, Hes1 exhibited a dosage-dependent effect on luciferase activity for the Stat3 promoter (Fig. 10B).
Figure 10.
Regulation of Stat3 expression by Hes1. A, Luciferase assay on the promoter activity of Stat3 using HEK293T cells. Expression of Notch1 ICD, AICD, TrkB ICD, P75 ICD, Lrp1 ICD, Dag ICD, EphA4 ICD, NG2 ICD, or N-cad ICD did not affect the luciferase activity of the Stat3 promoter (n = 4 replicates). Expression of Hes1 significantly repressed the luciferase activity of the Stat3 promoter (n = 4 replicates). ***p < 0.005. B, Hes1 repressed the luciferase activity of the Stat3 promoter in a dosage-dependent manner in HEK293T cells (n = 4 replicates). ***p < 0.005. C, Luciferase assay on various fragments of the Stat3 promoter. Hes1 repressed the luciferase activity for Fragments 1 and 2 but not for Fragments 3 and 4 (n = 4 replicates). *p < 0.05. **p < 0.01. D, Luciferase assay on the GFAP promoter. Expression of Notch1 ICD, AICD, TrkB ICD, P75 ICD, Lrp1 ICD, Dag ICD, EphA4 ICD, NG2-ICD, N-cad ICD, or Hes1 did not affect the luciferase activity of the GFAP promoter (n = 4 replicates). E, Luciferase assay on the GFAP promoter. Expression of activated Stat3, human Stat3, or mouse Stat3 significantly inhibited the luciferase activity of the GFAP promoter. Expression of inactive Stat3 did not affect (n = 4 replicates). *p < 0.05. ***p < 0.005. F, A schematic model for the fate switch of OPCs into astrocytes regulated by Pen-2. First, deletion of Pen-2 leads to downregulation of Hes1 by blocking the cleavage of Notch receptors. Second, Hes1 negatively regulates the expression of Stat3. Third, activation of Stat3 promotes GFAP expression. Stat3 and GFAP together trigger the differentiation of OPCs into astrocytes. NS, not significant.
To further explore molecular mechanisms, we performed fragment analysis on the Stat3 promoter (Fig. 10C). Hes1 is a bHLH transcription factor which binds to the N box (CACNAG) but not the E box (CANNTG) in the promoter of its targeted genes (Ohsako et al., 1994; Kageyama et al., 2007). We conducted luciferase experiments using four plasmids containing different regions of the Stat3 promoter. We found that expression of Hes1 repressed luciferase activities for Fragments 1 and 2, but not 3 and 4 (Fig. 10C). Therefore, the promoter region from –1338 to –834 bp upstream of the transcriptional start site (TSS) in Stat3 was critical for Hes1 to inhibit Stat3 expression.
To find out whether GFAP could be directly regulated by any γ-secretase cleavage product, HEK293T cells were cotransfected with plasmids expressing luciferase system driven by the GFAP promoter and one of the above ICDs. However, neither ICD nor Hes1 significantly inhibited the promoter activity of GFAP (Fig. 10D). We found that expression of activated Stat3, human Stat3, or mouse Stat3 enhanced the promoter activity of GFAP (Fig. 10E). In contrast, inactive Stat3 did not change the promoter activity of GFAP (Fig. 10E).
Discussion
Accumulating evidence has shown that astrocytes can be generated from OPCs and NPCs in the cortex (Belachew et al., 2003; Cai et al., 2007; Freeman, 2010; Namihira and Nakashima, 2013; Huang et al., 2014, 2018). Here we show that deletion of Pen-2 results in increased number of astrocytes. Whereas deletion of Pen-2 does not cause abnormal neuronal death or enhanced neuroinflammatory responses, it leads to increased number of OPCs but reduced population of OLs. Fate-mapping experiments reveal that abnormally generated astrocytes are derived from Cre-expressing OPCs in Pen-2 cKO and Pen-2 icKO mice. Mechanistically, we demonstrate that Stat3 is a key transcriptional factor to regulate Pen-2-dependent astrogliogenesis from OPCs in the cortex (Figs. 9, 10).
Although Notch1 is a major γ-secretase substrate (De Strooper, 2003), Notch1 cKO mice (Wang et al., 1998; Genoud et al., 2002; Woodhoo et al., 2009; Zhang et al., 2009) exhibit distinct phenotypes from those in Pen-2 cKO animals. Therefore, the role of γ-secretase in OPCs is different from that of Notch1. This discrepancy may be solved by the following explanations. Since γ-secretase cleaves all Notch receptors, phenotypes in Pen-2 cKO and Pen-2 icKO mice could be caused by complete inhibition of Notch function. In contrast, the remaining Notch receptors (e.g., Notch2/3/4) may produce partial compensatory effects on astrocyte development in Notch1 cKO mice (Wang et al., 1998; Genoud et al., 2002; Zhang et al., 2009). Although inhibition on γ-secretase activity in OLs enhances myelination (Watkins et al., 2008), it is unknown whether it also promotes astrogliogenesis. Nevertheless, the Watkins et al. (2008) study is not contradictory to this one, since γ-secretase activity is completely abolished in OPCs and OLs in Pen-2 cKO mice.
It has well been established that OPCs possess multipotency for differentiation in the cortex (Belachew et al., 2003; Cai et al., 2007; Huang et al., 2014; Zuo et al., 2018). Whereas mechanisms by which NPCs differentiate into astrocytes have well been defined (Freeman, 2010; Imayoshi and Kageyama, 2014), those governing OPCs to differentiate into astrocytes are not well understood. We report excessive astrogliogenesis in the cortices of Pen-2 cKO mice. Results on TUNEL, NeuN, and Iba1 have excluded the possibility that abnormal astrocyte generation in Pen-2 cKO mice is astrogliosis. The following observations have led us to conclude that deletion of Pen-2 causes enhanced OPC-to-astrocyte differentiation. First, the number of GFAP+/tdTomato+ cells is increased in Pen-2 cKO or Pen-2 icKO mice expressing tdTomato compared with controls. Second, there is larger quantity of Pdgfrα+/GFAP+ and Olig2+/GFAP+ cells in Pen-2 cKO cortices than in controls. Third, the number of Aldh1l1+/tdTomato+ cells is increased in Pen-2 icKO cortices.
Previous evidence has shown that Hdac3 and Olig2 play critical roles in the fate switch of OPCs into astrocytes (Zhang et al., 2016; Zuo et al., 2018). Since protein and mRNA levels of Hdac3 are not significantly changed in Pen-2 cKO cortices, Hdac3 may not be involved in Pen-2-dependent differentiation of OPCs into astrocytes. Although there are increased levels of Olig2 and Sox10 in Pen-2 cKO cortical samples, these changes may be because Pen-2 cKOs have larger amount of OPCs than controls do. Interestingly, we find that the expression of Olig2 and Sox10 is significantly decreased in OPCs, which undergo differentiating into astrocytes in Pen-2 cKO cortices. Thus, this observation is consistent with a widely accepted concept that downregulation of Olig2 and Sox10 is required for OPCs to exit the OL lineage program (Zhang et al., 2016; Zuo et al., 2018).
This study reveals that deletion of Pen-2 leads to increased number of OPCs but decreased number of OLs in the cortex. Since proliferation of OPCs is enhanced in the cortex of Pen-2 cKO mice, this may account for the increase in the OPC number. However, we cannot exclude the possibility that OPCs may compensate for the loss of OPCs that are becoming astrocytes in Pen-2 cKO mice. The following explanations may help answer why the number of OLs is decreased in Pen-2 cKOs. First, since deletion of Pen-2 promotes OPCs differentiating into astrocytes, this may diminish the pool of OPCs to take the route for OL differentiation, and eventually reduce the number of mature OLs. Second, at the molecular level, deletion of Pen-2 causes upregulation of STAT3 and GFAP, key transcriptional factors to control astrocyte fate. High levels of these molecules may inhibit the expression of Olig2 and Sox10 so that OL differentiation is impaired.
Stat3 is an important transcriptional factor to regulate astrocyte differentiation (Fan et al., 2005). We demonstrate that expression of Hes1 inhibits the promoter activity of Stat3 in a dosage-dependent manner, and that expression of Stat3 enhances the promoter activity of GFAP. We reason that deletion of Pen-2 sequentially induces activation of Stat3 and GFAP, which drives the OPC-to-astrocyte differentiation and may in turn inhibit the expression of Olig2. This explanation helps understand why Olig2 is downregulated in Olig2+/GFAP+ cells compared with Olig2+/GFAP– cells. Since this study has uncovered mechanisms by which Pen-2 regulates the fate switch of OPCs into astrocytes, a cellular model is proposed to highlight key molecular events related to this process (Fig. 10F). First, deletion of Pen-2 prevents the cleavage of Notch receptors, which leads to decreased Hes1 levels. Second, Hes1 regulates expression of Stat3 through binding to a specific region of the Stat3 promoter. Third, elevated Stat3 activates GFAP expression, which drives OPCs to differentiate into astrocytes.
Since mutations on γ-secretase subunits cause various brain disorders (Dermaut et al., 2002; Sala Frigerio et al., 2005; Shen and Kelleher, 2007; Zhong et al., 2009; Forzano et al., 2012; Gana et al., 2012), findings in this study may provide insights on the developmental etiology for abnormal astrogliogenesis in these diseases. Although this study has shown that Pen-2 plays a critical role in OPC development, mechanisms are unknown. It has recently been reported that inhibition of the Notch signaling results in increased mRNA levels of Olig2 (Sagner et al., 2018). Thus, there is a possibility that conditional deletion of Pen-2 in OL lineages causes increased number of OPCs via regulation of Olig2. Further investigations are warranted to dissect the underlying molecular mechanisms in near future.
Footnotes
This work was supported by National Natural Science Foundation of China Grants 91849113 and 31271123, Natural Science Foundation of Jiangsu Grant BK20201255, NJU-SKLPB Project Grant ZZYJ202112, Nanjing Medical University Science and Technology Development Key Project Grant 2018NJMUZD026, and Nanjing University Innovation Program for PhD Candidate. W.H. and F.K. were supported by Sino-German Deutsche Forschungsgemeinschaft Grant KI503/14-1.
The authors declare no competing financial interests.
References
- Acx H, Serneels L, Radaelli E, Muyldermans S, Vincke C, Pepermans E, Müller U, Chávez-Gutiérrez L, De Strooper B (2017) Inactivation of γ-secretases leads to accumulation of substrates and non-Alzheimer neurodegeneration. EMBO Mol Med 9:1088–1099. 10.15252/emmm.201707561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechler ME, Byrne L, Ffrench-Constant C (2015) CNS myelin sheath lengths are an intrinsic property of oligodendrocytes. Curr Biol 25:2411–2416. 10.1016/j.cub.2015.07.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belachew S, Chittajallu R, Aguirre AA, Yuan XQ, Kirby M, Anderson S, Gallo V (2003) Postnatal NG2 proteoglycan-expressing progenitor cells are intrinsically multipotent and generate functional neurons. J Cell Biol 161:169–186. 10.1083/jcb.200210110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi H, Zhou C, Zhang Y, Cai X, Ji M, Yang J, Chen G, Hu Y (2021) Neuron-specific deletion of presenilin enhancer2 causes progressive astrogliosis and age-related neurodegeneration in the cortex independent of the Notch signaling. CNS Neurosci Ther 27:174–185. 10.1111/cns.13454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Chen Y, Cai WH, Hurlock EC, Wu H, Kernie SG, Parada LF, Lu QR (2007) A crucial role for Olig2 in white matter astrocyte development. Development 134:1887–1899. 10.1242/dev.02847 [DOI] [PubMed] [Google Scholar]
- Cheng S, Liu T, Hu Y, Xia Y, Hou J, Huang C, Zou X, Liang J, Stone Shi Y, Zheng Y, Lu J, Chen G (2019) Conditional inactivation of Pen-2 in the developing neocortex leads to rapid switch of apical progenitors to basal progenitors. J Neurosci 39:2195–2207. 10.1523/JNEUROSCI.2523-18.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Strooper B (2003) Aph-1, Pen-2, and nicastrin with presenilin generate an active gamma-secretase complex. Neuron 38:9–12. 10.1016/S0896-6273(03)00205-8 [DOI] [PubMed] [Google Scholar]
- Dermaut B, Theuns J, Sleegers K, Hasegawa H, Van den Broeck M, Vennekens K, Corsmit E, St. George-Hyslop P, Cruts M, van Duijn CM, Van Broeckhoven C (2002) The gene encoding nicastrin, a major gamma-secretase component, modifies risk for familial early-onset Alzheimer disease in a Dutch population-based sample. Am J Hum Genet 70:1568–1574. 10.1086/340732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dries DR, Zhu Y, Brooks MM, Forero DA, Adachi M, Cenik B, West JM, Han YH, Yu C, Arbella J, Nordin A, Adolfsson R, Del-Favero J, Lu QR, Callaerts P, Birnbaum SG, Yu G (2016) Loss of nicastrin from oligodendrocytes results in hypomyelination and Schizophrenia with compulsive behavior. J Biol Chem 291:11647–11656. 10.1074/jbc.M116.715078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaz B, Popko B (2019) Molecular control of oligodendrocyte development. Trends Neurosci 42:263–277. 10.1016/j.tins.2019.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan GP, Martinowich K, Chin MH, He F, Fouse SD, Hutnick L, Hattori D, Ge WH, Shen Y, Hao W, ten Hoeve J, Shuai K, Sun YE (2005) DNA methylation controls the timing of astrogliogenesis through regulation of JAK-STAT signaling. Development 132:3345–3356. 10.1242/dev.01912 [DOI] [PubMed] [Google Scholar]
- Figlia G, Gerber D, Suter U (2018) Myelination and mTOR. Glia 66:693–707. 10.1002/glia.23273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forzano F, Napoli F, Uliana V, Malacarne M, Viaggi C, Bloise R, Coviello D, Di Maria E, Olivieri I, Di Iorgi N, Faravelli F (2012) 19q13 microdeletion syndrome: further refining the critical region. Eur J Med Genet 55:429–432. 10.1016/j.ejmg.2012.03.002 [DOI] [PubMed] [Google Scholar]
- Freeman MR (2010) Specification and morphogenesis of astrocytes. Science 330:774–778. 10.1126/science.1190928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gana S, Veggiotti P, Sciacca G, Fedeli C, Bersano A, Micieli G, Maghnie M, Ciccone R, Rossi E, Plunkett K, Bi W, Sutton VR, Zuffardi O (2012) 19q13.11 cryptic deletion: description of two new cases and indication for a role of WTIP haploinsufficiency in hypospadias. Eur J Hum Genet 20:852–856. 10.1038/ejhg.2012.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genoud S, Lappe-Siefke C, Goebbels S, Radtke F, Aguet M, Scherer SS, Suter U, Nave KA, Mantei N (2002) Notch1 control of oligodendrocyte differentiation in the spinal cord. J Cell Biol 158:709–718. 10.1083/jcb.200202002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J, Cheng S, Chen L, Wang Q, Shi Y, Xu Y, Yin Z, Chen G (2016) Astroglial activation and tau hyperphosphorylation precede to neuron loss in a neurodegenerative mouse model. CNS Neurosci Ther 22:244–247. 10.1111/cns.12509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang WH, Zhao N, Bai XS, Karram K, Trotter J, Goebbels S, Scheller A, Kirchhoff F (2014) Novel NG2-CreERT2 knock-in mice demonstrate heterogeneous differentiation potential of NG2 glia during development. Glia 62:896–913. 10.1002/glia.22648 [DOI] [PubMed] [Google Scholar]
- Huang WH, Bai XS, Stopper L, Catalin B, Cartarozzi LP, Scheller A, Kirchhoff F (2018) During development NG2 glial cells of the spinal cord are restricted to the oligodendrocyte lineage, but generate astrocytes upon acute injury. Neuroscience 385:154–165. 10.1016/j.neuroscience.2018.06.015 [DOI] [PubMed] [Google Scholar]
- Imayoshi I, Kageyama R (2014) bHLH factors in self-renewal, multipotency, and fate choice of neural progenitor cells. Neuron 82:9–23. 10.1016/j.neuron.2014.03.018 [DOI] [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T, Kobayashi T (2007) The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development 134:1243–1251. 10.1242/dev.000786 [DOI] [PubMed] [Google Scholar]
- Kelenis D, Hart E, Edwards-Fligner M, Johnson J, Vue TY (2018) ASCL1 regulates proliferation of NG2-glia in the embryonic and adult spinal cord. Glia 66:1862–1880. 10.1002/glia.23344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Shen J (2008) Presenilins are required for maintenance of neural stem cells in the developing brain. Mol Neurodegeneration 3:2. 10.1186/1750-1326-3-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberly WT, LaVoie MJ, Ostaszewski BL, Ye W, Wolfe MS, Selkoe DJ (2003) Gamma-secretase is a membrane protein complex comprised of presenilin, nicastrin, Aph-1, and Pen-2. Proc Natl Acad Sci USA 100:6382–6387. 10.1073/pnas.1037392100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Raff M (2000) Oligodendrocyte precursor cells reprogrammed to become multipotential CNS stem cells. Science 289:1754–1757. 10.1126/science.289.5485.1754 [DOI] [PubMed] [Google Scholar]
- Lee HJ, Jung KM, Huang YZ, Bennett LB, Lee JS, Mei L, Kim TW (2002) Presenilin-dependent gamma-secretase-like intramembrane cleavage of ErbB4. J Biol Chem 277:6318–6323. 10.1074/jbc.M110371200 [DOI] [PubMed] [Google Scholar]
- Li HL, Richardson WD (2016) Evolution of the CNS myelin gene regulatory program. Brain Res 1641:111–121. 10.1016/j.brainres.2015.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Xu P, Guan Z, Qian X, Dockery P, Fitzgerald U, O'Brien T, Shen S (2018) Ulk4 deficiency leads to hypomyelination in mice. Glia 66:175–190. 10.1002/glia.23236 [DOI] [PubMed] [Google Scholar]
- Liu T, Ye X, Zhang J, Yu T, Cheng S, Zou X, Xu Y, Chen G, Yin Z (2017) Increased adult neurogenesis associated with reactive astrocytosis occurs prior to neuron loss in a mouse model of neurodegenerative disease. CNS Neurosci Ther 23:885–893. 10.1111/cns.12763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie IA, Ohayon D, Li HL, de Faria JP, Emery B, Tohyama K, Richardson WD (2014) Motor skill learning requires active central myelination. Science 346:318–322. 10.1126/science.1254960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei L, Nave KA (2014) Neuregulin-ERBB signaling in the nervous system and neuropsychiatric diseases. Neuron 83:27–49. 10.1016/j.neuron.2014.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, Krenick R, Ullian E, Tsai HH, Deneen B, Richardson WD, Barres BA, Rowitch DH (2012) Astrocytes and disease: a neurodevelopmental perspective. Genes Dev 26:891–907. 10.1101/gad.188326.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L (2007) A global double-fluorescent Cre reporter mouse. Genesis 45:593–605. 10.1002/dvg.20335 [DOI] [PubMed] [Google Scholar]
- Namihira M, Nakashima K (2013) Mechanisms of astrocytogenesis in the mammalian brain. Curr Opin Neurobiol 23:921–927. 10.1016/j.conb.2013.06.002 [DOI] [PubMed] [Google Scholar]
- Nave KA, Trapp BD (2008) Axon-glial signaling and the glial support of axon function. Annu Rev Neurosci 31:535–561. 10.1146/annurev.neuro.30.051606.094309 [DOI] [PubMed] [Google Scholar]
- Ning F, Li X, Yu L, Zhang B, Zhao Y, Liu Y, Zhao B, Shang Y, Hu X (2019) Hes1 attenuates type I IFN responses via VEGF-C and WDFY1. J Exp Med 216:1396–1410. 10.1084/jem.20180861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsako S, Hyer J, Panganiban G, Oliver I, Caudy M (1994) Hairy function as a DNA-binding helix-loop-helix repressor of Drosophila sensory organ formation. Genes Dev 8:2743–2755. 10.1101/gad.8.22.2743 [DOI] [PubMed] [Google Scholar]
- Parras CM, Galli R, Britz O, Soares S, Galichet C, Battiste J, Johnson JE, Nakafuku M, Vescovi A, Guillemot F (2004) Mash1 specifies neurons and oligodendrocytes in the postnatal brain. EMBO J 23:4495–4505. 10.1038/sj.emboj.7600447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Sun J, Hon S, Nylander AN, Xia WM, Feng YP, Wang XL, Lemere CA (2010) L-3-n-butylphthalide improves cognitive impairment and reduces amyloid-beta in a transgenic model of Alzheimer's disease. J Neurosci 30:8180–8189. 10.1523/JNEUROSCI.0340-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff MC, Miller RH, Noble M (1983) A glial progenitor-cell that develops In vitro into an astrocyte or an oligodendrocyte depending on culture-medium. Nature 303:390–396. 10.1038/303390a0 [DOI] [PubMed] [Google Scholar]
- Rowitch DH, Kriegstein AR (2010) Developmental genetics of vertebrate glial-cell specification. Nature 468:214–222. 10.1038/nature09611 [DOI] [PubMed] [Google Scholar]
- Sagner A, Gaber ZB, Delile J, Kong JH, Rousso DL, Pearson CA, Weicksel SE, Melchionda M, Mousavy Gharavy SN, Briscoe J, Novitch BG (2018) Olig2 and Hes regulatory dynamics during motor neuron differentiation revealed by single cell transcriptomics. PLoS Biol 16:e2003127. 10.1371/journal.pbio.2003127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala Frigerio C, Piscopo P, Calabrese E, Crestini A, Malvezzi Campeggi L, Civita di Fava R, Fogliarino S, Albani D, Marcon G, Cherchi R, Piras R, Forloni G, Confaloni A (2005) PEN-2 gene mutation in a familial Alzheimer's disease case. J Neurol 252:1033–1036. 10.1007/s00415-005-0799-7 [DOI] [PubMed] [Google Scholar]
- Saura CA, Choi SY, Beglopoulos V, Malkani S, Zhang D, Shankaranarayana Rao BS, Chattarji S, Kelleher RJ 3rd, Kandel ER, Duff K, Kirkwood A, Shen J (2004) Loss of presenilin function causes impairments of memory and synaptic plasticity followed by age-dependent neurodegeneration. Neuron 42:23–36. 10.1016/S0896-6273(04)00182-5 [DOI] [PubMed] [Google Scholar]
- Sehgal R, Sheibani N, Rhodes SJ, Belecky AT (2009) BMP7 and SHH regulate Pax2 in mouse retinal astrocytes by relieving TLX repression. Dev Biol 332:429–443. 10.1016/j.ydbio.2009.05.579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Kelleher RJ 3rd (2007) The presenilin hypothesis of Alzheimer's disease: evidence for a loss-of-function pathogenic mechanism. Proc Natl Acad Sci USA 104:403–409. 10.1073/pnas.0608332104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SH, Zhu XJ, Huang H, Guo W, Tang T, Xie BH, Xu XF, Zhang ZY, Shen Y, Dai ZM, Qiu M (2019) WNT signaling represses astrogliogenesis via Ngn2-dependent direct suppression of astrocyte gene expression. Glia 67:1333–1343. 10.1002/glia.23608 [DOI] [PubMed] [Google Scholar]
- Tabuchi K, Chen G, Sudhof TC, Shen J (2009) Conditional forebrain inactivation of nicastrin causes progressive memory impairment and age-related neurodegeneration. J Neurosci 29:7290–7301. 10.1523/JNEUROSCI.1320-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari N, Pataskar A, Peron S, Thakurela S, Sahu SK, Figueres-Onate M, Marichal N, Lopez-Mascaraque L, Tiwari VK, Berninger B (2018) Stage-specific transcription factors drive astrogliogenesis by remodeling gene regulatory landscapes. Cell Stem Cell 23:557–571. 10.1016/j.stem.2018.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Liu M, Zou G, Wang L, Duan W, He X, Ji M, Zou X, Hu Y, Yang J, Chen G (2021) Deletion of PDK1 in oligodendrocyte lineage cells causes white matter abnormality and myelination defect in the central nervous system. Neurobiol Dis 148:105212. 10.1016/j.nbd.2020.105212 [DOI] [PubMed] [Google Scholar]
- Wang S, Sdrulla AD, diSibio G, Bush G, Nofziger D, Hicks C, Weinmaster G, Barres BA (1998) Notch receptor activation inhibits oligodendrocyte differentiation. Neuron 21:63–75. 10.1016/S0896-6273(00)80515-2 [DOI] [PubMed] [Google Scholar]
- Watkins TA, Emery B, Mulinyawe S, Barres BA (2008) Distinct stages of myelination regulated by gamma-secretase and astrocytes in a rapidly myelinating CNS coculture system. Neuron 60:555–569. 10.1016/j.neuron.2008.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhoo A, Alonso MB, Droggiti A, Turmaine M, D'Antonio M, Parkinson DB, Wilton DK, Al-Shawi R, Simons P, Shen J, Guillemot F, Radtke F, Meijer D, Feltri ML, Wrabetz L, Mirsky R, Jessen KR (2009) Notch controls embryonic Schwann cell differentiation, postnatal myelination and adult plasticity. Nat Neurosci 12:839–847. 10.1038/nn.2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M, Yue T, Ma ZY, Wu FF, Gow A, Lu QR (2005) Myelinogenesis and axonal recognition by oligodendrocytes in brain are uncoupled in Olig1-Null mice. J Neurosci 25:1354–1365. 10.1523/JNEUROSCI.3034-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang GH, Zhou R, Zhou Q, Guo XF, Yan CY, Ke M, Lei JL, Shi YG (2019) Structural basis of Notch recognition by human gamma-secretase. Nature 565:192–197. 10.1038/s41586-018-0813-8 [DOI] [PubMed] [Google Scholar]
- Zhang LG, He XL, Liu L, Jiang MQ, Zhao CT, Wang HB, He DY, Zheng T, Zhou XY, Hassan A, Ma ZX, Xin M, Sun Z, Lazar MA, Goldman SA, Olson EN, Lu QR (2016) Hdac3 interaction with p300 histone acetyltransferase regulates the oligodendrocyte and astrocyte lineage fate switch. Dev Cell 36:316–330. 10.1016/j.devcel.2016.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Argaw AT, Gurfein BT, Zameer A, Snyder BJ, Ge CH, Lu QR, Rowitch DH, Raine CS, Brosnan CF, John GR (2009) Notch1 signaling plays a role in regulating precursor differentiation during CNS remyelination. Proc Natl Acad Sci USA 106:19162–19167. 10.1073/pnas.0902834106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, Dong-hai Q, Hong-ying L, Qing-feng L (2009) Analysis of the nicastrin promoter rs10752637 polymorphism and its association with Alzheimer's disease. Eur J Neurosci 30:1831–1836. 10.1111/j.1460-9568.2009.06942.x [DOI] [PubMed] [Google Scholar]
- Zhou R, Yang GH, Guo XF, Zhou Q, Lei JL, Shi YG (2019) Recognition of the amyloid precursor protein by human gamma-secretase. Science 363:eaaw0930. 10.1126/science.aaw0930 [DOI] [PubMed] [Google Scholar]
- Zhu XQ, Zuo H, Maher BJ, Serwanski DR, LoTurco JJ, Lu QR, Nishiyama A (2012) Olig2-dependent developmental fate switch of NG2 cells. Development 139:2299–2307. 10.1242/dev.078873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo L, Sun B, Zhang CL, Fine A, Chiu SY, Messing A (1997) Live astrocytes visualized by green fluorescent protein in transgenic mice. Dev Biol 187:36–42. 10.1006/dbio.1997.8601 [DOI] [PubMed] [Google Scholar]
- Zuo H, Wood WM, Sherafat A, Hill RA, Lu QR, Nishiyama A (2018) Age-dependent decline in fate switch from NG2 cells to astrocytes after Olig2 deletion. J Neurosci 38:2359–2371. 10.1523/JNEUROSCI.0712-17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]