Abstract
Fetal growth is an important determinant of cardiometabolic disease risk during childhood and adulthood. The genetic architecture of fetal growth remains largely understudied in ancestrally diverse populations. We conducted genome-wide admixture mapping scan and analysis of genetic ancestry among Hispanic American, African American, European American, and Asian American pregnant women to identify genetic loci associated with fetal growth measures across 13 to 40 weeks gestation. Fetal growth measures were associated with genome-wide average African, European, Amerindigenous and East Asian ancestry proportions (P ranged from10−3 to 4.8×10−2). Admixture mapping analysis identified 10 African ancestry loci and 3 Amerindigenous ancestry loci significantly associated with fetal growth measures at Bonferroni-corrected levels of significance (P ranged from 2.18×10−8 to 3.71×10−6). At the chr2q23.3–24.2 locus in which higher African ancestry was associated with long bone (femur and humerus) lengths, the T allele of rs13030825 (GALNT13) was associated with longer humerus length in African Americans (β= 0.44, P= 6.25×10−6 at week 27; β= 0.39, P=7.72×10−5 at week 40). The rs13030825 SNP accounted for most of the admixture association at the chr2q23.3–24.2 locus and has substantial allele frequency difference between African and European reference samples (FST=0.55, P=0.03). Regulatory annotation shows that rs13030825 overlaps with the serum response factor (SRF) transcription factor previously implicated in postnatal bone development of mice. Overall, we identified ancestry-related maternal genetic loci that influence fetal growth, shedding light on molecular pathways that regulate fetal growth and potential effects on health across the lifespan.
Keywords: admixture mapping, fetal growth, early origins of disease, African ancestry, Amerindigenous ancestry
Introduction
Abnormal fetal growth has immediate and life-long consequences on health. Several studies found consistent associations of fetal growth measures with infant mortality and morbidity (Rossen and Schoendorf 2014), childhood morbidity (Gaskins et al. 2010; Mikkola et al. 2005), and adult cardiometabolic diseases and cognitive function (Barker et al. 1992; Godfrey and Barker 2000; Hales et al. 1991). Complex interactions between genetic and environmental factors contribute to variations in growth trajectories (Kiserud et al. 2018). The fetal genome directly influences fetal growth, whereas the maternal genome can contribute to offspring birthweight through maternally inherited offspring risk alleles or the influence of maternal genotypes on the intrauterine environment (Warrington et al. 2019).
In European ancestry populations, genome-wide association studies (GWAS) have identified several genetic markers associated with birthweight (Beaumont et al. 2018; Freathy et al. 2010; Horikoshi et al. 2016; Horikoshi et al. 2013; Warrington et al. 2019), and documented that 7.6% of the variance in birthweight could be attributed to maternal genetic variants on the genome-wide arrays (Warrington et al. 2019). However, the genetic contribution to fetal growth at different gestational ages may be different from that of size at birth (Roland et al. 2012; Workalemahu et al. 2018). Apart from a recent trans-ethnic GWAS meta-analysis that identified a maternal variant in the ITPR1 gene associated with lower fetal weight and head circumference during late second trimester and early third trimester of pregnancy (Tekola-Ayele et al. 2020), the genetic architecture of fetal growth is not clearly understood. Moreover, the genetic basis of early life growth remains understudied in non-European ancestry populations.
The majority of present-day human populations have mixed genetic ancestries that vary tremendously even among people from the same population group (Baker et al. 2017; Shriner et al. 2014), and the diversity of the United States (U.S.) population is increasing (Cooper et al. 2018). Genomes of present-day African Americans have admixed genomic segments derived from African and European ancestries (Reiner et al. 2005) and Hispanic Americans have genomic segments derived from European, African, and Amerindigenous ancestries (Bonilla et al. 2004). Ancestry of admixed individuals can be characterized as genome-wide averages of ancestral proportions namely global ancestry, and ancestral states at each locus namely local ancestry (Shriner 2017). This mosaic pattern of ancestry varies among individuals belonging to an admixed population, and admixture mapping analysis strategies can be used to identify ancestry-related genetic loci associated with a phenotype (Shriner 2017). Using an admixture mapping scan, one study has found an association between higher African ancestry at chr12q14 locus and lower birthweight (Ochs-Balcom et al. 2018). We hypothesized that differentially distributed genetic variants that vary with genetic ancestry may underlie fetal growth variations. Using a cohort of ancestrally diverse pregnant women in the U.S., we investigated the following: (1) identified local ancestry loci associated with fetal growth in African Americans and Hispanic Americans using genome-wide admixture mapping, and (2) examined association of global ancestry proportion with fetal growth among African American, Hispanic Americans, European Americans and Asian Americans.
Methods
Study participants
This analysis included 1,935 pregnant women (i.e., 531 Hispanic Americans, 587 African Americans, 601 European Americans, and 216 Asian Americans) from the NICHD Fetal Growth Studies – Singletons. Briefly, the NICHD Fetal Growth Studies – Singletons was a multi-ethnic prospective cohort study designed to establish standards for fetal growth in the U.S. population. A total of 2,802 pregnant women were recruited between 8–13 gestational weeks between July 2009 and January 2013 from 12 clinic sites in the U.S. Women self-identified themselves to be Hispanic (here after called Hispanic Americans), non-Hispanic Black (African Americans), non-Hispanic White (European Americans), and Asian (Asian American). Detailed recruitment and inclusion criteria of the study have been reported previously (Buck Louis et al. 2015; Grewal et al. 2018). The study implemented a standardized ultrasonography protocol with established quality control after intensive training and credentialing of sonographers (Hediger et al. 2016). After the first ultrasound to confirm gestational age, pregnant women underwent five standardized ultrasounds at a priori defined gestational ages.
Fetal growth measures
At each ultrasound visit, head circumference, biparietal diameter, abdominal circumference, humerus length, and femur length were measured. Estimated fetal weight was calculated from head circumference, abdominal circumference, and femur length using the Hadlock formula (Hadlock et al. 1985). Fetal growth trajectories were created using a linear mixed model with a cubic spline mean structure and a cubic polynomial random effect (Pinheiro and Bates 2000). The study was approved by the institutional review boards of NICHD and each of the participating clinic sites. Written informed consent was obtained from all study participants.
Genotyping and quality control
DNA was successfully extracted from stored buffy coat specimens obtained from 2,215 individuals (582 Hispanic Americans, 652 African Americans, 641 European Americans, and 340 Asian Americans). Single nucleotide polymorphisms (SNPs) were genotyped using the Infinium Multiethnic Global BeadChip microarray (Illumina) that has 1,710,785 autosomal SNPs. Genotype positions were aligned to the human reference genome build GRCh37. Quality control of genome-wide SNP data was carried out within each population group using PLINK version 1.9 (Purcell et al. 2007). First, we applied sample quality filters which removed samples with more than 5% missing SNP genotypes, high degree of relatedness (Pi hat ≥0.25), excess heterozygosity (≥3 S.D. from the mean), and outliers from the distribution of the Hispanic, African, European, and East Asian clusters of the 1000 Genomes reference population based on multi-dimensional scaling plots (Genomes Project et al. 2015). Next, we applied SNP quality filters which removed insertion-deletions, SNPs that are multi-allelic and duplicated, SNPs that had more than 5% missing values, minor allele frequency <0.5%, and not in Hardy-Weinberg equilibrium (P <10−4). Details of samples and SNPs removed based on these quality control filters has been previously described (Tekola-Ayele et al. 2020). After additional removal of individuals that did not have at least two fetal ultrasound measures and live birth data, a total of 1,935 women were included in subsequent analyses.
Estimation of global ancestry
We estimated global ancestry to determine the proportions of African, European, Amerindigenous, and East Asian ancestries of women. Reference samples for African, European and East Asian ancestries were obtained from genotype data of the 1000 Genomes samples: Yoruba in Ibadan, Nigeria (YRI), Utah residents with Northern and Western European ancestry from the CEPH collection (CEU), and Han Chinese in Beijing, China (CHB), respectively (Genomes Project et al. 2015). Representative samples for Amerindigenous ancestry were obtained by combining samples from the 1000 Genomes project and the Human Genome Diversity Project (HGDP) (n=938) (Li et al. 2008). First, we integrated genotype data for SNPs with a minor allele frequency ≥5% (n=148,335 SNPs) from samples in HGDP (n=938) (Li et al. 2008), and samples from the 1000 Genomes Project with at least one of the four continental ancestries (n=761; Peruvian from Lima, Peru (PEL), Mexican ancestry from Los Angeles USA (MXL), Puerto Ricans from Puerto Rico (PUR), and Colombians from Medellin, Colombia (CLM) for Amerindigenous ancestry; YRI for African ancestry; CEU for European ancestry; and CHB for East Asian ancestry) (Genomes Project et al. 2015). We removed 62 HGDP samples identified as second degree or closer relatives using the --genome function in PLINK version 1.9 (Purcell et al. 2007), three SNPs with a call rate <5%, and 494 SNPs in the HLA (chr6:29640000–33120000) and LCT (chr2:136545415–136594750) gene loci. We performed linkage disequilibrium pruning using an r2 threshold of 0.1 and a sliding window of 50 SNPs by skipping 10 SNPs between consecutive windows resulting in a genome-wide dataset of 1,637 individuals with genotypes of 42,523 SNPs that were not correlated with each other.
Next, unsupervised clustering analysis was performed using ADMIXTURE version 1.3 (Alexander et al. 2009) with 10-fold cross-validation for the number of ancestral components (K)=2 to K=7 on the combined HGDP and 1000 Genomes dataset (n=1,637). At K=4, the value of K in which cross-validation error was the lowest, we obtained 46 samples with percent Amerindigenous ancestry of 90% or higher (25 PEL, 12 Maya, 2 Colombian, 2 Suri, 2 Pima, 2 MXL, and 1 Karitiana). Finally, we combined the representative ancestral samples (i.e., 46 Amerindigenous, 108 YRI, 99 CEU, and 103 CHB) with our dataset that passed quality control and performed ADMIXTURE analysis at K=4 in order to estimate the respective percent global ancestries in our study samples. The admixture proportions were plotted using Genesis v 0.2.6b (http://www.bioinf.wits.ac.za/software/genesis/). The mean ± S.D. global ancestry percentages in each population group was as follows: 46.8% ± 19.8% European, 34.8% ± 27.0% Amerindigenous and 16.6% ± 16.6% African ancestries among Hispanic Americans; 79.5% ± 13.7% African and 17.5% ± 13.1% European ancestries among African Americans; 96.3% ± 3.3% European ancestry among European Americans; and 95.5% ± 3.4% East Asian ancestry among Asian Americans (Fig S1, Table S1). We evaluated whether the ancestry estimates were stable to reference sample size differences by downsizing each of the representative reference samples to 46. The ancestry percentages in the original and downsized analyses were highly correlated (Pearson correlation >0.99). There were also high correlations between global ancestry percentages estimated using ADMIXTURE and genome-wide average of local ancestry estimates using RFMix (Maples et al. 2013) (Pearson correlation =0.96 for African ancestry in African Americans, and 0.96, 0.97, and 0.98 for African, European, and Amerindigenous ancestry in Hispanic Americans. In each population group, an ancestry component was taken forward for analyses if the mean percent ancestry in the respective population group was at least 5%, minimizing inclusion of ancestries that are not well differentiated and artefacts in ancestry estimation (Alexander and Lange 2011; Alexander et al. 2009). Moreover, among African Americans, the proportions of European and African ancestries per sample mirror each other adding to ~100%; so only African ancestry was included in analysis to avoid duplication. Overall, the subsequent analyses included African, European, and Amerindigenous ancestries in Hispanic Americans; African ancestry in African Americans; European ancestry in European Americans; and East Asian ancestry in Asian Americans.
Local ancestry inference
Local ancestry was inferred separately for African Americans and Hispanic Americans, two ancestrally admixed populations in the U.S. with widely varying proportions of genetic ancestry (Bryc et al. 2015), using RFMix v.2.0 (Maples et al. 2013). Analysis was performed using imputed and phased genotype data. Imputation procedures have been described previously (Tekola-Ayele et al. 2020). The reference panels from the 1000 Genomes Project and HDGP were used for each population group as described above. The input parameters for RFMix were included per recommendation (Maples et al. 2013): 5 minimum number of haplotypes per node, 3 EM iterations, and 6 and 11 generations of admixture for African Americans and Hispanic Americans, respectively. To evaluate the local ancestry estimates, we repeated the analysis using raw (unimputed) genotypes and found high concordance between the estimate based on imputed and genotyped markers (Pearson correlation ranged from 0.98 to 0.99). Details of how RFMix defines local ancestry intervals (windows) for inferring local ancestry has previously been described (Maples et al. 2013). To account for local ancestry assignments that span multiple loci, one locus was selected for each 0.2 cM window as defined by RFMix v.2.0, resulting in a total of 23,150 local ancestry intervals in African Americans and 13,071 local ancestry windows in Hispanic Americans.
Statistical analysis
We evaluated the association between each percent increase in genetic ancestry and estimated fetal growth measure at each week of gestation from 13 to 40 weeks using linear regression models. Analyses were adjusted for maternal pre-pregnancy weight (continuous), height (continuous), education (some college or more, high school or less), employment (employed or student, unemployed), marital status (married, other), health insurance coverage (private or managed care, other), age (continuous), parity (continuous), and fetal sex (male, female).
Genome-wide admixture mapping analysis was performed in African Americans and Hispanic Americans to identify local ancestry signals associated with fetal growth measures at end of the first, second, and third trimester of pregnancy. In African Americans, we tested whether the number of African ancestry alleles at a locus was associated with fetal growth measures, and in Hispanic Americans we separately tested whether the number of African, European and Amerindigenous ancestry alleles at a locus was associated with fetal growth measures. Analyses were adjusted for fetal sex and the first five genetic principal components. The first five PCs were included based on visual inspection of principal components analysis and scree plots, as previously described (Tekola-Ayele et al. 2020). Statistical significance was established at the Bonferroni-corrected P value threshold of 2.16×10−6 in African Americans (i.e., 0.05/23150 local ancestry windows) and 3.83×10−6 in Hispanic Americans (i.e., 0.05/13071 local ancestry windows). All analyses, unless specified otherwise, were implemented using the software package R version 3.1.2 (R Development Core Team) or PLINK version 1.9 (Purcell et al. 2007). Genetic positions were aligned based on the human genome reference build GRCh37 (hg19).
Results
Distribution of genetic ancestry percentages vary by characteristics of participants. For African Americans for example, mean African ancestry percentage was higher among unmarried compared to married women (81% vs. 77.8%, P=0.004) and among women without compared to those with private or managed care health insurance (81.4% vs. 77.4%, P=0.0004). For Hispanic American women, mean Amerindigenous ancestry percentage was higher among married compared to unmarried (36.8% vs. 29%, P=3.3×10−3), among unemployed compared to employed (41.4% vs. 29.9%, P=1.7×10−6), and among high school or less educated compared to college or higher educated (42.8% vs.26.1%, P=1.6×10−13), and among women without compared to women with private or managed care health insurance (38.7% vs. 27.6%, P=2.0×10−6), and was inversely correlated with maternal pre-pregnancy weight (ρ=−0.14, P=0.001) and height (ρ=−0.35, P=1.3×10−13) (Table S2).
Associations of global ancestry with fetal growth
In covariate-adjusted models, fetal weight was inversely associated with percentage of African ancestry in Hispanic Americans at 23–40 weeks and East Asian ancestry in Asian Americans at 38–40 weeks, but was positively associated with percentage of Amerindigenous ancestry in Hispanic Americans at 27–40 weeks and European ancestry in European Americans at 13–40 weeks (P ranged from 0.001 to 0.048; Fig 1). Similar patterns were found for other growth measures, except long bones (humerus and femur lengths) which were positively associated with percentage of African ancestry in Hispanic Americans and African Americans at 24–40 weeks and inversely associated with percentage of East Asian ancestry at 31–40 weeks (Fig S2 and Table S3). The variance in fetal growth measures explained by the model showed a small increment (adjusted coefficient of determination (adjusted R2) ranging from 0.01% to 3.6%) in models that contained genetic ancestry and socio-demographic factors compared to models that contained only socio-demographic factors (Table S4).
Fig 1.
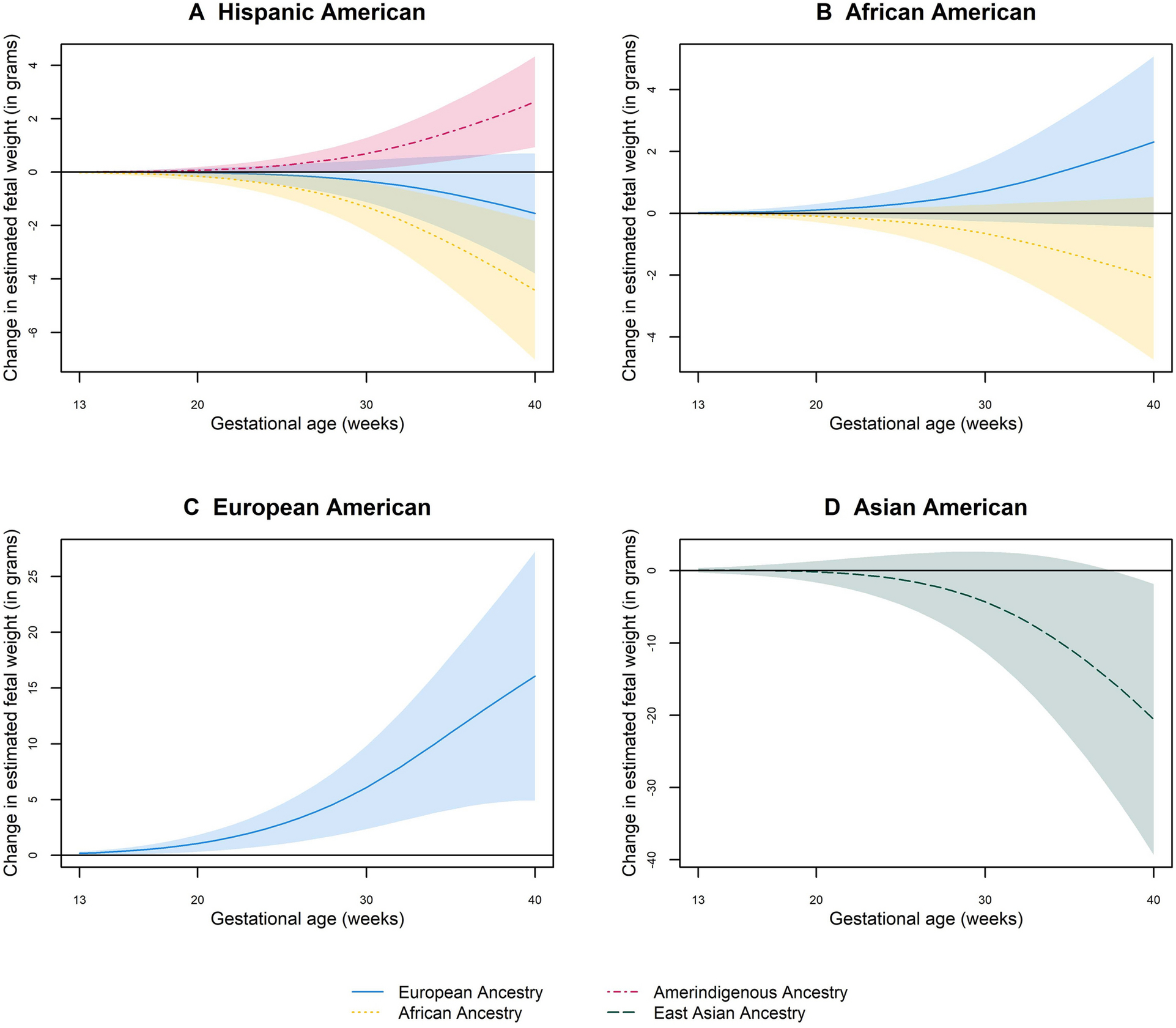
Change in fetal weight at 13–40 weeks’ gestation per each percent increase in African, European, Amerindigenous, and East Asian ancestry
Black horizontal line along the 0-mark on the vertical axis represents the null. Color shaded areas represent the lower and upper bounds of 95% Confidence Intervals. All models were adjusted for maternal age, education, insurance, employment, height, pre-pregnancy weight, parity, and marital status.
Novel admixture mapping loci associated with fetal growth
A genome-wide admixture mapping identified 13 local ancestry regions significantly associated with fetal growth measures (P ranged from 2.18×10−8 to 3.71×10−6). To evaluate robustness of the findings, empirical P values were obtained based on 1000 permutations and found no instances in which P values from the permutation tests were greater than those observed to be genome-wide significant (empirical P =0.000999). These local ancestry regions marked higher African ancestry at chr4.q26 associated with decreased fetal weight at 27 and 40 weeks’ gestation; higher African ancestry at chr2.q21.2, chr4.q25, chr7.q11.22, chr9.p22.1-p21.3, and chr10.q23.1 associated with decreased abdominal circumference at 27 and/or 40 weeks; higher Amerindigenous ancestry at chr2.p22.2, chr6.q21, and chr8.p22 associated with increased abdominal circumference at 27 and/or 40 weeks; higher African ancestry at chr6.p25.3 and chr14.q31.1 associated with decreased head circumference at 40 weeks; and higher African ancestry at chr2q14.3-q21.1, and chr2.q23.3-q24.2 associated with longer long bones (femur and humerus lengths at 27 and 40 weeks) (Table 1, Table S5 and Fig S3). Annotation of the local ancestry intervals using the National Human Genome Research Institute-European Bioinformatics Institute (NHGRI-EBI) Catalog of published GWAS (Welter et al. 2014) found that 9 out of the 13 local ancestry regions harbor genes previously found to be associated with anthropometry traits (Table 2).
Table 1.
Local ancestry signals significantly associated with fetal growth measures based on genome-wide admixture mapping
| Fetal growth measure | Ancestry | Locus | Number of significantly associated local ancestry locic | Effect on fetal growth measure | P | Genes within locus |
|---|---|---|---|---|---|---|
| Fetal weight at week 27 | African ancestrya | chr4.q26 | 4 | Decrease | 1.17×10−6–1.94×10−6 | C4orf32, AP1AR, TIFA |
| Fetal weight at week 40 | African ancestrya | chr4.q26 | 4 | Decrease | 1.59×lO−6–2.67×10−6 | C4orf32, AP1AR, TIFA |
| Abdominal circumference at week 27 | African ancestrya | chr4.q25 | 15 | Decrease | 1.55×10−7–3.71×10−6 | MIR297, AC024198.1, LYPLA1P2, AP1AR, NEUROG2, ZGRF1, LARP7, MIR367, MIR302, C4orf32, AP1AR, TIFA, NEUROG2, ZGRF1, LARP7, MIR367, MIR302, ANK2, MIR1243, MIR8082, CAMK2D, ARSJ |
| Amerindigenous ancestrya | chr6.q21 | 6 | Increase | 7.81×10−7–3.39×10−6 | PRDM1, ATG5, AIM1, RTN4IP1, QRSL1, MIR587, C6orf203, BEND3, PDSS2, SOBP, SCML4, SEC63, OSTM1, AS1 | |
| African ancestrya | chr7.q11.22 | 6 | Decrease | 7.36×10−7–3.63×10−6 | AUST2, CCDS5540.1, WBSCR17–001 | |
| African ancestrya | Chr10.q23.1 | 7 | Decrease | 1.35×10−6–2.47×10−6 | HMGN2P8, GHITM, C10orf99, CDHR1, RGR, CCSER2, LINC00858, TNPO1P1, RPS3AP5 | |
| African ancestrya | Chr14.q31.3-q32.1 | 4 | Decrease | 3.22×10−6–3.64×10−6 | FAM35CP, GPR65, GALC, LINC01146, KCNK10, EFCAB11, RAB42P1, TDP1 | |
| Abdominal circumference at week 40 | African ancestrya | chr2.q21.2 | 6 | Decrease | 1.32×10−6–3.17×10−6 | MIR3679, RMDN2, AS1, CYP1B1 |
| Amerindigenous ancestrya | chr2.p22.2 | 2 | Increase | 2.47×10−6–2.95×10−6 | RMDN2, CYP1B1 | |
| African ancestrya | chr4.q25 | 13 | Decrease | 1.24×10−7–2.98×10−6 | C4orf32, AP1AR, TIFA, NEUROG2, ZGRF1, LARP7, MIR367, MIR302, ANK2, MIR1243, MIR8082, CAMK2D, ARSJ | |
| Amerindigenous ancestrya | chr6.q21 | 16 | Increase | 2.32×10−7–2.78×10−6 | PRDM1, ATG5, AIM1, RTN4IP1, QRSL1, MIR587, C6orf203, BEND3, PDSS2, SOBP, SCML4, SEC63, OSTM1, AS1 | |
| Amerindigenous ancestrya | chr8.p22 | 20 | Increase | 2.52×10−7–3.53×10−6 | DEFB134, DEFB135, DEFB136, USP17L2, USP17L7, FAM90A2P, FAM86B1, DEFB130, FAM66A, DEFB109P1, FAM90A25P, FAM86B2, MIR5692A2, LONRF1, MIR3926–1, MIR3926–2, KIAA1456, DLC1, C8orf48, SGCZ, TUSC3, MSR1 | |
| African ancestrya | chr9.p22.1-p21.3 | 16 | Decrease | 2.59×10−7–3.57×10−6 | IFNA21, IFNA4, IFNA7, IFNA10, IFNA16, IFNA17, IFNA14, IFNA22P, IFNA5, KLHL9, IFNA6, IFNA13, IFNA2, IFNA8, SLC24A2, MLLT3, MIR4473, FOCAD, FOCAD-AS1, MIR491, HACD4, IFNB1, IFNW1, IFNA1, MIR31HG, IFNE, MIR31, MTAP | |
| Head circumference at week 40 | African ancestryb | chr6.p25.3 | 15 | Decrease | 3.00×l0−8–1.88×10−6 | EXOC2, EXOC3, FOXQ1, FOXF2, MIR6720 |
| African ancestryb | Chr14.q31.1 | 9 | Decrease | 5.24×10−7–1.93×10−6 | DIO, AS1, CEP128, TSHR | |
| Femur length at week 27 | African ancestryb | chr2.q24.2 | 1 | Increase | 2.13×10−6 | BAZ2B |
| Femur length at week 40 | African ancestryb | chr2.q14.3-q21.1 | 2 | Increase | 1.84×10−6–1.42×10−6 | |
| Humerus length at week 27 | African ancestryb | chr2.q23.3-q24.2 | 28 | Increase | 4.05×l0−8–2.11×10−6 | GALNT13, RPRM, KCNJ3, ACVR1, UPP2, TANC1, BAZ2B, MARCH7, CD302, LY75, CD302, PLA2R1, ITGB6 |
| Humerus length at week 40 | African ancestryb | chr2.q23.3-q24.2 | 4 | Increase | 1.56×10−7–1.74×10−6 | GALNT13 |
Hispanic Americans
African Americans
Bonferroni-corrected P < 2.16 × 10−6 in African Americans and < 3.83×10−6 in Hispanic Americans. For exact P per locus, see Table S5.
Table 2.
Overlaps between admixture mapping signals associated with fetal growth measures (present study) and loci associated with anthropometry traits in published genome-wide association studies.
| Locus | SNP | Position (hgl9) | Gene | Published genome-wide association studies | ||
|---|---|---|---|---|---|---|
| Disease/trait | P-value | PubMed ID | ||||
| 2p22.2 | rs6706045 | 37888051 | RMDN2 | Height | 6.00E-14 | 30595370 |
| rs162332 | 38095219 | CYP1B1-AS1 | Height | 2.00E-08 | 30595370 | |
| 2q24.1 | rs55920843 | 157556189 | ACVR1C | Waist-to-hip ratio adjusted for body mass index | 3.00E-20 | 30239722 |
| rs56188432 | 157550353 | ACVR1C | Birth weight | 1.00E-16 | 31043758 | |
| rs55920843 | 157556189 | ACVR1C | Waist-to-hip ratio adjusted for body mass index | 3.00E-14 | 30778226 | |
| rs7563664 | 157487943 | CYTIP | Birth weight | 2.00E-08 | 31043758 | |
| rs2444770 | 157647227 | ACVR1C | Waist-to-hip ratio adjusted for body mass index | 3.00E-16 | 30575882 | |
| rs2444770 | 157647227 | ACVR1C | Waist-hip ratio | 5.00E-08 | 30575882 | |
| rs55920843 | 157556189 | ACVR1C | Waist-hip ratio | 2.00E-11 | 30239722 | |
| rs55920843 | 157556189 | ACVR1C | Waist-hip ratio | 9.00E-19 | 30595370 | |
| rs72927479 | 157659825 | ACVR1C | Waist-hip ratio | 3.00E-13 | 30595370 | |
| rs12620249 | 154170934 | GALNT13 | Body mass index | 2.00E-08 | 30595370 | |
| 2q24.2 | rs10173538 | 159712765 | MARCH7 | Birth weight | 1.00E-08 | 31043758 |
| rs11674055 | 159023593 | TANC1 | Height | 4.00E-09 | 30595370 | |
| rs2124969 | 160132975 | ITGB6 | Waist circumference adjusted for body mass index | 7.00E-09 | 25673412 | |
| 4q25 | rs4833407 | 112390634 | ALPK1 | Body mass index | 4.00E-08 | 26426971 |
| rs7668738 | 113208714 | TIFA | Body mass index | 4.28E-08 | 17903300 | |
| 4q26 | rs7669672 | 113824425 | CAMK2D | Height | 8.00E-11 | 30595370 |
| rs7677674 | 113968218 | ARSJ | Height | 1.00E-15 | 30595370 | |
| 6q21 | rs803522 | 106394073 | CRYBG1 | Visceral fat | 1.00E-09 | 30942860 |
| rs803522 | 106394073 | CRYBG1 | Visceral fat | 4.00E-10 | 30942860 | |
| 8p22 | rs4123853 | 14233516 | SGCZ | Body mass index | 1.00E-15 | 30239722 |
| rs354508 | 15677717 | TUSC3 | Body mass index | 3.00E-10 | 30239722 | |
| rs74871039 | 12805555 | AC123777.1 | Height | 1.00E-13 | 30595370 | |
| rs59738387 | 13342703 | DLC1 | Height | 2.00E-20 | 30595370 | |
| rs76364830 | 13514611 | DLC1 | Height | 1.00E-43 | 30595370 | |
| rs6990042 | 14316465 | SGCZ | Body mass index | 1.00E-08 | 26426971 | |
| rs13263601 | 14238391 | SGCZ | Body mass index | 2.00E-14 | 30595370 | |
| rs12679528 | 15708655 | TUSC3 | Body mass index | 4.00E-11 | 30595370 | |
| 9p21.3 | rs143138242 | 21159039 | IFNW1 | Height | 4.00E-08 | 30595370 |
| 14q31.1 | rs935728 | 80491580 | CEP128 | Height | 2.00E-11 | 30595370 |
| rs10498603 | 86610956 | FLRT2, GALC | Body mass index | 1.56E-10 | 17903300 | |
Further analyses of associations between SNPs in the local ancestry regions and the respective fetal growth measures found that variants in the chr2q23.3-q24.2 locus were associated with humerus length among African Americans after correction for multiple testing (Table 3). All variants had strong LD (r2>0.8), with rs13030825 (intronic in GALNT13) having the strongest association with humerus length (T allele frequency = 0.90 among African Americans, β (s.e)= 0.44 (0.10), P= 6.25×10−6 at week 27; β (s.e.)= 0.39 (0.10), P=7.72×10−5 at week 40) (Fig 2). When the analysis was conditioned on the local African ancestry at the chr2q23.3-q24.2 locus, the association of rs13030825 with humerus length was attenuated, but was not eliminated (β (s.e.)= 0.37 (0.12), P= 3×10−3 at week 27; β (s.e.)= 0.33 (0.13), P=9×10−3 at week 40). This observation suggests that the SNP is not merely an ancestry informative marker. The frequency of rs13030825 T allele is 0.99 in the YRI (the parental African sample from the 1000 Genomes Project used in our admixture analysis) but 0.42 among the CEU (the parental European sample used in the admixture mapping). The Weir and Cockerham FST, which measures population differentiation based on allele frequency differences, for rs13030825 between YRI and CEU was 0.55 (P=0.03).
Table 3.
Single nucleotide polymorphisms associated with long bone length at the chr2q24.1-q24.2 locus in African Americans
| SNP | Chr: position (hgl9) | Effect allele | Non-effect allele | Humerus length z-score at week 27 | Humerus length z-score at week 40 | ||
|---|---|---|---|---|---|---|---|
| β (s.e.) | P | β (s.e.) | P | ||||
| rs13030825 | 2:154816802 | T | C | 0.44 (0.1) | 6.25×10−6 | 0.39 (0.1) | 7.72×10−5 |
| rs6714832 | 2:154808697 | T | C | 0.44 (0.1) | 9.32×10−6 | 0.39 (0.1) | 8.51×10−5 |
| rs6738659 | 2:154826521 | A | G | 0.44 (0.1) | 7.64×10−6 | 0.38 (0.1) | 1.07×10−4 |
| rs10205027 | 2:154828241 | T | G | 0.44 (0.1) | 7.95×10−6 | 0.38 (0.1) | 1.12×10−4 |
| rs6761233 | 2:154837081 | A | G | 0.44 (0.1) | 1.01×10−5 | 0.38 (0.1) | 1.46×10−4 |
| rs7603550 | 2:154849933 | G | A | 0.43 (0.1) | 9.22×10−6 | 0.38 (0.1) | 1.20×10−4 |
| rs7578311 | 2:154850187 | C | A | 0.43 (0.1) | 9.22×10−6 | 0.38 (0.1) | 1.20×10−4 |
| rs7578960 | 2:154851413 | T | C | 0.43 (0.1) | 9.21×10−6 | 0.38 (0.1) | 1.20×10−4 |
| rs11689769 | 2:154856671 | A | G | 0.43 (0.1) | 1.33×10−5 | 0.37 (0.1) | 1.94×10−4 |
| rs1596637 | 2:154858857 | A | C | 0.43 (0.1) | 1.33×10−5 | 0.37 (0.1) | 1.94×10−4 |
| rs6736399 | 2:154869895 | G | A | 0.43 (0.1) | 8.96×10−6 | 0.38 (0.1) | 1.18×10−4 |
| rs6435010 | 2:154873967 | A | C | 0.43 (0.1) | 1.25×10−5 | 0.37 (0.1) | 1.85×10−4 |
| rs7572016 | 2:154874149 | T | C | 0.43 (0.1) | 1.25×10−5 | 0.38 (0.1) | 1.85×10−4 |
| rs10195968 | 2:154880327 | T | G | 0.43 (0.1) | 1.09×10−5 | 0.38 (0.1) | 1.34×10−4 |
| rs7608978 | 2:154889809 | A | G | 0.44 (0.1) | 1.37×10−5 | 0.37 (0.1) | 1.98×10−4 |
Abbreviation: SNP, single nucleotide polymorphism, chr: chromosome, s.e: standard error
Fig 2.
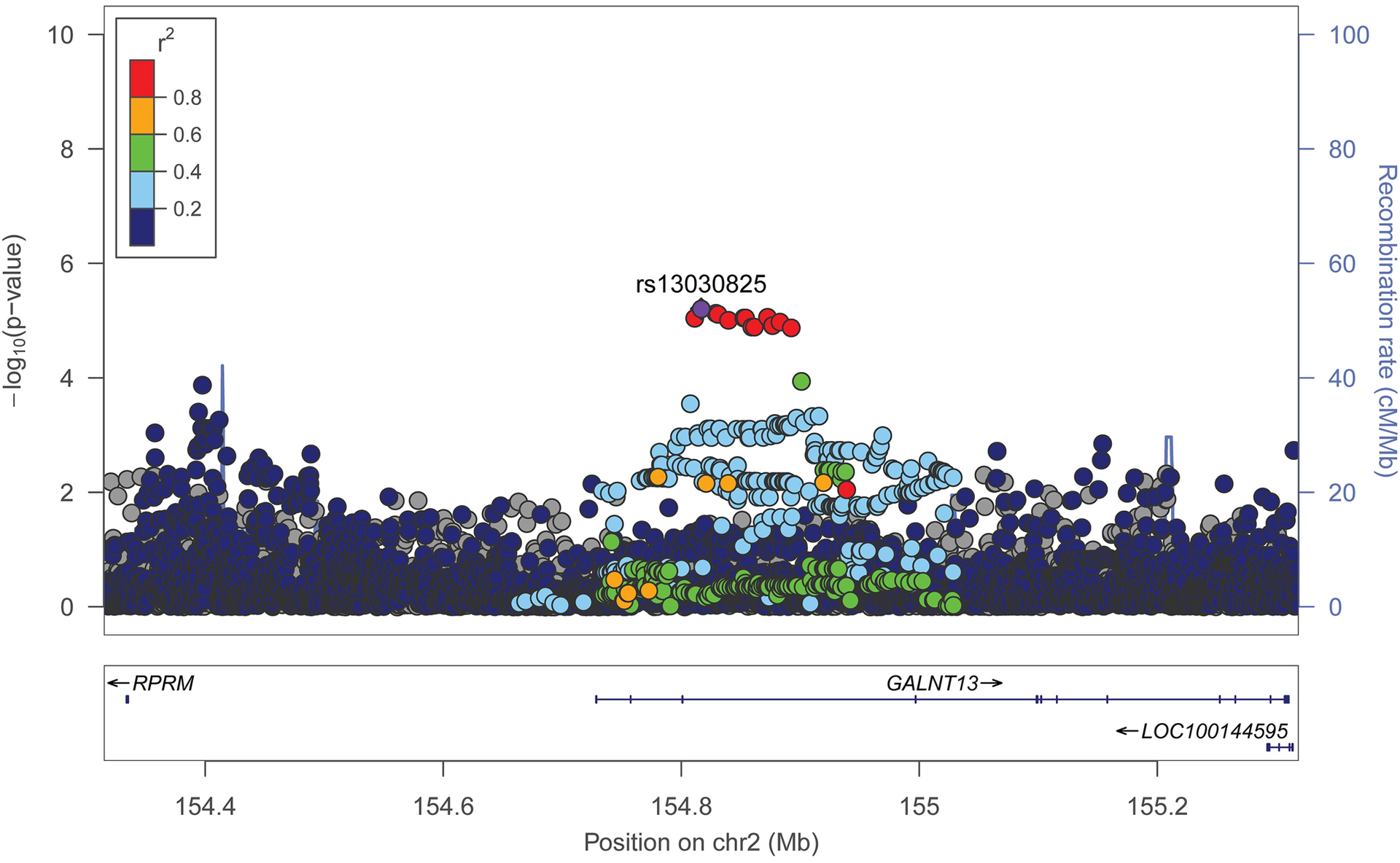
Regional plot of variants in the ch2q23.3-q24.4 African ancestry locus associated with longer bone lengths
Data span 500 kb centered at the top associated SNP (rs13030825). The x-axis denotes genomic position (hg19) and the y axis denotes the log10 P-value, and recombination rate (cM/Mb). The purple circle point represents the index SNP. The color of each data point indicates its linkage disequilibrium value (r2) with the index SNP based on HapMap2’s African reference genome.
To determine whether rs13030825 accounted for the association finding at the chr2q23.3-q24.2 locus, we repeated the admixture mapping analyses by adding rs13030825 as a covariate in the model. When adjusted for rs13030825, the association of African ancestry at the chr2q23.3-q24.2 locus with humerus length was no longer significant (β=0.17, P=0.08 at week 27 and β=0.14, P=0.15 at week 40). Evidence from Haploreg (Ward and Kellis 2012) shows that rs13030825 overlaps with enriched regulatory annotations for enhancer histone marks and altered regulatory motifs in multiple tissues including serum response factor (SRF) transcription factor.
Discussion
This is the first genome-wide admixture mapping scan and analysis of genetic ancestry on fetal growth. The genome-wide averages of African, European, Amerindigenous and East Asian ancestry composition were associated with fetal growth measures. Admixture mapping analysis identified several novel signals in genetic regions that differ in local ancestry. Specifically, we found ten loci at which higher African ancestry was associated with smaller fetal weight, smaller abdominal circumference, smaller head circumference and longer skeletal bones, and three loci at which higher Amerindigenous ancestry was associated with larger abdominal circumference. Notably, the rs13030825 (GALNT13) variant at the chr2q23.3–24.2 local ancestry locus was found to be strongly associated with long bone length and appears to explain most of the association between higher African ancestry at the locus and longer bone length in African Americans.
Most of the admixture mapping loci overlap with genetic loci found to be associated with anthropometric and cardiometabolic traits in previous GWASs (Welter et al. 2014). Notably, the chr2q.23.3-q24.2 locus at which African ancestry was found to be associated with long bones in the present study harbors the ACVR1C, GALNT13 and CYTIP loci that have been associated with birthweight in the most recent GWAS of birthweight (Warrington et al. 2019). ACVR1C is a receptor for transforming growth factor beta family of signaling molecules and is highly expressed in adipose tissues. Studies in mice have shown that ACVR1C down-regulates glucose-stimulated insulin release in pancreatic beta cells (Bertolino et al. 2008), and participates in the molecular regulation of adipose tissue homeostasis (Andersson et al. 2008).
We also observed that common SNPs within the chr2q.23.3-q24.2 locus were associated with long bone length in African Americans and the lead SNP explained most of the admixture signal. Particularly, SNP rs13030825 (GALNT13) most strongly associated with humerus and femur lengths overlaps with regulatory motifs including SRF, which is a widely expressed transcription factor that regulates expression of genes involved in cell proliferation, differentiation, migration, survival, and apoptosis (Miano 2003). In addition to its well documented role in embryonic organ development, a recent study in mice has demonstrated that SRF deletion downregulates insulin like growth factor 1(IGF-1), a growth factor crucial in skeletal development and bone remodeling (Chen et al. 2012). The same study also found that osteoblast-derived SRF deletion resulted in decreased postnatal bone development of adult mice (Chen et al. 2012). Altogether, the admixture signal identified in our study identified genetic variants that have a role in skeletal bone development.
Global ancestry was associated with fetal growth after adjustment for some socio-demographic factors. The ancestry effect direction on fetal growth was consistent with studies that found that adult height is inversely correlated with Amerindigenous ancestry (Asgari et al. 2020) and positively correlated with African and European ancestries (Ruiz-Linares et al. 2014). Fetal weight has been found to be smaller in African Americans and Asian Americans compared to European Americans (Buck Louis et al. 2015), and fetal long bones have been found to be longer in African Americans and shorter in Asian Americans compared to European Americans (Buck Louis et al. 2015; Chang et al. 2003; Harper et al. 2010; Kovac et al. 2002; Shipp et al. 2001). Genetic risk scores of birthweight-reducing variants exhibited population differences (Tekola-Ayele et al. 2018); however, the loci were discovered in GWASs involving predominantly European ancestry populations and may not tag the causal variants in non-Europeans (Martin et al. 2019). In our study, there was a small increment of the variance in fetal growth measures explained by models incorporating global ancestry and sociodemographic factors compared to models incorporating only sociodemographic factors. However, we observed that higher African and Amerindigenous ancestries are correlated with lower socioeconomic indicators such as unemployment and lower educational status. We did not have data to account for inter-generational and early life social experiences that could influence fetal growth (Astone et al. 2007; Burris and Hacker 2017; Geronimus 1996; Mutambudzi et al. 2017). Given possible relationships between ancestry and structural disadvantages (Krieger et al. 1998; Parra et al. 2004), it is important to be cautious that unaccounted social factors correlated with global ancestry may in part explain the observed associations between global ancestry and fetal growth.
We acknowledge that our study’s sample size may have limited our ability to identify admixture signals with modest effect. Replication of the local ancestry signals is needed in an independent dataset but was not possible within the scope of our study because similar cohorts with longitudinal fetal biometry measures in diverse populations are not common. We also observed that the signals detected in African Americans did not transfer to Hispanic Americans and vice versa. Absence of cross-population extrapolation is not unexpected because the causal variants may be outside the local ancestry interval boundaries which are predicted with some degree of uncertainty. In addition, local ancestry inference by RFMix depends on both the reference and admixed sample chromosomes to determine window interval size, model training and assignment of ancestry per window. Demographic differences between African Americans and Hispanic Americans in the number of admixture events, selective pressures, and their historical timing may also lead to widely different contributions of an ancestry tract (Bryc et al. 2015). Moreover, the loci may interact with environmental factors that differ between the two groups. The maternal local ancestry signals could impact fetal growth through their direct influence on the intrauterine environment or by contributing alleles to the fetus (Eaves et al. 2014; Warrington et al. 2019). Future large cohorts with maternal and paternal genotypes or maternal and offspring genotypes are needed to differentiate these effects and identify molecular pathways. Finally, sequence data is being increasingly generated for global and diverse population including the HGDP samples (Bergstrom et al. 2020). In the future, these improvements in coverage of denser genetic markers and rarer variants from genetic sequence data can be leveraged to identify local ancestry association signals at population- differentiated and clinically relevant loci such as the exome.
In conclusion, in a cohort of pregnant women in the U.S. with high quality repeated longitudinal measurement of fetal biometry, we found novel African and Amerindigenous local ancestry signals associated with fetal growth measures. Notably, the admixture mapping signal at the chr2q23.3-q24.2 locus associated with long bone lengths in African Americans may be driven by a difference in allele frequency between the African and European ancestry at rs13030825, a SNP that overlaps with the SRF transcription factor previously implicated in postnatal bone development. Future functional studies may give insights into the molecular pathways that underlie the relationships between the novel local ancestry markers and fetal growth.
Supplementary Material
Acknowledgements
The authors acknowledge the research teams at all participating clinical centers for the NICHD Fetal Growth Studies, including Christina Care Health Systems, Columbia University, Fountain Valley Hospital, California, Long Beach Memorial Medical Center, New York Hospital, Queens, Northwestern University, University of Alabama at Birmingham, University of California, Irvine, Medical University of South Carolina, Saint Peters University Hospital, Tufts University, and Women and Infants Hospital of Rhode Island. The authors also acknowledge C-TASC and The EMMES Corporations in providing data and imaging support. Genotyping was performed in the Department of Laboratory Medicine and Pathology, University of Minnesota. This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov).
Funding
This work was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), National Institutes of Health (NIH) including American Recovery and Reinvestment Act funding via contract numbers HHSN275200800013C; HHSN275200800002I; HHSN27500006; HHSN275200800003IC; HHSN275200800014C; HHSN275200800012C; HHSN275200800028C; HHSN275201000009C and HHSN27500008. Additional support was obtained from the NIH Office of the Director, the National Institute on Minority Health and Health Disparities (NIMHD) and the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK).
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ethics approval
The study was approved by the institutional review boards of NICHD and each of the participating clinic sites.
Consent to participate
Written informed consent was obtained from all study participants.
Data availability
The genotype data underlying the findings reported in this study are available for interested researchers upon request through the NICHD Division of Intramural Population Health Research Biospecimen Repository Access and Data Sharing platform.
Consent for publication
Not applicable
Code availability
Not applicable
Animal research
Not applicable
Plant reproducibility
Not applicable
Gels and blots/Image manipulation
Not applicable
Clinical trials registration
References
- Alexander DH, Lange K (2011) Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinformatics 12: 246. doi: 10.1186/1471-2105-12-246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19: 1655–64. doi: 10.1101/gr.094052.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson O, Korach-Andre M, Reissmann E, Ibanez CF, Bertolino P (2008) Growth/differentiation factor 3 signals through ALK7 and regulates accumulation of adipose tissue and diet-induced obesity. Proc Natl Acad Sci U S A 105: 7252–6. doi: 10.1073/pnas.0800272105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asgari S, Luo Y, Akbari A, Belbin GM, Li X, Harris DN, Selig M, Bartell E, Calderon R, Slowikowski K, Contreras C, Yataco R, Galea JT, Jimenez J, Coit JM, Farronay C, Nazarian RM, O’Connor TD, Dietz HC, Hirschhorn JN, Guio H, Lecca L, Kenny EE, Freeman EE, Murray MB, Raychaudhuri S (2020) A positively selected FBN1 missense variant reduces height in Peruvian individuals. Nature 582: 234–239. doi: 10.1038/s41586-020-2302-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astone NM, Misra D, Lynch C (2007) The effect of maternal socio-economic status throughout the lifespan on infant birthweight. Paediatr Perinat Epidemiol 21: 310–8. doi: 10.1111/j.1365-3016.2007.00821.x [DOI] [PubMed] [Google Scholar]
- Baker JL, Rotimi CN, Shriner D (2017) Human ancestry correlates with language and reveals that race is not an objective genomic classifier. Sci Rep 7: 1572. doi: 10.1038/s41598-017-01837-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DJ, Godfrey KM, Osmond C, Bull A (1992) The relation of fetal length, ponderal index and head circumference to blood pressure and the risk of hypertension in adult life. Paediatr Perinat Epidemiol 6: 35–44. [DOI] [PubMed] [Google Scholar]
- Beaumont RN, Warrington NM, Cavadino A, Tyrrell J, Nodzenski M, Horikoshi M, Geller F, Myhre R, Richmond RC, Paternoster L, Bradfield JP, Kreiner-Moller E, Huikari V, Metrustry S, Lunetta KL, Painter JN, Hottenga JJ, Allard C, Barton SJ, Espinosa A, Marsh JA, Potter C, Zhang G, Ang W, Berry DJ, Bouchard L, Das S, Early Growth Genetics C, Hakonarson H, Heikkinen J, Helgeland O, Hocher B, Hofman A, Inskip HM, Jones SE, Kogevinas M, Lind PA, Marullo L, Medland SE, Murray A, Murray JC, Njolstad PR, Nohr EA, Reichetzeder C, Ring SM, Ruth KS, Santa-Marina L, Scholtens DM, Sebert S, Sengpiel V, Tuke MA, Vaudel M, Weedon MN, Willemsen G, Wood AR, Yaghootkar H, Muglia LJ, Bartels M, Relton CL, Pennell CE, Chatzi L, Estivill X, Holloway JW, Boomsma DI, Montgomery GW, Murabito JM, Spector TD, Power C, Jarvelin MR, Bisgaard H, Grant SFA, Sorensen TIA, Jaddoe VW, Jacobsson B, Melbye M, McCarthy MI, Hattersley AT, Hayes MG, Frayling TM, Hivert MF, Felix JF, Hypponen E, Lowe WL Jr., Evans DM, Lawlor DA, Feenstra B, Freathy RM (2018) Genome-wide association study of offspring birth weight in 86 577 women identifies five novel loci and highlights maternal genetic effects that are independent of fetal genetics. Hum Mol Genet 27: 742–756. doi: 10.1093/hmg/ddx429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom A, McCarthy SA, Hui R, Almarri MA, Ayub Q, Danecek P, Chen Y, Felkel S, Hallast P, Kamm J, Blanche H, Deleuze JF, Cann H, Mallick S, Reich D, Sandhu MS, Skoglund P, Scally A, Xue Y, Durbin R, Tyler-Smith C (2020) Insights into human genetic variation and population history from 929 diverse genomes. Science 367. doi: 10.1126/science.aay5012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolino P, Holmberg R, Reissmann E, Andersson O, Berggren PO, Ibanez CF (2008) Activin B receptor ALK7 is a negative regulator of pancreatic beta-cell function. Proc Natl Acad Sci U S A 105: 7246–51. doi: 10.1073/pnas.0801285105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla C, Parra EJ, Pfaff CL, Dios S, Marshall JA, Hamman RF, Ferrell RE, Hoggart CL, McKeigue PM, Shriver MD (2004) Admixture in the Hispanics of the San Luis Valley, Colorado, and its implications for complex trait gene mapping. Ann Hum Genet 68: 139–53. doi: 10.1046/j.1529-8817.2003.00084.x [DOI] [PubMed] [Google Scholar]
- Bryc K, Durand EY, Macpherson JM, Reich D, Mountain JL (2015) The genetic ancestry of African Americans, Latinos, and European Americans across the United States. Am J Hum Genet 96: 37–53. doi: 10.1016/j.ajhg.2014.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck Louis GM, Grewal J, Albert PS, Sciscione A, Wing DA, Grobman WA, Newman RB, Wapner R, D’Alton ME, Skupski D, Nageotte MP, Ranzini AC, Owen J, Chien EK, Craigo S, Hediger ML, Kim S, Zhang C, Grantz KL (2015) Racial/ethnic standards for fetal growth: the NICHD Fetal Growth Studies. Am J Obstet Gynecol 213: 449 e1–449 e41. doi: 10.1016/j.ajog.2015.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burris HH, Hacker MR (2017) Birth outcome racial disparities: A result of intersecting social and environmental factors. Semin Perinatol 41: 360–366. doi: 10.1053/j.semperi.2017.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SC, O’Brien KO, Nathanson MS, Caulfield LE, Mancini J, Witter FR (2003) Fetal femur length is influenced by maternal dairy intake in pregnant African American adolescents. Am J Clin Nutr 77: 1248–54. doi: 10.1093/ajcn/77.5.1248 [DOI] [PubMed] [Google Scholar]
- Chen J, Yuan K, Mao X, Miano JM, Wu H, Chen Y (2012) Serum response factor regulates bone formation via IGF-1 and Runx2 signals. J Bone Miner Res 27: 1659–68. doi: 10.1002/jbmr.1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper RS, Nadkarni GN, Ogedegbe G (2018) Race, ancestry, and reporting in medical journals. JAMA. doi: 10.1001/jama.2018.10960 [DOI] [PubMed] [Google Scholar]
- Eaves LJ, Pourcain BS, Smith GD, York TP, Evans DM (2014) Resolving the effects of maternal and offspring genotype on dyadic outcomes in genome wide complex trait analysis (“M-GCTA”). Behav Genet 44: 445–55. doi: 10.1007/s10519-014-9666-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freathy RM, Mook-Kanamori DO, Sovio U, Prokopenko I, Timpson NJ, Berry DJ, Warrington NM, Widen E, Hottenga JJ, Kaakinen M, Lange LA, Bradfield JP, Kerkhof M, Marsh JA, Magi R, Chen CM, Lyon HN, Kirin M, Adair LS, Aulchenko YS, Bennett AJ, Borja JB, Bouatia-Naji N, Charoen P, Coin LJ, Cousminer DL, de Geus EJ, Deloukas P, Elliott P, Evans DM, Froguel P, Genetic Investigation of ATC, Glaser B, Groves CJ, Hartikainen AL, Hassanali N, Hirschhorn JN, Hofman A, Holly JM, Hypponen E, Kanoni S, Knight BA, Laitinen J, Lindgren CM, Meta-Analyses of G, Insulin-related traits C, McArdle WL, O’Reilly PF, Pennell CE, Postma DS, Pouta A, Ramasamy A, Rayner NW, Ring SM, Rivadeneira F, Shields BM, Strachan DP, Surakka I, Taanila A, Tiesler C, Uitterlinden AG, van Duijn CM, Wellcome Trust Case Control C, Wijga AH, Willemsen G, Zhang H, Zhao J, Wilson JF, Steegers EA, Hattersley AT, Eriksson JG, Peltonen L, Mohlke KL, Grant SF, Hakonarson H, Koppelman GH, Dedoussis GV, Heinrich J, Gillman MW, Palmer LJ, Frayling TM, Boomsma DI, Davey Smith G, Power C, Jaddoe VW, Jarvelin MR, Early Growth Genetics C, McCarthy MI(2010) Variants in ADCY5 and near CCNL1 are associated with fetal growth and birth weight. Nat Genet 42: 430–5. doi: 10.1038/ng.567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaskins RB, LaGasse LL, Liu J, Shankaran S, Lester BM, Bada HS, Bauer CR, Das A, Higgins RD, Roberts M (2010) Small for gestational age and higher birth weight predict childhood obesity in preterm infants. Am J Perinatol 27: 721–30. doi: 10.1055/s-0030-1253555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR (2015) A global reference for human genetic variation. Nature 526: 68–74. doi: 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geronimus AT (1996) Black/white differences in the relationship of maternal age to birthweight: a population-based test of the weathering hypothesis. Soc Sci Med 42: 589–97. [DOI] [PubMed] [Google Scholar]
- Godfrey KM, Barker DJ (2000) Fetal nutrition and adult disease. Am J Clin Nutr 71: 1344S–52S. doi: 10.1093/ajcn/71.5.1344s [DOI] [PubMed] [Google Scholar]
- Grewal J, Grantz KL, Zhang C, Sciscione A, Wing DA, Grobman WA, Newman RB, Wapner R, D’Alton ME, Skupski D, Nageotte MP, Ranzini AC, Owen J, Chien EK, Craigo S, Albert PS, Kim S, Hediger ML, Buck Louis GM (2018) Cohort Profile: NICHD Fetal Growth Studies-Singletons and Twins. Int J Epidemiol 47: 25–25l. doi: 10.1093/ije/dyx161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadlock FP, Harrist RB, Sharman RS, Deter RL, Park SK (1985) Estimation of fetal weight with the use of head, body, and femur measurements--a prospective study. American journal of obstetrics and gynecology 151: 333–337. [DOI] [PubMed] [Google Scholar]
- Hales CN, Barker DJ, Clark PM, Cox LJ, Fall C, Osmond C, Winter PD (1991) Fetal and infant growth and impaired glucose tolerance at age 64. BMJ 303: 1019–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper LM, Gray D, Dicke J, Stamilio DM, Macones GA, Odibo AO (2010) Do race-specific definitions of short long bones improve the detection of down syndrome on second-trimester genetic sonograms? J Ultrasound Med 29: 231–5. [DOI] [PubMed] [Google Scholar]
- Hediger ML, Fuchs KM, Grantz KL, Grewal J, Kim S, Gore-Langton RE, Buck Louis GM, D’Alton ME, Albert PS (2016) Ultrasound Quality Assurance for Singletons in the National Institute of Child Health and Human Development Fetal Growth Studies. J Ultrasound Med 35: 1725–33. doi: 10.7863/ultra.15.09087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikoshi M, Beaumont RN, Day FR, Warrington NM, Kooijman MN, Fernandez-Tajes J, Feenstra B, van Zuydam NR, Gaulton KJ, Grarup N, Bradfield JP, Strachan DP, Li-Gao R, Ahluwalia TS, Kreiner E, Rueedi R, Lyytikainen LP, Cousminer DL, Wu Y, Thiering E, Wang CA, Have CT, Hottenga JJ, Vilor-Tejedor N, Joshi PK, Boh ETH, Ntalla I, Pitkanen N, Mahajan A, van Leeuwen EM, Joro R, Lagou V, Nodzenski M, Diver LA, Zondervan KT, Bustamante M, Marques-Vidal P, Mercader JM, Bennett AJ, Rahmioglu N, Nyholt DR, Ma RCW, Tam CHT, Tam WH, Group CCHW, Ganesh SK, van Rooij FJ, Jones SE, Loh PR, Ruth KS, Tuke MA, Tyrrell J, Wood AR, Yaghootkar H, Scholtens DM, Paternoster L, Prokopenko I, Kovacs P, Atalay M, Willems SM, Panoutsopoulou K, Wang X, Carstensen L, Geller F, Schraut KE, Murcia M, van Beijsterveldt CE, Willemsen G, Appel EVR, Fonvig CE, Trier C, Tiesler CM, Standl M, Kutalik Z, Bonas-Guarch S, Hougaard DM, Sanchez F, Torrents D, Waage J, Hollegaard MV, de Haan HG, Rosendaal FR, Medina-Gomez C, Ring SM, Hemani G, McMahon G, Robertson NR, Groves CJ, Langenberg C, Luan J, Scott RA, Zhao JH, Mentch FD, MacKenzie SM, Reynolds RM, Early Growth Genetics C, Lowe WL Jr., Tonjes A, Stumvoll M, Lindi V, et al. (2016) Genome-wide associations for birth weight and correlations with adult disease. Nature 538: 248–252. doi: 10.1038/nature19806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikoshi M, Yaghootkar H, Mook-Kanamori DO, Sovio U, Taal HR, Hennig BJ, Bradfield JP, St Pourcain B, Evans DM, Charoen P, Kaakinen M, Cousminer DL, Lehtimaki T, Kreiner-Moller E, Warrington NM, Bustamante M, Feenstra B, Berry DJ, Thiering E, Pfab T, Barton SJ, Shields BM, Kerkhof M, van Leeuwen EM, Fulford AJ, Kutalik Z, Zhao JH, den Hoed M, Mahajan A, Lindi V, Goh LK, Hottenga JJ, Wu Y, Raitakari OT, Harder MN, Meirhaeghe A, Ntalla I, Salem RM, Jameson KA, Zhou K, Monies DM, Lagou V, Kirin M, Heikkinen J, Adair LS, Alkuraya FS, Al-Odaib A, Amouyel P, Andersson EA, Bennett AJ, Blakemore AI, Buxton JL, Dallongeville J, Das S, de Geus EJ, Estivill X, Flexeder C, Froguel P, Geller F, Godfrey KM, Gottrand F, Groves CJ, Hansen T, Hirschhorn JN, Hofman A, Hollegaard MV, Hougaard DM, Hypponen E, Inskip HM, Isaacs A, Jorgensen T, Kanaka-Gantenbein C, Kemp JP, Kiess W, Kilpelainen TO, Klopp N, Knight BA, Kuzawa CW, McMahon G, Newnham JP, Niinikoski H, Oostra BA, Pedersen L, Postma DS, Ring SM, Rivadeneira F, Robertson NR, Sebert S, Simell O, Slowinski T, Tiesler CM, Tonjes A, Vaag A, Viikari JS, Vink JM, Vissing NH, Wareham NJ, Willemsen G, Witte DR, Zhang H, et al. (2013) New loci associated with birth weight identify genetic links between intrauterine growth and adult height and metabolism. Nat Genet 45: 76–82. doi: 10.1038/ng.2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiserud T, Benachi A, Hecher K, Perez RG, Carvalho J, Piaggio G, Platt LD (2018) The World Health Organization fetal growth charts: concept, findings, interpretation, and application. Am J Obstet Gynecol 218: S619–S629. doi: 10.1016/j.ajog.2017.12.010 [DOI] [PubMed] [Google Scholar]
- Kovac CM, Brown JA, Apodaca CC, Napolitano PG, Pierce B, Patience T, Hume RF Jr., Calhoun BC (2002) Maternal ethnicity and variation of fetal femur length calculations when screening for Down syndrome. J Ultrasound Med 21: 719–22; quiz 724–5. [DOI] [PubMed] [Google Scholar]
- Krieger N, Sidney S, Coakley E (1998) Racial discrimination and skin color in the CARDIA study: implications for public health research. Coronary Artery Risk Development in Young Adults. Am J Public Health 88: 1308–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, Myers RM (2008) Worldwide human relationships inferred from genome-wide patterns of variation. Science 319: 1100–4. doi: 10.1126/science.1153717 [DOI] [PubMed] [Google Scholar]
- Maples BK, Gravel S, Kenny EE, Bustamante CD (2013) RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. Am J Hum Genet 93: 278–88. doi: 10.1016/j.ajhg.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ (2019) Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet 51: 584–591. doi: 10.1038/s41588-019-0379-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miano JM (2003) Serum response factor: toggling between disparate programs of gene expression. J Mol Cell Cardiol 35: 577–93. doi: 10.1016/s0022-2828(03)00110-x [DOI] [PubMed] [Google Scholar]
- Mikkola K, Ritari N, Tommiska V, Salokorpi T, Lehtonen L, Tammela O, Paakkonen L, Olsen P, Korkman M, Fellman V (2005) Neurodevelopmental outcome at 5 years of age of a national cohort of extremely low birth weight infants who were born in 1996–1997. Pediatrics 116: 1391–400. doi: 10.1542/peds.2005-0171 [DOI] [PubMed] [Google Scholar]
- Mutambudzi M, Meyer JD, Reisine S, Warren N (2017) A review of recent literature on materialist and psychosocial models for racial and ethnic disparities in birth outcomes in the US, 2000–2014. Ethn Health 22: 311–332. doi: 10.1080/13557858.2016.1247150 [DOI] [PubMed] [Google Scholar]
- Ochs-Balcom HM, Shaw H, Preus L, Palmer JR, Haddad SA, Rosenberg L, Ruiz-Narvaez EA (2018) Admixture mapping and fine-mapping of birth weight loci in the Black Women’s Health Study. Hum Genet 137: 535–542. doi: 10.1007/s00439-018-1908-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra EJ, Kittles RA, Shriver MD (2004) Implications of correlations between skin color and genetic ancestry for biomedical research. Nat Genet 36: S54–60. doi: 10.1038/ng1440 [DOI] [PubMed] [Google Scholar]
- Pinheiro J, Bates D (2000) Mixed-Effects Models in S and S-PLUS, 1 edn. Springer-Verlag New York, New York [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–75. doi: 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner AP, Ziv E, Lind DL, Nievergelt CM, Schork NJ, Cummings SR, Phong A, Burchard EG, Harris TB, Psaty BM, Kwok PY (2005) Population structure, admixture, and aging-related phenotypes in African American adults: the Cardiovascular Health Study. Am J Hum Genet 76: 463–77. doi: 10.1086/428654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roland MC, Friis CM, Voldner N, Godang K, Bollerslev J, Haugen G, Henriksen T (2012) Fetal growth versus birthweight: the role of placenta versus other determinants. PLoS One 7: e39324. doi: 10.1371/journal.pone.0039324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossen LM, Schoendorf KC (2014) Trends in racial and ethnic disparities in infant mortality rates in the United States, 1989–2006. Am J Public Health 104: 1549–56. doi: 10.2105/AJPH.2013.301272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Linares A, Adhikari K, Acuna-Alonzo V, Quinto-Sanchez M, Jaramillo C, Arias W, Fuentes M, Pizarro M, Everardo P, de Avila F, Gomez-Valdes J, Leon-Mimila P, Hunemeier T, Ramallo V, Silva de Cerqueira CC, Burley MW, Konca E, de Oliveira MZ, Veronez MR, Rubio-Codina M, Attanasio O, Gibbon S, Ray N, Gallo C, Poletti G, Rosique J, Schuler-Faccini L, Salzano FM, Bortolini MC, Canizales-Quinteros S, Rothhammer F, Bedoya G, Balding D, Gonzalez-Jose R (2014) Admixture in Latin America: geographic structure, phenotypic diversity and self-perception of ancestry based on 7,342 individuals. PLoS Genet 10: e1004572. doi: 10.1371/journal.pgen.1004572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipp TD, Bromley B, Mascola M, Benacerraf B (2001) Variation in fetal femur length with respect to maternal race. J Ultrasound Med 20: 141–4. [DOI] [PubMed] [Google Scholar]
- Shriner D (2017) Overview of Admixture Mapping. Curr Protoc Hum Genet 94: 1 23 1–1 23 8. doi: 10.1002/cphg.44 [DOI] [PubMed] [Google Scholar]
- Shriner D, Tekola-Ayele F, Adeyemo A, Rotimi CN (2014) Genome-wide genotype and sequence-based reconstruction of the 140,000 year history of modern human ancestry. Sci Rep 4: 6055. doi: 10.1038/srep06055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekola-Ayele F, Workalemahu T, Amare AT (2018) High burden of birthweight-lowering genetic variants in Africans and Asians. BMC Med 16: 70. doi: 10.1186/s12916-018-1061-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekola-Ayele F, Zhang C, Wu J, Grantz KL, Rahman ML, Shrestha D, Ouidir M, Workalemahu T, Tsai MY (2020) Trans-ethnic meta-analysis of genome-wide association studies identifies maternal ITPR1 as a novel locus influencing fetal growth during sensitive periods in pregnancy. PLoS Genet 16: e1008747. doi: 10.1371/journal.pgen.1008747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward LD, Kellis M (2012) HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 40: D930–4. doi: 10.1093/nar/gkr917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrington NM, Beaumont RN, Horikoshi M, Day FR, Helgeland O, Laurin C, Bacelis J, Peng S, Hao K, Feenstra B, Wood AR, Mahajan A, Tyrrell J, Robertson NR, Rayner NW, Qiao Z, Moen GH, Vaudel M, Marsit CJ, Chen J, Nodzenski M, Schnurr TM, Zafarmand MH, Bradfield JP, Grarup N, Kooijman MN, Li-Gao R, Geller F, Ahluwalia TS, Paternoster L, Rueedi R, Huikari V, Hottenga JJ, Lyytikainen LP, Cavadino A, Metrustry S, Cousminer DL, Wu Y, Thiering E, Wang CA, Have CT, Vilor-Tejedor N, Joshi PK, Painter JN, Ntalla I, Myhre R, Pitkanen N, van Leeuwen EM, Joro R, Lagou V, Richmond RC, Espinosa A, Barton SJ, Inskip HM, Holloway JW, Santa-Marina L, Estivill X, Ang W, Marsh JA, Reichetzeder C, Marullo L, Hocher B, Lunetta KL, Murabito JM, Relton CL, Kogevinas M, Chatzi L, Allard C, Bouchard L, Hivert MF, Zhang G, Muglia LJ, Heikkinen J, Consortium EGG, Morgen CS, van Kampen AHC, van Schaik BDC, Mentch FD, Langenberg C, Luan J, Scott RA, Zhao JH, Hemani G, Ring SM, Bennett AJ, Gaulton KJ, Fernandez-Tajes J, van Zuydam NR, Medina-Gomez C, de Haan HG, Rosendaal FR, Kutalik Z, Marques-Vidal P, Das S, Willemsen G, Mbarek H, Muller-Nurasyid M, Standl M, Appel EVR, Fonvig CE, et al. (2019) Maternal and fetal genetic effects on birth weight and their relevance to cardio-metabolic risk factors. Nat Genet 51: 804–814. doi: 10.1038/s41588-019-0403-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, Parkinson H (2014) The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res 42: D1001–6. doi: 10.1093/nar/gkt1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workalemahu T, Grantz KL, Grewal J, Zhang C, Louis GMB, Tekola-Ayele F (2018) Genetic and Environmental Influences on Fetal Growth Vary during Sensitive Periods in Pregnancy. Sci Rep 8: 7274. doi: 10.1038/s41598-018-25706-z [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


