Abstract
The aim of this study was exploration of the genetic background of conjunctival melanoma (CM) and correlation with recurrent and metastatic disease. Twenty-eight CM from the Rotterdam Ocular Melanoma Study group were collected and DNA was isolated from the formalin-fixed paraffin embedded tissue. Targeted next-generation sequencing was performed using a panel covering GNAQ, GNA11, EIF1AX, BAP1, BRAF, NRAS, c-KIT, PTEN, SF3B1, and TERT genes. Recurrences and metastasis were present in eight (29%) and nine (32%) CM cases, respectively. TERT promoter mutations were most common (54%), but BRAF (46%), NRAS (21%), BAP1 (18%), PTEN (14%), c-KIT (7%), and SF3B1 (4%) mutations were also observed. No mutations in GNAQ, GNA11, and EIF1AX were found. None of the mutations was significantly associated with recurrent disease. Presence of a TERT promoter mutation was associated with metastatic disease (p-value = 0.008). Based on our molecular findings, CM comprises a separate entity within melanoma, although there are overlapping molecular features with uveal melanoma, such as the presence of BAP1 and SF3B1 mutations. This warrants careful interpretation of molecular data, in the light of clinical findings. About three quarter of CM contain drug-targetable mutations, and TERT promoter mutations are correlated to metastatic disease in CM.
Keywords: conjunctiva, melanoma, molecular medicine, prognosis, TERT promotor mutation
1. Introduction
Conjunctival melanoma (CM) comprises 5–10% of all ocular melanoma [1,2,3]. The majority derives from primary acquired melanosis with atypia (PAM), but infrequently, CM develops from a pre-existing nevus or de novo [1,3,4,5,6]. CM has an incidence of 0.2–0.8 per million [3,6,7], with an increasing trend [3,8]. The 5- and 10-years cumulative incidence of CM-related mortality is 17–31% and 22–59%, respectively [5,7,9,10,11]. The prognosis of ocular melanoma, including CM and uveal melanoma (UM), depends on clinical and histopathological features, as well as the molecular genetic make-up [3,12,13]. During the past decade, the molecular make-up of UM has been well-characterized, with UM harboring recurrent mutations in guanine-nucleotide-binding protein-Q (GNAQ), guanine-nucleotide-binding protein-alpha 11 (GNA11), BRCA-associated protein 1 (BAP1), splicing factor 3 subunit 1 (SF3B1), and eukaryotic translation initiation factor 1A (EIF1AX). BAP1 and SF3B1 mutations are associated with the development of metastasis in UM. After the diagnosis of metastatic disease, patients with UM have a survival between 2–9 months [12]. When CM has metastasized, there are also very limited treatment options [1,13]. Yet, although CM as well as UM are ocular melanoma, CM certainly do show overlapping features, including molecular abnormalities with cutaneous melanoma [1,3,6,13,14]. For example, in 25–40% of the CM driver v-raf murine sarcoma, viral oncogene homolog B1 (BRAF) V600E/K mutations are described [1,2,6,13,15]. This incidence is higher as compared to other mucosal melanoma, which harbor a BRAF mutation in only 12% of cases. Although a correlation between BRAF mutations and poor prognostic factors has been described in cutaneous melanoma, no predictive value is yet reported for mucosal melanoma [16,17]. Other genes in which mutations have been identified in CM are the neuroblastoma RAS viral oncogene homolog (NRAS), Kirsten RAS oncogene homolog (KRAS), neurofibromin 1 (NF1), telomerase reverse transcriptase (TERT), tyrosine protein kinase (c-KIT), TP53, and BAP1 [3,6,15,18]. Mutations in GNAQ/GNA11 have also been described, but these are not the known activating hotspot mutations at amino acid Q209 or R183, which occur in UM [15,19]. The genetic background of the melanoma originating from these different locations, emphasizes the differences between UM and CM, and the similarities between CM and cutaneous melanoma. Furthermore, in contrast to UM, some of the mutations frequently found in CM are amenable to targeted therapies. However, the prognostic value of these molecular abnormalities in CM is largely unclear. The aim of this study was to further elucidate the genetic background of CM within the spectrum of melanoma and to correlate these findings with the development of recurrences and metastasis.
2. Results
2.1. Clinical and Histopathological Characteristics
Clinical and histopathological characteristics are listed in Table 1. Based on the availability of sufficient formalin-fixed paraffin-embedded (FFPE) tissue for DNA isolation, twenty-eight cases could be included. Gender was equally divided with 50% males and 50% females. The median age at the time of diagnosis was 64 years (range 16–89 years). Based on the clinical information, most tumors were (at least partly) located on the bulbar conjunctiva (16 cases, 57%) with involvement of the palpebral conjunctiva in 10 cases (36%), the fornix in 5 cases (18%), and the caruncle in 1 case (4%). The tumors had a median diameter of 0.7 cm (range 0.05–1.8 cm), with a median tumor thickness of 3.0 mm (range 0.18–7.70 mm). According to the Eighth Edition of the American Joint Committee on Cancer (AJCC) Cancer Staging [20], twelve cases (43%) were pathological tumor (pT) stage pT1, including six pT1a cases (21%) and five pT1b cases (18%), and thirteen cases were pT2 cases (46%), comprising one pT2a case (4%), eleven pT2b cases (39%), and two cases (7%) with unknown tumor thickness. In three cases (11%), the pT status was unknown. In eighteen cases (64%), the melanoma were derived from PAM, four melanoma (14%) developed from a nevus, and three melanoma (11%) were de novo lesions. In three cases (11%), the origin could not be reliably determined, based on the pathology reports and the available clinical information.
Table 1.
Clinical and histopathological characteristics of the included conjunctival melanoma (CM).
| Clinical Characteristics | |
|---|---|
| Median age at diagnosis (years) | 63 (16–89) |
| Gender | |
| Male | 14 (50%) |
| Female | 14 (50%) |
| Location | |
| Bulbar | 16 (57%) |
| Palpebral | 10 (36%) |
| Fornix | 5 (18%) |
| Caruncle | 1 (4%) |
| Metastasis | |
| No | 19 (68%) |
| Yes | 9 (32%) |
| Local recurrence | |
| No | 20 (71%) |
| Yes | 8 (29%) |
| Histopathological Characteristics | |
| Median diameter (cm) | 0.7 (0.05–1.8) |
| Median tumor thickness (mm) | 3.0 (0.18–7.70) |
| pT status | |
| pT1a | 6 (21%) |
| pT1b | 5 (18%) |
| pT2a | 1 (4%) |
| pT2b | 11 (39%) |
| pTx | 5 (18%) |
| Origin | |
| PAM | 18 (64%) |
| Nevus | 4 (14%) |
| De novo | 3 (11%) |
| Unknown | 3 (11%) |
Clinical and histopathological characteristics of the included conjunctival melanoma (CM) cases. PAM = primary acquired melanosis with atypia. pT status = pathological tumor status.
Local recurrent disease occurred in eight patients (29%), between 6.8–156.8 months (median 29.3 months) after treatment. Nine patients (32%) developed metastatic disease between 1.7–49.2 months (median 14.3 months). Metastatic sites included lymph nodes (solitary or within the parotid gland) in all patients (n = 9), with metastatic disease in the orbit (n = 1), thyroid (n = 1), breast (n = 1), lung (n = 1), brain (n = 1), and spleen (n = 1). The thyroid and breast metastases were present in one patient, and the orbit and brain metastases were identified in one patient as well. The spleen and brain metastases were not histologically confirmed. The mean overall survival was 77.4 months (range 3.85–257.2 months), with a median of 62.8 months.
2.2. Mutation Analysis
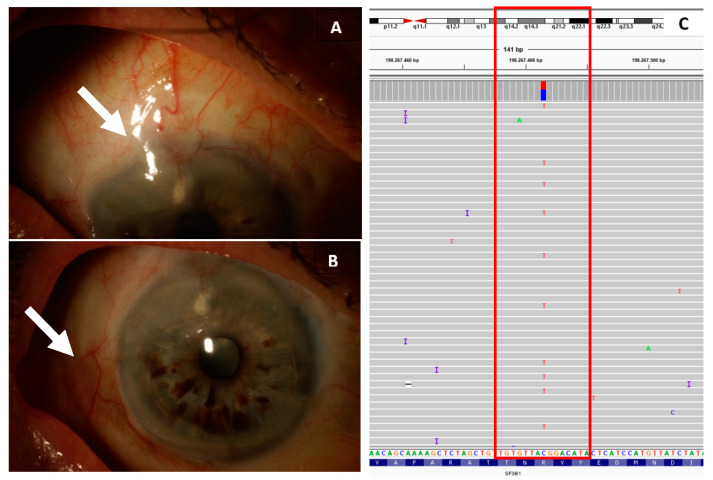
The specific mutations found per case are listed in Supplementary Table S1, with a summary of the mutations including correlation with metastatic and recurrent disease in Table 2. Fifteen CM cases (54%) showed a TERT promoter mutation. A mutation in the BRAF gene was identified in thirteen CM (46%), mostly affecting amino acid V600. NRAS mutations were seen in six cases (21%) and mutations in BAP1 were identified in five CM (18%). A PTEN mutation was found in four CM (14%), and in two CM (7%), a mutation in c-KIT was identified. Interestingly, a p.Arg625His mutation in SF3B1 was detected in one CM (4%). The diagnosis was unequivocally a CM in terms of both clinical and pathological reports. It was located in the nasal superior in the bulbar conjunctiva (Figure 1). None of the CM cases carried a mutation of GNAQ, GNA11, or EIF1AX.
Table 2.
Presence of a mutation versus metastasis-free survival (MFS) and recurrence-free survival (RFS).
| Gene | Presence of a Mutation | n (%) | Metastasis n (%) | MFS p-Value |
Recurrences n (%) | RFS p-Value |
|---|---|---|---|---|---|---|
| SF3B1 | 0.45 | 0.45 | ||||
| Yes | 1 (4) | 0 (0) | 0 (0) | |||
| No | 27 (96) | 9 (33) | 8 (30) | |||
| BAP1 | 0.46 | 0.69 | ||||
| Yes | 5 (18) | 1 (20) | 2 (40) | |||
| No | 23 (82) | 8 (35) | 6 (26) | |||
| TERT | 0.008 | 0.20 | ||||
| Yes | 15 (54) | 7 (47) | 2 (13) | |||
| No | 13 (46) | 2 (15) | 6 (46) | |||
| NRAS | 0.17 | 0.82 | ||||
| Yes | 6 (21) | 4 (67) | 2 (33) | |||
| No | 22 (79) | 5 (23) | 6 (27) | |||
| KIT | 0.26 | 0.88 | ||||
| Yes | 2 (7) | 0 (0) | 1 (50) | |||
| No | 26 (93) | 9 (35) | 7 (28) | |||
| PTEN | 0.53 | 0.25 | ||||
| Yes | 4 (14) | 1 (25) | 2 (50) | |||
| No | 24 (86) | 8 (33) | 6 (25) | |||
| BRAF | 0.052 | 0.76 | ||||
| Yes | 13 (46) | 5 (38) | 2 (15) | |||
| No | 15 (54) | 4 (27) | 6 (40) |
The total number of included conjunctival melanoma cases was twenty-eight. This table depicts the percentages of the specific mutations in the cohort, as well as the development of metastatic disease and recurrent disease within the group of a specific mutation. The statistically significant p-value is depicted in bold. MFS = metastasis-free survival. RFS = recurrence-free survival.
Figure 1.
Clinical pictures and molecular data concerning the conjunctival melanoma harboring a SF3B1 mutation. In (A) the macroscopic view of the melanoma located on the bulbar conjunctiva, within (B) the primary acquired melanomsis with atypia component (white arrow). Depicted in the red box in (C) is the molecular data concerning a p.Arg625His mutation in SF3B1, with an allele frequency of 42%, using the Integrative Genomics Viewer.
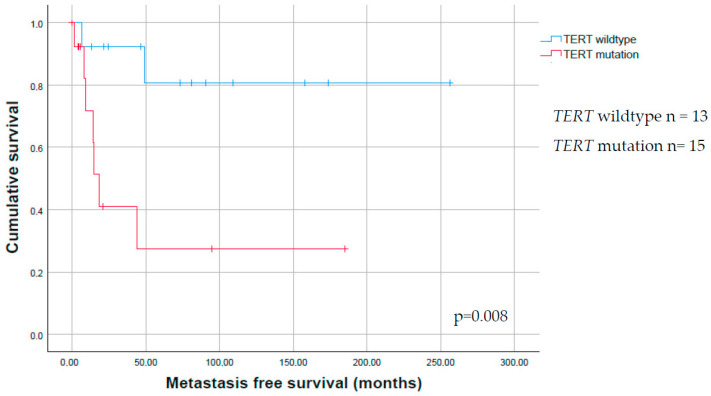
The metastasis-free survival (MFS) of patients with a TERT promoter mutation was significantly shorter as compared to patients without a TERT promoter mutation in the tumor (p = 0.008, Table 2, Figure 2). No correlation between metastasis-free survival and mutation status of BRAF, BAP1, SF3B1, NRAS, c-KIT, and PTEN could be observed.
Figure 2.
Kaplan–Meier survival estimate for the presence of a TERT promoter mutation in conjunctival melanoma. Kaplan–Meier survival estimate for the time to metastasis of conjunctival melanoma (CM), showing that patients with a CM with a TERT promoter mutation are more likely to develop metastatic disease.
No correlation was found between the presence of any mutations and the development of recurrences (Table 2). We also analyzed whether the mutations were correlated with sex, age, location (bulbar only versus involvement of the palpebral/caruncular/forniceal conjunctiva), pT status (pT1 versus pT2), tumor thickness, origin (PAM-derived melanoma versus non-PAM-derived melanoma). We did find an association between the presence of a TERT promoter mutation and the origin of the lesion (p-value = 0.005), with most cases (54%) developing either de novo or from a melanocytic nevus (Table 3).
Table 3.
Mutations versus clinical and histopathological parameters.
|
TERT n = 15 (%) |
P |
BRAF n = 13 (%) |
P |
BAP1 n = 5 (%) |
P |
NRAS n = 6 (%) |
p |
PTEN n = 4 (%) |
P |
c-KIT n= 2 (%) |
P |
SF3B1 n = 1 (%) |
P | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gender | 0.26 | 0.71 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | ||||||||
| Male | 6 (40) | 6 (46) | 3 (60) | 3 (50) | 3 (75) | 1 (50) | 1 (100) | ||||||||
| Female | 9 (60) | 7 (54) | 2 (40) | 3 (50) | 1 (25) | 1 (50) | 0 (0) | ||||||||
| Age | 0.91 | 0.91 | 0.52 | 0.32 | 0.92 | 0.24 | 0.50 | ||||||||
| <50y | 2 (13) | 2 (15) | 0 (0) | 2 (33) | 0 (0) | 0 (0) | 0 (0) | ||||||||
| 50–65y | 7 (47) | 6 (46) | 3 (60) | 2 (33) | 2 (50) | 2 (100) | 1 (100) | ||||||||
| >65y | 6 (40) | 5 (38) | 2 (40) | 2 (33) | 2 (50) | 0 (0) | 0 (0) | ||||||||
| Location | 0.16 | 0.85 | 1.00 | 1.00 | 0.59 | 1.00 | 0.48 | ||||||||
| Bulbar | 8 (53) | 6 (46) | 2 (40) | 2 (33) | 1 (25) | 1 (50) | 1 (100) | ||||||||
| Forniceal/ palpebral/ caruncular involvement |
5 (33) | 6 (46) | 2 (40) | 2 (33) | 2 (50) | 1 (50) | 0 (0) | ||||||||
| Tumor thick-ness | 0.67 | 0.68 | 1.00 | 0.63 | 0.56 | 0.53 | 0.31 | ||||||||
| Tumor thickness ≤2 mm | 5 (33) | 3 (23) | 1 (20) | 2 (33) | 2 (50) | 1 (50) | 1 (100) | ||||||||
| Tumor thickness >2mm | 8 (53) | 9 (69) | 4 (80) | 3 (50) | 2 (50) | 1 (50) | 0 (0) | ||||||||
| pT status | 0.16 | 0.85 | 1.00 | 1.00 | 0.59 | 1.00 | 0.48 | ||||||||
| pT1 | 8 (53) | 6 (46) | 2 (40) | 2 (33) | 1 (25) | 1 (50) | 1 (100) | ||||||||
| pT2 | 5 (33) | 6 (46) | 2 (40) | 2 (33) | 2 (50) | 1 (50) | 0 (0) | ||||||||
| Origin | 0.01 | 1.00 | 1.00 | 0.30 | 1.00 | 1.00 | 1.00 | ||||||||
| PAM | 6 (40) | 7 (54) | 3 (60) | 3 (50) | 3 (80) | 2 (100) | 1 (100) | ||||||||
| Non PAM (nevus/de novo) | 7 (47) | 3 (23) | 1 (20) | 3 (50) | 1 (25) | 0 (0) | 0 (0) |
P = p-value calculated with either the Pearson’s χ2 test or Fisher’s exact test. In bold, the association between the presence of a TERT promoter mutation and origin of the lesion (p-value = 0.01), with most cases (54%) developing either de novo or from a melanocytic nevus. None of the cases showed GNAQ, GNA11, or EIF1AX mutations; therefore, these mutations are not included in the table. pT status = pathological tumor status.
2.3. Immunohistochemistry
In five CM cases that revealed a BAP1 mutation using molecular testing, there was enough material available for testing the presence of a BAP1 mutation using immunohistochemistry. Four of these cases did not show loss of expression of BAP1 using immunohistochemistry, while one CM case did show loss of expression using BAP1 immunohistochemistry, with presence of positive (internal) control tissue.
3. Discussion
Pathways involved in the pathogenesis of CM included the MAPK/ERK pathway and the PI3K/AKT pathways, and these pathways overlap with the pathways involved in cutaneous melanoma [6].
The mutation that we found most frequent in CM is a TERT promoter mutation, congruent with other studies concerning ocular melanoma [6,13,14] and cancer originating from other sites. These mutations result in a new consensus binding site for E-twenty-six (ETS) transcription factors and this may contribute to increased TERT. The ETS transcription factors are downstream targets of the RAS-RAF-MAPK pathways, and TERT promoter mutations are suggested to have synergistic effects with activating BRAF or NRAS mutations to promote tumor cell proliferation [21]. TERT is involved in the AKT pathway, and plays an important role in cellular immortality [6]. TERT mRNA overexpression does not completely explain all effects of the TERT promoter mutations in tumorigenesis, and the role of immunohistochemistry in determining the TERT status is still a topic of debate [22]. Consequently, other undefined or epigenetic mechanisms of TERT-upregulating are expected to exist [21,23,24]. While a TERT promoter mutation is not found in conjunctival nevi, it is found in both PAM [14] and CM [6,14], with increased TERT expression leading to tumor progression [6]. In this context, the C>T or CC>TT nucleotide changes in these mutations are of interest, since this is the typical UV signature, in line with the UV-exposed location of most CM, as seen in our study and as compared to the molecular make up of cutaneous melanoma [6]. UM usually do not harbor mutations in or near the TERT gene [14,18,25]. It indicates that different pathways are involved in the development of CM and UM, as is also suggested by the differences in the presence of mutations in BRAF, NRAS, and GNAQ/GNA11. Since TERT promoter mutations are relatively common in CM, these mutations are of special interest with respect to clinical consequences. We did not find a correlation between the presence of any of the investigated mutations in this study and the well-known adverse histopathological parameters, as has been described for cutaneous melanoma, such as increasing tumor thickness and more advanced pT stage [26]. Previous studies reported an association between PAM with atypia and PAM-derived melanoma, with the presence of a TERT promoter mutation [13,14]. Remarkably, in the current study, we found a significant association with the presence of a TERT promoter mutation and non PAM-derived melanoma. This difference needs to be clarified by testing larger cohorts. The presence of a TERT promoter mutation in the tumor could have important clinical consequences, including the correlation of mutation status of this gene and follow-up. We found a correlation between the presence of a TERT promoter mutation and MFS, with a lower MFS in patients with a CM with a TERT promoter mutation, congruent with the findings in our previous study [13]. TERT promoter mutations have also been described as an independent prognostic factor in cutaneous melanoma. From this perspective, it is important to mention that most lesions in our cohort concerned relatively large tumors located at prognostic adverse locations (palpebra, fornix, or caruncle) [6], suggesting a bias. Patients with a TERT-promoter-mutated CM might benefit from an intensified follow-up program.
In addition to TERT promoter mutations, CM frequently harbors BRAF mutations, which are known to activate the downstream kinases MEK1/2 and ERK1/2, resulting in tumor proliferation [1,6]. In this study, we identified BRAF mutations in almost half the cases, almost all resulting in V600E mutations. This is in line with the literature in which 30–40% of all CM harbor mutations in BRAF, almost all being V600E mutations [3,6,13,27,28]. These mutations, and specifically the V600E mutation, are also present in about half of all patients with cutaneous melanoma [29], whereas this mutation is not frequently involved in other mucosal melanoma or UM [6].
In cutaneous melanoma, the presence of a TERT promoter mutation in addition to a BRAF mutation is associated with unfavorable clinicopathological characteristics, such as large tumor thickness and a high mitotic rate [26]. Unfortunately, the number of cases in the current cohort was too small to render any conclusions concerning these correlations in CM.
Determining the mutation status of the tumor could be useful with regards to therapeutic consequences, since several studies have shown an improved progression-free survival and overall survival, in patients with metastasized cutaneous melanoma harboring a BRAF mutation, using BRAF inhibitors [30]. BRAF mutations are also attractive as a target for adjuvant therapy in CM [6,31,32,33].
NRAS mutations are described in 27% of cutaneous melanoma, with a Q61K mutation as the most common mutation followed by Q61R [34]. NRAS-mutated cutaneous melanoma have an unfavorable prognosis as compared to BRAF mutated or wild-type melanoma [34]. We identified NRAS mutations in 21% of all CM in our cohort, which is in line with the 17% previously reported [15] and is somewhat lower compared to other literature [6]. Due to the small numbers of NRAS-mutated cases in our cohort, no correlations to prognosis could be determined. NRAS mutations are mutually exclusive with BRAF mutations [6]. NRAS mutations are amenable to MEK inhibitor therapy, as has been shown for cutaneous melanoma [35]. MEK inhibitors reduce the growth of NRAS mutant CM cell lines [1]. As yet, no cases of NRAS-mutated metastatic melanoma treated with MEK inhibitors have been published.
Interestingly, we detected an SF3B1 mutation at the hotspot R625, which is well-known in UM [3,28], and was reported in one CM case. The presence of a SF3B1 mutation was reported previously in CM, however, this concerned a p.C1123Y mutation and not a hotspot mutation [36], and another study reported a missense mutation [15]. Although R625 SF3B1 mutations are very rare in most melanoma, they have been identified in UM, including iris melanoma [19], and are less frequent in cutaneous melanoma as well as in vulvovaginal mucosal melanoma [36,37,38,39]. The occurrence of SF3B1 mutations in mucosal melanoma other than CM is higher, with a prevalence of 42% and hotspot mutations in 30–37% [39,40]. The clinical significance of this mutation in CM is unknown, whereas in UM, SF3B1 mutation is correlated to late metastatic disease [41]. The CM with this mutation was treated with excision. This case also included PAM and showed local recurrence, three and eight years after primary treatment. No metastasis developed in the follow-up period of 6.8 years. However, metastasis in SF3B1-mutated UM was described even after 10 years [41].
The CM cases in our cohort also harbored mutations in c-KIT, PTEN, and BAP1. These findings of mutations in c-KIT, NRAS, and PTEN are congruent with other literature [1,6], with c-KIT mutations reported in 39% of mucosal melanoma and being feasible for targeted therapy [42]. Of interest is the finding of mutations in BAP1, which is a common hemizygous mutation in UM [12,43]. BAP1 is a tumor suppressor gene and individuals with cutaneous melanocytic neoplasm with a germline BAP1 mutation, often have BRAF mutations, with these lesions reported to have a benign clinical course [43]. However, UM with somatic BAP1 mutations are correlated to loss of chromosome 3 and early metastatic disease. CM has also been described in a patient with the BAP1 tumor predisposition syndrome [44]. We identified heterozygous BAP1 mutations that can be explained as passenger mutations without consequences, due to expression of the remaining non-affected allele.
The genetic profile of CM differs from UM, another subtype of ocular melanoma, in which mutations in GNAQ/GNA11 are frequently described [45]. In this study, none of the CM harbored an activating hotspot mutation in GNAQ or GNA11. These findings are congruent with other studies analyzing mutations in CM [15,46]. BRAF and NRAS mutations are extremely rare in UM [37]. Therefore, these mutations can be useful in distinguishing CM from UM. This may be of interest in the identification of the primary tumor site in the case of metastatic melanoma with unknown primary. It also warrants the need for exploration of the genetic background of metastatic melanocytic lesions. However, such molecular results need to be interpreted with care, since we describe BAP1 and SF3B1 mutations in CM in the current cohort.
We did not find a correlation concerning the presence of any of the mutations and the development of recurrent disease. Cases with recurrent disease harbored the most frequently found mutations only in a (very) low number of cases. This may imply that recurrence and metastasis relate to different molecular or physical processes.
In conclusion, based on our molecular findings, CM comprises a separate entity within the ocular melanoma group, although there certainly are overlapping molecular features with UM, such as the presence of BAP1 and SF3B1 mutations. This warrants careful interpretation of molecular data in the light of clinical findings. About three-quarter of CM contain drug-targetable mutations in BRAF, NRAS, or c-KIT, supporting the relevance of molecular genetic testing in CM for therapeutic reasons. Within this study, we confirmed that TERT promoter mutations are frequently found in CM and are correlated to metastatic disease, supporting the relevance of molecular genetic testing for prognostic reasons.
4. Materials and Methods
4.1. Material Selection
We collected twenty-eight CM, diagnosed between 1987 and 2016 at the Erasmus MC—University Medical Center (Rotterdam, The Netherlands) and The Rotterdam Eye Hospital (Rotterdam, The Netherlands). Ethics Committee approval was obtained by the Medical Ethics Committee, Erasmus MC-University Medical Center, Rotterdam, The Netherlands (4 October 2018) and was registered with reference 67865. The study was performed according to the tenets of the Declaration of Helsinki. Samples were included when sufficient FFPE material was available for testing. Data regarding gender, age at the time of diagnosis, location, tumor thickness, the origin of the lesion, and information of development of recurrences and metastasis were collected from the patient records and information was obtained from the pathology reports and the nationwide-pathology network and registry system (Pathologisch-Anatomisch Landelijk Geautomatiseerd Archief). Recurrence was defined as histopathological proven CM at the same location, either after complete excision of the primary lesion or a tumor-free mapping biopsy, after a first incomplete excision of the primary tumor. Recurrence-free survival was defined as the time from the primary treatment to the date of recurrence or last date of follow-up. Metastasis-free survival was defined as time from the primary treatment to the date of metastatic disease or last date of follow-up.
4.2. DNA Isolation
DNA from FFPE tissue was isolated using lysis buffer (Promega, Madison, WI, USA) and 5% Chelex (Bio-Rad, Hercules, CA, USA), as described previously [27] and stored at −20 °C. DNA concentrations were measured with the Quant-iT™ PicoGreen™ ds DNA Assay Kit (Thermo Fisher Scientific, Inc., Waltham, MA, USA).
4.3. Targeted Next-Generation Sequencing
The Ion Personal Genome Machine and Torrent Server (Thermo Fisher Scientific, Waltham, MA, USA) was used for targeted next-generation sequencing (NGS), according to the manufacturer’s protocol. An input of DNA was used depending on the available amount of DNA. An extended gene panel covering GNAQ, GNA11, EIF1AX, SF3B1, BAP1, BRAF, NRAS, c-KIT, PTEN, and TERT was used, as described previously [27].
4.4. Mutation Analysis
Mutation analysis was performed independently by an ophthalmology resident (NvP) and a fellow in ophthalmic pathology (JvI), trained in the evaluation of NGS data. All data were analyzed manually using Integrative Genomics Viewer (IGV) Version 2.3.68 (97) (Broad Institute, Cambridge, MA). Furthermore, an automatic filtering of the variant calling files (vcf) was done according to the following criteria—inclusion of the hotspots at GNAQ/GNA11 (R183 and Q209) and SF3B1 (R625), and other variants meeting the following criteria—coverage of at least 50 reads and an allele frequency of at least 10%. Single nucleotide pleomorphisms (SNP’s), synonymous, intergenic, and intronic variants were excluded, but intronic variants with possible splice effects were scored. Subsequently, the filtered mutations were verified using IGV (Broad Institute, Cambridge, MA, USA), and compared to the mutations that were detected manually.
4.5. Immunohistochemistry
The presence of a mutation in the BAP1 gene was also evaluated using BAP1 immunohistochemistry, clone sc-28383, 1:50 dilution (Santa Cruz Biotechnology, Dallas, TX, USA). The samples were scored through masked screening, by an experienced ophthalmic pathologist (RVE).
4.6. Survival Analysis
All statistical analysis was performed using IBM SPSS Statistics Version 25 (IBM, Armonk, NY, USA). Kaplan Meier estimates were used to compare survival between groups. Log-rank test was used to test the null hypothesis that there was no difference in survival. A p-value < 0.05 was considered to be statistically significant. For the purpose of analyzing age related to the mutation, age was categorized into three groups: <50 years, 50–65 years, >65 years, analogous to other literature [28]. Fisher’s exact test was used to analyze whether a specific mutation was correlated with a specific clinical or histopathological parameter.
Acknowledgments
We would like to thank R. van Marion for his help performing next-generation sequencing.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms22115784/s1. Supplementary Table S1: Overview of mutations detected in conjunctival melanoma.
Author Contributions
Conceptualization, R.M.V., E.K. and A.d.K.; methodology, R.M.V., E.K., A.d.K., J.A.v.I., H.J.D. and N.M.v.P.; software, B.E. and T.B.; validation, J.A.v.I., N.M.v.P. and F.M.; formal analysis, J.A.v.I. and N.M.v.P.; investigation, J.A.v.I., N.M.v.P., J.V. and Q.v.d.B.; resources D.P., N.N., H.J.D. and E.K.; data curation, T.B., E.B., R.M.V., E.K., A.d.K., J.A.v.I. and N.M.v.P.; writing original draft preparation, J.A.v.I. and N.M.v.P.; writing review and editing, R.M.V., E.K., A.d.K., D.P., N.N., J.V., F.M., T.B., B.E., E.B. and Q.v.d.B., project administration, R.M.V., A.d.K. and E.K., funding acquisition, R.M.V. and E.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Stichting Nederlands Oogheelkundig Onderzoek (SNOO). Grant no: 2013-2021, Stichting Wetenschappelijk Oogheelkundig Onderzoek (SWO). Grant no: 2016-2017 and KWF Dutch Cancer Society. Grant no: 6905.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Medical Ethics Committee, Erasmus MC-University Medical Center, Rotterdam, The Netherlands (4 October 2018) and was registered with reference 67865.
Informed Consent Statement
Patient consent was waived due to the Code of Conduct for Responsible Use of Human Tissue and Medical Research” that applies to medical research in The Netherlands.
Data Availability Statement
Data can be provided upon request.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cao J., Heijkants R.C., Jochemsen A.G., Dogrusöz M., De Lange M.J., Van Der Velden P.A., Van Der Burg S.H., Jager M.J., Verdijk R.M. Targeting of the MAPK and AKT pathways in conjunctival melanoma shows potential synergy. Oncotarget. 2017;8:58021–58036. doi: 10.18632/oncotarget.10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larsen A.C., Dahmcke C.M., Dahl C., Siersma V.D., Toft P.B., Coupland S.E., Prause J.U., Guldberg P., Heegaard S. A retrospective review of conjunctival malignant melanoma presentation, treatment and outcome and an investigation of features associated with BRAF mutations. JAMA Ophthalmol. 2015;133:1295–1303. doi: 10.1001/jamaophthalmol.2015.3200. [DOI] [PubMed] [Google Scholar]
- 3.Griewank K.G., Westekemper H., Murali R., Mach M., Schilling B., Wiesner T., Schimming T., Livingstone E., Sucker A., Grabellus F., et al. Conjunctival Melanomas Harbor BRAF and NRAS Mutations and Copy Number Changes Similar to Cutaneous and Mucosal Melanomas. Clin. Cancer Res. 2013;19:3143–3152. doi: 10.1158/1078-0432.CCR-13-0163. [DOI] [PubMed] [Google Scholar]
- 4.Dalvin L.A., Salomão D.R., Patel S.V. Population-based incidence of conjunctival tumours in Olmsted County, Minnesota. Br. J. Ophthalmol. 2018;102:1728–1734. doi: 10.1136/bjophthalmol-2017-311530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Missotten G.S., Keijser S., De Keizer R.J.W., De Wolff-Rouendaal D. Conjunctival Melanoma in The Netherlands: A Nationwide Study. Investig. Opthalmol. Vis. Sci. 2005;46:75–82. doi: 10.1167/iovs.04-0344. [DOI] [PubMed] [Google Scholar]
- 6.Rossi E., Schinzari G., Maiorano B.A., Pagliara M.M., Di Stefani A., Bria E., Peris K., Blasi M.A., Tortora G. Conjunctival Melanoma: Genetic and Epigenetic Insights of a Distinct Type of Melanoma. Int. J. Mol. Sci. 2019;20:5447. doi: 10.3390/ijms20215447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou C., Wang Y., Jia R., Fan X. Conjunctival Melanoma in Chinese Patients: Local Recurrence, Metastasis, Mortality, and Comparisons with Caucasian Patients. Investig. Opthalmol. Vis. Sci. 2017;58:5452–5459. doi: 10.1167/iovs.17-22376. [DOI] [PubMed] [Google Scholar]
- 8.Wong J.R., Nanji A.A., Galor A., Karp C.L. Management of conjunctival malignant melanoma: A review and update. Expert Rev. Oph-Thalmol. 2014;9:185–204. doi: 10.1586/17469899.2014.921119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kujala E., Tuomaala S., Eskelin S., Kivelä T. Mortality after uveal and conjunctival melanoma: Which tumour is more deadly? Acta Ophthalmol. 2009;87:149–153. doi: 10.1111/j.1755-3768.2008.01369.x. [DOI] [PubMed] [Google Scholar]
- 10.Esmaeli B., Wang X., Youssef A., Gershenwald J.E. Patterns of regional and distant metastasis in patients with conjunctival melanoma: Experience at a cancer center over four decades. Ophthalmology. 2001;108:2101–2105. doi: 10.1016/S0161-6420(01)00782-5. [DOI] [PubMed] [Google Scholar]
- 11.Paridaens A.D., Minassian D.C., McCartney A.C., Hungerford J.L. Prognostic factors in primary malignant melanoma of the conjunctiva: A clinicopathological study of 256 cases. Br. J. Ophthalmol. 1994;78:252–259. doi: 10.1136/bjo.78.4.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smit K.N., Van Poppelen N.M., Vaarwater J., Verdijk R., van Marion R., Kalirai H., Coupland S.E., Thornton S., Farquhar N., Dubbink H.J., et al. Combined mutation and copy-number variation detection by targeted next-generation sequencing in uveal melanoma. Mod. Pathol. 2018;31:763–771. doi: 10.1038/modpathol.2017.187. [DOI] [PubMed] [Google Scholar]
- 13.van Ipenburg J.A., Naus N.C., Dubbink H.J., van Ginderdeuren R., Missotten G.S., Paridaens D., Verdijk R.M. Prognostic value of TERT promoter mutations in conjunctival melanomas in addition to clinicopathological features. Br. J. Ophthalmol. 2020 doi: 10.1136/bjophthalmol-2020-317405. [DOI] [PubMed] [Google Scholar]
- 14.Koopmans A.E., Ober K., Dubbink H.J., Paridaens D., Naus N.C., Belunek S., Krist B., Post E., Zwarthoff E.C., de Klein A., et al. Prevalence and Implications of TERT Promoter Mutation in Uveal and Conjunc-tival Melanoma and in Benign and Premalignant Conjunctival Melanocytic Lesions. Invest. Ophthalmol. Vis. Sci. 2014;55:6024–6030. doi: 10.1167/iovs.14-14901. [DOI] [PubMed] [Google Scholar]
- 15.Scholz S.L., Cosgarea I., Süßkind D., Murali R., Möller I., Reis H., Leonardelli S., Schilling B., Schimming T., Hadaschik E., et al. NF1 mutations in conjunctival melanoma. Br. J. Cancer. 2018;118:1243–1247. doi: 10.1038/s41416-018-0046-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui C., Lian B., Zhou L., Song X., Zhang X., Wu D., Chi Z., Si L., Sheng X., Kong Y., et al. Multifactorial Analysis of Prognostic Factors and Survival Rates Among 706 Mucosal Melanoma Patients. Ann. Surg. Oncol. 2018;25:2184–2192. doi: 10.1245/s10434-018-6503-9. [DOI] [PubMed] [Google Scholar]
- 17.Ellerhorst J.A., Greene V.R., Ekmekcioglu S., Warneke C.L., Johnson M.M., Cooke C.P., Wang L.E., Prieto V.G., Gershenwald J.E., Wei Q., et al. Clinical correlates of NRAS and BRAF mutations in primary human melanoma. Clin. Cancer Res. 2011;17:229–235. doi: 10.1158/1078-0432.CCR-10-2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griewank K.G., Murali R., Schilling B.E., Scholz S., Sucker A., Song M., Susskind D., Grabellus F., Zimmer L.O., Hillen U., et al. TERT promoter mutations in ocular melanoma distinguish between conjunctival and uveal tumours. Br. J. Cancer. 2013;109:497–501. doi: 10.1038/bjc.2013.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Raamsdonk C.D., Griewank K.G., Crosby M.B., Garrido M.C., Vemula S., Wiesner T., Obenauf A.C., Wackernagel W., Green G., Bouvier N., et al. Mutations in GNA11 in uveal melanoma. N. Engl. J. Med. 2010;363:2191–2199. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jain P., Finger P.T., Damato B., Coupland S.E., Heimann H., Kenawy N., Brouwer N.J., Marinkovic M., Van Duinen S.G., Caujolle J.P., et al. Multicenter, International Assessment of the Eighth Edition of the American Joint Committee on Cancer Cancer Staging Manual for Conjunctival Melanoma. JAMA Ophthalmol. 2019;137:905–911. doi: 10.1001/jamaophthalmol.2019.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dratwa M., Wysoczańska B., Lacina P., Kubik T., Bogunia-Kubik K. TERT-Regulation and Roles in Cancer Formation. Front. Immunol. 2020;11:589929. doi: 10.3389/fimmu.2020.589929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Torres-Cabala C., Li-Ning-Tapia E., Hwu W.-J. Pathology-based Biomarkers Useful for Clinical Decisions in Melanoma. Arch. Med Res. 2020;51:827–838. doi: 10.1016/j.arcmed.2020.09.008. [DOI] [PubMed] [Google Scholar]
- 23.Akincilar S.C., Chan C.H.T., Ng Q.F., Fidan K., Tergaonkar V. Non-canonical roles of canonical telomere binding proteins in cancers. Cell. Mol. Life Sci. 2021:1–23. doi: 10.1007/s00018-021-03783-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colebatch A.J., Dobrovic A., Cooper W.A. TERT gene: Its function and dysregulation in cancer. J. Clin. Pathol. 2019;72:281–284. doi: 10.1136/jclinpath-2018-205653. [DOI] [PubMed] [Google Scholar]
- 25.Dono M., Angelini G., Cecconi M., Amaro A., Esposito A.I., Mirisola V., Maric I., Lanza F., Nasciuti F., Viaggi S., et al. Mutation frequencies of GNAQ, GNA11, BAP1, SF3B1, EIF1AX and TERT in uveal melanoma: Detection of an activating mutation in the TERT gene promoter in a single case of uveal melanoma. Br. J. Cancer. 2014;110:1058–1065. doi: 10.1038/bjc.2013.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macerola E., Loggini B., Giannini R., Garavello G., Giordano M., Proietti A., Niccoli C., Basolo F., Fontanini G. Coexistence of TERT promoter and BRAF mutations in cutaneous melanoma is associated with more clinicopathological features of aggressiveness. Virchows Archiv. 2015;467:177–184. doi: 10.1007/s00428-015-1784-x. [DOI] [PubMed] [Google Scholar]
- 27.Triay E., Bergman L., Nilsson B., All-Ericsson C., Seregard S. Time trends in the incidence of conjunctival melanoma in Sweden. Br. J. Ophthalmol. 2009;93:1524–1528. doi: 10.1136/bjo.2009.157933. [DOI] [PubMed] [Google Scholar]
- 28.Larsen A.C., Dahl C., Dahmcke C.M., Lade-Keller J., Siersma V.D., Toft P.B., Coupland S.E., Prause J.U., Guldberg P., Heegaard S. BRAF mutations in conjunctival melanoma: Investigation of incidence, clinico-pathological features, prognosis and paired premalignant lesions. Acta Ophthalmol. 2016;94:463–470. doi: 10.1111/aos.13007. [DOI] [PubMed] [Google Scholar]
- 29.Ascierto P.A., Kirkwood J.M., Grob J.-J., Simeone E., Grimaldi A.M., Maio M., Palmieri G., Testori A., Marincola F.M., Mozzillo N. The role of BRAF V600 mutation in melanoma. J. Transl. Med. 2012;10:1–9. doi: 10.1186/1479-5876-10-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pasquali S., Hadjinicolaou A.V., Sileni V.C., Rossi C.R., Mocellin S. Systemic treatments for metastatic cutaneous melanoma. Cochrane Database Syst. Rev. 2018;2:CD011123. doi: 10.1002/14651858.CD011123.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kenawy N., Kalirai H., Sacco J.J., Lake S.L., Heegaard S., Larsen A.-C., Finger P.T., Milman T., Chin K., Mosci C., et al. Conjunctival melanoma copy number alterations and correlation with mutation status, tumor features, and clinical outcome. Pigment. Cell Melanoma Res. 2019;32:564–575. doi: 10.1111/pcmr.12767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mor J.M., Heindl L.M. Systemic BRAF/MEK Inhibitors as a Potential Treatment Option in Metastatic Conjunctival Melanoma. Ocul. Oncol. Pathol. 2017;3:133–141. doi: 10.1159/000452473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shields C.L., Chang M., Lally S.E., Dalvin L.A., Orloff M.M. Conjunctival melanoma with orbital invasion and liver metastasis managed with systemic immune checkpoint inhibitor therapy. Indian J. Ophthalmol. 2019;67:2071–2073. doi: 10.4103/ijo.IJO_663_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heppt M.V., Siepmann T., Engel J., Schubert-Fritschle G., Eckel R., Mirlach L., Kirchner T., Jung A., Gesierich A., Ruzicka T., et al. Prognostic significance of BRAF and NRAS mutations in melanoma: A German study from routine care. BMC Cancer. 2017;17:536. doi: 10.1186/s12885-017-3529-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adam C., Fusi L., Weiss N., Goller S.G., Meder K., Frings V.G., Kneitz H., Goebeler M., Houben R., Schrama D., et al. Efficient Suppression of NRAS-Driven Melanoma by Co-Inhibition of ERK1/2 and ERK5 MAPK Pathways. J. Investig. Dermatol. 2020;140:2455–2465. doi: 10.1016/j.jid.2020.03.972. [DOI] [PubMed] [Google Scholar]
- 36.Quek C., Rawson R.V., Ferguson P.M., Shang P., Silva I., Saw R.P., Shannon K., Thompson J.F., Hayward N.K., Long G.V., et al. Recurrent hotspot SF3B1 mutations at codon 625 in vulvovaginal muco-sal melanoma identified in a study of 27 Australian mucosal melanomas. Oncotarget. 2019;10:930–941. doi: 10.18632/oncotarget.26584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Poppelen N.M., Vaarwater J., Mudhar H.S., Sisley K., Rennie I.G., Rundle P., Brands T., Van Den Bosch Q.C., Mensink H.W., de Klein A., et al. Genetic Background of Iris Melanomas and Iris Melanocytic Tumors of Uncertain Malignant Potential. Ophthalmology. 2018;125:904–912. doi: 10.1016/j.ophtha.2017.12.022. [DOI] [PubMed] [Google Scholar]
- 38.Kong Y., Krauthammer M., Halaban R. Rare SF3B1 R625 mutations in cutaneous melanoma. Melanoma Res. 2014;24:332–334. doi: 10.1097/CMR.0000000000000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hintzsche J.D., Gorden N.T., Amato C.M., Kim J., Wuensch K.E., Robinson S.E., Applegate A.J., Couts K.L., Medina T.M., Wells K.R., et al. Whole-exome sequencing identifies recurrent SF3B1 R625 mutation and comutation of NF1 and KIT in mucosal melanoma. Melanoma Res. 2017;27:189–199. doi: 10.1097/CMR.0000000000000345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang H.M., Hsiao S.J., Schaeffer D.F., Lai C., Remotti H.E., Horst D., Mansukhani M.M., Horst B.A. Identification of recurrent mutational events in anorectal melanoma. Mod. Pathol. 2016;30:286–296. doi: 10.1038/modpathol.2016.179. [DOI] [PubMed] [Google Scholar]
- 41.Yavuzyigitoglu S., Koopmans A.E., Verdijk R.M., Vaarwater J., Eussen B., Van Bodegom A., Paridaens D., Kiliç E., de Klein A. Uveal Melanomas with SF3B1 Mutations: A Distinct Subclass Associated with Late-Onset Metastases. Ophthalmology. 2016;123:1118–1128. doi: 10.1016/j.ophtha.2016.01.023. [DOI] [PubMed] [Google Scholar]
- 42.Pham D.M., Guhan S., Tsao H. KIT and Melanoma: Biological Insights and Clinical Implications. Yonsei Med. J. 2020;61:562–571. doi: 10.3349/ymj.2020.61.7.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piris A., Mihm M.C., Jr., Hoang M.P. BAP1 and BRAFV600E expression in benign and malignant melanocytic proliferations. Hum. Pathol. 2015;46:239–245. doi: 10.1016/j.humpath.2014.10.015. [DOI] [PubMed] [Google Scholar]
- 44.Chau C., van Doorn R., van Poppelen N.M., van der Stoep N., Mensenkamp A.R., Sijmons R.H., van Paassen B.W., van den Ouweland A.M., Naus N.C., van der Hout A.H., et al. Families with BAP1-Tumor Predisposition Syndrome in The Nether-lands: Path to Identification and a Proposal for Genetic Screening Guidelines. Cancers. 2019;11:1114. doi: 10.3390/cancers11081114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koopmans A.E., Vaarwater J., Paridaens D., Naus N.C., Kiliç E., De Klein A. Patient survival in uveal melanoma is not affected by oncogenic mutations in GNAQ and GNA11. Br. J. Cancer. 2013;109:493–496. doi: 10.1038/bjc.2013.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bol K.F., Donia M., Heegaard S., Kiilgaard J.F., Svane I.M. Genetic Biomarkers in Melanoma of the Ocular Region: What the Medical Oncologist Should Know. Int. J. Mol. Sci. 2020;21:5231. doi: 10.3390/ijms21155231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data can be provided upon request.