Abstract
The Negative on TATA-less (NOT) 2_3_5 domain proteins play key roles in mRNA metabolism and transcription regulation, but few comprehensive studies have focused on this protein family in plants. In our study, a total of 30 NOT2_3_5 genes were identified in four cotton genomes: Gossypium. arboretum, G. raimondii, G. hirsutum and G. barbadense. Phylogenetic analysis showed that all the NOT2_3_5 domain proteins were divided into two classes. The NOT2_3_5 genes were expanded frequently, and segmental duplication had significant effects in their expansion process. The cis-regulatory elements analysis of NOT2_3_5 promoter regions indicated that NOT2_3_5 domain proteins might participate in plant growth and development processes and responds to exogenous stimuli. Expression patterns demonstrated that all of the GhNOT2_3_5 genes were expressed in the majority of tissues and fiber development stages, and that these genes were induced by multiple stresses. Quantitative real-time PCR showed that GbNOT2_3_5 genes were up-regulated in response to verticillium wilt and the silencing of GbNOT2_3_5-3/8 and GbNOT2_3_5-4/9 led to more susceptibility to verticillium wilt than controls. Identification and analysis of the NOT2_3_5 protein family will be beneficial for further research on their biological functions.
Keywords: NOT2_3_5 domain protein, Verticillium wilt, cotton
1. Introduction
Negative on TATA-less (NOT) proteins are important constitutive components of the carbon catabolite repression 4 (CCR4)-NOT complex, which exists in all eukaryotes and plays a key role in mRNA metabolism and transcription regulation [1]. The NOT genes of yeast were isolated from mutations in which transcription of the HIS3 gene was increased [2,3]. Collart et al. indicated that mutations of the NOT genes cause up-regulated expression of many genes [4]. To date, several kinds of NOT proteins have been isolated from yeasts, flies, humans and Arabidopsis thaliana, including NOT1, NOT2, NOT3, NOT4, NOT5, NOT9, NOT10 and NOT11 [5]. The NOT proteins function in a complex. Among them, NOT1, as a >200 kDa subunit, functions as a scaffold for the CCR4-NOT complex and other components of the CCR4-NOT complex dock on the NOT1 protein [6]. As core proteins, NOT2, NOT3 and NOT4 docked on the C-terminal domain of the NOT1 scaffold [7,8], while NOT10 and NOT11 dock onto the N-terminal domain of the NOT1 scaffold [9,10].
Genetic and biochemical analysis indicated that NOT2, NOT3 and NOT5 shared a conserved NOT2_3_5 domain and were closely associated [11]. Additionally, NOT3 and NOT5 from Saccharomyces cerevisiae were regarded as paralogous genes recently [12], and NOT5 had up to 44% sequence identity with NOT3 protein in the N-terminal region of [13]. Many previous studies indicated that NOT2, NOT3 and NOT5 function together and play key roles in the vegetative growth and transcription machinery of yeast [12]. Until now, there have been more studies on NOT proteins containing a NOT2_3_5 domain than on other NOT proteins. Russell et al. identified four not2 mutations of Saccharomyces cerevisiae and suggested that there was no 1.9 MDa CCR4-NOT complex in the not2:L9 mutant, and stability of the CCR4-NOT complex was decreased slightly in the not2-4 mutant [14]. Additionally, NOT2 protein had a significant influence on the integrity of the CCR4-NOT complex and the associations with other NOT proteins. Proteins NOT2 and NOT3/5 were also essential for assembly of the proteasome, and together with some chaperones and proteasomes strongly regulated protein synthesis and degradation in the cell [12]. Additionally, NOT2 was found to be involved in transcription elongation and mutation of NOT2 decreased the interaction between RNA polymerase II and NOT proteins [15]. Sohn et al. [16] found that CNOT2 facilitated the differentiation and lipogenesis of 3T3-L1 adipocyte through regulating expression of PPARγ, CEBPα, GSK3α/β and β-catenin. The expression level of CNOT2 was positively correlated with the expression level of PPARγ and CEBPα and negatively correlated with that of GSK3α/β and β-catenin [16]. Besides studies on NOT2_3_5 proteins from yeast and human, there has been some research focused on NOT2_3_5 proteins from phytopathogenic fungi and plants. Gene FonNot2 was expressed higher in conidia and germinating conidia than in mycelia and played important roles in vegetative growth and conidia production and the conidial morphology of watermelon fusarium wilt pathogen (Fusarium oxysporum f. sp.) [17]. The FonNot2 was significantly up-regulated during infection of watermelon roots compared to mycelia under axenic conditions and FonNot2 increased virulence in the infection process through regulating cell wall integrity, oxidative stress response, ROS production and fatty acids (FA) biosynthesis [17]. In previous studies, two highly similar NOT2 homologs in Arabidopsis were identified including AtNOT2a and AtNOT2b; AtNOT2b was previously known as VIRE2-INTERACTING PROTEIN2 (VIP2) and was isolated using a yeast two-hybrid system. Anand et al. [18] suggested that AtVIP2 contains a NOT domain, which is conserved between plants and animals and that AtVIP2, likely as a transcriptional regulator, was involved in Agrobacterium-mediated stable transformation through promoting the integration of T-DNA. Wang et al. [19] reported that NOT2 proteins were vital for plant development and inactivation of Os-NOT2 in rice and At-NOT2 in Arabidopsis thaliana caused seedling death and defect in male gametogenesis, respectively. Protein NOT2 interacted with RNA polymerase II and several microRNA (miRNA) biogenesis factors containing DCL1, CBP20, CBP80 and SE and acted as general factors to regulate the level of pri-miRNAs and mature miRNAs involved in miRNA biogenesis. Zhao et al. cloned the TaVIP2 gene, which encoded a NOT2 protein from wheat and indicated that overexpression of TaNOT2 in tobacco increased the efficiency of Agrobacterium-mediated transformation and resistance to powdery mildew of tobacco [20].
Cotton is one of the most important economic crops in the world because its fibers are the main natural source for textiles [21]. With the deterioration of global climate, cotton cultivation faces a number of biotic and abiotic stresses [22]. The genomes of diploid and allotetraploid cotton including Gossypium arboretum, G. raimondii, G. hirsutum and G. barbadense have been successfully sequenced and released, which helps in excavating functional genes and genome-wide analysis of gene families [23,24]. Although the structure and function of the NOT2_3_5 proteins have been studied in yeasts, human, phytopathogenic fungi and some model organisms, little is known about the NOT2_3_5 protein in cotton. Previous studies indicated that NOT2 played a key role in miRNA biogenesis and miRNA regulated expression level of target genes involving in plant growth, development and response to abiotic and biotic stresses [18]. Thus, we wanted to research the NOT2_3_5 family and attempt to utilize the transcriptional regulation of NOT2_3_5 proteins to increase resistance in cotton.
Here, we characterize NOT2_3_5 domain proteins from diploid and tetraploid cotton species and examine their expression patterns during normal growth and development and after abiotic and biotic stresses. Two of these genes were induced in a verticillium-resistant cotton variety after pathogen inoculation. Verticillium wilt, a soil-borne vascular disease, causes devastating losses of cotton yield and quality every year [25,26].The results will provide new approaches to improve resistance to verticillium wilt in cotton and provide new inspiration on further research on transcriptional regulation of NOT proteins.
2. Results
2.1. Identification of NOT2_3_5 Domain Protein Family in Cotton
In order to determine the NOT2_3_5 domain protein family in cotton, we conducted an HMMER alignment search on the protein sequences of the genomes of the four cotton species, with the hidden Markov model profile of the NOT2_3_5 domain (PF04153). After filtration of proteins without the NOT2_3_5 domain and non-full-length segments, a total of 5, 5, 10 and 10 NOT2_3_5 domain protein genes were identified in G. arboretum, G. raimondii, G. hirsutum and G. barbadense, respectively. All the above genes were numbered consecutively from GaNOT2_3_5-1 to GaNOT2_3_5-5, GrNOT2_3_5-1 to GrNOT2_3_5-5, GhNOT2_3_5-1 to GhNOT2_3_5-10 and GbNOT2_3_5-1 to GbNOT2_3_5-11, respectively, according to their localization on chromosomes (Table S1). The protein length of obtained NOT2_3_5 domain proteins ranged within 563-1048 amino acids and mean length was 737 amino acids. The MW and pI were also predicted, with ranges of 59019.22 to 115638.5 kDa and 5.27–9.45, respectively, and corresponding means of 81715.43 kDa and 6.75, respectively. Subcellular localization prediction results suggested all of the NOT2_3_5 domain proteins were predicted to be located in the nucleus (Table S1).
2.2. Phylogenetic and Conserved Amino Residue Analysis of NOT2_3_5 Domain Proteins
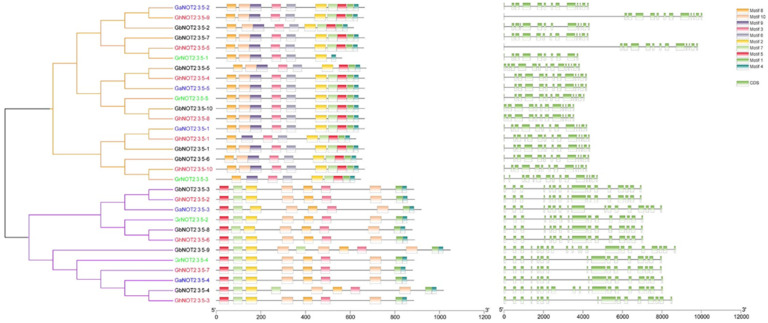
To analyze the phylogenetic relationship of NOT2_3_5 domain proteins from four cotton and other plant species, we also obtained three and five NOT2_3_5 domain proteins from Arabidopsis and rice, respectively, using the same method. All of the 38 NOT2_3_5 domain protein sequences were aligned using Clustal X. The alignment results were used to construct a phylogenetic tree by MEGA7 software using the NJ method with a bootstrap value set as 500 (Figure 1). All of the NOT2_3_5 domain proteins were divided into two classes: Class I and Class II, with 16 and 22 members. Using the WebLogo online software, the conserved domain-sequence analysis of the two classes of NOT2_3_5 domain proteins was performed. The conserved NOT2_3_5 domains were likely located in the C-terminal and the conservation degrees of amino acid residue from each class of NOT2_3_5 domains were very high (Figure S1). Several amino acid residues showed no variations. However, there were significant differences in the sequences of NOT2_3_5 domains from Classes I and II (Figure S1).
Figure 1.
Phylogenetic analysis of NOT2_3_5 genes from cotton and other plants. Prefixes At, Os, Gr, Ga, Gh and Gb represented Arabidopsis thaliana, Oryza sativa, G. raimondii, G. arboretum, G. hirsutum and G. barbadense, respectively. NOT 2_3_5 domain protein from G. hirsutum, G. barbadense, G. raimondii, G. arboretum, Arabidopsis thaliana and Oryza sativa were marked red, black, green, blue, turquoise and pink, respectively.
Structures of NOT2_3_5 domain protein genes were analyzed by aligning the genomic DNA sequences and coding sequences using the GSDS online software. Genes from the same class had similar gene structures with conserved size and number of exons and variable length of introns. The exon number of NOT2_3_5 domain protein genes from Class I ranged within 10–15. Among them, GrNOT2_3_5-1 and GrNOT2_3_5-3 had the lowest and highest number of exons, respectively. Most NOT2_3_5 domain protein genes from Class II had 16 exons, but GaNOT2_3_5-3 had 17, GbNOT2_3_5-4 had 21 and GbNOT2_3_5-4 had 19. The conserved motifs of the 30 NOT2_3_5 domain protein sequences were predicted by MEME software. The NOT2_3_5 domain proteins from Class I had 7–10 conserved motifs and most had 10. Most NOT2_3_5 domain proteins from Class II contained nine motifs except for GhNOT2_3_5-6 and GbNOT2_3_5-4 with 11 and 9, respectively (Figure 2). All the conserved motif sequences were uploaded to InterPro online tools to analyze their functions. Motifs 1 2, 3, 5, 6, 7, 9 and 10 were classified into NOT2/NOT3/NOT5 protein family membership (IPR040168). Motifs 4 and 8 were not classified into any membership (Table S2).
Figure 2.
Analysis of conserved motif and gene structure of NOT2_3_5 domain proteins. NOT2_3_5 domain protein from G. hirsutum, G. barbadense, G. raimondii and G. arboretum were marked red, black, green and blue, respectively.
2.3. Chromosome Location and Gene Collinearity Analysis
All NOT2_3_5 domain protein genes from the four cotton species were mapped onto chromosomes or scaffolds. Of 30 NOT2_3_5 domain protein genes, 28 were located on chromosomes and 2 were mapped to scaffolds (Figure S2). Five GrNOT2_3_5 genes were found in D04, D07, D09 and D13; and five GaNOT2_3_5 genes were mapped to A05, A08, A11 and A13. Ten GbNOT2_3_5 genes were mapped to AD2_A05, AD2_A08, AD2_A11 and AD2_A13 and the corresponding D genome. Two of GhNOT2_3_5 genes were mapped to scaffolds, and the chromosome locations of the remaining GhNOT2_3_5 genes were similar to those of GbNOT2_3_5 genes. Loci of GhNOT2_3_5 and GbNOT2_3_5 genes were deeply conserved between At and Dt sub-genomes in allotetraploid cotton, indicating that G. hirsutum and G. barbadense were typical allotetraploid species and were applicable to natural polyploidization study.
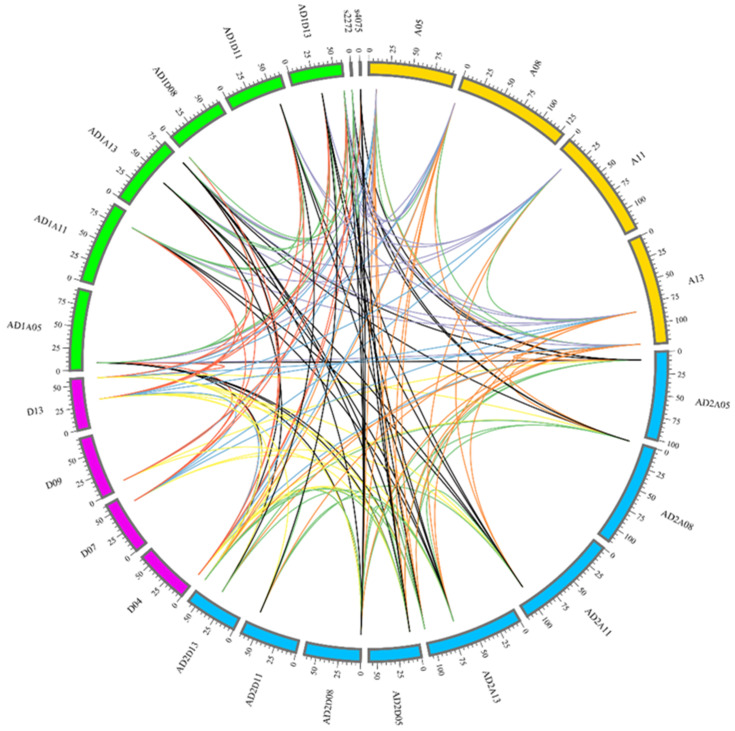
Gene families are created by duplications and variations from a common ancestral gene. Gene duplication events play key roles in species evolution and environmental adaptation. Segmental duplication and tandem repeat are the main types of gene duplications. MCSCAN software was used to analyze the expansion and synteny of NOT2_3_5 domain protein genes from G. arboretum, G. raimondii, G. hirsutum and G. barbadense. The duplication events were selected according to the standard described in the methods. Results indicated that all gene family members of NOT2_3_5 domain proteins were produced by duplication events and segmental duplication events, but no tandem duplication events were observed in their expansion process. There were 155 pairs of duplicated genes of NOT2_3_5 domain proteins identified among the four cotton species. There were 4, 17, 12 and 2 duplicated gene pairs observed in G. arboretum, G. barbadense, G. hirsutum and G. raimondii, respectively. There were 19, 11 and 22 pairs of segmental duplication events detected between G. arboretum and G. hirsutum, G. arboretum and G. raimondii and G. arboretum and G. barbadense, respectively. There were 18, 18 and 32 pairs of segmental duplication events obtained between G. barbadense and G. raimondii, G. hirsutum and G. raimondii and G. barbadense and G. hirsutum, respectively (Figure 3). Results suggested that genes of NOT2_3_5 domain proteins were expanded frequently, and segmental duplication had a significant effect on their expansion.
Figure 3.
Gene collinearity analysis among cotton NOT2_3_5 genes. AD1AX and AD1DX (green blocks) indicated chromosomes of A sub-genome and D sub-genome of G. hirsutum. AD2AX and AD2DX (blue blocks) indicated chromosomes of A sub-genome and D sub-genome of G. barbadense. AX (yellow blocks) and DX (pink blocks) represented chromosomes of G. arboretum and G. raimondii, respectively. Green lines exhibited duplicated gene pairs among NOT2_3_5 genes in each G. arboretum, G. barbadense, G. hirsutum, G. raimondii cotton species. Orange lines, blue lines, red lines, yellow lines, purple lines and black lines separately exhibited duplicated gene pairs among NOT2_3_5 genes from and G. arboretum and G. barbadense, Gossypium arboretum and G. raimondii, G. hirsutum and G. raimondii, G. raimondii and G. barbadense, G. hirsutum and G. arboretum, G. barbadense and Gossypium arboretum.
The Ka/Ks ratio was applied to predict the selection pressure on gene pairs in the four cotton species, with Ka/Ks = 1, >1 and <1 representing neutral, positive and purifying selections, respectively. Results suggested a Ka/Ks < 1 for 153 NOT2_3_5 domain protein gene pairs indicating that the duplicated genes in the four species had evolved mainly by purifying selection (Figure S3). Additionally, only one gene pair (GbNOT2_3_5-3 and GaNOT2_3_5-3) processed a Ka/Ks > 1, which suggested that that gene pair experienced positive selection pressure. Detailed information for the Ka/Ks ratio of duplicated gene pairs of NOT2_3_5 domain proteins is given in Table S3.
2.4. Cis-Regulatory Elements Analysis of NOT 2_3_5 Promoter Regions
The cis-regulatory elements of promoter regions play vital roles in regulating gene expressions in serious conditions and plant developmental stages. Through identifying and analyzing cis-regulatory elements, the expression patterns, regulatory network and potential functions of NOT2_3_5 domain protein genes were studied deeply. The 2.5-kb upstream genomic sequences from the start codon were considered as putative promoter regions and applied to predict cis-regulatory elements. There were 198 cis-elements (15 types) obtained from 10 putative GhNOT2_3_5 promoters, which were classified into three groups: Cellular development and metabolism, phytohormone regulation and abiotic and biotic stress response (Figure 4 and Table S4). The cis-elements mentioned above, involved in cellular development and metabolism, included O2-site, CAT-box, HD-Zip 1, circadian and MBSI, which represented zein metabolism, meristem expression, palisade mesophyll cell differentiation, circadian control and flavonoid biosynthetic gene regulation, respectively. Additionally, more than half of the cis-elements (107, 18 different kinds) were associated with light response. In the phytohormone regulation group, five types of cis-elements were detected: P-box (gibberellin-responsive element), ABRE (abscisic acid responsiveness), TCA-element (SA responsiveness), CGTCA-motif (MeJA-responsiveness) and AuxRR-core (auxin-responsive element). There were five types of cis-elements in the abiotic and biotic stress response group: WUN-motif, MBS, ARE, TC-rich repeats and LTR, which responded to wound, drought, anaerobic environment, pathogenic bacteria and low temperature, respectively. All the results indicated that NOT2_3_5 domain proteins might participate in plant growth and development processes and respond to exogenous stimuli.
Figure 4.
Cis-Regulatory elements analysis of NOT 2_3_5 promoter regions. Different colors represented different cis-regulatory elements and numbers in abscissa were the numbers of cis-regulatory element.
2.5. Prediction of MiRNAs Targeting GhNOT2_3_5 Domain Protein Genes
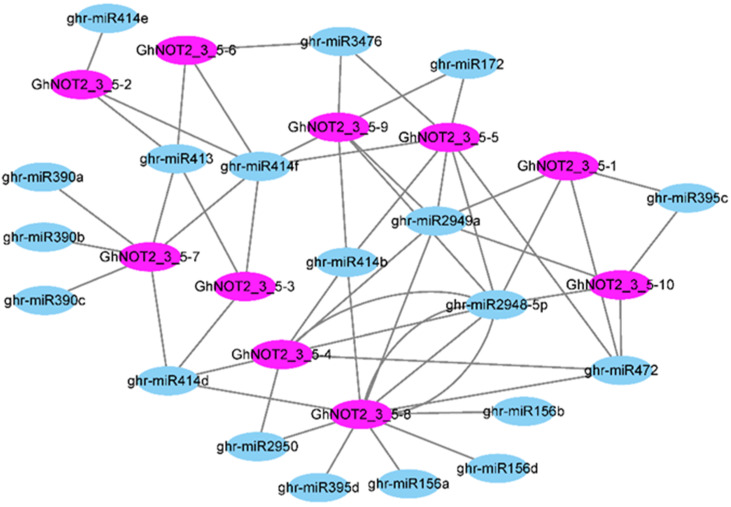
Plant regulatory small RNAs, especially miRNAs, play key roles in regulating gene expression and are involved in plant responses to abiotic and biotic stress processes. The prediction of miRNAs targeting genes would shed new light on understanding the crosstalk between miRNAs from upland cotton and NOT2_3_5 domain protein genes. There were 55 interaction relationships obtained, including 19 ghr-miRNAs targeting 10 GhNOT2_3_5 genes (Figure 5). Among them, ghr-miR2949a targeted six GhNOT2_3_5 genes (GhNOT2_3_5-1, GhNOT2_3_5-4, GhNOT2_3_5-5, GhNOT2_3_5-8, GhNOT2_3_5-9 and GhNOT2_3_5-10) indicating that ghr-miR2949a was closely related to the expression regulation of GhNOT2_3_5 genes. Gene GhNOT2_3_5-8 was potentially targeted by nine ghr-miRNAs (ghr-miR2950, ghr-miR395d, ghr-miR156a, ghr-miR156d, ghr-miR2948-5p, ghr-miR472, ghr-miR2949a, ghr-miR414b and ghr-miR414d), making it the most targeted GhNOT2_3_5 gene. Especially, GhNOT2_3_5-8 was targeted by ghr-miR2948-5p at three different sites. Additionally, other GhNOT2_3_5 genes were also targeted by several ghr-miRNAs: GhNOT2_3_5-4, GhNOT2_3_5-5, GhNOT2_3_5-7 and GhNOT2_3_5-9 were predicted to be targeted by 5–6 ghr-miRNAs.
Figure 5.
Prediction of miRNAs targeting GhNOT2_3_5 genes. Connections among nodes and lines indicate the targeting relationship of miRNA and GhNOT2_3_5 genes.
2.6. Expression Patterns of NOT2_3_5 Genes in Different Tissues and Responding to Stresses and Phytohormones
Gene functions have a close relationship with their expression profiles including expression level and expression location. To better and further study the function of NOT2_3_5 domain protein, the expression profiles of GhNOT2_3_5 genes in different tissues of root, stem, leaf, petal, pistil, calycle, stamen, torus and in different fiber and ovule development stages 5, 10, 20 and 25 days post-anthesis (DPA) for fiber and -3, -1, 0, 1, 3, 5, 10, 20, 25 and 35 DPA for ovules were analyzed using G. hirsutum RNA-seq data [27]. Results demonstrated that all GhNOT2_3_5 genes were expressed in all tissues and development stages mentioned above, but their expression levels varied. However, the expression patterns of GhNOT2_3_5 genes from the A sub-genome were similar to genes from the corresponding D sub-genome. We observed that GhNOT2_3_5-1-3/7 were abundantly expressed with mean values of log2 FPKM > 3.5, while GhNOT2_3_5-1-2/6 were slightly expressed with values of log2 FPKM < 2.0, suggesting that duplicated GhNOT2_3_5 genes might have diverse biological functions. Genes GhNOT2_3_5-1-7 and GhNOT2_3_5-8 were up-regulated in petal, pistil, calycle, stamen and torus, indicating significant roles in floral development. Gene GhNOT2_3_5-10 showed a continuous increase in expression level during fiber and ovule development processes. Gene GhNOT2_3_5-1 had higher expression levels in root, stem and leaf than other tissues suggesting a function associated with stress resistance. The expression profile of GhNOT2_3_5 genes indicated that the NOT2_3_5 domain protein family might be involved in all cotton growth processes and in fiber and ovule development (Figure 6A). Meanwhile, the expression profiles of GhNOT2_3_5 genes responding to stresses including cold, hot, polyethene glycol (PEG) and salt were also investigated using the RNA-seq data. The heat map suggested that GhNOT2_3_5-1/10/3/7 genes were expressed highly during all stress treatments but GhNOT2_3_5-2/6 genes were barely expressed. Under hot treatment, GhNOT2_3_5-5 were up-regulated at an early treatment stage compared to the control. The expression level of GhNOT2_3_5-7 was induced by PEG and salt treatment, with peak expression at 12 h after PEG treatment and 6 h after salt treatment. Gene GhNOT2_3_5-9 was down-regulated after 12h of cold treatment (Figure 6B).
Figure 6.
Heat maps of expression patterns of NOT2_3_5 genes. (A) Expression patterns of NOT2_3_5 genes in different tissues of G. hirsutum. (B) Expression patterns of NOT2_3_5 genes under different stresses of G. hirsutum.
The predicted results of cis-regulatory elements from GhNOT2_3_5 putative promoter regions suggested that GhNOT2_3_5 genes might respond to exogenous hormones and pathogenic bacteria. Thus, expression patterns of GhNOT2_3_5 members in G. hirsutum var TM-1 and G. barbadense var Hai 7124 under exogenous hormone treatment (SA, MeJA and ET), t and after V. dahliae infection were analyzed using qPCR. The expression levels of GhNOT2_3_5 genes did not significantly change under MeJA treatment indicating that they were not involved in MeJA response pathways. All of the GhNOT2_3_5 genes were down-regulated at 12h after ET treatment. However, gene GhNOT2_3_5-1/10 was slightly down-regulated at 6h after treatment and up-regulated after experiencing a long period of SA treatment. Gene GhNOT2_3_5-5/9 showed a higher expression level at the early SA treatment time point (Figure 7A). Expression of each gene pair between 0 time point for each exogenous hormones treatment experiment did not vary more than 1.339 fold. Genes of NOT2_3_5 from G. hirsutum and G. barbadense exhibited different expression patterns following V. dahlia infection. None of the GhNOT2_3_5 genes were induced by V. dahliae infection, suggesting that they had no roles in cotton resistance against verticillium wilt. Notably, expression levels of GbNOT2_3_5-5/10, GbNOT2_3_5-3/8 and GbNOT2_3_5-4/9 were up-regulated under V. dahliae treatment. Among them, GbNOT2_3_5-4/9 showed significantly higher expression at four treatment time points, with expression peaking at 12 h after inoculation. The expression level of GbNOT2_3_5-5/10 gene increased during the inoculation process. Gene GbNOT2_3_5-3/8 was induced early, with expression peaking at 6 h after inoculation. The expression profiles of NOT2_3_5 under V. dahliae inoculation was in accordance with disease resistance of cotton varieties. The G. barbadense var Hai 7124 is V. dahliae resistant and some GbNOT2_3_5 genes were up-regulated during V. dahliae inoculation; however, G. hirsutum var TM-1 is V. dahliae susceptible and all of its GhNOT2_3_5 genes were not induced under treatment (Figure 7B).
Figure 7.
Expression patterns of NOT2_3_5 genes under exogenous hormone and after infection of V. dahlia using qPCR. (A) Expression patterns of NOT2_3_5 genes in G. hirsutum var TM-1 under exogenous hormone, including SA, MeJA and ET, treatment. (B) Expression patterns of NOT2_3_5 genes in G. barbadense var Hai 7124 after infection of V. dahlia.
2.7. Silencing of GbNOT2_3_5 Genes Reduced Verticillium Wilt Resistance in Cotton
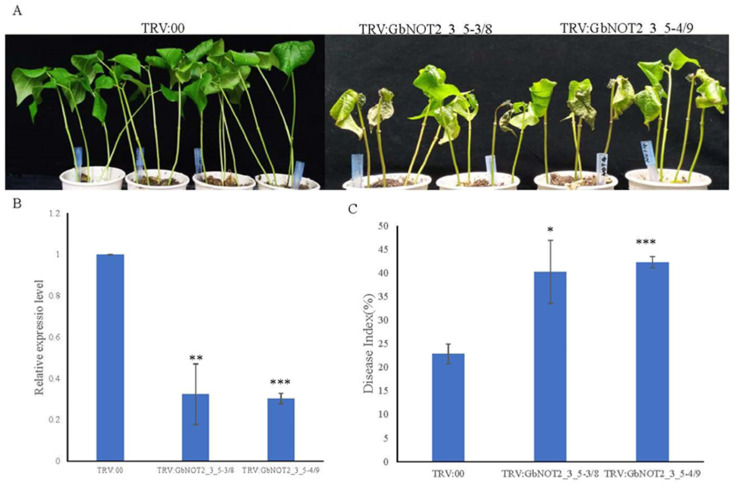
Tobacco rattle virus (TRV)-based VIGS systems were used to further study whether GbNOT2_3_5 genes played roles in cotton responses to verticillium wilt. Genes GbNOT2_3_5-3/8 and GbNOT2_3_5-4/9 were selected because their expression levels were significantly up-regulated during V. dahliae inoculation. About 300 bp of GbNOT2_3_5-3/8 and GbNOT2_3_5-4/9 genes were amplified using suitable primers and were inserted into the pTRV2 vector, forming recombinant vector pTRV2–GbNOT2_3_5-3/8 and pTRV2–GbNOT2_3_5-4/9. An Agrobacterium strain carrying the above recombinant plasmids was injected into cotyledons. After VIGS, the expression level of GbNOT2_3_5-3/8 and GbNOT2_3_5-4/9 genes were significantly reduced (Figure 8B). After the TRV:PDS cotton seedlings showed a photobleaching phenotype across all true leaves, the remaining cotton seedlings infiltrated with recombinant vectors and empty vectors were inoculated with V. dahliae. After 25 d, GbNOT2_3_5 silenced cotton displayed obvious symptoms of verticillium wilt compared to the control, including wilting of true leaves and yellowing and falling cotyledons and true leaves (Figure 8A). The DI values of TRV: GbNOT2_3_5-3/8, TRV: GbNOT2_3_5-4/9 were remarkably higher than of the control (TRV:00) after inoculation, with values of 40.27 and 42.32 versus 22.90, respectively (Figure 8C). Stems of silenced cotton seedlings and controls were extracted and cut through the middle of the vascular bundle. The xylem of TRV: GbNOT2_3_5 plants showed significantly more browning than TRV:00 plants (Figure S4). Trypan blue assay showed that leaves from silenced cotton were severely dyed blue, indicating more dead cells in the leaves of TRV:GbNOT2_3_5 plants after V. dahliae infection (Figure S4). All of the above results showed that silencing GbNOT2_3_5-3/8 and GbNOT2_3_5-4/9 genes compromised cotton resistance during V. dahliae inoculation.
Figure 8.
Silencing GbNOT2_3_5 genes reduced Verticillium wilt resistance in cotton. (A) The phenotypes of silencing plants and control after infection of V. dahlia. TRV:00 represented empty vector, TRV:GbNOT2_3_5 represented GbNOT2_3_5 genes silenced. (B) Expression levels of GbNOT2_3_5 genes in silencing plants and control. (C) The DI values of silencing plants and control after infection of V. dahlia. * p < 0.1 by Student ’s t test, ** p < 0.05 and *** p < 0.01.
3. Discussion
The NOT2_3_5 domain proteins are important constitutive components of the CCR4-NOT complex and are evolutionarily conserved proteins in eukaryotes. Research on NOT2_3_5 domain proteins has mainly focused on yeast and mammals and those in plants have not been well studied. To further examine NOT2_3_5 domain proteins, we comprehensively analyzed the NOT2_3_5 protein subfamily in four diploid and allotetraploid cotton species. The structural, conserved domain, evolution direction, chromosome location and expression features of the NOT2_3_5 domain proteins were characterized.
3.1. Evolution and Duplication of NOT2_3_5 Genes
During evolution, whole-genome duplication (WGD)/polyploidization has occurred in numerous organisms, accompanied by gene movement, gene loss, chromosome structural changes and gene functional divergence [28]. Thus, WGD is one of the main driving forces of genome evolution and formation of new species [29]. Cotton is an ideal model for exploring the mechanism of natural polyploidy, and understanding polyploid evolution in cotton might improve our insight concerning other polyploid plants [30]. The Gossypium contains about 45 diploid species grouped into eight monophyletic groups: A, B, C, D, E, F, G and K genomes. According to their locations, diploid cotton species are divided into three main lineages: the C, G and K genomes (Sturita subgenus) derived from Australia; the D genome (Houzingenia subgenus) originated from America; and the A, B, E and F genomes derived from Africa and Asia [30]. The formation of the allotetraploid cotton species occurred 1–2 million years ago and the closest extant progenitors of tetraploid cotton are G. herbaceum L.(A1) and G. raimondii Ulbrich (D5) [31,32]. Because of polyploidization and the formation of allotetraploid cotton species, the number of NOT2_3_5 family members from allotetraploid cotton species is double that in diploid species. Meanwhile, WGD resulted in expansion of gene families of cotton species and NOT2_3_5 domain protein genes were duplicated remarkably. The results of collinearity analysis indicated that segmental duplication was the main approach during duplication of the NOT2_3_5 gene family. In tetraploid cotton species, the chromosome location of some of NOT2_3_5 family members did not correspond with that in diploid cotton species, indicating that during evolution of the NOT2_3_5 gene family, chromosome structural changes and chromosome rearrangement may have occurred. Genes of NOT2_3_5 domain proteins in the four cotton species mainly experienced purifying selection, suggesting that differentiation of gene function was partly inhibited.
3.2. Relationship between MiRNA and NOT2_3_5 Genes
As a class of non-coding RNA, miRNA are the key molecular device of RNA silencing, which is involved in regulating gene expression at either the posttranscriptional or transcriptional levels. The miRNAs regulate expression levels of target genes involved in plant growth, development and response to abiotic and biotic stresses [33]. Previous studies have shown that the AtNOT2b protein is involved in regulating the level of pri-miRNAs and mature miRNAs via interacting with RNA polymerase II and several miRNA biogenesis factors containing DCL1, CBP20, CBP80 and SE. In our study, the prediction of miRNAs targeting GhNOT2_3_5 domain protein genes produced 55 interaction relationships including 19 ghr-miRNAs targeting 10 GhNOT2_3_5 genes, which indicated that NOT2_3_5 domain proteins had a close relationship with miRNAs. The NOT2_3_5 domain protein regulated the biogenesis of miRNA and, in turn, the expression level of NOT2_3_5 genes was regulated by some miRNAs. Previous studies have shown that many plant miRNAs are involved in stress response for example miR414 and miR390 are involved in salinity stress [34,35]; miR471, miR472 and miR395 are involved in immune response [36]; miR156 is involved in drought stress [37] and miR2949 is involved in high temperature response [38]. Thus, the prediction results of miRNAs targeting GhNOT2_3_5 genes inspired us to study the mechanism underlying GhNOT2_3_5 genes responding to various stresses in depth.
3.3. Differences between NOT2_3_5 Domain Proteins from Two Classes
Phylogenetic analysis showed that all of the NOT2_3_5 domain proteins were divided into two classes: Class I and Class II. There were significant differences among the NOT2_3_5 domain proteins from different classes, including: sequences, gene structure and conserved motifs. However, the biological functions of NOT2_3_5 domain proteins were uncertain in cotton. In our study, we found three pairs of the GbNOT2_3_5 gene, referred to as GbNOT2_3_5-3/8 and GbNOT2_3_5-4/9, were significantly up-regulated after inoculation with V. dahliae. Another three gene pairs of GbNOT2_3_5 gene did not remarkably respond to Verticillium wilt. Gene pairs of GbNOT2_3_5-3/8 and GbNOT2_3_5-4/9 were from Class I and another gene pair was from Class II. Thus, these previously mentioned differences between different classes partly resulted in the function division among the protein family. The cis-regulatory elements of promoter regions among GhNOT2_3_5 members were analyzed, and response elements related to light, environmental stress, plant hormones and metabolism were detected. The distribution and kind of cis-regulatory elements among GhNOT2_3_5 members from the same class were discriminatory. Because cis-regulatory DNA sequences of promoter region were the key factors for controlling the accurate spatiotemporal gene expression process, this was essential for plant development and response to external stimulus [39]. Various cis-regulatory elements in promoter regions promote the diversification of genes function and make plants more adaptable to environmental changes.
3.4. Expression Patterns and Putative Disease Resistant Functions of NOT2_3_5 Domain Protein Genes
Expression patterns of GhNOT2_3_5 genes in various tissues were studied and showed that all GhNOT2_3_5 genes were expressed in all tissues and fiber development stages, suggesting that they were involved in the whole cotton growth process and that NOT2_3_5 proteins were important constitutive components of the CCR4-NOT complex and played critical roles in transcription regulation. Similarly, expression profiles of GhNOT2_3_5 genes under abiotic and biotic stresses were investigated. It is well known that abiotic and biotic stresses including verticillium wilt, drought, salt and heat seriously restrict cotton production all over the world. However, few studies have focused on NOT2_3_5 domain proteins involved in plant response to abiotic and biotic stresses. In our study, some NOT2_3_5 domain proteins were differentially expressed under various stresses and might demonstrate that GhNOT2_3_5 genes were involved in responding to exogenous stimulus. Especially, some GbNOT2_3_5 genes (including GbNOT2_3_5-5/10, GbNOT2_3_5-3/8 and GbNOT2_3_5-4/9) were significantly up-regulated after infection of verticillium wilt. To further study the function of GbNOT2_3_5 genes in resistance to verticillium wilt, silencing of GbNOT2_3_5 genes using VIGS technology compromised resistance to V. dahliae in G. barbadense var Hai 7124. Moreover, the cis-regulatory elements analysis of promoter regions suggested that several NOT2_3_5 genes contained a TCA element (SA responsiveness) and were up-regulated under SA treatment. There is growing evidence that SA is involved in plant disease resistance and regulates expression of pathogenesis-related protein [40]. Thus, we speculate that GbNOT2_3_5-3/8 and GbNOT2_3_5-4/9 genes participated in the SA signal pathway and enhanced resistance to verticillium wilt in cotton.
4. Materials and Methods
4.1. Identification of the NOT2_3_5 Domain Protein Family
The genome sequences of four cotton species were downloaded from the CottonFGD website (https://cottonfgd.org/ (accessed on 14 November 2019)): G. arboretum (CRI), G. raimondii (JGI), G. hirsutum (NAU) and G. barbadense (NAU). [27]. The genome sequences of Arabidopsis, Brachypodium sylvaticum and rice were downloaded from the JGI website (https://phytozome.jgi.doe.gov/pz/portal.html (accessed on 14 November 2019)). The Pfam protein family databases with the NOT2_3_5 domain (PF04153) were used with HMMER software version 3.0 to identify the NOT2_3_5 domain protein in these four cotton and other plant species mentioned above (E < 10−10) [41]. The obtained protein sequences were further verified using the SMART database [42]. After selection, sequences with no NOT2_3_5 domain were deleted and the remaining NOT2_3_5 domain protein sequences were recognized as belonging to NOT2_3_5 domain protein family in cotton and other plant species. The ExPASy database (http://www.expasy.org/ (accessed on 8 December 2019)) was used to analyze the protein sequence length, molecular weight and isoelectric point (pI) [43]. The CELLO v2.5 server was applied in predicting the subcellular localization information [44].
4.2. Construction of Phylogenetic Tree and Chromosome Map
Clustal X software was used to perform the multiple sequence alignments of NOT2_3_5 domain proteins from four cotton and other plant species [45]. The phylogenetic tree was constructed by MEGA7 software using the neighbor-joining (NJ) method with bootstrap value set as 500 [46]. MapChart 2.2 software was used to analyze the respective chromosome distributions of genes encoding the NOT2_3_5 domain proteins in four cotton species [47].
4.3. Gene Collinearity and Structural Analysis
The protein sequences of four cotton species were aligned with each other using the Basic Local Alignment Search Tool (BLAST). The collinearity analysis was performed using MCscan software according to the above blast results. Collinear pairs in the NOT2_3_5 domain protein family were selected, and CIRCS software was used to draw the collinearity map [48]. Additionally, to analyze the divergence of NOT2_3_5 families, their coding sequences were aligned using MEGA6.0 software and the non-synonymous substitution rate (Ka) and synonymous substitution rate (Ks) values of the orthologous gene pairs were calculated using DnaSP5.0 software. The Ka/Ks ratio was applied to estimate the selection pressure for the duplication events of NOT2_3_5 families. The exon and intron positions were obtained by aligning the genomic DNA sequences and coding sequences using the Gene Structure Display Server (GSDS) online software. The Multiple Em for Motif Elicitation (MEME) software was used to analyze the protein sequences from four cotton species.
4.4. Target Prediction between MiRNAs and GhNOT2_3_5 Members
The miRNA sequences of upland cotton (G. hirsutum) were downloaded from the plant microRNA database. The transcript sequences of GhNOT2_3_5 families were uploaded to the psRNATarget server (2017 update) and predicted candidate target transcripts against G. hirsutum miRNA sequences [49]. The target prediction results of G. hirsutum miRNA and GhNOT2_3_5 families were displayed using Cytoscape software [50].
4.5. Prediction of Upstream cis-Elements
The 2.5-kb upstream genomic sequences from the start codon of each NOT2_3_5 gene were extracted from the genome files of the four cotton species. The above-2.5-kb promoter regions were submitted to the Plant CARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/ (accessed on 13 Feburary 2020)) to predict cis-elements of NOT2_3_5 gene promoters [51].
4.6. Expression Pattern Analysis
The expression patterns of GhNOT2_3_5 members in G. hirsutum var TM-1 in various tissues and under different stresses were analyzed using the RNA-seq data, which were downloaded from the CottonFGD website (https://cottonfgd.org/(accessed on 15 May 2020)) [27]. The heat maps were generated using the log2FPKM values by MeV software. Expression patterns of GhNOT2_3_5 members in G. hirsutum var TM-1 under exogenous hormone treatment and after infection of verticillium wilt were analyzed using quantitative real-time PCR (qPCR) data. Selected primer pairs were predicted to be specific to each gene pair based on the sequence alignment. Seedlings of G. hirsutum var TM-1 were grown in plant growth chambers at 25 °C during the day and 23 °C at night, 16/8 h light/dark photoperiod, and 70% relative humidity. For the exogenous hormone treatment, exogenous hormones: salicylic (SA), ethylene (ET) and methyl jasmonate (MeJA) were sprayed onto leaves at 1 μmol/mL when the cotton seedlings possessed two true leaves. Each time point represents RNA isolated from the leaves of 4 different plants harvested 0, 6, 12, 24, and 48 h after treatment (total of 20 plants/treatment) and then stored at −80 °C. For inoculation of verticillium wilt, roots of cotton seedlings with two true leaves were uprooted gently, dipped in the 10 mL of conidial suspensions of Verticillium dahliae (1 × 107 conidia/mL, Vd991) for 5 min and the plants replanted in the pots. Mixed roots, isolated from the roots of 4 different plants, were harvested 0, 6, 12 and 24 h after treatment and then stored at −80 °C. Total RNA of the above plant samples were isolated using RNAprep Pure Plant Plus Kit (Polysaccharides & Polyphenolics-rich) (TIANGEN, Beijing, China) according to the manufacturer’s instructions, and cDNA was synthesized using PrimeScriptTM II 1st strand cDNA Synthesis Kit (TaKaRa, Dalian, China). The qPCR was performed on QuantStudio TM 6 Flex Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA) using TB Green Premix Ex TaqTM (Tli RNaseH Plus), Bulk (TaKaRa, Dalian, China). Cotton actin gene was used as an internal control gene for normalization of expression values. All primers used in this study are listed in Table S5.
4.7. Vector Construction and Development of Virus-Induced Gene Silencing (VIGS) in Cotton
About 300 bp of GbNOT2_3_5-3/8 and GbNOT2_3_5-4/9 gene fragments were amplified by corresponding primers. The above fragments were inserted into pYL156 vector by ClonExpressTM II One Step Cloning Kit (Vazyme, Nanjing, China). Plasmids containing the pYL156–GbNOT2_3_5-3/8, pYL156–GbNOT2_3_5-4/9, pYL156–GhPDS, pYL156 and pYL192 were transformed into Agrobacterium tumefaciens GV3101 by the freeze–thaw method, respectively [52]. The A. tumefaciens containing recombinant plasmids were grown overnight at 28 °C and collected. The A. tumefaciens carrying pYL156–GbNOT2_3_5-3/8 and pYL156–GbNOT2_3_5-4/9 were respectively mixed with A. tumefaciens carrying pYL192 in a 1:1 ratio. Mixed A. tumefaciens were injected into two fully expanded cotyledons of cotton seedlings using sterile needleless injectors [52]. When the positive control plants appeared as albino phenotypes, expression level of above GbNOT2_3_5 gene were tested. Inoculation with V. dahliae was performed as described above until purposed GbNOT2_3_5 gene were silenced. VIGS experiments were performed with at least three biological repeats and for each repeat, there were more than 10 plants per constructed vector. Morbidity situation was investigated 25 d after inoculation with V.dahliae.
Acknowledgments
We would like to thank Zhen Chen at Henan Agricultural University for support of the data analysis in this work.
Abbreviations
| NOT | Negative on TATA-less |
| CCR4-NOT | Carbon Catabolite Repression 4-NOT |
| SA | Salicylic Acid |
| ET | Ethylene |
| MeJA | Methyl Jasmonate |
| VIGS | Virus-Induced Gene Silencing |
| pI | isoelectric point |
| MW | molecular weight |
| Dpa | Days Post Anthesis |
| TRV | Tobacco Rattle Virus |
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms22115634/s1, Figure: S1: Conserved domain-sequence and amino residues analysis of NOT2_3_5 domain proteins. A: Conserved domain-sequence analysis. NOT2_3_5 domain were located in the C-terminal. B: Conserved amino residues analysis. The conservation degrees of amino acid residue from each class of NOT2_3_5 domains were very high. Figure: S2 Chromosome Location of NOT2_3_5 genes from four cotton species. Abbreviated A2, D5, AD1, AD2 represented G. arboretum, G. raimondii, G. hirsutum and G. barbadense, respectively. Figure S3: Distribution of Ka/Ks ratios of NOT2_3_5 gene pairs in tetraploid cotton. A: Ka/Ks ratios between A and D sub-genomes of G. hirsutum. B: Ka/Ks ratios between G. hirsutum and G. barbadense. C: Ka/Ks ratios between A and D sub-genomes of G. barbadense. Circles represented outlier and triangles represented average value. Figure S4: The stems and leaves of silencing GbNOT2_3_5 genes plants showed more severe symptoms after infection of V. dahlia. A: The stems from silencing GbNOT2_3_5 genes plants and control. B: Trypan blue array of leaves from silencing GbNOT2_3_5 genes plants and control. Table S1: The detailed information of NOT2_3_5 domain protein family from four cotton species. Table S2: The top ten conserved motifs and the functional searches on InterProScan database. Table S3: The details information for the Ka/Ks ratio of duplicated genes pairs of NOT2_3_5 domain proteins. Table S4: Cis-elements from 10 putative GhNOT2_3_5 promoter regions. Table S5: Primer sequences used in the study.
Author Contributions
H.W. and Y.Z. conceived and designed the experiments, P.Z. and T.Q. performed the experiments and analyzed the data, P.Z. and T.Q. wrote the paper, T.Q. and Y.Z. revised the manuscript, W.C. and X.S. prepared the materials. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Natural Science Foundation of Henan Province of China (No. 202300410551) and the Central Public-interest Scientific Institution Basal Research Fund (No. 1610162021032).
Data Availability Statement
The genome sequences of plant species and RNA-seq data used in our manuscript were downloaded from the CottonFGD website (https://cottonfgd.org/).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gupta I., Villanyi Z., Kassem S., Hughes C., Panasenko O.O., Steinmetz L.M., Collart M.A. Translational capacity of a cell is determined during transcription elongation via the ccr4-not complex. Cell Rep. 2016;15:1782–1794. doi: 10.1016/j.celrep.2016.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collart M.A., Struhl K. Cdc39, an essential nuclear protein that negatively regulates transcription and differentially affects the constitutive and inducible his3 promoters. EMBO J. 1993;12:177–186. doi: 10.1002/j.1460-2075.1993.tb05643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collart M.A., Struhl K. Not1(cdc39), not2(cdc36), not3, and not4 encode a global-negative regulator of transcription that differentially affects tata-element utilization. Genes Dev. 1994;8:525–537. doi: 10.1101/gad.8.5.525. [DOI] [PubMed] [Google Scholar]
- 4.Collart M.A. The not, spt3, and mot1 genes functionally interact to regulate transcription at core promoters. Mol. Cell Biol. 1996;16:6668–6676. doi: 10.1128/MCB.16.12.6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collart M.A. The ccr4-not complex is a key regulator of eukaryotic gene expression. Wiley Interdiscip. Rev. RNA. 2016;7:438–454. doi: 10.1002/wrna.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collart M.A., Panasenko O.O. The ccr4-not complex: Architecture and structural insights. Subcell. Biochem. 2017;83:349–379. doi: 10.1007/978-3-319-46503-6_13. [DOI] [PubMed] [Google Scholar]
- 7.Bhaskar V., Roudko V., Basquin J., Sharma K., Urlaub H., Seraphin B., Conti E. Structure and rna-binding properties of the not1-not2-not5 module of the yeast ccr4-not complex. Nat. Struct. Mol. Biol. 2013;20:1281–1288. doi: 10.1038/nsmb.2686. [DOI] [PubMed] [Google Scholar]
- 8.Boland A., Chen Y., Raisch T., Jonas S., Kuzuoglu-Ozturk D., Wohlbold L., Weichenrieder O., Izaurralde E. Structure and assembly of the not module of the human ccr4-not complex. Nat. Struct. Mol. Biol. 2013;20:1289–1297. doi: 10.1038/nsmb.2681. [DOI] [PubMed] [Google Scholar]
- 9.Bawankar P., Loh B., Wohlbold L., Schmidt S., Izaurralde E. Not10 and c2orf29/not11 form a conserved module of the ccr4-not complex that docks onto the not1 n-terminal domain. RNA Biol. 2013;10:228–244. doi: 10.4161/rna.23018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mauxion F., Preve B., Seraphin B. C2orf29/cnot11 and cnot10 form a new module of the ccr4-not complex. RNA Biol. 2013;10:267–276. doi: 10.4161/rna.23065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bai Y., Salvadore C., Chiang Y.C., Collart M.A., Liu H.Y., Denis C.L. The ccr4 and caf1 proteins of the ccr4-not complex are physically and functionally separated from not2, not4, and not5. Mol. Cell Biol. 1999;19:6642–6651. doi: 10.1128/MCB.19.10.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collart M.A., Panasenko O.O., Nikolaev S.I. The not3/5 subunit of the ccr4-not complex: A central regulator of gene expression that integrates signals between the cytoplasm and the nucleus in eukaryotic cells. Cell Signal. 2013;25:743–751. doi: 10.1016/j.cellsig.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 13.Oberholzer U., Collart M.A. Characterization of not5 that encodes a new component of the not protein complex. Gene. 1998;207:61–69. doi: 10.1016/S0378-1119(97)00605-7. [DOI] [PubMed] [Google Scholar]
- 14.Russell P., Benson J.D., Denis C.L. Characterization of mutations in not2 indicates that it plays an important role in maintaining the integrity of the ccr4-not complex. J. Mol. Biol. 2002;322:27–39. doi: 10.1016/S0022-2836(02)00707-6. [DOI] [PubMed] [Google Scholar]
- 15.Kruk J.A., Dutta A., Fu J., Gilmour D.S., Reese J.C. The multifunctional ccr4-not complex directly promotes transcription elongation. Genes Dev. 2011;25:581–593. doi: 10.1101/gad.2020911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sohn E.J., Jung D.B., Lee J., Yoon S.W., Won G.H., Ko H.S., Kim S.H. Ccr4-not2 promotes the differentiation and lipogenesis of 3t3-l1 adipocytes via upregulation of pparxγ, cebpalpha and inhibition of p-gsk3alpha/beta and beta-catenin. Cell Physiol. Biochem. 2015;37:1881–1889. doi: 10.1159/000438549. [DOI] [PubMed] [Google Scholar]
- 17.Dai Y., Cao Z., Huang L., Liu S., Shen Z., Wang Y., Wang H., Zhang H., Li D., Song F. Ccr4-not complex subunit not2 plays critical roles in vegetative growth, conidiation and virulence in watermelon fusarium wilt pathogen fusarium oxysporum f. Sp. Niveum. Front. Microbiol. 2016;7:1449. doi: 10.3389/fmicb.2016.01449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anand A., Krichevsky A., Schornack S., Lahaye T., Tzfira T., Tang Y., Citovsky V., Mysore K.S. Arabidopsis vire2 interacting protein2 is required for agrobacterium t-DNA integration in plants. Plant Cell. 2007;19:1695–1708. doi: 10.1105/tpc.106.042903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L., Song X., Gu L., Li X., Cao S., Chu C., Cui X., Chen X., Cao X. Not2 proteins promote polymerase ii-dependent transcription and interact with multiple microrna biogenesis factors in arabidopsis. Plant Cell. 2013;25:715–727. doi: 10.1105/tpc.112.105882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao P., Wang K., Lin Z., Zhang W., Du L., Zhang Y., Ye X. Cloning and characterization of tavip2 gene from triticum aestivum and functional analysis in nicotiana tabacum. Sci. Rep. 2016;6:37602. doi: 10.1038/srep37602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paterson A.H., Wendel J.F., Gundlach H., Guo H., Jenkins J., Jin D., Llewellyn D., Showmaker K.C., Shu S., Udall J., et al. Repeated polyploidization of gossypium genomes and the evolution of spinnable cotton fibres. Nature. 2012;492:423–427. doi: 10.1038/nature11798. [DOI] [PubMed] [Google Scholar]
- 22.Lu P., Magwanga R.O., Kirungu J.N., Dong Q., Cai X., Zhou Z., Wang X., Xu Y., Hou Y., Peng R., et al. Genome-wide analysis of the cotton g-coupled receptor proteins (gpcr) and functional analysis of gtom1, a novel cotton gpcr gene under drought and cold stress. BMC Genom. 2019;20:651. doi: 10.1186/s12864-019-5972-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du X., Huang G., He S., Yang Z., Sun G., Ma X., Li N., Zhang X., Sun J., Liu M., et al. Resequencing of 243 diploid cotton accessions based on an updated a genome identifies the genetic basis of key agronomic traits. Nat. Genet. 2018;50:796–802. doi: 10.1038/s41588-018-0116-x. [DOI] [PubMed] [Google Scholar]
- 24.Wang M., Tu L., Yuan D., Zhu D., Shen C., Li J., Liu F., Pei L., Wang P., Zhao G., et al. Reference genome sequences of two cultivated allotetraploid cottons, gossypium hirsutum and gossypium barbadense. Nat. Genet. 2019;51:224–229. doi: 10.1038/s41588-018-0282-x. [DOI] [PubMed] [Google Scholar]
- 25.Li X., Zhang Y., Ding C., Xu W., Wang X. Temporal patterns of cotton fusarium and verticillium wilt in jiangsu coastal areas of china. Sci. Rep. 2017;7:12581. doi: 10.1038/s41598-017-12985-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J., Hu H.L., Wang X.N., Yang Y.H., Zhang C.J., Zhu H.Q., Shi L., Tang C.M., Zhao M.W. Dynamic infection of verticillium dahliae in upland cotton. Plant Biol. 2020;22:90–105. doi: 10.1111/plb.13037. [DOI] [PubMed] [Google Scholar]
- 27.Zhu T., Liang C., Meng Z., Sun G., Meng Z., Guo S., Zhang R. Cottonfgd: An integrated functional genomics database for cotton. BMC Plant Biol. 2017;17:101. doi: 10.1186/s12870-017-1039-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rong J., Feltus F.A., Liu L., Lin L., Paterson A.H. Gene copy number evolution during tetraploid cotton radiation. Heredity. 2010;105:463–472. doi: 10.1038/hdy.2009.192. [DOI] [PubMed] [Google Scholar]
- 29.Adams K.L., Wendel J.F. Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 2005;8:135–141. doi: 10.1016/j.pbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 30.Wendel J.E., Cronn R.C. Polyploidy and the evolutionary history of cotton. Adv. Agron. 2003;78:139–186. [Google Scholar]
- 31.Fang L., Guan X.Y., Zhang T.Z. Asymmetric evolution and domestication in allotetraploid cotton (Gossypium hirsutum L.) Crop J. 2017;5:159–165. doi: 10.1016/j.cj.2016.07.001. [DOI] [Google Scholar]
- 32.Zhang T.Z., Hu Y., Jiang W.K., Fang L., Guan X.Y., Chen J.D., Zhang J.B., Saski C.A., Scheffler B.E., Stelly D.M., et al. Sequencing of allotetraploid cotton (gossypium hirsutum l. Acc. Tm-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015;33:531–537. doi: 10.1038/nbt.3207. [DOI] [PubMed] [Google Scholar]
- 33.Xie Z., Johansen L.K., Gustafson A.M., Kasschau K.D., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C. Genetic and functional diversification of small rna pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Macovei A., Tuteja N. Micrornas targeting dead-box helicases are involved in salinity stress response in rice (Oryza sativa L.) BMC Plant Biol. 2012;12:183. doi: 10.1186/1471-2229-12-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He F., Xu C., Fu X., Shen Y., Guo L., Leng M., Luo K. The microrna390/trans-acting short interfering rna3 module mediates lateral root growth under salt stress via the auxin pathway. Plant Physiol. 2018;177:775–791. doi: 10.1104/pp.17.01559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boccara M., Sarazin A., Thiebeauld O., Jay F., Voinnet O., Navarro L., Colot V. The arabidopsis mir472-rdr6 silencing pathway modulates pamp- and effector-triggered immunity through the post-transcriptional control of disease resistance genes. PLoS Pathog. 2014;10:e1003883. doi: 10.1371/journal.ppat.1003883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feyissa B.A., Arshad M., Gruber M.Y., Kohalmi S.E., Hannoufa A. The interplay between mir156/spl13 and dfr/wd40-1 regulate drought tolerance in alfalfa. BMC Plant Biol. 2019;19:434. doi: 10.1186/s12870-019-2059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen J., Pan A., He S., Su P., Yuan X., Zhu S., Liu Z. Different microrna families involved in regulating high temperature stress response during cotton (Gossypium hirsutum L.) anther development. Int. J. Mol. Sci. 2020;21:1280. doi: 10.3390/ijms21041280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Franco-Zorrilla J.M., Solano R. Identification of plant transcription factor target sequences. Biochim. Biophys. Acta Gene Regul. Mech. 2017;1860:21–30. doi: 10.1016/j.bbagrm.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 40.Yang Y.X., Ahammed G.J., Wu C., Fan S.Y., Zhou Y.H. Crosstalk among jasmonate, salicylate and ethylene signaling pathways in plant disease and immune responses. Curr. Protein. Pept. Sci. 2015;16:450–461. doi: 10.2174/1389203716666150330141638. [DOI] [PubMed] [Google Scholar]
- 41.Finn R.D., Bateman A., Clements J., Coggill P., Eberhardt R.Y., Eddy S.R., Heger A., Hetherington K., Holm L., Mistry J., et al. Pfam: The protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Letunic I., Doerks T., Bork P. Smart 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Artimo P., Jonnalagedda M., Arnold K., Baratin D., Csardi G., de Castro E., Duvaud S., Flegel V., Fortier A., Gasteiger E., et al. Expasy: Sib bioinformatics resource portal. Nucleic Acids Res. 2012;40:W597–W603. doi: 10.1093/nar/gks400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu C.S., Lin C.J., Hwang J.K. Predicting subcellular localization of proteins for gram-negative bacteria by support vector machines based on n-peptide compositions. Protein. Sci. 2004;13:1402–1406. doi: 10.1110/ps.03479604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. Clustal w and clustal x version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 46.Kumar S., Stecher G., Tamura K. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Voorrips R.E. Mapchart: Software for the graphical presentation of linkage maps and qtls. J. Hered. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- 48.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dai X., Zhuang Z., Zhao P.X. Psrnatarget: A plant small rna target analysis server (2017 release) Nucleic Acids Res. 2018;46:W49–W54. doi: 10.1093/nar/gky316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lescot M., Dehais P., Thijs G., Marchal K., Moreau Y., Van de Peer Y., Rouze P., Rombauts S. Plantcare, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao X., Shan L. Functional genomic analysis of cotton genes with agrobacterium-mediated virus-induced gene silencing. Methods Mol. Biol. 2013;975:157–165. doi: 10.1007/978-1-62703-278-0_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome sequences of plant species and RNA-seq data used in our manuscript were downloaded from the CottonFGD website (https://cottonfgd.org/).