Abstract
Expansion microscopy (ExM) physically magnifies biological specimens to enable nanoscale-resolution imaging on conventional microscopes. Current ExM methods permeate specimens with free-radical-chain-growth-polymerized polyacrylate hydrogels, whose network structure limits the local isotropy of expansion, and the preservation of morphology and shape at the nanoscale. Here we report that ExM is possible using hydrogels with more homogeneous network structure, assembled via non-radical terminal linking of tetrahedral monomers. As with earlier forms of ExM, such “tetra-gel”-embedded specimens can be iteratively expanded for greater physical magnification. Iterative tetra-gel expansion of HSV-1 virions by ~10x in linear dimension results in a median spatial error of 9.2 nm for localizing the viral envelope layer, rather than 14.3 nm from earlier versions of ExM. Moreover, tetra-gel-based expansion better preserved virion spherical shape. Thus, tetra-gels may support ExM with reduced spatial errors and improved local isotropy, pointing the way towards single biomolecule precision ExM.
Expansion microscopy (ExM) is in increasingly widespread use for biological imaging because it enables, via physical magnification of specimens1–3, nanoscale imaging on diffraction-limited optical microscopes. In ExM, biological specimens are densely permeated by a swellable hydrogel. Biomolecules and/or fluorescent tags are covalently anchored to the hydrogel network, the specimen chemically softened, and the hydrogel expanded upon immersion in water (typically ~4.5x in linear dimension). Protocols using off-the-shelf-chemicals4 have helped ExM find utility in a wide variety of contexts, ranging from the mapping of ribosome components and RNAs in synaptic compartments5, to the analysis of circadian rhythm neural circuitry in the Drosophila brain6, to the analysis of cancer in human biopsies7. Variants of ExM have been developed that achieve nanoscale localization of proteins and RNAs in preserved cells and tissues on diffraction-limited microscopes4,7–13. Additionally, multiple strategies have been introduced to expand specimens ~10x-20x, including applying the expansion process repeatedly (iterative expansion microscopy, or iExM)14, or by using superabsorbent hydrogels (X10 expansion microscopy)15. However, all ExM variants to date form the hydrogel mesh via free-radical chain-growth polymerization, a process that results in nanoscale structural heterogeneity in the hydrogel network16–20. This raises the question of whether another polymer might provide better structural homogeneity, and thus less spatial error, during the expansion process.
We describe two earlier ExM protocols to provide context. One popular version of ExM is protein-retention expansion microscopy (proExM)8. First, fixed biological specimens labelled with fluorophore-conjugated antibodies are exposed to a small molecule (acryloyl-X, SE, or AcX) that equips primary amine groups on antibodies and endogenous proteins with a polymerizable acryloyl group. The specimen is then immersed in a solution containing sodium acrylate and acrylamide, as well as the crosslinker N,N’-methylenebisacrylamide, to form a dense network of crosslinked sodium polyacrylate/polyacrylamide hydrogel throughout the specimen. During the polymerization, antibodies and proteins are covalently anchored to the hydrogel via AcX. Finally, proteinase K treatment cleaves most of the proteins, largely sparing the antibody-conjugated fluorophores, and allowing for expansion of the polymer-specimen composite in water. As a second example, in iExM14, fixed specimens are first labelled with primary antibodies and then DNA oligo-conjugated secondary antibodies. Next, DNA oligos bearing a gel-anchorable moiety are hybridized to the secondary antibody-conjugated DNA oligos. Then a first hydrogel is formed as above, which anchors the gel-anchorable DNA oligos to the hydrogel at the locations of the immunostained proteins; this gel is made with a cleavable cross-linker. Then, this gel is expanded as described above. Next, another round of hybridization of DNA oligos complementary to those anchored to the first hydrogel, and again bearing a gel-anchorable moiety, is performed. Then, a second hydrogel is formed throughout the expanded first hydrogel, so that the new DNA oligos are anchored to the second hydrogel at the original sites of immunostained proteins. Finally, the first hydrogel is cleaved, allowing the second hydrogel to expand upon immersion in water. The protein locations can be imaged after the polymer-anchored oligos are labelled by applying complementary oligos, potentially equipped with branched DNA for amplification purposes, bearing fluorescent dyes. The oligos, in short, provide a molecular mechanism (based on complementary strand hybridization) for signal transfer between the first and second hydrogels, as well as a scaffold for amplification and fluorescence readout.
In both cases, the free-radical chain-growth polymerization process that forms the sodium polyacrylate/polyacrylamide hydrogel has nanoscale structural heterogeneity. The size of local cross-linker density variations (Fig. 1a, “1”) can amount to 15–25 nm for thus-synthesized polyacrylamide gels16. Topological defects, such as dangling ends (Fig. 1a, “2”) and loops (Fig. 1a, “3”), can introduce deviations from uniform polymer meshes at the 1–10 nm length scale19,20. To date, many attempts have been made to obtain a more ideal hydrogel matrix19–24. One strategy was to adopt step-growth polymerization. For example, photoinitiated thiol-ene step-growth polymerization formed a more homogenous network structure vs. that yielded by chain-growth polymerization25–27. In another strategy, terminal linking of two kinds of tetrahedral polyethylene glycol (PEG) monomer was utilized to form a diamond lattice-like polymer network19,20,24. The hydrogel thus formed, via non-radical step-growth polymerization, known as a tetra-PEG gel, approximated an ideal polymer network, nearly void of structural defects, both as-synthesized and when slightly swollen (<1.5x linear expansion in water)28,29. The tetra-PEG gel structure was further extended to create hydrogels with versatile chemical and mechanical properties, including polyelectrolyte hydrogels30 and highly compressible and stretchable hydrogels31.
Figure 1. Design and synthesis of tetra-gel (TG) for expansion microscopy.
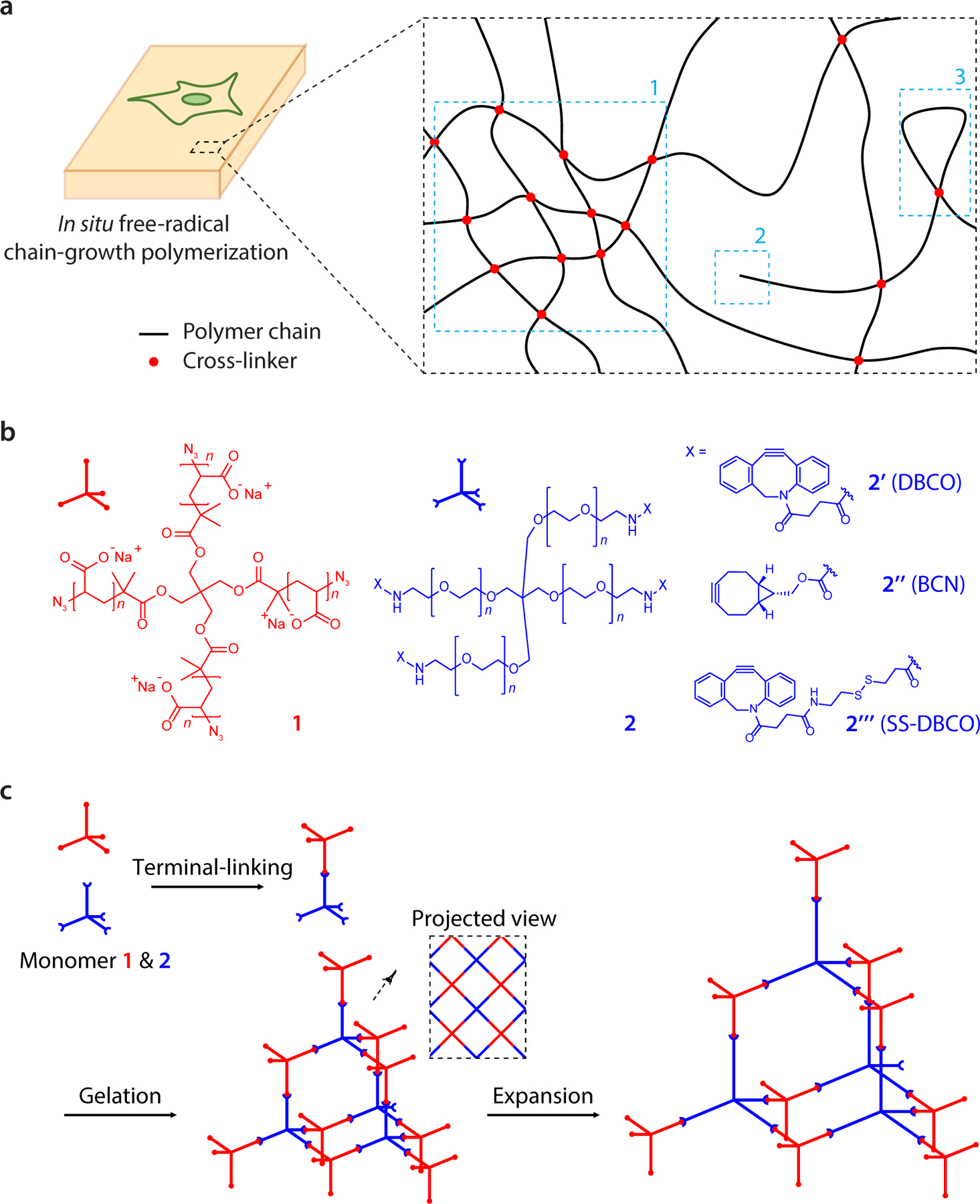
a, Cell/tissue-hydrogel composites formed by in situ free-radical chain-growth polymerization are known to have structural inhomogeneities in the range of tens of nanometres due to (1) local fluctuations of monomer and cross-linking densities, (2) dangling ends, and (3) loops formed within the polymer network. b, Design of TG monomer 1 and 2 with tetrahedral symmetry and reactive terminal groups. Specific monomer 2 terminal groups (2’, 2’’ and 2’’’) enable, for example, control of the reaction rate between the monomers and addition of functionalities to the polymer network. c, Formation and expansion of TG via click-chemistry-based terminal-linking of monomer 1 and 2. Inset, projected view of the TG polymer network.
We here report an ExM methodology using swellable hydrogels assembled by click-chemistry-based, non-radical linking of two complementary, tetrahedral monomers comprising backbones of polyacrylate and PEG respectively, which we call a tetra-gel (TG). TG-expanded herpes simplex virus type 1 (HSV-1) virions exhibited envelope shapes with significantly smaller spatial errors (9.2 nm of deviation) compared to classical sodium polyacrylate/polyacrylamide gel (PAAG)-expanded virions (14.3 nm). As a result of these reduced spatial errors, TG-based ExM significantly better preserved the spherical shapes of virions, compared to PAAG-based ExM. Thus, the TG serves as a candidate for a more structurally homogeneous expansion microscopy matrix compared to that synthesized by free-radical chain-growth polymerization, and points towards new ways to improve and extend the expansion microscopy toolbox.
Design of tetra-gel (TG) structure and assembly mechanism
We designed and synthesized tetrahedral monomers closely related to those used for tetra-PEG gels (Fig. 1b)24,28–30. One monomer (1) had a tetra-arm polyacrylate backbone with a clickable terminal group (azide) (synthesis and NMR spectra in Supplementary Fig. 1, 2 respectively), and the other (2) had a tetra-arm PEG backbone with a complementary terminal group (alkyne). The alkyne was varied to tune reactivity of terminal linking and functionality of the hydrogel (Fig 1b, monomers 2’, 2’’, 2’’’). The monomers had comparable molecular weights of ~10–20 kDa and an arm length of ~3–6 nm at the gelation step (at an ionic strength of ~0.150 M). In the solution phase, monomer 1 had four negatively charged polyacrylate arms (n = ~21 acrylate units), each arm of which is estimated to extend ~4.0–5.7 nm, based on the previously characterized persistence length of ≥ 4.0 nm32 and the fully stretched length of ~5.7 nm. Monomer 2 had four uncharged PEG arms (n = either ~57 or ~114 ethylene glycol units, depending on which version is utilized), each of which has a fully stretched length of ~21.7 or ~43.3 nm, respectively, which greatly exceeds its persistence length (0.38 nm)33. The PEG arm can thus be modelled as a freely jointed chain in solution34,35, whose root-mean-square arm length can be calculated as ~2.9 or ~4.1 nm, respectively. For monomer 2 synthesis, the n = ~57 backbone was used for variants 2’ and 2’’ and the n = ~114 backbone was used for variant 2’’’. The increased number of ethylene glycol units in 2’’’ enhanced its solubility, compensating for the hydrophobic nature of the disulfide-dibenzocyclooctyne (SS-DBCO) moiety. Finally, when monomer 1 and a selected monomer 2 are mixed, a hydrogel forms via click-chemistry-based terminal linking (Fig. 1c, left). Then, in water or aqueous buffers with ionic strength <~0.05M (concentration estimated from the Debye-Huckel theory for electrostatic potential energy in electrolyte solutions)36, the hydrogel would swell due to the reduced electrostatic screening of salt ions between the mutually repelling monomer 1 units (~84 negative charges per monomer), which in turn would elongate the originally unstructured PEG arms of the interconnecting monomer 2 (Fig. 1c, right).
Of monomer 2 variants in Fig. 1b, the bicyclononyne (BCN) version (monomer 2’’) was designed to support expansion of thicker specimens, because its slower click-reaction kinetics (by ~55% vs. standard DBCO)37 would provide additional time for monomers to equilibrate in concentration throughout tissues during pre-gelation incubation. In addition, a slower gelation reaction38 has been demonstrated to increase polymer network homogeneity. Indeed, gelation occurred in ~1 hour and ~2.5 hours at 4 °C for monomers 2’ and 2’’, respectively. The SS-DBCO version (monomer 2’’’) would support post-expansion cleavage of the polymer network into individual monomers, rendering it compatible with iterative expansion14. Monomer 2’’’-based gelation took ~4 hours at 4 °C, again suggesting utility for thicker specimens.
TG expansion of cells and tissues
We mixed monomer 1 and 2 in a 1:1 ratio (although small deviations may result depending on the exact fraction of monomer 1 whose t-butyl group was removed in the final step of monomer 1 synthesis, Supplementary Fig. 1, bottom row), and cast the gelling solution into a circular mold. Notably, deviation from a 1:1 ratio would result in additional defects in the polymer network and thus an increase in structural heterogeneity28. Using a virion-based local expansion isotropy assay (described later), we observed growing discrepancies vs. ground-truth virion size, when the monomer ratio deviated from 1:1 (Supplementary Fig. 3). After 1–2 hours of incubation at 37 °C, the gelling solution solidified into an optically transparent and mechanically elastic hydrogel (Fig. 2a, left). Similar to the PAAG gel and other types of hydrogel used in ExM that were formed by free-radical chain-growth polymerization7–13, the gel swelled after salt elution in water (~3-fold in linear dimension) (Fig. 2a, right). Fluorescently labelled gels (Fig. 2b) showed the linear expansion factor to be ~3.0–3.5x.
Figure 2. TG-mediated expansion of cells and tissues.

a, Image of TG (using monomer 2’’’) as synthesized (left, pre-expansion) and after swelling in deionized water (right, post-expansion). The two gels were cast in circular molds with identical dimensions. Grid size, 5 mm. b, TG labelled with fluorescein in pre- (left, 2’; same sizes and shapes when 2’’ and 2’’’ were used) and post-expansion (right, 2’, 2’’ and 2’’’) states. Irregular boundaries on the post-expansion images reflect the meniscus of water used to expand the gels. Scale bar, 5 mm. c, Left, HEK293 cells with alpha-tubulin immunostaining in pre- (inset) and post-expansion states. Expansion factor, 2.85x. Scale bars, 20 µm. Right, magnified views of the boxed regions 1 (top) and 2 (bottom). Scale bars, 1 µm [bottom, 2.85 µm; here and after, unless otherwise noted, scale bar sizes are provided at pre-expansion scale (i.e., biological scale) with the corresponding post-expansion size (i.e., physical size) indicated in parentheses]. Results are representative of 5 cells from the same cell culture. d, Root-mean-square (RMS) error curve for HEK293 cell expansion (blue line, mean; shaded, standard deviation; n = 8 cells from one culture). Inset, non-rigidly registered and overlaid pre- (green) and post-expansion (magenta) images used for the RMS error analysis. Scale bar, 20 µm (57 µm). e, Pre- (left top) and post-expansion (left bottom and right) Thy1-YFP mouse brain slices. Expansion factor, 3.00x. Scale bars, 5 mm (left) and 10 µm (right, 30 µm). The gelled brain slice on the left bottom panel was immunostained against YFP after the proteolysis step to enhance fluorescence. Inset, magnified view of the boxed region. Scale bar, 1 µm (3 µm). Results are representative of 2 brain slices from the same single batch of brain slice preparation for each panel. f, Pre-expansion (left column) and post-expansion (right column) Thy1-YFP mouse brain slices immunostained with Homer1 primary antibody and Alexa Fluor 647 (AF647)-conjugated secondary antibody, using sodium polyacrylate/polyacrylamide gel (PAAG, top row) and TG (bottom row). Expansion factor, 3.93x (PAAG) and 2.72x (TG). Scale bars, 300 µm (top right, 1.18 mm; bottom right, 815 µm). Results are representative of 3 brain slices from the same single batch of brain slice preparation for each gel type. g, Fluorescence retention of AF647, Cy5, and AF680 with TG and PAAG in immunostained mouse brain slices processed as in f (bar height, mean; black dots, individual data points; error bar, standard error of the mean; n = 3 brain slices from one mouse).
Next, we implemented proExM using TG. We infused antibody-stained cells and tissue slices with NHS-azide to link proteins and antibodies (via primary amines) to the polymer network through click-chemistry (Fig. 1b). We then formed the TG in situ, followed by adding proteinase K, and swelling in water. In one implementation, HEK cells were immunostained with antibodies against microtubules and then processed via the DBCO version (monomer 2’) of TG. Water swelled the HEK cell-TG composite ~2.9-fold, resulting in more sharply resolved microtubules than before expansion, on the same diffraction-limited confocal microscope (Fig. 2c). We aligned pre- and post- expansion confocal microscopy images of the microtubules via non-rigid registration, and quantified the amount of distortion in the expansion process (Fig. 2d), finding ~2.5% error over lengths of ~20 µm, comparable to that of PAAG-based proExM1,8. In another implementation, mouse brain slices expressing yellow fluorescent protein (YFP), processed using the BCN version (monomer 2’’) of TG, after immunostaining against YFP to enhance fluorescence (Fig. 2e), expanded ~3.0-fold and showed excellent detail in structures such as dendritic spines, as seen with PAAG-based proExM8.
Since TG forms via non-radical polymerization, TG-based expansion could potentially preserve molecules that are inactivated during free-radical chain-growth polymerization. We found that the fluorescence of dyes known to be inactivated by PAAG-based proExM8 (Fig. 2f), including Alexa Fluor 647 (AF647), Cy5, and AF 680, was largely retained after TG-based expansion (Fig. 2f, 2g). Thus TG-based ExM may preserve molecules not compatible with PAAG-based ExM.
TG-based iterative expansion
The SS-DBCO version of TG (using monomer 2’’’) would allow the TG polymer network to be cleaved at each network node (Fig. 3a), as appropriate for iExM14. Indeed, we found that HeLa cells, thus TG-expanded, could be iteratively expanded (Fig. 3b). We used a free-radical-chain-growth-polymerized gel for the second round of iExM, reasoning that the spatial errors introduced by second-round expansion would be negligible compared to those introduced by the first round (since, when considered in biological units, i.e., in terms of the relative spacing of biomolecules with respect to each other, second-round errors are effectively divided by the expansion factor of the first round). TG-based iExM expanded cells by ~16-fold, revealing the hollow of microtubules (Fig. 3c), with a sidewall separation comparable to that seen previously with iExM of BS-C-1 cells using PAAG-based iExM (Fig. 3d). We note that the larger sidewall distance than that seen by, for example, STORM, can be explained by the label size effect described previously – classical iExM requires larger labels than used in other super-resolution methods14. Finally, multiple DNA oligo-conjugated antibodies could be used at once, e.g., to label tubulin and clathrin in TG-expanded HeLa cells (Fig. 3e)14.
Figure 3. TG-based iterative expansion.

a, Monomeric cleaving of TG (monomer 2’’’) after re-embedding in a second hydrogel. A reducing agent, tris(2-carboxyethyl)phosphine) (TCEP), was applied to cleave the disulfide bonds in the TG polymer network. b, HeLa cell with beta-tubulin immunostaining, expanded by cleavable TG-based 2-round iterative expansion. Expansion factor, 15.6x. Scale bar, 5 µm (78.2 µm). Results are representative of 10 cells from the same cell culture. c, Top, magnified view of the boxed region in b. Scale bar, 100 nm (1.56 µm). Bottom, transverse line intensity profile of the microtubule in a single xy-plane in the dotted box (circle) and the fitted sum of two Gaussians (red line). The line intensity profile was averaged in the direction parallel to the microtubule axis over a length of 200 nm. d, Histogram of peak-to-peak distance between microtubule sidewalls in HeLa cells (n = 336 segments of a length of 200 nm from 10 cells in one culture). e, Left, HeLa cell with two-colour labelling of clathrin coated pits/vesicles and microtubules, expanded by TG-based 2-round iterative expansion. Expansion factor, 15.6x. Scale bar, 10 µm (156 µm). Right, magnified view of the boxed region for each colour channel. Scale bars, 1 µm (15.6 µm). Results are from 1 cell from the same cell culture preparation as in b.
Spatial analysis of TG-based ExM expansion error
To assess spatial errors introduced by TG-based iExM, we imaged the nanoscale arrangement of virion envelope proteins. To minimize the label size effect mentioned above, we developed a direct, nonspecific labelling strategy that targets primary amines of virus envelope proteins and reduces the label size from ~21 nm14 to that of a single DNA oligo (~7 nm). First, we directly conjugated 22-bp oligonucleotides to the envelope proteins on virions via a hydrazone-based DNA-to-peptide conjugation (Fig. 4a). We were careful not to expose viruses to any buffers containing detergents, for all steps from fixation to gelation, aiming to preserve the virus’s lipid bilayer envelope integrity, aiming to direct the majority of the labelling to accessible envelope proteins on the outside of the membrane. Herpes simplex virus type 1 (HSV-1) virions have a well-defined envelope protein layer that has been characterized by methods such as electron microscopy (EM)39–41, electron tomography (ET)42,43, and super-resolution microscopy44, and have appropriate length scale (170–190 nm diameter)44, with features in the tens of nanometres, to characterize the local homogeneity of TG. After direct labelling of envelope proteins, we then expanded HSV-1 virions with TG-based iExM vs. PAAG-based iExM (in both cases using sodium polyacrylate/polyacrylamide second-round gels) (Fig. 4b). We note that this protocol was compatible with dual-colour labelling and imaging of envelope proteins and DNA (Supplementary Fig. 4), and could be applied to other types of enveloped viruses, such as vesicular stomatitis virus (VSV), an RNA virus (Supplementary Fig. 5), and human immunodeficiency virus (HIV), a retrovirus (Supplementary Fig. 6). For quantitative analysis of envelope shapes, to gauge TG-based iExM spatial error, we focused on viruses of spherical geometry (i.e., HSV-142,43 and HIV45).
Figure 4. Spatial errors introduced by TG-based vs. classical PAAG-based iterative expansion microscopy.

a, Short DNA oligos (22 bp) were covalently conjugated to the envelope proteins of herpes simplex virus type 1 (HSV-1) virions via hydrazone formation, which allows labelling transfer across multiple hydrogels, amplification based on branched DNA, and fluorescence readout based on hybridization of fluorescent oligos. b, Schematic illustration of TG- (top) and PAAG-based (bottom) iterative expansion of HSV-1 virions with the direct oligo-conjugation as in a. PAAG-based re-embedding and expansion was used for all expansion rounds after the first round, reasoning that most of the error of an iterative expansion protocol is introduced in the first round of expansion (see text for details). c, HSV-1 virions with directly labelled envelope proteins, expanded by TG- (left) and PAAG-based (right) 2-round iterative expansion. Scale bar, 1 µm (TG, 10.3 µm; PAAG, 15.3 µm). Expansion factor, 10.3x (TG) and 15.3x (PAAG). Results are representative of 3 separately acquired fields of view from a single hydrogel preparation for each gel type, using virions from the same single batch of live HSV-1 preparation. d, Single-particle averaged images of HSV-1 virions after TG (top)- and PAAG-based (bottom) 2-round iterative expansion (TG, n = 396; PAAG, n = 362 virions; virions from the same single batch of live HSV-1 preparation). Full width half maximum (FWHM) of the radial intensity profile (the line intensity profile from the particle centre to the edge of the particle), 73.6 nm (TG) and 91.0 nm (PAAG). Scale bars, 100 nm. e, Single-particle averaged images of HIV virions expanded by TG- (top) and PAAG-based (bottom) 2-round iterative expansion (TG, n = 61; PAAG, n = 73 virions; virions from the same single batch of live HIV preparation). FWHM of the radial intensity profile, 49.1 nm (TG) and 119.0 nm (PAAG). Scale bars, 100 nm. f, Top left and middle, representative single xy-plane images of HSV-1 virions expanded by TG- and PAAG-based 2-round iterative expansion. White lines indicate the 8 directions along which the virion’s radii (R) were measured. Scale bars, 100 nm. Top right, representative line intensity profile (circles) along a single direction (Direction 1) of the TG-expanded virion and the fitted Gaussian (red line). Distance from the centre of the Gaussian to the centre of the virion was defined as R along that direction. Bottom, histogram of the particle-mean-normalized R (Rn) for all the measured line profiles [TG, n = 1977; PAAG, n =1889 profiles; from 352 (TG) and 330 (PAAG) virions from the same single batch of live HSV-1 preparation]. g, Standard deviation (σ) of HSV-1 virion radii (R) within individual virions for TG- and PAAG-based iterative expansion (p = 2.4 × 10−24, 2-sided Wilcoxon rank sum test; TG, n = 352; PAAG, n = 330 virions; virions from the same single batch of live HSV-1 preparation). Data are presented as box plots with ends of whiskers representing maximum and minimum values of the distribution after outlying points (values above 75th percentile + 1.5 × interquartile range, or below 25th percentile – 1.5 × interquartile range) are excluded, upper line of box representing 75th percentile, middle line of box representing 50th percentile (median), lower line of box representing 25th percentile, and plus signs representing individual values for the outlying points.
The expansion factor for TG-based iExM of HSV-1 virions was ~10.3–13.3, and for PAAG-based iExM, it was ~15.3–18.7. TG-based iExM appeared to result in more spherical virions than PAAG-based iExM (Fig. 4c). From single-particle averaged virion images, we found that the envelope protein profile of TG-expanded HSV-1 virions was substantially sharper than that of PAAG-expanded ones (Fig. 4d). Similarly, TG-expanded HIV virions had sharper envelope profiles than PAAG-expanded ones (Fig. 4e).
To quantify spatial errors associated with each expansion process, we measured the standard deviation of virion radii (σ) within the midplane of individual HSV-1 virions (Fig. 4f; computed in biological units, i.e., normalized by the expansion factor, to compensate for the different expansion factors for TG- and PAAG-based iExM protocols). TG-expanded HSV-1 virions [σm (median σ across virions) = 9.2 nm; n = 352 virions; virions from a single batch of live HSV-1 preparation] had a significantly smaller σm value compared to PAAG-expanded HSV-1 virions [σm = 14.3 nm; n = 330 virions; virions from the same single batch of live HSV-1 preparation] (p = 2.4 × 10−24, 2-sided Wilcoxon rank sum test) (Fig. 4g). We calculated the same metric of σ for the single-particle averaged images of HSV-1 virions (Fig. 4d), and saw smaller spatial errors for TG-expanded virions [σa (σ of the averaged virion image) = 3.4 nm] compared to the PAAG-expanded ones (σa = 4.0 nm); note that the absolute difference between the two σa values was numerically small because of population averaging. HIV virions had much sparser envelope proteins per virion than HSV-1 virions, which precluded computing σ at the individual particle level, but for single-particle averaged images, TG-expanded HIV virions (σa = 8.9 nm) had a smaller σa value than the PAAG-expanded ones (σa = 12.9 nm), showing again reduced spatial errors for TG-based iExM (Fig. 4e).
We made a unifying computational model so that different measures of hydrogel-based variability could be compared. We took the spatial errors calculated across individual particles (Fig. 4g) as gold-standard metrics of spatial error, and sought to examine whether other metrics, such as those derived from single-particle averaged images (Fig. 4d, 4e), were consistent. For the model, we considered both chemical variability (e.g., hydrogel-associated local anisotropy) and biological variability (e.g., biological distribution of virion size). We developed a computational model that simulated the averaged images from individual simulated virions (Supplementary Methods, Supplementary Fig. 7). When single-particle averaged images were simulated based on this model, we observed sharper envelope profiles for TG than PAAG, consistent with the experimental results (Supplementary Fig. 7). Moreover, the metrics derived from the simulated images were comparable to the experimental values to a great extent, despite the highly simplified assumptions and designs of the model.
To assess how the improved accuracy of TG- vs. PAAG-based expansion helps preserve the shapes of nanoscale objects like viruses, we characterized the expanded HSV-1 virion cross-sectional shapes with key shape descriptors, such as eccentricity, the ratio of the minor axis to the major axis (e; e = 1 for a perfect circle; see examples in Fig. 5a, top, for sketches of shapes with different e), sphericity, the ratio of the radius of the inscribing circle to the radius of the circumscribing circle (s; s = 1 for a perfect circle; see examples in Fig. 5b, top, for sketches of shapes with different s), and normalized circular standard deviation, the standard deviation of the 8 radii within each particle divided by the mean for that particle (σn; σn = 0 for a perfect circle; see examples in Fig. 5c, top, for sketches of shapes with different σn). We found that the TG-expanded virions were significantly closer to circular compared to PAAG-expanded virions (p < 10−5 for all the descriptors, 2-sided Wilcoxon rank sum test) (Fig. 5). The changes were not small – to wit, the median deviation from a perfect circle was substantially reduced for TG compared to PAAG, i.e. by 62% (e), 64% (s), and 59% (σn) of the PAAG deviation. We applied the same shape analysis to HIV virions, and obtained data consistent with the HSV-1 data (Supplementary Fig. 6). Going forward, even higher expansion factors, potentially revealing individual proteins on virions, may be possible by expanding ~40x via 3 rounds of iterative expansion, with the initial expansion round TG-based and the final two rounds sodium polyacrylate/polyacrylamide-based (Supplementary Fig. 8, Supplementary Video 1).
Figure 5. Shape analysis of TG- vs PAAG-expanded HSV-1 virions.
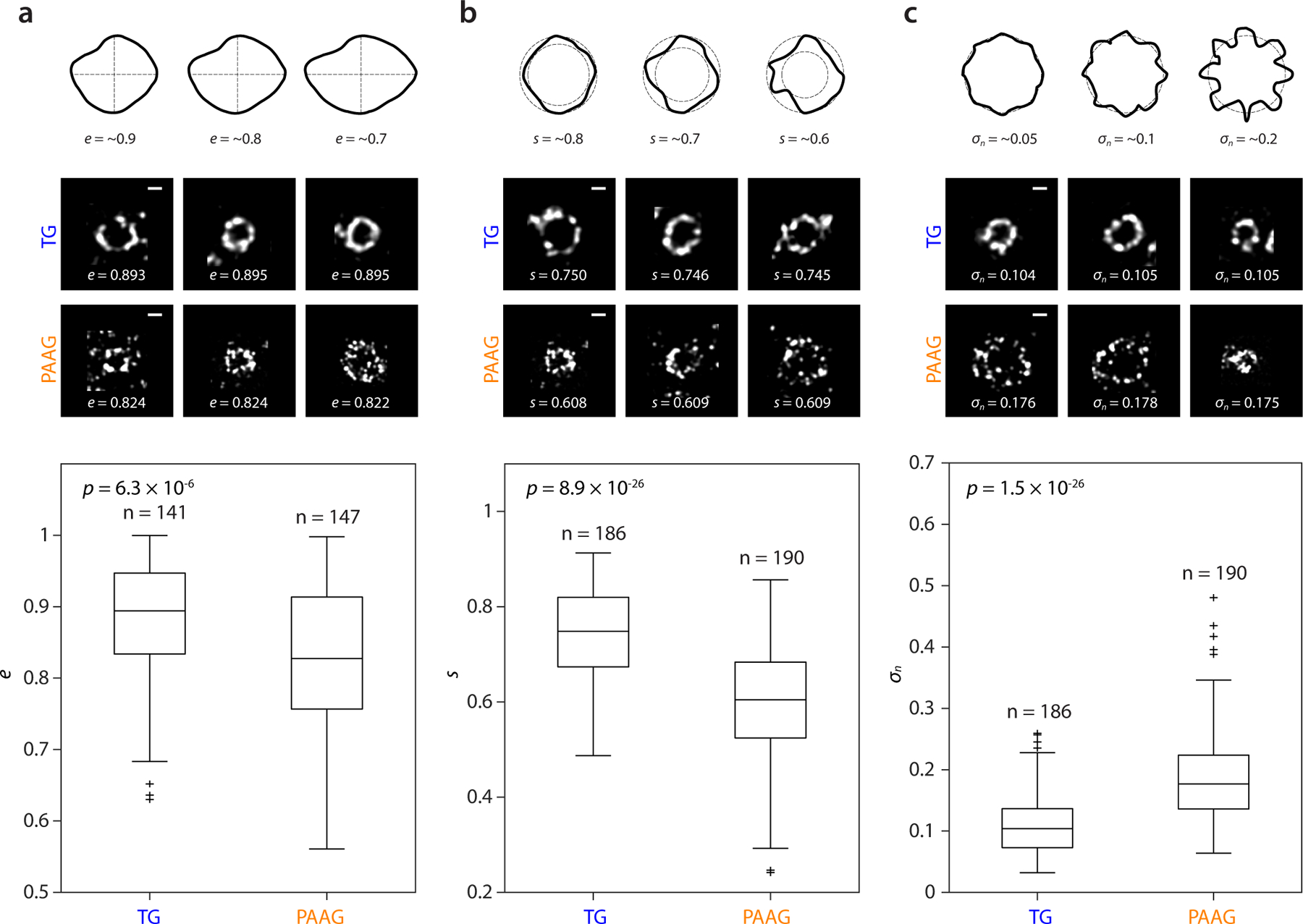
a. Top, schematic examples of shapes with ~0.9, 0.8, and 0.7 eccentricity (e). Middle, HSV-1 virions expanded by TG- (top row) and PAAG-based (bottom row) 2-round iterative expansion, with the z-midplane cross-sectional shapes scoring close to the median value of e within each hydrogel group. Bottom, eccentricity (e) of HSV-1 virion z-midplane cross-sections for TG- and PAAG-based iterative expansion (p = 6.3 × 10−6, 2-sided Wilcoxon rank sum test; TG, n = 141; PAAG, n = 147 virions; virions from the same single batch of live HSV-1 preparation). Scale bars, 100 nm. b, Top, schematic examples of shapes with ~0.8, 0.7, and 0.6 sphericity (s). Middle, HSV-1 virions expanded by TG- (top row) and PAAG-based (bottom row) 2-round iterative expansion, with the z-midplane cross-sectional shapes scoring close to the median value of s within each hydrogel group. Bottom, sphericity (s) of HSV-1 virion z-midplane cross-sections for TG- and PAAG-based iterative expansion (p = 8.9 × 10−26, 2-sided Wilcoxon rank sum test; TG, n = 186; PAAG, n = 190 virions; virions from the same single batch of live HSV-1 preparation). Scale bars, 100 nm. c, Top, schematic examples of shapes with ~0.05, 0.1, and 0.2 normalized circular standard deviation (σn). Middle, HSV-1 virions expanded by TG- (top row) and PAAG-based (bottom row) 2-round iterative expansion, with the z-midplane cross-sectional shapes scoring close to the median value of σn within each hydrogel group. Bottom, normalized circular standard deviation (σn) of HSV-1 virion z-midplane cross-sections for TG- and PAAG-based iterative expansion (p = 1.5 × 10−26, 2-sided Wilcoxon rank sum test; TG, n = 186; PAAG, n = 190 virions; virions from the same single batch of live HSV-1 preparation). Scale bars, 100 nm. In a–c, data are presented as box plots with ends of whiskers representing maximum and minimum values of the distribution after outlying points (values above 75th percentile + 1.5 × interquartile range, or below 25th percentile – 1.5 × interquartile range) are excluded, upper line of box representing 75th percentile, middle line of box representing 50th percentile (median), lower line of box representing 25th percentile, and plus signs representing individual values for the outlying points.
Conclusions
We have found that tetra-gel (TG) polymer assembled via non-radical step-growth polymerization of tetrahedral monomers is capable of mediating ExM. When TG-based iterative expansion protocols were compared to classical PAAG-based protocols, using HSV-1 virion envelope proteins as an assay, we saw a reduction in spatial errors introduced by the expansion process, from 14.3 nm to 9.2 nm at the individual particle level. We also found that TG-based iExM better retained the nanoscale shape of the HSV-1 virion envelope, using multiple classical shape descriptors such as ellipticity and sphericity to capture the improvements of TG over PAAG. Thus, TG-based ExM may overcome local anisotropy of the expansion process inherent in previous methods, and support expansion with better preserved nanoscale morphology and shape. The single-digit distortions introduced by TG-based ExM raises the tantalizing question of whether new hydrogel chemistries might be able to achieve expansion precision approaching that of the size of individual biomolecules such as proteins.
Our current study is focused on the design principles of the hydrogel matrices for ExM, and is not yet a protocol for general scientific use. For TG-based ExM to be useful in everyday scientific investigations, a number of improvements will be required. The chemicals must be made broadly available, through commercialization or other arrangements. Biomolecules of interest are currently anchored to the ends of monomers when incorporated into the hydrogel network, meaning that every anchored biomolecule results in a “defect” in the polymer network because it terminates one of the monomer’s arms; replacing such terminal-based anchoring with sidechain-based anchoring may improve TG structural homogeneity for ExM still further. The ideal size of the monomer has not yet been established; current TG-based ExM monomers are much larger than the sodium acrylate monomers used in classical ExM protocols, raising the question of whether smaller TG monomers would better permeate dense cells and tissues. Finally, appreciation of the improvements offered by TG over PAAG most likely will require post-expansion antibody staining, or the use of very small tags such as nanobodies for pre-expansion staining, since the improvements offered by TG are smaller than the sizes of antibodies. To enable post-expansion antibody staining, the disulfide in the monomer will need to be replaced by a cleavable moiety compatible with the high-temperature treatments, basic pHs, and other conditions typically used to denature proteins and expand them away from each other4,8,11. For this approach, intramolecular epoxide cross-linking might be helpful for epitope preservation46. Such a second-generation TG-based ExM process, in the future, may be useful for investigating the detailed nanoscale spatial arrangement of molecular species in complexes, in cells and tissues, in healthy and disease states.
Methods
Synthesis of tetra-gel (TG) monomers
Monomer 1 was synthesized using a procedure modified from a previously described synthesis (Supplementary Fig. 1)30. First, tetra-arm poly(t-butyl acrylate) with bromo terminal groups (4) was synthesized by atom transfer radical polymerization (ATRP). Next, tetra-arm poly(t-butyl acrylate) with azide terminal groups (5) was synthesized by replacing bromines of 4 with azides (Supplementary Fig. 2). Finally, monomer 1 was synthesized by hydrolysis and neutralization of 5 to a final pH of ~7. Monomer 2’, 2’’, and 2’’’ were synthesized by N-hydroxysuccinimide (NHS) ester-based conjugation of the alkynes (DBCO-NHS, BCN-NHS, or DBCO-SS-NHS) to the terminal primary amines of tetra-arm polyethylene glycols (PEGs). A detailed procedure for the synthesis can be found in the Supplementary Methods.
Cell culture
HEK293FT cells (Thermo Fisher) were cultured in chambered coverglasses (CultureWell, Thermo Fisher) to a confluency of 60–80%1, fixed, and immunostained1,4,8. Briefly, the cells were treated with 3% (w/v) formaldehyde and 0.1% (w/v) glutaraldehyde in phosphate buffered saline (PBS, 1x unless otherwise noted) for 10 min at room temperature before the subsequent quenching, blocking, immunostaining, and expansion procedure. HeLa cells (ATCC CCL-2) were plated on coverglasses coated with Matrigel (BD Sciences) to a confluency of 50–90% and fixed1,4,8,14. The cells were treated with PBS + 3% (w/v) formaldehyde + 0.1% (w/v) glutaraldehyde for 10 min at room temperature before the subsequent quenching, blocking, and expansion procedure. All the cells were not subjected to additional authentication and were not tested for mycoplasma contamination. A detailed procedure for the immunostaining can be found in the Supplementary Methods.
Thy1-YFP mouse brain slice
All procedures involving Thy1-YFP-H transgenic mice (Jackson Laboratory) were carried out in accordance with the US National Institutes of Health Guide for the Care and Use of Laboratory Animals and approved by the MIT Committee on Animal Care. All the animals were housed in group in standardized cages (temperature: 20–22 °C, humidity: 30–70%) with a 12-hour light/12-hour dark cycle with unrestricted access to food and water. 50–100 μm coronal brain slices of Thy1-YFP-H mice of 2–4 months old, both male and female, were prepared and immunostained for expansion4,8,13. A detailed procedure for the immunostaining can be found in the Supplementary Methods.
General procedure for gelation, digestion, and expansion
Fixed (and immunostained) cells and tissues were incubated in ~0.1–0.2 mg/mL NHS-azide in PBS overnight at room temperature and washed with PBS twice. To form the gelling solution, the two monomer solutions were mixed at a close to 1:1 molar ratio, and an additional amount of water was added to adjust the final concentration of monomer 1 to ~3.3% (w/v). For example, 10 µL each of monomer 1 and monomer 2’ (both ~200 mg/mL) and 40 µl of water were mixed to yield the gelling solution. Gelation was carried out for 1–2 hours at 37 °C (blank gels) or overnight at 4 °C (cell and tissue samples) in a gelation chamber4,8. The gelled cell and tissue samples were incubated in digestion buffer with proteinase K (8 units/mL) (New England BioLabs; 1:100 dilution) overnight at room temperature4,8 and expanded in an excess amount of water three times, each time for 20 min.
Expansion of HeLa cells (pre-expansion immunostaining and iterative expansion)
Fixed HeLa cells were stained with primary antibodies, oligo-conjugated secondary antibodies, and azide-modified tertiary oligos as previously described1,14. The cells were gelled with a cleavable TG gelling solution prepared by mixing monomer 1, monomer 2’’’, and water. The gelled samples were incubated in digestion buffer with Proteinase K at 8 units/mL overnight at room temperature with gentle shaking before de-hybridization of the oligos from the gel-anchored oligos. The expanded samples were re-embedded in N,N′-bis(acryloyl)cystamine (BAC)-crosslinked non-expanding gel, hybridized with 1st linker oligos, re-embedded in N,N′-diallyl L-tartardiamide (DATD)-crosslinked expanding gel, and incubated in BAC-cleaving buffer. For fluorescence readout, the samples were incubated with fluorophore-conjugated locked nucleic acid (LNA) oligos and expanded in water. A detailed procedure for the expansion can be found in the Supplementary Methods.
Expansion of HSV-1 virions (direct-labelling and iterative expansion)
Purified HSV-1 virion stock47 was diluted before being drop-casted onto a plasma cleaned #0 circular 12-mm coverslip. After 15 min of incubation at room temperature, the virions were fixed in 4% PFA in PBS for 10 min. Azide-modified oligos were directly conjugated to the virion envelope proteins via SoluLink bioconjugation chemistry as previously described1. The virions were gelled, digested, expanded, and hybridized with fluorophore-conjugated LNA oligos using a similar procedure to that of the HeLa cell expansion. For 3-round iterative expansion, BAC-cleaved samples were re-embedded in DATD-crosslinked non-expanding gel, hybridized with 2nd linker oligos, re-embedded in bis-crosslinked expanding gel, and incubated in DATD-cleaving buffer before the LNA oligo-conjugation, expansion, and imaging. A detailed procedure for the expansion and for the sodium polyacrylate/polyacrylamide gel (PAAG) control can be found in the Supplementary Methods.
Expansion of HIV and VSV virions (direct labelling and iterative expansion)
Purified HIV and VSV virions were immobilized, fixed, conjugated with oligos, gelled, and expanded following the 2-round iterative expansion protocol of HSV-1 virions.
Imaging and visualization
All the expanded samples were imaged with a diffraction-limited spinning disk confocal microscope (CSU-W1, Yokogawa on Eclipse Ti-E microscope body, Nikon) with a CFI Apo LambdaS LWD 40x, 1.15 NA water-immersion objective (Nikon), controlled by NIS-Elements AR v4.60.00 (Nikon). The two-colour HeLa cell images and all the virion images were deconvolved with theoretical point-spread-functions (PSFs) (Huygens Essential for MacX11 built on Feb 7, 2013, SVI) before visualization and image analysis. Unless otherwise noted, all the 3D renderings were generated using Imaris x64 8.3 (Oxford Instruments). Part of the image datasets were analysed and visualized using ImageJ (Fiji) 2.10/1.53c. Part of the plots were generated using OriginPro8.1 (Origin).
Image analysis
For virion envelope protein analysis, first, single-particle averaged virion images were generated using a semi-automated image analysis pipeline implemented on MATLAB R2018a-R2020b (“Virus Particle Analysis”)48. Each virion was manually aligned, automatically cropped, calibrated with the expansion factor, and arithmetically averaged to generate the single-particle averaged images. Next, using Virus Particle Analysis, spatial arrangements of the virion envelope proteins were quantified. Radii in 8 directions (45 degrees apart) were measured for each virion as the distance from the particle centroid to the Gaussian-fitted centre of the envelope profile. After inspection to remove unfitted profiles, standard deviations of all the accepted radii within the same particle (σ) were reported as population statistics for virions with >= 3 accepted radii. For HSV-1 virion shape analysis, virions with >= 6 accepted radii (for eccentricity, virions with >= 6 accepted radii and with the accepted radii constituting at least 2 mutually perpendicular diameters) were used to assure an accurate approximation of the particle shapes. Eccentricity (e) was defined as the ratio of the minor axis to the major axis. Sphericity (s) was defined as the ratio of the radius of the inscribing circle to the radius of the circumscribing circle. The normalized circular standard deviation (σn) was defined as the standard deviation of normalized radii (Rn) within each particle. For microtubule analysis, the peak-to-peak distance between microtubule sidewalls was measured using a semi-automated algorithm implemented on MATLAB R2018a-R2020b (“Microtubule Peak-to-Peak Distance Analysis”)48. On maximum intensity z-projection images of beta-tubulin-stained HeLa cells, two points were manually selected along the centreline of a microtubule segment, and a 200 nm (in biological length) segment was cropped out from the selected segment. The line intensity profile along the 200 nm segment was fitted with two Gaussian functions to detect the two peaks in the fluorescence intensity, between which the distance was measured as the peak-to-peak distance of the microtubule sidewalls. A detailed description of image analysis can be found in the Supplementary Methods.
Modelling and simulation of single-particle averaged HSV-1 virion images
A model of spherical virions was developed on MATLAB R2020a-R2020b based on experimental parameters derived from individual HSV-1 virions, in order to simulate the single-particle averaged HSV-1 virion images and their quantitative metrics (“HSV-1 Averaged Particle Image Simulation”)48. A detailed description of the modelling can be found in the Supplementary Methods.
Supplementary Material
Acknowledgements
We thank Y.-Y. Chou and T. Kirchhausen at HMS for help with VSV stock preparation and virion immobilization; C. Linghu, and O. Shemesh for help with HSV-1 stock preparation; P. Valdes and C. Zhang for helpful discussion about sample staining and expansion; M.J. Kauke for helpful discussion about DNA staining; S.M. Asano for helpful discussion about image analysis; G.H. Huynh for help with mouse brain slice preparation; F. Chen for helpful discussion about monomer and polymerization chemistry design; R. Herlo at HMS for helpful discussion about virion envelope proteins. E.S.B. acknowledges Lisa Yang and Y. Eva Tan, John Doerr, the Open Philanthropy Project, NIH 1R01NS087950, NIH 1RM1HG008525, NIH 1R01DA045549, NIH 2R01DA029639, NIH 1R01NS102727, NIH 1R01EB024261, NIH 1R01MH110932, the HHMI-Simons Faculty Scholars Program, IARPA D16PC00008, U.S. Army Research Laboratory and the U.S. Army Research Office under contract/grant W911NF1510548, U.S.-Israel Binational Science Foundation Grant 2014509, NSF Grant 1734870, and NIH Director’s Pioneer Award 1DP1NS087724. C.-C.Y. acknowledges the McGovern Institute for Brain Research at MIT for the Friends of the McGovern Fellowship. S.U. was supported by Biogen and NIH 5R01GM075252–13S grants awarded to T. Kirchhausen.
Footnotes
Competing interests
R.G., C.-C.Y., L.G., and E.S.B. have filed for patent protection on a subset of the technologies here described. E.S.B. is a cofounder of a company that aims to commercialize ExM for medical purposes. R.G., C.-C.Y., L.G., and E.S.B. are a co-inventor on multiple patents related to ExM. The authors declare no other competing interests.
Additional information
Supplementary information is available in the online version of the paper. Reprints and permission information is available online at www.nature.com/reprints. Correspondence and requests for materials should be addressed to E.S.B.
Code availability
Analysis code used in this study, including “Virus Particle Analysis”, “HSV-1 Averaged Particle Image Simulation”, and “Microtubule Peak-to-Peak Distance Analysis”, are available at https://github.com/jayyu0528/.
Data availability
The total raw data size of the study exceeds 350 GB. The data that support this study are available from the authors upon reasonable request.
References
- 1.Chen F, Tillberg PW & Boyden ES Expansion microscopy. Science 347, 543–548 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao R, Asano SM & Boyden ES Q&A: Expansion microscopy. BMC Biol 15, 50 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wassie AT, Zhao Y & Boyden ES Expansion microscopy: Principles and uses in biological research. Nat. Methods 16, 33–41 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asano SM et al. Expansion microscopy: Protocols for imaging proteins and RNA in cells and tissues. Curr. Protoc. Cell Biol 80, e56 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hafner AS, Donlin-Asp PG, Leitch B, Herzog E & Schuman EM Local protein synthesis is a ubiquitous feature of neuronal pre- And postsynaptic compartments. Science 364, eaau3644 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Schlichting M et al. Light-mediated circuit switching in the Drosophila neuronal clock network. Curr. Biol 29, 3266–3276.e3 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Zhao Y et al. Nanoscale imaging of clinical specimens using pathology-optimized expansion microscopy. Nat. Biotechnol 35, 757–764 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tillberg PW et al. Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nat. Biotechnol 34, 987–992 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chozinski TJ et al. Expansion microscopy with conventional antibodies and fluorescent proteins. Nat. Methods 13, 485–488 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen F et al. Nanoscale imaging of RNA with expansion microscopy. Nat. Methods 13, 679–84 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ku T et al. Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nat. Biotechnol 34, 973–981 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gambarotto D et al. Imaging cellular ultrastructures using expansion microscopy (U-ExM). Nat. Methods 16, 71–74 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao R et al. Cortical column and whole-brain imaging with molecular contrast and nanoscale resolution. Science 363, eaau8302 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang JB et al. Iterative expansion microscopy. Nat. Methods 14, 593–599 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Truckenbrodt S et al. X10 Expansion Microscopy enables 25 nm resolution on conventional microscopes. EMBO Rep 19, e45836 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen Y, Ramon O, Kopelman IJ & Mizrahi S Characterization of inhomogeneous polyacrylamide hydrogels. J. Polym. Sci. Part B Polym. Phys 30, 1055–1067 (1992). [Google Scholar]
- 17.Yazici I & Okay O Spatial inhomogeneity in poly(acrylic acid) hydrogels. Polymer (Guildf) 46, 2595–2602 (2005). [Google Scholar]
- 18.Orakdogen N & Okay O Correlation between crosslinking efficiency and spatial inhomogeneity in poly(acrylamide) hydrogels. Polym. Bull 57, 631–641 (2006). [Google Scholar]
- 19.Di Lorenzo F & Seiffert S Nanostructural heterogeneity in polymer networks and gels. Polym. Chem 6, 5515–5528 (2015). [Google Scholar]
- 20.Gu Y, Zhao J & Johnson JAA (macro)molecular-level understanding of polymer network topology. Trends Chem 1, 318–334 (2019). [Google Scholar]
- 21.Martens P & Anseth KS Characterization of hydrogels formed from acrylate modified poly(vinyl alcohol) macromers. Polymer (Guildf) 41, 7715–7722 (2000). [Google Scholar]
- 22.Lutolf MP & Hubbell JA Synthesis and physicochemical characterization of end-linked poly(ethylene glycol)-co-peptide hydrogels formed by Michael-type addition. Biomacromolecules 4, 713–722 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Malkoch M et al. Synthesis of well-defined hydrogel networks using Click chemistry. Chem. Commun 2006, 2774–2776 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Sakai T et al. Design and fabrication of a high-strength hydrogel with ideally homogeneous network structure from tetrahedron-like macromonomers. Macromolecules 41, 5379–5384 (2008). [Google Scholar]
- 25.Fairbanks BD et al. A versatile synthetic extracellular matrix mimic via thiol-norbornene photopolymerization. Adv. Mater 21, 5005–5010 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cui J et al. Synthetically simple, highly resilient hydrogels. Biomacromolecules 13, 584–588 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saffer EM et al. SANS study of highly resilient poly(ethylene glycol) hydrogels. Soft Matter 10, 1905–1916 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsunaga T, Sakai T, Akagi Y, Chung U & Shibayama M Structure characterization of tetra-PEG gel by small-angle neutron scattering. Macromolecules 42, 1344–1351 (2009). [Google Scholar]
- 29.Matsunaga T, Sakai T, Akagi Y, Chung U Il & Shibayama, M. SANS and SLS studies on tetra-arm PEG gels in as-prepared and swollen states. Macromolecules 42, 6245–6252 (2009). [Google Scholar]
- 30.Oshima K, Fujimoto T, Minami E & Mitsukami Y Model polyelectrolyte gels synthesized by end-linking of tetra-arm polymers with click chemistry: Synthesis and mechanical properties. Macromolecules 47, 7573–7580 (2014). [Google Scholar]
- 31.Kamata H, Akagi Y, Kayasuga-Kariya Y, Chung U Il & Sakai T ‘Nonswellable’ hydrogel without mechanical hysteresis. Science 343, 873–875 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Tricot M Comparison of experimental and theoretical persistence length of some polyelectrolytes at various ionic strengths. Macromolecules 17, 1698–1704 (1984). [Google Scholar]
- 33.Kienberger F et al. Static and dynamical properties of single poly(ethylene glycol) molecules investigated by force spectroscopy. Single Mol 1, 123–128 (2000). [Google Scholar]
- 34.Kuhn W Über die Gestalt fadenförmiger Moleküle in Lösungen. Kolloid-Zeitschrift 68, 2–15 (1934). [Google Scholar]
- 35.Kuhn W & Kuhn H Die Frage nach der Aufrollung von Fadenmolekeln in strömenden Lösungen. Helv. Chim. Acta 26, 1394–1465 (1943). [Google Scholar]
- 36.Debye P & Huckel E The theory of electrolytes. I. Lowering of freezing point and related phenomena. Phys. Zeitschrift 24, 185–206 (1923). [Google Scholar]
- 37.Dommerholt J, Rutjes FPJT & van Delft FL Strain-promoted 1,3-dipolar cycloaddition of cycloalkynes and organic azides. Top. Curr. Chem 374, 16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zander ZK, Hua G, Wiener CG, Vogt BD & Becker ML Control of mesh size and modulus by kinetically dependent cross-linking in hydrogels. Adv. Mater 27, 6283–6288 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang XT, Unnerstall JR, Ibuki T, Wang H & Pappas GD Dielectrophoretic manipulation and characterization of herpes simplex virus-1 capsids. Eur. Biophys. J 30, 268–272 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Liu F & Zhou ZH Comparative virion structures of human herpesviruses. in Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis (eds. Arvin A et al. ) (Cambridge University Press, 2007). [PubMed] [Google Scholar]
- 41.Brown JC & Newcomb WW Herpesvirus capsid assembly: Insights from structural analysis. Curr. Opin. Virol 1, 142–149 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grünewald K et al. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science 302, 1396–1398 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Maurer UE, Sodeik B & Grunewald K Native 3D intermediates of membrane fusion in herpes simplex virus 1 entry. Proc. Natl. Acad. Sci 105, 10559–10564 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laine RF et al. Structural analysis of herpes simplex virus by optical super-resolution imaging. Nat. Commun 6, 5980 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu J, Wright ER & Winkler H Chapter thirteen - 3D visualization of HIV virions by cryoelectron tomography. in Cryo-EM, Part C: Analyses, Interpretation, and Case studies (ed. Jensen GJBT-M in E.) vol. 483 267–290 (Academic Press, 2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park YG et al. Protection of tissue physicochemical properties using polyfunctional crosslinkers. Nat. Biotechnol 37, 73–83 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neve RL, Neve KA, Nestler EJ & Carlezon WA Use of herpes virus amplicon vectors to study brain disorders. Biotechniques 39, 381–391 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Yu C-C Virus Particle Analysis, HSV-1 Averaged Particle Image Simulation, and Microtubule Peak-to-Peak Distance Analysis https://github.com/jayyu0528/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The total raw data size of the study exceeds 350 GB. The data that support this study are available from the authors upon reasonable request.


