Abstract
Large-scale RNA sequencing and genome-wide profiling data revealed the identification of a heterogeneous group of noncoding RNAs, known as long noncoding RNAs (lncRNAs). These lncRNAs play central roles in health and disease processes in diabetes and cancer. The critical association between aberrant expression of lncRNAs in diabetes and diabetic kidney disease have been reported. LncRNAs regulate diverse targets and can function as sponges for regulatory microRNAs, which influence disease phenotype in the kidneys. Importantly, lncRNAs and microRNAs may regulate bidirectional or crosstalk mechanisms, which need to be further investigated. These studies offer the novel possibility that lncRNAs may be used as potential therapeutic targets for diabetes and diabetic kidney diseases. Here, we discuss the functions and mechanisms of actions of lncRNAs, and their crosstalk interactions with microRNAs, which provide insight and promise as therapeutic targets, emphasizing their role in the pathogenesis of diabetes and diabetic kidney disease
Keywords: long noncoding RNAs, microRNAs in kidney, kidney fibrosis, EMT, EndMT, diabetes mellitus, diabetic kidney disease
1. Long Non-Coding RNA (lncRNA)
Long non-coding RNAs (LncRNAs) account for the major class of the noncoding RNAs in the genome and are linear transcripts longer than 200 nucleotide sequences that share similar characteristics with mRNAs [1]. Most LncRNAs are transcribed by RNA polymerase II and have capping at the 5′ end and splicing and the polyadenylated tail at the 3′ end. LncRNAs have defined promoter regions [1]. However, compared to mRNA, lncRNAs do not have open reading frames (ORFs) and have lesser exons (lncRNAs contain around 2.8 exons whereas mRNA contains 11 exons). LncRNAs can be transcribed as a whole or partial natural antisense transcripts (NAT) to coding genes, or located between genes or within introns [1]. Some lncRNAs originate from pseudogenes [2]. LncRNAs can be divided into several subtypes according to their position (such as antisense, intergenic, overlapping, intronic, bidirectional and recessed) and transcriptional direction in relation to other genes [3,4].
2. Synthesis Procedure and Location
Gene expression profiling and in situ hybridization studies have shown that the expression of lncRNAs can be tissue- and cell-specific, and may vary spatially, temporally or in response to stimuli [5]. Many lncRNAs are located exclusively in the nucleus, however, some are cytoplasmic or are located in both nucleus and cytoplasm. LncRNAs are critical regulators of gene expression and have functions in a wide range of cellular and developmental processes [5]. LncRNAs function through both inhibition and activation of genes [6].
LncRNAs are classified into four groups based on their location in the genome: (1) the intergenic lncRNAs, (2) the sense or antisense lncRNAs, (3) the intronic lncRNAs and (4) the processed transcripts; these lncRNAs reside in a gene-loci that has no ORF [6,7].
Based on their functions, lncRNAs have been characterized as signal, decoy, scaffold, guide, enhancer RNAs and short peptides [8,9]. Signal lncRNA acts as a molecular signal that regulates transcription processes [10]. Decoy lncRNAs act by reducing the availability of key molecules that are involved in gene regulation. These lncRNAs alter the transcription level by sequestering regulatory factors, and microRNAs, hence minimizing their expression level [11]. The scaffold class of lncRNAs provides structural support for complex proteins [12] and transcriptional activation or repression is conferred depending on the types of regulatory proteins and RNAs that exist [13]. Guide lncRNAs interact with ribonucleoproteins complex and influence the gene transcriptional level [14].
3. LNCs in Diabetic Kidney Disease
Available evidence has indicated the important roles of lncRNAs in the pathophysiology of diabetic kidney disease (DKD), and the crosstalk between lncRNAs and DKD were reported in recent years [15,16,17,18,19]. Altered expression levels of lncRNAs play key roles in the development of proteinuria and associated diabetic nephropathy (DN) [15,20]. LncRNAs are involved in the progression of kidney disease through regulation of many important factors, such as pathologic processes in mesangial cells, podocytes, reactive oxidative species, epithelial-to-mesenchymal transition (EMT), endothelial-to-mesenchymal transitions (EndMT) and actions on microRNAs [21,22,23].
Several lncRNAs participate in the regulation of renal disease (Table 1). For example, plasmacytoma variant translocation (PVT1) participates in the development of DN by regulating ECM accumulation. PVTI is the first non-coding RNA reported to be associated with kidney disease, which is highly expressed in human renal mesangial cells under high-glucose conditions and significantly promotes the expression of fibronectin protein, type IV collagen, TGF-β1 and type I plasminogen activator inhibitor [20,24,25]. Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is aberrantly upregulated in early DN [26,27,28]. MALAT1 initiates inflammation and oxidative stress; these pathogenic pathways regulate glucose-stimulated induction of the proinflammatory cytokines IL-6 and TNF-α by activating serum amyloid antigen 3. These changes alter endothelial cell stability in DN [20,29]. Gm4419 is located in chromosome 12 and is a regulator of the nuclear factor kappa light-chain enhancer of activated B cells (NF-κB), which is a crucial inflammatory factor for DN [20,30]. Gm4419 interacts with p50 and induces the NF-κB/NLRP3 inflammasome signal transduction pathway in mesangial cells, which is associated with inflammation, fibrosis and proliferation in high-glucose conditions [30]. NR_033515 is significantly upregulated in serum of DN patients [31]. Overexpression of NR_033515 promotes mesangial cell proliferation and inhibits apoptosis [31]. NR_033515 has been shown to upregulate the gene expression levels of proliferation-related genes, fibrosis-associated genes and EMT markers by targeting miR-743b-5p [31]. Kidney-specific deletion of Erbb4-IR has been shown to confer protective effects against DN complications [32]. Erbb4-IR inhibits the expression level of reno-protective miR-29b. Therefore, the level of fibrosis was enhanced by Erbb4-IR i diabetic kidneys [32]. Antisense mitochondrial noncoding RNA-2 (ASncmtRNA-2) is a mitochondrial lncRNA [33]. ASncmtRNA-2 is upregulated in ageing and senescence in endothelial cells [33]. ASncmtRNA-2 induces oxidative stress and causes tubular injury through (i) accelerated lipid peroxidation and protein crosslinking, (ii) damage to DNA and (iii) promoting inflammatory pathways, such as NF-κB and transforming growth factor-β1 (TGFβ1) [33]. Lnc-MGC is regulated by an ER stress-related transcription factor, CHOP (C/EBP homologous protein), and by TGFβ1-dependent and independent mechanisms [34]. ER stress increases in patients with progressive DN [34]. Nuclear enriched abundant transcript-1 (NEAT1) is highly expressed in high-glucose conditions, and interacts with AKT/mTOR pathways [35,36]. NEAT1 inhibition leads to suppression in levels of TGFβ1, FN and COL4A1 in DN [36]. NEAT1 promotes high glucose-stimulated mesangial cell hypertrophy by targeting the miR-222-3p/CDKN1B axis [37]. Similarly, lncRNA ERBB4-IR is involved in the development of renal fibrosis in diabetes and its silencing in diabetic mice protects against albuminuria and fibrogenic processes [32,38].
Table 1.
LncRNAs and circular RNAs in diabetes and diabetic kidney disease.
| lncRNAs | Expression | Samples | Targets | Functions | References |
|---|---|---|---|---|---|
| Plasmacytoma variant translocation (PVT1) |
Up | High-glucose stimulated mesangial cells | Fibronectin, collagen IV, TGF-β1, and PAI-1 | DN, ECM accumulation. | [20,24,25] |
| Metastasis associated lung adenocarcinoma transcript 1 (MALAT-1) | Up | Endothelial cells, STZ mice Podocytes, HEK-293 cells Renal tissues, proximal tubular epithelial cells Serum |
IL-6, TNF-α, SAA3, miR-29b CTNNBIP1, SRSF1 miR-23c, ELAVL1, NLRP3 miR-499a |
renal fibrosis, disrupts endothelial cell stability Podocytes cell damage Injuries in tubular cells DN phenotypes |
[26,27,28,29] |
| Gm4419 | Up | High-glucose stimulated mesangial cells | NF-κB/NLRP3 | Fibrosis, cell proliferation | [30] |
| GM5524 | Up | Diabetic tissues, High-glucose stimulated podocytes | Bcl2 and Bax protein LC3/ATG autophagy pathway |
DN, Podocytes cells damage | [50,51] |
| NR_033515 | Up | Serum, HEK293 T cells, mesangial cells | PCNA, cyclin D1, P38, ASK1, fibronectin, and α-SMA, E-cadherin and vimentin and miR-743b-5p | DN phenotypes, EMT and cell proliferation | [31] |
| Erbb4-IR | Up | Renal tissue | miR-29b, TGF-β/Smad3 | Renal fibrosis | [32,38] |
| Antisense mitochondrial noncoding RNA-2 (ASncmtRNA-2) | Up | Endothelial cells | ROS, (i) inducing lipid peroxidation, protein crosslinking, and the formation of DNA adducts; (ii) inducing direct damage to cellular DNA; and (iii) activating multiple cellular signaling pathways, including NF-κB and TGF-β1. | Damage to endothelial cells, Ageing, replicative senescence and fibrosis | [33] |
| Lnc-MGC | Up | Renal tissues Podocytes Mesangial cells |
Endoplasmic reticulum (ER) stress-related transcription factor, CHOP (C/EBP homologous protein), TGF-β1. | ER stress, renal fibrosis, glomerular hypertrophy, and podocyte cells injury EMT and DN. |
[34] |
| GAS5 | Up | Human tubular epithelial cells |
miR-27, P53, CASP3, NF-κB, BNIP3 | Tubular cell apoptosis | [133] |
| GM6135 | Up | Glucose-stimulated-mesangial cells | TLR4, miR-203 | Renal inflammation and fibrosis | [134] |
| LnC-H19 | Up | Diabetic mice UUO mice Endothelial cells |
TGF-β/Smad3, miR-29a | Renal inflammation and fibrosis | [22,96,97,98,99] |
| CJ241444- miR-192 | Up | Renal cortex and mesangial cells | TGF-β, Akt, Col1a2, Col4A1, Smad, Ets1, miR-192 |
Glomerular fibrosis | [81] |
| NEAT1 | Up | Renal tissues | Akt, Mtor, collagen IV, Fibronectin, TGF-β1. Zeb1, miR-27b-3p, Ask1, fibronectin |
Glomerular fibrosis Mesangial cell proliferation |
[36,37] |
| Circular noncoding RNAs circRNA_15698 |
Up | Renal cortical cells mesangial cells | collagen IV, collagen I, Fibronectin, TGF-β1. miR-185 |
Renal fibrosis | [135] |
| circLRP6 | Up | Renal cells | miR-205, HMGB1 | DN progression | [136] |
| circACTR2 | Up | Tubular cells | interleukin (IL)-1β, collagen IV and fibronectin | Pyroptosis, Fibrosis in Renal Tubular Cells | [137] |
| circHIPK3 | Up | DN tissues, Glucose-stimulated mesangial cells | Cyclin D1, PCNA, TGF-β1, Collagen I, Fibronectin and miR-185 | DN progression | [138] |
| circ_0000491 | Up | Glucose-stimulated-mesangial cells | TGFβR1, miR-101b | Glomerular fibrosis | [139] |
| circRNA_010383 | down | Kidneys of db/db mice mesangial cells |
Sponges for miR-135a | DN | [140] |
| Taurine up-regulated 1 (TUG1) | down | Glucose-stimulated mesangial cells, renal cortex, mesangial cells | endogenous sponge of miR-377, PGC-1α, PAI-1, TGF-β1, FN, collagen IV | Mesangial cells damage, podocyte cell death | [42] |
| Myocardial infarction-associated transcript (MIAT) | down | HK-2 cells | Nuclear factor erythroid 2-related factor 2 (Nrf2), Acta2 | tubular cells damage | [20,35] |
| Cancer susceptibility candidate 2 (CASC2) | down | serum and renal tissues | JNK pathway | renal failure, podocyte cell death | [45,46,47,48] |
| ENSMUST00000147869 | down | mesangial cells, renal cortex | ECM synthesis, fibronectin and Collagen IV | Mesangial cells damage | [41] |
| 1700020I14Rik | down | mesangial cells, Renal tissues | miR-34a-5p, Sirt1, HIF-1α | renal fibrosis | [49] |
| CYP4B1-PS1-001 | down | mesangial cells, renal tissues | nucleolin (NCL), ubiquitin proteasome-dependent pathway | mesangial cells proliferation and fibrosis | [39,40] |
| Gm15645 | down | Kidneys of Db/db mice and high-glucose-stimulated podocytes | Bcl2/Bax and LC3/ATG pathways | DN, podocyte cell apoptosis | [50] |
| LINC01619 | down | DN tissues, podocytes | miR-27a, FoxO1, ROS, CHOP, GRP78 | DN | [51] |
| LncRIAN | down | Renal biopsy, podocyte cell | Acta2, Smad2, Smad3, miR-150 | Myofibroblasts formation | [81] |
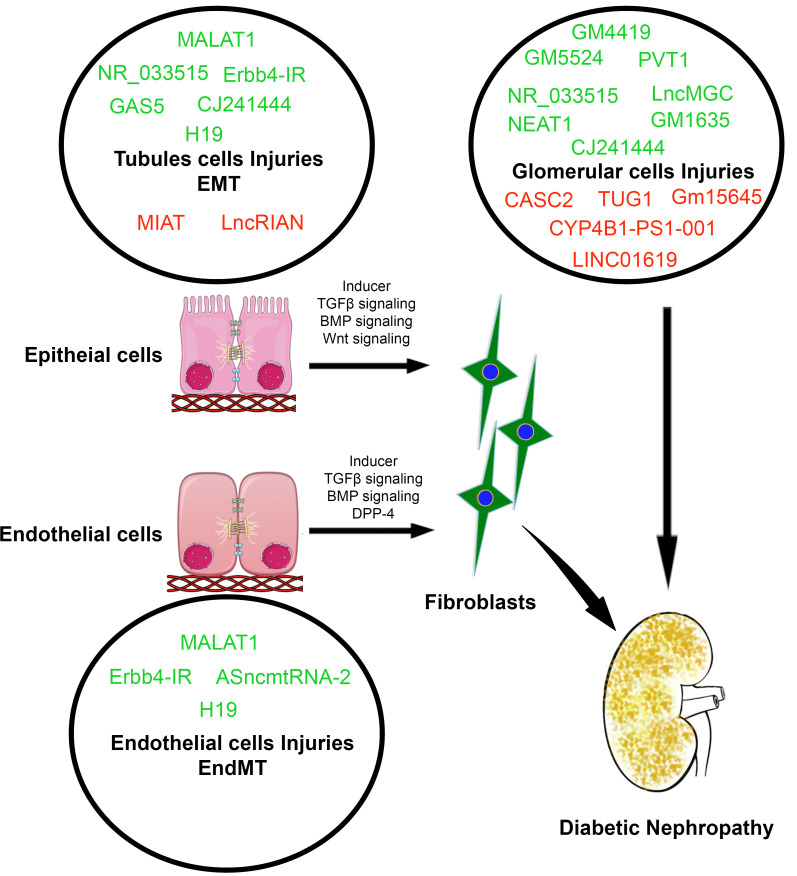
Conversely, the expression of CYP4B1-PS1-001, which upregulates the nucleolin protein level, is suppressed in high-glucose conditions [39,40]. CYP4B1-PS1-001 overexpression suppresses the levels of FN, COL4A1 and proliferation markers in diabetic mice [40]. Another example of a reno-protective lncRNA is the lncRNA ENSMUST00000147869, which target ECM production in kidneys of diabetic mice [41]. ENSMUST00000147869 affects ECM synthesis and dramatically decreases the levels of fibronectin and collagen IV in mesangial cells under high-glucose conditions [41], though the exact role of this lncRNA is unknown. TUG1 functions as a repressor of miR-377. miR-377 directly targets the 3′UTR of PGC-1α and fibrosis markers. Therefore, TUG1 upregulates the level of PGC-1α and alleviates ECM production and downregulates the expression levels of proinflammatory cytokines in high-glucose stimulated mesangial cells [42]. Myocardial infarction-associated transcript (MIAT), also known as retinal noncoding RNA 2 (RNCR2), has been known to associate with myocardial infarction [35]. MIAT regulates cell viability through stabilizing nuclear factor erythroid 2-related factor 2 (NRF2) expression in renal tubules [20]. NRF2 pathologically and functionally protects the kidneys against diabetic damage [43]. Interestingly, expression of Nrf2 can be enhanced by MIAT overexpression in glucose-treated renal tubular epithelial cell lines [44]. Cancer susceptibility candidate 2 (CASC2) has critical functions in tumorigenesis [45]. Downregulated expression of CASC2 has been observed in serum and kidney tissues in diabetic kidneys and is predictive of diabetic complications [46]. Low-plasma levels of CASC2 are associated with higher risk of renal failure in DN patients [47,48]. Another lncRNA, 1700020I14Rik, which is located in chromosome 2 (Chr2: 119594296–119600744), functions as an endogenous RNA and regulates the expression levels of microRNAs in diabetes [20,49]. Overexpression of 1700020I14Rik suppresses the expression level of miR-34a-5p by Sirt1/HIF-1α signal pathway and accelerates fibrosis in mesangial cells [49]. CYP4B1-PS1-001 is downregulated in early DN [40]. Its overexpression inhibits fibrosis of mesangial cells by interacting with nucleolin [40]. Gm15645 is downregulated in DN and high-glucose-stimulated, cultured podocytes [50]. The mechanism of Gm15645 is contrary to that of Gm5524, which affects podocyte cell death and autophagy regulation in DN [50]. LINC01619 regulates miR-27a/FoxO1 (forkhead box protein O1) and ER stress-associated podocyte cell injury in diabetes [51]. Downregulated expression levels of LINC01619 are associated with proteinuria and declines in kidney function in DN patients; hence, targeting LINC01619 is one of the potential therapeutic options for treatment of DN [51]. Figure 1 demonstrates the lncRNAs involvement in influencing EMT, EndMT and glomerular injury in diabetic nephropathy.
Figure 1.
Involvement of lncRNAs in the regulation of diverse cell types in kidneys. Green color indicates upregulated expression levels whereas red color indicates downregulated expression levels during injury in cell types.
4. LncRNAs Involvement in the Regulation of EMT
EMT involves a series of processes by which epithelial cells lose their epithelial characteristics and acquire properties of mesenchymal cells [52,53,54,55,56,57]. Figure 1 depicts the involvement of lncRNAs in the regulation of EMT, EndMT and mesenchymal cells. Epithelial cells are normally associated tightly with their neighbor cells. In contrast, mesenchymal cells do not form intercellular adhesion complexes [58]. Mesenchymal cells are elongated in shape and exhibit end-to-end polarity and focal adhesions, allowing for increased migratory capacity [58]. The main function of fibroblasts, which are prototypical mesenchymal cells that are found in several tissues, is to maintain structural integrity by secreting extracellular matrix (ECM). Fibroblast-specific protein 1 (FSP-1), alpha-smooth muscle actin (SMA), vimentin, fibronectin and collagen I are the markers that characterize mesenchymal products in diabetic kidneys [58,59,60]. Inflammation results in the recruitment of multiple types of cells that are involved in the induction of EMT processes. Elevated levels of TGFβ1, platelet-derived growth factor (PDGF), epidermal growth factor (EGF) and fibroblast growth factor-2 (FGF-2) contribute to EMT processes [59,60,61]. MALAT1, NR_033515, Erbb4-IR, GAS5 and CJ241444 are involved in tubular injury and contribute to EMT processes whereas MIAT and LncRIAN have shown tubular protective activity and may have regulate EMT processes in diabetic kidneys (Figure 1).
5. LncRNAs Involvement in the Regulation of EndMT
Endothelial cells form fibroblasts by undergoing a transition, referred as EndMT [57,58,62,63,64,65]. EndMT is characterized by the loss of endothelial cell phenotypes, and gain of mesenchymal proteins [58,62,64,65,66,67]. EndMT participates in fibrogenic processes in kidneys and, in diabetic kidneys, can alter the physiology and function of other neighboring cells [58,62,65,68]. Pathological stimuli such as inflammation, diabetes and ageing influence EndMT events in the kidneys [69]. Endothelial SIRT3, the nuclear receptor glucocorticoid receptor (GR) and cell surface FGFR1 are critical regulators of TGFβ signaling and EndMT in diabetic kidneys [70,71,72,73]. The kidneys of diabetic mice showed both progressive glomerular sclerosis and tubulointerstitial fibrosis, which was associated with approximately 40% of all FSP-1-positive cells and 50% of αSMA-positive stromal cells were CD31-positive [74]. Similarly, in the kidneys of COL4A3 knockout mice, 45% of all αSMA-positive fibroblasts and 60% of all FSP-1-positive fibroblasts were CD31-positive, suggesting that these fibroblasts are of endothelial origin and that EndMT might contribute critically to the development and progression of renal fibrosis [74]. During the process of EndMT, biochemical changes lead to the decreased expression of endothelial markers and the gain of mesenchymal markers such as FSP-1, αSMA, smooth muscle 22-alpha (SM22α), N-cadherin, fibronectin, vimentin, types I and III collagen, nestin, cluster of differentiation, 73 (CD73), matrix metalloproteinase-2 (MMP-2) and matrix metalloproteinase-9 (MMP-9) [58,75,76]. MALAT1, Erbb4-IR and ASncmtRNA2 cause endothelial cell injury and may involve EndMT-associated renal fibrosis (Figure 1). LncRNA H19 is associated with kidney fibrosis by activating EndMT processes in diabetes (Figure 1).
6. LNCs Interaction with microRNA
The miRNA and lncRNA interaction is one of mechanisms for regulation of gene expression [77]. This multilevel regulation is involved in almost all the physiological and cellular processes at the transcriptional, post-transcriptional and post-translational levels [77,78]. In some studies, it has been reported that miRNA triggers lncRNA decay [77]. On the contrary, lncRNAs generate miRNAs, act as miRNA sponges and miRNA decoys, and compete with miRNA for binding at mRNAs [77].
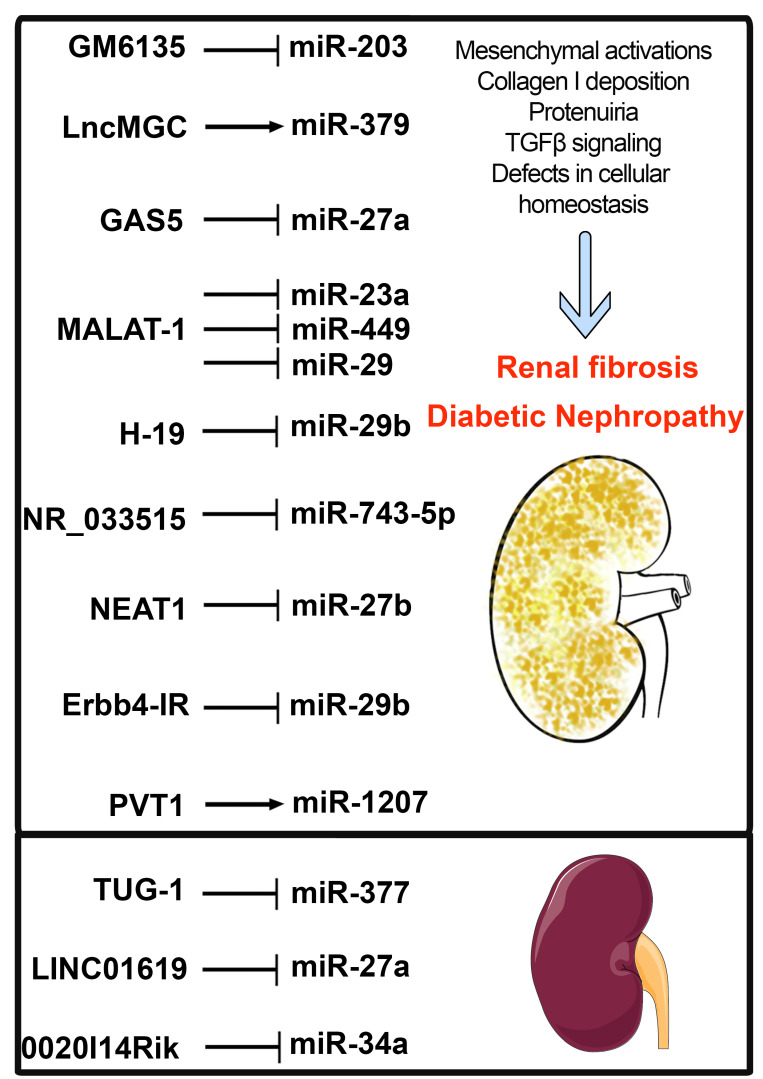
LncRNA genes can harbor microRNAs and these microRNAs can be released by post-transcriptional processing. For example, the lncRNA PVT1 serves as a host of miR-1207-5p and has been implicated in DN [79]. microRNAs are often present in clusters, having been localized to the PVT1 locus, and are upregulated by high-glucose and affect extracellular matrix accumulation [80]. MiRNA clusters in lncRNAs can get very large as demonstrated by a megacluster of more than 40 miRNAs harbored in lnc-MGC [34]. This cluster is induced in diabetic glomeruli through endoplasmic reticulum stress signaling, which responds to high glucose and TGFβ-activation as well [34].
The interactions between microRNAs and lncRNAs are important to study the key steps in DN progression. DN mice exhibit interactions between lncRNA CJ241444-miR-192 that induces TGFβ1/Smad3 signaling [81] and lncRNA Erbb4- IR-miR-129b activates collagen genes and ECM genes and, hence, kidney injury [82]. These lncRNAs can act as miRNA sponges [32,81]. Similarly, lncRNA PVT-1 participates in ECM accumulation via the actions of its-derived miRNAs, miR-1207-5p and miR-1207-3p [25]. Under high-glucose conditions, higher expression of both PVT-1 and its miRNAs cause an increase of TGFβ1/Smad3 signaling and ECM accumulation [25]. Similarly, miR-379 clusters that are regulated by ER stress in DN and lncMGC are also hosted in this same cluster [34]. LncMGC regulates the expression of the miR-379 clusters, and the upregulation of the miR-379 clusters induces ECM accumulation and renal hypertrophy [34]. Thus, antagonism of lncMGC expression can be used as a potential therapeutic for DN to reduce the effects of the miR-379 cluster, following ER stress [34]. Besides that, lncRNA NEAT1 antagonism is also a potential therapy, since NEAT1 antagonism leads to the suppression of ECM deposition via the reduction of ASK1, FN and TGFβ1 production [83]. This NEAT1-associated ECM suppression is due to its interaction with miR-27b-3p, and its target, the TGFβ and Zeb1 [83]. Administration of the antiapoptotic lncRNA, TUG-1, suppresses miR-377 expression and its target gene PPARγ and thus prevents ECM accumulation in DN mice [42]. Therefore, treatment to increase TUG-1 expression could be beneficial to treat the DN phenotype and restore kidney structure, although further studies are needed to understand its potential [42]. These findings will allow the development of an understanding of the interactions between lncRNAs and their target miRNAs, that can be useful for therapeutic target selection to prevent ECM deposition and for the management of DN progression. Figure 2 demonstrates LncRNAs and microRNAs interactions in the regulation of diabetic nephropathy.
Figure 2.
Interactions of LncRNAs and microRNAs. Arrow  —Upregulates whereas
—Upregulates whereas  —downregulates expression level.
—downregulates expression level.
7. LNCs in the Regulations of Antifibrotic microRNAs Crosstalk
TGFβ suppresses antifibrotic miRNAs such as miR-29 clusters and miR-let-7 clusters [84]. Suppression of such TGFβ1-regulated crosstalk of miRNAs was reported in type I diabetic subjects who had higher rates of ESRD progression [85]. The data from our laboratory reveal that clusters of the miR-29 family and the miR-let-7 family showed a protective effect against endothelial-to-mesenchymal transition (EndMT) and demonstrate bidirectional regulation under physiological conditions [86,87,88,89]. This bidirectional regulation is essential for endothelial cell homeostasis and protects against EndMT in diabetic kidneys [76]. Targeting EndMT is one of the potential therapeutic options for the treatment of diabetic kidney fibrosis [56,58]. miR-29 clusters show negative, bidirectional regulation with TGFβ receptors [76]. miRNAs regulate gene expression of each other directly or indirectly. This crosstalk phenomenon is linked with maintenance of antifibrotic activity in the kidney and its disruption results in accelerated renal fibrosis [76]. Interventions that prevent the disruption of this crosstalk are beneficial in protecting against kidney diseases [56,86]. DPP-4 inhibition shows suppression in TGFβ signaling-driven EndMT in diabetic kidneys by elevating miR-29 clusters [67,88]. miR-29 clusters target the profibrotic molecule DPP-4, and its inhibition elevates the miR-29 level; therefore, DPP-4 inhibitors are potential leads for the treatment of diabetic nephropathy [88].
MiR-let-7 inhibits TGFβ receptor 1 [90], and TGFβ-smad3 signaling has been demonstrated as an inhibitory pathway for miR-29 gene expression [84,88,91,92]; therefore, as expected, miR-let-7 induces the expression of miR-29 in endothelial cells. An alternative mechanism of miR-29-linked-miR-let-7 expression was explained by the interferon-gamma (IFNγ)-FGFR1 axis. miR-29 targets IFN-γ [93] and moreover, IFN-γ inhibits FGFR1. FGFR1 plays crucial roles in the expression of miR-let-7 family clusters [90]. Downregulation of miR-29 clusters causes a rise in IFN-γ levels, which subsequently discourage FGFR1 and FGFR1-associated expression of miR-let-7 clusters. This suppression of miR-let-7 expression causes activation of TGFβR1 protein expression. Triggering TGF-β/smad3 signaling in turn inhibits the expression of miR-29 family clusters [88].
AcSDKP is a key peptide that is partially synthesized in the distal tubular regions from the enzymatic action of polyoligopeptidase on thymosinβ4 and is degraded by angiotensin converting enzyme. Hence, angiotensin converting enzyme inhibitors have been shown to elevate the level of AcSDKP in the plasma of mice and diabetic subjects [86,89]. Several studies have been analyzed for renal protective abilities of AcSDKP and ACE inhibitors can perform antifibrotic activities by partially elevating AcSDKP levels [70,89,94]. Most importantly AcSDKP is a key endogenous peptide that restores kidney structure and suppresses renal fibrosis by counteracting DPP-4-associated EndMT through elevating microRNA crosstalk regulations between miR-29 and miR-let-7 [86]. Moreover, inhibition of ACE elevates the level of AcSDKP and cause upregulation of antifibrotic microRNAs and restores the antifibrotic cross-talk in cultured endothelial cells, while angiotensin receptor blockers have minimal effects [76,86,89]. These events control crosstalk regulation between miR-29s and miR-let-7s in fibrotic kidneys of diabetic mice [86]. AcSDKP maintains kidney homeostasis partly by elevating the bidirectional regulation between miR-29s and miR-let-7s [76,86].
Lnc-H19 expression is upregulated in TGFβ2-induced endothelial cells and in fibrotic kidneys of diabetic mice [22]. H19 suppression significantly reduces EndMT and kidney fibrosis [22]. The upregulated H19 expression in diabetic kidneys is associated with downregulated levels of miR-29a [22]. H19 and miR-29 association contributes to a regulatory network involved in EndMT [22]. Similar H19 regulatory mechanisms have previously been reported, such as the H19/miR675 pathway, which inhibits cell growth and Igf1r expression [95]; H19/Let-7-mediated inhibition of HMGA2-mediated epithelial-to-mesenchymal transition [96] and the H19/miR-675 axis inhibits prostate cancer metastasis via TGFβ1 [97]. Xie et al. (2016) also found that H19 interaction with miR17 contributed to a regulatory network involved in renal fibrosis [98]. H19 acts as a competitive endogenous RNA. The regulatory network integrates the transcriptional and post-transcriptional regulatory network of EndMT and renal fibrosis [22]. Interestingly, inhibition of H19 only altered miR-29a levels, not miR-29b or miR-29-c, and suppressed TGF-β/Smad signaling in order to regulate EndMT and renal fibrosis in diabetes [22].
Figure 3 demonstrates the involvement of LncRNAs in the regulation of microRNAs crosstalk and its implications in the mesenchymal activation in diabetic kidneys.
Figure 3.
Crosstalk regulation between LncRNAs and antifibrotic microRNAs. Arrow  —Upregulates whereas
—Upregulates whereas  —downregulates expression level.
—downregulates expression level.
8. LncRNA-miRNA-Based Treatment for DKD, Future directions and Perspectives
Many non-coding RNAs (miRNAs, lncRNAs and circRNAs) regulate the expression of critical genes involved in DN phenotypes. These non-coding RNAs (nc RNAs) are stable in biological fluids and can offer potential biomarkers in a diverse array of diseases. Non-coding RNAs are involved in the disease processes of hypertrophy, ECM synthesis, apoptosis and renal fibrosis. Moreover, some research has advanced to synthesize ncRNAs-based treatments, with a few of these ncRNAs already in the clinical trial phase. Therefore, these ncRNAs-based therapeutics would be an alternative approach for the treatment of DN [99].
MiRNA-based therapeutics can be used as alternative therapy for treatment of several diseases including diabetic nephropathy. The application of artificially synthesized oligonucleotides to mimics (miRNA mimics) or knockdown microRNAs (antagomiRs) has evolved [99,100]. In this series, locked nucleic acid (LNA) inhibitor was developed to suppress a specific miRNA expression or action [99,100]. Dramatically, LNA-miR-192 treatment improves the DN phenotype, and thus can be utilized as a potential DN therapy [101].
Other work has showed that subcutaneous injection of anti-miR-21 suppressed the fibrosis level in chronic kidney disease mice [102]. miR-29 family significantly improves renal structure and fibrosis in DN mice [103], thus anti-miR-29-based therapy could be potentially used as an alternative option for DN treatment. miRNA-based treatment is gaining momentum over the last decade. However, the problem lies within the delivery methods. miRNAs regulate several targets at the same time; thus, they may affect other pathways. Therefore, research in miRNA-based therapies is now switching to focus on delivery methods and efficacy and safety to target a specific route and tissue localization [104,105,106].
Moreover, the size of the therapeutic molecule should be small enough to cross the endothelium to the organ or site of interest and should not be filtered out by the kidney [107]. Interestingly, this filtration problem is an advantage for miRNA-based treatment, since the epithelial cells reabsorb the therapeutic agents from the ultrafiltrate, thus reducing the loss [107,108]. Therefore, it is believed that miRNA-based treatments could be safely used for DN subjects, although advanced work or large clinical trials are still needed. Several miRNA-based treatments have advanced to clinical trials, though none for the treatment of DN. Miravirsen (LNA based miR-122 inhibitor) already entered phase II clinical trials to treat HCV infection in patients [109]. Many miRNA-based therapeutics are currently in development for several other diseases; therefore, the use of miRNA-based treatment for DN is a new hope. Another possible treatment option is lncRNAs-mediated treatment for DN. It is comparatively favorable to targeting the lncRNA expression when compared to miRNAs, because of its functional role in transcriptional control, tissue-specific expression and disease-specific alterations. LncRNAs are mainly present in the nucleus; synthetic antisense oligonucleotides (ASOs) are widely suggested to silence the lncRNA expression in the nucleus by commencing RNase H-dependent degradation [110,111]. Design of ASOs is very important as it should bind to the LncRNA-specific site and target a single lncRNA. Furthermore, the real challenge is to treat with ASO in vivo. Similar to miRNA-based treatments, the problems lie within the delivery efficiency and efficacy. Another problem related to lncRNAs based therapeutics is the heterogeneous nature and unconserved intron sequence of lncRNAs [1,112]. Further studies are needed to identify small molecules that induce the expression of renal protective non-coding RNAs. There is a need to search for compounds that induce the expression of antifibrotic non-coding RNAs in diabetic kidneys, such as flavonoids, chalcones, polyhydroquinolines, propiophenone derivatives, deoxyandrographolides, 2-methoxy-estradiol and thiazolidin-4-one derivatives; these synthetic or plant-based compounds have shown protective effects in mouse models of diabetes mellitus [113,114,115,116,117,118,119,120,121,122,123,124,125], and could be further tested and used in the management of DN. ncRNAs play crucial roles in the pathogenesis of type II DM and diabetic complications; despite their limitations, tissue-specific microRNAs expression should be further studied [56,126,127]. Physiological dysfunction, metabolic alterations, ER-stress and inflammation are observed before later features such as proteinuria, which is a major contributor to the development of DKD [20]. Proteinuria determines the cardio-renal outcomes of patients with DKD [128,129,130]. Higher proteinuria leads to tubular damage and is associated with renal inflammation and interstitial fibrosis in diabetes [129,130,131]. Minutolo et al. studied the crucial role of proteinuria in patients who have chronic diabetic kidney disease (DM-CKD), and discussed new information on cardio-renal prognosis in DM-CKD patients [128]. In the absence of proteinuria, DM-CKD patients did not have increased cardio-renal risk when compared with non-diabetic CKD patients [128]. However, in CKD patients with proteinuria, the risk of end-stage renal disease was mainly due to the proteinuria level independent of diabetes [20,132]. The physiological and cellular roles of altered sets of microRNAs and lncRNAs are relevant to study proteinuria and associated DN. In addition, lncRNAs such as GAS5 and GM6135, which are upregulated during renal inflammation, might be addressed by a Lnc-inhibitor [133,134]. Similarly, research on circular RNAs and their role in the health and disease of diabetic kidneys is gaining momentum as well. circRNA_15698, circLRP6, circACTR2, circHIPK3 and circ_0000491 are associated with renal inflammation and fibrosis whereas circRNA_010383 is reno-protective [135,136,137,138,139,140]. Therefore, better understanding of the role of these regulatory circular RNAs in the physiology of diverse kidney cell types is needed. Table 1 presents the list of lncRNAs and circular RNAs, and their targets in kidney disease.
The role of lncRNAs should be analyzed in preclinical settings before utilizing their therapeutic potential in the management of diabetic nephropathy. Hence, extensive researches demonstrating the role of miRNAs and LncRNAs interaction are needed to validate the possibility of using these miRNAs/lncRNAs-based treatments in proteinuria and associated DN.
9. Conclusions
miRNAs and lncRNAs interactions influence DKD progression by targeting genes related to fibrogenesis, ER stress, inflammation, oxidative stress and metabolic dysfunction [8,49,110]. Identification of pathways regulating early-stage (physiological dysfunction, metabolic alteration, ER-stress and inflammation) and late-stage (proteinuria) features are of key importance in studies of DN pathogenesis. miRNAs and LncRNAs interactions open a wide area for basic research and for the development of new therapeutic options against diabetic complications including DKD.
Abbreviations
| AcSDKP | N-acetyl-seryl-lysyl-proline |
| ACE | Angiotensin-converting enzyme |
| AKT | protein kinase B |
| ASK1 | Apoptosis signal-regulating kinase 1 |
| ASncmtRNA | Antisense mitochondrial noncoding RNA-2 |
| ASO | Antisense oligonucleotide |
| CASC2 | Cancer susceptibility candidate 2 |
| CD73 | Cluster of differentiation 73 |
| CD31 | Cluster of differentiation 31 |
| CDKN1B | Cyclin Dependent Kinase Inhibitor 1B |
| CHOP | C/EBP homologous protein |
| CKD | Chronic kidney disease |
| COL4A1 | Collagen Type IV Alpha 1 Chain |
| DPP-4 | Dipeptidyl transferase-4 |
| DKD | Diabetic kidney disease |
| DM | Diabetes mellitus |
| DN | Diabetic nephropathy |
| ECM | extracellular matrix |
| EGF | Epidermal growth factor |
| EMT | epithelial-to-mesenchymal transition |
| EndMT | endothelial-to-mesenchymal transition |
| ER | endoplasmic reticulum stress |
| ESRD | End-stage renal disease |
| FSP-1 | Fibroblast-specific protein-1 |
| FGF-2 | Fibroblasts specific growth factor-2 |
| FN | Fibronectin |
| FoxO1 | Forkhead box protein O1 |
| FGFR1 | Fibroblast growth factor receptor 1 |
| GR | Glucocorticoid receptor |
| HIF1 α | Hypoxia Inducible Factor 1 Subunit Alpha |
| IGF1R | Insulin-like growth factor 1 receptor |
| IL-6 | Interleukins-6 |
| IFN-γ | Interferon-gamma |
| LncRNA | Long non-coding RNAs |
| LNA | Locked nucleic acid |
| MALAT1 | Metastasis-associated lung adenocarcinoma transcript 1 |
| MiRNA | MicroRNA |
| MIAT | Myocardial infarction-associated transcript |
| MMP-2 | Matrix metalloproteinase-2 |
| MMP-9 | Matrix metalloproteinase-9 |
| mTOR | Mammalian target of rapamycin |
| NAT | Natural antisense transcript |
| ncRNA | Non-coding RNAs |
| NEAT1 | Nuclear enriched abundant Transcript-1 |
| NF-κB | Nuclear factor kappa light-chain enhancer of activated B cells |
| NRF2 | nuclear factor erythroid 2-related factor 2 |
| NLRP3 | NOD-, LRR- and pyrin domain-containing protein 3 |
| ORF | Open reading frame |
| PGC-1α | Peroxisome proliferator-activated receptor gamma coactivator 1-alpha |
| PPAR-γ | Peroxisome proliferator-activated receptor gamma PPAR-γ |
| PVT1 | Plasmacytoma variant translocation 1 |
| RNCR2 | Retinal non-coding RNA 2 |
| SIRT1 | Sirtuin 1 |
| SIRT3 | Sirtuin 3 |
| SM22α | Smooth muscle 22-alpha |
| αSMA | Alpha smooth muscle actin |
| TGFβ1 | Transforming growth factor β1 |
| TNF α | Tumor necrosis factor α |
| TUG1 | Taurine Up-Regulated 1 |
| UTR | Untranslated region |
Author Contributions
Conceptualization, S.P.S.; software, S.P.S.; resources, S.P.S. and J.E.G.; writing original draft, S.P.S.; writing review and editing, S.P.S., P.T. and J.E.G.; supervision, S.P.S. and D.K.; Project administration, K.K. and D.K. Funding, J.E.G. and D.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by research grant from Kanazawa Medical University to D.K. (C2011-4, C2012-1), the Grant for Promoted Research awarded to K.K. (S2015-3, S2016-3, S2017-1) and a grant from the National Institutes of Health (R01HL131952) to J.E.G.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Statello L., Guo C.J., Chen L.L., Huarte M. Gene regulation by long non-coding rnas and its biological functions. Nat. Rev. Mol. Cell Biol. 2021;22:96–118. doi: 10.1038/s41580-020-00315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Milligan M.J., Lipovich L. Pseudogene-derived lncrnas: Emerging regulators of gene expression. Front. Genet. 2015;5:476. doi: 10.3389/fgene.2014.00476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peschansky V.J., Wahlestedt C. Non-coding rnas as direct and indirect modulators of epigenetic regulation. Epigenetics. 2013;9:3–12. doi: 10.4161/epi.27473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mattick J.S., Rinn J.L. Discovery and annotation of long noncoding rnas. Nat. Struct. Mol. Biol. 2015;22:5–7. doi: 10.1038/nsmb.2942. [DOI] [PubMed] [Google Scholar]
- 5.Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G., et al. The gencode v7 catalog of human long noncoding rnas: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma L., Bajic V.B., Zhang Z. On the classification of long non-coding rnas. RNA Biol. 2014;10:924–933. doi: 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mercer T.R., Dinger M.E., Mattick J.S. Long non-coding rnas: Insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 8.Li X., Wu Z., Fu X., Han W. Lncrnas: Insights into their function and mechanics in underlying disorders. Mutat. Res. Rev. Mutat. Res. 2014;762:1–21. doi: 10.1016/j.mrrev.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 9.Moran V.A., Perera R.J., Khalil A.M. Emerging functional and mechanistic paradigms of mammalian long non-coding rnas. Nucleic Acids Res. 2012;40:6391–6400. doi: 10.1093/nar/gks296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pandey R.R., Mondal T., Mohammad F., Enroth S., Redrup L., Komorowski J., Nagano T., Mancini-DiNardo D., Kanduri C. Kcnq1ot1 antisense noncoding rna mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 11.Kallen A.N., Zhou X.-B., Xu J., Qiao C., Ma J., Yan L., Lu L., Liu C., Yi J.-S., Zhang H., et al. The imprinted h19 lncrna antagonizes let-7 micrornas. Mol. Cell. 2013;52:101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang L., Froberg J.E., Lee J.T. Long noncoding rnas: Fresh perspectives into the rna world. Trends Biochem. Sci. 2014;39:35–43. doi: 10.1016/j.tibs.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aguilo F., Zhou M.-M., Walsh M.J. Long noncoding rna, polycomb, and the ghosts haunting ink4b-arf-ink4a expression. Cancer Res. 2011;71:5365–5369. doi: 10.1158/0008-5472.CAN-10-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang Y., Fullwood M.J. Roles, functions, and mechanisms of long non-coding rnas in cancer. Genom. Proteom. Bioinform. 2016;14:42–54. doi: 10.1016/j.gpb.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu Y.-Y., Lu F.-H., Huang X.-R., Zhang L., Mao W., Yu X.-Q., Liu X.-S., Lan H.-Y. Non-coding rnas as biomarkers and therapeutic targets for diabetic kidney disease. Front. Pharmacol. 2021;11:583528. doi: 10.3389/fphar.2020.583528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo J., Liu Z., Gong R. Long noncoding rna: An emerging player in diabetes and diabetic kidney disease. Clin. Sci. 2019;133:1321–1339. doi: 10.1042/CS20190372. [DOI] [PubMed] [Google Scholar]
- 17.Lv J., Wu Y., Mai Y., Bu S. Noncoding rnas in diabetic nephropathy: Pathogenesis, biomarkers, and therapy. J. Diabetes Res. 2020;2020:3960857. doi: 10.1155/2020/3960857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou Q., Chen W., Yu X.-Q. Long non-coding rnas as novel diagnostic and therapeutic targets in kidney disease. Chronic Dis. Transl. Med. 2019;5:252–257. doi: 10.1016/j.cdtm.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coellar J.D., Long J., Danesh F.R. Long noncoding rnas and their therapeutic promise in diabetic nephropathy. Nephron. 2021:1–11. doi: 10.1159/000515422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y., Xu K., Xu K., Chen S., Cao Y., Zhan H. Roles of identified long noncoding rna in diabetic nephropathy. J. Diabetes Res. 2019;2019:1–8. doi: 10.1155/2019/5383010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin J., Jiang Z., Liu C., Zhou D., Song J., Liao Y., Chen J. Emerging roles of long non-coding rnas in renal fibrosis. Life. 2020;10:131. doi: 10.3390/life10080131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi S., Song L., Yu H., Feng S., He J., Liu Y., He Y. Knockdown of lncrna-h19 ameliorates kidney fibrosis in diabetic mice by suppressing mir-29a-mediated endmt. Front Pharm. 2020;11:586895. doi: 10.3389/fphar.2020.586895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zoja C., Xinaris C., Macconi D. Diabetic nephropathy: Novel molecular mechanisms and therapeutic targets. Front. Pharmacol. 2020;11:586892. doi: 10.3389/fphar.2020.586892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanson R.L., Craig D.W., Millis M.P., Yeatts K.A., Kobes S., Pearson J.V., Lee A.M., Knowler W.C., Nelson R.G., Wolford J.K. Identification of pvt1 as a candidate gene for end-stage renal disease in type 2 diabetes using a pooling-based genome-wide single nucleotide polymorphism association study. Diabetes. 2007;56:975–983. doi: 10.2337/db06-1072. [DOI] [PubMed] [Google Scholar]
- 25.Lee L.T.O., Alvarez M.L., DiStefano J.K. Functional characterization of the plasmacytoma variant translocation 1 gene (pvt1) in diabetic nephropathy. PLoS ONE. 2011;6:e18671. doi: 10.1371/journal.pone.0018671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou L.-J., Yang D.-W., Ou L.-N., Guo X.-R., Wu B.-L. Circulating expression level of lncrna malat1 in diabetic kidney disease patients and its clinical significance. J. Diabetes Res. 2020;2020:4729019. doi: 10.1155/2020/4729019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu M., Wang R., Li X., Fan M., Lin J., Zhen J., Chen L., Lv Z. Lncrna malat1 is dysregulated in diabetic nephropathy and involved in high glucose-induced podocyte injuryviaits interplay with β-catenin. J. Cell. Mol. Med. 2017;21:2732–2747. doi: 10.1111/jcmm.13189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X., Zeng L., Cao C., Lu C., Lian W., Han J., Zhang X., Zhang J., Tang T., Li M. Long noncoding rna malat1 regulates renal tubular epithelial pyroptosis by modulated mir-23c targeting of elavl1 in diabetic nephropathy. Exp. Cell Res. 2017;350:327–335. doi: 10.1016/j.yexcr.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 29.Shang J., Wang S., Jiang Y., Duan Y., Cheng G., Liu D., Xiao J., Zhao Z. Identification of key lncrnas contributing to diabetic nephropathy by gene co-expression network analysis. Sci. Rep. 2019;9:3328. doi: 10.1038/s41598-019-39298-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yi H., Peng R., Zhang L., Sun Y., Peng H., Liu H., Yu L., Li A., Zhang Y., Jiang W., et al. Lincrna-gm4419 knockdown ameliorates nf-κb/nlrp3 inflammasome-mediated inflammation in diabetic nephropathy. Cell Death Dis. 2017;8:e2583. doi: 10.1038/cddis.2016.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao J., Wang W., Wang F., Guo C. Lncrna-nr_033515 promotes proliferation, fibrogenesis and epithelial-to-mesenchymal transition by targeting mir-743b-5p in diabetic nephropathy. Biomed. Pharmacother. 2018;106:543–552. doi: 10.1016/j.biopha.2018.06.104. [DOI] [PubMed] [Google Scholar]
- 32.Sun S.F., Tang P.M.K., Feng M., Xiao J., Huang X.R., Li P., Ma R.C.W., Lan H.Y. Novel lncrna erbb4-ir promotes diabetic kidney injury in db/db mice by targeting mir-29b. Diabetes. 2018;67:731–744. doi: 10.2337/db17-0816. [DOI] [PubMed] [Google Scholar]
- 33.Gao Y., Chen Z.-Y., Wang Y., Liu Y., Ma J.-X., Li Y.-K. Long non-coding rna asncmtrna-2 is upregulated in diabetic kidneys and high glucose-treated mesangial cells. Exp. Ther. Med. 2017;13:581–587. doi: 10.3892/etm.2017.4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kato M., Wang M., Chen Z., Bhatt K., Oh H.J., Lanting L., Deshpande S., Jia Y., Lai J.Y., O’Connor C.L., et al. An endoplasmic reticulum stress-regulated lncrna hosting a microrna megacluster induces early features of diabetic nephropathy. Nat. Commun. 2016;7:12864. doi: 10.1038/ncomms12864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alfaifi M., Ali Beg M.M., Alshahrani M.Y., Ahmad I., Alkhathami A.G., Joshi P.C., Alshehri O.M., Alamri A.M., Verma A.K. Circulating long non-coding rnas nkila, neat1, malat1, and miat expression and their association in type 2 diabetes mellitus. BMJ Open Diabetes Res. Care. 2021;9:e001821. doi: 10.1136/bmjdrc-2020-001821. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Huang S., Xu Y., Ge X., Xu B., Peng W., Jiang X., Shen L., Xia L. Long noncoding rna neat1 accelerates the proliferation and fibrosis in diabetic nephropathy through activating akt/mtor signaling pathway. J. Cell. Physiol. 2018;234:11200–11207. doi: 10.1002/jcp.27770. [DOI] [PubMed] [Google Scholar]
- 37.Liao L., Chen J., Zhang C., Guo Y., Liu W., Liu W., Duan L., Liu Z., Hu J., Lu J. Lncrna neat1 promotes high glucose-induced mesangial cell hypertrophy by targeting mir-222-3p/cdkn1b axis. Front. Mol. Biosci. 2021;7:627827. doi: 10.3389/fmolb.2020.627827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng M., Tang P.M.-K., Huang X.-R., Sun S.-F., You Y.-K., Xiao J., Lv L.-L., Xu A.-P., Lan H.-Y. Tgf-β mediates renal fibrosis via the smad3-erbb4-ir long noncoding rna axis. Mol. Ther. 2018;26:148–161. doi: 10.1016/j.ymthe.2017.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang M., Wang S., Yao D., Yan Q., Lu W. A novel long non-coding rna cyp4b1-ps1-001 regulates proliferation and fibrosis in diabetic nephropathy. Mol. Cell. Endocrinol. 2016;426:136–145. doi: 10.1016/j.mce.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 40.Wang S., Chen X., Wang M., Yao D., Chen T., Yan Q., Lu W. Long non-coding rna cyp4b1-ps1-001 inhibits proliferation and fibrosis in diabetic nephropathy by interacting with nucleolin. Cell. Physiol. Biochem. 2018;49:2174–2187. doi: 10.1159/000493821. [DOI] [PubMed] [Google Scholar]
- 41.Wang M., Yao D., Wang S., Yan Q., Lu W. Long non-coding rna ensmust00000147869 protects mesangial cells from proliferation and fibrosis induced by diabetic nephropathy. Endocrine. 2016;54:81–92. doi: 10.1007/s12020-016-0950-5. [DOI] [PubMed] [Google Scholar]
- 42.Duan L.-J., Ding M., Hou L.-J., Cui Y.-T., Li C.-J., Yu D.-M. Long noncoding rna tug1 alleviates extracellular matrix accumulation via mediating microrna-377 targeting of pparγ in diabetic nephropathy. Biochem. Biophys. Res. Commun. 2017;484:598–604. doi: 10.1016/j.bbrc.2017.01.145. [DOI] [PubMed] [Google Scholar]
- 43.Nezu M., Suzuki N. Roles of nrf2 in protecting the kidney from oxidative damage. Int. J. Mol. Sci. 2020;21:2951. doi: 10.3390/ijms21082951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou L., Xu D.Y., Sha W.G., Shen L., Lu G.Y., Yin X. Long non-coding miat mediates high glucose-induced renal tubular epithelial injury. Biochem. Biophys. Res. Commun. 2015;468:726–732. doi: 10.1016/j.bbrc.2015.11.023. [DOI] [PubMed] [Google Scholar]
- 45.Ghafouri-Fard S., Dashti S., Taheri M. The role of long non-coding rna casc2 in the carcinogenesis process. Biomed. Pharmacother. 2020;127:110202. doi: 10.1016/j.biopha.2020.110202. [DOI] [PubMed] [Google Scholar]
- 46.Wang L., Su N., Zhang Y., Wang G. Clinical significance of serum lncrna cancer susceptibility candidate 2 (casc2) for chronic renal failure in patients with type 2 diabetes. Med. Sci. Monit. 2018;24:6079–6084. doi: 10.12659/MSM.909510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lian H., Cheng Y., Wu X. Tmem16a exacerbates renal injury by activating p38/jnk signaling pathway to promote podocyte apoptosis in diabetic nephropathy mice. Biochem. Biophys. Res. Commun. 2017;487:201–208. doi: 10.1016/j.bbrc.2017.04.021. [DOI] [PubMed] [Google Scholar]
- 48.Yang H., Kan Q.e., Su Y., Man H. Long non-coding rna casc2 improves diabetic nephropathy by inhibiting jnk pathway. Exp. Clin. Endocrinol. Diabetes. 2018;127:533–537. doi: 10.1055/a-0629-9958. [DOI] [PubMed] [Google Scholar]
- 49.Li A., Peng R., Sun Y., Liu H., Peng H., Zhang Z. Lincrna 1700020i14rik alleviates cell proliferation and fibrosis in diabetic nephropathy via mir-34a-5p/sirt1/hif-1α signaling. Cell Death Dis. 2018;9:461. doi: 10.1038/s41419-018-0527-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feng Y., Chen S., Xu J., Zhu Q., Ye X., Ding D., Yao W., Lu Y. Dysregulation of lncrnas gm5524 and gm15645 involved in high-glucose-induced podocyte apoptosis and autophagy in diabetic nephropathy. Mol. Med. Rep. 2018;18:3657–3664. doi: 10.3892/mmr.2018.9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bai X., Geng J., Li X., Wan J., Liu J., Zhou Z., Liu X. Long noncoding rna linc01619 regulates microrna-27a/forkhead box protein o1 and endoplasmic reticulum stress-mediated podocyte injury in diabetic nephropathy. Antioxid. Redox Signal. 2018;29:355–376. doi: 10.1089/ars.2017.7278. [DOI] [PubMed] [Google Scholar]
- 52.Hills C.E., Squires P.E. The role of tgf-beta and epithelial-to mesenchymal transition in diabetic nephropathy. Cytokine Growth Factor Rev. 2011;22:131–139. doi: 10.1016/j.cytogfr.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 53.Grande M.T., Sanchez-Laorden B., Lopez-Blau C., De Frutos C.A., Boutet A., Arevalo M., Rowe R.G., Weiss S.J., Lopez-Novoa J.M., Nieto M.A. Snail1-induced partial epithelial-to-mesenchymal transition drives renal fibrosis in mice and can be targeted to reverse established disease. Nat. Med. 2015;21:989–997. doi: 10.1038/nm.3901. [DOI] [PubMed] [Google Scholar]
- 54.Lovisa S., LeBleu V.S., Tampe B., Sugimoto H., Vadnagara K., Carstens J.L., Wu C.C., Hagos Y., Burckhardt B.C., Pentcheva-Hoang T., et al. Epithelial-to-mesenchymal transition induces cell cycle arrest and parenchymal damage in renal fibrosis. Nat. Med. 2015;21:998–1009. doi: 10.1038/nm.3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marquez-Exposito L., Lavoz C., Rodrigues-Diez R.R., Rayego-Mateos S., Orejudo M., Cantero-Navarro E., Ortiz A., Egido J., Selgas R., Mezzano S., et al. Gremlin regulates tubular epithelial to mesenchymal transition via vegfr2: Potential role in renal fibrosis. Front Pharm. 2018;9:1195. doi: 10.3389/fphar.2018.01195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Srivastava S.P., Hedayat A.F., Kanasaki K., Goodwin J.E. Microrna crosstalk influences epithelial-to-mesenchymal, endothelial-to-mesenchymal, and macrophage-to-mesenchymal transitions in the kidney. Front Pharm. 2019;10:904. doi: 10.3389/fphar.2019.00904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Amar S.K., Srivastav A.K., Srivastava S.P. Current Advances in Breast Cancer Research: A Molecular Approach. Springer; Berlin/Heidelberg, Germany: 2020. Advances of the current therapeutic approach for the management of breast cancer; pp. 328–345. [Google Scholar]
- 58.Srivastava S.P., Koya D., Kanasaki K. Micrornas in kidney fibrosis and diabetic nephropathy: Roles on emt and endmt. Biomed. Res. Int. 2013;2013:125469. doi: 10.1155/2013/125469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kalluri R., Weinberg R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alidadiani N., Ghaderi S., Dilaver N., Bakhshamin S., Bayat M. Epithelial mesenchymal transition transcription factor (tf): The structure, function and microrna feedback loop. Gene. 2018;674:115–120. doi: 10.1016/j.gene.2018.06.049. [DOI] [PubMed] [Google Scholar]
- 61.Liu X., Sun N., Mo N., Lu S., Song E., Ren C., Li Z. Quercetin inhibits kidney fibrosis and the epithelial to mesenchymal transition of the renal tubular system involving suppression of the sonic hedgehog signaling pathway. Food Funct. 2019;10:3782–3797. doi: 10.1039/C9FO00373H. [DOI] [PubMed] [Google Scholar]
- 62.Glover E.K., Jordan N., Sheerin N.S., Ali S. Regulation of endothelial-to-mesenchymal transition by micrornas in chronic allograft dysfunction. Transplantation. 2019;103:e64–e73. doi: 10.1097/TP.0000000000002589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li J., Shi S., Srivastava S.P., Kitada M., Nagai T., Nitta K., Kohno M., Kanasaki K., Koya D. Fgfr1 is critical for the anti-endothelial mesenchymal transition effect of n-acetyl-seryl-aspartyl-lysyl-proline via induction of the map4k4 pathway. Cell Death Dis. 2017;8:e2965. doi: 10.1038/cddis.2017.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lovisa S., Kalluri R. Fatty acid oxidation regulates the activation of endothelial-to-mesenchymal transition. Trends Mol. Med. 2018;24:432–434. doi: 10.1016/j.molmed.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 65.Curci C., Castellano G., Stasi A., Divella C., Loverre A., Gigante M., Simone S., Cariello M., Montinaro V., Lucarelli G., et al. Endothelial-to-mesenchymal transition and renal fibrosis in ischaemia/reperfusion injury are mediated by complement anaphylatoxins and akt pathway. Nephrol. Dial Transpl. 2014;29:799–808. doi: 10.1093/ndt/gft516. [DOI] [PubMed] [Google Scholar]
- 66.Zeisberg E.M., Tarnavski O., Zeisberg M., Dorfman A.L., McMullen J.R., Gustafsson E., Chandraker A., Yuan X., Pu W.T., Roberts A.B., et al. Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nat. Med. 2007;13:952–961. doi: 10.1038/nm1613. [DOI] [PubMed] [Google Scholar]
- 67.Shi S., Srivastava S.P., Kanasaki M., He J., Kitada M., Nagai T., Nitta K., Takagi S., Kanasaki K., Koya D. Interactions of dpp-4 and integrin beta1 influences endothelial-to-mesenchymal transition. Kidney Int. 2015;88:479–489. doi: 10.1038/ki.2015.103. [DOI] [PubMed] [Google Scholar]
- 68.Medici D. Endothelial-mesenchymal transition in regenerative medicine. Stem. Cells Int. 2016;2016:6962801. doi: 10.1155/2016/6962801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cho J.G., Lee A., Chang W., Lee M.S., Kim J. Endothelial to mesenchymal transition represents a key link in the interaction between inflammation and endothelial dysfunction. Front. Immunol. 2018;9:294. doi: 10.3389/fimmu.2018.00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li J., Liu H., Srivastava S.P., Hu Q., Gao R., Li S., Kitada M., Wu G., Koya D., Kanasaki K. Endothelial fgfr1 (fibroblast growth factor receptor 1) deficiency contributes differential fibrogenic effects in kidney and heart of diabetic mice. Hypertension. 2020;76:1935–1944. doi: 10.1161/HYPERTENSIONAHA.120.15587. [DOI] [PubMed] [Google Scholar]
- 71.Zhou H., Mehta S., Srivastava S.P., Grabinska K., Zhang X., Wong C., Hedayat A., Perrotta P., Fernandez-Hernando C., Sessa W.C., et al. Endothelial cell-glucocorticoid receptor interactions and regulation of wnt signaling. JCI Insight. 2020;5:131384. doi: 10.1172/jci.insight.131384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Srivastava S.P., Zhou H., Setia O., Liu B., Kanasaki K., Koya D., Dardik A., Fernandez-Hernando C., Goodwin J. Loss of endothelial glucocorticoid receptor accelerate diabetic nephropathy. Nat. Commun. 2021;12:2368. doi: 10.1038/s41467-021-22617-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Srivastava S.P., Li J., Takagaki Y., Kitada M., Goodwin J., Kanasaki K., Koya D. Endothelial SIRT3 regulates myofibroblast metabolic shifts in diabetic kidneys. iScience. 2021;24:102390. doi: 10.1016/j.isci.2021.102390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zeisberg E.M., Potenta S.E., Sugimoto H., Zeisberg M., Kalluri R. Fibroblasts in kidney fibrosis emerge via endothelial-to-mesenchymal transition. J. Am. Soc. Nephrol. 2008;19:2282–2287. doi: 10.1681/ASN.2008050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Medici D., Kalluri R. Endothelial-mesenchymal transition and its contribution to the emergence of stem cell phenotype. Semin. Cancer Biol. 2012;22:379–384. doi: 10.1016/j.semcancer.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Srivastava S.P., Shi S., Kanasaki M., Nagai T., Kitada M., He J., Nakamura Y., Ishigaki Y., Kanasaki K., Koya D. Effect of antifibrotic micrornas crosstalk on the action of n-acetyl-seryl-aspartyl-lysyl-proline in diabetes-related kidney fibrosis. Sci. Rep. 2016;6:29884. doi: 10.1038/srep29884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fernandes J., Acuña S., Aoki J., Floeter-Winter L., Muxel S. Long non-coding rnas in the regulation of gene expression: Physiology and disease. Non-Coding RNA. 2019;5:17. doi: 10.3390/ncrna5010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shi X., Sun M., Liu H., Yao Y., Song Y. Long non-coding rnas: A new frontier in the study of human diseases. Cancer Lett. 2013;339:159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 79.Lee L.T.O., Alvarez M.L., Khosroheidari M., Eddy E., Kiefer J. Role of microrna 1207-5p and its host gene, the long non-coding rna pvt1, as mediators of extracellular matrix accumulation in the kidney: Implications for diabetic nephropathy. PLoS ONE. 2013;8:e77468. doi: 10.1371/journal.pone.0077468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huppi K., Volfovsky N., Runfola T., Jones T.L., Mackiewicz M., Martin S.E., Mushinski J.F., Stephens R., Caplen N.J. The identification of micrornas in a genomically unstable region of human chromosome 8q24. Mol. Cancer Res. 2008;6:212–221. doi: 10.1158/1541-7786.MCR-07-0105. [DOI] [PubMed] [Google Scholar]
- 81.Loganathan T.S., Sulaiman S.A., Abdul Murad N.A., Shah S.A., Abdul Gafor A.H., Jamal R., Abdullah N. Interactions among non-coding rnas in diabetic nephropathy. Front. Pharmacol. 2020;11:191. doi: 10.3389/fphar.2020.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sun Z., Ma Y., Chen F., Wang S., Chen B., Shi J. Mir-133b and mir-199b knockdown attenuate tgf-beta1-induced epithelial to mesenchymal transition and renal fibrosis by targeting sirt1 in diabetic nephropathy. Eur. J. Pharm. 2018;837:96–104. doi: 10.1016/j.ejphar.2018.08.022. [DOI] [PubMed] [Google Scholar]
- 83.Wang X., Xu Y., Zhu Y.C., Wang Y.K., Li J., Li X.Y., Ji T., Bai S.J. Lncrna neat1 promotes extracellular matrix accumulation and epithelial-to-mesenchymal transition by targeting mir-27b-3p and zeb1 in diabetic nephropathy. J. Cell. Physiol. 2018;234:12926–12933. doi: 10.1002/jcp.27959. [DOI] [PubMed] [Google Scholar]
- 84.Wang B., Komers R., Carew R., Winbanks C.E., Xu B., Herman-Edelstein M., Koh P., Thomas M., Jandeleit-Dahm K., Gregorevic P., et al. Suppression of microrna-29 expression by tgf-beta1 promotes collagen expression and renal fibrosis. J. Am. Soc. Nephrol. 2012;23:252–265. doi: 10.1681/ASN.2011010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pezzolesi M.G., Satake E., McDonnell K.P., Major M., Smiles A.M., Krolewski A.S. Circulating tgf-beta1-regulated mirnas and the risk of rapid progression to esrd in type 1 diabetes. Diabetes. 2015;64:3285–3293. doi: 10.2337/db15-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Srivastava S.P., Goodwin J.E., Kanasaki K., Koya D. Inhibition of angiotensin-converting enzyme ameliorates renal fibrosis by mitigating dpp-4 level and restoring antifibrotic micrornas. Genes. 2020;11:211. doi: 10.3390/genes11020211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nitta K., Shi S., Nagai T., Kanasaki M., Kitada M., Srivastava S.P., Haneda M., Kanasaki K., Koya D. Oral administration of n-acetyl-seryl-aspartyl-lysyl-proline ameliorates kidney disease in both type 1 and type 2 diabetic mice via a therapeutic regimen. Biomed. Res. Int. 2016;2016:9172157. doi: 10.1155/2016/9172157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kanasaki K., Shi S., Kanasaki M., He J., Nagai T., Nakamura Y., Ishigaki Y., Kitada M., Srivastava S.P., Koya D. Linagliptin-mediated dpp-4 inhibition ameliorates kidney fibrosis in streptozotocin-induced diabetic mice by inhibiting endothelial-to-mesenchymal transition in a therapeutic regimen. Diabetes. 2014;63:2120–2131. doi: 10.2337/db13-1029. [DOI] [PubMed] [Google Scholar]
- 89.Nagai T., Kanasaki M., Srivastava S.P., Nakamura Y., Ishigaki Y., Kitada M., Shi S., Kanasaki K., Koya D. N-acetyl-seryl-aspartyl-lysyl-proline inhibits diabetes-associated kidney fibrosis and endothelial-mesenchymal transition. Biomed. Res. Int. 2014;2014:696475. doi: 10.1155/2014/696475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen P.Y., Qin L., Barnes C., Charisse K., Yi T., Zhang X., Ali R., Medina P.P., Yu J., Slack F.J., et al. Fgf regulates tgf-beta signaling and endothelial-to-mesenchymal transition via control of let-7 mirna expression. Cell Rep. 2012;2:1684–1696. doi: 10.1016/j.celrep.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Blahna M.T., Hata A. Smad-mediated regulation of microrna biosynthesis. FEBS Lett. 2012;586:1906–1912. doi: 10.1016/j.febslet.2012.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Qin W., Chung A.C., Huang X.R., Meng X.M., Hui D.S., Yu C.M., Sung J.J., Lan H.Y. Tgf-beta/smad3 signaling promotes renal fibrosis by inhibiting mir-29. J. Am. Soc. Nephrol. 2011;22:1462–1474. doi: 10.1681/ASN.2010121308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ma F., Xu S., Liu X., Zhang Q., Xu X., Liu M., Hua M., Li N., Yao H., Cao X. The microrna mir-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-gamma. Nat. Immunol. 2011;12:861–869. doi: 10.1038/ni.2073. [DOI] [PubMed] [Google Scholar]
- 94.Srivastava S.P., Goodwin J.E., Kanasaki K., Koya D. Metabolic reprogramming by n-acetyl-seryl-aspartyl-lysyl-proline protects against diabetic kidney disease. Br. J. Pharm. 2020;177:3691–3711. doi: 10.1111/bph.15087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Keniry A., Oxley D., Monnier P., Kyba M., Dandolo L., Smits G., Reik W. The h19 lincrna is a developmental reservoir of mir-675 that suppresses growth and igf1r. Nat. Cell Biol. 2012;14:659–665. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ma C., Nong K., Zhu H., Wang W., Huang X., Yuan Z., Ai K. H19 promotes pancreatic cancer metastasis by derepressing let-7’s suppression on its target hmga2-mediated emt. Tumor Biol. 2014;35:9163–9169. doi: 10.1007/s13277-014-2185-5. [DOI] [PubMed] [Google Scholar]
- 97.Zou T., Jaladanki S.K., Liu L., Xiao L., Chung H.K., Wang J.-Y., Xu Y., Gorospe M., Wang J.-Y. H19long noncoding rna regulates intestinal epithelial barrier function via microrna 675 by interacting with rna-binding protein hur. Mol. Cell. Biol. 2016;36:1332–1341. doi: 10.1128/MCB.01030-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xie H., Xue J.-D., Chao F., Jin Y.-F., Fu Q. Long non-coding rna-h19 antagonism protects against renal fibrosis. Oncotarget. 2016;7:51473–51481. doi: 10.18632/oncotarget.10444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Baumann V., Winkler J. Mirna-based therapies: Strategies and delivery platforms for oligonucleotide and non-oligonucleotide agents. Future Med. Chem. 2014;6:1967–1984. doi: 10.4155/fmc.14.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fu Y., Chen J., Huang Z. Recent progress in microrna-based delivery systems for the treatment of human disease. ExRNA. 2019;1:1–14. doi: 10.1186/s41544-019-0024-y. [DOI] [Google Scholar]
- 101.Putta S., Lanting L., Sun G., Lawson G., Kato M., Natarajan R. Inhibiting microrna-192 ameliorates renal fibrosis in diabetic nephropathy. J. Am. Soc. Nephrol. 2012;23:458–469. doi: 10.1681/ASN.2011050485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gomez I.G., MacKenna D.A., Johnson B.G., Kaimal V., Roach A.M., Ren S., Nakagawa N., Xin C., Newitt R., Pandya S., et al. Anti-microrna-21 oligonucleotides prevent alport nephropathy progression by stimulating metabolic pathways. J. Clin. Investig. 2015;125:141–156. doi: 10.1172/JCI75852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chen H.Y., Zhong X., Huang X.R., Meng X.M., You Y., Chung A.C., Lan H.Y. Microrna-29b inhibits diabetic nephropathy in db/db mice. Mol. Ther. 2014;22:842–853. doi: 10.1038/mt.2013.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Beltrami C., Simpson K., Jesky M., Wonnacott A., Carrington C., Holmans P., Newbury L., Jenkins R., Ashdown T., Dayan C., et al. Association of elevated urinary mir-126, mir-155, and mir-29b with diabetic kidney disease. Am. J. Pathol. 2018;188:1982–1992. doi: 10.1016/j.ajpath.2018.06.006. [DOI] [PubMed] [Google Scholar]
- 105.Simpson K., Wonnacott A., Fraser D.J., Bowen T. Micrornas in diabetic nephropathy: From biomarkers to therapy. Curr. Diabetes Rep. 2016;16:35. doi: 10.1007/s11892-016-0724-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Assmann T.S., Recamonde-Mendoza M., Costa A.R., Punales M., Tschiedel B., Canani L.H., Bauer A.C., Crispim D. Circulating mirnas in diabetic kidney disease: Case-control study and in silico analyses. Acta Diabetol. 2019;56:55–65. doi: 10.1007/s00592-018-1216-x. [DOI] [PubMed] [Google Scholar]
- 107.Schena F.P., Serino G., Sallustio F. Micrornas in kidney diseases: New promising biomarkers for diagnosis and monitoring. Nephrol. Dial Transpl. 2014;29:755–763. doi: 10.1093/ndt/gft223. [DOI] [PubMed] [Google Scholar]
- 108.Schena F.P., Sallustio F., Serino G. Micrornas in glomerular diseases from pathophysiology to potential treatment target. Clin. Sci. 2015;128:775–788. doi: 10.1042/CS20140733. [DOI] [PubMed] [Google Scholar]
- 109.Janssen H.L., Kauppinen S., Hodges M.R. Hcv infection and miravirsen. N. Engl. J. Med. 2013;369:878. doi: 10.1056/NEJMc1307787. [DOI] [PubMed] [Google Scholar]
- 110.Maruyama R., Yokota T. Gapmers. Springer; Berlin/Heidelberg, Germany: 2020. Knocking down long noncoding rnas using antisense oligonucleotide gapmers; pp. 49–56. [DOI] [PubMed] [Google Scholar]
- 111.Lennox K.A., Behlke M.A. Cellular localization of long non-coding rnas affects silencing by rnai more than by antisense oligonucleotides. Nucleic Acids Res. 2016;44:863–877. doi: 10.1093/nar/gkv1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Garitano-Trojaola A., Agirre X., Prósper F., Fortes P. Long non-coding rnas in haematological malignancies. Int. J. Mol. Sci. 2013;14:15386–15422. doi: 10.3390/ijms140815386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Verma A.K., Singh H., Satyanarayana M., Srivastava S.P., Tiwari P., Singh A.B., Dwivedi A.K., Singh S.K., Srivastava M., Nath C., et al. Flavone-based novel antidiabetic and antidyslipidemic agents. J. Med. Chem. 2012;55:4551–4567. doi: 10.1021/jm201107g. [DOI] [PubMed] [Google Scholar]
- 114.Raza S., Srivastava S.P., Srivastava D.S., Srivastava A.K., Haq W., Katti S.B. Thiazolidin-4-one and thiazinan-4-one derivatives analogous to rosiglitazone as potential antihyperglycemic and antidyslipidemic agents. Eur. J. Med. Chem. 2013;63:611–620. doi: 10.1016/j.ejmech.2013.01.054. [DOI] [PubMed] [Google Scholar]
- 115.Arha D., Pandeti S., Mishra A., Srivastava S.P., Srivastava A.K., Narender T., Tamrakar A.K. Deoxyandrographolide promotes glucose uptake through glucose transporter-4 translocation to plasma membrane in l6 myotubes and exerts antihyperglycemic effect in vivo. Eur. J. Pharm. 2015;768:207–216. doi: 10.1016/j.ejphar.2015.10.055. [DOI] [PubMed] [Google Scholar]
- 116.Kumar A., Sharma S., Tripathi V.D., Maurya R.A., Srivastava S.P., Bhatia G., Tamrakar A.K., Srivastava A.K. Design and synthesis of 2,4-disubstituted polyhydroquinolines as prospective antihyperglycemic and lipid modulating agents. Bioorgan. Med. Chem. 2010;18:4138–4148. doi: 10.1016/j.bmc.2009.11.061. [DOI] [PubMed] [Google Scholar]
- 117.Kumar A., Sharma S., Gupta L.P., Ahmad P., Srivastava S.P., Rahuja N., Tamrakar A.K., Srivastava A.K. Synthesis of propiophenone derivatives as new class of antidiabetic agents reducing body weight in db/db mice. Bioorgan. Med. Chem. 2012;20:2172–2179. doi: 10.1016/j.bmc.2011.12.027. [DOI] [PubMed] [Google Scholar]
- 118.Kanasaki M., Srivastava S.P., Yang F., Xu L., Kudoh S., Kitada M., Ueki N., Kim H., Li J., Takeda S., et al. Deficiency in catechol-o-methyltransferase is linked to a disruption of glucose homeostasis in mice. Sci. Rep. 2017;7:7927. doi: 10.1038/s41598-017-08513-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shukla P., Srivastava S.P., Srivastava R., Rawat A.K., Srivastava A.K., Pratap R. Synthesis and antidyslipidemic activity of chalcone fibrates. Bioorgan. Med. Chem. Lett. 2011;21:3475–3478. doi: 10.1016/j.bmcl.2011.03.057. [DOI] [PubMed] [Google Scholar]
- 120.Balaramnavar V.M., Srivastava R., Rahuja N., Gupta S., Rawat A.K., Varshney S., Chandasana H., Chhonker Y.S., Doharey P.K., Kumar S., et al. Identification of novel ptp1b inhibitors by pharmacophore based virtual screening, scaffold hopping and docking. Eur. J. Med. Chem. 2014;87:578–594. doi: 10.1016/j.ejmech.2014.09.097. [DOI] [PubMed] [Google Scholar]
- 121.Mishra A., Srivastava R., Srivastava S.P., Gautam S., Tamrakar A.K., Maurya R., Srivastava A.K. Antidiabetic activity of heart wood of pterocarpus marsupium roxb. And analysis of phytoconstituents. Indian J. Exp. Biol. 2013;51:363–374. [PubMed] [Google Scholar]
- 122.Srivastava R., Srivastava S.P., Jaiswal N., Mishra A., Maurya R., Srivastava A.K. Antidiabetic and antidyslipidemic activities of cuminum cyminum l. In validated animal models. Med. Chem. Res. 2010;20:1656–1666. doi: 10.1007/s00044-010-9483-2. [DOI] [Google Scholar]
- 123.Srivastava S.P., Mishra A., Bhatia V., Narender T., Srivastava A.K. Acacia catechu hard wood: Potential anti-diabetic cum anti-dyslipidemic. Med. Chem. Res. 2010;20:1732–1739. doi: 10.1007/s00044-010-9479-y. [DOI] [Google Scholar]
- 124.Jaiswal N. Inhibition of alpha-glucosidase by acacia nilotica prevents hyperglycemia along with improvement of diabetic complications via aldose reductase inhibition. J. Diabetes Metab. 2012;6:9. doi: 10.4172/2155-6156.S6-004. [DOI] [Google Scholar]
- 125.Jaiswal N., Bhatia V., Srivastava S.P., Srivastava A.K., Tamrakar A.K. Antidiabetic effect of eclipta alba associated with the inhibition of alpha-glucosidase and aldose reductase. Nat. Prod. Res. 2012;26:2363–2367. doi: 10.1080/14786419.2012.662648. [DOI] [PubMed] [Google Scholar]
- 126.Pandey A.K., Verma G., Vig S., Srivastava S., Srivastava A.K., Datta M. Mir-29a levels are elevated in the db/db mice liver and its overexpression leads to attenuation of insulin action on pepck gene expression in hepg2 cells. Mol. Cell Endocrinol. 2011;332:125–133. doi: 10.1016/j.mce.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 127.Kaur K., Pandey A.K., Srivastava S., Srivastava A.K., Datta M. Comprehensive mirnome and in silico analyses identify the wnt signaling pathway to be altered in the diabetic liver. Mol. Biosyst. 2011;7:3234–3244. doi: 10.1039/c1mb05041a. [DOI] [PubMed] [Google Scholar]
- 128.Minutolo R., Gabbai F.B., Provenzano M., Chiodini P., Borrelli S., Garofalo C., Sasso F.C., Santoro D., Bellizzi V., Conte G., et al. Cardiorenal prognosis by residual proteinuria level in diabetic chronic kidney disease: Pooled analysis of four cohort studies. Nephrol Dial Transpl. 2018;33:1942–1949. doi: 10.1093/ndt/gfy032. [DOI] [PubMed] [Google Scholar]
- 129.Yamanouchi M., Furuichi K., Hoshino J., Toyama T., Hara A., Shimizu M., Kinowaki K., Fujii T., Ohashi K., Yuzawa Y., et al. Nonproteinuric versus proteinuric phenotypes in diabetic kidney disease: A propensity score-matched analysis of a nationwide, biopsy-based cohort study. Diabetes Care. 2019;42:891–902. doi: 10.2337/dc18-1320. [DOI] [PubMed] [Google Scholar]
- 130.Lewis E.J., Xu X. Abnormal glomerular permeability characteristics in diabetic nephropathy: Implications for the therapeutic use of low-molecular weight heparin. Diabetes Care. 2008;31:S202–S207. doi: 10.2337/dc08-s251. [DOI] [PubMed] [Google Scholar]
- 131.Jermendy G., Ruggenenti P. Preventing microalbuminuria in patients with type 2 diabetes. Diabetes Metab. Res. Rev. 2007;23:100–110. doi: 10.1002/dmrr.693. [DOI] [PubMed] [Google Scholar]
- 132.Long J., Badal S.S., Ye Z., Wang Y., Ayanga B.A., Galvan D.L., Green N.H., Chang B.H., Overbeek P.A., Danesh F.R. Long noncoding rna tug1 regulates mitochondrial bioenergetics in diabetic nephropathy. J. Clin. Investig. 2016;126:4205–4218. doi: 10.1172/JCI87927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lv L., Li D., Ye Z., Tian F., Li X., Zhang J., Yu X. Silence of lncRNA GAS5 alleviates high glucose toxicity to human renal tubular epithelial HK-2 cells through regulation of miR-27a. Artif. Cells Nanomed. Biotechnol. 2019;47:2205–2212. doi: 10.1080/21691401.2019.1616552. [DOI] [PubMed] [Google Scholar]
- 134.Ji T., Wang Y., Zhu Y., Gao C., Li X., Li J., Bai F., Bai S. Long noncoding RNA Gm6135 functions as a competitive endogenous RNA to regulate toll-like receptor 4 expression by sponging miR-203-3p in diabetic nephropathy. J. Cell. Physiol. 2019;234:6633–6641. doi: 10.1002/jcp.27412. [DOI] [PubMed] [Google Scholar]
- 135.Hu W., Han Q., Zhao L., Wang L. Circular RNA circRNA_15698 aggravates the extracellular matrix of diabetic nephropathy mesangial cells via miR-185/TGF-β1. J. Cell. Physiol. 2019;234:1469–1476. doi: 10.1002/jcp.26959. [DOI] [PubMed] [Google Scholar]
- 136.Chen B., Li Y., Liu Y., Xu Z. circLRP6 regulates high glucose-induced proliferation, oxidative stress, ECM accumulation, and inflammation in mesangial cells. J. Cell. Physiol. 2019;234:21149–21259. doi: 10.1002/jcp.28730. [DOI] [PubMed] [Google Scholar]
- 137.Wen S., Li S., Li L., Fan Q. circACTR2: A Novel Mechanism Regulating High Glucose-Induced Fibrosis in Renal Tubular Cells via Pyroptosis. Biol. Pharm. Bull. 2020;43:558–564. doi: 10.1248/bpb.b19-00901. [DOI] [PubMed] [Google Scholar]
- 138.Zhuang L., Wang Z., Hu X., Yang Q., Pei X., Jin X. CircHIPK3 Alleviates High Glucose Toxicity to Human Renal Tubular Epithelial HK-2 Cells Through Regulation of miR-326/miR-487a-3p/SIRT1. Diabetes Metab. Syndr. Obes. 2021;14:729–740. doi: 10.2147/DMSO.S289624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Mou X., Chenv J.W., Zhou D.Y., Liu K., Chen L.J., Zhou D., Hu Y.B. A novel identified circular RNA, circ_0000491, aggravates the extracellular matrix of diabetic nephropathy glomerular mesangial cells through suppressing miR-101b by targeting TGFβRI. Mol. Med. Rep. 2020;22:3785–3794. doi: 10.3892/mmr.2020.11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Peng F., Gong W., Li S., Yin B., Zhao C., Liu W., Chen X., Luo C., Huang Q., Chen T., et al. circRNA_010383 Acts as a Sponge for miR-135a and its Downregulated Expression Contributes to Renal Fibrosis in Diabetic Nephropathy. Diabetes. 2020:db200203. doi: 10.2337/db200203. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.