Abstract
In recent years, much progress has been made in elucidating the functional roles of plant glycine-rich RNA-binding proteins (GR-RBPs) during development and stress responses. Canonical GR-RBPs contain an RNA recognition motif (RRM) or a cold-shock domain (CSD) at the N-terminus and a glycine-rich domain at the C-terminus, which have been associated with several different RNA processes, such as alternative splicing, mRNA export and RNA editing. However, many aspects of GR-RBP function, the targeting of their RNAs, interacting proteins and the consequences of the RNA target process are not well understood. Here, we discuss recent findings in the field, newly defined roles for GR-RBPs and the actions of GR-RBPs on target RNA metabolism.
Keywords: glycine-rich RNA-binding protein, plant growth and development, stress responses, RNA immunoprecipitation, RNA post-transcriptional regulation
1. Introduction
An increasing occurrence of extreme weather events (heatwaves, drought, torrential rains), together with dire projections regarding climate change, makes the improvement of crop resilience to environmental and pathogen stress of paramount importance for feeding a growing global population [1]. To this end, a greater understanding of molecular mechanisms involving stress tolerance genes is essential for genetic improvement of crop species.
One protein family, which has been linked to stresses such as cold, wounding, UV radiation, salinity and pathogen infection, is the glycine-rich RNA-binding proteins (GR-RBPs), which is a sub-family within the larger glycine-rich protein (GRP) superfamily [2]. GR-RBPs are widely distributed in organisms ranging from prokaryotes to eukaryotes, and in the plant kingdom, they have been identified in Arabidopsis thaliana, tobacco (Nicotiana tabacum), rice (Oryza sativa), maize (Zea mays), sweet potato (Ipomoea batatas), Camelina sativa and others [3,4,5,6,7,8]. GR-RBPs are critical for RNA processing and metabolism. In the last two years, more and more evidence that GR-RBPs can affect growth and development of different plant species has also been published. Processes of crucial importance for growth and development, such as the regulation of transcription and post-transcriptional modifications, are known to require the assistance of RNA-binding proteins (RBPs), however, in which capacity is not yet clear. GR-RBPs contain a canonical RNA recognition motif (RRM) or a cold-shock domain (CSD) at the N-terminus and a glycine-rich region at the C-terminus, and are involved in several processes, including pre-mRNA splicing, nucleocytoplasmic transport of RNA and RNA editing. GR-RBPs shuttling between the nucleus and the cytoplasm have been shown to bind their target transcripts in the nucleus to influence their processing, and so to understand the biological roles of GRPs in plants, the key is to find their direct target RNA and RNA-binding sites [9]. Recently, new technologies such as RNA immunoprecipitation (RIP) and UV crosslinking immunoprecipitation (CLIP) combined with high-throughput sequencing have been used to find target RBP RNAs and RNA-binding sites [10].
In summary, the biological functions of GR-RBPs have been described in various species, with special focus on regulation of gene expression, and RBPs are known to be involved in RNA mechanisms and several defense pathways; despite this, little is known about their structure/function relationship. To our knowledge, this is the first compilation of available data regarding the participation of plant GR-RBPs in various biological functions, such as development and stress responses, using different key technologies.
2. The GR-RBP Family
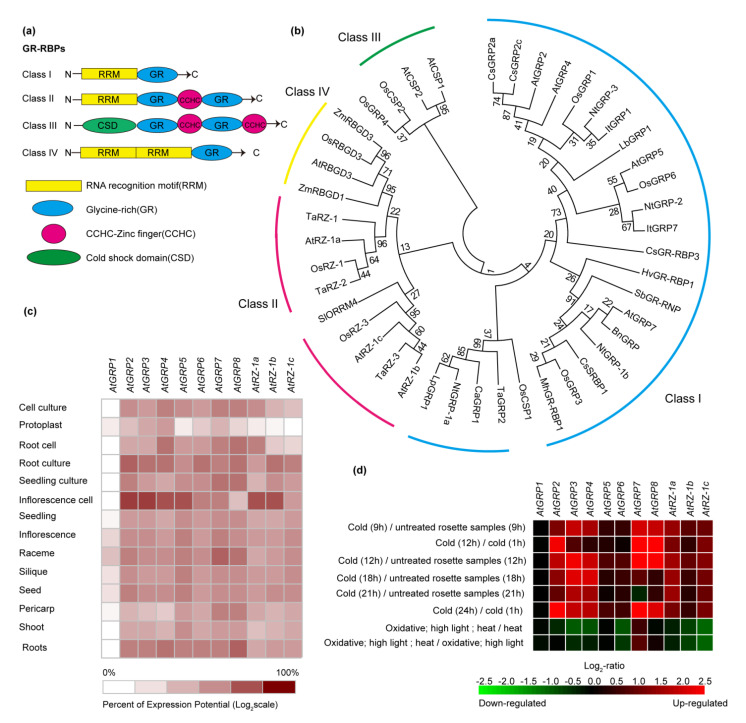
Plant glycine-rich proteins (GRPs) are characterized by a high content of glycine (Gly, 20–70%) and have been categorized into five classes, based on the presence of additional motifs and the arrangement of Gly repeats [11]. Among them, members of class IV, known as GR-RBPs, are particularly abundant in plants [12]. A canonical GR-RBP structure is shown in Figure 1a. This class is further divided into four subclasses (denoted I–IV). Subclass I contains one RNA recognition motif (RRM) domain as well as a glycine-rich (GR) motif, while subclass IV contains two RRM domains as well as the GR motif. The other two classes (II, III) both contain CCHC zinc-finger motifs, and are referred to as zinc finger-containing glycine-rich RNA-binding proteins (RZs). These have been found in Arabidopsis thaliana and rice (Oryza sativa) [5,11,13]. The biggest difference between these groups lies in the cold-shock domain (CSD), which is present only in subclass III. According to Table 1, the amount of subclass I in genomes of various plant species was much bigger than any other subclass (Figure 1b). Furthermore, we analyzed 11 GR-RBPs in different tissues and organs of Arabidopsis thaliana. As can be seen from Figure 1c, AtGRP1 was weakly expressed in all organs. However, the expression levels of the other genes were the highest in inflorescence cells and the lowest in protoplast.
Figure 1.
(a) Schematic representation of domain structures in plant glycine-rich RNA-binding proteins (GR-RBPs). Members of class I have an RNA recognition motif (RRM) at the N-terminus and a glycine-rich region at the C-terminus. Class II proteins, which are also called zinc finger-containing glycine-rich RNA-binding proteins (RZs), contain an RRM and a glycine-rich region separated by a CCHC-type zinc finger motif. Members of class III have an N-terminal cold shock domain (CSD) and a C-terminal glycine-rich region with two or more zinc finger motifs while class IV proteins have two RRMs and a glycine-rich region at the C-terminus. (b) Phylogenetic tree based on the alignment of protein sequences of GR-RBPs from sixteen plant species. These GR-RBPs are divided into four subclasses (denoted I–IV). Phylogenetic analyses were conducted in MEGA5. Species names are abbreviated as follows: Sl, Solanum lycopersicum; Bn, Brassica napus; Nt, Nicotiana tabacum; At, Arabidopsis thaliana; Cs, Camelina sativa; Ca, Capsicum annuum; Sb, Sorghum bicolor; Mh, Malus hupehensis; Hv, Hordeum vulgare; Cs, Cucumis sativa; Ca, Capsicum annuum; Os, Oryza sativa; Lp, Lolium perenne; Lb, Limonium bicolor; Ta, Triticum aestivum; It, Ipomoea trifida. (c) Expression profile of GR-RBPs in different tissues of Arabidopsis thaliana. (d) Expression profile of GR-RBPs from Arabidopsis thaliana under different treatments.
Table 1.
Number of predicted genes for each GR-RBP subclass in various plants.
| Plant | Subclass I | Subclass II | Subclass III | Subclass IV | Total |
|---|---|---|---|---|---|
| Arabidopsis thaliana | 8 | 3 | 2 | 5 | 18 |
| Oryza sativa | 6 | 3 | 2 | 4 | 15 |
| Zea mays | 6 | 6 | 2 | 9 | 23 |
| Theobroma cacao | 6 | 0 | 0 | 9 | 15 |
|
Brassica rapan L. ss P. pekinensis |
15 | 4 | 5 | 6 | 30 |
| Gossypium raimondii | 11 | 6 | 6 | 9 | 32 |
| Gossypium arboreum | 14 | 4 | 7 | 12 | 37 |
3. Roles of GR-RBPs in Plant Growth and Development
Three distinct GR-RBP functions or development-associated expression patterns have been distinguished: stress-related seed germination, vegetative growth; and those related to flowering time and fruit ripening.
3.1. GR-RBPs in Stress-Related Seed Germination
Many GR-RBPs are known to play roles in stress-related seed germination (Table 2). The Arabidopsis genome encodes eight GR-RBPs in subclass I, and it includes three members which are involved in stress-related seed germination [14,15,16,17]. For example, AtGRP7 and AtGRP2 from A. thaliana have been shown to accelerate seed germination and seedling growth under low-temperature conditions [14,18]. In contrast, AtGRP4-overexpressing seeds were reported to have delayed germination during high-salt or dehydration stress [19]. Similarly, the rice genome encodes six GR-RBPs, in which only two members (OsGRP1 and OsGRP4) promoted seed germination and seedling growth under low temperatures [5,19]. The Arabidopsis AtRZ-1a, a zinc finger-containing glycine-rich RNA-binding protein, has a different function in the modulation of seed germination from AtRZ-1b and AtRZ-1c [20]. Overexpression of AtRZ-1a seeds were reported to have delayed germination, while the AtRZ-1a mutant resulted in accelerated seed germination under salt and freezing treatments [21]. In addition, AtRZ-1a suppressed seed germination through an abscisic acid (ABA)-dependent pathway [21]. By contrast, knocking out AtRZ-1b and AtRZ-1c expression suppressed the seed germination rate compared to WT [22]. Interestingly, among the three RZ family members present in wheat genomes, all three TaRZ-overexpressing transgenic Arabidopsis seeds showed delayed germination compared to WT under salt stress conditions. However, overexpression of Triticum aestivum TaRZ-2 and TaRZ-3 in A. thaliana resulted in retarded seed germination under dehydration stress conditions [23]. Furthermore, overexpression of Malus prunifolia MpGR-RBP1 in A. thaliana resulted in accelerated seed germination under NaCl treatment [24]. Organelle RNA recognition motif-containing (ORRM) proteins are RNA editing factors in plants. In A. thaliana, ORRM3, ORRM4 and ORRM5 were previously characterized as GR-RBP3, GR-RBP5 and GR-RBP2 respectively, based on the presence of an RRM domain as well as the GR motif [25]. GR-RBP2 (ORRM5) was reported to accelerate seed germination under cold stress conditions, and Kwak et al. (2005) demonstrated that overexpressing GR-RBP5 (ORRM4) in A. thaliana suppressed seed germination compared to WT plants under salt or dehydration stress conditions [19]. In summary, these studies demonstrate diverse functional roles for plant GR-RBPs in stress-related seed germination.
Table 2.
Description of the identified plant glycine-rich RNA binding proteins.
| Gene | Subclass | Gene Source | Growth Phenotype | Roles | References |
|---|---|---|---|---|---|
|
AtGRP2
ORRM5 |
I | Arabidopsis thaliana | Under cold stress accelerate seed germination and seedling growth, under high salt and dehydration stress conditions affect plants growth and stress tolerance, slow growth and late flowering |
Overexpressing enhances freezing tolerance of Arabidopsis plants, AtGRP2 in inhibition of the early stages of ZYMV infection, mitochondrial RNA editing | [14,25,26,27,28,29] |
| AtGRP4 | I | Arabidopsis thaliana | Overexpressing seeds delayed germination during high salt or dehydration stress | The transcripts increase under cold stress, and downregulated by high salinity and dehydration stress | [18,19] |
| AtGRP7 | I | Arabidopsis thaliana | Affect the growth and stress tolerance of A. thaliana plants, influence flowering time, overexpressing in rice shows higher recovery rates and grain yields |
The transcripts increase significantly under cold stress, influence AS or polyadenylation, influence mRNA export from the nucleus to the cytoplasm under cold stress conditions, |
[18,27,28,30,31,32,33,34,35] |
| involvement in plant defenses, such as Pseudomonas syringae and TMV | |||||
|
ORRM4
GR-RBP5 |
I | Arabidopsis thaliana | Slow growth and late flowering | Mitochondrial RNA editing | [19,36] |
| SlORRM4 | I | Solanum lycopersicum | Delayed tomato fruit ripening | Mitochondrial RNA editing | [37,38] |
| AtRZ-1a | III | Arabidopsis thaliana | Enhances tolerance to cold stress in A. thaliana | Overexpressing in salt and cold stress retards seed germination | [21,22] |
| AtRZ-1b | III | Arabidopsis thaliana | Enhances tolerance to cold stress, AtRZ-1b and AtRZ-1c knockout mutants delayed seed germination, reduced stature and serrated leaves | Promote efficient splicing of FLC introns and repress FLC transcription | [22,39] |
| AtRZ-1c | III | Arabidopsis thaliana | AtRZ-1b and AtRZ-1c knockout mutants delayed seed germination, reduced stature and serrated leaves | Promote efficient splicing of FLC introns and repress FLC transcription | [22,39] |
| TaRZ - 2 | III | Triticum aestivum | Overexpressing retards seed germination under dehydration stress condition | Not determined | [23] |
| TaRZ - 3 | III | Triticum aestivum | Overexpressing retards seed germination under dehydration stress condition | Not determined | [23] |
| OsGRP1 | I | Oryza sativa | Overexpressing suppresses the dwarf phenotype of Arabidopsis bri1-5 mutant, under low temperatures promotes seed germination and seedling growth | Promotes cell expansion and elongation, enhances freezing tolerance | [5,40] |
| OsGRP4 | I | Oryza sativa | Under low temperatures promotes seed germination and seedling growth | Enhances tolerance to cold stress | [5] |
| OsGRP6 | I | Oryza sativa | Not determined | Enhances tolerance to cold stress | [5] |
| OsRZ-2 | III | Oryza sativa | Under low temperatures rescues grp7- knockout plants | Not determined | [13] |
| NtGRP-1a | I | Nicotiana tabacum | Not determined | Upregulation of abundance during heat and drought stress | [4] |
| NtGRP-3 | I | Nicotiana tabacum | Not determined | Upregulation of abundance during heat and drought stress | [4] |
| BnGRP1 | I | Brassica napus | Under cold stress accelerates seed germination | Enhances tolerance to cold stress | [41] |
| CsGRP2 | I | Camelina sativa | Complement cold-sensitive mutants at low temperatures | Upregulation of cold stress | [8] |
| CsGRP3 | I | Cucumis sativ a | Overexpressing contributes to cold and freezing stress tolerance | Not determined | [42] |
| LpGRP1 | I | Lolium perenne | Not determined | Upregulation of cold stress | [43] |
|
ItGRP1
ItGRP5 ItGRP7 |
I | Ipomoea trifida | Not determined | Upregulation of heat stress | [7] |
| LbGRP1 | I | Limonium bicolo r | Not determined | Improves tolerance to salt stress | [44] |
| SbGR-RNP | I | Sorghum bicolor | Not determined | Upregulation of salt stress | [45] |
| ZjGRP | I | Zoysia japonica | Overexpressing increases salt sensitivity in A. thaliana | Not determined | [46] |
| MhGR-RBP1 | I | Malus hupehensis | Not determined | Upregulation of salt stress | [47] |
| CsSRBP1 | I | Cucumis sativus | Not determined | Inhibits the initial stages of Zucchini yellow mosaic virus (ZYMV) infection | [29] |
| TaGRP2 | I | Triticum aestivum | Not determined | Inhibits transcript accumulation of TaVRN1 | [48] |
| HvGR-RBP1 | I | Hordeum vulgare | Not determined | Involvement in the timing of anthesis, senescence and levels of grain protein | [49] |
| CaGRP1 | I | Capsicum annuum | Not determined | Resistance to Xanthomonas campestris pv vesicatoria (Xcv) infection | [50] |
3.2. GR-RBP Function during Vegetative Growth
Studies have identified a role for RBPs in rice, where partial loss of RBP-P, a protein with two RRMs and a GR domain at the C-terminus, was reported to cause a broad range of phenotypes, including dwarfism, chlorophyll deficiency, sterility, late flowering and low spikelet fertility [51]. Similarly, a report by Tian et al. (2019) highlighted that the RBP-L knockout mutant also caused dwarfism, late flowering and smaller seeds in rice [52]. In addition, loss of function of AtRZ-1b and AtRZ-1c conferred defective phenotypes, including delayed seed germination, reduced stature and serrated leaves [39]. Notably, OsGRP1 was shown to promote cell expansion and elongation, and Wang et al. (2010) demonstrated that when overexpressing OsGRP1 in the A. thaliana brassinosteroid-insensitive mutant, bri1-5 partially suppressed its dwarf phenotype [40]. Furthermore, Staszak and Pawlowski (2014) suggested that a glycine-rich RNA-binding protein from Norway maple (Acer platanoides) may be directly involved in seed dormancy acquisition control [53].
3.3. GR-RBPs and Reproductive Growth
GR-RBPs have also been linked to the control of flowering time. In A. thaliana, partial loss of AtGRP7 function resulted in late flowering and more rosette leaves when bolting compared to the wild-type control [30]. Additionally, in barley (Hordeum vulgare), HvGR-RBP1 participates in the control of the timing of anthesis and senescence and levels of grain protein [49]. Moreover, in wheat (Triticum aestivum), overexpression of TaGRP2 resulted in late flowering compared with the wild type (WT) [48]. Furthermore, ORRM4 (GR-RBP5) and ORRM5 (GR-RBP2) are required for normal development, as A. thaliana loss-of-function mutants exhibit slower growth and delayed flowering time owing to defective mitochondrial editing [26,36,54], Finally, loss of tomato SlORRM4 was reported to cause a delay in fruit ripening and defects in mitochondrial RNA editing, presumably as a consequence of significant changes in the editing of target RNAs [37].
4. GR-RBPs and Stress Responses
4.1. Temperature Treatments
GRP expression is known to be affected by different environmental stresses (e.g., cold, drought and salinity) in a number of plant species. Examples of studies describing their responses to altered temperatures are presented below. Notably, numbers of GR-RBPs responded sensitively to cold treatment in Arabidopsis leaves and roots. In subclass I, the level of AtGRP2, AtGRP4 and AtGRP7 transcripts increased significantly due to a decrease in the temperature. It can be seen from Figure 1d that in the cold treatment of multiple groups of experiments, the expression of AtGRP2, AtGRP4 and AtGRP7 was significantly upregulated. In addition, Kim et al. (2007) reported that overexpressing AtGRP2 enhanced freezing tolerance of Arabidopsis plants compared with the WT and grp2-knockout mutants [18]. Interestingly, heterologous expression of AtGRP7 was seen to enhance growth in a cold-sensitive Escherichia coli mutant under low-temperature conditions [31]. Kim et al. (2010) observed that two members of the zinc finger-containing GR-RBPs, AtRZ-1a and AtRZ-1b, enhanced cold and freezing tolerance in A. thaliana [22]. However, AtRZ-1c appeared unable to complement cold sensitivity in the E.coli BX04 mutant [22]. Furthermore, overexpression of two cold-shock domain proteins (AtCSDP1 and AtCSDP2) was shown to rescue cold-sensitive AtGRP7 mutant plants from freezing damage [32]. Kim et al. (2010) reported that rice GRPs (OsGRP1, OsGRP4 and OsGRP6) contribute to the enhancement of cold and freezing tolerance, while three OsRZ proteins (OsRZ-1, OsRZ-2 and OsRZ-3) of OsRZ-2 have the ability to rescue grp7-knockout plants from cold conditions, but not OsRZ-1 and OsRZ-2 [13]. Furthermore, expression of BnGRP1 from Brassica napus in A. thaliana was shown to result in accelerated seed germination and enhanced freezing tolerance grown under cold or freezing conditions [41]. Finally, the expression of three CsGRP2 genes (CsGRP2a, CsGRP2b and CsGRP2c) from Camelina sativa L. was highly upregulated under cold stress and when heterologously expressed in Escherichia coli and had the ability to complement cold-sensitive mutants at low temperatures [8]. Other insights into roles of GR-RBPs in chilling responses came from studies of harvested cucumber (Cucumis sativus) fruit, where overexpressing CsGR-RBP3 in Arabidopsis lines was shown to contribute to cold and freezing stress tolerance [42]. A role in cold acclimation has also been shown through studies of perennial ryegrass (Lolium perenne) in which GRP1 transcription was significantly increased [43]. GR-RBPs are also thought to contribute to heat tolerance: SbGRBP from Sorghum bicolor expression is modulated by heat stress in seedlings [55] and the sweet potato genes ItGRP1, ItGRP5 and ItGRP7 are upregulated by heat stress [7]. The expression of NtGRP-1a and NtGRP-3 in tobacco was strongly upregulated, while the expression of NtRGP-1b was unaffected by high-temperature treatment [4].
4.2. Salinity Stress
Salinity stress is one of the most significant factors limiting agricultural crop productivity [56] and improving crop salt tolerance is recognized as being essential for future sustainable food production. GR-RBPs have also been associated with salinity stress responses. As examples, overexpressing Limonium bicolor LbGRP1 in tobacco improved its salt stress tolerance [44], and under 500 mM NaCl and 10 µm ABA treatments, the transcript levels of S. bicolor SbGR-RNP increased between four- and seven-fold [45]. A Zoysia japonica salt-induced glycine-rich RNA-binding protein, ZjGRP, resulted in increased salt sensitivity in A. thaliana when overexpressed [46]. Additionally, both AtGRP7 and AtGRP2 affect the growth and stress tolerance of A. thaliana plants under high salt and dehydration stress conditions [27,33], while the expression of AtGRP4 was downregulation by high salinity or dehydration stress. Finally, MhGR-RBP1 from Malus hupehensis was reported to show a doubling of transcript levels in leaves 2 days after salt treatment [47]. Kwak et al. (2013) reported that the expression of CsGRP2a was highly upregulated, whereas the transcript levels of CsGRP2b and CsGRP2c were decreased under salt stress conditions [8].
4.3. Drought Stress
Drought stress is one of the major abiotic stresses having a direct impact on plant growth and limiting plant productivity [57,58]. Many scientific studies have focused on the development of drought-tolerant crop plants. As examples, overexpressing AtGRP2 or AtGRP7 in rice showed much higher recovery rates and grain yields compared with WT under drought stress conditions [28]. The expression of NtGRP-1a and NtGRP-3 in tobacco was weakly upregulated under drought stress [4].
4.4. Biotic Stress
Plant virus-induced diseases of crops are of great importance to the agricultural industry due to the deleterious effects on product quality and yield [59]. Genetic resistance represents one approach to protect crops from viral infections [60] and this has been a motivating factor for studies of resistance genes.
Numerous studies have suggested that GR-RBP genes are likely involved in plant defenses. A GR-RBP from cucumber, CsRBP1, has been shown to inhibit the initial stages of Zucchini yellow mosaic virus (ZYMV) infection [29]. Another GRP, AtGRP7 from A. thaliana, has been shown to respond to challenge by Pseudomonas syringae, a necrotrophic bacterium (Pectobacterium carotovorum SCC1; formerly Erwinia carotovora SCC1), a necrotrophic fungus (Botrytis cinerea) and a biotrophic virus (Tobacco mosaic virus, TMV) [61]. In addition, a report by Yan et al. (2019) highlighted the importance of AtGRP7, AtGRP8 and AtGRP2 in inhibition of the early stages of ZYMV infection, and in particular the importance of the GR domain, which was shown to be both necessary and sufficient to block the early stages of infection [29].
An inverse correlation between expression of genes encoding GR-RBPs and virus resistance has also been reported. Huang et al. (2019) demonstrated that knockdown of GR-RBP NgRBP in Nicotiana glutinosa by virus-induced gene silencing enhanced Potato virus X (PVX) and Cucumber mosaic virus resistance [62]. In addition, a gene encoding GR-RBP2 was found to be highly expressed in a susceptible maize line associated with host hypersensitivity and susceptibility [63]. Finally, a study by Kim et al. (2015) supported silencing CaGRP1 in pepper (Capsicum annuum) for enhanced resistance to Xanthomonas campestris pv vesicatoria (Xcv) infection [50]. Furthermore, CaGRP1 interacts with RECEPTOR-LIKE CYTOPLASMIC PROTEIN KINASE 1 (CaPIK1). These phosphorylated CaGRP1 and CaGRP1/CaPIK1 complexes suppress the expression of pathogen-related genes and negatively regulate CaPIK1-triggered cell death and defense responses.
5. Technologies Used to Investigate RNA–GR-RBP Interactions
The interaction between RNA-binding proteins and their target RNAs is a key factor in their fate and an important challenge in understanding the biological function of RNA-binding proteins is identifying the RNA targets in a cellular context. To date, the predominant methods for identifying unknown RNA targets have included protein immunoprecipitation, including RNA immunoprecipitation (RIP), individual nucleotide resolution crosslinking and immunoprecipitation (iCLIP), and high-resolution RIP-seq. Here, we describe three strategies for identifying RNA targets of GR-RBPs in plants.
5.1. RIP-qPCR
RIP is an antibody-based technique used to identify interactions between proteins and RNA targets in vivo. Specific RNA–protein complexes can be immunoprecipitated from a cellular lysate with an antibody raised against the protein of interest. RNA immunoprecipitation can be divided into two main classes: native and crosslinked.
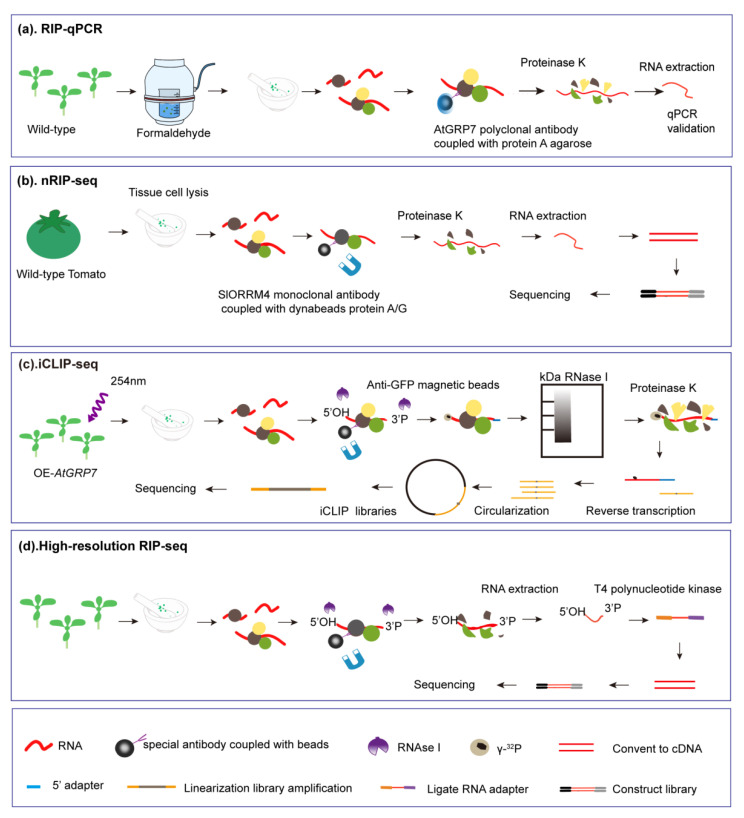
There are two methods for crosslinked RNA immunoprecipitation: irreversible UV crosslinking and reversible formaldehyde crosslinking [64]. Xiao et al. (2015) demonstrated that AtGRP7 binds to antisense FLC premRNA in vivo by RIP-qPCR using the crosslinkage of Arabidopsis seedling tissue (Figure 2a) [30]. Furthermore, Xiao et al. (2014) reported that TaGRP2 directly binds TaVRN1 pre-mRNA with conserved binding sites through RIP [48].
Figure 2.
Workflow of RIP-qPCR, native RIP-seq, iCLIP and high-resolution RIP-seq analyses of plants. (a) RIP with a AtGRP7-specific antibody followed by qPCR to identify the binding status of AtGRP7 at the FLC locus. Point mutation of R49Q (Arg to Gln at the 49th amino acid) in the RRM motif was used as a negative control. Arabidopsis seedling tissue was cross-linked by 0.5% (v/v) formaldehyde. After tissue lysis, immunoprecipitation was performed using anti-AtGRP7 antibodies coupled with protein A agarose. Then, the protein was degraded by proteinase K, and RNA was extracted by acidic phenol/chloroform. The percentage of RIP-enriched RNA relative to that of input sample was determined by qRT-PCR. (b) For native RIP, tomato fruit (36 days post-anthesis (DPA)) cells were directly lysed and immunoprecipitation was performed using anti-SlORRM4 antibodies coupled with dynabeads protein A/G and proteins were digested using proteinase K. RNA extraction was performed and analyzed by next-generation sequencing. The negative control was IP from slorrm4 mutant with anti-SlORRM4. (c) The GFP-tagged AtGRP7 was expressed in the grp7 mutant. Plant materials used for iCLIP were subjected to crosslinking with UV light 254 nm wavelength. After sample homogenization in liquid N2, a lysate was prepared and RNA–protein complexes were precipitated using specific antibodies coupled with magnetic beads. RNAs were fragmented by treatment with RNAse I and the fragments were radioactively labeled at the 5′ end only. After proteins were digested and RNAs were isolated, iCLIP sequencing libraries were prepared. In parallel, the negative control libraries were immunoprecipitated for GFP-only transgene plants and AtGRP7 R49Q-GFP plants. (d) For high-resolution RIP-seq, tissues extracts were pre-treated with RNAse I. Immunoprecipitation was then performed using a specific antibody that recognized the protein of interest. After the proteins were digested, the RNA was phosphorylated using a T4 polynucleotide kinase and processed for sequencing using a Small RNA-seq Kit.
Native RIP assays involve identification of RNA targets directly bound in the immunoprecipitated sample, while crosslinked RIP assays involve detection of direct and indirect binding sites on RNA targets (Figure 2b). RNA immunoprecipitation under native conditions without crosslinking combined with high-throughput sequencing (nRIP-seq) is a powerful technique that facilitates the direct target coding and non-coding RNAs associated with a particular protein [65]. Recently, nRIP-Seq analysis revealed that SlORRM4 is directly associated with 31 transcripts, 21 coding genes and 10 tRNAs, which were dramatically enriched in the immunoprecipitation of SlORRM4 (Figure 2b) [38]. The nRIP-Seq technique can find the RNAs targeted to GR-RBPs directly, but it cannot verify the interface between GR-RBPs and target RNAs.
5.2. ICLIP-Seq
The identification of RNA-binding sites facilitates the understanding of the molecular function of RNA-binding proteins. The iCLIP protocol starts with UV irradiation and then RNA–protein complexes are immunoprecipitated. After RNAse I digestion, the RNA 5′ ends are labeled with [γ-32P] ATP using a polynucleotide kinase and RNA–protein complexes are transferred to nitrocellulose membranes [9]. Finally, the associated RNAs are reverse transcribed into cDNAs, which are then PCR-amplified to generate libraries that are subjected to high-throughput sequencing (Figure 2c). In one study, the iCLIP approach revealed 858 transcripts, showing changes due to alternative splicing (AS) or polyadenylation in atgrp7 knockout and atgrp8 knockdown mutants, compared to AtGRP7-overexpressing plants [34]. FATTY ACID DESATURASE 2 (FAD2), which has been shown to contribute to salt tolerance, was identified as a target RNA for AtGRP7 by iCLIP, and it was reported that overexpression of AtGRP7 in A. thaliana had a negative effect on germination and seedling growth under salt stress conditions due to FAD2 downregulation [34]. The disadvantage of this technique is that the irreversible UV crosslinking leads identification of RNA-binding sites to include both direct and indirect binding sites on RNA targets. In summary, these studies demonstrate that identification of the RNA target is critical for demonstration of the molecular regulatory mechanism underlying the effect of GR-RBPs on plant growth and development.
5.3. High-Resolution RIP-Seq
Recently, it was reported that modifying the standard RIP-seq protocol by adding a ribonuclease I digestion step after the chloroplast extraction step, but prior to antibody addition, led to the identification of an RNA-binding protein (HCF173) interaction with the target mRNA-binding site in A. thaliana (Figure 2d) [66]. In this case, the protein binding site was too short or had too low affinity to be identified with this method. However, this technology can be used to effectively explore the functional roles of GR-RBPs in gene regulation in the future.
6. Post-Transcriptional RNA Regulation by GR-RBPs
RBPs are chaperones that bind RNA via one or multiple RRM domains, thereby changing the function or fate of the RNA targets. Here, we provide examples of the involvement of GR-RBPs in plant RNA metabolism, which collectively underscore the complexity of in vivo RNA-mediated processes (Figure 3).
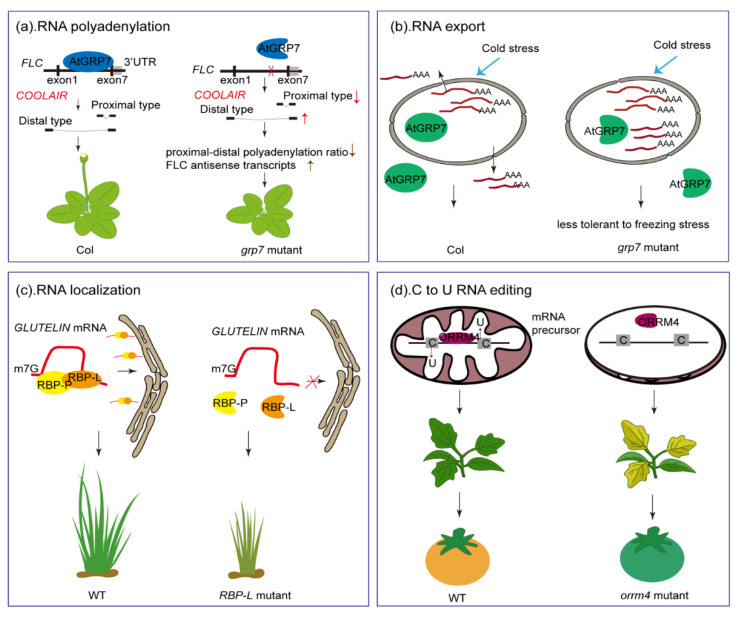
Figure 3.
Cellular functions of diverse GR-RBPs involved in RNA metabolism during plant growth, development and stress responses. (a) AtGRP7 affects the polyadenylation site usage of COOLAIR transcripts, leading to an altered ratio of proximally–distally spliced variants. Loss of AtGRP7 function leads to increased abundance of FLC antisense transcripts and a reduced proximal–distal polyadenylation ratio, resulting in late flowering compared with the wild type. (b) AtGRP7 is located in the nucleus and cytoplasm and is involved in mRNA export from the nucleus to the cytoplasm under cold stress conditions. In grp7 mutants, the export of mRNA is impaired, leading to its accumulation in the nucleus, while mRNAs transcribed in the nucleus are efficiently exported to the cytoplasm in wild-type plants subjected to cold stress. (c) An insertional allele in rice, RBP-L, causes growth defects due to the loss of RBP and the consequent mislocalization of GLUTELIN target RNA. (d) Knocking out the mitochondrial RNA editing factor ORRM4 in tomato causes defective mitochondrial editing, leading to yellowish seedlings and delayed fruit ripening.
6.1. GR-RBP Function in AS and Polyadenylation
AtGRP7 has been shown to affect AS and regulate pre-mRNA splicing in A. thaliana by binding to the target pre-mRNA and generating an intron-terminating alternatively spliced transcript, which is unstable and degraded through the nonsense-mediated decay pathway [35]. In a subsequent study, overexpression of AtGRP7 was found to result in significant changes in the ratios of AS isoforms in 59 out of 288 analyzed AS events [67].
Cold-induced long antisense intragenic RNAs (COOLAIR) originate from the promoter adjacent to the FLOWERING LOCUS C (FLC) 3′UTR. Two structural variants of COOLAIR have been identified, terminating at proximal (sense intron 6) or distal (sense promoter) sites to repress FLC expression following cold exposure [68]. AtGRP7 has been shown to bind to FLC antisense pre-mRNA and the binding site is close to the polyadenylation site of the proximal COOLAIR structural type. Loss of AtGRP7 function was observed to cause decreased levels of proximal type ASI (type I/total) and increased levels of distal type ASII (type II/total) COOLAIR to be expressed. The reduced proximal–distal polyadenylation ratio resulted in an increase in the total abundance of the functional sense FLC transcript and consequently influenced flowering time (Figure 3a) [30]. Wu et al. (2016) demonstrated that AtRZ-1c bound to FLC, promoting efficient splicing of FLC introns and repressing FLC transcription [39]
6.2. The Role of GR-RBPs in RNA Export
To experimentally validate mRNA subcellular localization, poly (A) in situ hybridization assays were performed in wild-type and grp7 mutant plants. Strong fluorescence signals were observed in the nuclei of leaf cells in grp7 plants subjected to cold stress while no noticeable fluorescence signals were present in the wild type [69], indicating thatAtGRP7 plays a role in mRNA export from the nucleus to the cytoplasm under cold stress conditions (Figure 3b) [69]. Through Northern blot analysis, Yan et al. (2019) showed that AtGRP7 binds ZYMV viral siRNA (vsiRNA), thus implicating it in cell-to-cell delivery of siRNA, leading to activation of the RNAi pathway in neighboring cells [29].
6.3. RNA Localization
RNA-binding proteins have been shown to play critical roles in transporting mRNA to specific subcellular locations for localized translation. Recent studies identified two RBPs, RBP-P and RBP-L, containing two and three RRM domains and a GR domain at the C-terminal, respectively [51,52], which specifically bound to the glutelin zipcode mRNA sequences, thereby regulating glutelin mRNA localization (Figure 3c) [51,52]. Glutelin RNAs are asymmetrically distributed on distinct endoplasmic reticulum (ER) subdomains of the cortical–ER complex [52].
6.4. The Role of GR-RBPs in C-to-U RNA Editing
RNA editing allows modification of genetic information at the transcriptional level by inserting, knocking out and replacing RNA base sequences, and C (cytidine) to U (uridine) RNA editing in plants occurs in chloroplast and mitochondrial transcripts [70]. It was reported that the knockout of ORRM4 (glycine-rich RNA-binding protein 5), which functions as a major mitochondrial editing factor, in A. thaliana caused defective mitochondrial editing in 44% of the mitochondrial sites surveyed [36]. In addition, knocking out ORRM5 expression resulted in an increase of the editing extent in 14% of the mitochondrial sites [26]. The knockout orrm5 mutant reduced the splicing efficiency of the first nad5 intron, causing slower growth [26]. Furthermore, SlORRM4 from tomato was also shown to be a key RNA editing factor and responsible for 61% of the 552 C-to-U editing events in mitochondria. SlORRM4-mediated RNA editing events normally cause about 56% of the expressed mitochondrial genes to develop missense mutations, and defects in RNA editing in the slorrm4 mutant had a significant effect on the expression levels of these mitochondrial genes [38]. Moreover, using nRIP-seq, 19 SlORRM4 RNA targets were identified, which were mostly subunits of mitochondrial respiratory chain complexes [38]. Defective RNA editing in the slorrm4 mutant resulted in yellowish seedlings and a major delay in the initiation of tomato fruit ripening (Figure 3d) [38].
7. Conclusions and Discussion
GR-RBPs are involved in a range of abiotic and biotic stress responses, including cold adaptation and responses to pathogens. Czolpinska et al. (2018) demonstrated that GRPs play important roles in responses to diverse stress conditions [71]. In this review, we systematically summarized GR-RBPs that are involved in RNA metabolism during plant growth and development. Importantly, identification of the RNA target is critical for demonstration of the molecular regulatory mechanism underlying the function of GR-RBPs. The RRM domain confers RNA-binding (and potentially DNA-binding) activity, allowing the regulation of genes with multiple roles. Additional regulatory roles for GR-RBPs include the regulation of pre-mRNA alternative splicing, mRNA export and mRNA localization. Importantly, members of the GR-RBP family have been shown in mammals to be extensively involved in various forms of neurodegenerative diseases and neurodevelopmental disorders. Unlike in plants, it has been reported that in mammals, GRPs not only regulate the transcription of target genes but also affect the translation efficiency of proteins. For instance, expression of RNA-binding motif 3 (Rbm3), a cold-induced member of the GRP family, can increase total protein synthesis under both physiological and mild hypothermic temperatures in N2a cells [72]. RNA-binding motif single-stranded interacting protein 3 (Rbms3), a glycine-rich RNA-binding protein, can bind to the 3′ untranslated region (UTR) of pancreas-associated transcription factor 1a (Ptf1a) mRNA, regulating the accumulation of the encoded Ptf1a protein to maintain pancreas development [73]. Among the many important unanswered questions regarding the role of GR-RBPs in plants is whether they are associated with translational regulation of their RNA targets. Biomacromolecules contain between tens and several thousands of protein and RNA species which aggregate to form membraneless organelles [74]. These condensates often contain RBPs with large intrinsically disordered regions (IDRs) that can promote phase separation [75]. Liquid–liquid phase separation occurs in diverse biological processes, including signal transduction, gene expression regulation, higher-order chromatin organization, cell division, etc. [76]. The study of protein phase separation is an emerging field in animal and yeast research but is still not common in plants [77]. Xie et al. (2020) showed that nuclear dicing bodies (D-bodies, containing DICER-LIKE 1, HYPONASTIC LEAVES 1 and SERRATE) are phase-separated condensates and essential for efficient miRNA processing [78]. Furthermore, Huang et al. (2021) reported that reactive oxygen species (ROS) regulate reversible protein phase separation to direct stem cell fate for flowering transition. Proteins contain large IDRs that are enriched in amino acids, such as glycine, serine, glutamine, asparagine, phenylalanine and tyrosine [79]. Therefore, many GR-RBPs are predicted by the presence of IDR sequences to be involved in phase separation under specific physiological conditions. Functional characterization of newly identified GR-RBPs and their RNA targets will pave the way for a better understanding of post-transcriptional and translational regulation of plant growth and development.
Author Contributions
L.M. and H.Z. designed the review and wrote the manuscript. C.Z. generated Table 1. L.M. generated Figure 1, Figure 2 and Figure 3 and Table 2. K.C., J.L. and Z.D. revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31972472 and 31672208) to H.Z.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bailey-Serres J., Parker J.E., Ainsworth E.A., Oldroyd E.G., Schroeder J.I. Genetic strategies for improving crop yields. Nature. 2019;575:109–118. doi: 10.1038/s41586-019-1679-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sachetto-Martins G., Franco L.O., Oliveira D.E.D. Plant glycine-rich proteins: A family or just proteins with a common motif? Biochimica. Biophys. Acta. 2000;1492:1–14. doi: 10.1016/S0167-4781(00)00064-6. [DOI] [PubMed] [Google Scholar]
- 3.Nocker S.V., Vierstra R.D. Two cDNAs from Arabidopsis thaliana encode putative RNA binding proteins containing glycine-rich domains. Plant Mol. Biol. 1993;21:695–699. doi: 10.1007/BF00014552. [DOI] [PubMed] [Google Scholar]
- 4.Chen X., Zeng Q.C., Lu X.P., Yu D.Q., Li W.Z. Characterization and Expression Analysis of Four Glycine-Rich RNA-Binding Proteins Involved in Osmotic Response in Tobacco (Nicotiana tabacum cv. Xanthi) Agric. Sci. China. 2010;9:1577–1587. doi: 10.1016/S1671-2927(09)60254-6. [DOI] [Google Scholar]
- 5.Kim J.Y., Kim W.Y., Kwak K.J., Oh S.H., Han Y.S., Kang H. Glycine-rich RNA-binding proteins are functionally conserved in Arabidopsis thaliana and Oryza sativa during cold adaptation process. J. Exp. Bot. 2010;61:2317–2325. doi: 10.1093/jxb/erq058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J., Zhao Y., Xiao H., Zheng Y., Yue B. Genome-wide identification, evolution, and expression analysis of RNA-binding glycine-rich protein family in maize. J. Integr. Plant Biol. 2014;56:1020–1031. doi: 10.1111/jipb.12210. [DOI] [PubMed] [Google Scholar]
- 7.Lu Y., Sun J., Yang Z., Zhao C., Xu T. Genome-wide identification and expression analysis of glycine-rich RNA-binding protein family in sweet potato wild relative Ipomoea trifida. Gene. 2018;686:177–186. doi: 10.1016/j.gene.2018.11.044. [DOI] [PubMed] [Google Scholar]
- 8.Kwak K.J., Kang H., Han K.H., Ahn S.J. Molecular cloning, characterization, and stress-responsive expression of genes encoding glycine-rich RNA-binding proteins in Camelina sativa L. Plant Physiol. Biochem. 2013;68:44–51. doi: 10.1016/j.plaphy.2013.03.023. [DOI] [PubMed] [Google Scholar]
- 9.Köster T., Reichel M., Staiger D. CLIP and RNA interactome studies to unravel genome-wide RNA-protein interactions in vivo in Arabidopsis thaliana. Methods. 2020;178:63–71. doi: 10.1016/j.ymeth.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 10.Huppertz I., Attig J., D’Ambrogio A., Easton L.E., Sibley C.R., Sugimoto Y., Tajnik M., Kônig J., Ule J. iCLIP: Protein–RNA interactions at nucleotide resolution. Methods. 2014;65:274–287. doi: 10.1016/j.ymeth.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mangeon A., Junqueira R.M., Sachetto-Martins G. Functional diversity of the plant glycine-rich proteins superfamily. Plant Signal. Behav. 2010;5:99–104. doi: 10.4161/psb.5.2.10336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krishnamurthy P., Kim J.A., Jeong M.J., Kang C.H., Lee S.I. Defining the RNA-binding glycine-rich (RBG) gene superfamily: New insights into nomenclature, phylogeny, and evolutionary trends obtained by genome-wide comparative analysis of Arabidopsis, Chinese cabbage, rice and maize genomes. Mol. Genet. Genom. 2015;290:2279–2295. doi: 10.1007/s00438-015-1080-0. [DOI] [PubMed] [Google Scholar]
- 13.Kim J.Y., Kim W.Y., Kwak K.J., Oh S.H., Kang H. Zinc finger-containing glycine-rich RNA-binding protein in Oryza sativa has an RNA chaperone activity under cold stress conditions. Plant Cell Environ. 2010;33:759–768. doi: 10.1111/j.1365-3040.2009.02101.x. [DOI] [PubMed] [Google Scholar]
- 14.Fusaro A.F., Bocca S.N., Ramos R.L.B., Barrôco R.M., Magioli C., Jorge V.C., Coutinho T.C., Rangel-Lima C.M., De Rycke R., Inzé D., et al. AtGRP2, a cold-induced nucleo-cytoplasmic RNA-binding protein, has a role in flower and seed development. Planta. 2007;225:1339–1351. doi: 10.1007/s00425-006-0444-4. [DOI] [PubMed] [Google Scholar]
- 15.Juntawong P., Sorenson R., Bailey-Serres J. Cold shock protein 1 chaperones mRNAs during translation in Arabidopsis thaliana. Plant J. 2013;74:1016–1028. doi: 10.1111/tpj.12187. [DOI] [PubMed] [Google Scholar]
- 16.Karlson D., Imai R. Conservation of the cold shock domain protein family in plants. Plant Physiol. 2003;131:12–15. doi: 10.1104/pp.014472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Y., Karlson D.T. Overexpression of AtCSP4 affects late stages of embryo development in Arabidopsis. J. Exp. Bot. 2011;62:2079–2091. doi: 10.1093/jxb/erq400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J.Y., Park S.J., Jang B., Jung C.H., Ahn S.J., Goh C.H., Cho K., Han O., Kang H. Functional characterization of a glycine-rich RNA-binding protein 2 in Arabidopsis thaliana under abiotic stress conditions. Plant J. 2007;50:439–451. doi: 10.1111/j.1365-313X.2007.03057.x. [DOI] [PubMed] [Google Scholar]
- 19.Kwak K.J., Kim Y.O., Kang H. Characterization of transgenic Arabidopsis plants overexpressing GR-RBP4 under high salinity, dehydration, or cold stress. J. Exp. Bot. 2005;56:3007–3016. doi: 10.1093/jxb/eri298. [DOI] [PubMed] [Google Scholar]
- 20.Kim Y.O., Kim J.S., Kang H. Cold-inducible zinc finger-containing glycine-rich RNA-binding protein contributes to the enhancement of freezing tolerance in Arabidopsis thaliana. Plant J. 2005;42:890–900. doi: 10.1111/j.1365-313X.2005.02420.x. [DOI] [PubMed] [Google Scholar]
- 21.Kim Y.O., Pan S., Jung C.H., Kang H. A zinc finger-containing glycine-rich RNA-binding protein, atRZ-1a, has a negative impact on seed germination and seedling growth of Arabidopsis thaliana under salt or drought stress conditions. Plant Cell Physiol. 2007;48:1170–1181. doi: 10.1093/pcp/pcm087. [DOI] [PubMed] [Google Scholar]
- 22.Kim W.Y., Kim J.Y., Jung H.J., Oh S.H., Han Y.S., Kang H. Comparative analysis of Arabidopsis zinc finger-containing glycine-rich RNA-binding proteins during cold adaptation. Plant Physiol. Biochem. 2010;48:866–872. doi: 10.1016/j.plaphy.2010.08.013. [DOI] [PubMed] [Google Scholar]
- 23.Xu T., Gu L., Choi M.J., Kim R.J., Suh M.C., Kang H. Comparative functional analysis of wheat (Triticum aestivum). zinc finger-containing glycine-rich RNA-binding proteins in response to abiotic stresses. PLoS ONE. 2014;9:e96877. doi: 10.1371/journal.pone.0096877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan Y., Qin Y., Li Y., Li M., Ma F. Overexpression of MpGR-RBP1, a glycine-rich RNA-binding protein gene from Malus prunifolia (Willd.) Borkh., confers salt stress tolerance and protects against oxidative stress in Arabidopsis. Plant Cell Tissue Organ Cult. 2014;119:635–646. doi: 10.1007/s11240-014-0563-8. [DOI] [Google Scholar]
- 25.Shi X.W., Bentolila S., Hanson M.R. Organelle RNA recognition motif-containing (ORRM) proteins are plastid and mitochondrial editing factors in Arabidopsis. Plant Signal. Behav. 2016;11:e1167299. doi: 10.1080/15592324.2016.1167299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi X.W., Castandet B., Germain A., Hanson M.R., Bentolil B. ORRM5, an RNA recognition motif-containing protein, has a unique effect on mitochondrial RNA editing. J. Exp. Bot. 2017;11:2833–2847. doi: 10.1093/jxb/erx139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ortega-Amaro M.A., Rodríguez-Hernández A.A., Rodríguez-Kessler M., Hernández-Lucero E., Rosales-Mendoza S., Ibáñez-Salazar A., Delagado-Sánchez P., Jiménez-Bremont J.F. Overexpression of AtGRDP2, a novel glycine-rich domain protein, accelerates plant growth and improves stress tolerance. Front. Plant Sci. 2015;5:782. doi: 10.3389/fpls.2014.00782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang D.H., Kwak K.J., Kim M.K., Park S.J., Yang K.Y., Kang H. Expression of Arabidopsis glycine-rich RNA-binding protein AtGRP2 or AtGRP7 improves grain yield of rice (Oryza sativa) under drought stress conditions. Plant Sci. Int. J. Exp. Plant Biol. 2014;214:106–112. doi: 10.1016/j.plantsci.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 29.Yan Y., Ham B.K., Chong Y.H., Yeh S.D., Lucas W.J. A Plant SMALL RNA-BINDING PROTEIN 1 Family Mediates Cell-to-Cell Trafficking of RNAi Signals. Mol. Plant. 2019;13:321–335. doi: 10.1016/j.molp.2019.12.001. [DOI] [PubMed] [Google Scholar]
- 30.Xiao J., Li C.H., Xu S.J., Xing L.J., Chong K. Jacalin-Lectin Like1 Regulates the Nuclear Accumulation of Glycine-Rich RNA-Binding Protein7, influencing the RNA Processing of Flowering Locus C Antisense Transcripts and Flowering Time in Arabidopsis. Plant Physiol. 2015;69:2102–2117. doi: 10.1104/pp.15.00801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim J.S., Park S.J., Kwak J.K., Kim Y.O., Kim J.Y., Song J.Y., Jang B., Jung C.H., Kang H. Cold shock domain proteins and glycine-rich RNA-binding proteins from Arabidopsis thaliana can promote the cold adaptation process in Escherichia coli. Nucleic Acids Res. 2007;35:506–516. doi: 10.1093/nar/gkl1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park S.J., Kwak K.J., Oh T.R., Kim Y.O., Kang H. Cold shock domain proteins affect seed germination and growth of Arabidopsis thaliana under abiotic stress conditions. Plant Cell Physiol. 2009;50:869–878. doi: 10.1093/pcp/pcp037. [DOI] [PubMed] [Google Scholar]
- 33.Cao S.Q., Jiang L., Song S.Y., Jing R., Xu G.S. AtGRP7 is involved in the regulation of abscisic acid and stress responses in Arabidopsis. Cell. Mol. Biol. Lett. 2006;11:526–535. doi: 10.2478/s11658-006-0042-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meyer K., KoSter T., Nolte C., Weinholdt C., Lewinski M., Grosse I., Staiger D. Adaptation of iCLIP to plants determines the binding landscape of the clock-regulated RNA-binding protein AtGRP7. Genome Biol. 2017;18:204. doi: 10.1186/s13059-017-1332-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Staiger D., Zecca L., Wieczorek-Kirk D.A., Apel K., Eckstein L. The circadian clock regulated RNA-binding protein AtGRP7 autoregulates its expression by infuencing alternative splicing of its own pre-mRNA. Plant J. 2003;33:361–371. doi: 10.1046/j.1365-313X.2003.01629.x. [DOI] [PubMed] [Google Scholar]
- 36.Shi X.W., Germain A., Hanso M.R., Bentolil B. RNA Recognition Motif-Containing Protein ORRM4 Broadly Affects Mitochondrial RNA Editing and Impacts Plant Development and Flowering. Plant Physiol. 2015;17:294–309. doi: 10.1104/pp.15.01280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Y.F., Zhu G.N., Li R., Yan S.J., Fu D.Q., Zhu B.Z., Tian H.Q., Luo Y.B., Zhu H.L. The RNA editing factor SlORRM4 is required for normal fruit ripening in tomato. Plant Physiol. 2017;175:1690–1702. doi: 10.1104/pp.17.01265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Y.F., Liu X.Y., Wang K.R., Li J.Y., Zhu G.N., Ren S., Deng Z.P., Zhu B.Z., Fu D.Q., Luo Y.B., et al. Molecular and functional diversity of organelle RNA editing mediated by RNA recognition motif-containing protein ORRM4 in tomato. New Phytol. 2020;228:570–585. doi: 10.1111/nph.16714. [DOI] [PubMed] [Google Scholar]
- 39.Wu Z., Zhu D.L., Lin X.Y., Miao J., Gu L.F., Deng X., Yang Q., Sun K.T., Zhu D.M., Cao X.F., et al. RNA Binding Proteins RZ-1B and RZ-1C play critical roles in regulating pre-mRNA splicing and gene expression during development in Arabidopsis. Plant Cell. 2016;28:55–73. doi: 10.1105/tpc.15.00949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang F.R., Bai M.Y., Deng Z.P., Oses-Prieto J.A., Burlingame A.L., Lu T.L., Chong K., Wang Z.Y. Proteomic Study Identifies Proteins Involved in Brassinosteroid Regulation of Rice Growth. J. Integr. Plant Biol. 2010;12:1075–1085. doi: 10.1111/j.1744-7909.2010.00992.x. [DOI] [PubMed] [Google Scholar]
- 41.Kim K.M., Jung H.J., Kim D.H., Kang H. Characterization of glycine-rich RNA-binding proteins in Brassica napus under stress conditions. Physiol. Plant. 2012;146:297–307. doi: 10.1111/j.1399-3054.2012.01628.x. [DOI] [PubMed] [Google Scholar]
- 42.Wang B., Wang G., Shen F., Zhu S.J. A Glycine-Rich RNA-Binding Protein, CsGR-RBP3, Is Involved in Defense Responses Against Cold Stress in Harvested Cucumber (Cucumis sativus L.) Fruit. Front. Plant Sci. 2018;9:540. doi: 10.3389/fpls.2018.00540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shinozuka H., Hisano H., Yoneyama S., Shimamoto Y., Jones E.S., Forster J.W., Yamada T., Kanazawa A. Gene expression and genetic mapping analyses of a perennial ryegrass glycine-rich RNA-binding protein gene suggest a role in cold adaptation. Mol. Genet. Genet. 2006;275:399–408. doi: 10.1007/s00438-005-0095-3. [DOI] [PubMed] [Google Scholar]
- 44.Wang C., Zhang D.W., Wang Y.C., Zheng L., Yang C.P. A glycine-rich RNA-binding protein can mediate physiological responses in transgenic plants under salt stress. Mol. Biol. Rep. 2012;39:1047–1053. doi: 10.1007/s11033-011-0830-2. [DOI] [PubMed] [Google Scholar]
- 45.Aneeta, Sanan-Mishra N., Tuteja N., Sopory S.K. Salinity- and ABA-induced up-regulation and light-mediated modulation of mRNA encoding glycine-rich RNA-binding protein from Sorghum bicolor. Biochem. Biophys. Res. Commun. 2002;296:1063–1068. doi: 10.1016/S0006-291X(02)02050-8. [DOI] [PubMed] [Google Scholar]
- 46.Teng K., Tan P., Xiao G., Han L., Chang Z., Chao Y. Heterologous expression of a novel Zoysia japonica salt-induced glycine-rich RNA-binding protein gene, ZjGRP, caused salt sensitivity in Arabidopsis. Plant Cell Rep. 2016;36:179–191. doi: 10.1007/s00299-016-2068-x. [DOI] [PubMed] [Google Scholar]
- 47.Wang S.C., Wang R.C., Liang D., Ma F.W., Shu H.R. Molecular characterization and expression analysis of a glycine-rich rna-binding protein gene from malus hupehensis rehd. Mol. Biol. Rep. 2012;39:4145–4153. doi: 10.1007/s11033-011-1197-0. [DOI] [PubMed] [Google Scholar]
- 48.Xiao J., Xu S.J., Li C.H., Xu Y.Y., Chong K. O-GlcNAc-mediated interaction between VER2 and TaGRP2 elicits TaVRN1 mRNA accumulation during vernalization in winter wheat. Nat. Commun. 2014;5:4572. doi: 10.1038/ncomms5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alptekin B., Mangel D., Pauli D., Blake T., Lachowiec J., Hoogland T., Fischer A., Sherman J. Combined effects of a glycine-rich RNA-binding protein and a NAC transcription factor extend grain fill duration and improve malt barley agronomic performance. Theor. Appl. Genet. 2021;134:351–366. doi: 10.1007/s00122-020-03701-1. [DOI] [PubMed] [Google Scholar]
- 50.Kim D.S., Kim N.H., Hwang B.K. Glycine-rich RNA-binding protein1 interacts with receptor-like cytoplasmic protein kinase1 and suppresses cell death and defense responses in pepper (Capsicum annuum) New Phytol. 2015;205:786–800. doi: 10.1111/nph.13105. [DOI] [PubMed] [Google Scholar]
- 51.Tian L., Chou H.L., Zhang L., Hwang S.K., Starkenburg S.R., Doroshenk K.A., Kumamaru T., Okita T.W. RNA-binding protein RBP-P is required for glutelin and prolamine mRNA localization in rice endosperm cells. Plant Cell. 2018;30:2529–2552. doi: 10.1105/tpc.18.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tian L., Chou H.L., Zhang L., Okita T.W. Targeted endoplasmic reticulum localization of storage protein mRNAs requires the RNA-binding protein RBP-L. Plant Physiol. 2019;179:1111–1131. doi: 10.1104/pp.18.01434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Staszak A., Pawlowski T. Proteomic Analysis of Embryogenesis and the Acquisition of Seed Dormancy in Norway Maple (Acer platanoides L.) Int. J. Mol. Sci. 2014;15:10868–10891. doi: 10.3390/ijms150610868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun T., Germain A., Giloteaux L., Hammani K., Barkan A., Hanson M.R., Bentolila S. An RNA recognition motif-containing protein is required for plastid RNA editing in Arabidopsis and maize. Proc. Natl. Acad. Sci. USA. 2013;110:E1169–E1178. doi: 10.1073/pnas.1220162110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singh S., Virdi A.S., Jaswal R., Chawla M., Kapoor S., Mohapatra S.B., Manoj N., Pareek A., Kumar S., Singh P. A temperature-responsive gene in sorghum encodes a glycine-rich protein that interacts with calmodulin. Biochimie. 2017;137:115–123. doi: 10.1016/j.biochi.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 56.Boyer J.S. Plant productivity and environment. Science. 1982;218:443–448. doi: 10.1126/science.218.4571.443. [DOI] [PubMed] [Google Scholar]
- 57.Zhao Y., Chan Z.L., Gao J.H., Xing L., Cao M.J., Yu C.M., Hu Y.L., You J., Shi H.T., Zhu Y.F., et al. BA receptor PYL9 promotes drought resistance and leaf senescence. Proc. Natl. Acad. Sci. USA. 2016;113:1949–1954. doi: 10.1073/pnas.1522840113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zargar S.M., Nagar P., Deshmukh R., Nazir M., Wani A.A., Masoodi K.Z., Agrawal G.K., Rakwal R. Aquaporins as potential drought tolerance inducing proteins: Towards instigating stress tolerance. J. Proteom. 2017;169:233–238. doi: 10.1016/j.jprot.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 59.Zhao L., Feng C., Wu K., Chen W., Wu Y. Advances and prospects in biogenic substances against plant virus: A review. Pestic. Biochem. Physiol. 2015;135:15–26. doi: 10.1016/j.pestbp.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 60.Zhao Y., Yang X., Zhou G., Zhang T. Engineering plant virus resistance: From RNA silencing to genome editing strategies. Plant Biotechnol. J. 2020;18:328–336. doi: 10.1111/pbi.13278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee H.J., Kim J.S., Yoo S.J., Kang E.Y., Han S.H. Different roles of glycine-rich RNA-binding protein7 in plant defense against Pectobacterium carotovorum, Botrytis cinerea, and tobacco mosaic viruses. Plant Physiol. Biochem. 2012;60:46–52. doi: 10.1016/j.plaphy.2012.07.020. [DOI] [PubMed] [Google Scholar]
- 62.Huang X., Yu R.W., Li R., Geng L., Jing X., Zhu C., Liu H. Identification and characterisation of a glycine-rich RNA-binding protein as an endogenous suppressor of RNA silencing from Nicotiana glutinosa. Planta. 2019;249:1811–1822. doi: 10.1007/s00425-019-03122-5. [DOI] [PubMed] [Google Scholar]
- 63.Kelley R.Y., Williams W.P., Mylroie J.E., Boykin D.L., Harper J.W., Windham G.L., Ankala A., Shan X.Y. Identification of Maize Genes Associated with Host Plant Resistance or Susceptibility to Aspergillus flavus Infection and Aflatoxin Accumulation. PLoS ONE. 2012;7:e36892. doi: 10.1371/journal.pone.0036892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gagliardi M., Matarazzo M.R. RIP: RNA Immunoprecipitation. Methods Mol. Boil. 2016;1480:73–86. doi: 10.1007/978-1-4939-6380-5_7. [DOI] [PubMed] [Google Scholar]
- 65.Zhao J.C. nRIP-seq: A technique to identify RNA targets of an RNA binding protein on a genome-wide scale. Methods Mol. Med. 2015;1206:97–106. doi: 10.1007/978-1-4939-1369-5_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mcdermott J.J., Watkins K.P., Williams-Carrier R., Barkan A. Ribonucleoprotein capture by in vivo expression of a designer pentatricopeptide repeat protein in Arabidopsis. Plant Cell. 2019;31:1723–1733. doi: 10.1105/tpc.19.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Streitner C., Köster T., Simpson C.G., Shaw P., Danisman S., Brown J.W.S., Staiger D. An hnRNP-like RNA-binding protein affects alternative splicing by in vivo interaction with transcripts in Arabidopsis thaliana. Nucleic Acids Res. 2012;40:11240–11255. doi: 10.1093/nar/gks873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qi H.D., Lin Y., Ren Q.P., Wang Y.Y., Xiong F., Wang X.L. RNA Splicing of FLC Modulates the Transition to Flowering. Front. Plant Sci. 2019;10:1625. doi: 10.3389/fpls.2019.01625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim J.S., Jung H.J., Lee H.J., Kim K.A., Goh C.H., Woo Y., Oh S.H., Han Y.S., Kang H. Glycine-rich RNA-binding protein7 affects abiotic stress responses by regulating stomata opening and closing in Arabidopsis thaliana. Plant J. 2008;55:455–466. doi: 10.1111/j.1365-313X.2008.03518.x. [DOI] [PubMed] [Google Scholar]
- 70.He P., Xiao G.H., Liu H., Zhang L.H., Zhao L., Tang M.J., Huang S., An Y.J., Yu J.N. Two pivotal RNA editing sites in the mitochondrial atp1mRNA are required for ATP synthase to produce sufficient ATP for cotton fiber cell elongation. New Phytol. 2018;218:167–182. doi: 10.1111/nph.14999. [DOI] [PubMed] [Google Scholar]
- 71.Czolpinska M., Rurek M. Plant Glycine-Rich Proteins in Stress Response: An Emerging, Still Prospective Story. Front. Plant Sci. 2018;9:302. doi: 10.3389/fpls.2018.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dresios J., Aschrafi A., Owens G.C., Vanderklish P.W., Edelman G.M., Mauro V.P. Cold stress-induced protein Rbm3 binds 60S ribosomal subunits, alters microRNA levels, and enhances global protein synthesis. Proc. Natl. Acad. Sci. USA. 2005;102:1865–1870. doi: 10.1073/pnas.0409764102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lu C.K., Lai Y.C., Chen H.R., Chiang M.K. Rbms3, an RNA-Binding Protein, Mediates the Expression of Ptf1a by Binding to Its 3′UTR during Mouse Pancreas Development. DNA Cell Biol. 2012;31:1245–1251. doi: 10.1089/dna.2012.1619. [DOI] [PubMed] [Google Scholar]
- 74.Hennig S., Kong G., Mannen T., Sadowska A., Kobelke S., Blythe A., Knott G.J., Lyer K.S., Ho D.W., Newcombe E.A., et al. Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles. J. Cell Biol. 2015;210:529–539. doi: 10.1083/jcb.201504117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Maharana S., Wang J., Papadopoulos D.K., Richter D., Pozniakovsky A., Poser I., Poser I., Bickle M., Rizk S., Guillen-Boixét J., et al. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science. 2018;370:918–921. doi: 10.1126/science.aar7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Banani S.F., Lee H.O., Hyman A.A., Rosen M.K. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Emenecker R.J., Holehouse A.S., Strader L.C. Emerging roles for phase separation in plants. Dev. Cell. 2020;55:69–83. doi: 10.1016/j.devcel.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xie D.Q., Chen M., Niu J.R., Wang L., Li Y., Fang X.F., Li P.L., Qi Y. Phase separation of SERRATE drives dicing body assembly and promotes miRNA processing in Arabidopsis. Nat. Cell Biol. 2020;23:32–39. doi: 10.1038/s41556-020-00606-5. [DOI] [PubMed] [Google Scholar]
- 79.Huang X.Z., Chen S.D., Li W.P., Tang L.L., Xu C. ROS regulated reversible protein phase separation synchronizes plant flowering. Nat. Chem. Biol. 2021;17:549–557. doi: 10.1038/s41589-021-00739-0. [DOI] [PubMed] [Google Scholar]