Abstract
The increase in the use of antimicrobials such as colistin for the treatment of infectious diseases has led to the appearance of Aeromonas strains resistant to this drug. However, resistance to colistin not only occurs in the clinical area but has also been determined in Aeromonas isolates from the environment or animals, which has been determined by the detection of mcr genes that confer a resistance mechanism to colistin. The variants mcr-1, mcr-3, and mcr-5 have been detected in the genus Aeromonas in animal, environmental, and human fluids samples. In this article, an overview of the resistance to colistin in Aeromonas is shown, as well as the generalities of this molecule and the recommended methods to determine colistin resistance to be used in some of the genus Aeromonas.
Keywords: colistin, antimicrobial resistance, Aeromonas
1. Introduction
Colistin is a lipopeptide antibiotic from the group of polymyxins. It has a cyclic peptide chain that is linked to a fatty acid. Colistin is used in the medical field, since it is an extended-spectrum antimicrobial, and it is used as a last line of treatment in human infections that are caused by Gram-negative bacilli [1]. Until 2016, resistance to colistin was reported in some genera of bacteria intrinsically and contained in the bacterial genophore, until the presence of a gene called mcr, present in a plasmid that confers resistance to this antimicrobial, was detected in an Escherichia coli strain [2]. After this report, the number of isolates of various origins with mcr genes and a colistin resistance phenotype was increased; in addition, it was found that resistance to this molecule could be transferred horizontally [3,4].
The use of colistin as a treatment for infections increased after the appearance of multidrug resistance phenotypes (MDR) in Gram-negative bacilli and the appearance of carbapenemase-producing enterobacteria type KPC (Klebsiella pneumoniae carbapenemase) or NDM (New Delhi metallo-β-lactamase), in addition to Gram-negative bacilli classified as XDR (extensively drug resistant) that continue to appear, especially in bacteria such as Klebsiella pneumoniae, Pseudomonas aeruginosa, and other Gram-negative bacilli, such as Aeromonas, which is also of medical and veterinary importance and is isolated from environmental samples [5,6].
Aeromonas is a Gram-negative bacillus, positive for oxidase and catalase tests, a glucose fermenter, and it is resistant to vibriostatic O/129 (2,4-diamino-6,7-diisopropylpteridine) [7]. In humans, it can cause intestinal and extra-intestinal infections. It is important in the medical area, mainly in patients with diarrhea, or with infections in the skin and soft tissue; moreover, it can cause bacteremia, which progresses to sepsis, or endocarditis [8,9].
The genus Aeromonas is widely distributed in diverse ecosystems; however, it is a bacterium native to aquatic systems, hence the largest number of isolates are from water. Isolates have been obtained from drinking water, wastewater, bottled water, seawater, and deep and surface water samples. Food isolates have been obtained from vegetables, fruits, pork, poultry, and beef, as well as seafood and fish. In animals, it is considered a pathogen, especially in fish, in which it can cause furunculosis, ulcers, and hemorrhages, among other diseases. This pathogen has also been isolated from infections in rabbits, dogs, cats, chickens, horses, and crustaceans [8,9].
2. The Colistin Situation
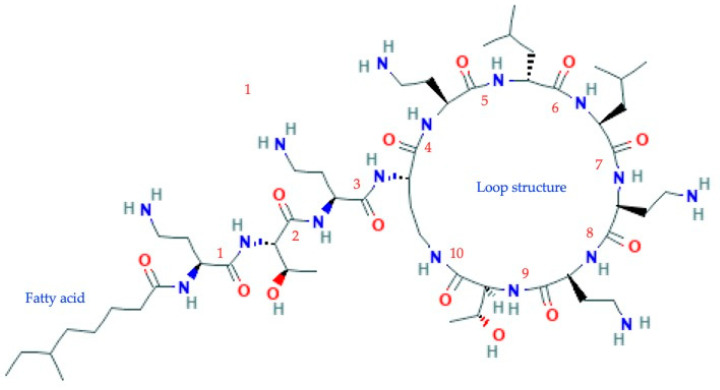
Colistin is a lipopeptide molecule that is included in the polymyxin family. This molecule has a peptide chain that is made up of 10 amino acids and is linked to a fatty acid. It has surface-active properties that alter the permeability of the Gram-negative membrane in contact with lipopolysaccharides (LPS), resulting in cell death due to the electrostatic attraction that is exerted between the positive charge of the amino groups and the anions, phosphate, and carboxylate (Figure 1) [1,3,4].
Figure 1.
Polymyxin or colistin structure. Basic structure of polymyxin E composed of a chain of 6-methyl-octanoic acid (fatty acid), linked to a cyclic lipopeptide of 10 amino acids. 1, diaminobutyric acid (L-Dab); 2, threonine (L-Thr); 3, diaminobutyric acid (L-Dab); 4–5, diaminobutyric acid (L-Dab); 6, leucine (D-Leu); 7, leucine (L-Leu); 8–9, diaminobutyric acid (L-Dab); 10, threonine (L-Thr). The residues in position 6 of D-Leu and L-Leu in position 7 determine polymyxin E. Amino acid substitution at positions 6 and 7 determines the colistin subtype. The loop is formed by the amino acids from position 4 to 10. This cyclic peptide is composed of non-proteinogenic amino acids (D-Leu; L-Dab) and contains an isopeptide bond linking amino acids 4 and 10. These characteristics make the peptide resistant to the action of most peptidases/proteases and indicate that the compound originates from a non-ribosomal protein synthesis (NRPS) biosynthetic pathway. Modified from PubChem (https://pubchem.ncbi.nlm.nih.gov/compound/Colistin#section=Names-and-Identifiers, access date: 21 May 2021).
This molecule contains a structure called a cyclic loop, formed by the seven amino acids that are among the amino group of the diaminobutyric acid side chain in position 4 and the carboxyl group of the terminal carbon of residue number 10 of threonine (Thr). This loop is linked by a chain of three amino acids to the amino terminal fatty acid [1].
Colistin’s mechanism of action is focused on the membrane structure of Gram-negative bacteria; they have the outer membrane that is a barrier of the passage of various substances, including antimicrobials. This membrane has a phospholipid layer inside and LPS on the outside. The LPS structure has three domains: lipid A, which is the domain that is anchored to the membrane and the hydrophobic and endotoxic portions of the structure; the O antigen, an oligosaccharide polymer; and the nucleus or core, which is also an oligosaccharide, and is the domain that connects lipid A to antigen O and is divided into the inner core and the outer core [1,3,10,11,12].
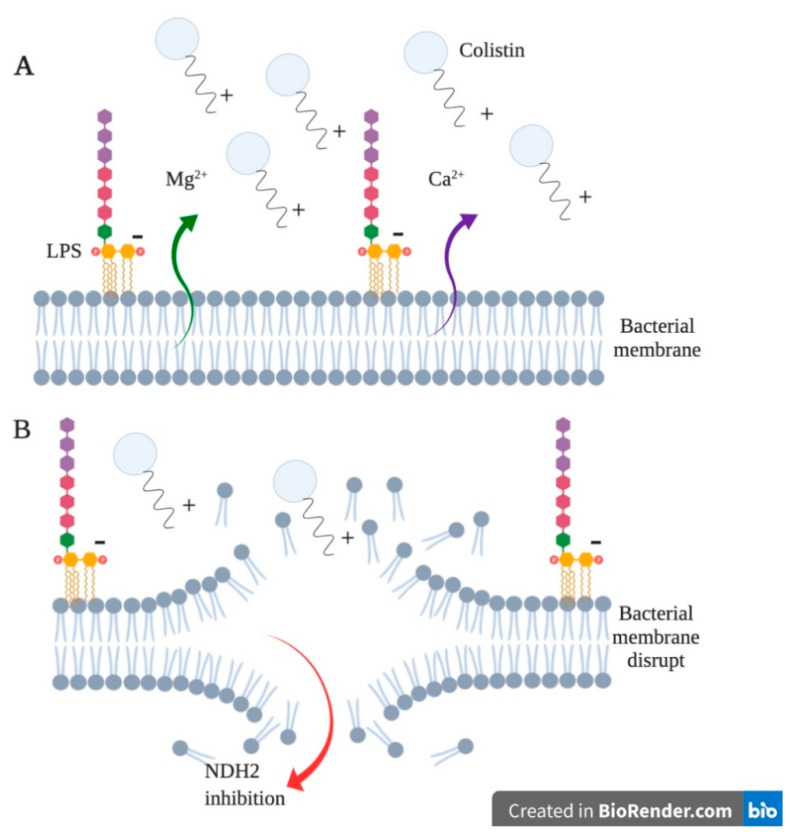
Lipid A is a glucosamine disaccharide linked by a β-1,6 bond, esterified at four positions with saturated fatty acids, and phosphorylated at ends 1 and 4. Two divalent cations participate in the mechanism of action of colistin, Mg2+ and Ca2+, associated with the phosphoesters of lipid A. These cations keep the adjacent LPS molecules together and stable, in such a way that the membrane of the Gram-negative bacterium exerts a dual function, as a mechanical barrier and at the same time an electrostatic barrier with a high repulsive anionic charge that is generated by the phosphoesters of lipid A, and by the phosphates and carboxylates of the core sugars and the O antigen (Figure 2) [1,3,10,11,12].
Figure 2.
Colistin mechanism of action. (A) The positively charged colistin molecule interacts with the negative charges of the LPS membrane, resulting in the release of Ca2+/Mg2+ ions. (B) This interaction results in a membrane disruption, therefore, the output of Ca2+/Mg2+ ions is generated. Finally, colistin enters the cytoplasm and inhibits the activity of the oxidoreductases enzyme (NDH2). Generated from this work.
2.1. Colistin Activity Spectrum
Colistin resistance rates are generally low, in contrast to carbapenems, which are constantly increasing. The limited therapeutic options against MDR microorganisms such as A. baumanni or P. aeruginosa lead to an increased use of colistin in the treatment of this type of microorganism. For this reason, it is controlled in the hospital area and the use of other antimicrobials or the combination of two is preferred, before resorting to colistin [13].
The use of colistin is focused mainly on the family Enterobacteriaceae and non-fermenting bacilli. Within these genera, there are groups with intrinsic resistance to colistin, such as Neisseria spp., Stenotrophomonas maltophilia, Brucella spp., Proteus spp., Providencia spp., Serratia spp., and Burkholderia spp., which are outside the spectrum of this antimicrobial, and likewise, Gram-positive cocci and strict anaerobes, such as Bacteroides fragilis (Table 1) [3].
Table 1.
Colistin activity spectrum on Gram-negative bacteria.
| Sensitive | Resistance | Variables |
|---|---|---|
|
Escherichia coli * Klebsiella pneumoniae * Pseudomonas aeruginosa * Acinetobacter baumannii * Salmonella spp. Shigella spp. Legionella pneumophila Haemophilus influenzae Bordetella pertussis Prevotella spp.a Fusobacterium spp.a |
Proteus spp. Burkholderia spp. Helicobacter pylori Neisseria meningitidis Bacteroides fragilis a |
Stenotrophomonas maltophilia Moraxella catarrhalis Vibrio spp. Aeromonas spp. |
* Sensitive Gram-negative bacilli with reported colistin resistance. a Strict anaerobic bacteria. Modified from the literature [6].
In addition to the bacterial groups of medical importance with colistin resistance that have been isolated from clinical samples and other environmental sources, from animals or from food, there are other Gram-negative bacilli that are important, such as human and animal pathogens, which have been isolated from various sources, such as the genus Aeromonas, from which their resistance to this drug has been determined and their probable responsibility that mcr genes are resistant to it. Resistance to colistin is also present in isolates from environmental samples, from which antimicrobial resistance has been documented [14,15,16,17,18].
2.2. Colistin, Pharmacology and Application
Currently, the use of colistin has increased in combination with other drugs, such as phosphomycin and tigecycline, as has the emergence of resistance to carbapenems, in combination therapy, double or triple, due to the shortage of active pharmacological molecules. This was derived from the presentation of resistant phenotypes of KPC or NDM, and those Gram-negative rods classified as XDR that were increased to include Gram-negative rods and non-fermenter bacilli [1,2,3].
Among the drugs that are part of the treatment strategy against MDR microorganisms are polymyxins B and E (colistin). Although these antibiotics have adverse effects, such as nephrotoxicity and neurotoxicity, they are one of the last alternatives available for human infections caused by multidrug-resistant Gram-negative bacilli. In contrast, genes such as mcr have been found, whose products are related to colistin resistance [11,12].
3. Antimicrobial Resistance in Aeromonas
The molecular basis of antimicrobial resistance in Aeromonas spp. has been widely studied, but their importance in the hospital area as a cause of outbreaks is not fully established. Likewise, reports on resistance are varied regarding the origin of isolation and the type of antimicrobials tested in vitro [7]. The Aeromonas resistance profile has not changed significantly; until now, the mechanism of action of inducible chromosomal β-lactamases and carbapenemase expression has been suggested as being the main resistance mechanism for Aeromonas species. Three classes of β-lactamases are recognized in Aeromonas; one of class C cephalosporinase, one of class D penicillinase, and one of class B metallo-β-lactamase (MBL) [9,19].
The occurrence of MDR-type Aeromonas spp. isolates has been increasing. Different authors have suggested that antimicrobial resistance in the clinical setting is closely related to resistance mechanisms detected in environmental isolates [20]. In the genus Aeromonas, the occurrence of MDR strains is equivalent, due to their origin in aquatic environments, which is attributed to the extensive use of antibiotics in aquaculture. Therefore, this environment becomes an ideal setting for the acquisition of these mechanisms of resistance to antimicrobials and other toxic agents [21].
Colistin Resistance in Aeromonas
Since the report in 2016, where it was shown that colistin resistance can be encoded by the mcr genes detected within a plasmid, it was determined that these genes are not only in bacterial genophores but can also be present in mobile genetic elements as plasmids [2]. From this report, attention was paid to the search for and detection of these genes in different bacterial genera, mainly those of medical importance, but also isolated from other sources, such as the environment or animals. In K. pneumoniae, P. aeruginosa, or others, such as Aeromonas genus, mcr gene variants have been detected, and the reports are increasing [22,23].
Colistin resistance in Aeromonas has been reported in several regions of the world, mainly in Europe and Asia. Resistance to this antibiotic has been reported in Latin America in other genera, but not in Aeromonas. The detection of colistin resistance is more common in the clinical area; however, colistin-resistant strains of Aeromonas have been isolated from other origins that have been detected, from which investigations and reports have emerged in the world. The extensive use of antibiotics in aquaculture and in human treatment has led to an increase in the resistance of this genus to antimicrobial drugs [20,21].
The species A. dhakensis, A. hydrophila, A. caviae, and A. veronii are considered the main causes of human infections that can cause infection in wounds, diarrheal syndromes, and other clinical presentations [9]. Commonly, the isolates do not present resistance to antimicrobials; however, MDR isolates have still appeared, and in recent years the report of Aeromonas isolates from clinical samples and from various sources with resistance to colistin has increased [24]. This resistance has been investigated in Aeromonas spp. isolates, by means of disk diffusion test and by minimum inhibitory concentration (MIC), showing the MIC method to be more effective. Induction of colistin resistance in the strains showed an 85% increase after overnight incubation in a tube with Müller–Hinton broth and a 50 µL colistin disk. This result allowed the establishment of a phenotypic marker in the Aeromonas isolates [25].
In an Aeromonas veronii isolate from chicken meat, two adjacent genes with colistin resistance markers, called mcr-3.3 and mcr-3-like, were detected in the genophore. The result had 95.2 and 84.19% identity in the nucleotide sequence, when compared to the mcr-3 gene of an E. coli of porcine origin [26].
The evidence of the mcr genes in Aeromonas was demonstrated by a group of scientists who analyzed a total of 6497 strains that were collected in 13 provinces of China between 2016 and 2017. In these samples, the presence of the mcr-3 genes was detected by PCR. The mcr-3 gene was detected in 49 strains only, of which eight strains corresponded to the genus Aeromonas, two A. hydrophila strains, one with mcr-3.8 variant, and one with mcr-3.9 variant, one A. caviae with mcr-3.1 variant, and one A. media with mcr-3.6 variant. Of the four remaining strains, one each were of A. veronii, A. media, and A. caviae, and one was Aeromonas spp. with mcr-3 without variant. All the strains were grouped into a subclade, after the phylogenetic analysis of the sequences of the mcr-3 genes detected in the strains [18].
Another group of researchers found four Aeromonas isolates with the presence of the mcr-3 gene through PCR, while the mcr-1 or mcr-2 genes were not detected. Each of the four isolates with mcr-3 genes presented a different variant each; these presented identities in the amino acid chain were of 95 to 98% compared to the original protein MCR-3. These variants of the protein were designated as MCR-3.6 obtained from the A. allosaccharophila strain isolated from Leuciscus idus, MCR-3.7 for the protein detected in the A. media strain isolated from Meleagris gallopavo, MCR-3.8 for that detected in the A. jandaei strain isolated from a Cyprinus carpio carp, and MCR-3.9 for the protein of the A. hydrophila strain of Cyprinus carpio. The isolate with the mcr-3.9 gene also contained an additional mcr-3.8 gene in the MIC test, with colistin showing an MIC ≥ 128 mg/L higher compared to the other isolates [27].
The reports include a new variant of the mcr-3 gene in A. caviae, also detected in Proteus mirabilis and E. coli that were isolated from a domestic duck. These strains were obtained from sewage samples from free-range ducks, which were raised near a river in the suburban area of Qingdao, Shangdon Province, in China. The presence of the mcr-3 gene was demonstrated in 1 of 15 samples processed in this study. The result was obtained by detection of the mcr gene directly in the sample; the positive sample was seeded in a CHROMagar plate from Biomerieux®, France, to which 2 mg/L of colistin was added. Based on the above, three positive strains were detected for mcr-3 gene. A. caviae 17AC, P. mirabilis 17PM, and E. coli 17EC strains were identified by MALDI-TOF (Matrix-Assisted Laser Desorption/Ionization-Time of Flight) technology, and by 16S rRNA gene sequencing [27]. In another study, the prevalence was determined, complemented by a genetic analysis of the mcr-3 gene in Aeromonas species. These isolates were obtained from human rectal exudates, meat for human consumption, and environmental water samples [28].
The variant mcr-5 gene of colistin resistance was detected in an A. hydrophila strain isolated from a fecal sample from a backyard pig. In this case, the mcr-5 gene was detected in a plasmid with 7915 base pairs (bp) named pI064-2. Additionally, they analyzed the possibility of transforming the A. hydrophila strains susceptible to colistin into a resistant strain [29].
Various mechanisms of resistance to colistin, in addition to the mechanism mediated by mcr genes, have been described in some bacteria, including P. aeruginosa, A. baumannii, members of the Enterobacteriaceae family, such as E. coli, Salmonella spp., and K. pneumoniae—they have an acquired resistance against colistin. However, the possibility of the appearance of strains resistant to this antibiotic should be monitored, due to the presence of mutations, new mechanisms, or adaptations [11].
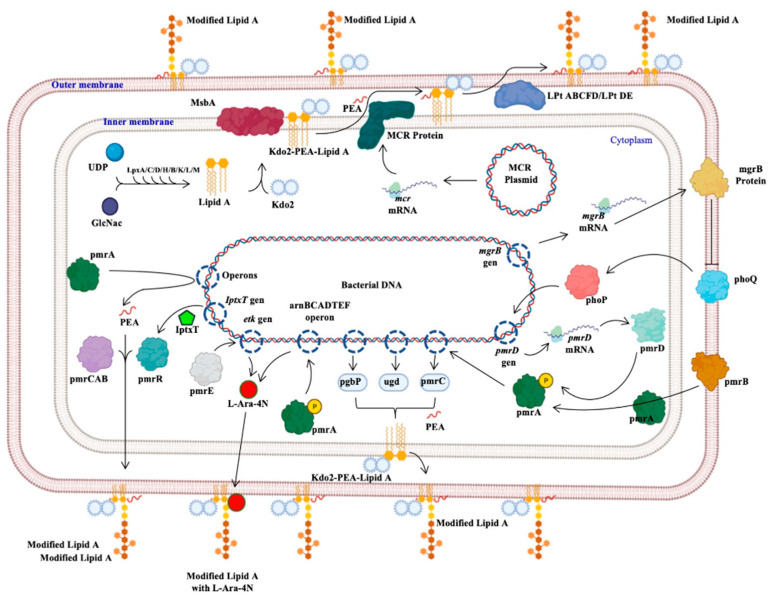
The protein generated by the mcr genes confers resistance to polymyxins; this protein, called MCR, from the inner membrane, adds a molecule of phosphoethanolamine (PEA) to lipid A of lipopolysaccharide (Kdo2-PEA-Lipid A), synthesized by the binding of uridine diphosphate (UDP) and N-acetyl glucosamine (GlcNac) mediated by Lpx proteins (C, D, H, B, K, L, M). The product binds to 2-keto deoxioctanate acid (Kdo2), which is transported by the MsbA protein to the inner membrane, where the PEA molecule is added. The Lpt protein complex (ABCFD/DE) transports the modified LPS that generates resistance to colistin, since the negative charge with which it interacts was modified (Figure 3) [30,31,32].
Figure 3.
Bacterial mechanisms of resistance to colistin. The different mechanisms of resistance to colistin described in Gram-negative bacilli and other bacteria are presented: the mechanism mediated by the mcr genes, those regulated by the phoP and phoQ proteins, others regulated by the pmr proteins or by the iptxT and etk genes, and operons that encode activation proteins of pmr proteins [11,30,31,32]. Generated from this work.
Another mechanism of resistance to colistin is the two-component system composed of the membrane proteins phoP and phoQ. This mechanism is regulated by the expression of the mgrB gene, and when the phoQ protein is activated, it acts on phoP, that functions as an inducer of the prmD gene, which phosphorylates the prmA protein; additionally, the prmA protein can be activated by the prmB protein. The phosphorylated prmA protein activates the expression of the pgbP, ugd, and prmC genes, which independently add a PEA molecule to the LPS, generating resistance to colistin in the bacteria. Likewise, the prmA protein can induce the expression of operons, whose composition has not been determined, and which adds a PEA molecule mediated by the prmC, A, and B proteins. Furthermore, the lptx protein, with the help of the prmR protein, can also add a PEA to the LPS (Figure 3) [11,30].
The last colistin resistance mechanism described in this article is the one that adds a 4-amino-4-deoxy-L-arabinose (L-Ara-4N) to the LPS, in which the prmA protein induces the expression of the arnBCADTEF operon; additionally, the prmE protein induces the expression of the etk gene, which is also capable of adding an L-Ara-4N to the LPS. This modification generates a different structure in the LPS that prevents colistin from interacting with Lipid A of LPS; therefore, colistin is unable to perform its function (Figure 3) [11,30,31,32].
Until now, only the mcr-1, mcr-3, and mcr-5 gene variants have been reported in the genus Aeromonas. The variability in the presence of these genes in Gram-negative bacilli may be due to the diversity of their origin, the plasticity of the genus to receive external genetic material, or the selective pressure exerted by both the environment and exposure to antimicrobials on the health environment [33].
4. Detection Methods of Colistin Resistance
In the 2021 version of the CLSI, methods for the determination of resistance or susceptibility to colistin by MIC are described. The accepted methods are microdilution in broth, elution in broth, and diffusion in agar. These methods are indicated for the Enterobacteriaceae family and for P. aeruginosa; however, there are no indications for other Gram-negative bacilli, such as Aeromonas [34].
4.1. Elution Method in Broth
The CLSI 2021 (M-100) indicates the use of four tubes with 10 mL of Müeller–Hinton broth with cation adjustment for Ca2+ and Mg2+ in each one. Aseptically, one disc of colistin (10 μg/mL) to tube “1”, two discs to tube “2”, and four to tube “4” are added; the control tube will not have a disc. The tubes are completely closed after adding the colistin discs and left to stand for no more than an hour, so that the colistin elutes from the discs in the broth, at room temperature. These tubes are considered as the stock or stock solution for use with each strain [6,22,35].
From growth of 24 h in a plate of the strains to be tested, an inoculum will be adjusted with sterile physiological solution. From the adjusted inoculum, 50 μL are placed in each of the tubes marked “1”, “2”, “4”, and the control, to reach a final concentration of approximately 7.5 × 105 CFU/mL. Additionally, it is recommended to inoculate blood agar plates with 5% sheep blood, to check viability and purity. The inoculum is mixed with each tube with colistin softly, so that the colistin does not adhere to the walls of the tube or the cap. The tubes are incubated between 33 and 35 °C for 16–20 h, taking time to observe the test [6,36].
For results interpretation, bacterial growth is expected in the control tube to validate the test. The MIC will be that of the tube with no growth and with the lowest concentration of colistin. If the MIC is ≤2 μg/mL it is intermediate, or resistant if the MIC is ≥4 μg/mL. If inconsistency is shown in the results, for example, bacterial growth in tube 1 and no bacterial growth in tube 2, the method is repeated. These results may be due to the presence of contamination in the inoculum, presence of heteroresistance, errors in tube inoculation, or inappropriate concentrations in elution [6,36,37].
4.2. Plate Test Method
Initially, the strain purity is analyzed in a plate incubated for 24 h on plates with colistin agar, and the concentration is adjusted to 0, 1, 2, and 4 μg/mL. The tube of the McFarland Nephelometer scale is adjusted to 0.5 in 4–5 mL of sterile saline solution, then diluted 1:10. Up to 10 strains of the dilution are seeded on colistin agar plates with a bacteriological loop adjusted to 10 μL. Then, they are incubated between 33 and 35 °C for 16–20 h. The growth in the four plates is checked and interpreted for whether or not there is growth, so that it is intermediate if it grows in the plate with 2 μg/mL, or resistant if it grows in the plate with 4 μg/mL [34].
5. Conclusions
The genus Aeromonas is a bacterium widely distributed in the environment. Its presence in various ecosystems and ability to cause infections in animals and humans makes it a bacterium of interest in the study related to the appearance of strains resistant to antimicrobials. The presence of antimicrobial-resistant isolates from the first line of care, and those of the last alternative, such as colistin, detected in the clinical area, in animals or in the environment, demonstrates that the resistance comes from the exposure of bacteria to antimicrobials in clinical care, but it is also suggested that resistance originates in the environment. In addition, it is important to study Aeromonas isolates with colistin resistance from different sources, and also with the detection of the mcr genes.
Acknowledgments
L.U.G.-A. and M.Á.L.C. received grant-aided support from “Consejo Nacional de Ciencia y Tecnología” (CONACyT, México), while C.H.-C., J.M.B.-L., and G.C.-E. received support from the “Sistema Nacional de Investigadores (SNI)” from CONACyT, Mexico. G.C.-E. received support from Estímulos al Desempeño en Investigación, Comisión y Fomento de Actividades Académicas (Instituto Politécnico Nacional). This study was approved by Secretaría de Investigación y Posgrado del Instituto Politécnico Nacional (SIP 20200675, 20211351). The authors are also grateful to Sofia Mulia for correcting the style of the manuscript.
Author Contributions
L.U.G.-A. and G.C.-E. developed the structural design of the review and drafted the manuscript. M.A.L.-C., C.H.-C., and J.M.B.-L. reviewed the manuscript critically for important intellectual content and appropriate academic content. L.U.G.-A. designed Figure 2 and Figure 3 for this manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interests.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Upert G., Luther A., Obrecht D., Ermert P. Emerging peptide antibiotics with therapeutic potential. Med. Drug Discov. 2021;9:100078. doi: 10.1016/j.medidd.2020.100078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Y.Y., Wang Y., Walsh T.R., Yi L.X., Zhang R., Spencer J., Yu L.F. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 3.Medina J., Paciel D., Noceti O., Rieppi G. Actualización acerca de colistina (polimixina E): Aspectos clínicos, PK/PD y equivalencias. Rev. Méd. Uru. 2017;33:79–114. doi: 10.29193/RMU.33.3.5. [DOI] [Google Scholar]
- 4.Karaiskos I., Souli M., Galani I., Giamarellou H. Colistin: Still a lifesaver for the 21st century? Expert Opin. Drug. Met. 2017;13:59–71. doi: 10.1080/17425255.2017.1230200. [DOI] [PubMed] [Google Scholar]
- 5.Anandan S., Gopi R., Ragupathi N.K.D., Sethuvel D.P.M., Gunasekaran P., Walia K., Veeraraghavan B. First report of blaOXA-181-mediated carbapenem resistance in Aeromonas caviae in association with pKP3-A: Threat for rapid dissemination. J. Glob. Antimicrob. Resist. 2017;10:310–314. doi: 10.1016/j.jgar.2017.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Jiménez-Pearson M.A., Galas M., Corso A., Hormazábal J.C., Duarte-Valderrama C., Salgado-Marcano N., Melano R.G. Consenso latinoamericano para definir, categorizar y notificar patógenos multirresistentes, con resistencia extendida o panresistentes. Rev. Panam. Salud Púb. 2019;43:e65. doi: 10.26633/RPSP.2019.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janda J.M., Abbott S.L. The genus Aeromonas: Taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 2010;23:35–73. doi: 10.1128/CMR.00039-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Figueras M.J., Beaz-Hidalgo R. Aeromonas infections in humans. In: Graf J., editor. Aeromonas. 1st ed. Caister Academic Press; Pole, UK: 2015. pp. 65–108. Chapter 4. [Google Scholar]
- 9.Fernández-Bravo A., Figueras M.J. An Update on the Genus Aeromonas: Taxonomy, Epidemiology, and Pathogenicity. Microorganisms. 2020;8:129. doi: 10.3390/microorganisms8010129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deris Z.Z., Swarbrick J.D., Roberts K.D., Azad M.A., Akter J., Horne A.S., Nation R.L., Rogers K.L., Thompson P.E., Velkov T., et al. Probing the penetration of antimicrobial polymyxin lipopeptides into gram-negative bacteria. Bioconjugate Chem. 2014;25:750–760. doi: 10.1021/bc500094d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olaitan A.O., Morand S., Rolain J.M. Mechanisms of polymyxin resistance: Acquired and intrinsic resistance in bacteria. Front. Microbiol. 2014;5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu Z., Qin W., Lin J., Fang S., Qiu J. Antibacterial mechanisms of polymyxin and bacterial resistance. BioMed Res. Int. 2015;679109 doi: 10.1155/2015/679109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tietgen M., Semmler T., Riedel-Christ S., Kempf V.A., Molinaro A., Ewers C., Göttig S. Impact of the colistin resistance gene mcr-1 on bacterial fitness. Internat. J. Antimicrob. Ag. 2018;51:554–561. doi: 10.1016/j.ijantimicag.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 14.Kaskhedikar M., Chhabra D. Multiple drug resistance of Aeromonas hydrophila isolates from chicken samples collected from Mhow and Indore city of Madhyapradesh. World. 2009;2:31–32. [Google Scholar]
- 15.Kaskhedikar M., Chhabra D. Multiple drug resistance in Aeromonas hydrophila isolates of fish. Food Microbiol. 2010;28:157–168. [Google Scholar]
- 16.Bialvaei A.Z., Samadi-Kafil H. Colistin, mechanisms and prevalence of resistance. Curr. Med. Res. Opin. 2015;31:707–721. doi: 10.1185/03007995.2015.1018989. [DOI] [PubMed] [Google Scholar]
- 17.Cabello F.C., Godfrey H.P. Aquaculture, exaptation, and the origin of mcr-positive colistin resistance. Antimicrob. Agents Chemother. 2018;62:e01903-18. doi: 10.1128/AAC.01903-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Y., Zhong L.L., Srinivas S., Sun J., Huang M., Paterson D.L., Lei S., Lin J., Li X., Tang Z., et al. Spread of MCR-3 colistin resistance in China: An epidemiological, genomic and mechanistic study. EbioMedicine. 2018;34:139–157. doi: 10.1016/j.ebiom.2018.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tekedar H.C., Kumru S., Blom J., Perkins A.D., Griffin M.J., Abdelhamed H., Karsi A., Lawrence M.L. Comparative genomics of Aeromonas veronii: Identification of a pathotype impacting aquaculture globally. PLoS ONE. 2019;14:e0221018. doi: 10.1371/journal.pone.0221018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Esteve C., Alcaide E., Giménez M.J. Multidrug-resistant (MDR) Aeromonas recovered from the metropolitan area of Valencia (Spain): Diseases spectrum and prevalence in the environment. Eur. J. Clin. Microbiol. 2015;34:137–145. doi: 10.1007/s10096-014-2210-z. [DOI] [PubMed] [Google Scholar]
- 21.Zhou Y., Yu L., Nan Z., Zhang P., Kan B., Yan D., Su J. Taxonomy, virulence genes and antimicrobial resistance of Aeromonas isolated from extra-intestinal and intestinal infections. BMC Infect. Dis. 2019;19:158. doi: 10.1186/s12879-019-3766-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tansarli G.S., Papaparaskevas J., Balaska M., Samarkos M., Pantazatou A., Markogiannakis A., Daikos G.L. Colistin resistance in carbapenemase-producing Klebsiella pneumoniae bloodstream isolates: Evolution over 15 years and temporal association with colistin use by time series analysis. Int. J. Antimicrob. Agents. 2018;52:397–403. doi: 10.1016/j.ijantimicag.2018.06.012. [DOI] [PubMed] [Google Scholar]
- 23.Jorgensen J.H., Hindler J.F., Reller L.B., Weinstein M.P. New consensus guidelines from the Clinical and Laboratory Standards Institute for antimicrobial susceptibility testing of infrequently isolated or fastidious bacteria. Clin. Infect. Dis. 2007;44:280–286. doi: 10.1086/510431. [DOI] [PubMed] [Google Scholar]
- 24.Bravo-Fariñas L., Cabrera-Rodríguez L.E., Margarita-Ramírez M., Llop-Hernández A., Verdecía-Pérez J., Borrego-Hernández G., Fernández-Abreu A. Resistencia antimicrobiana en cepas de Aeromonas spp. aisladas de pacientes con bacteriemia. Rev. Biomédica. 2007;18:176–181. [Google Scholar]
- 25.Fosse T., Giraud-Morin C., Madinier I. Induced colistin resistance as an identifying marker for Aeromonas phenospecies groups. Lett. Appl. Microbiol. 2003;36:25–29. doi: 10.1046/j.1472-765X.2003.01257.x. [DOI] [PubMed] [Google Scholar]
- 26.Ling Z., Yin W., Li H., Zhang Q., Wang X., Wang Z., Shen J. Chromosome-mediated mcr-3 variants in Aeromonas veronii from chicken meat. Antimicrob. Agents Chemother. 2017;61:e01272-17. doi: 10.1128/AAC.01272-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eichhorn I., Feudi C., Wang Y., Kaspar H., Feßler A.T., Lübke-Becker A., Michael G.B., Shen J., Schwarz S. Identification of novel variants of the colistin resistance gene mcr-3 in Aeromonas spp. from the national resistance monitoring programme GE RM-Vet and from diagnostic submissions. J. Antimicrob. Chemother. 2018;73:1217–1221. doi: 10.1093/jac/dkx538. [DOI] [PubMed] [Google Scholar]
- 28.Shen Y., Xu C., Sun Q., Schwarz S., Ou Y., Yang L., Zhang R. Prevalence and genetic analysis of mcr-3-positive Aeromonas species from humans, retail meat, and environmental water samples. Antimicrob. Agents Chemother. 2018;62:e00404-18. doi: 10.1128/AAC.00404-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma S., Sun C., Hulth A., Li J., Nilsson L.E., Zhou Y., Wang Y. Mobile colistin resistance gene mcr-5 in porcine Aeromonas hydrophila. J. Antimicrob. Chemother. 2018;73:1777–1780. doi: 10.1093/jac/dky110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aghapour Z., Gholizadeh P., Ganbarov K., Bialvaei A.Z., Mahmood S.S., Tanomand A., Yousefi M., Asgharzadeh M., Yousefi B., Kafil H.S. Molecular mechanisms related to colistin resistance in Enterobacteriaceae. Infect. Drug Resist. 2019;12:965–975. doi: 10.2147/IDR.S199844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trebosc V., Gartenmann S., Tötzl M., Lucchini V., Schellhorn B., Pieren M., Lociuro S., Gitzinger M., Tigges M., Bumann D., et al. Dissecting Colistin Resistance Mechanisms in Extensively Drug-Resistant Acinetobacter baumannii Clinical Isolates. mBio. 2019;10:e01083-19. doi: 10.1128/mBio.01083-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venter H., Henningsen M.L., Begg S.L. Antimicrobial resistance in healthcare, agriculture and the environment: The biochemistry behind the headlines. Essays Biochem. 2017;61:1–10. doi: 10.1042/EBC20160053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anyanwu M.U., Jaja I.F., Nwobi O.C. Occurrence and Characteristics of Mobile Colistin Resistance (mcr) Gene-Containing Isolates from the Environment: A Review. Int. J. Environ. Res. Public Health. 2020;17:1028. doi: 10.3390/ijerph17031028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clinical and Laboratory Standars Institute (CLSI) Performance Standards for Antimicrobial Susceptibility Testing. 29th ed. Clinical and Laboratory Standars Institute; Wayne, PA, USA: 2021. CLSI supplement M100. [Google Scholar]
- 35.Kavanagh A., Ramu S., Gong Y., Cooper M.A., Blaskovich M.A. Effects of microplate type and broth additives on microdilution MIC susceptibility assays. Antimicrob. Agents Chemother. 2019;63:e01760-18. doi: 10.1128/AAC.01760-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simner P.J., Bergman Y., Trejo M., Roberts A.A., Marayan R., Tekle T., Tamma P.D. Two-site evaluation of the colistin broth disk elution test to determine colistin in vitro activity against Gram-negative bacilli. J. Clin. Microbiol. 2019;57:e01163-18. doi: 10.1128/JCM.01163-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dalmolin T.V., Mazzetti A., Ávila H., Kranich J., Carneiro G.I.B., Arend L.N.V.S., Pillonetto M. Elution methods to evaluate colistin susceptibility of Gram-negative rods. Diagn. Microbiol. Infect. Dis. 2020;96:114910. doi: 10.1016/j.diagmicrobio.2019.114910. [DOI] [PubMed] [Google Scholar]