Abstract
The early detection of cancer favors a greater chance of curative treatment and long-term survival. Exciting new technologies have been developed that can help to catch the disease early. Liquid biopsy is a promising non-invasive tool to detect cancer, even at an early stage, as well as to continuously monitor disease progression and treatment efficacy. Various methods have been implemented to isolate and purify bio-analytes in liquid biopsy specimens. Aptamers are short oligonucleotides consisting of either DNA or RNA that are capable of binding to target molecules with high specificity. Due to their unique properties, they are considered promising recognition ligands for the early detection of cancer by liquid biopsy. A variety of circulating targets have been isolated with high affinity and specificity by facile modification and affinity regulation of the aptamers. In this review, we discuss recent progress in aptamer-mediated liquid biopsy for cancer detection, its associated challenges, and its future potential for clinical applications.
Keywords: early cancer detection, circulating biomarkers, liquid biopsy, aptamer, extracellular vesicles, non-invasive diagnosis, circulating tumor cells, DNA-nanostructures
1. Introduction
The early detection of cancer increases the chances for a cure and the long-term survival of cancer patients [1,2] and is therefore of the utmost importance for optimized cancer treatment. Currently, most cancers lack effective non-invasive screening tests and are therefore detected too late. Liquid biopsy is a promising tool for the early detection of cancer [3,4], and significant advancement of the technique shows potential as a diagnostic tool for a variety of cancer types, including melanoma [5,6,7,8], breast [9,10,11], colorectal [12,13,14], lung [15], liver [16,17,18], ovarian [19], pancreatic [20], and gastric cancers [21]. The most common analytical targets of liquid biopsy are circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), microRNA, and extracellular vesicles (EVs) [22,23,24,25,26]. Various biological fluids including peripheral blood, urine, pleural fluid, ascites, seminal fluid, and cerebrospinal fluid (CSF) are used to isolate circulating targets for diagnostic applications. However, it is challenging to preserve the biological functions and viability of the analytes isolated from liquid biopsy specimens.
Therefore, there is an urgent need for efficient procedures to specifically and reliably detect targets in the analyte of choice. In particular, the affinity-based capture of analytes from liquid biopsy relies on the specificity and binding affinity of the ligands toward the surface receptors of the analytes. Recently, nucleic acid aptamers have been extensively studied as a powerful detection method. Aptamers are single-stranded oligonucleotides with unique tertiary structures capable of specifically binding to a wide range of targets, including proteins [27], small molecules [28], metal ions [29], viruses [30], bacteria [31], and whole cells [32].
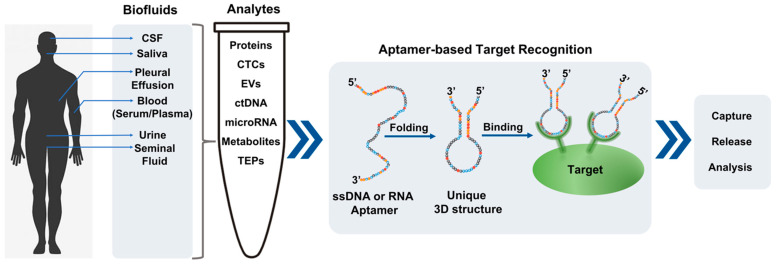
In the recent past, nucleic acid aptamers have emerged as attractive alternatives to antibodies and small molecules in liquid biopsy-based diagnostic, imaging, and targeting applications [33,34,35]. Unlike antibodies, aptamers are easily evolved, synthesized, tailored, and engineered by in vitro methods against the target to find the tightest binding candidates with prominent characteristics for liquid biopsy. In addition, their versatility in structural design and stability greatly the improves sensitivity and high-accuracy large-scale production of aptamers for liquid biopsy applications. In this review, we discuss the recent advances and challenges in the development of aptamer-based liquid biopsy (Figure 1), with a particular focus on the isolation and detection of CTCs, EVs, and other analytes.
Figure 1.
Schematic diagram for common aptamer selection and their applications for circulating tumor targets in liquid biopsy. CSF: Cerebrospinal fluids; CTCs: Circulating tumor cells; ctDNA: Circulating tumor DNA; TEPs: Tumor-educated platelets; EVs: Extracellular vesicles; ssDNA: Single-stranded DNA.
2. SELEX for Aptamer Discovery
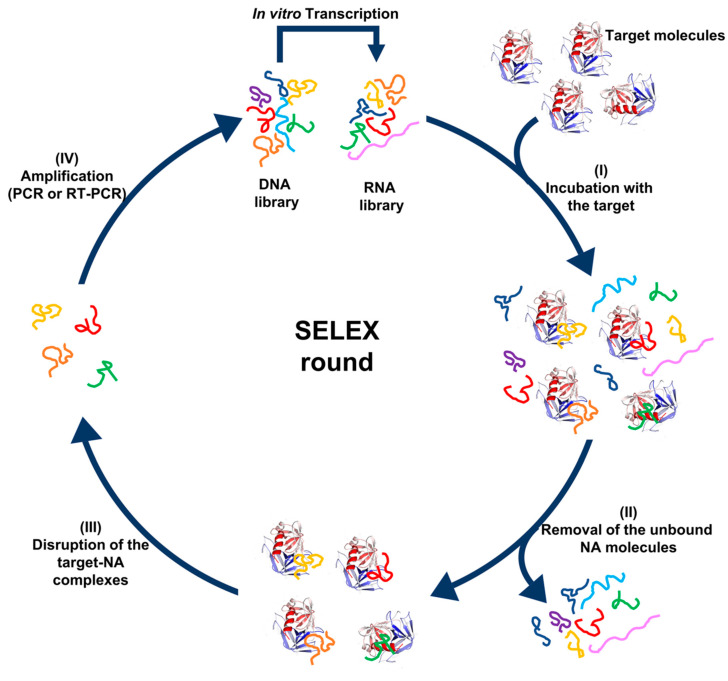
SELEX—systematic evolution of ligands by exponential enrichment—has been developed as a method to select aptamers from liquid biopsy specimens [36]. SELEX was first reported by Tuerk et al. and Ellington et al. in 1990 [27,37]. The general procedure consists of the following steps: (1) the desired selection targets, traditionally proteins, are incubated with a random library consisting of 1012 to 1014 unique single-stranded oligonucleotides, 20 to 80 bases in length. (2) The target-binding oligonucleotides are separated from the unbound ones. (3) The target-binding oligonucleotides are eluted and amplified by PCR to create the library for the next round of selection. (4) Several rounds of selection are performed with increasing selection pressure to obtain target specific aptamers. (5) The obtained aptamer candidates are sequenced and further characterized (Figure 2).
Figure 2.
Schematic diagram of the SELEX method using DNA and RNA libraries.
Existing SELEX methods can be categorized by target types, which include purified proteins, whole cells, and tissues [38,39,40,41]. SELEX methods are onerous and time-consuming procedures, and the selected aptamers might display an unsatisfactory recognition performance despite successful selection. In the following, we will discuss certain modifications that have been implemented to improve selection efficiency and optimize aptamer properties for use with liquid biopsy.
2.1. High-Throughput SELEX
The selection efficiency of SELEX has been improved by a set of strategies [42,43]. In order to isolate aptamers from random sequences with high specificity and affinity, it is critical to have a diverse library during the selection process and to minimize technical bias. The multiple cycles of conventional PCR used for traditional SELEX run the risk of amplifying nonspecific byproducts, thus introducing bias during the amplification steps [44]. On one hand, some sequences may be preferentially processed by DNA polymerase and thus be over-enriched during PCR. On the other hand, sequences with complex structures may be more difficult to amplify and therefore be eliminated over the course of the procedure. Novel PCR technologies such as droplet digital [45] and emulsion PCR [46,47,48] have helped to optimize amplification and preserve library diversity by decreasing the accumulation of byproducts and minimizing PCR bias.
After the completion of amplification, the final PCR products are usually cloned into Escherichia coli for identification via sequencing. This step is lengthy and tedious, and the resulting clones do not necessarily represent the entire population of aptamers. Inefficient cloning strategies or a small number of clones can miss infrequent aptamers despite excellent affinity. Cloning bias can be bypassed by utilizing high-throughput sequencing and bioinformatics analysis in combination with SELEX (HT-SELEX). HT-SELEX allows for the visualization of dynamic changes among millions of sequence reads throughout the selection process. This reduces cloning bias and enables the identification of high-affinity aptamers much earlier in the selection.
2.2. Cell SELEX
Traditional in vitro SELEX uses purified proteins as targets for selecting aptamers. Yet, aptamers binding to purified proteins might not necessarily be able to bind to the same proteins within the cell due to varying protein levels and specific cellular conditions. Cell SELEX, however, which was firstly developed by Daniels et al. [49], uses the whole cell as a target. This increases the chance that the selected aptamers will be able to bind to the target within the physiological context and can be directly applied for diagnostic and therapeutic purposes. Cell SELEX has several advantages over in vitro SELEX: first, targets on the cell surface are presented in their natural conformation and the selected aptamers represent the final results; second, there is no need for time-consuming protein purification— there is actually not even a need for prior identification or characterization of the molecular targets on the cell surface; third, cell SELEX represents an unbiased method for the discovery of new biomarkers and even unknown surface molecules.
Currently, several cell SELEX modifications have been developed to enhance successful aptamer screening. Hicke et al. developed a hybrid system linking the advantages of cell SELEX and traditional SELEX based on purified proteins [50]. In the first step, aptamer candidates were selected against native tenascin-C by incubating the random library with tenascin-C-bearing cells. The following selection rounds were performed using purified tenascin-C protein as the target.
In order to overcome the tedious and time-consuming task of the purification of the recombinant proteins, Ohuchi et al. developed the so-called target expressed on cell surface-SELEX (TECS SELEX) method [51]. TECS SELEX is a SELEX method that directly uses a cell-surface displaying recombinant proteins as the selection target. Using this approach, they successfully identified RNA aptamers binding to TGFBR3 (transforming growth factor-β receptor III) expressed on Chinese hamster ovary cells. Chen et al. used this approach to successfully select aptamers against DC-SIGN (dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin) on the surface of NIH3T3 cells [52].
Another recent hybrid method by Soldevilla et al. combines cell SELEX and peptide SELEX. Using this method, they were able to target chemotherapy-resistant tumors expressing MRP1 [53]. Further, cell SELEX-based modifications and hybrid methods include FACS-SELEX [54,55], 3D cell SELEX [56], and cell-internalization SELEX [57,58].
2.3. Microfluidic SELEX
In 2006, Hybarger et al. combined traditional SELEX with a microfluidic system introducing microfluidic SELEX or M-SELEX [59]. The prototype consists of microlines that are loaded with reagents, a pressurized reagent reservoir and distributor, a PCR cycler, and operable valves for selection and sample routing. Using this prototype, they successfully selected an RNA aptamer-binding lysozyme. In 2009, Luo et al. developed an automated, more rapid aptamer selection system [60]. For this approach, a magnetic bead-based SELEX is combined with microfluidic technology, and a continuous-flow magnetically activated chip is used as a separation device. Following this strategy, the enriched aptamer pool, which was obtained after only a single round of selection, was able to bind to recombinant botulinum neurotoxin type A with high affinity. However, several technical challenges were encountered. Aggregation of the magnetic beads in the microchannels influenced the purity and yield of the recovery. In addition, the flow streams could be distorted by microbubbles in the microchannels. To overcome these challenges, Soh and coworkers further advanced the M-SELEX method by furnishing the microchannel with magnetic materials, thus successfully isolating aptamers targeting streptavidin with a Kd value of 25 nM in only three selection rounds [60].
Another novel microfluidic SELEX, developed by Park et al. in 2009, incorporates nanoporous sol-gel material from protein microarrays [61]. Sol-gels can hold a large number of target molecules due to their nanoporous structure, which facilitates the selection of aptamers against multiple targets. Several target molecules with Kd values in the low nanomolar range have been used with sol-gel SELEX. Using this approach, the efficiency of aptamer selection is improved, and only five to eight rounds of selection are usually needed [62,63,64]. However, some of the concerns remain in sol-gel SELEX, such as molecule integrity and stability throughout the multiple selection cycles.
Other modifications of microfluidic techniques aiming at enhancing the efficiency of selecting aptamers include capillary electrophoresis (CE) microfluidic SELEX [65], bead-based microfluidic SELEX [66,67], and protein microarray-microfluidic chip SELEX [68].
3. Development of Cancer-Specific Aptamers for Diagnosis and Treatment
The majority of traditional cancer drugs are neither specific nor selective and can cause serious side effects and toxicity within the body during and after treatment [69,70]. The use of aptamers in cancer treatment opens an exciting new avenue. Aptamers are a class of small nucleic acid ligands with high affinity and specificity for their targets [27,37]. Such targets can be either cancer-specific molecules or biomarkers that are related to the development of cancer. Aptamers selected against these targets can be used as drugs themselves or as vehicles to deliver drugs to a certain target after coupling the aptamers with drugs, siRNA, nanoparticles, etc. This creates a powerful targeted drug delivery system that is specific for the selected target and minimizes toxicity to normal cells. This can potentially reduce the dose required for treatment and, thus, enhances therapeutic efficacy.
Thus far, a number of aptamers that target tumor cells have been identified by SELEX: A10, an anti-prostate-specific membrane antigen (PSMA) aptamer [71]; AS1411, an anti-nucleolin aptamer [72,73]; EpCAM, anti-epithelial cell adhesion molecule aptamer [74,75]; Sgc8, an anti-protein tyrosine kinase 7 (PTK7) aptamer [76,77,78] and MUC1, anti-mucin1 aptamer [79]. In addition, a variety of drug delivery systems have been developed using the aptamers for the targeted treatment of specific cancer cells.
Early and exact cancer diagnosis is of great clinical significance as it can help doctors to implement the best treatment strategies in a timely manner, evaluate treatment response, and monitor for recurrence or metastasis, as well as to provide an accurate prognosis. At present, mostly antibodies are used in clinical applications and cancer diagnostics, such as immunohistochemistry, flow cytometry, the detection of tumor markers, in vivo imaging, etc. [80,81,82]. However, antibodies come with certain disadvantages, such as immunogenicity, poor stability, and high production costs due to limited and time-consuming production methods, which limit their usefulness to a certain extent. While aptamers can bind targets with high affinity and specificity comparable to antibodies, aptamers have obvious advantages when it comes to stability, chemical modifications, and production cost [83]. Aptamers have therefore found vast applications in the field of cancer diagnosis, such as immunohistochemical analysis, in vivo imaging, and the detection of CTCs.
In addition, numerous aptamers against specific cancer biomarkers have been utilized for the capture and analysis of targets from liquid biopsy specimens, including aptamers against PSMA, prostate-specific antigen (PSA), mucin 1 (MUC1), and epidermal growth factor receptor 2 (HER2) [84,85,86]. Molecular markers on the surface of exosomes, such as CD63, and the extracellular domain of HSP70 have been targeted for the development of specific aptamers to isolate and analyze EVs [87,88]. Herein, we summarize some of the existing aptamers successfully applied in liquid biopsy and their applications for various cancer types (Table 1).
Table 1.
A List of Selected Aptamers Used in Liquid Biopsy by Cancer Types.
| Aptamer | Tumor Type | Application |
|---|---|---|
| A8 | Breast, lung, and ovarian cancer | Quantification of tumor-derived exosomes [89] |
| MO-1, MO-2 | Cervical cancer | Detection of cancer-specific EVs [90] |
| H2 and SYL3C | Breast cancer | Detection of cancer-specific EVs [91] |
| LZH8 | Hepatocellular carcinoma | Detection of cancer-derived exosomes [92] |
| MUC_3 | Gastric cancer | Detection of cancer-derived exosomes [93] |
| EpCAM/Ep114 | Breast cancer, colorectal cancer | Detection of cancer-specific EVs [94,95] |
| CD63 | Gastric, lung, and breast cancer | Detection and quantification of cancer-derived EVs [94,96,97,98] |
| H2, CEA, and PSMA | Breast, colorectal, and prostate cancer | Detection of cancer-derived exosomes [99] |
| PL1-8 | Pancreatic ductal adenocarcinoma | Biomarkers identification, drug delivery [100] |
| Aptamers 1 and 146 | Pancreatic ductal adenocarcinoma | Detection of CTC [101] |
| AS1411 | Breast cancer | Drug-targeting nanovesicles [102] |
| PSMA and EGFR | Breast, colorectal, and prostate cancer | siRNA delivery via aptamer-functionalized EVs [103] |
| sgc8 | Leukemia, lymphoma | Aptamer-functionalized exosomes for drug delivery [104] |
| CTLA-4 | Melanoma, lymphoma, colon cancer, renal cell cancer, fibrosarcoma |
Immune-checkpoint blockade [105]; targeting STAT3 siRNA [106] |
| PD1 | Colon cancer | Immune-checkpoint blockade [107] |
| TIM3 | Colon cancer | Immune-checkpoint blockade [108] |
| IL10R | Colon cancer | Immune-checkpoint blockade [109] |
| IL6 | Glioma and hepatoma | In vitro growth inhibition [110] |
| IL4R | Breast cancer | Targeting myeloid-derived suppressor cells (MDSC) and tumor-associated macrophages (TAM) [111] |
| 4-1BB | Mastocytoma, melanoma, colon cancer, breast cancer, oncogene-induced high-grade glioma, MCA fibrosarcomas |
Costimulatory receptor agonist [112]; targeting costimulation to the tumor [113,114] |
| OX40 | Melanoma | Costimulatory receptor agonist [115] |
| CD28 | Lymphoma, melanoma | Costimulatory receptor agonist [116]; targeting costimulation to the tumor [53] |
| CD40 | Lymphoma | Stimulatory receptor agonist [117] |
| CD16α | Lysis of human gastric and lung cancer cell lines in vitro | Antibody-dependent cell-mediated cytotoxicity (ADCC) [118] |
| BAFF-R | Mantle cell lymphoma | Targeted STAT-3 inhibition [119] |
3.1. CTCs from Liquid Biopsy
CTCs are cells detached from solid cancer tissues circulating in the bloodstream. They are believed to be responsible for the hematogenous spread of cancer cells essential for the development of distant metastasis [120]. CTCs retain unique cellular structures and functions and can provide real-time information about the originating cancer issues. This makes them an ideal target for diagnostics as well as for guiding therapy.
Over the past years, CTC-based liquid biopsy has found applications in a variety of clinical settings including early cancer diagnosis, treatment monitoring, prognosis, and personalized cancer therapy [121]. Aptamers, with their unique properties and advantages, are an exciting new tool for the isolation and detection of CTCs.
3.1.1. Capture
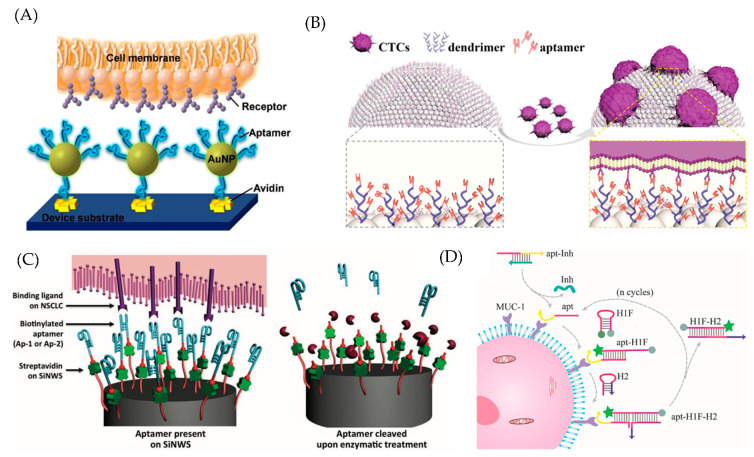
CTCs from different cancers have been successfully isolated with aptamer-based magnetic separation [122,123,124]. For this approach, a magnetic field is applied to separate magnetically tagged target cells from their respective pool [125]. The disadvantage of this method is the use of a single aptamer, which runs the risk of losing some CTC subpopulations due to the unequal expression of CTC biomarkers. Sheng et al. utilized gold nanoparticles (AuNPs) as scaffolds to assemble multivalent aptamer nanospheres, which were further coated into micro- channels for isolation of leukemia cells in whole blood (Figure 3A) [126]. Up to 95 aptamer ligands were attached onto each gold nanoparticle (approximately 14 nm). When the microchannel of a microfluidic chip was modified by gold nanoparticles (AuNPs), a 39-fold increase in binding affinity was developed compared to a flat surface coated with aptamer alone. The results showed that the efficiency of cell capture increased from 49% by using aptamer alone to 92% by using AuNP aptamer, indicating the strong potential of AuNP-based microfluidic chip devices for the analysis of CTCs. Zheng et al. employed barcode particles of spherical colloidal crystal clusters decorated with dendrimer-amplified aptamer probes to capture, detect, and release multiple types of CTCs (Figure 3B) [127]. This barcode-particle technology can simultaneously capture, detect, and release multiple types of CTCs from a complex sample with a high capture efficiency of 92.83%. Shen and co-workers developed a new-generation NanoVelcro chip (Figure 3C) [128]. Two aptamers were coated on silicon nanowire substrates (SiNWS), which were utilized to immobilize CTCs in a stationary device setting. Then, SiNWS was integrated with a polydimethylsiloxane (PDMS)-based chaotic mixer that enhanced the contact frequency between flow-through cancer cells and the substrate, thereby improving the capture efficiency of the CTCs. At optimized conditions, the capture efficiency exceeded 80% for A549 cells in artificial blood samples, and the release efficiency was more than 85%. This SiNW-based microfluidic chip platform can not only improve CTC capture efficiency, but can also realize controllable CTC release via nuclease treatment. In another report, Liu et al. developed a homogeneous fluorescent method to detect cancer cells based on catalytic hairpin assembly (CHA) and bifunctional aptamers (Figure 3D) [129]. The strategy showed high specificity for discriminating normal cells and leukocytes, and the detection limit was 10 cells/mL. Zamay et al. used a targeted selection of DNA aptamers and applied two different aptamer clones for the isolation and detection of CTCs in peripheral blood samples from patients with different lung cancer types and benign lung tumors [130]. In addition, these aptamers were further utilized for a bioluminescent solid-phase sandwich-type microassay to detect lung tumor elements circulating in the blood [131]. Fang’s group introduced a cocktail of aptamer-modified magnetic nanoparticles (apt-MNPs) to capture heterogeneous CTC populations [132]. A microwell chip was used to improve the CTC purity for downstream applications. Eventually, platforms were developed to simultaneously capture and analyze CTCs in situ. Another example is the magnetic nanoparticle-quantum dot (QD)-aptamer copolymers by Li et al., which isolated CTCs with about 80% efficiency and simultaneously phenotyped them using a fluorescent approach [133]. Magnetic-based affinity isolation is easy to perform, can be conveniently integrated into various platforms, and has a high capture efficiency; however, CTC purity can be a concern due to the nonspecific binding of magnetic tags to other cells in the bloodstream. In some devices, a uniform multiscale TiO2 nanorod array is fabricated to provide a “multi-scale interacting platform” for cell capture by Sun et al. [134]. The platform showed up to 85–95% capture yield of the target cancer cells on the BSA-aptamer TiO2 nanorod substrates, revealing the potential application of the TiO2 nanorods for the efficient and sensitive capture of rare CTCs.
Figure 3.
Schematic illustration of aptamer-based techniques for the capture and detection of circulating tumor cells (CTCs). (A) Multivalent AuNP-aptamer for the enhanced capture of tumor cells on microdevices. Reprinted with permission from ref. [126]. (B) SiNW-based platform for CTC capture and release with temperature stimulation. Reprinted with permission from ref. [127]. (C) NanoVelcro Chip consisted of aptamer-coated silicon nanowire substrate (SiNWS) and an overlaid PDMS chaotic mixer. Reprinted with permission from ref. [128]. (D) Bi-functional aptamer-mediated catalytic hairpin assembly for the fluorescence turn-on detection of rare cancer cells. Reproduced with permission from ref. [129].
3.1.2. Release
After the successful capture of CTCs, it is critical to release them from whatever matrix they are bound to so as to allow for downstream processing and analysis. Different from other recognition ligands, aptamers can be easily manipulated to allow for the convenient release of CTCs. The aptamer-CTC bond can be disrupted either by the denaturation of the aptamer or by the detachment of the aptamer from the capture interface. Disruption of the aptamer-CTC interaction can be achieved by physical or chemical strategies such as adsorption, covalent binding, or the use of linker molecules to allow for the release of the CTCs [135].
Au−S bonds, for example, can be easily disrupted by electrochemical reduction and competitive ligand binding, which leads to the release of the CTCs bound to aptamers by Au substrates [136,137,138,139]. Yang’s group created an AP-Octopus-Chip using thiol-terminated aptamer-functionalized AuNPs for capturing CTCs. After capture, glutathione (GSH) with three thiol moieties was utilized to release the CTCs through competitive binding to AuNPs with more than 80% efficiency and 96% viability [139]. Aside from the direct disruption, different approaches have been developed to alter the aptamer-interface interaction. Wang’s group developed an elegant approach using UV light to release CTCs. They used azobenzene-tagged aptamers that interact with a cyclodextrin (CD)-modified surface. Exposure to UV light leads to a switch from trans- to cis-isomers, which then leads to an unmatching of the host-guest pairs, thereby releasing the captured CTCs [140].
3.1.3. Analysis
Downstream detection and analysis have been at the core of CTC research over the past decade. Clinical studies have suggested a correlation between the detection and quantification of CTCs and clinical outcomes, proposing CTCs not only as a diagnostic, but also as a prognostic tool. The opportunity to monitor treatment response via the detection and quantification of CTCs can be used to directly and efficiently guide treatment strategies. As CTCs are believed to be the origin of distant metastasis, the elimination of CTCs might even become useful as an additional cancer therapy option, potentially preventing metastatic cancer spread.
Obtaining CTCs from blood samples is far less invasive than obtaining a surgical specimen or even performing an interventional biopsy. Nonetheless, it provides vast genotypic and phenotypic information on the original cancer, potentially replacing histologic confirmation—which is generally required before initiating cancer treatment—in the future. In addition, CTCs are easy to analyze and manipulate in the laboratory, making for an ideal research platform to gain insights into the mechanism of tumorigenesis and cancer progression, as well as guiding personalized cancer therapy.
Fluorescence assays are widely used for the identification and detection of CTCs. They are highly sensitive, easy to manipulate, and can be conveniently combined with various different isolation methods. However, autofluorescence background is a major issue, interfering with the sensitive and reliable detection of CTCs [129,141,142,143,144]. To overcome this disadvantage, Liu’s group labeled tumor cells with aptamer-conjugated upconversion nanoparticles (UCNPs). In a second step, magnetic nanoparticles were used to further isolate the labeled tumor cells. The resulting upconversion luminescence imaging was free of autofluorescence and allowed for the sensitive and specific detection of CTCs in blood samples [129]. Near-infrared (NIR) fluorescent probes can also be useful by offering reduced light scattering and minimal autofluorescence from living tissues when detecting CTCs from the complex blood matrix. Ding et al. were able to detect as little as 5 CTCs per mL of sample using NIR fluorescent nanoprobes based on aptamer-modified Ag2S nanodots [143]. The turn-on method is another strategy to work around fluorescence interference. Fluorescence detection is limited by the performance of fluorescent tags, including fluorescence intensity, labeling specificity, and stability, among others. The refinement and optimization of fluorescent tags will be needed to further improve the sensitive and reliable detection of CTCs.
3.2. EVs from Liquid Biopsy
EVs are lipid bilayer-delimited particles that are secreted from most cell types and can be found in all biological fluids, such as blood, saliva, pleura fluid, and urine [87,145,146]. EVs contain a unique set of proteins, DNA, and RNA from the originating cells and act as mediators of intercellular communication [147]. EVs are grouped by size and origin into exosomes (30−200 nm), microvesicles (200−1000 nm), and apoptotic bodies (>1000 nm) [148]. Tumor-derived EVs carry information about cancer cells and are believed to be important players in cancer progression and metastasis, as well as the development of drug resistance. Tumor-derived EVs are therefore promising biomarkers not only for the early detection of cancer, but also for the evaluation of therapeutic response, the assessment of prognosis, and the monitoring of cancer recurrence and/or metastasis [87,146]. Despite the exciting potential, there are still many technical challenges to overcome with regard to the isolation and detection of EVs [149]. Currently, size, density, surface charge, membrane proteins, and lipid membranes have been explored for the isolation of EVs from various body fluids. Among these approaches, bioaffinity-based EV isolation targeting unique EV membrane antigens such as EpCAM, EGFR, HER2, PSA and Tim4 requires specific capture molecules with high affinity [150,151,152]. Aptamer-based detection of EVs has emerged as an advantageous strategy for the efficient and specific isolation of EVs utilizing affinity-based isolation and quantitative detection using signal transduction, as well as signal amplification.
3.2.1. Capture
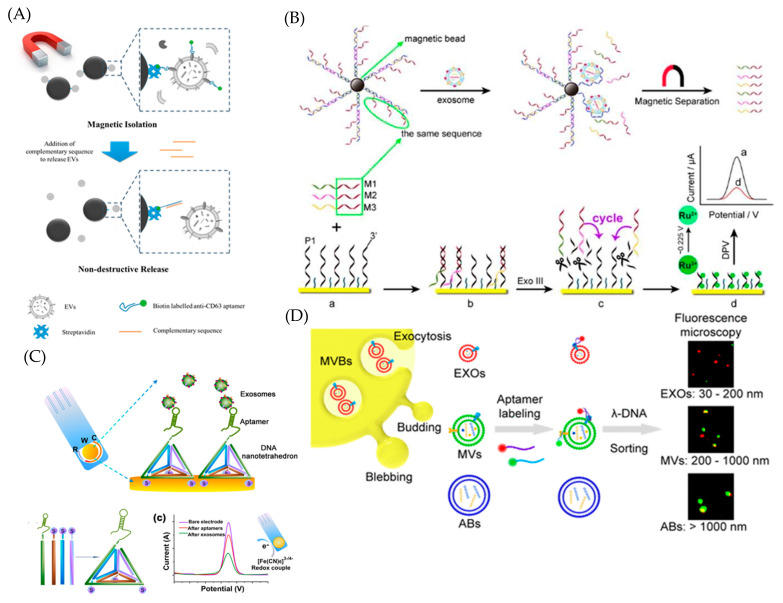
EVs can be isolated utilizing antibodies or aptamers as recognition ligands. The advantage of aptamer-based isolation is the fast and efficient release of EVs, low immunogenicity, and an easy incorporation of downstream processing such as signal transformation and amplification for the further analysis of EVs. Zhang et al. could isolate EVs in as little as 1.5 h with an efficiency of ~78% (Figure 4A) [97]. By using complementary sequences to release the EVs, the isolated EVs remained stable and functional, allowing for further analysis. Aptamer-anchored DNA nanostructures can be designed according to specific substrates for capture and detection, and can easily be controlled and manipulated. The more efficient and thorough the capture and isolation of the EVs, the better the conditions for the downstream analysis, biofunctional studies, and biomedical applications of the EVs. Dong et al. used aptamer magnetic bead bioconjugates to capture tumor exosomes (Figure 4B) [153]. A strategy based on aptamer recognition-induced multi-DNA release was implemented, where the recognition of the exosomes causes the release of the detected complementary strands and the recognition event produces a decrease in the voltammetric signal of the marker in response to the exosome target.
Figure 4.
Schematic illustration of aptamer-based techniques for the capture and detection of EVs. (A) DNA aptamer-based magnetic isolation system (AMI) for the rapid capture and nondestructive release of EVs. Reprinted with permission from ref. [97]. (B) Electrochemical detection of tumor exosomes based on aptamer recognition-induced multi-DNA release. Reprinted with permission from ref. [153]. (C) Nanotetrahedron-assisted aptasensor for the electrochemical detection of cancerous exosomes. Reproduced with permission from ref. [92]. (D) λ-DNA-mediated sorting of EV subpopulations and aptamer-mediated fluorescence analysis of EVs. Reproduced with permission from ref. [91].
Xue et al. reported a multivalent, long single-stranded aptamer with repeated units for EV enrichment and retrieval [154]. EVs were secured by biotin-labeled multivalent aptamers (MAs), and 45% of EVs were isolated from the spiked samples in 40 min with a depletion of 84.7% of albumin contamination. Furthermore, 93.1% of the isolated EVs were retrieved via DNase-mediated aptamer degradation in 10 min for downstream molecular analyses.
3.2.2. Detection and Quantification of EVs
The detection and quantification of cancer-derived EVs can aid in early cancer diagnosis and the assessment of therapeutic response. A large variety of approaches and technologies utilizing novel aptamer-based methods have been implemented to detect and quantify cancer-derived EVs in complex biological or clinical samples.
Kelly’s group used an elegant electrochemical approach, wherein they constructed an on-chip multiplexed electrochemical sensor and used aptamer-modified metal nanoparticles as probes to detect and quantify EVs [155]. Zhou et al. developed an aptamer-based electrochemical biosensor, which could detect as few as 1 × 106 particles/mL of exosomes by integrating Au electrode arrays in microchannels [156]. EVs were targeted via aptamers specific to transmembrane protein CD63, which were also hybridized to redox-labeled probing chains. Once bound, conformational change resulted in the release of redox reporters and, thus, in a decreased electrochemical signal. A nanotetrahedron (NTH)-assisted aptasensor was developed by Tan’s group. They linked DNA-based nanostructure and portable electrochemical devices to directly capture and detect hepatocellular exosomes. The NTH-assisted aptasensor was 100 times more sensitive for the detection of EVs than the single-stranded aptamer-functionalized aptasensor (Figure 4C) [92]. While electrochemical detection comes with excellent sensitivity, its practical application is limited by the rather low stability of EVs within body fluids. Another easy and convenient method for detecting EVs is a colorimetric approach. Colorimetric tools can be used to quantify and profile exosomal protein information in a simple manner. Aptamers have the ability to enhance nanozymes, nanoparticles with peroxidase-like activity, which can be used for colorimetric experiments [85,157,158]. Wang et al. attached aptamers to graphitic carbon nitride nanosheets (g-C3N4 NSs) [158], which detached from the sheets once bound to EVs. This resulted in a decrease in the peroxidase activity of the nanozymes, leading to a color change, which could be quantified using spectrometry. A third detection strategy utilizes fluorescence. Target proteins on the surface of EVs are labeled with small aptamers (2–3 mm) without significantly changing the overall size. Sun’s group used two kinds of fluorescent dye-labeled aptamers to label EVs. This was followed by on-chip λ-DNA-mediated separation by size (Figure 4D) [91]. Fluorescent analysis demonstrated a heterogeneous marker expression even within single EV subpopulations. This approach was further advanced by the same group to profile EV surface proteins via a size-dependent thermophoretic strategy. With a thermophoretic aptasensor (TAS) using a panel of seven fluorescent aptamers to profile EV surface proteins and then separating them by size, they were able to classify cancer types with an accuracy of 68%, a sensitivity of 95%, and a specificity of 100% [148].
However, especially in early stage cancer, the efficient detection and quantification of EVs with low protein expression levels remains a challenge. Several signal amplification strategies, including direct and competitive amplifications, have been utilized. For direct amplification, aptamers bind to EVs to directly trigger signal amplification reactions, such as rolling circle amplification (RCA) or hybridization chain reaction (HCR) [93,159,160]. In the competitive amplification assay, aptamers are released from their carrier once bound to EVs, thus starting a signal amplification reaction [151].
3.2.3. Analysis
EV secretion has been found to be increased in response to various stimuli, such as inflammation, hypoxia, and acidic microenvironments, among others. In cancer, EV secretion levels have been shown to correlate with the invasiveness of the cancer [161,162]. The heterogeneity of EV subpopulations can be accurately captured by the multiplex analysis of EV biomarkers, which can be performed at a single EV level [147,163,164]. Lee et al. used this approach to characterize single EVs by multiplex protein analysis with eleven markers. For this, EVs were first immobilized in a microfluidic chamber, then fluorescently stained in several rounds and analyzed [163]. Lu’s group used a multiplex barcode technique to profile single-cell EV secretion from thousands of human oral squamous cell cancer cells [147]. They demonstrated an association between the downregulation of certain EV phenotypes and the aggressiveness of the originating cancer cell. Apart from protein analysis approaches, RNA analysis approaches of EVs have been developed to facilitate the detection of early stage cancer and to screen for drug resistance [165,166]. Weissleder’s group, for example, identified mRNA markers associated with drug resistance using a microfluidic chip. This chip combined immunomagnetic capture, on chip RNA isolation, and the real-time RNA analysis of EVs [166]. Furthermore, the detection of DNA mutations and microRNAs in EVs was found to facilitate cancer diagnosis [167,168]. Various miRNAs, as well as associated proteins, have been identified within cancer derived EVs which have the potential to be used as biomarkers, such as CPNE3 in colorectal cancer EVs [169], miR-451a in non-small cell lung cancer EVs [170], and miR-451a in pancreatic ductal adenocarcinoma EVs [171].
4. Conclusions and Future Perspective
In this review, we discussed recent advances in aptamer-mediated liquid biopsy with a focus on CTC and EVs, their isolation, release, and analysis. Clinical application of liquid biopsy is already paving the way for precision medicine, and the main advantage of the liquid biopsy approach stands with the possibility of capturing tumor heterogeneity as a whole. Liquid biopsy is an easy, minimally invasive, and reproducible way to obtain CTCs and cancer-derived EVs, which are powerful platforms for cancer diagnosis; the real-time monitoring of treatment response; and, ultimately, personalized cancer therapy. Aptamers themselves are an ideal tool for the isolation and detection of CTCs and EVs. They are small in size; fast and cheap to synthesize; can be easily modified and integrated in downstream analysis and various applications; and—most importantly—detect CTCs and EVs with high efficiency, sensitivity, and specificity.
Despite their clinical potential, there are certain challenges and limitations of aptamer-based liquid biopsies, and their excellent features and versatility need to be reinforced with reliable clinical applications. Aptamers may behave differently within the context of complex body fluids. They can become degraded by nuclease activity and suffer decreased affinity compared to their in vitro function. Most reports of aptamer-based liquid biopsy approaches lack information on clinical standards, such as sampling, preservation, the processing of samples, detection, and readout, limiting the actual clinical utility at this time. To reach the goal of clinical applications of aptamer-based liquid biopsy, clinical standards, reagent kits, and standard instruments have to be developed to streamline the process and allow them for use as part of the clinical routine. In addition, new selection and modification strategies have to be developed to generate reliably and highly performing aptamers.
In comparison to antibodies, aptamer-based technologies are still at an early stage of development, and further research is needed to ensure their widespread adoption. Recent advances in next-generation sequencing (NGS) and the use of high-throughput NGS can be expected to have a huge impact on the aptamer field. Interestingly, therapeutic applications of aptamers have been slow to catch on, despite many clinical trials. The U.S. Food and Drug Administration (FDA) has, so far, only approved Pegaptanib (Macugen), a 28-base ribonucleic acid aptamer, against all isoforms of human vascular endothelial growth factor (VEGF) for clinical use [172]. However, with the recent expiration of the patent on SELEX technology, we can expect more aptamer-based drugs in the near future. Due to their unique three-dimensional structure, aptamers can bind targets with high affinity and specificity. They can be easily coupled to various platforms due to their small size and ease of manipulation. Combining these advantages, aptamer technology has the potential for innumerable clinical applications, from cancer detection and therapy to the treatment of bacterial or viral infections and targeted drug delivery. Aptamer-based approaches can offer an extremely high resolution and will therefore be an invaluable tool for shaping the future landscape of personalized medicine.
Author Contributions
Writing—original draft preparation, D.R., J.C.S., M.A.J.; writing—review and editing, D.R., J.C.S., M.A.J., A.P.; visualization, D.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Huang A.C., Postow M.A., Orlowski R.J., Mick R., Bengsch B., Manne S., Xu W., Harmon S., Giles J.R., Wenz B., et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature. 2017;545:60–65. doi: 10.1038/nature22079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prigerson H.G., Bao Y., Shah M.A., Paulk M.E., LeBlanc T.W., Schneider B.J., Garrido M.M., Reid M.C., Berlin D.A., Adelson K.B., et al. Chemotherapy Use, Performance Status, and Quality of Life at the End of Life. JAMA Oncol. 2015;1:778–784. doi: 10.1001/jamaoncol.2015.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng J., Li B., Ying J., Pan W., Liu C., Luo T., Lin H., Zheng L. Liquid Biopsy: Application in Early Diagnosis and Monitoring of Cancer. Small Struct. 2020;1:2000063. doi: 10.1002/sstr.202000063. [DOI] [Google Scholar]
- 4.Cohen J.D., Li L., Wang Y., Thoburn C., Afsari B., Danilova L., Douville C., Javed A.A., Wong F., Mattox A., et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926–930. doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galanzha E.I., Menyaev Y.A., Yadem A.C., Sarimollaoglu M., Juratli M.A., Nedosekin D.A., Foster S.R., Jamshidi-Parsian A., Siegel E.R., Makhoul I., et al. In vivo liquid biopsy using Cytophone platform for photoacoustic detection of circulating tumor cells in patients with melanoma. Sci. Transl. Med. 2019;11 doi: 10.1126/scitranslmed.aat5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Juratli M.A., Sarimollaoglu M., Siegel E.R., Nedosekin D.A., Galanzha E.I., Suen J.Y., Zharov V.P. Real-time monitoring of circulating tumor cell release during tumor manipulation using in vivo photoacoustic and fluorescent flow cytometry. Head Neck. 2014;36:1207–1215. doi: 10.1002/hed.23439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lucci A., Hall C.S., Patel S.P., Narendran B., Bauldry J.B., Royal R.E., Karhade M., Upshaw J.R., Wargo J.A., Glitza I.C., et al. Circulating Tumor Cells and Early Relapse in Node-positive Melanoma. Clin. Cancer Res. 2020;26:1886–1895. doi: 10.1158/1078-0432.CCR-19-2670. [DOI] [PubMed] [Google Scholar]
- 8.Gandini S., Zanna I., De Angelis S.P., Cocorocchio E., Queirolo P., Lee J.H., Carlino M.S., Mazzarella L., Achutti Duso B., Palli D., et al. Circulating tumour DNA and melanoma survival: A systematic literature review and meta-analysis. Crit. Rev. Oncol. Hematol. 2021;157:103187. doi: 10.1016/j.critrevonc.2020.103187. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Murillas I., Chopra N., Comino-Mendez I., Beaney M., Tovey H., Cutts R.J., Swift C., Kriplani D., Afentakis M., Hrebien S., et al. Assessment of Molecular Relapse Detection in Early-Stage Breast Cancer. JAMA Oncol. 2019;5:1473–1478. doi: 10.1001/jamaoncol.2019.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Juratli M.A., Sarimollaoglu M., Nedosekin D.A., Melerzanov A.V., Zharov V.P., Galanzha E.I. Dynamic Fluctuation of Circulating Tumor Cells during Cancer Progression. Cancers. 2014;6:128–142. doi: 10.3390/cancers6010128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tian F., Zhang S., Liu C., Han Z., Liu Y., Deng J., Li Y., Wu X., Cai L., Qin L., et al. Protein analysis of extracellular vesicles to monitor and predict therapeutic response in metastatic breast cancer. Nat. Commun. 2021;12:2536. doi: 10.1038/s41467-021-22913-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vacante M., Ciuni R., Basile F., Biondi A. The Liquid Biopsy in the Management of Colorectal Cancer: An Overview. Biomedicines. 2020;8:308. doi: 10.3390/biomedicines8090308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li W.M., Bing T., Wei J.Y., Chen Z.Z., Shangguan D.H., Fang J. Cell-SELEX-based selection of aptamers that recognize distinct targets on metastatic colorectal cancer cells. Biomaterials. 2014;35:6998–7007. doi: 10.1016/j.biomaterials.2014.04.112. [DOI] [PubMed] [Google Scholar]
- 14.Li X., An Y., Jin J., Zhu Z., Hao L., Liu L., Shi Y., Fan D., Ji T., Yang C.J. Evolution of DNA aptamers through in vitro metastatic-cell-based systematic evolution of ligands by exponential enrichment for metastatic cancer recognition and imaging. Anal. Chem. 2015;87:4941–4948. doi: 10.1021/acs.analchem.5b00637. [DOI] [PubMed] [Google Scholar]
- 15.Li K., Xiu C.-L., Gao L.-M., Liang H.-G., Xu S.-F., Shi M., Li J., Liu Z.W. Screening of specific nucleic acid aptamers binding tumor markers in the serum of the lung cancer patients and identification of their activities. Tumor Biol. 2017;39:1010428317717123. doi: 10.1177/1010428317717123. [DOI] [PubMed] [Google Scholar]
- 16.Wang F.-B., Rong Y., Fang M., Yuan J.-P., Peng C.-W., Liu S.-P., Li Y. Recognition and capture of metastatic hepatocellular carcinoma cells using aptamer-conjugated quantum dots and magnetic particles. Biomaterials. 2013;34:3816–3827. doi: 10.1016/j.biomaterials.2013.02.018. [DOI] [PubMed] [Google Scholar]
- 17.Chen H., Yuan C.H., Yang Y.F., Yin C.Q., Guan Q., Wang F.B., Tu J.C. Subtractive Cell-SELEX Selection of DNA Aptamers Binding Specifically and Selectively to Hepatocellular Carcinoma Cells with High Metastatic Potential. Biomed Res. Int. 2016;2016:5735869. doi: 10.1155/2016/5735869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vogl T.J., Riegelbauer L.J., Oppermann E., Kostantin M., Ackermann H., Trzmiel A., Stein S., Eichler K., Zharov V.P., Roy D., et al. Early dynamic changes in circulating tumor cells and prognostic relevance following interventional radiological treatments in patients with hepatocellular carcinoma. PLoS ONE. 2021;16:e0246527. doi: 10.1371/journal.pone.0246527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benedetto G., Hamp T.J., Wesselman P.J., Richardson C. Identification of epithelial ovarian tumor-specific aptamers. Nucleic Acid Ther. 2015;25:162–172. doi: 10.1089/nat.2014.0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu X., Zhao Z., Bai H., Fu T., Yang C., Hu X., Liu Q., Champanhac C., Teng I.T., Ye M., et al. DNA Aptamer Selected against Pancreatic Ductal Adenocarcinoma for in vivo Imaging and Clinical Tissue Recognition. Theranostics. 2015;5:985–994. doi: 10.7150/thno.11938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao H.-Y., Yuan A.-H., Shi X.-S., Chen W., Miao Y. Evolution of a gastric carcinoma cell-specific DNA aptamer by live cell-SELEX. Oncol. Rep. 2014;32:2054–2060. doi: 10.3892/or.2014.3411. [DOI] [PubMed] [Google Scholar]
- 22.Siravegna G., Marsoni S., Siena S., Bardelli A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017;14:531–548. doi: 10.1038/nrclinonc.2017.14. [DOI] [PubMed] [Google Scholar]
- 23.Roy D., Tiirikainen M. Diagnostic Power of DNA Methylation Classifiers for Early Detection of Cancer. Trends Cancer. 2020;6:78–81. doi: 10.1016/j.trecan.2019.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roy D., Taggart D., Zheng L., Liu D., Li G., Li M., Zhang K., Etten R.A.V. Abstract 837: Circulating cell-free DNA methylation assay: Towards early detection of multiple cancer types. Cancer Res. 2019;79:837. doi: 10.1158/1538-7445.Am2019-837. [DOI] [Google Scholar]
- 25.Juratli M.A., Siegel E.R., Nedosekin D.A., Sarimollaoglu M., Jamshidi-Parsian A., Cai C., Menyaev Y.A., Suen J.Y., Galanzha E.I., Zharov V.P. In Vivo Long-Term Monitoring of Circulating Tumor Cells Fluctuation during Medical Interventions. PLoS ONE. 2015;10:e0137613. doi: 10.1371/journal.pone.0137613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koonce N.A., Juratli M.A., Cai C., Sarimollaoglu M., Menyaev Y.A., Dent J., Quick C.M., Dings R.P.M., Nedosekin D., Zharov V., et al. Real-time monitoring of circulating tumor cell (CTC) release after nanodrug or tumor radiotherapy using in vivo flow cytometry. Biochem. Biophys. Res. Commun. 2017;492:507–512. doi: 10.1016/j.bbrc.2017.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 28.Hermann T., Patel D.J. Adaptive Recognition by Nucleic Acid Aptamers. Science. 2000;287:820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- 29.Rajendran M., Ellington A.D. Selection of fluorescent aptamer beacons that light up in the presence of zinc. Anal. Bioanal. Chem. 2008;390:1067–1075. doi: 10.1007/s00216-007-1735-8. [DOI] [PubMed] [Google Scholar]
- 30.Tang Z., Parekh P., Turner P., Moyer R.W., Tan W. Generating Aptamers for Recognition of Virus-Infected Cells. Clin. Chem. 2009;55:813–822. doi: 10.1373/clinchem.2008.113514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bruno J.G., Kiel J.L. In vitro selection of DNA aptamers to anthrax spores with electrochemiluminescence detection. Biosens. Bioelectron. 1999;14:457–464. doi: 10.1016/S0956-5663(99)00028-7. [DOI] [PubMed] [Google Scholar]
- 32.Tang Z., Shangguan D., Wang K., Shi H., Sefah K., Mallikratchy P., Chen H.W., Li Y., Tan W. Selection of aptamers for molecular recognition and characterization of cancer cells. Anal. Chem. 2007;79:4900–4907. doi: 10.1021/ac070189y. [DOI] [PubMed] [Google Scholar]
- 33.Jayasena S.D. Aptamers: An emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999;45:1628–1650. doi: 10.1093/clinchem/45.9.1628. [DOI] [PubMed] [Google Scholar]
- 34.Hong H., Goel S., Zhang Y., Cai W. Molecular imaging with nucleic acid aptamers. Curr. Med. Chem. 2011;18:4195–4205. doi: 10.2174/092986711797189691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bouchard P.R., Hutabarat R.M., Thompson K.M. Discovery and development of therapeutic aptamers. Annu. Rev. Pharm. Toxicol. 2010;50:237–257. doi: 10.1146/annurev.pharmtox.010909.105547. [DOI] [PubMed] [Google Scholar]
- 36.Ulrich H., Martins A.H., Pesquero J.B. RNA and DNA aptamers in cytomics analysis. Cytom. A. 2004;59:220–231. doi: 10.1002/cyto.a.20056. [DOI] [PubMed] [Google Scholar]
- 37.Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 38.Mercier M.C., Dontenwill M., Choulier L. Selection of Nucleic Acid Aptamers Targeting Tumor Cell-Surface Protein Biomarkers. Cancers. 2017;9:69. doi: 10.3390/cancers9060069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu M., Yu X., Chen Z., Yang T., Yang D., Liu Q., Du K., Li B., Wang Z., Li S., et al. Aptamer selection and applications for breast cancer diagnostics and therapy. J. Nanobiotechnol. 2017;15:81. doi: 10.1186/s12951-017-0311-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pang X., Cui C., Wan S., Jiang Y., Zhang L., Xia L., Li L., Li X., Tan W. Bioapplications of Cell-SELEX-Generated Aptamers in Cancer Diagnostics, Therapeutics, Theranostics and Biomarker Discovery: A Comprehensive Review. Cancers. 2018;10:47. doi: 10.3390/cancers10020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y., Lai B.S., Juhas M. Recent Advances in Aptamer Discovery and Applications. Molecules. 2019;24:941. doi: 10.3390/molecules24050941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ozer A., Pagano J.M., Lis J.T. New Technologies Provide Quantum Changes in the Scale, Speed, and Success of SELEX Methods and Aptamer Characterization. Mol. Ther. Nucleic Acids. 2014;3:e183. doi: 10.1038/mtna.2014.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Darmostuk M., Rimpelova S., Gbelcova H., Ruml T. Current approaches in SELEX: An update to aptamer selection technology. Biotechnol. Adv. 2015;33:1141–1161. doi: 10.1016/j.biotechadv.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 44.Musheev M.U., Krylov S.N. Selection of aptamers by systematic evolution of ligands by exponential enrichment: Addressing the polymerase chain reaction issue. Anal. Chim. Acta. 2006;564:91–96. doi: 10.1016/j.aca.2005.09.069. [DOI] [PubMed] [Google Scholar]
- 45.Ouellet E., Foley J.H., Conway E.M., Haynes C. Hi-Fi SELEX: A High-Fidelity Digital-PCR Based Therapeutic Aptamer Discovery Platform. Biotechnol. Bioeng. 2015;112:1506–1522. doi: 10.1002/bit.25581. [DOI] [PubMed] [Google Scholar]
- 46.Shao K., Ding W., Wang F., Li H., Ma D., Wang H. Emulsion PCR: A high efficient way of PCR amplification of random DNA libraries in aptamer selection. PLoS ONE. 2011;6:e24910. doi: 10.1371/journal.pone.0024910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Levay A., Brenneman R., Hoinka J., Sant D., Cardone M., Trinchieri G., Przytycka T.M., Berezhnoy A. Identifying high-affinity aptamer ligands with defined cross-reactivity using high-throughput guided systematic evolution of ligands by exponential enrichment. Nucleic. Acids Res. 2015;43:e82. doi: 10.1093/nar/gkv534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yufa R., Krylova S.M., Bruce C., Bagg E.A., Schofield C.J., Krylov S.N. Emulsion PCR significantly improves nonequilibrium capillary electrophoresis of equilibrium mixtures-based aptamer selection: Allowing for efficient and rapid selection of aptamer to unmodified ABH2 protein. Anal. Chem. 2015;87:1411–1419. doi: 10.1021/ac5044187. [DOI] [PubMed] [Google Scholar]
- 49.Daniels D.A., Chen H., Hicke B.J., Swiderek K.M., Gold L. A tenascin-C aptamer identified by tumor cell SELEX: Systematic evolution of ligands by exponential enrichment. Proc. Natl. Acad. Sci. USA. 2003;100:15416–15421. doi: 10.1073/pnas.2136683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hicke B.J., Marion C., Chang Y.F., Gould T., Lynott C.K., Parma D., Schmidt P.G., Warren S. Tenascin-C aptamers are generated using tumor cells and purified protein. J. Biol. Chem. 2001;276:48644–48654. doi: 10.1074/jbc.M104651200. [DOI] [PubMed] [Google Scholar]
- 51.Ohuchi S.P., Ohtsu T., Nakamura Y. Selection of RNA aptamers against recombinant transforming growth factor-beta type III receptor displayed on cell surface. Biochimie. 2006;88:897–904. doi: 10.1016/j.biochi.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 52.Chen F., Zeng J., Sun P., Pan Q., Zhang X.L. [Selection and identification of DNA aptamers against DC-SIGN] Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi Chin. J. Cell. Mol. Immunol. 2008;24:1133–1136. [PubMed] [Google Scholar]
- 53.Soldevilla M.M., Villanueva H., Casares N., Lasarte J.J., Bendandi M., Inoges S., Lopez-Diaz de Cerio A., Pastor F. MRP1-CD28 bi-specific oligonucleotide aptamers: Target costimulation to drug-resistant melanoma cancer stem cells. Oncotarget. 2016;7:23182–23196. doi: 10.18632/oncotarget.8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mayer G., Ahmed M.S., Dolf A., Endl E., Knolle P.A., Famulok M. Fluorescence-activated cell sorting for aptamer SELEX with cell mixtures. Nat. Protoc. 2010;5:1993–2004. doi: 10.1038/nprot.2010.163. [DOI] [PubMed] [Google Scholar]
- 55.Kim J.W., Kim E.Y., Kim S.Y., Byun S.K., Lee D., Oh K.J., Kim W.K., Han B.S., Chi S.W., Lee S.C., et al. Identification of DNA aptamers toward epithelial cell adhesion molecule via cell-SELEX. Mol. Cells. 2014;37:742–746. doi: 10.14348/molcells.2014.0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Souza A.G., Marangoni K., Fujimura P.T., Alves P.T., Silva M.J., Bastos V.A., Goulart L.R., Goulart V.A. 3D Cell-SELEX: Development of RNA aptamers as molecular probes for PC-3 tumor cell line. Exp. Cell Res. 2016;341:147–156. doi: 10.1016/j.yexcr.2016.01.015. [DOI] [PubMed] [Google Scholar]
- 57.Thiel W.H., Thiel K.W., Flenker K.S., Bair T., Dupuy A.J., McNamara J.O., 2nd, Miller F.J., Giangrande P.H. Cell-internalization SELEX: Method for identifying cell-internalizing RNA aptamers for delivering siRNAs to target cells. Methods Mol. Biol. 2015;1218:187–199. doi: 10.1007/978-1-4939-1538-5_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yan A., Levy M. Cell internalization SELEX: In vitro selection for molecules that internalize into cells. Methods Mol. Biol. 2014;1103:241–265. doi: 10.1007/978-1-62703-730-3_18. [DOI] [PubMed] [Google Scholar]
- 59.Hybarger G., Bynum J., Williams R.F., Valdes J.J., Chambers J.P. A microfluidic SELEX prototype. Anal Bioanal. Chem. 2006;384:191–198. doi: 10.1007/s00216-005-0089-3. [DOI] [PubMed] [Google Scholar]
- 60.Lou X., Qian J., Xiao Y., Viel L., Gerdon A.E., Lagally E.T., Atzberger P., Tarasow T.M., Heeger A.J., Soh H.T. Micromagnetic selection of aptamers in microfluidic channels. Proc. Natl. Acad. Sci. USA. 2009;106:2989–2994. doi: 10.1073/pnas.0813135106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park S.M., Ahn J.Y., Jo M., Lee D.K., Lis J.T., Craighead H.G., Kim S. Selection and elution of aptamers using nanoporous sol-gel arrays with integrated microheaters. Lab Chip. 2009;9:1206–1212. doi: 10.1039/b814993c. [DOI] [PubMed] [Google Scholar]
- 62.Ahn J.Y., Jo M., Dua P., Lee D.K., Kim S. A sol-gel-based microfluidics system enhances the efficiency of RNA aptamer selection. Oligonucleotides. 2011;21:93–100. doi: 10.1089/oli.2010.0263. [DOI] [PubMed] [Google Scholar]
- 63.Ahn J.Y., Lee S., Jo M., Kang J., Kim E., Jeong O.C., Laurell T., Kim S. Sol-gel derived nanoporous compositions for entrapping small molecules and their outlook toward aptamer screening. Anal. Chem. 2012;84:2647–2653. doi: 10.1021/ac202559w. [DOI] [PubMed] [Google Scholar]
- 64.Bae H., Ren S., Kang J., Kim M., Jiang Y., Jin M.M., Min I.M., Kim S. Sol-gel SELEX circumventing chemical conjugation of low molecular weight metabolites discovers aptamers selective to xanthine. Nucleic. Acid. Ther. 2013;23:443–449. doi: 10.1089/nat.2013.0437. [DOI] [PubMed] [Google Scholar]
- 65.Nie H., Chen Y., Lu C., Liu Z. Efficient selection of glycoprotein-binding DNA aptamers via boronate affinity monolithic capillary. Anal. Chem. 2013;85:8277–8283. doi: 10.1021/ac4015353. [DOI] [PubMed] [Google Scholar]
- 66.Wang Q., Liu W., Xing Y., Yang X., Wang K., Jiang R., Wang P., Zhao Q. Screening of DNA aptamers against myoglobin using a positive and negative selection units integrated microfluidic chip and its biosensing application. Anal. Chem. 2014;86:6572–6579. doi: 10.1021/ac501088q. [DOI] [PubMed] [Google Scholar]
- 67.Park J.W., Lee S.J., Ren S., Lee S., Kim S., Laurell T. Acousto-microfluidics for screening of ssDNA aptamer. Sci. Rep. 2016;6:27121. doi: 10.1038/srep27121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu X., Li H., Jia W., Chen Z., Xu D. Selection of aptamers based on a protein microarray integrated with a microfluidic chip. Lab Chip. 2016;17:178–185. doi: 10.1039/C6LC01208F. [DOI] [PubMed] [Google Scholar]
- 69.Zugazagoitia J., Guedes C., Ponce S., Ferrer I., Molina-Pinelo S., Paz-Ares L. Current Challenges in Cancer Treatment. Clin. Ther. 2016;38:1551–1566. doi: 10.1016/j.clinthera.2016.03.026. [DOI] [PubMed] [Google Scholar]
- 70.Soldevilla M.M., Villanueva H., Pastor F. Aptamers: A Feasible Technology in Cancer Immunotherapy. J. Immunol. Res. 2016;2016:1083738. doi: 10.1155/2016/1083738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lupold S.E., Hicke B.J., Lin Y., Coffey D.S. Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res. 2002;62:4029–4033. [PubMed] [Google Scholar]
- 72.Cao Z., Tong R., Mishra A., Xu W., Wong G.C., Cheng J., Lu Y. Reversible cell-specific drug delivery with aptamer-functionalized liposomes. Angew. Chem. Int. Ed. Engl. 2009;48:6494–6498. doi: 10.1002/anie.200901452. [DOI] [PubMed] [Google Scholar]
- 73.Shieh Y.A., Yang S.J., Wei M.F., Shieh M.J. Aptamer-based tumor-targeted drug delivery for photodynamic therapy. ACS Nano. 2010;4:1433–1442. doi: 10.1021/nn901374b. [DOI] [PubMed] [Google Scholar]
- 74.Xiang D., Shigdar S., Qiao G., Wang T., Kouzani A.Z., Zhou S.F., Kong L., Li Y., Pu C., Duan W. Nucleic acid aptamer-guided cancer therapeutics and diagnostics: The next generation of cancer medicine. Theranostics. 2015;5:23–42. doi: 10.7150/thno.10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Song Y., Zhu Z., An Y., Zhang W., Zhang H., Liu D., Yu C., Duan W., Yang C.J. Selection of DNA aptamers against epithelial cell adhesion molecule for cancer cell imaging and circulating tumor cell capture. Anal. Chem. 2013;85:4141–4149. doi: 10.1021/ac400366b. [DOI] [PubMed] [Google Scholar]
- 76.An Y., Jin T., Zhu Y., Zhang F., He P. An ultrasensitive electrochemical aptasensor for the determination of tumor exosomes based on click chemistry. Biosens. Bioelectron. 2019;142:111503. doi: 10.1016/j.bios.2019.111503. [DOI] [PubMed] [Google Scholar]
- 77.Zhu G., Hu R., Zhao Z., Chen Z., Zhang X., Tan W. Noncanonical self-assembly of multifunctional DNA nanoflowers for biomedical applications. J. Am. Chem. Soc. 2013;135:16438–16445. doi: 10.1021/ja406115e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhu G., Zheng J., Song E., Donovan M., Zhang K., Liu C., Tan W. Self-assembled, aptamer-tethered DNA nanotrains for targeted transport of molecular drugs in cancer theranostics. Proc. Natl. Acad. Sci. USA. 2013;110:7998–8003. doi: 10.1073/pnas.1220817110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ferreira C.S., Matthews C.S., Missailidis S. DNA aptamers that bind to MUC1 tumour marker: Design and characterization of MUC1-binding single-stranded DNA aptamers. Tumour Biol. 2006;27:289–301. doi: 10.1159/000096085. [DOI] [PubMed] [Google Scholar]
- 80.Alix-Panabières C. EPISPOT assay: Detection of viable DTCs/CTCs in solid tumor patients. Recent Results Cancer Res. 2012;195:69–76. doi: 10.1007/978-3-642-28160-0_6. [DOI] [PubMed] [Google Scholar]
- 81.Zhang X., Soori G., Dobleman T.J., Xiao G.G. The application of monoclonal antibodies in cancer diagnosis. Expert Rev. Mol. Diagn. 2014;14:97–106. doi: 10.1586/14737159.2014.866039. [DOI] [PubMed] [Google Scholar]
- 82.De Rubis G., Rajeev Krishnan S., Bebawy M. Liquid Biopsies in Cancer Diagnosis, Monitoring, and Prognosis. Trends Pharmacol. Sci. 2019;40:172–186. doi: 10.1016/j.tips.2019.01.006. [DOI] [PubMed] [Google Scholar]
- 83.Keefe A.D., Pai S., Ellington A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jiang Y., Shi M., Liu Y., Wan S., Cui C., Zhang L., Tan W. Aptamer/AuNP Biosensor for Colorimetric Profiling of Exosomal Proteins. Angew. Chem. Int. Ed. Engl. 2017;56:11916–11920. doi: 10.1002/anie.201703807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen J., Xu Y., Lu Y., Xing W. Isolation and Visible Detection of Tumor-Derived Exosomes from Plasma. Anal. Chem. 2018;90:14207–14215. doi: 10.1021/acs.analchem.8b03031. [DOI] [PubMed] [Google Scholar]
- 86.Labib M., Green B., Mohamadi R.M., Mepham A., Ahmed S.U., Mahmoudian L., Chang I.H., Sargent E.H., Kelley S.O. Aptamer and Antisense-Mediated Two-Dimensional Isolation of Specific Cancer Cell Subpopulations. J. Am. Chem. Soc. 2016;138:2476–2479. doi: 10.1021/jacs.5b10939. [DOI] [PubMed] [Google Scholar]
- 87.Boriachek K., Islam M.N., Möller A., Salomon C., Nguyen N.T., Hossain M.S.A., Yamauchi Y., Shiddiky M.J.A. Biological Functions and Current Advances in Isolation and Detection Strategies for Exosome Nanovesicles. Small. 2018;14 doi: 10.1002/smll.201702153. [DOI] [PubMed] [Google Scholar]
- 88.Joy A.P., Ayre D.C., Chute I.C., Beauregard A.P., Wajnberg G., Ghosh A., Lewis S.M., Ouellette R.J., Barnett D.A. Proteome profiling of extracellular vesicles captured with the affinity peptide Vn96: Comparison of Laemmli and TRIzol© protein-extraction methods. J. Extracell Vesicles. 2018;7:1438727. doi: 10.1080/20013078.2018.1438727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gobbo J., Marcion G., Cordonnier M., Dias A.M.M., Pernet N., Hammann A., Richaud S., Mjahed H., Isambert N., Clausse V., et al. Restoring Anticancer Immune Response by Targeting Tumor-Derived Exosomes With a HSP70 Peptide Aptamer. J. Natl. Cancer Inst. 2016:108. doi: 10.1093/jnci/djv330. [DOI] [PubMed] [Google Scholar]
- 90.Murakami K., Zhao J., Yamasaki K., Miyagishi M. Biochemical and structural features of extracellular vesicle-binding RNA aptamers. Biomed. Rep. 2017;6:615–626. doi: 10.3892/br.2017.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu C., Zhao J., Tian F., Chang J., Zhang W., Sun J. λ-DNA- and Aptamer-Mediated Sorting and Analysis of Extracellular Vesicles. J. Am. Chem. Soc. 2019;141:3817–3821. doi: 10.1021/jacs.9b00007. [DOI] [PubMed] [Google Scholar]
- 92.Wang S., Zhang L., Wan S., Cansiz S., Cui C., Liu Y., Cai R., Hong C., Teng I.T., Shi M., et al. Aptasensor with Expanded Nucleotide Using DNA Nanotetrahedra for Electrochemical Detection of Cancerous Exosomes. ACS Nano. 2017;11:3943–3949. doi: 10.1021/acsnano.7b00373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Huang R., He L., Xia Y., Xu H., Liu C., Xie H., Wang S., Peng L., Liu Y., Liu Y., et al. A Sensitive Aptasensor Based on a Hemin/G-Quadruplex-Assisted Signal Amplification Strategy for Electrochemical Detection of Gastric Cancer Exosomes. Small. 2019;15:1900735. doi: 10.1002/smll.201900735. [DOI] [PubMed] [Google Scholar]
- 94.Jin D., Yang F., Zhang Y., Liu L., Zhou Y., Wang F., Zhang G.-J. ExoAPP: Exosome-Oriented, Aptamer Nanoprobe-Enabled Surface Proteins Profiling and Detection. Anal. Chem. 2018;90:14402–14411. doi: 10.1021/acs.analchem.8b03959. [DOI] [PubMed] [Google Scholar]
- 95.Yoshida M., Hibino K., Yamamoto S., Matsumura S., Yajima Y., Shiba K. Preferential capture of EpCAM-expressing extracellular vesicles on solid surfaces coated with an aptamer-conjugated zwitterionic polymer. Biotechnol. Bioeng. 2018;115:536–544. doi: 10.1002/bit.26489. [DOI] [PubMed] [Google Scholar]
- 96.Xu H., Liao C., Zuo P., Liu Z., Ye B.-C. Magnetic-Based Microfluidic Device for On-Chip Isolation and Detection of Tumor-Derived Exosomes. Anal. Chem. 2018;90:13451–13458. doi: 10.1021/acs.analchem.8b03272. [DOI] [PubMed] [Google Scholar]
- 97.Zhang K., Yue Y., Wu S., Liu W., Shi J., Zhang Z. Rapid Capture and Nondestructive Release of Extracellular Vesicles Using Aptamer-Based Magnetic Isolation. ACS Sens. 2019;4:1245–1251. doi: 10.1021/acssensors.9b00060. [DOI] [PubMed] [Google Scholar]
- 98.Zhang Z., Tang C., Zhao L., Xu L., Zhou W., Dong Z., Yang Y., Xie Q., Fang X. Aptamer-based fluorescence polarization assay for separation-free exosome quantification. Nanoscale. 2019;11:10106–10113. doi: 10.1039/C9NR01589B. [DOI] [PubMed] [Google Scholar]
- 99.Wang Z., Zong S., Wang Y., Li N., Li L., Lu J., Wang Z., Chen B., Cui Y. Screening and multiple detection of cancer exosomes using an SERS-based method. Nanoscale. 2018;10:9053–9062. doi: 10.1039/C7NR09162A. [DOI] [PubMed] [Google Scholar]
- 100.Champanhac C., Teng I.T., Cansiz S., Zhang L., Wu X., Zhoa Z., Fu T., Tan W. Development of a panel of DNA Aptamers with High Affinity for Pancreatic Ductal Adenocarcinoma. Sci. Rep. 2015;5:16788. doi: 10.1038/srep16788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kim Y.J., Lee H.S., Jung D.E., Kim J.M., Song S.Y. The DNA aptamer binds stemness-enriched cancer cells in pancreatic cancer. J. Mol. Recognit. 2017;30 doi: 10.1002/jmr.2591. [DOI] [PubMed] [Google Scholar]
- 102.Wan Y., Wang L., Zhu C., Zheng Q., Wang G., Tong J., Fang Y., Xia Y., Cheng G., He X., et al. Aptamer-Conjugated Extracellular Nanovesicles for Targeted Drug Delivery. Cancer Res. 2018;78:798. doi: 10.1158/0008-5472.CAN-17-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pi F., Binzel D.W., Lee T.J., Li Z., Sun M., Rychahou P., Li H., Haque F., Wang S., Croce C.M., et al. Nanoparticle orientation to control RNA loading and ligand display on extracellular vesicles for cancer regression. Nat. Nanotechnol. 2018;13:82–89. doi: 10.1038/s41565-017-0012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zou J., Shi M., Liu X., Jin C., Xing X., Qiu L., Tan W. Aptamer-Functionalized Exosomes: Elucidating the Cellular Uptake Mechanism and the Potential for Cancer-Targeted Chemotherapy. Anal. Chem. 2019;91:2425–2430. doi: 10.1021/acs.analchem.8b05204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Santulli-Marotto S., Nair S.K., Rusconi C., Sullenger B., Gilboa E. Multivalent RNA aptamers that inhibit CTLA-4 and enhance tumor immunity. Cancer Res. 2003;63:7483–7489. [PubMed] [Google Scholar]
- 106.Herrmann A., Priceman S.J., Kujawski M., Xin H., Cherryholmes G.A., Zhang W., Zhang C., Lahtz C., Kowolik C., Forman S.J., et al. CTLA4 aptamer delivers STAT3 siRNA to tumor-associated and malignant T cells. J. Clin. Investig. 2014;124:2977–2987. doi: 10.1172/JCI73174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Prodeus A., Abdul-Wahid A., Fischer N.W., Huang E.H., Cydzik M., Gariépy J. Targeting the PD-1/PD-L1 Immune Evasion Axis With DNA Aptamers as a Novel Therapeutic Strategy for the Treatment of Disseminated Cancers. Mol. Ther. Nucleic Acids. 2015;4:e237. doi: 10.1038/mtna.2015.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hervas-Stubbs S., Soldevilla M.M., Villanueva H., Mancheño U., Bendandi M., Pastor F. Identification of TIM3 2’-fluoro oligonucleotide aptamer by HT-SELEX for cancer immunotherapy. Oncotarget. 2016;7:4522–4530. doi: 10.18632/oncotarget.6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Berezhnoy A., Stewart C.A., McNamara J.O., 2nd, Thiel W., Giangrande P., Trinchieri G., Gilboa E. Isolation and optimization of murine IL-10 receptor blocking oligonucleotide aptamers using high-throughput sequencing. Mol. Ther. 2012;20:1242–1250. doi: 10.1038/mt.2012.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gupta S., Hirota M., Waugh S.M., Murakami I., Suzuki T., Muraguchi M., Shibamori M., Ishikawa Y., Jarvis T.C., Carter J.D., et al. Chemically modified DNA aptamers bind interleukin-6 with high affinity and inhibit signaling by blocking its interaction with interleukin-6 receptor. J. Biol. Chem. 2014;289:8706–8719. doi: 10.1074/jbc.M113.532580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Roth F., De La Fuente A.C., Vella J.L., Zoso A., Inverardi L., Serafini P. Aptamer-mediated blockade of IL4Rα triggers apoptosis of MDSCs and limits tumor progression. Cancer Res. 2012;72:1373–1383. doi: 10.1158/0008-5472.CAN-11-2772. [DOI] [PubMed] [Google Scholar]
- 112.McNamara J.O., Kolonias D., Pastor F., Mittler R.S., Chen L., Giangrande P.H., Sullenger B., Gilboa E. Multivalent 4-1BB binding aptamers costimulate CD8+ T cells and inhibit tumor growth in mice. J. Clin. Investig. 2008;118:376–386. doi: 10.1172/JCI33365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pastor F., Kolonias D., McNamara J.O., 2nd, Gilboa E. Targeting 4-1BB costimulation to disseminated tumor lesions with bi-specific oligonucleotide aptamers. Mol. Ther. 2011;19:1878–1886. doi: 10.1038/mt.2011.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schrand B., Berezhnoy A., Brenneman R., Williams A., Levay A., Kong L.Y., Rao G., Zhou S., Heimberger A.B., Gilboa E. Targeting 4-1BB costimulation to the tumor stroma with bispecific aptamer conjugates enhances the therapeutic index of tumor immunotherapy. Cancer Immunol. Res. 2014;2:867–877. doi: 10.1158/2326-6066.CIR-14-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dollins C.M., Nair S., Boczkowski D., Lee J., Layzer J.M., Gilboa E., Sullenger B.A. Assembling OX40 aptamers on a molecular scaffold to create a receptor-activating aptamer. Chem. Biol. 2008;15:675–682. doi: 10.1016/j.chembiol.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pastor F., Soldevilla M.M., Villanueva H., Kolonias D., Inoges S., de Cerio A.L., Kandzia R., Klimyuk V., Gleba Y., Gilboa E., et al. CD28 aptamers as powerful immune response modulators. Mol. Ther. Nucleic Acids. 2013;2:e98. doi: 10.1038/mtna.2013.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Soldevilla M.M., Villanueva H., Bendandi M., Inoges S., López-Díaz de Cerio A., Pastor F. 2-fluoro-RNA oligonucleotide CD40 targeted aptamers for the control of B lymphoma and bone-marrow aplasia. Biomaterials. 2015;67:274–285. doi: 10.1016/j.biomaterials.2015.07.020. [DOI] [PubMed] [Google Scholar]
- 118.Boltz A., Piater B., Toleikis L., Guenther R., Kolmar H., Hock B. Bi-specific aptamers mediating tumor cell lysis. J. Biol. Chem. 2011;286:21896–21905. doi: 10.1074/jbc.M111.238261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhou J., Tiemann K., Chomchan P., Alluin J., Swiderski P., Burnett J., Zhang X., Forman S., Chen R., Rossi J. Dual functional BAFF receptor aptamers inhibit ligand-induced proliferation and deliver siRNAs to NHL cells. Nucleic Acids Res. 2013;41:4266–4283. doi: 10.1093/nar/gkt125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Williams S.C. Circulating tumor cells. Proc. Natl. Acad. Sci. USA. 2013;110:4861. doi: 10.1073/pnas.1304186110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Joosse S.A., Gorges T.M., Pantel K. Biology, detection, and clinical implications of circulating tumor cells. EMBO Mol. Med. 2015;7:1–11. doi: 10.15252/emmm.201303698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sun C., Zhang R., Gao M., Zhang X. A rapid and simple method for efficient capture and accurate discrimination of circulating tumor cells using aptamer conjugated magnetic beads and surface-enhanced Raman scattering imaging. Anal. Bioanal. Chem. 2015;407:8883–8892. doi: 10.1007/s00216-015-9049-8. [DOI] [PubMed] [Google Scholar]
- 123.Ding L., Wu Y., Liu W., Liu L., Yu F., Yu S., Tian Y., Feng J., He L. Magnetic-assisted self-assembled aptamer/protein hybrid probes for efficient capture and rapid detection of cancer cells in whole blood. Talanta. 2019;205:120129. doi: 10.1016/j.talanta.2019.120129. [DOI] [PubMed] [Google Scholar]
- 124.Huang M., Song J., Huang P., Chen X., Wang W., Zhu Z., Song Y., Yang C. Molecular Crowding Evolution for Enabling Discovery of Enthalpy-Driven Aptamers for Robust Biomedical Applications. Anal. Chem. 2019;91:10879–10886. doi: 10.1021/acs.analchem.9b02697. [DOI] [PubMed] [Google Scholar]
- 125.Wen C.Y., Wu L.L., Zhang Z.L., Liu Y.L., Wei S.Z., Hu J., Tang M., Sun E.Z., Gong Y.P., Yu J., et al. Quick-response magnetic nanospheres for rapid, efficient capture and sensitive detection of circulating tumor cells. ACS Nano. 2014;8:941–949. doi: 10.1021/nn405744f. [DOI] [PubMed] [Google Scholar]
- 126.Sheng W., Chen T., Tan W., Fan Z.H. Multivalent DNA nanospheres for enhanced capture of cancer cells in microfluidic devices. ACS Nano. 2013;7:7067–7076. doi: 10.1021/nn4023747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zheng F., Cheng Y., Wang J., Lu J., Zhang B., Zhao Y., Gu Z. Aptamer-functionalized barcode particles for the capture and detection of multiple types of circulating tumor cells. Adv. Mater. 2014;26:7333–7338. doi: 10.1002/adma.201403530. [DOI] [PubMed] [Google Scholar]
- 128.Shen Q., Xu L., Zhao L., Wu D., Fan Y., Zhou Y., Ouyang W.H., Xu X., Zhang Z., Song M., et al. Specific capture and release of circulating tumor cells using aptamer-modified nanosubstrates. Adv. Mater. 2013;25:2368–2373. doi: 10.1002/adma.201300082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Liu J., Zhang Y., Zhao Q., Situ B., Zhao J., Luo S., Li B., Yan X., Vadgama P., Su L., et al. Bifunctional aptamer-mediated catalytic hairpin assembly for the sensitive and homogenous detection of rare cancer cells. Anal. Chim. Acta. 2018;1029:58–64. doi: 10.1016/j.aca.2018.04.068. [DOI] [PubMed] [Google Scholar]
- 130.Zamay G.S., Kolovskaya O.S., Ivanchenko T.I., Zamay T.N., Veprintsev D.V., Grigorieva V.L., Garanzha I.I., Krat A.V., Glazyrin Y.E., Gargaun A. Development of DNA aptamers to native EpCAM for isolation of lung circulating tumor cells from human blood. Cancers. 2019;11:351. doi: 10.3390/cancers11030351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bashmakova E.E., Krasitskaya V.V., Zamay G.S., Zamay T.N., Frank L.A. Bioluminescent aptamer-based solid-phase microassay to detect lung tumor cells in plasma. Talanta. 2019;199:674–678. doi: 10.1016/j.talanta.2019.03.030. [DOI] [PubMed] [Google Scholar]
- 132.Dong Z., Tang C., Zhao L., Xu J., Wu Y., Tang X., Zhou W., He R., Zhao R., Xu L., et al. A Microwell-Assisted Multiaptamer Immunomagnetic Platform for Capture and Genetic Analysis of Circulating Tumor Cells. Adv. Healthc. Mater. 2018;7:e1801231. doi: 10.1002/adhm.201801231. [DOI] [PubMed] [Google Scholar]
- 133.Li Z., Wang G., Shen Y., Guo N., Ma N. DNA-Templated Magnetic Nanoparticle-Quantum Dot Polymers for Ultrasensitive Capture and Detection of Circulating Tumor Cells. Adv. Funct. Mater. 2018;28:1707152. doi: 10.1002/adfm.201707152. [DOI] [Google Scholar]
- 134.Sun N., Li X., Wang Z., Zhang R., Wang J., Wang K., Pei R. A Multiscale TiO2 Nanorod Array for Ultrasensitive Capture of Circulating Tumor Cells. ACS Appl. Mater. Interfaces. 2016;8:12638–12643. doi: 10.1021/acsami.6b02178. [DOI] [PubMed] [Google Scholar]
- 135.Wu L., Zhu L., Huang M., Song J., Zhang H., Song Y., Wang W., Yang C. Aptamer-based microfluidics for isolation, release and analysis of circulating tumor cells. TrAC Trends Anal. Chem. 2019;117:69–77. doi: 10.1016/j.trac.2019.05.003. [DOI] [Google Scholar]
- 136.Zhai T.-T., Ye D., Zhang Q.-W., Wu Z.-Q., Xia X.-H. Highly Efficient Capture and Electrochemical Release of Circulating Tumor Cells by Using Aptamers Modified Gold Nanowire Arrays. ACS Appl. Mater. Interfaces. 2017;9:34706–34714. doi: 10.1021/acsami.7b11107. [DOI] [PubMed] [Google Scholar]
- 137.Sun D., Lu J., Chen D., Jiang Y., Wang Z., Qin W., Yu Y., Chen Z., Zhang Y. Label-free electrochemical detection of HepG2 tumor cells with a self-assembled DNA nanostructure-based aptasensor. Sens. Actuators B Chem. 2018;268:359–367. doi: 10.1016/j.snb.2018.04.142. [DOI] [Google Scholar]
- 138.Ou D., Sun D., Liang Z., Chen B., Lin X., Chen Z. A novel cytosensor for capture, detection and release of breast cancer cells based on metal organic framework PCN-224 and DNA tetrahedron linked dual-aptamer. Sens. Actuators B Chem. 2019;285:398–404. doi: 10.1016/j.snb.2019.01.079. [DOI] [Google Scholar]
- 139.Song Y., Shi Y., Huang M., Wang W., Wang Y., Cheng J., Lei Z., Zhu Z., Yang C. Bioinspired Engineering of a Multivalent Aptamer-Functionalized Nanointerface to Enhance the Capture and Release of Circulating Tumor Cells. Angew. Chem. Int. Ed. 2019;58:2236–2240. doi: 10.1002/anie.201809337. [DOI] [PubMed] [Google Scholar]
- 140.Bian Q., Wang W., Wang S., Wang G. Light-Triggered Specific Cancer Cell Release from Cyclodextrin/Azobenzene and Aptamer-Modified Substrate. ACS Appl. Mater. Interfaces. 2016;8:27360–27367. doi: 10.1021/acsami.6b09734. [DOI] [PubMed] [Google Scholar]
- 141.Fang S., Wang C., Xiang J., Cheng L., Song X., Xu L., Peng R., Liu Z. Aptamer-conjugated upconversion nanoprobes assisted by magnetic separation for effective isolation and sensitive detection of circulating tumor cells. Nano Res. 2014;7:1327–1336. doi: 10.1007/s12274-014-0497-9. [DOI] [Google Scholar]
- 142.Yang H., Li H., Xu D. High-density micro-well array with aptamer-silver conjugates for cell sorting and imaging at single cells. Anal. Chim. Acta. 2019;1063:127–135. doi: 10.1016/j.aca.2019.02.034. [DOI] [PubMed] [Google Scholar]
- 143.Ding C., Zhang C., Yin X., Cao X., Cai M., Xian Y. Near-Infrared Fluorescent Ag2S Nanodot-Based Signal Amplification for Efficient Detection of Circulating Tumor Cells. Anal. Chem. 2018;90:6702–6709. doi: 10.1021/acs.analchem.8b00514. [DOI] [PubMed] [Google Scholar]
- 144.Zeng Z., Tung C.-H., Zu Y. A Cancer Cell-Activatable Aptamer-Reporter System for One-Step Assay of Circulating Tumor Cells. Mol. Ther. Nucleic Acids. 2014;3 doi: 10.1038/mtna.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wang W., Luo J., Wang S. Recent Progress in Isolation and Detection of Extracellular Vesicles for Cancer Diagnostics. Adv. Healthc. Mater. 2018;7:1800484. doi: 10.1002/adhm.201800484. [DOI] [PubMed] [Google Scholar]
- 146.Tian F., Liu C., Lin L., Chen Q., Sun J. Microfluidic analysis of circulating tumor cells and tumor-derived extracellular vesicles. TrAC Trends Anal. Chem. 2019;117:128–145. doi: 10.1016/j.trac.2019.05.013. [DOI] [Google Scholar]
- 147.Ji Y., Qi D., Li L., Su H., Li X., Luo Y., Sun B., Zhang F., Lin B., Liu T., et al. Multiplexed profiling of single-cell extracellular vesicles secretion. Proc. Natl. Acad. Sci. USA. 2019;116:5979. doi: 10.1073/pnas.1814348116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Liu C., Zhao J., Tian F., Cai L., Zhang W., Feng Q., Chang J., Wan F., Yang Y., Dai B., et al. Low-cost thermophoretic profiling of extracellular-vesicle surface proteins for the early detection and classification of cancers. Nat. Biomed. Eng. 2019;3:183–193. doi: 10.1038/s41551-018-0343-6. [DOI] [PubMed] [Google Scholar]
- 149.Contreras-Naranjo J.C., Wu H.-J., Ugaz V.M. Microfluidics for exosome isolation and analysis: Enabling liquid biopsy for personalized medicine. Lab Chip. 2017;17:3558–3577. doi: 10.1039/C7LC00592J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Balaj L., Atai N.A., Chen W., Mu D., Tannous B.A., Breakefield X.O., Skog J., Maguire C.A. Heparin affinity purification of extracellular vesicles. Sci. Rep. 2015;5:10266. doi: 10.1038/srep10266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kang Y.T., Purcell E., Palacios-Rolston C., Lo T.W., Ramnath N., Jolly S., Nagrath S. Isolation and Profiling of Circulating Tumor-Associated Exosomes Using Extracellular Vesicular Lipid–Protein Binding Affinity Based Microfluidic Device. Small. 2019;15:1903600. doi: 10.1002/smll.201903600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Nakai W., Yoshida T., Diez D., Miyatake Y., Nishibu T., Imawaka N., Naruse K., Sadamura Y., Hanayama R. A novel affinity-based method for the isolation of highly purified extracellular vesicles. Sci. Rep. 2016;6:1–11. doi: 10.1038/srep33935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dong H., Chen H., Jiang J., Zhang H., Cai C., Shen Q. Highly Sensitive Electrochemical Detection of Tumor Exosomes Based on Aptamer Recognition-Induced Multi-DNA Release and Cyclic Enzymatic Amplification. Anal. Chem. 2018;90:4507–4513. doi: 10.1021/acs.analchem.7b04863. [DOI] [PubMed] [Google Scholar]
- 154.Xue F., Chen Y., Wen Y., Abhange K., Zhang W., Cheng G., Quinn Z., Mao W., Wan Y. Isolation of extracellular vesicles with multivalent aptamers. Analyst. 2021;146:253–261. doi: 10.1039/D0AN01420F. [DOI] [PubMed] [Google Scholar]
- 155.Zhou Y.-G., Mohamadi R.M., Poudineh M., Kermanshah L., Ahmed S., Safaei T.S., Stojcic J., Nam R.K., Sargent E.H., Kelley S.O. Interrogating Circulating Microsomes and Exosomes Using Metal Nanoparticles. Small. 2016;12:727–732. doi: 10.1002/smll.201502365. [DOI] [PubMed] [Google Scholar]
- 156.Zhou Q., Rahimian A., Son K., Shin D.-S., Patel T., Revzin A. Development of an aptasensor for electrochemical detection of exosomes. Methods. 2016;97:88–93. doi: 10.1016/j.ymeth.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 157.Xia Y., Liu M., Wang L., Yan A., He W., Chen M., Lan J., Xu J., Guan L., Chen J. A visible and colorimetric aptasensor based on DNA-capped single-walled carbon nanotubes for detection of exosomes. Biosens. Bioelectron. 2017;92:8–15. doi: 10.1016/j.bios.2017.01.063. [DOI] [PubMed] [Google Scholar]
- 158.Wang Y.-M., Liu J.-W., Adkins G.B., Shen W., Trinh M.P., Duan L.-Y., Jiang J.-H., Zhong W. Enhancement of the Intrinsic Peroxidase-Like Activity of Graphitic Carbon Nitride Nanosheets by ssDNAs and Its Application for Detection of Exosomes. Anal. Chem. 2017;89:12327–12333. doi: 10.1021/acs.analchem.7b03335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wan S., Zhang L., Wang S., Liu Y., Wu C., Cui C., Sun H., Shi M., Jiang Y., Li L., et al. Molecular Recognition-Based DNA Nanoassemblies on the Surfaces of Nanosized Exosomes. J. Am. Chem. Soc. 2017;139:5289–5292. doi: 10.1021/jacs.7b00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Shen W., Guo K., Adkins G.B., Jiang Q., Liu Y., Sedano S., Duan Y., Yan W., Wang S.E., Bergersen K., et al. A Single Extracellular Vesicle (EV) Flow Cytometry Approach to Reveal EV Heterogeneity. Angew. Chem. Int. Ed. 2018;57:15675–15680. doi: 10.1002/anie.201806901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Boussadia Z., Lamberti J., Mattei F., Pizzi E., Puglisi R., Zanetti C., Pasquini L., Fratini F., Fantozzi L., Felicetti F., et al. Acidic microenvironment plays a key role in human melanoma progression through a sustained exosome mediated transfer of clinically relevant metastatic molecules. J. Exp. Clin. Cancer Res. 2018;37:245. doi: 10.1186/s13046-018-0915-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Brennan K., Martin K., FitzGerald S.P., O’Sullivan J., Wu Y., Blanco A., Richardson C., Mc Gee M.M. A comparison of methods for the isolation and separation of extracellular vesicles from protein and lipid particles in human serum. Sci. Rep. 2020;10:1039. doi: 10.1038/s41598-020-57497-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lee K., Fraser K., Ghaddar B., Yang K., Kim E., Balaj L., Chiocca E.A., Breakefield X.O., Lee H., Weissleder R. Multiplexed Profiling of Single Extracellular Vesicles. ACS Nano. 2018;12:494–503. doi: 10.1021/acsnano.7b07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Chen C., Zong S., Liu Y., Wang Z., Zhang Y., Chen B., Cui Y. Profiling of Exosomal Biomarkers for Accurate Cancer Identification: Combining DNA-PAINT with Machine- Learning-Based Classification. Small. 2019;15:1901014. doi: 10.1002/smll.201901014. [DOI] [PubMed] [Google Scholar]
- 165.Hu J., Sheng Y., Kwak K.J., Shi J., Yu B., Lee L.J. A signal-amplifiable biochip quantifies extracellular vesicle-associated RNAs for early cancer detection. Nat. Commun. 2017;8:1683. doi: 10.1038/s41467-017-01942-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Shao H., Chung J., Lee K., Balaj L., Min C., Carter B.S., Hochberg F.H., Breakefield X.O., Lee H., Weissleder R. Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nat. Commun. 2015;6:6999. doi: 10.1038/ncomms7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wan Y., Cheng G., Liu X., Hao S.-J., Nisic M., Zhu C.-D., Xia Y.-Q., Li W.-Q., Wang Z.-G., Zhang W.-L., et al. Rapid magnetic isolation of extracellular vesicles via lipid-based nanoprobes. Nature Biomed. Eng. 2017;1:0058. doi: 10.1038/s41551-017-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Gao X., Li S., Ding F., Fan H., Shi L., Zhu L., Li J., Feng J., Zhu X., Zhang C. Rapid Detection of Exosomal MicroRNAs Using Virus-Mimicking Fusogenic Vesicles. Angew. Chem. Int. Ed. 2019;58:8719–8723. doi: 10.1002/anie.201901997. [DOI] [PubMed] [Google Scholar]
- 169.Sun B., Li Y., Zhou Y., Ng T.K., Zhao C., Gan Q., Gu X., Xiang J. Circulating exosomal CPNE3 as a diagnostic and prognostic biomarker for colorectal cancer. J. Cell. Physiol. 2019;234:1416–1425. doi: 10.1002/jcp.26936. [DOI] [PubMed] [Google Scholar]
- 170.Kanaoka R., Iinuma H., Dejima H., Sakai T., Uehara H., Matsutani N., Kawamura M. Usefulness of Plasma Exosomal MicroRNA-451a as a Noninvasive Biomarker for Early Prediction of Recurrence and Prognosis of Non-Small Cell Lung Cancer. Oncology. 2018;94:311–323. doi: 10.1159/000487006. [DOI] [PubMed] [Google Scholar]
- 171.Takahasi K., Iinuma H., Wada K., Minezaki S., Kawamura S., Kainuma M., Ikeda Y., Shibuya M., Miura F., Sano K. Usefulness of exosome-encapsulated microRNA-451a as a minimally invasive biomarker for prediction of recurrence and prognosis in pancreatic ductal adenocarcinoma. J. Hepatobiliary Pancreat Sci. 2018;25:155–161. doi: 10.1002/jhbp.524. [DOI] [PubMed] [Google Scholar]
- 172.Ng E.W., Shima D.T., Calias P., Cunningham E.T., Jr., Guyer D.R., Adamis A.P. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006;5:123–132. doi: 10.1038/nrd1955. [DOI] [PubMed] [Google Scholar]