Abstract
Simple Summary
Here, we monitored the course of the disease and treatment of sinonasal melanoma patients. Since treatment options are rare, immunotherapy is often the treatment of choice. However, intrinsic or acquired resistance to treatment may occur. We assessed the mutational status of the tumors and metastases during the course of therapy and recognized a switch of the oncogenic drivers to mutant NRAS in progressing disease. As a switch of drivers (other than the addition of a second driver) has not been reported yet, longitudinal molecular testing and the awareness of molecular heterogeneity of sinonasal melanoma is crucial.
Abstract
Mucosal melanoma can be driven by various driver mutations in genes such as NRAS, KIT, or KRAS. However, some cases present with only weak drivers, or lacking known oncogenic drivers, suggesting immunotherapy over targeted therapy. While resistance mechanisms to immunotherapy in cutaneous melanoma have been uncovered, including alterations in JAK1/2, B2M, or STK11, a switch of oncogenic drivers under immunotherapy has not yet been observed. We report three cases of metastatic sinonasal melanoma that switched oncogenic drivers from KRAS, KIT, or no driver to NRAS during or after immunotherapy, thereby showing progressive disease. One of the cases presented with three spatially separate driver mutations in the primary tumor, whereas the NRAS clone persisted under immunotherapy. In comparison, three different control cases receiving radiotherapy only did not show a change of the detectable molecular drivers in their respective recurrences or metastases. In summary, these data provide an important rationale for longitudinal molecular testing, based on evidence for an unforeseen recurrent event of molecular driver switch to NRAS in progressing sinonasal melanoma. These findings provide the basis for further studies on a potential causal relation of emerging NRAS mutant clones and immunotherapy.
Keywords: mucosal melanoma, sinonasal cancer, oncogenic driver, immunotherapy, tumor heterogeneity, disease monitoring
1. Introduction
Sinonasal melanoma is a rare melanoma subtype (0.5 cases per Mio/year) [1]. While treatment options for cutaneous melanoma emerged in recent years due to the successful implementation of targeted therapy and immunotherapy (IT) [2], the treatment of metastatic sinonasal melanoma remains challenging. Molecular targets for therapy are infrequent [3], and little is known about responses to immunotherapy [1]. Therefore, metastatic sinonasal melanoma patients are currently treated according to the guidelines for cutaneous melanoma [4]. Besides immunotherapy, targeted therapy with imatinib or nilotinib are options in patients with KIT mutations [5,6] or binimetinib for patients harboring NRAS mutations [7]. Zaretsky et al. recently reported the acquirement of loss-of-function mutations in JAK1/2 or B2M, causing an interruption of the interferon gamma signaling or loss of MHC-I surface expression, respectively, leading to resistance to anti-PD-1 treatment [8]. As resistance mechanisms to targeted therapy with BRAF/MEK inhibitors, mutations in MAP2K1/2, alternative splice variants of BRAF, BRAF amplifications or a gain of NRAS mutations were detected [9,10]. However, the emergence or selection of NRAS-mutant clones in progressing disease has not been shown yet. We report three cases of sinonasal melanoma, initially harboring KRAS, KIT, or no detectable driver mutation. For the first time, we describe a switch from the initial distinct oncogenic drivers (or no detectable driver) to solely NRAS in the course of therapy.
2. Materials and Methods
Collection of patient material: The local ethics review board approved this study (BASEC 2020-01663, approved: 30 July 2020) and written informed consent was obtained (BASEC PB_2017-00494, amendment approved: 25 July 2017). Molecular data from the primary tumor of patient 2 and of RT-patient 2 were previously published [3].
Collection of clinical data: All patients were presented regularly at the multidisciplinary tumor board. Clinical workup included molecular analysis to search for driver mutations and possible therapeutic targets. All patients were re-sequenced during the course of metastatic disease.
Molecular analysis: For Next Generation Sequencing, patient material was reviewed by an attending pathologist and the area of interest was marked on a hematoxylin and eosin (H&E) slide. Punch biopsies were then taken from the corresponding FFPE tissue block and DNA was isolated using the Maxwell 16 FFPE Tissue LEV DNA Purification Kit (Promega, Madison, WI, USA). For detection of low frequency mutations with the Oncomine™ Colon cfDNA assay (see below), DNA was isolated from whole sections of the FFPE tumor blocks. DNA was quantified using a fluorometric assay (Qubit, Thermo Fisher Scientific, Waltham, MA, USA). Library preparation was performed using the KAPA HyperPlus Kit (Roche, Basel, Switzerland) according to the manufacturer’s manual. Target capture was performed according to the protocol using a customized probe set (KAPA Hyper Choice, Roche) covering 190 melanoma-typical genes (Table S1) and heterozygous SNPs across the genome to detect copy number changes. HLA-A, HLA-B, and HLA-C loci were covered to conduct HLA typing. The tumor mutational burden (TMB) was calculated using all non-synonymous coding mutations divided by the size of the coding target region of the panel. All libraries were sequenced paired-end (100 bp) on an Illumina NextSeq550 sequencer. Data analysis was performed using a customized pipeline and open-source software [3].
For liquid biopsy, cell-free DNA (cfDNA) was isolated from 2 mL of plasma using the QIAamp circulating nucleic acid kit (Qiagen, Hilden, Germany). The DNA was quantified using a fluorometric assay (Qubit, Thermo Fisher Scientific). NGS on liquid biopsies, as well as NGS on whole sections of the tumors was performed using the Oncomine™ Colon cfDNA Assay (Thermo Fisher Scientific). The libraries were templated and loaded on a 540er chip using the Ion Chef™ instrument (Thermo Fisher Scientific) and sequenced on the Ion S5™ sequencer (Thermo Fisher Scientific). The data were analyzed using the Ion Reporter software version 5.12 (Thermo Fisher Scientific).
Variant reporting focused on main melanoma drivers (MAPK pathway) and potential resistance mutations to approved therapies (e.g., PIK3CA/mTOR pathway).
Histology/Immunohistochemistry: FFPE specimen of 2 µm were stained with hematoxylin and eosin for morphological assessment. Immunohistochemistry was performed using the Bond III automated staining system (Leica), and the monoclonal anti-NRAS (mutated Q61R) antibody SP174 (Abcam, Cambridge, UK). For NRAS staining, pretreatment with H2 was done for 60 min. The recombinant anti-rabbit monoclonal anti-NRAS (mutated Q61R) antibody SP174 (Abcam) was used in a 1:50 dilution. Detection was performed using the Bond Polymer Refine Red Detection Kit (Leica, Wetzlar, Germany). Staining of CD8 and PD-L1 was performed as described previously [3]. The morphological features were evaluated by an experienced head and neck pathologist (NJR). PD-L1 immunohistochemistry was scored as described previously [3]. Numbers of CD8+ T cells were semi-quantitatively assessed (no, low, moderate, high infiltration) in two locations: intratumoral and tumor margin.
FDG-PET/CT: Patients underwent several FDG-PET/CT examinations on different scanners (all manufactured by GE Healthcare, Waukesha, WI, USA).
Dissimilarity and co-ancestry analysis: Dissimilarities were calculated using biallelic SNPs, usually used for CNV analysis, in addition to the 190 MelArray genes. In total, we genotyped 22,246 loci across all chromosomes.
3. Results
3.1. Patient History Shows Emergence and Selection of an NRAS-Mutant Clone during the Course of Immunotherapy
3.1.1. Case 1
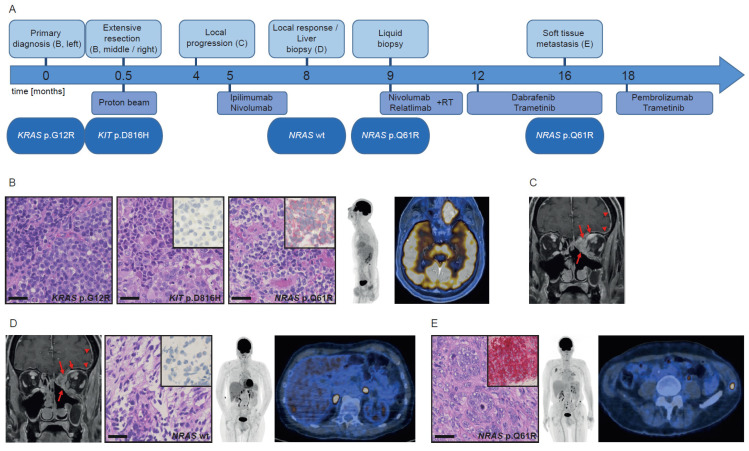
This 68-year-old male patient (Figure 1A) presented with progressing pressure sensation in the left cheek and orbita. The staging FDG-PET/CT revealed a large, FDG-avid mass in the left ethmoid infiltrating in the cribriform plate and showed absence of regional or distant metastases (Figure 1B). The resection specimen (Figure 1B, left) revealed a KRAS p.G12R mutation. Interestingly, the dural part of the tumor (Figure 1B, middle) revealed a KIT p.D816H mutation. The multidisciplinary tumor board opted for adjuvant proton beam therapy with 72 Gy. Due to local progression three months later (Figure 1C), immunotherapy with ipilimumab/nivolumab was initiated. Follow-up MRI and FDG-PET/CT after three months showed local tumor response (Figure 1D, left), but liver metastases, which were confirmed by biopsy (Figure 1D, middle and right). One month later, NGS of a liquid biopsy revealed an NRAS p.Q61R mutation. Retrospectively, the NRAS mutation had not been present in the liver biopsy, which was corroborated by negative staining of the NRAS Q61R antibody (Figure 1D, insert). However, when staining all available blocks from the primary tumor, a minute portion of cells were positive for the NRAS Q61R antibody (Figure 1B, right, insert). Systemic treatment was changed to nivolumab/relatlimab. Subsequent metastases in the bone and lung were additionally treated with stereotactic radiation. Owing to local tumor progression and distant metastases, treatment was changed to dabrafenib/trametinib three months later. An excised soft tissue metastasis of the left flank confirmed the previously detected NRAS mutation. Thus, the patient is currently being treated with pembrolizumab/trametinib.
Figure 1.
Course of disease, treatment, and morpho-molecular workup of patient 1. (A) Timeline. (B) Histology of the primary tumor, including NRAS Q61R immunohistochemistry (inserts); left: KRAS mutant region, middle: KIT mutant region, right: NRAS mutant region; PET MIP display and fused PET/CT image at primary diagnosis. (C) T1-weighted contrast-enhanced and fat-suppressed MR image at local progression (long arrows: sinonasal tumor mass, short arrows: intraorbital extraconal tumor mass, arrowheads: dural enhancement. (D) MR image at local regression (arrows/arrowheads: see above), histology of the liver biopsy, including NRAS Q61R immunohistochemistry (insert); PET MIP display and fused PET/CT image before liver biopsy. (E) Histology of the soft tissue metastasis, including NRAS Q61R immunohistochemistry (insert); PET MIP display and fused PET/CT image at progression under kinase inhibitors. Scale bar: 40 µm. wt: wild type.
3.1.2. Case 2
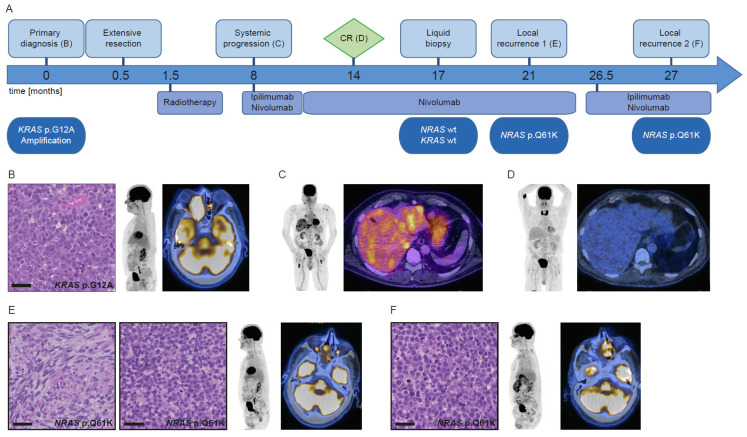
This 73-year-old male patient (Figure 2A) presented with watery discharge from the right nostril, pressure on the right eye, and double vision. An MRI scan revealed a mass in the right-sided nasal cavity and maxillary sinus, extending into the orbita and anterior skull base. Biopsy revealed a sinonasal melanoma without regional or distant metastases on FDG-PET/CT (Figure 2B). NGS showed a KRAS p.G12A mutation and amplification. Transnasal-transcribriform resection was carried out, followed by intensity-modulated radiation therapy with 70 Gy. On follow-up PET/CT, there was no evidence of local tumor recurrence, but multiple new neck lymph node metastases and distant metastases were found (Figure 2C). A combination of ipilimumab/nivolumab was initiated and after four cycles switched to nivolumab monotherapy (11 cycles). Follow-up PET/CT four months after the start of monotherapy showed complete remission (CR) (Figure 2D). A liquid biopsy taken three months after CR revealed no KRAS or NRAS mutation. This result can either indicate sustained remission or absence of tumor DNA in the blood for other reasons. More than one year after initiation of immunotherapy, the patient developed a mass in the right nasal cavity, visible both clinically and on PET/CT. No other tumor manifestation was found. Tumor recurrence was confirmed (Figure 2E) and nivolumab treatment was continued. NGS of the biopsy revealed an NRAS p.Q61K mutation. Six months later, a subtotal tumor removal was anticipated due to progression. Treatment with trametinib will be started as the NRAS mutation is still evident.
Figure 2.
Course of disease, treatment, and morpho-molecular workup of patient 2. (A) Timeline. (B) Histology of the primary tumor; PET MIP display and fused PET/CT image at primary diagnosis. (C) PET MIP display and fused PET/CT image at the start of immunotherapy. (D) PET MIP display and fused PET/CT image at CR. (E) Histology of the first local recurrence, left: spindle cell shaped morphology, right: monomorphic epithelioid morphology; PET MIP display and fused PET/CT image at first local recurrence. (F) Histology of the second local recurrence; PET MIP display and fused PET/CT image at second local recurrence. Scale bar: 40 µm. wt: wild type, CR: complete remission.
3.1.3. Case 3
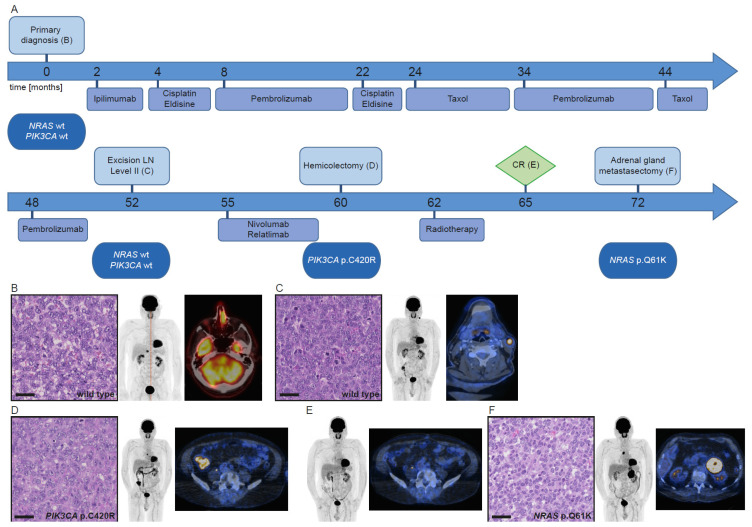
This 72-year-old male patient (Figure 3A) underwent surgery of a left-sided endonasal tumor. Histological workup revealed an amelanotic melanoma (Figure 3B), which had no evident driver mutation, but mutations in JAK2 and MITF. For the next two years, the patient underwent various treatment lines that were continuously adjusted depending on disease progression monitored by FDG-PET/CT (Figure 3A).
Figure 3.
Course of disease, treatment, and morpho-molecular workup of patient 3. (A) Timeline. (B) Histology of the primary tumor; PET MIP display and fused PET/CT image at primary diagnosis. (C) Histology of the lymph node metastasis; PET MIP display and fused PET/CT image at progression after 34 cycles of pembrolizumab. (D) Histology of the colon metastasis; PET MIP display and fused PET/CT image at progression under nivolumab/relatlimab. (E) PET MIP display and fused PET/CT image at CR. (F) Histology of the adrenal gland metastasis; PET MIP display and fused PET/CT image at progression without therapy. Scale bar: 40 µm. wt: wild type, LN: lymph node, CR: complete remission.
Forty-eight months after initial diagnosis, a left-sided cervical lymph node metastasis was resected (Figure 3C) and sequenced, which revealed a mutational profile similar to the primary tumor. Due to a progressing metastasis in the cecum, treatment with nivolumab/relatlimab was started. After five months under immunotherapy this lesion was removed surgically (Figure 3D), and molecular analysis revealed a PIK3CA p.C420R mutation together with the known JAK2 and MITF mutations. Systemic treatment was discontinued, and another mesenteric lymph node metastasis was irradiated. Thereafter, the patient was considered to be in CR (Figure 3E). Seven months later, a left-sided adrenal gland metastasis was removed surgically (Figure 3F) and confirmed by immunohistochemistry. Molecular analysis revealed an NRAS p.Q61K mutation and the known MITF mutation, but no JAK2 mutation. Owing to disease progression, the patient is currently under treatment with ipilimumab/nivolumab.
3.2. Mutational Analysis of Different Tumors Reveals Both Intra-Patient Similarities and Molecular Differences
We analyzed heterozygous SNPs across the genome of all tumors, which revealed the relationship of all tumors within each patient. Of note, intra-patient heterogeneity could also be observed (Figure S1).
For each patient we intersected the set of all mutations found in the tumors. Tumors of patient 1 had an overlap of 46.2% (six mutations; Figure S2A). Tumors of patient 2 had an overlap of 33.3% (four mutations; Figure S2B), whereas additional 25.0% (three mutations; Figure S2B) overlapped between the two NRAS-mutant tumors. The four tumors of patient 3 had an overlap of 48.1% (13 mutations; Figure S2C). When overlapping the mutations of NRAS-mutant tumors from all patients, there were no similarities of the mutational profile between the different patients (Figure S2D). The mutational profile of all tumors is provided in Tables S2–S4. The mutation frequency in synopsis with the tumor cell content revealed the presence of the NRAS mutation on one allele, excluding subclonal events.
3.3. NRAS Clones either Preexisted or Emerged during the Course of Disease and Therapy
To investigate whether the NRAS mutations were already present in the tumor before immunotherapy on a subclonal level, we searched for a low frequency presence of the NRAS mutations in the sequencing data from the investigated tissue punches (mean coverage of coding target region: patient 1: 168x (primary diagnosis) and 791x (extensive resection); patient 2: 757x; patient 3: 1371x) but did not find any. Conversely, NRAS-mutant tumors did not show any low-frequency traces of the previously detected KRAS, KIT or PIK3CA mutations in the NRAS mutant tumors of all patients (mean coverage of coding target region: patient 1: 710x; patient 2: 290x and 1077x; patient 3: 1720x). For the detection of the NRAS mutation in the melanoma of patient 1 (NRAS p.Q61R), a specific antibody is available. This NRAS Q61R antibody revealed no positivity in the KRAS-mutant or KIT-mutant part of the primary tumor of patient 1 (Figure S3). However, a minute region was positive in another part, indicative for the NRAS mutation. To further investigate this molecular heterogeneity, whole sections of the tumor tissue before immunotherapy were selected and sequenced with a small but highly sensitive NGS panel (Oncomine™ Colon cfDNA Assay) that includes HotSpot regions of NRAS and KRAS (Table 1).
Table 1.
High sensitivity NGS analysis of whole tumor sections before immunotherapy.
| Mutations | Frequency | Molecular Depth | Tumor Cell Content | Comment | |
|---|---|---|---|---|---|
| Patient 1 |
KRAS p.G12R NRAS p.Q61R NRAS p.G12V |
4.6% 3.27% 7.04% |
913x 765x 270x |
40% |
KIT not covered by the assay |
| Patient 2 | KRAS p.G12A | 64.21% | 992x | 70% | |
| Patient 3 | WT | 30% |
WT = wild type.
The tumor of patient 1 revealed the previously detected KRAS p.G12R mutation (frequency: 4.6%), confirmed the NRAS p.Q61R mutation, previously detected using the corresponding antibody (frequency: 3.3%), and revealed an additional NRAS p.G12V mutation (frequency: 7.0%). These findings indicate a molecularly heterogeneous tumor with different subclones (tumor cell content of the whole section: 40%). The previously detected KIT mutation is not covered by the target region of the selected panel. The tumor of patient 2 revealed a high proportion of KRAS p.G12A mutated cells (64.2%), concordant with our previous results and reflecting an amplification of the mutant allele, but no NRAS mutation (tumor cell content of the whole section: 70%). The tumor of patient 3 showed no mutation in NRAS (tumor cell content of the whole section: 30%). MITF and JAK2 are not covered by the selected panel.
As all patients underwent additional radiotherapy (RT) during their course of disease, we investigated whether the emergence of the NRAS mutation could be an effect of this therapy. We retrospectively identified a control group of sinonasal melanoma patients who underwent solely radiotherapy (n = 3). Primary tumors and tumor recurrences or metastases of these three patients before and after radiotherapy were sequenced, whereas no emerging NRAS mutation was detected (Table S5).
3.4. Tumor Morphology Revealed No Association with Mutational Status or Response to Therapy
Furthermore, we compared the morphology of the different tumors (Figure 1, Figure 2 and Figure 3 histological pictures, Table 2). The initial KRAS-mutated melanoma of patient 1 (Figure 1B, left) showed only minor differences to the KIT-mutated and NRAS-mutated part of the tumor (Figure 1B, middle, right). The NRAS-mutated soft tissue metastasis showed a similar morphology, however with obvious chromatin clearing of the nuclei (Figure 1E), while the liver biopsy showed only few vital cells (Figure 1D).
Table 2.
Comparison of tumor histology.
| Size | Morphology | Nuclei | Other Features |
Main Molecular Alteration |
|
|---|---|---|---|---|---|
| Patient 1 | |||||
| Primary diagnosis |
medium/large | pleomorphic, atypical | variable size, prominent nucleoli, eccentric nuclei | eosinophilic cytoplasm, abundant mitoses |
KRAS p.G12R |
| Extensive resection |
medium/large | pleomorphic, atypical | convoluted nuclei, variable size, prominent nucleoli, eccentric nuclei | eosinophilic cytoplasm, abundant mitoses |
KIT p.D816H |
| Liver biopsy |
few vital cells, myxoid stromal changes | WT | |||
| Soft tissue metastasis |
medium/large | pleomorphic, atypical | chromatin clearing of the nuclei | eosinophilic cytoplasm, abundant mitoses |
NRAS p.Q61R |
| Patient 2 | |||||
| Primary diagnosis |
medium/large | monomorphic, atypical | variously distinct nucleoli | eosinophilic cytoplasm |
KRAS p.G12A/amplification |
| Local recurrence 1 |
spindle cells: medium/large; epithelioid cells: small |
biphasic appearance (spindle cell shaped, monomorphic epithelioid) | NRAS p.Q61K | ||
| Local recurrence 2 |
small/ medium |
monomorphic, atypical | NRAS p.Q61K | ||
| Patient 3 | |||||
| Primary diagnosis |
medium/large | atypical, epithelioid |
prominent nucleoli |
eosinophilic cytoplasm, abundant mitoses |
WT |
| LN level II | medium/large | atypical, epithelioid |
prominent nucleoli |
eosinophilic cytoplasm, abundant mitoses |
WT |
| Hemi- colectomy |
medium/large | atypical, epithelioid |
prominent nucleoli |
eosinophilic cytoplasm, abundant mitoses |
PIK3CA p.C420R |
| Adrenal gland metastasis |
small/
medium |
atypical, epithelioid |
lacking prominent nucleoli | eosinophilic cytoplasm, abundant mitoses |
NRAS p.Q61K |
LN = lymph node, WT = wild type, major differences are printed in bold.
The initial KRAS-mutated melanoma of patient 2 (Figure 2B) showed differences in cell size and morphology compared to the first local recurrence, which had a biphasic appearance (Figure 2E), while the second local recurrence (Figure 2F) was similar to the initial tumor.
For patient 3, all manifestations presented a similar morphology (Figure 3B–D), with the NRAS-mutant tumor showing smaller cells and lacking the initial prominent nucleoli (Figure 3F).
When comparing the NRAS-mutant tumors of the three patients, their morphology did not reveal a distinctive mark to identify this molecular subtype.
3.5. CD8+ T Cell Infiltration, PD-L1 Expression and Tumor Mutational Burden (TMB) Do Not Correlate with Therapy Outcome
The tumors before immunotherapy of the responding patients (patients 1 and 2) showed no or only low intratumoral infiltration of CD8+ cells with low to moderate accumulation at the tumor margins, while NRAS mutant tumors emerging under therapy showed moderate intratumoral infiltration and moderate to high accumulation of CD8+ cells at the tumor margins. PD-L1 expression was constantly negative or weak in tumors from both patients before and after therapy. The tumor before IT of the intrinsic resistant patient (patient 3) showed a moderate intratumoral infiltration of CD8+ cells with a high number of cells at the tumor margin, while the NRAS mutant tumor showed only single intratumoral CD8+ infiltrating cells with moderate presence at the tumor margin. PD-L1 expression was high in both tumor and immune cells before and after therapy (Figure S4). In summary, NRAS mutant tumors of the initially responding patients showed a slightly higher CD8+ accumulation in the tumor and at its margins, while the NRAS mutant tumor of the intrinsic resistant patient showed a decreased infiltration compared to the WT tumors (Table S6).
Moreover, we calculated tumor mutational burden (TMB) based on our MelArray NGS panel (Table S7). Patients 1 and 2 that initially responded to the treatment had a lower TMB than the intrinsic resistant patient 3. Thus, higher TMB does not seem to correlate with response to IT in our cases.
4. Discussion
We have documented the evolution of metastatic sinonasal melanoma in three patients receiving combinational immunotherapy with nivolumab. While two patients (patients 2 and 3) lacked molecular intra-patient heterogeneity before immunotherapy, harboring only a KRAS mutation or no evident driver mutation, the primary tumor of patient 1 showed a heterogeneous molecular profile with several different clones and different driver mutations. Both patients 1 and 2 initially responded to immunotherapy, whereas the relapse showed presence of mutant NRAS. While the mutation was not detectable in the pre-treatment lesion of patient 2 performing a highly sensitive assay, it seemed to be the dominant, and therefore metastasizing, clone in patient 1. In patient 2, the NRAS mutant local recurrence appeared one year after the start of IT with ipilimumab/nivolumab, while the patient was in CR for already 6 months but still under nivolumab treatment. In patient 1 the NRAS mutant clone was present in the liquid biopsy four months after treatment initiation and in the later occurring soft tissue metastasis, while the other initially detected drivers were missing. Retrospectively, the NRAS mutation was already present on a subclonal level in the primary tumor. Patient 3 had intrinsic resistance to immunotherapy and CR was only reached due to surgery and RT. However, no NRAS clone was detectable in the tumor prior to IT, suggesting its emergence after therapy. While the NRAS mutation appeared under IT or in close proximity to the end of IT in the first two patients, it occurred late in patient 3 with one year after stop of IT without any further treatment. As this patient had an intrinsic resistance to the IT, it cannot be clarified whether the NRAS mutation is directly related to the treatment.
To investigate whether the NRAS mutation occurred as a potential effect of radiotherapy (RT), that all patients received at one point during their course of disease, we sequenced tumors before and after RT, but did not find an occurring NRAS mutation. Given the rarity of sinonasal melanoma, it is difficult to find a larger group of patients with archived samples before and after a certain treatment. Therefore, our results indicate the occurrence of the NRAS mutation rather in the context of immunotherapy than of RT. However, statistical evidence is lacking due to the small cohort size. Nevertheless, we can state that this phenomenon occurred recurrently under real world conditions and is therefore important to know. Further, this finding is very unlikely to occur recurrently in three independent patients of this rare tumor entity just by chance.
Neither clinically nor on FDG-PET/CT did we find lesions suspicious for a second primary tumor that would possibly explain the detection of different oncogenic drivers. While tumor heterogeneity in melanoma is well known, a switch of the driver mutation or a selection for a specific driver mutation in the course of immunotherapy as a possible mechanism of therapy resistance has not been reported yet. It was shown for several cancer types, that intra-patient tumor heterogeneity is limited to passenger mutations, with a consistent main driver in all metastases [11,12,13]. Moreover, when acquiring an additional driver mutation, the initial driver remains present, even within the same tumor cell [14]. Only one study compared primary cutaneous melanoma and corresponding metastases and showed the detection of emerging NRAS mutations in two patients. However, no information about treatment of these two patients is available [15]. Due to known intratumoral molecular heterogeneity in melanoma, it can be difficult to assess the mutational status of the whole tumor. NGS results on punch biopsies are potentially biased in tumors with high intratumoral heterogeneity. This was especially evident in patient 1, where we only found a KRAS mutation in a punch from the primary resection of the tumor, and afterwards a KIT mutation in the extensive resection of the same tumor. Moreover, an NRAS mutation became evident in another part of this tumor after using a specific antibody for this mutation. To reduce the bias, whole sections of a tumor or even pooled whole sections of different FFPE blocks of a tumor, instead of punches, can be sequenced. In this way, we were able to detect several clones within the primary tumor of patient 1 (Table 1). Still, it is impossible to sequence every single part of a tumor, especially in a diagnostic setting.
Liquid biopsy is another option to detect different clones. However, liquid biopsy highly depends on the amount of tumor cells in the blood. If this amount is low, the liquid biopsy is likely to remain inconclusive. Moreover, not all clones of a heterogeneous primary tumor may metastasize. Furthermore, it is possible to pick up clones in the blood that originate from a completely different type of cancer (e.g., in a patient with both melanoma and lung cancer). To find an optimal cost-benefit and effort-benefit ratio in the clinical setting, it might be best to do NGS on whole sections of the tumor and disease monitoring using liquid biopsy.
Several reports on the correlation of mutation status and immunotherapy outcome in cutaneous melanoma have been published, but the results remain inconclusive. Kirchberger et al. reported a similar response rate of both NRAS-mutant and wild type melanoma to immunotherapy. However, median overall survival was significantly lower in the NRAS-mutant group. Moreover, a treatment with MEK inhibitors could lead to a survival benefit of NRAS-mutant patients [16]. Shoushtari et al. showed an inferior response of NRAS p.Q61 mutant melanoma to anti-PD-1 monotherapy and a trend towards inferior response to a combination of ipilimumab and nivolumab in cutaneous melanoma [17].
In contrast, Johnson et al. found a superior outcome of NRAS-mutant patients [18]. Since KRAS mutations are rare in melanoma, no results are available for immunotherapy outcome in this group.
The effect of immunotherapy in mucosal melanoma was only investigated in a few reports. However, it seems to be a promising treatment in this type of cancer with very limited therapeutic alternatives. The overall response rate (ORR) to pembrolizumab in mucosal melanoma was shown to be slightly lower than in cutaneous melanoma. However, even CRs were reported [19] similar to our patient 2. When comparing treatment with ipilimumab plus nivolumab in mucosal and cutaneous melanoma, the ORR rate was 37% in mucosal and 60% in cutaneous melanoma, respectively, showing that mucosal melanoma patients may also benefit from this therapy [20]. Several studies reported a correlation of response to immunotherapy and PD-L1 status in various types of cancer [21,22]. Nevertheless, in cutaneous melanoma also patients with negative PD-L1 status can respond to IT, as shown by a previous study with an ORR of 54% and 72% in patients with negative or positive PD-L1 status, respectively [23]. PD-L1 expression in mucosal melanoma is less common than in cutaneous melanoma (44% positivity) as previously demonstrated by Kaunitz et al. [24]. It even seems to be particularly low in the subtype of sinonasal melanoma (20% positivity) [25], which is consistent with our previously published data [3] and the observation in the present study. Still, two of our three patients responded to this treatment.
In all three cases, CD8+ cells were present in the proximity of the tumor before IT and infiltration was shown in resistant tumors. However, a slight increase of CD8+ cells in the initially responding patients was evident in the NRAS mutant tumors, while the intrinsic resistant patient showed a slight decrease of CD8+ cells. Whether the CD8+ T cells in the NRAS mutant, IT-resistant tumors are exhausted, cannot be concluded from our study. Moreover, with only three patients, it is not possible to draw statistically reliable conclusions about CD8+ cell infiltration and response to IT.
Some associations of activated MAPK signaling and response to IT were recently made, such as reduced T cell infiltration and activity, as shown in breast cancer [26]. Moreover, NRAS is upstream of both MAPK and PIK3CA signaling, the latter being involved in reduced CD8+ T cell infiltration and function [27].
Recent studies have shown a correlation of TMB and ORR to immune checkpoint inhibitors. Nevertheless, mucosal melanoma patients, that generally have a lower TMB [28], are able to respond to the treatment [20]. Similar to these findings, the TMB in our two initially responding patients was low, while the intrinsic resistant patient had a slightly higher TMB. Therefore, additional markers to TMB and PD-L1 may play a role in mucosal melanoma.
Resistance to immunotherapy in melanoma is associated with alterations in the interferon gamma pathway or a lack of MHC-I surface expression [8]. We did not detect any alteration affecting the mentioned pathways with our custom NGS panel. While all tumors within one patient shared several passenger mutations, no overlap of passenger mutations among NRAS-mutant tumors was evident. Moreover, the morphology of the NRAS-mutant tumors differed. This result is consistent with our previously reported lack of morpho-molecular correlation in primary sinonasal melanoma [3].
Our study is limited by the lack of fresh material and patient-derived cell lines for functional testing. Thus, we cannot prove a causal association between the emergence of NRAS mutated clones and immunotherapy. In general, activating NRAS mutations could be a survival advantage for sinonasal melanoma cells independent of therapy. However, our finding can set the scene for future investigations to clarify the significance of the here detected emerging NRAS mutations in the course of immunotherapy.
5. Conclusions
Our case series shows the importance of longitudinal testing, preferably by non-invasive liquid biopsy, to monitor not only the presence, but also the change of driver mutations in melanoma patients during the course of disease. Moreover, molecular testing of larger tissue amounts (e.g., whole slide tissue) is preferable over tissue punches in this entity to increase the chances of capturing heterogeneous tumors harboring different relevant driver mutations. Our study also stresses the importance of regular whole-body FDG-PET/CT imaging in the follow-up of sinonasal melanoma patients.
The awareness of the heterogeneity of primary sinonasal melanoma and the fact that a unique driver switch may occur will help to adjust the treatment regimen in time by combining immunotherapy and targeted therapy before the emergence of multiple treatment-resistant metastases. Due to a lack of awareness, the occurrence of this issue can lead to significant discomfiture for pathologists and clinicians in charge and may have dire consequences for patients.
Acknowledgments
We thank Susanne Dettwiler and Fabiola Prutek from the Biobank, Marion Bawohl, Annette Bohnert, André Fitsche and the immunohistochemistry laboratory, the IT division, and the Department of Pathology and Molecular Pathology for technical support.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/cancers13112725/s1, Figure S1: Individual dissimilarity and co-ancestry coefficient of all resected tumors from all patients, Figure S2: Overlap of mutations of the different tumor manifestations within each patient and of the NRAS mutant tumors, Figure S3: NRAS Q61R staining of the primary tumor of patient 1, Figure S4: Examples for CD8 and PD-L1 staining, Table S1: List of genes included in the MelArray NGS panel, Table S2: Mutational profile of all tumors of patient 1 acquired by MelArray NGS panel, Table S3: Mutational profile of all tumors of patient 2 acquired by MelArray NGS panel, Table S4: Mutational profile of all tumors of patient 3 acquired by MelArray NGS panel, Table S5: Mutational analysis of sinonasal melanoma before and after immunotherapy, Table S6: Distribution of CD8+ cells and PD-L1 status in all assessed tumors, Table S7: Mutational burden of all samples as calculated by MelArray.
Author Contributions
Conceptualization, S.N.F., D.H. and N.J.R.; data curation, S.N.F. and P.T.; formal analysis, S.N.F., P.T., M.H., G.B.M. and N.J.R.; funding acquisition, S.N.F., G.B.M. and N.J.R.; investigation, S.N.F., P.T., M.H., R.D., D.H. and N.J.R.; methodology, S.N.F., P.T., M.H., R.D., M.P.L., D.H. and N.J.R.; project administration, S.N.F.; resources, S.N.F., P.T., M.H., R.D., M.P.L., D.H. and N.J.R.; software, P.T.; supervision, S.N.F. and N.J.R.; validation, S.N.F. and N.J.R.; visualization, S.N.F., P.T., M.H. and N.J.R.; writing—Original draft, S.N.F., D.H. and N.J.R.; writing—Review & editing, P.T., M.H., R.D., G.B.M. and M.P.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Zurich Cancer League (grant recipient: S.N.F., co-recipient: N.J.R., G.B.M.)
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the local Ethics Committee of the Canton Zurich (BASEC 2020-01663 approved: 30 July 2020; BASEC PB_2017-00494 amendment approved: 25 July 2017).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to ethical/privacy restrictions.
Conflicts of Interest
S.N.F. receives honoraria from Oncobit. M.H. is a recipient of grants from GE Healthcare, grants for translational and clinical cardiac and oncological research from the Alfred and Annemarie von Sick legacy, and grants from the Artificial Intelligence in oncological Imaging Network by the University of Zurich outside the submitted work. RD has intermittent, project focused consulting and/or advisory relationships with Novartis, Merck Sharp & Dhome (MSD), Bristol-Myers Squibb (BMS), Roche, Amgen, Takeda, Pierre Fabre, Sun Pharma, Sanofi, Catalym, Second Genome, Regeneron, Alligator, MaxiVAX SA and touchIME outside the submitted work. M.P.L. is a founder of Oncobit, which holds a commercial license for the software used to analyze the melanoma target panel. N.J.R. discloses an advisory board function including receipt of honoraria from F. Hoffmann—La Roche AG outside the submitted work. P.T., G.B.M. and D.H. have no funding and conflicts of interest to disclose.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ascierto P.A., Accorona R., Botti G., Farina D., Fossati P., Gatta G., Gogas H., Lombardi D., Maroldi R., Nicolai P., et al. Mucosal melanoma of the head and neck. Crit. Rev. Oncol. Hematol. 2017;112:136–152. doi: 10.1016/j.critrevonc.2017.01.019. [DOI] [PubMed] [Google Scholar]
- 2.Leonardi G.C., Falzone L., Salemi R., Zanghì A., Spandidos D.A., McCubrey J.A., Candido S., Libra M. Cutaneous melanoma: From pathogenesis to therapy (review) Int. J. Oncol. 2018;52:1071. doi: 10.3892/ijo.2018.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freiberger S.N., Morand G.B., Turko P., Wager U., Dummer R., Hüllner M., Holzmann D., Rupp N.J., Levesque M.P. Morpho-molecular assessment indicates new prognostic aspects and personalized therapeutic options in sinonasal melanoma. Cancers. 2019;11:1329. doi: 10.3390/cancers11091329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dummer R., Keilholz U. Appendix 2: Cutaneous Melanoma (2): EUpdate Published Online September 2016. Ann. Oncol. 2016;27:v136–v137. doi: 10.1093/annonc/mdw432. [DOI] [PubMed] [Google Scholar]
- 5.Hodi F.S., Corless C.L., Giobbie-Hurder A., Fletcher J.A., Zhu M., Marino-Enriquez A., Friedlander P., Gonzalez R., Weber J.S., Gajewski T.F., et al. Imatinib for melanomas harboring mutationally activated or amplified kit arising on mucosal, acral, and chronically sun-damaged skin. J. Clin. Oncol. 2013;31:3182. doi: 10.1200/JCO.2012.47.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lebbe C., Chevret S., Jouary T., Dalac S., Dalle S., Guillot B., Arnault J.-P., Avril M.-F., Bedane C., Bens G., et al. Phase II multicentric uncontrolled national trial assessing the efficacy of nilotinib in the treatment of advanced melanomas with c-kit mutation or amplification. JCO. 2014;32:9032. doi: 10.1200/jco.2014.32.15_suppl.9032. [DOI] [Google Scholar]
- 7.Ascierto P.A., Schadendorf D., Berking C., Agarwala S.S., van Herpen C.M., Queirolo P., Blank C.U., Hauschild A., Beck J.T., St-Pierre A., et al. MEK162 for patients with advanced melanoma harbouring NRAS or Val600 BRAF mutations: A non-randomised, open-label phase 2 study. Lancet Oncol. 2013;14:249–256. doi: 10.1016/S1470-2045(13)70024-X. [DOI] [PubMed] [Google Scholar]
- 8.Zaretsky J.M., Garcia-Diaz A., Shin D.S., Escuin-Ordinas H., Hugo W., Hu-Lieskovan S., Torrejon D.Y., Abril-Rodriguez G., Sandoval S., Barthly L., et al. Mutations associated with acquired resistance to pd-1 blockade in melanoma. N. Engl. J. Med. 2016;375:819. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wagle N., Allen E.M.V., Treacy D.J., Frederick D.T., Cooper Z.A., Taylor-Weiner A., Rosenberg M., Goetz E.M., Sullivan R.J., Farlow D.N., et al. MAP kinase pathway alterations in BRAF-mutant melanoma patients with acquired resistance to combined RAF/MEK inhibition. Cancer Discov. 2014;4:61. doi: 10.1158/2159-8290.CD-13-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Long G.V., Fung C., Menzies A.M., Pupo G.M., Carlino M.S., Hyman J., Shahheydari H., Tembe V., Thompson J.F., Saw R.P., et al. Increased MAPK reactivation in early resistance to dabrafenib/trametinib combination therapy of BRAF-mutant metastatic melanoma. Nat. Commun. 2014;5:5694. doi: 10.1038/ncomms6694. [DOI] [PubMed] [Google Scholar]
- 11.Yates L.R., Gerstung M., Knappskog S., Desmedt C., Gundem G., Loo P.V., Aas T., Alexandrov L.B., Larsimont D., Davies H., et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat. Med. 2015;21:751. doi: 10.1038/nm.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hao J.-J., Lin D.-C., Dinh H.Q., Mayakonda A., Jiang Y.-Y., Chang C., Jiang Y., Lu C.-C., Shi Z.-Z., Xu X., et al. Spatial intratumor heterogeneity of genetic, epigenetic alterations and temporal clonal evolution in esophageal squamous cell carcinoma. Nat. Genet. 2016;48:1500. doi: 10.1038/ng.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rabbie R., Ansari-Pour N., Cast O., Lau D., Scott F., Welsh S.J., Parkinson C., Khoja L., Moore L., Tullett M., et al. Multi-site clonality analysis uncovers pervasive heterogeneity across melanoma metastases. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-18060-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raaijmakers M.I.G., Widmer D.S., Narechania A., Eichhoff O., Freiberger S.N., Wenzina J., Cheng P.F., Mihic-Probst D., Desalle R., Dummer R., et al. Co-Existence of BRAF and NRAS driver mutations in the same melanoma cells results in heterogeneity of targeted therapy resistance. Oncotarget. 2016;7:77163. doi: 10.18632/oncotarget.12848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pellegrini C., Cardelli L., Padova M., Nardo L., Ciciarelli V., Rocco T., Cipolloni G., Clementi M., Cortellini A., Ventura A., et al. Intra-patient heterogeneity of BRAF and NRAS molecular alterations in primary melanoma and metastases. Acta Derm. Venerol. 2020;100:1–8. doi: 10.2340/00015555-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirchberger M.C., Ugurel S., Mangana J., Heppt M.V., Eigentler T.K., Berking C., Schadendorf D., Schuler G., Dummer R., Heinzerling L. MEK inhibition may increase survival of NRAS-mutated melanoma patients treated with checkpoint blockade: Results of a retrospective multicentre analysis of 364 patients. Eur. J. Cancer. 2018;98:10–16. doi: 10.1016/j.ejca.2018.04.010. [DOI] [PubMed] [Google Scholar]
- 17.Shoushtari A.N., Chatila W.K., Arora A., Sanchez-Vega F., Kantheti H.S., Rojas Zamalloa J.A., Krieger P., Callahan M.K., Betof Warner A., Postow M.A., et al. Therapeutic implications of detecting MAPK-activating alterations in cutaneous and unknown primary melanomas. Clin. Cancer Res. 2021 doi: 10.1158/1078-0432.CCR-20-4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson D.B., Lovly C.M., Flavin M., Panageas K.S., Ayers G.D., Zhao Z., Iams W.T., Colgan M., DeNoble S., Terry C.R., et al. Impact of NRAS mutations for patients with advanced melanoma treated with immune therapies. Cancer Immunol. Res. 2015;3:288–295. doi: 10.1158/2326-6066.CIR-14-0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moya-Plana A., Herrera Gómez R.G., Rossoni C., Dercle L., Ammari S., Girault I., Roy S., Scoazec J.-Y., Vagner S., Janot F., et al. Evaluation of the efficacy of immunotherapy for non-resectable mucosal melanoma. Cancer Immunol. Immunother. 2019;68:1171–1178. doi: 10.1007/s00262-019-02351-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D’Angelo S.P., Larkin J., Sosman J.A., Lebbé C., Brady B., Neyns B., Schmidt H., Hassel J.C., Hodi F.S., Lorigan P., et al. Efficacy and safety of nivolumab alone or in combination with ipilimumab in patients with mucosal melanoma: A pooled analysis. JCO. 2016;35:226–235. doi: 10.1200/JCO.2016.67.9258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Topalian S.L., Hodi F.S., Brahmer J.R., Gettinger S.N., Smith D.C., McDermott D.F., Powderly J.D., Carvajal R.D., Sosman J.A., Atkins M.B., et al. Safety, activity, and immune correlates of anti–pd-1 antibody in cancer. N. Engl. J. Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garon E.B., Rizvi N.A., Hui R., Leighl N., Balmanoukian A.S., Eder J.P., Patnaik A., Aggarwal C., Gubens M., Horn L., et al. Pembrolizumab for the treatment of non–small-cell lung cancer. N. Engl. J. Med. 2015;372:2018–2028. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 23.Larkin J., Chiarion-Sileni V., Gonzalez R., Grob J.J., Cowey C.L., Lao C.D., Schadendorf D., Dummer R., Smylie M., Rutkowski P., et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N. Engl. J. Med. 2015;373:23–34. doi: 10.1056/NEJMoa1504030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaunitz G.J., Cottrell T.R., Lilo M., Muthappan V., Esandrio J., Berry S., Xu H., Ogurtsova A., Anders R.A., Fischer A.H., et al. Melanoma subtypes demonstrate distinct PD-L1 expression profiles. Lab. Investig. 2017;97:1063–1071. doi: 10.1038/labinvest.2017.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thierauf J., Veit J.A., Affolter A., Bergmann C., Grünow J., Laban S., Lennerz J.K., Grünmüller L., Mauch C., Plinkert P.K., et al. Identification and clinical relevance of pd-l1 expression in primary mucosal malignant melanoma of the head and neck. Melanoma Res. 2015;25:503–509. doi: 10.1097/CMR.0000000000000197. [DOI] [PubMed] [Google Scholar]
- 26.Loi S., Dushyanthen S., Beavis P.A., Salgado R., Denkert C., Savas P., Combs S., Rimm D.L., Giltnane J.M., Estrada M.V., et al. RAS/MAPK activation is associated with reduced tumor-infiltrating lymphocytes in triple-negative breast cancer: Therapeutic cooperation between MEK and PD-1/PD-L1 immune checkpoint inhibitors. Clin. Cancer Res. 2016;22:1499–1509. doi: 10.1158/1078-0432.CCR-15-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng W., Chen J.Q., Liu C., Malu S., Creasy C., Tetzlaff M.T., Xu C., McKenzie J.A., Zhang C., Liang X., et al. Loss of PTEN promotes resistance to t cell–mediated immunotherapy. Cancer Discov. 2016;6:202–216. doi: 10.1158/2159-8290.CD-15-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayward N.K., Wilmott J.S., Waddell N., Johansson P.A., Field M.A., Nones K., Patch A., Kakavand H., Alexandrov L.B., Mann G.J., et al. Whole-genome landscapes of major melanoma subtypes. Nature. 2017;545:175–180. doi: 10.1038/nature22071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to ethical/privacy restrictions.