Abstract
Objective
To develop a computer model to predict patients with nonalcoholic steatohepatitis (NASH) using machine learning (ML).
Materials and Methods
This retrospective study utilized two databases: a) the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) nonalcoholic fatty liver disease (NAFLD) adult database (2004-2009), and b) the Optum® de-identified Electronic Health Record dataset (2007-2018), a real-world dataset representative of common electronic health records in the United States. We developed an ML model to predict NASH, using confirmed NASH and non-NASH based on liver histology results in the NIDDK dataset to train the model.
Results
Models were trained and tested on NIDDK NAFLD data (704 patients) and the best-performing models evaluated on Optum data (~3,000,000 patients). An eXtreme Gradient Boosting model (XGBoost) consisting of 14 features exhibited high performance as measured by area under the curve (0.82), sensitivity (81%), and precision (81%) in predicting NASH. Slightly reduced performance was observed with an abbreviated feature set of 5 variables (0.79, 80%, 80%, respectively). The full model demonstrated good performance (AUC 0.76) to predict NASH in Optum data.
Discussion
The proposed model, named NASHmap, is the first ML model developed with confirmed NASH and non-NASH cases as determined through liver biopsy and validated on a large, real-world patient dataset. Both the 14 and 5-feature versions exhibit high performance.
Conclusion
The NASHmap model is a convenient and high performing tool that could be used to identify patients likely to have NASH in clinical settings, allowing better patient management and optimal allocation of clinical resources.
Keywords: artificial intelligence, machine learning, non-alcoholic fatty liver disease, NASH, NAFLD
INTRODUCTION
Nonalcoholic fatty liver disease (NAFLD) is a significant public health concern affecting 30% of the adult population in the United States.1 Characterized by excess fat accumulation in the liver, NALFD is associated with other metabolic comorbidities, such as arterial hypertension, dyslipidemia, obesity, and type 2 diabetes mellitus (T2DM).2,3 NAFLD is classified histologically as non-alcoholic fatty liver (NAFL) or non-alcoholic steatohepatitis (NASH).4 NASH is defined by steatosis and inflammation with hepatocyte injury (ballooning) with or without fibrosis2,4 and ultimately can lead to the development of end-stage liver disease, cirrhosis, or hepatocellular carcinoma (HCC). NASH is difficult to diagnose as symptoms are not specific and may not be readily overt on clinical examination or routine laboratory tests.3 The current reference standard for diagnosis and staging of NASH in clinical practice and investigational trials is liver biopsy,4 which is an invasive procedure with associated risks, such as post-procedural pain and bleeding5 as well as additional costs. There remains a need to develop a noninvasive, accurate, easy-to-use tool to identify patients with a high probability of NASH.6
The use of machine learning (ML) in healthcare has increased commensurately with the increase in electronic health records (EHR) and the advancement of big data analytics.7,8 While use cases for ML are broad, one interesting application is to predict the presence of disease for individual patients or in large databases. ML models vary in complexity from artificial neural networks to boosting techniques to more classic decision tree-based models. Prior work has shown promise for applying ML to predict NASH, but many of these approaches do not use readily available inputs or are not yet validated in a large cohort.
The successful identification of patients at risk of having NASH will allow for specific risk stratification, counseling, and identification of patients outside of specialized clinics (eg, in the primary care setting). This is of great interest to support physicians in overcoming the low awareness of the disease.9 As such, an ML model that utilizes common variables regularly collected in clinical practice could help facilitate suspicion of NASH at its earliest stages and allow the preselection of patients for more specific testing. Here, we present our approach and outcomes for the development and validation of a convenient and accurate ML model for predicting probable NASH in electronic health records.
MATERIAL AND METHODS
Data sources
Data were derived from two sources: The National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) NAFLD dataset and the Optum® de-identified Electronic Health Record dataset.
NIDDK NAFLD data consist of adult patients with NAFLD observed over a 4-year period (2004-2009). The patients were classified based on their liver status as NASH or non-NASH as confirmed through liver biopsy and histological assessment. Available data include demographic, histological, clinical biomarkers, and imaging variables. Patients with other liver diseases (eg, viral hepatitis, alcoholic cirrhosis) or excessive alcohol consumption were excluded. Complete inclusion and exclusion criteria as well as patient characteristics captured from NIDDK data are provided in the Supplementary Methods and Supplementary Table I.
The Optum dataset is a real-world dataset consisting of ∼86 million adult patient records collected by 150,000 providers, 2,000 hospitals, and 7,000 clinics in the United States from 2007-2018. This heterogeneous dataset contains patients with NAFLD based on International Classification of Disease (ICD) 9 and 10 codes and potential, yet undiagnosed or unconfirmed, NAFL and NASH patients. Patient records consist of demographics, diagnoses, procedures, medications, labs, and physician notes.
Cohort selection
Patients from the NIDDK database with confirmed NASH or non-NASH status by biopsy were included in this study. The index date was the date of liver biopsy. Patient data closest to the index date were used for analyses. For Optum EHR, true NASH and potential NASH patients were selected based on presence of liver biopsy reading and ICD-9 and 10 codes. Broad inclusion criteria containing both NAFLD and associated comorbidities, such as T2DM, were used to construct an initial cohort. This cohort was then classified as NASH or non-NASH. Patients with a biopsy and an ICD-10 diagnosis of NASH (K75.81) were identified as true NASH patients. Patients with the NAFLD-associated comorbidities used for inclusion but no ICD-9 or 10 diagnosis for any form of NAFLD were considered as non-NASH diagnosed (though they may be undiagnosed NASH). Patients with other diagnosed liver diseases (eg, hepatitis C infection), diagnosed alcohol dependence, or positive tests or treatment for other liver diseases were excluded from the analysis. The complete patient inclusion and exclusion criteria for the Optum EHR dataset are provided in the Supplementary Methods.
Statistical methods
Statistical applications for data analyses and model development included R and the packages eXtreme Gradient Boosting (XGBoost) and caret. The following methods were used:
Correlation, t-tests, and chi-squared tests were performed to determine the degree of association between patient data and NASH diagnosis. These were used to supplement manual review of data and not for variable selection.
K-nearest neighbor (kNN) was used for data imputation in cases of missing NIDDK data. kNN replaces missing values with the mean value of the feature from the k most similar neighbors for data imputation.10 We determined ∼26% of patients to have at least one missing feature value requiring imputation. Multiple values of k were tested ranging from 1 to 15 with a final selected value of k=5 yielding the best performance.
Random forest (RF) was tested as a potential model. Random forest is a tree-based ensemble method that utilizes parallel decision trees built on subsets of the data to develop an optimized predictive model.11 This idea of building multiple models from randomly selected subsets of the data is sometimes referred to as bagging or bootstrap aggregation. When predicting, each individual tree in the random forest votes based on its prediction, and the classification with the most votes becomes the overall model’s prediction. In this way, random forest uses a large set of uncorrelated trees to make a prediction as an ensemble that’s more accurate than a single tree.
XGBoost was also tested as a potential model. XGBoost is a popular ML approach that implements gradient boosting with decision trees as the underlying learners. Whereas random forest employs its individual trees in parallel to solve the same problem, XGBoost builds its individual trees sequentially. Each tree is trained to resolve the prediction error remaining after the prior tree and therefore improves the prediction.12 This provides another approach to building more complex and accurate models with trees, while controlling individual tree depth and complexity. XGBoost has shown very good performance across a wide variety of ML problems.
Model building
Step 1: Developing the model with NIDDK data
The target study population with confirmed NASH and non-NASH patients was split into train and test datasets. The following two-class models were trained: logistic regression, classification and regression trees (CART), random forest, and XGBoost.11–14 We started with 24 demographic variables and clinical biomarkers as features and then refined our models with recursive feature elimination (RFE) for feature selection. In RFE, all original features are ranked according to importance to the model, and each iteration results in the (backward) elimination of the weakest feature(s).15 RFE produces a feature subset that can improve generalization performance by reducing overfitting and improving efficiency. Applicable model hyperparameters were optimized for each model (eg, number of trees, maximum tree depth, minimum leaf size, observation sampling, feature sampling, and gamma) using model appropriate techniques (eg, cross validation and grid search).
Step 2: Evaluating the model on NIDDK data and selecting a model
The test dataset was used to evaluate model performance. Models were compared on key performance criteria that included sensitivity, specificity, precision, accuracy, and area under the curve (AUC). AUC was considered the primary criteria for overall model performance comparison and model selection as it is not dependent on the cut-off value. Of the remaining metrics, sensitivity and precision were considered first, given the focus on the positive class and potential for a very large negative class in real-world application of the model. Sensitivity-precision thresholds for each model were calculated to balance classifier sensitivity (classifying as NASH all true NASH patients) against precision (classifying as NASH only true NASH patients).
We conducted two further performance analyses using the NIDDK data. We repeated the training process 1,000 times with different random samples to ensure our performance is reproducible and generate an AUC confidence interval. We also analyzed performance including AUC, sensitivity, and specificity for T2DM patients and non-T2DM patients. T2DM is a common comorbidity in NAFLD, and these are two of the most important types of patients to understand model performance.
Step 3: Evaluating the model with Optum® EHR
Next, Optum data were used to evaluate model performance. For each patient, the best 6-month analysis window based on data completeness was identified, preferring the latest such window when multiple data were available. Complete records over a 6-month period were included in the analysis with 22% of patient records having all required features. Outliers were identified based on interquartile range (IQR, Q1 − 1.5*IQR and Q3 + 1.5*IQR). Outliers were capped or floored at the 99th and 1st percentiles respectively with thresholds calculated for both NIDDK and Optum data then the more permissive value used. The top performing classifier from step 2 was then applied to Optum data. Standard techniques for determining an appropriate prediction score cut-off were limited by the uncertainty of true NASH status in patients without a NASH diagnosis. Therefore, models were calibrated at a positive prediction rate (PPR) of 30% among these undiagnosed patients, which was approximately the same cut-off as used in the NIDDK dataset.
RESULTS
A total of 453 patients with NASH and 251 without NASH (non-NASH) from NIDDK were split into train (422) and test (282) datasets. The ratio of NASH to non-NASH (64:36) was maintained when splitting the train and test datasets. After splitting, demographics of the datasets were similar. The mean ± standard deviation (SD) age in the train and test datasets was 49.9 ± 9.8 and 48.6 ± 10.3 respectively. The gender distribution of the train dataset was 43% male, 57% female and 37% male, 63% female in the test dataset. Descriptive statistics for the NIDDK NAFLD cohort are provided in Supplementary Table II.
RFE led to a set of 14 features important for NASH classification. Table 1 shows these features in order of importance along with their relative feature importance in the final model. Class means for continuous variables are provided in Table 2.
Table 1.
NIDDK feature rank
| Feature | Rank | Relative Feature Importance |
|---|---|---|
| HbA1c | 1 | 100% |
| AST (units/L) | 2 | 86% |
| ALT (units/L) | 3 | 75% |
| Total protein (g/dl) | 4 | 71% |
| AST/ALT | 5 | 69% |
| BMI (kg/m2) | 6 | 66% |
| Triglycerides (mg/dl) | 7 | 64% |
| Height (cm) | 8 | 61% |
| Platelets (cell/μl) | 9 | 58% |
| WBC (1000 cells/μl) | 10 | 55% |
| Hematocrit (%) | 11 | 49% |
| Albumin (g/dl) | 12 | 42% |
| Hypertension | 13 | 16% |
| Gender | 14 | 12% |
Abbreviations: Hemoglobin A1C (HbA1c), alanine transaminase (ALT), aspartate transaminase (AST), white blood cell count (WBC), body mass index (BMI).
Table 2.
NIDDK feature values
|
Mean Value ± Standard Deviation |
T-Test | ||
|---|---|---|---|
| Laboratory test | NASH (N = 453) | Non-NASH (N = 251) | P-value |
| HbA1C (%) | 6.3 ± 1.4 | 5.8 ± 1.1 | <.01 |
| AST (units/L) | 67.1 ± 44.3 | 44.9 ± 29.9 | <.01 |
| ALT (units/L) | 88.7 ± 60.2 | 57.3 ± 41.3 | <.01 |
| Total Protein (g/dl) | 7.4 ± 0.5 | 7.1 ± 0.6 | <.01 |
| AST/ALT | 0.8 ± 0.3 | 0.9 ± 0.5 | .13 |
| BMI (kg/m2) | 34.0 ± 5.4 | 33.4 ± 5.8 | .16 |
| Triglycerides (mg/dl) | 189 ± 112 | 155 ± 86 | <.01 |
| Height (cm) | 167 ± 9 | 169 ± 9 | .01 |
| Platelets (cell/µl) | 233 877 ± 65 296 | 239 905 ± 70 449 | .24 |
| WBC (1000 cells/µl) | 7.1 ± 1.8 | 6.7 ± 1.7 | .01 |
| Hematocrit (%) | 41.8 ± 3.6 | 42.0 ± 3.7 | .45 |
| Albumin (g/dl) | 4.3 ± 0.4 | 4.2 ± 0.4 | <.01 |
Abbreviations: Hemoglobin A1C (HbA1c), alanine transaminase (ALT), aspartate transaminase (AST), white blood cell count (WBC), body mass index (BMI).
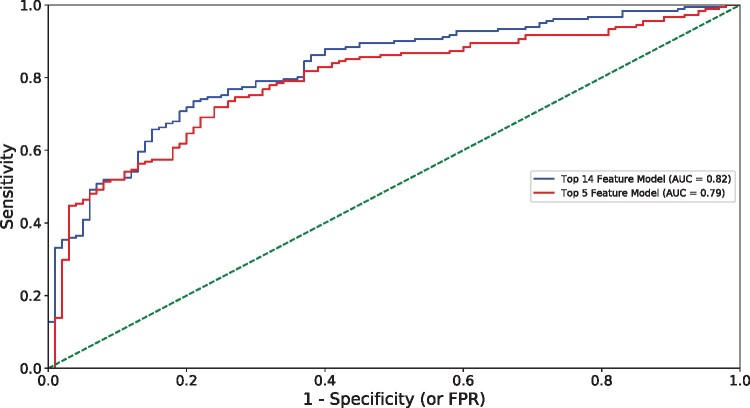
The performance of various ML models was evaluated on the NIDDK test dataset (Table 3). A 14-feature XGBoost model exhibited superior performance as determined by AUC (0.82) when compared to logistic regression (0.77) and CART (0.72) and comparable performance to random forest (0.82) with the same features. This XGBoost model exhibited 81% sensitivity and 81% precision to predict NASH. A larger 24-feature XGBoost model was the best performing model at 24 features and had equivalent AUC (0.82) but higher accuracy (78% vs. 75%), sensitivity (83% vs. 81%) and precision (83% vs. 81%) compared to the 14-feature XGBoost model. The slight reduction in performance at 14 features was considered acceptable, and the 14-feature XGBoost was selected as the preferred model. For this model, the Brier score was 0.19 and the area under the precision recall curve was 0.90. The final hyperparameters for this model are provided in Supplementary Table III. A reduced model was created with only five top performing predictive features (HbA1c, AST, ALT, total protein, and triglycerides). In the reduced model, XGBoost demonstrated slightly lower performance (AUC 0.79), sensitivity (80%), and precision (80%) for NASH prediction as compared to the 14-feature model (Table 3). Figure 1 compares the receiver operating characteristic (ROC) curve of the two XGBoost models. These two XGBoost models were further tested with 1,000 random repetitions of the training process on different train-test splits. The mean AUC of these was 0.82 indicating our results are not an outlier (standard deviation 0.02, median 0.82, 5th percentile 0.78, 95th percentile 0.85). For the 5-feature XGBoost model, the performance as measured by AUC was again consistent across repetitions (mean 0.80, standard deviation 0.02, median 0.80, 5th percentile 0.76, 95th percentile 0.83). These data indicate that the XGBoost models using either 14 features or five features exhibit high performance for NASH classification.
Table 3.
Model performance based on number of features (14 and 5 features) and method used (NIDDK test dataset)
| Performance | Logistic Regression | CART | Random Forest | XGBoost |
|---|---|---|---|---|
| 14-Feature Model | ||||
| AUC | 77% | 72% | 82% | 82% |
| Accuracy | 73% | 70% | 75% | 75% |
| Precision | 79% | 76% | 80% | 81% |
| Sensitivity | 79% | 78% | 82% | 81% |
| 5-Feature Model | ||||
| AUC | 75% | 73% | 78% | 79% |
| Accuracy | 73% | 69% | 70% | 74% |
| Precision | 78% | 77% | 77% | 80% |
| Sensitivity | 80% | 75% | 77% | 80% |
Abbreviations: Classification and regression trees (CART), area under the curve (AUC).
Figure 1.
Model performance in NASH prediction using NIDDK data. Area under the curve (AUC), false positive rate (FPR).
We examined the performance of our 14-feature model in T2DM (112) and non-T2DM (169) patients in the NIDDK test dataset. Among T2DM patients, 75% had NASH while 57% of the non-T2DM patients had NASH. The AUC was 0.79 in T2DM patients and 0.82 in non-T2DM patients. Sensitivity and precision were higher in T2DM (86% and 88% respectively) compared to non-T2DM (77% and 74%). While HbA1c is the most important feature in our model, NASHmap is able to predict NASH for both T2DM patients and non-T2DM patients.
The Optum dataset was analyzed to further evaluate model performance. Of the patients remaining following application of inclusion and exclusion criteria, ∼3 million had data for all 14 features in NASHmap and 22,946 of those had an ICD-10 code for NASH. Model validation was done with the classified NASH (1,016 patients with ICD-10 NASH and biopsy) and non-NASH patients (2,886,653 patients with no ICD-9 or 10 NASH/NAFLD code). Descriptive characteristics of Optum patients used for analyses are provided in Supplementary Table IV.
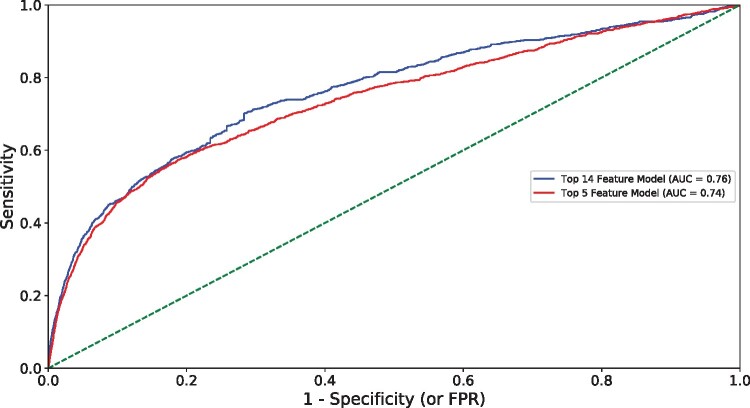
The full model exhibited good performance (AUC: 0.76), sensitivity (72%), and precision (80%) in NASH classification (Figure 2). The reduced 5-feature model demonstrated slightly lower performance (AUC: 0.74) and sensitivity (66%) when compared to the full model. Taken together, these data indicate that the proposed model, built on common clinical variables, exhibits good NASH classification and can maintain performance with diverse patient data.
Figure 2.
Model performance in NASH prediction using Optum data. Area under the curve (AUC), false positive rate (FPR).
DISCUSSION
Here, we report on the development of NASHmap an ML model for the prediction of NASH. The model was trained and tested in NASH and non-NASH patients as confirmed through liver biopsy and further validated in a real-world dataset (Optum EHR). Both versions of our model, with either the full 14 features or the reduced five features, achieved high performance, sensitivity, and precision to detect NASH in both the NIDDK and Optum datasets. This model represents a performant, non-invasive method for NASH screening, which could improve risk stratification measures and clinical management.
There are several key design choices and results that make NASHmap valuable. First, the model was developed and validated in two different databases. It was trained on the NIDDK NAFLD adult database with confirmed NASH status. While the dataset is not large by ML standards, it is one of the largest biopsy-confirmed datasets distinguishing NASH and non-NASH within the NAFLD disease spectrum. Biopsy-confirmed diagnoses ensured we trained the model on the best available input. Second, a broad set of features comprising demographic, laboratory and clinical variables were included and RFE was used to identify the optimal subset for NASH prediction. Similar to other variable selection approaches, RFE reduces the number of inputs required to apply the model while aiming to retain generalization performance. Unlike manual feature selection, RFE places the data first and clinical review second. As such, we relied less on existing clinical expectations in deriving the final list of features. Finally, the model exhibited high performance when tested on a holdout NIDDK dataset. As expected, model performance was slightly reduced when evaluated using the Optum dataset. A drop is inevitable as the Optum dataset contains heterogeneous patient data and the biases of today’s mis- or underdiagnosis of NASH in clinical practice. Nevertheless, the model performed well when applied to this real-world dataset.
The utilization of ML to predict disease allows for wider recognition, timely intervention, and targeted treatments to improve or mitigate disease progression. Other published studies have examined the utility of ML to predict or diagnose various forms of NAFLD.6,16–20 Perakakis et al. (2020) have compared a wide variety of models using different types of omics on a variety of NAFLD prediction problems. We compare a few such models and their differences (objectives, target cohort, methods, type of variables used, outcomes and applicability) to our study (Table 4).
Table 4.
Comparison of reported model performance
| Performance | NASHmap Model a | Cheng et al. 16 ,b | Atabaki-Pasdar et al. 17 ,c | Canbay et al. 18 | Fialoke et al. 19 | Perakakis et al. 20 ,d |
|---|---|---|---|---|---|---|
| Task | NASH vs. non-NASH | NAFLD vs. non-NAFLD | NAFLD vs. non-NAFLD | NASH vs. non-NASH | NASH vs. Healthy | NASH vs NAFL vs. Healthy |
| Cohort | NAFLD | Taiwanese high-tech workers | T2DM and non- T2DM at high risk | Obese with NAFLD | NAFLD and Healthy | Greek NAFLD and Healthy |
| Number of features | 14 | 8 | 9 | 5 | 23 | 29 |
| AUC | 82% | – | 82% | 70% | 88% | 95% |
| Accuracy | 75% | 87% | 74% | – | 80% | 88% |
| Precision | 81% | – | – | – | 81% | – |
| Sensitivity | 81% | 90% | 74% | – | 77% | 89% |
| Specificity | 66% | 81% | 73% | – | – | 94% |
| F1 Score | 81% | 70% | 79% |
Abbreviation: Type 2 Diabetes Mellitus (T2DM).
14-feature model on NIDDK data,
male data,
model 3 in IMI DIRECT,
29 lipid non-linear SVM OvR model with healthy >27.5 BMI.
Cheng et al. (2017) developed support vector machine (SVM) and random forest (RF) classifiers to identify presence of NAFLD in a Taiwanese high-tech industry worker cohort.16 Model performance was high with accuracy ranging from ∼80% in females (RF) to ∼87% in males (SVM). However, the models were built on a homogenous cohort that depended solely on ultrasound imaging for NALFD diagnosis. The models were not tested on a broader population. Atabaki-Pasdar et al. (2020) applied a least absolute shrinkage and selection operator (LASSO) model for feature selection and developed a series of random forest models with the objective of predicting if liver fat was <5% or ≥5%, consistent with non-NAFLD vs. NAFLD.17 Multi-omics and clinical variables were used as predictors while the target liver fat variable was quantified by magnetic resonance imaging (MRI). Multiple models were developed; one of the models (labeled model 3) achieved an AUC of 0.82 with nine clinically available features while a larger model with all omics and clinical features achieved an AUC of 0.84.
Canbay et al. (2019) utilized an ensemble method for feature identification (Ensemble Feature Selection21) and logistic regression for the classification of NAFL or NASH using an obese cohort with confirmed NAFL/NASH.18 The model, consisting of five features, exhibited moderate to good performance (AUC: 0.70) with an independent validation cohort. While the model was also trained on NAFL/NASH patients as confirmed through liver biopsy, the selection of obese patients for the model limits its application in other, non-obese populations.
Fialoke et al. (2018) developed a NASH classifier (NASH vs. healthy) using XGBoost in the Optum Integrated Claims-Clinical dataset (2007–2017). The model was trained and tested using ICD-9 and ICD-10 codes for NASH classification.19 The model's good performance (AUC: 0.88) could be attributed, in part, to the degree of expected separation between the two classes. NASH and healthy patients may be more differentiated than NASH and non-NASH NAFLD. Perakakis et al. (2020) note this is a complication in interpreting performance for models that include healthy patients.6 While Fialoke et al. applied their model to other NAFLD patients, they did not measure performance on that cohort given the limitations of diagnoses in Optum. The model also made use of longitudinal data; performance was significantly improved by inclusion of statistical summary features for AST, ALT, AST/ALT, and platelet data. The model highlights some of the advantages (easy availability of longitudinal data for features) as well as some of the limitations (bias in baseline diagnosis, difficulty defining the negative or non-NASH class) of training directly on real-world EHR data.
Perakakis et al. (2019) also developed an ML model for NASH. They included NASH, NAFL, and healthy patients, treating it as a 3-class problem and applying a one-vs-rest (OvR) approach.20 They collected serum samples from 31 NAFLD patients (16 NASH) and 49 healthy patients. These samples allowed them to measure 365 lipid species along with glycans and hormones. Their best performing models were support vector machines (SVMs) with 29 lipid features or 20 total features including lipids and glycans or hormones. This work supports the conclusion that NASH can be accurately identified using lipids and other serum markers. While the authors achieved very high performance, the large number of laboratory markers they use are not commonly captured in clinical care. This model would likely require additional, specific testing to be performed and could not be readily applied to existing EHR data. The authors note availability of this testing is currently limited and the cost of testing was $605 per patient in their study though it could be reduced in the future.
There are limitations to our approach. The ability of ML to predict NASH effectively is dependent on the quality of features within the model and the data used for training. The best set of features may also depend on the population under study as the incidence of NASH can vary across different ethnic groups.22 Our model could be improved by training on a larger dataset that includes other ethnic groups and broader, more diverse populations. Such a model could even reveal more about the pathophysiological processes associated with the development and progression of NASH. Additionally, while we tested the applicability of the model on Optum data, measurement of performance on that data is limited by the constraints of real-world data such as the inability to obtain confirmatory biopsies for non-NASH patients. Non-invasive ML models to predict NASH from clinical and lab data have yet to be prospectively validated in a large cohort.6 Finally, a further development of the model could be focused on predicting the specific NAFLD activity score (NAS) and fibrosis stage. However, the data requirements for training such a model are substantial. While ML and more specifically NASHmap will not be able to overcome the inherent limitations that liver biopsy has with regards to staging of NASH,23 it holds the promise to support physicians and other health care providers even outside of specialist practices.
CONCLUSION
The NASHmap model with 14 features is a robust model with 72-81% sensitivity at predicting NASH in patients from large, real-world datasets. Given NASH is perceived as a silent and greatly underdiagnosed disease, this model could be utilized as an initial screening tool to select patients with potential NASH for further confirmatory diagnostic steps and clinical management. Because it uses commonly available features, it could be automated through EHR systems and integrated into physicians’ workflows leveraging laboratory tests already being performed.
FUNDING
This study was funded by Novartis Pharma AG.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
AUTHOR CONTRIBUTIONS
All authors designed the study. M.D., S.A.R., G.C., and M-M.B. executed and refined the analysis. M.D., S.A.R., G.C., M-M.B., Q.Y., A.T., J.L., J.C., M.C.P., and J.M.S. interpreted results and developed the manuscript. All authors approved the final version of the manuscript.
DATA AVAILABILITY
No new data were collected in support of this research. The data underlying this article were provided by NIDDK and Optum, respectively, under license. Interested parties should reach out to the data owners to license this data: https://repository.niddk.nih.gov/studies/nafld_adult/ and https://www.optum.com/business.html.
CONFLICT OF INTEREST STATEMENT
J.M.S. reports consultancy: BMS, Boehringer Ingelheim, Echosens, Galmed, Genfit, Gilead Sciences, Intercept Pharmaceuticals, Madrigal, Nordic Bioscience, Novartis, Pfizer, Roche, Sanofi, and Siemens Healthineers. J.M.S. reports research funding: Gilead Sciences. J.M.S. is a speaker for Falk Foundation and MSD. M.D. and Q.Y. are employees of ZS Associates. S.A.R., G.C., M-M.B., N.J., A.T., J.L., and M.C.P. are employees and own stocks of Novartis Pharma AG. J.C. is an employee and owns stocks of Novartis Pharmaceuticals Corp.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Joanna Huang for her contributions to the study while at ZS, and Kirk Evanson and Superior Medical Experts for research and drafting assistance.
References
- 1. Younossi ZM, Marchesini G, Pinto-Cortez H, Petta S.. Epidemiology of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis: implications for liver transplantation. Transplantation 2019; 103 (1): 22–7. [DOI] [PubMed] [Google Scholar]
- 2. Suzuki A, Diehl AM.. Nonalcoholic steatohepatitis. Annu Rev Med 2017; 68 (1): 85–98. [DOI] [PubMed] [Google Scholar]
- 3. Brunt EM, Wong VW, Nobili V, et al. Nonalcoholic fatty liver disease. Nat Rev Dis Primers 2015; 1 (1): 15080. [DOI] [PubMed] [Google Scholar]
- 4. Chalasani N, Younossi Z, Lavine JE, et al. The diagnosis and management of nonalcoholic fatty liver disease: practice guidance from the American Association for the Study of Liver Diseases. Hepatology 2018; 67 (1): 328–57. [DOI] [PubMed] [Google Scholar]
- 5. Rockey DC, Caldwell SH, Goodman ZD, Nelson RC, Smith AD.. American Association for the Study of Liver D. Liver biopsy. Hepatology 2009; 49 (3): 1017–44. [DOI] [PubMed] [Google Scholar]
- 6. Perakakis N, Stefanakis K, Mantzoros CS.. The role of omics in the pathophysiology, diagnosis and treatment of non-alcoholic fatty liver disease. Metabolism 2020; 111: 154320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kononenko I. Machine learning for medical diagnosis: history, state of the art and perspective. Artif Intell Med 2001; 23 (1): 89–109. [DOI] [PubMed] [Google Scholar]
- 8. Beam AL, Kohane IS.. Big data and machine learning in health care. JAMA 2018; 319 (13): 1317–8. [DOI] [PubMed] [Google Scholar]
- 9. Lazarus JV, Colombo M, Cortez-Pinto H, et al. NAFLD - sounding the alarm on a silent epidemic. Nat Rev Gastroenterol Hepatol 2020; 17 (7): 377–9. [DOI] [PubMed] [Google Scholar]
- 10. Beretta L, Santaniello A.. Nearest neighbor imputation algorithms: a critical evaluation. BMC Med Inform Decis Mak 2016; 16 (S3): 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Breiman L. Random forests. Mach Learn 2001; 45 (1): 5–32. [Google Scholar]
- 12. Chen T, Guestrin C. XGBoost: A scalable tree boosting system. In: KDD '16: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining2016: 10.
- 13. Kotsiantis SB. Supervised machine learning: a review of classification techniques. In: Proceedings of the 2007 conference on Emerging Artificial Intelligence Applications in Computer Engineering: Real World AI Systems with Applications in eHealth, HCI, Information Retrieval and Pervasive Technologies2007: 22.
- 14. Safavian SR, Landgrebe DA, Landgrebe D. United States. National Aeronautics and Space Administration. A Survey of Decision Tree Classifier Methodology. West Lafayatte, IN; Washington DC, Springfield, VA: School of Electrical Engineering, National Aeronautics and Space Administration; National Technical Information Service, Distributor; 1990.
- 15. Svetnik V, Liaw A, Tong C, Culberson JC, Sheridan RP, Feuston BP.. Random forest: a classification and regression tool for compound classification and QSAR modeling. J Chem Inf Comput Sci 2003; 43 (6): 1947–58. [DOI] [PubMed] [Google Scholar]
- 16. Cheng Y, Chou C, Hsiung Y.. Application of machine learning methods to predict non-alcohol fatty liver disease in Taiwanese high-tech industry workers. In: International Conference on Data Mining 2017: 118–23. [Google Scholar]
- 17. Atabaki-Pasdar N, Ohlsson M, Viñuela A, et al. Predicting and elucidating the etiology of fatty liver disease: a machine learning modeling and validation study in the IMI DIRECT cohorts. PLoS Med 2020; 17 (6): e1003149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Canbay A, Kälsch J, Neumann U, et al. Non-invasive assessment of NAFLD as systemic disease-A machine learning perspective. PLoS One 2019; 14 (3): e0214436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fialoke S, Malarstig A, Miller MR, Dumitriu A.. Application of machine learning methods to predict Non-Alcoholic Steatohepatitis (NASH) in Non-Alcoholic Fatty Liver (NAFL) patients. AMIA Annu Symp Proc 2018; 2018: 430–9. [PMC free article] [PubMed] [Google Scholar]
- 20. Perakakis N, Polyzos SA, Yazdani A, et al. Non-invasive diagnosis of non-alcoholic steatohepatitis and fibrosis with the use of omics and supervised learning: a proof of concept study. Metabolism 2019; 101: 154005. [DOI] [PubMed] [Google Scholar]
- 21. Neumann U, Genze N, Heider D.. EFS: an ensemble feature selection tool implemented as R-package and web-application. BioData Min 2017; 10: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Danford CJ, Yao ZM, Jiang ZG.. Non-alcoholic fatty liver disease: a narrative review of genetics. J Biomed Res 2018; 32 (5): 389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schattenberg JM, Straub BK.. On the value and limitations of liver histology in assessing non-alcoholic steatohepatitis. J Hepatol 2020; 73 (6): 1592–3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No new data were collected in support of this research. The data underlying this article were provided by NIDDK and Optum, respectively, under license. Interested parties should reach out to the data owners to license this data: https://repository.niddk.nih.gov/studies/nafld_adult/ and https://www.optum.com/business.html.