Abstract
Proteomic analysis of cerebrospinal fluid (CSF) holds great promise in understanding the progression of neurodegenerative diseases, including Alzheimer’s disease (AD). As one of the primary reservoirs of neuronal biomolecules, CSF provides a window into the biochemical and cellular aspects of the neurological environment. CSF can be drawn from living participants allowing the potential alignment of clinical changes with these biochemical markers. Using cutting-edge mass spectrometry technologies, we perform a streamlined proteomic analysis of CSF. We quantify greater than 700 proteins across 10 pairs of age- and sex-matched participants in approximately one hour of analysis time each. Using the paired participant study structure, we identify a small group of biologically relevant proteins that show substantial changes in abundance between cognitive normal and AD participants, which were then analyzed at the peptide level using parallel reaction monitoring experiments. Our findings suggest the utility of fractionating a single sample and using matching to increase proteomic depth in cerebrospinal fluid, as well as the potential power of an expanded study.
Keywords: Alzheimer’s disease, cerebrospinal fluid, neurodegeneration, FAIMS
Introduction
Alzheimer’s disease (AD) is the sixth leading cause of death in the United States[1] and affects tens of millions worldwide[2]. Much remains to be understood about the onset and progression of AD, and no effective therapeutics to significantly alter its course currently exist[3]. Proteomic analysis of brain tissue across age[4], cell type[5], and brain region[6-8] has been extensive, but brain-focused studies require post-mortem tissue samples and thus offer limited insight into the molecular timeline of disease progression.
Proteomic analysis of cerebrospinal fluid (CSF), in contrast, allows for detection of molecular changes that occur during pathological decline. This approach holds great potential to discover additional biomarkers for AD and to increase understanding of the biological factors that lead to the diverse neurological effects observed across individuals. The unique benefits of CSF may be tempered by two competing objectives: targeting specific AD proteins[9-11] at the expense of discovery capacity vs. generating extensive catalogues of human CSF proteins[12-14] at the expense of preparation and analysis speed, which can impede the ability to make large-scale comparisons. In humans, large scale comparisons are often required to overcome population heterogeneity due to factors such as age and sex, both of which have been shown to have substantial effects on the protein abundances in CSF[15-19].
In this pilot study, we sought to determine the most effective solution for larger analyses of global protein expression in CSF by comparing three different proteomics methodologies: single-shot experiments, experiments with addition of a high-field asymmetric waveform ion mobility spectrometry (FAIMS) interface, and experiments analyzed in parallel with commercial CSF fractions. The capacity of these three strategies to quantify proteins and to capture variation in protein abundances was compared using a cohort of 20 individual samples. This sample set was comprised of ten age- and sex-matched AD case-control pairs, evenly split between males and females in an attempt to control for these sources of variability. We avoided individual sample fractionation and high-abundance protein depletion (common steps in CSF proteomics) to increase precision and ease of preparation while decreasing preparation time[20]. Despite the statistical limits of our sample size, this analysis quantified over 700 proteins that were detected across all 20 participants, including several found to be significantly associated with AD. These proteins included multiple 14-3-3 proteins, which have been previously colocalized to the neurofibrillary tangles and hypothesized to function in sequestering misfolded proteins, representing a potentially beneficial reaction to pathological protein aggregation. Our analysis also identifies more than 30 AD-associated proteins at lower significance, that have been previously observed in large-scale studies[15,16,21] and metareviews[22,23]. We believe this analysis can provide a valuable framework for large-scale global proteomic analysis of CSF, both in AD and in other neurodegenerative diseases.
Materials and Methods
Participant selection
Subjects came from the Wisconsin Alzheimer’s Disease Research Center (WADRC) clinical core, a longitudinal cohort study of middle-aged and older adults. Ten AD case/control pairs of subjects, matched for sex and age at lumbar puncture, were selected from the WADRC cohort for this study. The mean age of the AD case group was 72.1 with a maximum of 80.4 and a minimum of 62.1 while the control group ages ranged from 62.1 to 80.4 with a mean of 72.1 as well (Table 1).
Table 1. Sample Characteristics.
Mean and standard deviation of age, AB42 concentration, amyloid ratio and phosphorylated tau concentration for each sex-diagnosis group
| Age | CSF AB42 (pg/mL) | CSF AB42:AB40 | CSF ptau (pg/mL) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Diagnosis | Sex | Mean | SD | Mean | SD | Mean | SD | Mean | SD |
| AD | Female | 72.0 | 7.2 | 395.0 | 58.3 | 0.06 | 0.006 | 76.6 | 16.0 |
| Control | Female | 72.1 | 7.3 | 757.2 | 134.0 | 0.11 | 0.012 | 38.8 | 11.5 |
| AD | Male | 72.1 | 7.3 | 505.8 | 287.1 | 0.06 | 0.012 | 86.0 | 28.2 |
| Control | Male | 72.0 | 7.0 | 773.8 | 150.2 | 0.11 | 0.013 | 42.2 | 12.5 |
AD cases were defined as subjects who met all three of the following criteria: 1) diagnosed with dementia due to AD by consensus conference using the National Institute on Aging—Alzheimer’s Association criteria[24]; 2) amyloid positive status, defined here as having either a CSF Aβ42 measurement less than 471.54 pg/mL (Innotest method) or having a CSF Aβ42:Aβ40 ratio less than 0.09 (Triplex method)[25]; and 3) tau positive status, defined as having either a total CSF tau level greater than 461.26 pg/mL or a phosphorylated tau greater than 59.5 pg/mL[25]. Controls were defined as subjects meeting the following criteria: 1) cognitively normal according to the consensus conference; 2) amyloid negative status (defined as above); 3) tau negative status. For each subject, the clinical diagnosis used was the diagnosis closest in time to the date of the lumbar puncture that generated the CSF sample used in this study, and no diagnosis was more than six months removed from that date.
CSF Sample Collection
CSF was collected via lumbar puncture in the morning after a 12-hour fast[26]. Briefly, after gentle extraction, mixing, and centrifugation, supernatants were flash frozen and stored at −80 degrees until the time of preparation.
Extraction and Digestion
Samples were brought to 90% methanol and centrifuged, with the precipitate pellet then resuspended in 8M urea, 10mM TCEP, 40mM CAA, 100mM Tris pH 8. The solution was then diluted to 25% concentration with 100mM Tris, pH 8, and trypsin was added at a ratio of 50:1 w/w and digested overnight at ambient temperature. Digested peptides were desalted using Strata-X Polymeric Reverse Phase column (Phenomenex).
Chromatographic Columns
Online reverse-phase columns were prepared in-house using a high-pressure packing apparatus[27]. In brief, 1.5 μm Bridged Ethylene Hybrid C18 particles were packed at 30,000 psi into a New Objective PicoTip™ emitter (Stock# PF360-75-10-N-5) with an inner diameter of 75 μm and an outer diameter of 360 μm.
Experimental strategies
Strategy 1: Single-Shot Experiments
Mobile phase A consisted of 0.2% formic acid in water and mobile phase B consisted of 0.2% formic acid in 70% acetonitrile. Samples were loaded onto the column for 8.6 minutes at 300 nL/min. Mobile phase B was increased to 5% in the first 8.6 min then increased to 50% by 51 min. The method finished with a wash stage of 100% B from 52-54 minutes and an equilibration step of 1% B from 55-60 minutes.
Eluting ions were analyzed on a Thermo Orbitrap Fusion Lumos in a data-dependent manner with survey scans taken in the orbitrap at 240,000 resolution and MS2 scans taken in the ion trap using the “rapid” setting. Samples were analyzed in duplicate.
Strategy 2: FAIMS Experiments
Identical chromatographic methods were used for the FAIMS experiments as detailed above for the single-shot experiments.
Peptide ions passing through the FAIMS interface are destabilized by an alternating electric field, the effect of which is countered by a peptide-specific compensating voltage (CV). This stabilization allows for the selection of a specific subpopulation of peptide ions when using a specific compensation voltage setting. We performed three analyses on each participant sample, one for each of three CV settings: −60, −75, and −90.
Survey scans of precursors were taken in the orbitrap at 120,000 resolution while MS2 scans were taken in the ion trap using the “rapid” setting.
Strategy 3: Experiments Run with Fractions
Non-designated BioIVT cerebrospinal fluid was denatured, digested, and desalted as detailed above. Dried samples were resuspended in 0.2% formic acid and fractionated using an HPLC (Agilent, Infinity 2000) with a 150 mm C18 reverse phase column (Waters, XBridge Peptide BEH, particle size 3.5 μm). Mobile phase buffer A was a freshly prepared mixture of 10mM ammonium bicarbonate, pH 9.5; mobile phase B was composed of 10mM ammonium bicarbonate, 80% methanol, pH 9.5. The gradient method was 20 minutes in length with 32 fractions collected from 5 to 20 minutes and a flow rate of 800 nL/min throughout. Mobile phase B was increased from 5% to 35% in the first 2 minutes before increasing to 100% B by 13 minutes. 32 fractions were concatenated into 16 fractions by combining every other column in the sample collection plate.
The chromatographic method was lengthened slightly for fractions and participant samples run alongside fractions in order to accommodate the lower peptide concentration of fractions. Samples were loaded onto the column for 12 minutes at 350 nL/min. Mobile phase B increased to 12% in the first 12 min then increased to 65% by 55 min. The method finished with a wash stage of 100% B from 56-59 minutes and an equilibration step of 0% B from 60-70 minutes.
These experiments used the same instrument acquisition method as the single-shot experiments.
Protein Quantification
The resulting spectra were searched in MaxQuant (1.6.0.13) using fast LFQ against a full human proteome with isoforms downloaded from Uniprot (October 29, 2018). Each set of experimental strategies was searched separately. Carbamidomethylation of cysteine was set as fixed modification. Matching between runs was used with a retention time window of 0.7 min. Searches were performed using a protein FDR of 1%, a minimum peptide length of 7, and a 0.5 Da MS2 match tolerance. Protein data were then extracted from the “ProteinGroups.txt” file of the MaxQuant output after decoy, contaminants, and reverse sequences were removed. The protein counts were based on protein groups with an LFQ Intensity > 0.
Protein Variation, Pairwise Correlations, and Differential Abundance in AD
Single-shot analysis of participant samples alongside a fractionated commercial CSF sample searched using match-between-runs, was found to yield the greatest number of proteins quantified across all participant samples. This experimental strategy was then used to generate pairwise correlations between samples and to test the association with AD (paired t-test, unequal variance). A paired t-test was used due to the matched nature of the study design. Data analysis was performed in R (3.6.1) using the base package. Plots were generated using lattice plotting and ggplot2 in R.
Parallel Reaction Monitoring Experiments
The MaxQuant (1.6.0.13) output tables for the fractionated CSF were used to build a spectral library in Skyline (19.1.0.193). Proteins of interest were then selected from the library based on significance (t-test, p-value < 0.02) in the data-dependent analysis. Scheduled PRM experiments monitored 27 peptide ions from seven proteins using 4-min retention time windows. Peptides were isolated with a 1.6 m/z window before being scanned in the orbitrap at 60,000 resolution. Peak areas and spectral traces for parallel reaction monitoring experiments were extracted from Skyline and processed in R.
The MS proteomics files have been deposited to the publicly available Chorus Project repository (chorusproject.org) under the project title “Proteomic Analysis of Cerebrospinal Fluid in Alzheimer’s Disease”.
Results
Protein Quantification and Variation
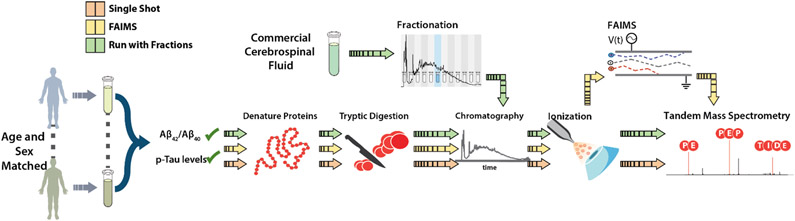
Proteins in the CSF samples were extracted, denatured, desalted, and then digested with trypsin[28]. Peptides resulting from trypsin digestion of the commercially available CSF were fractionated using high-pH reverse phase fractionation. All samples were injected separately onto an online liquid chromatography system and analyzed with a quadrupole-orbitrap dual-cell linear ion trap hybrid mass spectrometer (Figure 1) for all three strategies described above: single shot (40 total hours of instrument time), single shot with FAIMS added (60 hours), and single-shot analysis of participant samples run in parallel with a fractionated commercial CSF sample (65 hours).
Figure 1. Experimental Design.
20 individuals were age- and sex-matched to form 10 AD case-control pairs. CSF samples were collected by lumbar puncture, after which proteins were extracted, denatured, and digested before being analyzed by tandem mass spectrometry. Three different analysis strategies were utilized: single-shot (orange), addition of FAIMS (yellow), and single-shot analyses run in parallel with a fractionated commercial CSF sample (green).
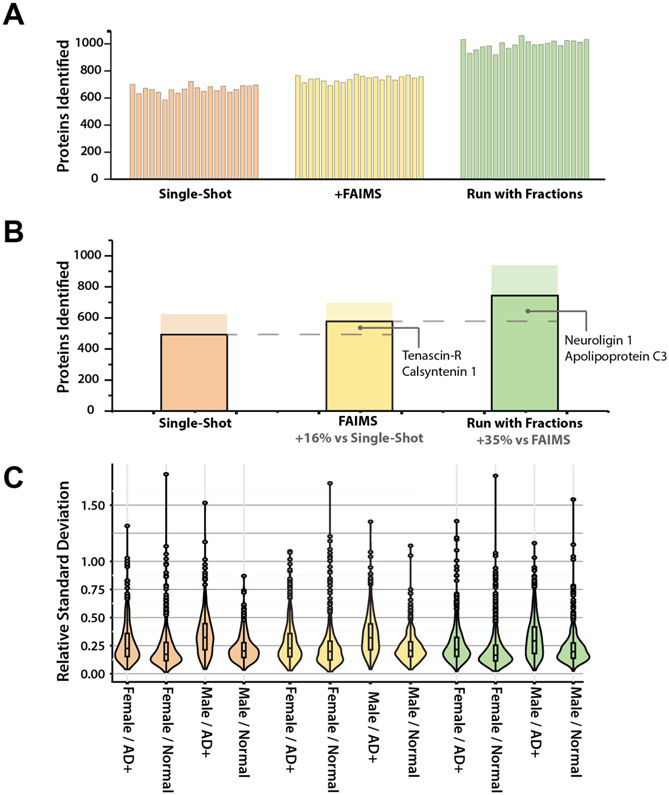
The latter strategy led to the highest number of identified proteins with a total of 2,118. Of these proteins, 939 were quantified in greater than 50% of each sex-disease group and 776 were quantified across all 20 participants (Supplemental Table 3). These numbers are comparable to those in recent DIA analyses of CSF, which relied on substantially more fractionation and longer chromatographic gradients[15,29]. Analyses that included fractions showed a 35% increase in the number of proteins quantified in all participant samples over the FAIMS analysis (Figure 2b) and a 56% increase over the single-shot analyses alone. These additional proteins included several well-characterized AD-related proteins, including neuroligin[30]. While FAIMS also increased the number of proteins quantified compared to single-shot, non-FAIMS experiments, the increase was not proportional to the additional analysis time, unlike the experiments with fractions (~50% increase in instrument time, ~50% increase in proteins quantified).
Figure 2. Distribution of proteins quantified across three methods.
(A) Number of proteins quantified in each sample for each analysis method. (B) Proteins quantified overall for each analysis method. Solid bar shows proteins quantified in all 20 samples. Translucent bar shows number of proteins quantified in at least 50% of each of the four sex-disease groups. Additional proteins quantified include several previously implicated in AD. (C) Relative standard deviation (RSD) distribution for each sex-disease group. AD groups exhibit higher median RSDs than healthy normal counterparts for all sexes and analysis strategies.
The label-free abundances (LFQ intensity) from experiments run alongside the commercial fractions were used for the remaining analyses in this study. To eliminate erroneous characterization[31], the following analyses were performed using only proteins for which an LFQ intensity was produced across all 20 participant samples, with no missing values(776 proteins referenced above).
Like many other body fluids, CSF is highly dynamic in molecular content[32,33], leading to higher relative standard deviations (RSDs) for protein abundance than would be expected for other tissues[34]. Indeed, large variations in protein levels have been one of bottlenecks of proteomic analysis in CSF, with relative standard deviations > 1.00 previously reported[35]. We observed median RSDs in AD samples for both sexes that were higher than those of the healthy controls (21% vs. 17% for females and 29 % vs. 20% for males in our experiments with fractions). Due to the consistency of this RSD pattern across all three methodologies (Figure 2c), we infer that these differences stemmed from the innate characteristics of the samples rather than artifacts of the analysis method.
Pairwise Correlation
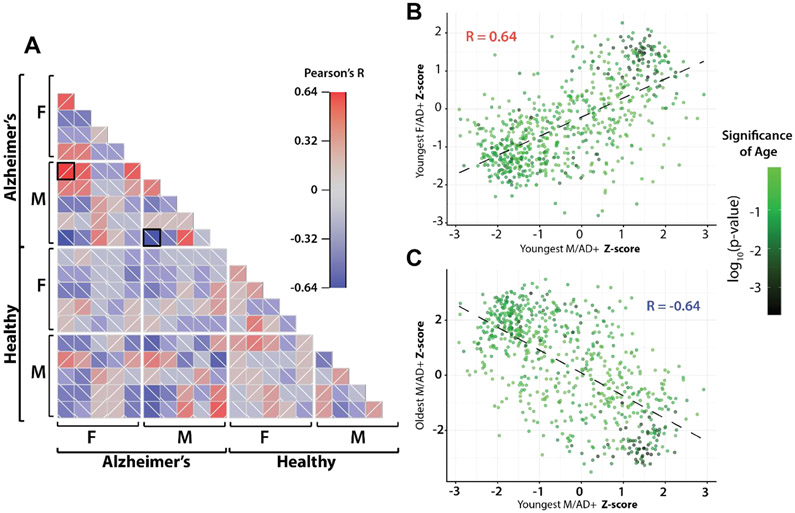
When comparing protein abundance across all samples using a pairwise Pearson correlation, we observe patterns that vary in direction and magnitude even within sex and disease state groups, with several unexpected correlations (Figure 3a). The strongest positive correlation (R=0.64) compares the protein abundances of the youngest samples from both the male and female AD+ group. The strongest anticorrelation (R = −0.64) compares two male, AD+ samples, the youngest and the oldest. These two correlations would initially seem counterintuitive given the misalignment of sex and the alignment of disease state, respectively.
Figure 3. Pairwise Pearson correlations.
(A) Pairwise Pearson correlation across all samples. Samples are separated by sex and disease state. Within disease/sex groups, samples are ordered by age with youngest samples at left on the x-axis and at top on y-axis. Black boxes indicate comparisons expanded in B and C. (B) Scatterplot of z-score normalized protein expression between youngest male AD+ sample and youngest female AD+ sample. Points are colored by significance when including age as explanatory variable in linear model (C) Scatterplot of z-score normalized expression between youngest male AD+ sample and oldest male AD+ sample. A high number of age-related proteins are present in the quadrants driving protein correlations in both B and C.
Although our sample population is not explicitly structured to examine age, we explored it as a possible explanation of these unexpected correlations by building two regression models: one relied on only disease state and sex as explanatory variables; the other also included age. Protein abundances were fit to each model, and the strength of the two models were compared using an analysis of variance (ANOVA). The p-value associated with that ANOVA was used to establish the significance of age in the expression profile for our sample set (Supplemental Table 1).
We plotted normalized protein abundances for the two comparisons described above (Figure 3a) with proteins colored by significance of age as an explanatory variable in the linear model (Figure 3b and 3c). In the positive correlation(Figure 3b), we see a dense clustering of age-associated proteins in quadrant I, strengthening the correlation by increasing the slope; while in the anticorrelated comparison (Figure3c), we see these same proteins in quadrant IV, decreasing the slope, and in turn magnifying the negative correlation. Previous work has shown a substantial effect of age on protein abundance in CSF[15], as well as in plasma[21] and the brain in Alzheimer’s[16]. Significant protein expression shifts have also been identified in CSF in the normal aging process [17,36]. These patterns indicate the importance of age in studying the human CSF proteome and neurodegeneration.
Differentially Expressed Disease Proteins
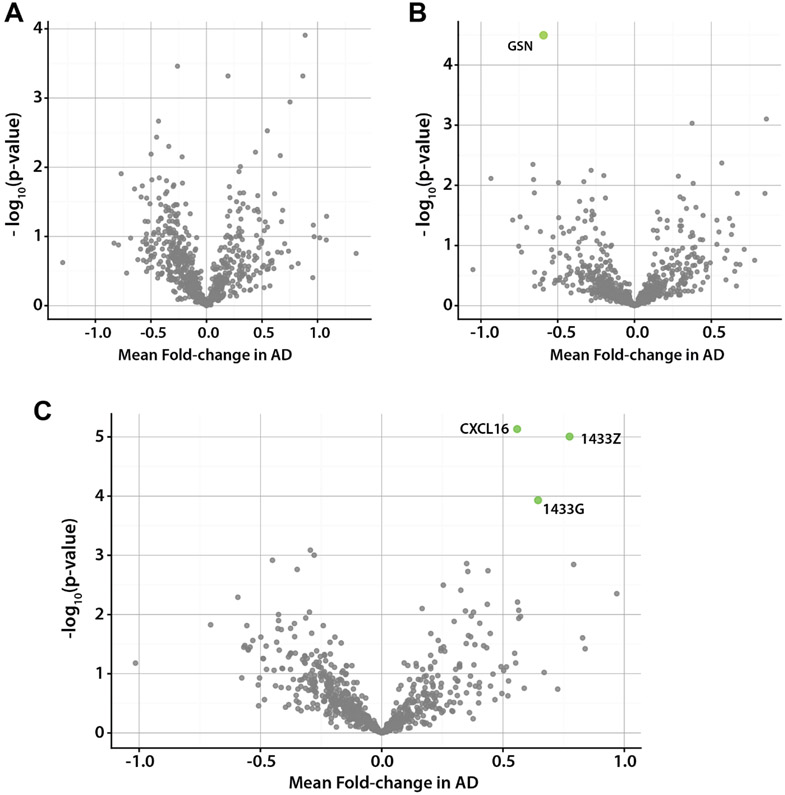
We next examined protein expression differences in CSF between AD and healthy samples using a paired t-test, accounting for unequal variance. Although several proteins in the female cohort exhibited high statistical significance (p-value < 0.001), they failed to meet our significance threshold after correction for multiple comparisons (Benjamini-Hochberg, 5% FDR)[37]. When examining the male sample group, one protein was significantly different after correction, N-acetylglucosamine-6-sulfatase (GSN), which decreased in the presence of AD (Figure 4b). GSN plays an important role in the regulation of the extracellular matrix in the brain by hydrolyzing heparan sulfate[38,39]. Decreased activity of GSN can lead to mucopolysaccharidosis, a condition associated with neurodegeneration[40]. Although previous work has shown alterations in the CSF[41] and plasma[42] proteome between sexes in mammals, differences observed may reflect limited statistical power due to small sample size, and a larger analysis would be required to fully disentangle these factors.
Figure 4. Differentially expressed proteins in disease.
(A) Volcano plot showing average fold change and significance for protein expression differences between female AD+/normal pairs. No proteins were found to be significant after correction for multiple comparisons. (B) Volcano plot showing fold change and significance for protein expression differences between male AD+/normal pairs. Only N-acetylglucosamine-6-sulfatase (GNS) was found to be significant after correction for multiple comparisons. (C) Volcano plot showing fold change and significance for protein expression across all AD+/normal pairs. Three proteins were identified to be significantly associated with disease 1433G, 1433Z and CXCL16.
When examining all ten sample pairs together, our analysis identified three proteins as significantly associated with disease: 14-3-3 protein zeta (1433Z), 14-3-3 protein gamma (1433G), C-X-C motif chemokine 16 (CXCL16)(Figure 4c, Supplemental Table 2). These three proteins were also found to be significant when using an unpaired t-test with the same correction. When comparing linear models with the addition of disease as an explanatory variable, similar to the above age analysis, only 1433Z and 1433G were found to be significant after correction. 14-3-3 proteins co-localize with neurofibrillary tangles within the brain[43]. One hypothesis for the role of these proteins in AD suggests that the proteins may operate in a similar capacity to their role as chaperones[44] by sequestering aggregated tau protein to reduce cytotoxicity[45]. 14-3-3 proteins are also associated with the development and maturation of synapses, where they function as signal transducers and recognition molecules[45-47]. Synaptic degradation is a sign of the neurodegeneration in AD[48]. We also observed differential expression to a lesser degree of significance (p-value < 0.05) of hypoxanthine phosphoribosyltransferase 1(HPRT1) and myristoylated alanine-rich protein C-kinase substrate (MARCKS), which play a role in the development of neurons[49] and synapses[50,51], respectively. These proteins as well as the 14-3-3 proteins, and CXCL16, have been identified as biomarkers of AD in several large-scale studies[15,16,21,23,52].
The upregulation of CXCL16 among the AD cases may reflect cellular signaling events occurring due to general neuroinflammation. This general neuroinflammation can occur as an effect of both normal aging and AD. CXCL16 functions as a chemokine signaler, helping to promote macrophage chemotaxis and endocytosis[53,54]. CXCL16 has both a transmembrane and soluble form, potentially increasing the chances of detecting elevated CXCL16 levels in a CSF analysis[55]. Two recent proteomic studies of CSF identified CXCL16 as a candidate biomarker for AD, observing increased abundance with disease[52,56].
Although only three proteins met our stringent significance cutoff, more than 80 proteins exhibited p-values < 0.05 (Supplemental Table 2), representing multiple biological processes previously associated with AD in CSF, including several proteins recently identified as part of a 40-protein AD signature[15]. We observe substantial upregulation of proteins involved in glycolysis and sugar metabolism including PML-RARA-regulated adapter molecule 1 (PRAM1), lactate dehydrogenase A (LDHA), aldolase fructose bisphosphate (ALDOA), malate dehydrogenase (MDH1), and hypoxanthine phosphoribosyltransferase 1(HPRT1) which have found to be elevated in the cerebrospinal fluid of Alzheimer’s patients[15,16,57-59]. Altered abundances were also observed for proteins involved in development and regulation of the extracellular matrix including serine protease HTRA1 (HTRA1), Pastin-2 (LCP1), and collagen alpha-2(IV) chain (COL4A). Previous work had grouped these proteins together into a co-expression module significantly correlated with AD[16].
We also assessed the role of known AD-related proteins in our data set, including amyloid precursor protein (APP), chitinase 3-like 1 protein (CHI3L1 or YKL-40), triggering receptor expressed on myeloid cells 2 protein (TREM-2), amyloid-like protein 1 (APLP1), and amyloid-like protein 2 (APLP2). Although these analyses did not find statistically significant differences between cases and controls, the method was still able to quantify these proteins across our samples. In future studies with larger sample sizes, the changes in these proteins may be better identified. Although other biomarkers such as NfL were sequenced in our fractionated samples, the associated peptides were not quantified in the participant samples, potentially reflecting the limitations of foregoing high-abundance depletion.
Targeted Analysis using Parallel Reaction Monitoring
The relatively low abundance of cells in the CSF poses a challenge to proteomic analysis, as in vivo deficiency leads to low protein content and diversity in our decellularized samples. Many of the peptides measured come from extracellular proteins[60] or those released by apoptosis[61-63]. Previous work has shown the potential utility of quantifying endogenous peptides processed by CSF native proteases[64,65]. These peptide subpopulations may experience abundance changes independent from overall protein expression shifts. To allow relative quantitation of specific peptides in a sensitive and accurate manner, we performed parallel reaction monitoring (PRM)[66], a targeted mass spectrometry technique that samples and quantifies a specific list of peptides using the peptides’ fragmentation spectra. Our PRM experiment targeted 26 peptides that included 31 precursor ions derived from seven proteins. Proteins were chosen due to their differential abundance in the untargeted study and functional similarity to identified disease-associated proteins.
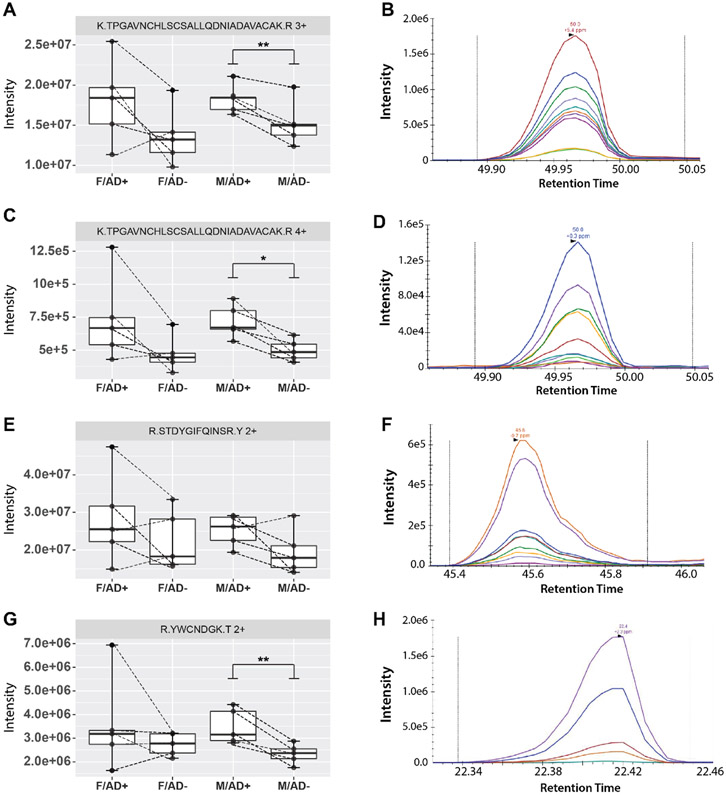
Summed fragment intensities were compared between case/control sample pairs. Of the 31 peptide ions, 24 had good quality spectral transitions across all 20 samples. Of those peptides, eight had p-values < 0.05 in at least one sex, four derived from the 14-3-3 proteins, three derived from Lysozyme C(LYZ), and one derived from heat shock cognate protein (HSPA8). Three out of the four precursor ions monitored from lysozyme C had p-values < 0.05 in male participants (Figure 5). Although previous proteomic studies in CSF have observed different magnitude fold changes of disease-associated proteins between males and females[15], here this difference may reflect statistical power limitations due to samples size. Lysozyme C plays a bacteriolytic role in humans, stemming from macrophages, which supports the continued theme of immune activation and inflammation signals being elevated in participants with AD. Evidence for the coincidence of AD with bacterial infection of the brain[67,68] and other tissues exists[69,70], which could promote upregulation of bactericidal pathways. Lysozyme C also increases the activity of other inflammatory signaling molecules [71].
Figure 5. Intensities and example transitions for Lysozyme C peptides targeted in parallel reaction monitoring experiments.
(A, B) TPGAVNCHLSCSALLQDNIADAVACAK, 3+ charge state (C,D) TPGAVNCHLSCSALLQDNIADAVACAK, 4+ charge state (E,F) STDYGIFQINSR, 2+ charge state (G,H) YWCNDGK, 2+ charge state. Dotted lines connect sample pairs. All four peptide ions exhibited increased intensity with addition of AD across a majority of disease/normal pairs. P-values were determined by paired sample t-test. * = p-value <0.05, ** = p-value <0.01. M = male, F = female, AD+ = Alzheimer’s disease, AD− = cognitively healthy control.
Discussion
Using a streamlined approach, we quantify more than 700 proteins across all 20 samples of our participant cohort, laying the groundwork for future large-scale proteomic analyses of CSF. This protein number encompasses > 20% of all proteins identified in the deepest CSF proteome analysis to date using a fraction of the preparation and analysis time [13]. A large collection of the proteins quantified have been previously associated with AD and neurodegeneration. Increased depth was achieved both with the addition of FAIMS and a parallel analysis of fractionated CSF. This proteomic depth was acquired without the use of immune depletion of high-abundance proteins or fractionation of the participant samples, allowing for efficient sample preparation and accurate reflection of protein abundances. Methods ran nearly one hour in total, providing a technique with excellent scaling potential to larger sample sizes.
This rapid analysis proved both efficient and effective, identifying multiple disease-associated proteins despite statistical limitations of sample size and large variations in protein abundance that occur in CSF. When comparing proteomic profiles of individual samples, several strong correlations arose not completely explained by sex or disease state groups. Upon closer examination we observed a possible effect of age, although an altered study would be required to confirm these findings. When comparing across case-control pairs, proteins were identified as significantly disease-associated in a single sex (GSN in males) and both sexes (1433G, 1433Z and CXCL16), showing the interconnected effects of sex and disease state on CSF protein composition.
PRM allowed for comparison of the relative abundance of specific peptides with high significance or AD-related functions. Several peptide ions from Lysozyme C exhibited significant shifts in abundance in males, with an increase in AD+ samples. Identification of Lysozyme C in follow-up analyses using PRM indicates the potential of this global profiling followed by targeted analysis to identify additional disease-associated proteins when equipped with greater statistical power. Although we controlled here for distribution of age and sex using our paired study design, these are only two of the myriad sources of between-individual variation in protein abundances in CSF. Alternative study structure would be required to identify the interaction between age, sex and disease in an expanded study.
This study highlights the advantages of CSF as an incredibly reactive and dynamic fluid that undergoes significant changes with neurodegeneration. Here, we present a next step in utilizing proteomic analysis of this tissue to gain a better understanding of the biochemical and cellular environment in AD. Given the promising findings detailed in this small pilot study, we expect that a coming study with expanded statistical power utilizing the same approach will identify more nuanced changes in this biological fluid.
Supplementary Material
Statement of Significance.
We combine here a well-structured age- and sex-matched sample cohort with a collection of strategies to increase proteomic depth, including the addition of ion mobility and parallel analysis of a fractionated sample. The study shows the capacity of this matched experimental design and streamlined approach, without high-abundance protein depletion or individual fractionation, to identify proteins and peptides significantly associated with Alzheimer’s disease in cerebrospinal fluid.
Acknowledgements
Research reported in this publication was supported by the National Institute on Aging of the National Institutes of Health under Award Number R01AG054047 and National Institute of General Medical Sciences under Award Number P41 GM108538. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
JJC is a consultant for Thermo Fisher Scientific
Abbreviations:
- CSF
cerebrospinal fluid
- FAIMS
high-field asymmetric waveform ion mobility spectrometry
- AD
Alzheimer’s disease
- PRM
parallel reaction monitoring
References
- [1].Association, A. s. (2019). 2019 Alzheimer's disease facts and figures. Alzheimer's & Dementia, 15(3), 321–387. [Google Scholar]
- [2].Prince M (2015). World Alzheimer Report 2015: the global impact of dimentia: an analysis of prevalence, incidence, cost and trends. Alzheimer's Disease International. [Google Scholar]
- [3].Evaluation, O. o. t. A. S. f. P. a. (2017). National Plan to Address Alzheimer's Disease: 2017 Update. Washington DC: Retrieved from https://aspe.hhs.gov/report/national-plan-address-alzheimers-disease-2017-update. [Google Scholar]
- [4].Carlyle BC, Kitchen RR, Kanyo JE, Voss EZ, Pletikos M, Sousa AMM, … Nairn AC (2017). A multiregional proteomic survey of the postnatal human brain. Nature neuroscience, 20(12), 1787–1795. doi: 10.1038/s41593-017-0011-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Seyfried NT, Dammer EB, Swarup V, Nandakumar D, Duong DM, Yin LM, … Levey AI (2017). A Multi-network Approach Identifies Protein-Specific Co-expression in Asymptomatic and Symptomatic Alzheimer's Disease. Cell Systems, 4(1), 60-+. doi: 10.1016/j.cels.2016.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].McKetney J, Runde RM, Hebert AS, Salamat S, Roy S, & Coon JJ (2019). Proteomic Atlas of the Human Brain in Alzheimer's Disease. Journal of Proteome Research, 18(3), 1380–1391. doi: 10.1021/acs.jproteome.9b00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mendonca CF, Kuras M, Nogueira FCS, Pla I, Hortobagyi T, Csiba L, … Rezeli M (2019). Proteomic signatures of brain regions affected by tau pathology in early and late stages of Alzheimer's disease. Neurobiol Dis, 130, 104509. doi: S0969-9961(19)30169-X [pii] 10.1016/j.nbd.2019.104509 [DOI] [PubMed] [Google Scholar]
- [8].Xu JS, Patassini S, Rustogi N, Riba-Garcia I, Hale BD, Phillips AM, … Unwin RD (2019). Regional protein expression in human Alzheimer's brain correlates with disease severity. Communications Biology, 2. doi: ARTN 43 10.1038/s42003-018-0254-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Andersson A, Remnestal J, Nellgard B, Vunk H, Kotol D, Edfors F, … Fredolini C (2019). Development of parallel reaction monitoring assays for cerebrospinal fluid proteins associated with Alzheimer's disease. Clin Chim Acta, 494, 79–93. doi: S0009-8981(19)30348-1 [pii] 10.1016/j.cca.2019.03.243 [DOI] [PubMed] [Google Scholar]
- [10].Brosseron F, Traschutz A, Widmann CN, Kummer MP, Tacik P, Santarelli F, … Heneka MT (2018). Characterization and clinical use of inflammatory cerebrospinal fluid protein markers in Alzheimer's disease. Alzheimers Research & Therapy, 10. doi: ARTN 25 10.1186/s13195-018-0353-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Quaranta A, Karlsson I, Ndreu L, Marini F, Ingelsson M, & Thorsen G (2019). Glycosylation profiling of selected proteins in cerebrospinal fluid from Alzheimer's disease and healthy subjects. Analytical Methods, 11(26), 3331–3340. doi: 10.1039/c9ay00381a [DOI] [Google Scholar]
- [12].Kroksveen AC, Guldbrandsen A, Vaudel M, Lereim RR, Barsnes H, Myhr KM, … Berven FS (2017). In-Depth Cerebrospinal Fluid Quantitative Proteome and Deglycoproteome Analysis: Presenting a Comprehensive Picture of Pathways and Processes Affected by Multiple Sclerosis. Journal of Proteome Research, 16(1), 179–194. doi: 10.1021/acs.jproteome.6b00659 [DOI] [PubMed] [Google Scholar]
- [13].Macron C, Lane L, Galindo AN, & Dayon L (2018). Deep Dive on the Proteome of Human Cerebrospinal Fluid: A Valuable Data Resource for Biomarker Discovery and Missing Protein Identification. Journal of Proteome Research, 17(12), 4113–4126. doi: 10.1021/acs.jproteome.8b00300 [DOI] [PubMed] [Google Scholar]
- [14].Guldbrandsen A, Vethe H, Farag Y, Oveland E, Garberg H, Berle M, … Berven FS (2014). In-depth Characterization of the Cerebrospinal Fluid (CSF) Proteome Displayed Through the CSF Proteome Resource (CSF-PR). Molecular & Cellular Proteomics, 13(11), 3152–3163. doi: 10.1074/mcp.M114.038554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Bader JM, Geyer PE, Müller JB, Strauss MT, Koch M, Leypoldt F, … Incesoy EI (2020). Proteome profiling in cerebrospinal fluid reveals novel biomarkers of Alzheimer's disease. Molecular systems biology, 16(6), e9356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Johnson EC, Dammer EB, Duong DM, Ping L, Zhou M, Yin L, … Troncoso JC (2020). Large-scale proteomic analysis of Alzheimer’s disease brain and cerebrospinal fluid reveals early changes in energy metabolism associated with microglia and astrocyte activation. Nature Medicine, 26(5), 769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Baird GS, Nelson SK, Keeney TR, Stewart A, Williams S, Kraemer S, … Montine TJ (2012). Age-Dependent Changes in the Cerebrospinal Fluid Proteome by Slow Off-Rate Modified Aptamer Array. American Journal of Pathology, 180(2), 446–456. doi: 10.1016/j.ajpath.2011.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Parrado-Fernández C, Blennow K, Hansson M, Leoni V, Cedazo-Minguez A, & Björkhem I (2018). Evidence for sex difference in the CSF/plasma albumin ratio in ~20 000 patients and 335 healthy volunteers. J Cell Mol Med, 22(10), 5151–5154. doi: 10.1111/jcmm.13767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lehallier B, Gate D, Schaum N, Nanasi T, Lee SE, Yousef H, … Wyss-Coray T (2019). Undulating changes in human plasma proteome profiles across the lifespan. Nat Med, 25(12), 1843–1850. doi: 10.1038/s41591-019-0673-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Galindo AN, Kussmann M, & Dayon L (2015). Proteomics of Cerebrospinal Fluid: Throughput and Robustness Using a Scalable Automated Analysis Pipeline for Biomarker Discovery. Analytical Chemistry, 87(21), 10755–10761. doi: 10.1021/acs.analchem.5b02748 [DOI] [PubMed] [Google Scholar]
- [21].Whelan CD, Mattsson N, Nagle MW, Vijayaraghavan S, Hyde C, Janelidze S, … Samad TA (2019). Multiplex proteomics identifies novel CSF and plasma biomarkers of early Alzheimer’s disease. Acta neuropathologica communications, 7(1), 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wesenhagen KE, Teunissen CE, Visser PJ, & Tijms BM (2020). Cerebrospinal fluid proteomics and biological heterogeneity in Alzheimer’s disease: a literature review. Critical reviews in clinical laboratory sciences, 57(2), 86–98. [DOI] [PubMed] [Google Scholar]
- [23].Pedrero-Prieto CM, García-Carpintero S, Frontiñán-Rubio J, Llanos-González E, Aguilera García C, Alcaín FJ, … Rabanal-Ruiz Y (2020). A comprehensive systematic review of CSF proteins and peptides that define Alzheimer’s disease. Clinical Proteomics, 17(1), 21. doi: 10.1186/s12014-020-09276-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR, Kawas CH, … Phelps CH (2011). The diagnosis of dementia due to Alzheimer's disease: Recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers & Dementia, 7(3), 263–269. doi: 10.1016/j.jalz.2011.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Clark LR, Berman SE, Norton D, Koscik RL, Jonaitis E, Blennow K, … Carlsson CM (2018). Age-accelerated cognitive decline in asymptomatic adults with CSF beta-amyloid. Neurology, 90(15), E1306–E1315. doi: 10.1212/Wnl.0000000000005291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Darst BF, Koscik RL, Racine AM, Oh JM, Krause RA, Carlsson CM, … Engelman CD (2017). Pathway-Specific Polygenic Risk Scores as Predictors of Amyloid-beta Deposition and Cognitive Function in a Sample at Increased Risk for Alzheimer's Disease. Journal of Alzheimers Disease, 55(2), 473–484. doi: 10.3233/Jad-160195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Shishkova E, Hebert AS, Westphall MS, & Coon JJ (2018). Ultra-High Pressure (>30,000 psi) Packing of Capillary Columns Enhancing Depth of Shotgun Proteomic Analyses. Analytical Chemistry, 90(19), 11503–11508. doi: 10.1021/acs.analchem.8b02766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Stefely JA, Kwiecien NW, Freiberger EC, Richards AL, Jochem A, Rush MJP, … Coon JJ (2016). Mitochondrial protein functions elucidated by multi-omic mass spectrometry profiling. Nature Biotechnology, 34(11), 1191-+. doi: 10.1038/nbt.3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Barkovits K, Linden A, Galozzi S, Schilde L, Pacharra S, Mollenhauer B, … Marcus K (2018). Characterization of Cerebrospinal Fluid via Data-Independent Acquisition Mass Spectrometry. Journal of Proteome Research, 17(10), 3418–3430. doi: 10.1021/acs.jproteome.8b00308 [DOI] [PubMed] [Google Scholar]
- [30].Sindi IA, Tannenberg RK, & Dodd PR (2014). A role for the neurexin-neuroligin complex in Alzheimer's disease. Neurobiology of Aging, 35(4), 746–756. doi: 10.1016/j.neurobiolaging.2013.09.032 [DOI] [PubMed] [Google Scholar]
- [31].Lim MY, Paulo JA, & Gygi SP (2019). Evaluating False Transfer Rates from the Match-between-Runs Algorithm with a Two-Proteome Model. Journal of Proteome Research, 18(11), 4020–4026. doi: 10.1021/acs.jproteome.9b00492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lin ZF, Gong Q, Wu CM, Yu JW, Lu TT, Pan XB, … Li XK (2012). Dynamic Change of Serum FGF21 Levels in Response to Glucose Challenge in Human. Journal of Clinical Endocrinology & Metabolism, 97(7), E1224–E1228. doi: 10.1210/jc.2012-1132 [DOI] [PubMed] [Google Scholar]
- [33].Yasui Y, Pepe M, Thompson ML, Adam BL, Wright GL, Qu YS, … Feng ZD (2003). A data-analytic strategy for protein biomarker discovery: profiling of high-dimensional proteomic data for cancer detection. Biostatistics, 4(3), 449–463. doi: DOI 10.1093/biostatistics/4.3.449 [DOI] [PubMed] [Google Scholar]
- [34].Trombetta BA, Carlyle BC, Koenig AM, Shaw LM, Trojanowski JQ, Wolk DA, … Arnold SE (2018). The technical reliability and biotemporal stability of cerebrospinal fluid biomarkers for profiling multiple pathophysiologies in Alzheimer's disease. Plos One, 13(3). doi: ARTN e0193707 10.1371/journal.pone.0193707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Schilde LM, Kosters S, Steinbach S, Schork K, Eisenacher M, Galozzi S, … May C (2018). Protein variability in cerebrospinal fluid and its possible implications for neurological protein biomarker research. Plos One, 13(11). doi: ARTN e0206478 10.1371/journal.pone.0206478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Jing Z, Goodlett DR, Peskind ER, Quinn JF, Yong Z, Qin W, … Montine TJ (2005). Quantitative proteomic analysis of age-related changes in human cerebrospinal fluid. Neurobiology of Aging, 26(2), 207–227. doi: 10.1016/j.neurobiolaging.2004.03.012 [DOI] [PubMed] [Google Scholar]
- [37].Benjamini Y, & Hochberg Y (1995). Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Statistical Methodology, 57(1), 289–300. [Google Scholar]
- [38].Hanson SR, Best MD, & Wong CH (2004). Sulfatases: structure, mechanism, biological activity, inhibition, and synthetic utility. Angew Chem Int Ed Engl, 43(43), 5736–5763. doi: 10.1002/anie.200300632 [DOI] [PubMed] [Google Scholar]
- [39].Parenti G, Meroni G, & Ballabio A (1997). The sulfatase gene family. Current Opinion in Genetics & Development, 7(3), 386–391. doi: Doi 10.1016/S0959-437x(97)80153-0 [DOI] [PubMed] [Google Scholar]
- [40].Mok A, Cao H, & Hegele RA (2003). Genomic basis of mucopolysaccharidosis type IIID (MIM 252940) revealed by sequencing of GNS encoding N-acetylglucosamine-6-sulfatase. Genomics, 81(1), 1–5. doi: 10.1016/s0888-7543(02)00014-9 [DOI] [PubMed] [Google Scholar]
- [41].Quintela T, Marcelino H, Deery MJ, Feret R, Howard J, Lilley KS, … Santos CRA (2016). Sex-Related Differences in Rat Choroid Plexus and Cerebrospinal Fluid: A cDNA Microarray and Proteomic Analysis. Journal of Neuroendocrinology, 28(1). doi: 10.1111/jne.12340 [DOI] [PubMed] [Google Scholar]
- [42].Lehallier B, Gate D, Schaum N, Nanasi T, Lee SE, Yousef H, … Wyss-Coray T (2019). Undulating changes in human plasma proteome profiles across the lifespan. Nature Medicine, 25(12), 1843–1850. doi: 10.1038/s41591-019-0673-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Umahara T, Uchihara T, Tsuchiya K, Nakamura A, Iwamoto T, Ikeda K, & Takasaki M (2004). 14-3-3 proteins and zeta isoform containing neurofibrillary tangles in patients with Alzheimer's disease. Acta Neuropathologica, 108(4), 279–286. doi: 10.1007/s00401-004-0885-4 [DOI] [PubMed] [Google Scholar]
- [44].Xu Z, Graham K, Foote M, Liang FS, Rizkallah R, Hurt M, … Zhou Y (2013). 14-3-3 protein targets misfolded chaperone-associated proteins to aggresomes. Journal of Cell Science, 126(18), 4173–4186. doi: 10.1242/jcs.126102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kaneko K, & Hachiya NS (2006). The alternative role of 14-3-3 zeta as a sweeper of misfolded proteins in disease conditions. Medical Hypotheses, 67(1), 169–171. doi: 10.1016/j.mehy.2006.01.019 [DOI] [PubMed] [Google Scholar]
- [46].Mackintosh C (2004). Dynamic interactions between 14-3-3 proteins and phosphoproteins regulate diverse cellular processes. Biochemical Journal, 381, 329–342. doi: Doi 10.1042/Bj20031332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Strochlic L, Cartaud A, Mejat A, Grailhe R, Schaeffer L, Changeux JP, & Cartaud J (2004). 14-3-3 gamma associates with muscle specific kinase and regulates synaptic gene transcription at vertebrate neuromuscular synapse. Proceedings of the National Academy of Sciences of the United States of America, 101(52), 18189–18194. doi: DOI 10.1073/pnas.0406905102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Preische O, Schultz SA, Apel A, Kuhle J, Kaeser SA, Barro C, … et al. (2019). Serum neurofilament dynamics predicts neurodegeneration and clinical progression in presymptomatic Alzheimer's disease. Nature Medicine, 25(2), 277-+. doi: 10.1038/s41591-018-0304-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Guibinga G-H, Hsu S, & Friedmann T (2010). Deficiency of the housekeeping gene hypoxanthine–guanine phosphoribosyltransferase (hprt) dysregulates neurogenesis. Molecular Therapy, 18(1), 54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Liu Y, Zhang P, Zheng Y, Yang C, Du T, Ge M, … Ma G (2018). Effects of NMDAR Antagonist on the Regulation of P-MARCKS Protein to Aβ 1− 42 Oligomers Induced Neurotoxicity. Neurochemical Research, 43(10), 2008–2015. [DOI] [PubMed] [Google Scholar]
- [51].McNamara RK, Hussain RJ, Simon EJ, Stumpo DJ, Blackshear PJ, Abel T, & Lenox RH (2005). Effect of myristoylated alanine-rich C kinase substrate (MARCKS) overexpression on hippocampus-dependent learning and hippocampal synaptic plasticity in MARCKS transgenic mice. Hippocampus, 15(5), 675–683. [DOI] [PubMed] [Google Scholar]
- [52].Sathe G, Na CH, Renuse S, Madugundu AK, Albert M, Moghekar A, & Pandey A (2019). Quantitative Proteomic Profiling of Cerebrospinal Fluid to Identify Candidate Biomarkers for Alzheimer's Disease. PROTEOMICS – Clinical Applications, 13(4), 1800105. doi: 10.1002/prca.201800105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Matloubian M, David A, Engel S, Ryan JE, & Cyster JG (2000). A transmembrane CXC chemokine is a ligand for HIV-coreceptor Bonzo. Nature Immunology, 1(4), 298–304. doi: Doi 10.1038/79738 [DOI] [PubMed] [Google Scholar]
- [54].Lleo A, Nuuez-Llaves R, Alcolea D, Chiva C, Balateu-Panos D, Colom-Cadena M, … Belbin O (2019). Changes in Synaptic Proteins Precede Neurodegeneration Markers in Preclinical Alzheimer's Disease Cerebrospinal Fluid. Molecular & Cellular Proteomics, 18(3), 546–560. doi: 10.1074/mcp.RA118.001290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Wilbanks A, Zondlo SC, Murphy K, Mak S, Soler D, Langdon P, … Briskin M (2001). Expression cloning of the STRL33/BONZO/TYMSTR ligand reveals elements of CC, CXC, and CX3C chemokines. Journal of Immunology, 166(8), 5145–5154. doi: DOI 10.4049/jimmunol.166.8.5145 [DOI] [PubMed] [Google Scholar]
- [56].Skillbäck T, Mattsson N, Hansson K, Mirgorodskaya E, Dahlén R, van der Flier W, … Gobom J (2017). A novel quantification-driven proteomic strategy identifies an endogenous peptide of pleiotrophin as a new biomarker of Alzheimer’s disease. Scientific Reports, 7(1), 13333. doi: 10.1038/s41598-017-13831-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Liguori C, Stefani A, Sancesario G, Sancesario G, Marciani M, & Pierantozzi M (2015). CSF lactate levels, τ proteins, cognitive decline: a dynamic relationship in Alzheimer's disease. Journal of Neurology, Neurosurgery & Psychiatry, 86(6), 655–659. [DOI] [PubMed] [Google Scholar]
- [58].Bélanger M, Allaman I, & Magistretti PJ (2011). Brain energy metabolism: focus on astrocyte-neuron metabolic cooperation. Cell metabolism, 14(6), 724–738. [DOI] [PubMed] [Google Scholar]
- [59].Higginbotham L, Ping L, Dammer EB, Duong DM, Zhou M, Gearing M, … Johnson EC (2020). Integrated proteomics reveals brain-based cerebrospinal fluid biomarkers in asymptomatic and symptomatic Alzheimer’s disease. Science Advances, 6(43), eaaz9360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Lun MP, Monuki ES, & Lehtinen MK (2015). Development and functions of the choroid plexus-cerebrospinal fluid system. Nature Reviews Neuroscience, 16(8), 445–457. doi: 10.1038/nrn3921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Uzan M, Erman H, Tanriverdi T, Sanus GZ, Kafadar A, & Uzun H (2006). Evaluation of apoptosis in cerebrospinal fluid of patients with severe head injury. Acta Neurochirurgica, 148(11), 1157–1164. doi: 10.1007/s00701-006-0887-1 [DOI] [PubMed] [Google Scholar]
- [62].Nemes Z, Fesus L, Egerhazi A, Keszthelyi A, & Degrell IM (2001). N-epsilon(gamma-glutamyl)lysine in cerebrospinal fluid marks Alzheimer type and vascular dementia. Neurobiology of Aging, 22(3), 403–406. doi: Doi 10.1016/S0197-4580(01)00224-X [DOI] [PubMed] [Google Scholar]
- [63].Vermes I, Steur ENHJ, Jirikowski GF, & Haanen C (2004). Elevated concentration of cerebrospinal fluid tissue transgrlutaminase in Parkinson's disease indicating apoptosis. Movement Disorders, 19(10), 1252–1254. doi: 10.1002/mds.20197 [DOI] [PubMed] [Google Scholar]
- [64].Hansson KT, Skillback T, Pernevik E, Kern S, Portelius E, Hoglund K, … Gobom J (2017). Expanding the cerebrospinal fluid endopeptidome. Proteomics, 17(5). doi: Artn 1600384 10.1002/Pmic.201600384 [DOI] [PubMed] [Google Scholar]
- [65].Hansson K, Dahlén R, Hansson O, Pernevik E, Paterson R, Schott JM, … Gobom J (2019). Use of the tau protein-to-peptide ratio in CSF to improve diagnostic classification of Alzheimer’s disease. Clinical Mass Spectrometry. doi: 10.1016/j.clinms.2019.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Peterson AC, Russell JD, Bailey DJ, Westphall MS, & Coon JJ (2012). Parallel Reaction Monitoring for High Resolution and High Mass Accuracy Quantitative, Targeted Proteomics. Molecular & Cellular Proteomics, 11(11), 1475–1488. doi: 10.1074/mcp.O112.020131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Maheshwari P, & Eslick GD (2015). Bacterial Infection and Alzheimer's Disease: A Meta-Analysis. Journal of Alzheimers Disease, 43(3), 957–966. doi: 10.3233/Jad-140621 [DOI] [PubMed] [Google Scholar]
- [68].Maheshwari P, & Eslick GD (2017). Bacterial Infection Increases the Risk of Alzheimer's Disease: An Evidence-Based Assessment. Handbook of Infection and Alzheimer's Disease, 5, 151–160. doi: 10.3233/978-1-61499-706-151 [DOI] [Google Scholar]
- [69].Ishida N, Ishihara Y, Ishida K, Tada H, Funaki-Kato Y, Hagiwara M, … Matsushita K (2017). Periodontitis induced by bacterial infection exacerbates features of Alzheimer's disease in transgenic mice. Npj Aging and Mechanisms of Disease, 3. doi: UNSP 15 10.1038/s41514-017-0015-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Dominy SS, Lynch C, Ermini F, Benedyk M, Marczyk A, Konradi A, … Potempa J (2019). Porphyromonas gingivalis in Alzheimer's disease brains: Evidence for disease causation and treatment with small-molecule inhibitors. Science Advances, 5(1). doi: ARTN eaau3333 10.1126/sciadv.aau3333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Reitamo S, Klockars M, Adinolfi M, & Osserman EF (1978). Human lysozyme (origin and distribution in health and disease). La Ricerca in clinica e in laboratorio, 8(4), 211–231. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.