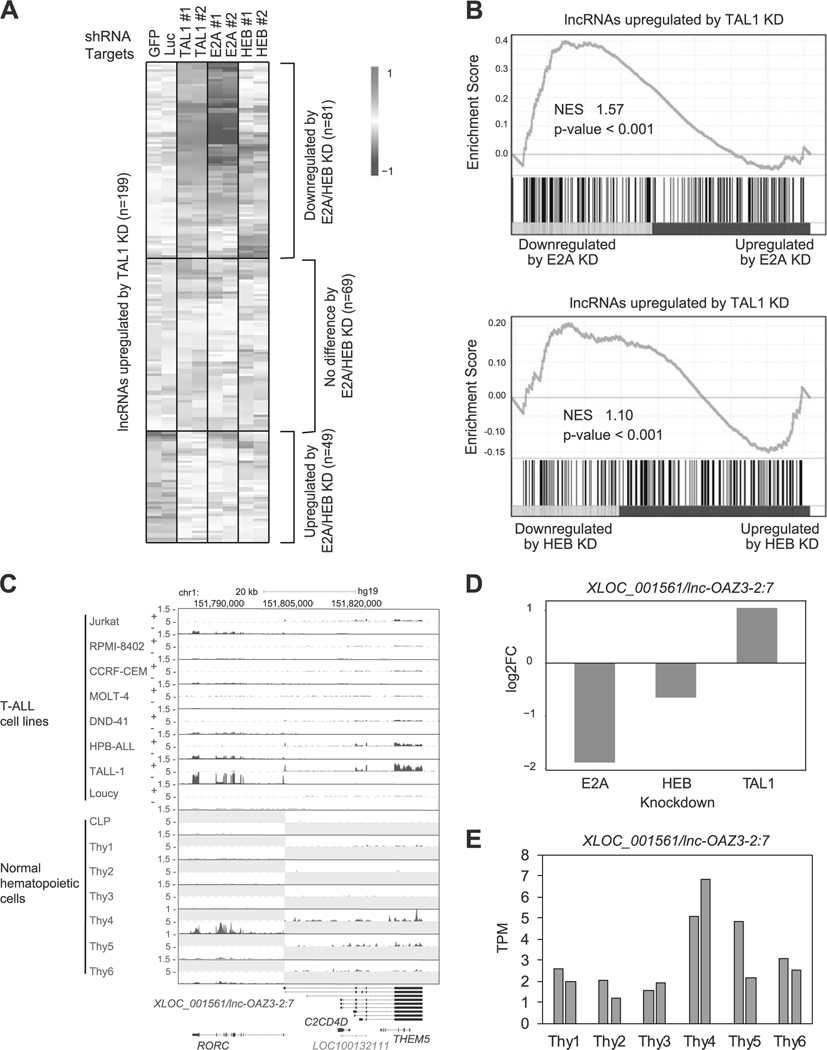
Fig. 5.
lncRNAs differentially controlled by TAL1 and E-proteins in T-ALL cells. a Jurkat cells were transduced with control shRNA (shGFP or shLuc) or shRNA targeting a transcription factor gene (TAL1, E2A, or HEB) by lentivirus infection. Two different shRNAs (1 and 2) were used for each gene. Heatmap images represent expression levels of 199 selected putative lncRNAs in two controls and two knockdown (KD) samples. b GSEA was performed to determine the correlation of lncRNAs negatively regulated by TAL1 (n = 199) and gene expression changes on KD of E2A and HEB. See Fig. 2c legend for details. c RNA-seq gene track showing expression and genomic loci of XLOC_001561/lnc-OAZ3–2:7 and RORC gene in T-ALL cell lines and normal hematopoietic cells. See Fig. 1b legend for details. d RNA expression changes of XLOC_001561/lnc-OAZ3–2:7 after knockdown of TAL1, E2A, and HEB in Jurkat cells. e RNA expression levels of XLOC_001561/lnc-OAZ3–2:7 in different stages of normal hematopoietic cells. See Fig. 4a legend for details.