Graphical abstract
Keywords: SARS-CoV-2, Responsive polymers, Misdiagnosis, Enrichment, Sensitivity
Abstract
The current commercially available SARS-CoV-2 diagnostic approaches including nucleic acid molecular assaying using polymerase chain reaction (PCR) have many limitations and drawbacks. SARS-CoV-2 diagnostic strategies were reported to have a high false-negative rate and low sensitivity due to low viral antibodies or antigenic load in the specimens, that is why even PCR test is recommended to be repeated to overcome this problem. Thus, in anticipation of COVID-19 current wave and the upcoming waves, we should have an accurate and rapid diagnostic tool to control this pandemic. In this study, we developed a novel preanalytical strategy to be used for SARS-CoV-2 specimen enrichment to avoid misdiagnosis. This method depends on the immuno-affinity trapping of the viral target followed by in situ thermal precipitation and enrichment. We designed, synthesized, and characterized a thermal-responsive polymer poly (N-isopropylacrylamide-co-2-hydroxyisopropylacrylamide-co-strained alkyne isopropylacrylamide) followed by decoration with SARS-CoV-2 antibody. Different investigations approved the successful synthesis of the polymeric antibody conjugate. This conjugate was shown to enrich recombinant SARS-CoV-2 nucleocapsid protein samples to about 6 folds. This developed system succeeded in avoiding the misdiagnosis of low viral load specimens using the lateral flow immunoassay test. The strength of this work is that, to the best of our knowledge, this report may be the first to functionalize SARS-CoV-2 antibody to a thermo-responsive polymer for increasing its screening sensitivity during the current pandemic.
1. Introduction
In anticipation of SARS-CoV-2 current wave and the upcoming waves, we should have an accurate and rapid diagnostic tool that will be critical in controlling the pandemic, which is expected to cause severe attacks, especially by changing whether temperature [1]. The current coronavirus disease 2019 (COVID-19) commercially available diagnostic approaches include two categories, the first one depends on viral RNA molecular assaying using polymerase chain reaction (PCR) based techniques and the second is the serological and immunological assays that detect antibodies or antigenic proteins in patient's specimens after viral exposure, as discussed recently by our group [2].
Unfortunately, most of the currently available diagnostic approaches have many limitations and drawbacks, even SARS-CoV-2 nucleic acid molecular assaying using polymerase chain reaction (PCR) based techniques that are considered as the gold standard methods for assaying the virus was reported to have a high false-negative rate and low sensitivity [3], [4]. The current SARS-CoV-2 RT-PCR positive rate of detection is only 30%-50% and the viral RNA detectability is very limited between days 15 to 39 after the onset of infection [5], [6].
Different factors contribute to SARS-CoV-2 RT-PCR false-negative results, but the sampling techniques and sample type are the common factors where nasopharyngeal and oropharyngeal (NP/OP) specimens are the common samples used in this assay despite the fluctuation in viral load according to the stage of infection where SARS-CoV-2 colonize in the lower respiratory tract [7], [8], [9], [10]. Most COVID-19 patients cannot be diagnosed before premorbid because of the low viral load in the specimens [11], [12], [9], [13].
SARS-CoV-2 viral load change is similar to influenza, but completely different from the change in SARS and MERS, comparable to chest CT scan, SARS-CoV-2 viral load peaked approximately at day 10 after the onset of infection [14], [15], [16]. Therefore, the low number of viral copies in the specimen increases the rate of false-negative PCR assay and virus misdiagnosis, that is why even the PCR test is recommended to be repeated several times to overcome this problem.
On the other hand, the presence of SARS-CoV-2 antibodies and antigenic proteins in plasma was reported to be <40% among the individuals after the first week of infection onset [17]. Viral antibody immune response may not be enhanced in some patients until the 4th week after the onset of infection, especially in asymptomatic patients, and that means SARS-CoV-2 negative results should not affect patient management and infection control decisions especially, these misdiagnosed peoples are active carriers of the virus and failure to quarantine them would be a great setback in SARS-CoV-2 transmission [18], [19], [20], [21].
Thus, it is now clear that our great challenge regarding the current pandemic diagnosis is how to avoid the false-negative diagnosis that may be caused by low viral antibodies or antigenic loads in the analyzed specimens regardless of the variations of the currently available diagnosis techniques, specimen type, sampling technique, and infection stage. In other words, we are in big need of a novel preanalytical strategy to be used for specimen enrichment to avoid misdiagnosis.
Immuno-affinity trapping strategies that mainly depend on antigen-antibody interaction have been confirmed as effective methods in concentrating trace biomarkers in various body fluids and biological samples [22]. Stimuli-responsive polymers decorated with specific antibodies have been used for the immune perception of different biomolecules in different biomedical applications, especially immunoassaying, and others depending on the reversible aggregation after induction of temperature, pH, light, etc. [23], [24], [25]. Our group possesses many achievements in this trend [26], [27], [28], [29], [30], [31].
In this study, we designed, synthesized, and characterized a temperature-responsive polymer, poly(N-isopropylacrylamide-co-2-hydroxyisopropylacrylamide-co-strained alkyne isopropylacrylamide) P(NIPAAm-co-HIPAAm-co-SAKIPAAm) followed by decoration using anti-SARS-CoV-2 nucleocapsid antibody for enabling affinity enrichment of recombinant SARS-CoV-2 nucleocapsid protein through in situ precipitation process as an initial model to be used as a pre-analytical technique to increase targets concentrations in SARS-CoV-2 assays to overcome misdiagnosis and improving its diagnostic sensitivity as shown in Fig. 1.
Fig. 1.
We developed a portable, fast, easy, and sensitive strategy based on the concepts of immune-affinity and responsive smart polymeric materials to be used as a preanalytical step for SARS-CoV-2 specimens enrichment to avoid the misdiagnosis that usually happens during SARS-CoV-2 diagnosis due to the low viral load in the tested samples.
2. Materials and methods
2.1. Materials
N-isopropylacrylamide (NIPAAm, Wako Pure Chemical, 97%) was recrystallized from hexane and dried under vacuum before use. chloroform (Wako Pure Chemical, 99.0%), D,L-2-amino-1-propanol (Tokyo Kasei, 98.0%), triethylamine (Wako Pure Chemical), acryloyl chloride (Tokyo Kasei, 95.0%), 2-propanol (Wako Pure Chemical, 99.7%), ethyl acetate (Kishida chemical, 99.5%), cyanomethyl dodecyl trithiocarbonate (CDT,Aldrich), 2,2′-azobis(isobutyronitrile) (AIBN, Wako Pure Chemical), tetrahydrofuran (THF, Kanto chemical, 99.0%,), diethyl either (Wako Pure Chemical, 99.5%), deuterium oxide containing 0.05 v/v% sodium 3-(trimethylsilyl)-1-propanesulfonate-d6 (TMSP) (D2O, Wako Pure Chemical, 99.9%), dimethyl sulfoxide-d6 containing 0.05 v/v% tetramethylsilane (TMS) (DMSO‑d6, Wako Pure Chemical, 99.9%), N,N-dimethylformamide (DMF, Wako Pure Chemical, 99.7%), dibenzylcyclooctynic acid (DBCO acid, Click Chemistry Tools, 95.0%), dichloromethane (DCM, Wako Pure Chemical, 99.0%), goat antibody to mouse IgG used for experimental trials (1.96 mg/mL, abcum), SARS-CoV-2 nucleocapsid humanized antibodies, >95% (MBS355887, Clone #5B, MyBioSource, Inc, San Diego, USA), Recombinant SARS-CoV-2 nucleocapsid Protein, >95% (MBS355892, MyBioSource, Inc, San Diego, USA), pierce™ modified lowry protein assay kit (thermo fisher scientific),4-dimethylaminopyridine (DMAP, Wako Pure Chemical), N′,N′-dicyclohexylcarbodiimide (DCC, Tokyo Kasei, 98.0%), ethanol (Wako Pure Chemical, 99.5%), Dulbecco’s phosphate buffered saline (PBS, Aldrich), azido-ethylene glycol (EG4)-NHS ester (Tokyo Kasei), dimethyl sulfoxide (DMSO, Wako Pure Chemical, 99.0%), copper(Ⅱ) sulfate, 5-hydrate (Aldrich), DL-2-aminobutyric acid (Tokyo Kasei, 99.0%), fluorescamine (Tokyo Kasei)、methanol (99.8%、Wako Pure Chemical), 10 × tris/glycine/SDS buffer (BIO-RAD), Coomassie brilliant blue R-250 (CBB, BIO-RAD), Laemmli sample buffer (BIO-RAD), 2-mercaptoethanol (Wako Pure Chemical, 99%), Precision plus protein unstained standards (BIO-RAD).
2.2. Synthesis and characterization of HIPAAm monomers
HIPAAm was synthesized according to the previous report [32], where d,l-2-amino-1-propanol (0.15 mol), and triethylamine (0.15 mol) were dissolved in anhydrous chloroform solvent followed by stirring at-5 °C for 20 min then acryloyl chloride (0.15 mol) was added to the prepared solution very slowly, followed by stirring at-5 °C for 2 h. The obtained solvent was evaporated using the Rota evaporator followed by re-dissolving in 2-propanol and keeping at −20 °C for more than 24 h. Finally, filtration was performed for salts removal followed by concentration and purification using column chromatography. Synthesis of HIPAAm monomer was confirmed using thin-layer chromatography (TLC) followed by 1H NMR (Solvent D2O).
2.3. Synthesis of P(NIPAAm-co-HIPAAm)
P(NIPAAm-co-HIPAAm) was synthesized by Reversible Addition-Fragmentation Chain Transfer Polymerization (RAFT polymerization) of HIPAAm hydroxyl derivative with NIPAAm (temperature-responsive polymer) as shown in Scheme 1(I). Reaction conditions were as the following (NIPAAm 1.89 g, HIPAAm 0.11 g, AIBN 1.31 mg, CDT 12.7 mg, ethanol 17.6 ml) followed by stirring at 20 °C for 20 h, evaporation, and vacuum drying.
Scheme 1.
(I) Synthesis of P(NIPAAm-co-HIPAAm). (II) Synthesis of P(NIPAAm-co-HIPAAm-co-SAKIPAAm). (III) Synthesis of azido-Anti- SARS-CoV-2 Antibody. (IV) Conjugation of azido-Anti- SARS-CoV-2 Antibody with the temperature responsive P(NIPAAm-co-HIPAAm-co-SAKIPAAm) using click chemistry.
2.4. Synthesis of P(NIPAAm-co-HIPAAm-co-SAKIPAAm)
Dibenzylcyclooctynic (DBCO) acid that contains an alkyne group was introduced into HIPAAm hydroxyl group by dehydration, condensation to obtain our clickable responsive polymer P(NIPAAm-co-HIPAAm-co-SAKIPAAm) as shown in Scheme 1(II). Reaction conditions were as the following (DCM 30 ml, DBCO acid 35.8 mg, DMAP 12 mg, P(NIPAAm-co-HIPAAm) 100 mg, DCCD 20 mg) followed by stirring overnight, evaporation, and vacuum drying.
2.5. Characterization of the polymeric synthesis
Both P(NIPAAm-co-HIPAAm) & P(NIPAAm-co-HIPAAm-co-SAKIPAAm) were confirmed by 1H NMR (Solvent:D2O, DMSO), Gel permeation chromatography (GPC) (Solvent:DMF, Standard: PS), Lower critical solution temperature (LCST) (Solvent: PBS, pH: 7.4, conc.: 2.0 mg/mL, temp.: 10–40 °C).
2.6. Introduction of azido group to the antibody
Azido-PEG4-NHS ester powder was dissolved in DMSO (10 mg/ml) and different concentrations of Azido-PEG4-NHS ester were prepared using carbonate buffer (pH 8.6). Azido-PEG4-NHS ester was added to the antibody with different feeding ratios followed by rotation at 4 °C for 5 h as shown in Scheme 1(III).
2.7. Evaluation of azido group introduction to the antibody
After conjugation of the azido group to the antibody, we measured the fluorescence of the antibody samples using Infinite® 200 PRO plate reader as following: Azido modified antibody samples were diluted to 0.5 mg/ml and then applied to a 96 microwell plate beside different concentrations of d,l-2-Aminobutyric acid that were used as standards (50 µL/well then 5 µL of fluorescamine (50 mg/ml) was added to each well followed by incubation in dark for 15 min and fluorescence investigation (395 nm/495 nm). All samples were applied as triplicates.
2.8. Preparation of SARS-CoV-2 antibody-temperature-responsive polymer conjugate via click reaction
The azido-modified antibody was conjugated to the alkyne group modified temperature-responsive polymer using click chemistry as following: 75 µL of P(NIPAAm-co-HIPAAm-co-SAKIPAAm) was mixed with 50 µL of azido-modified antibody (2.5 mg/mL in PBS) and stirred at 4 °C for 12 h as shown in Scheme 1(IV). The polymer concentration was adjusted for the following antibody: polymer ratios – 1:1, 1:2, 1:4, 1:8, 1:15, and 1:30. The succeeded conjugates were characterized by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) compared with the antibody and polymer.
2.9. Evaluation of SARS-CoV-2 antibody-temperature-responsive polymer enrichment efficacy
The previously prepared SARS-CoV-2 antibody-polymer conjugate (1:1) ratio enrichment efficacy with free polymer was evaluated by thermal precipitation. Purified SARS-CoV-2 -polymer conjugates (1.3 mg/mL, 100 µL in PBS with 15 equivalents of free polymer) were added to microtubes and centrifuged (13000×g) for 5 min at 37 °C. Then the supernatant (90 µL) and precipitate (10 µL) were collected, and the absorbance at 280 nm was measured by UV–vis. The molar ratios of SARS-CoV-2 antibodies in the liquid phase and the solid phase were estimated, and the enrichment ratios were calculated.
2.10. Evaluation of SARS-CoV-2 antibody-temperature-responsive polymer antigenic enrichment efficacy
Recombinant SARS-CoV-2 nucleocapsid protein enrichment capacity was evaluated against different concentrations of purified SARS-CoV-2 antibody polymer conjugates (1.3, 0.7, 0.35, 0.17, 0.08, 0 mg/mL, 50 µL in PBS) with 15 equivalent of the free polymer, where antibody-polymer conjugate (different concentrations, 50 µL in PBS) were mixed with the SARS-CoV-2 recombinant protein (1.0 mg/mL, 50 µL in PBS), and incubated for 1 h followed by addition of the free polymer to the solution and centrifugation in microtubes at 37 °C, 13000×g for 5 min. The supernatant (90 µL) and precipitate (10 µL) were collected, and the SARS-CoV-2 recombinant protein in the supernatant was measured using the modified Lowery protein assay kit according to the manufacturer instructions and finally, the enrichment ratios were calculated.
2.11. Evaluation of SARS-CoV-2 antibody-temperature-responsive polymer antigenic enrichment efficacy using lateral flow immunoassay strip
Different concentrations of purified SARS-CoV-2 recombinant protein (1.04 × 10−15–20.83 × 10−15 mol/mL, 20 µL in PBS) was tested using lateral flow immunoassay strips (that were obtained from Prof. Gamal Shiha, Egyptian Liver Research Institute and Hospital, Mansoura, Egypt) for the determination of the detection limit of SARS-CoV-2 lateral flow-immunoassay strip (LFIA) then the samples that were detected negative using LFIA were retested after using our devolved system with free polymers as following: purified SARS-CoV-2 recombinant protein (1.04 × 10−15 and 2.08 × 10−15 mol/ml,100 µL in PBS were mixed with the purified SARS-CoV-2 antibody-polymer conjugate, and incubated for 1 h followed by addition of 15 equivalent of the free polymer and centrifugation in microtubes at 37 °C, 13000×g for 5 min. The supernatant (180 µL) and precipitate (20 µL) were collected, the enriched part was assayed using LFIA.
2.12. Validation of the designed material strategy and polymeric conjugation in real settings
P(NIPAAm-co-HIPAAm-co-SAKIPAAm) and azido modified antibodies were mixed and allowed to react according to previous conditions using different real biological samples compared with PBS to evaluate the limitations that may be happened due to sample complexity and heterogenicity. PBS, nasopharyngeal samples, oropharyngeal samples, and mid-stream urine were used as the solvents. Nasopharyngeal, oropharyngeal, and urine samples were obtained from a healthy volunteer, samples were collected in sterile containers and used freshly in our experiments. Handling of these samples was performed according to the International Ethical Guidelines for Biomedical Research Involving Human Subjects (CIOMS/WHO, 1993). Finally, conjugates conducted in different samples were characterized using SDS-PAGE compared to the antibody and polymer.
2.13. Statistical analysis
Data were analyzed using the Statistical Package of Social Science (SPSS) program for Windows (Standard version 21). The descriptive statistics were presented as mean ± SD (standard deviation) for parametric data. ANOVA test was used to compare more than 2 means, and paired t-test was used to compare paired data. The threshold of significance was fixed at the 5% level (P-value). Results were considered significant when the probability of error was<5% (P < 0.05).
3. Results
3.1. Confirmation of HIPAAm synthesis
The purification of HIPAAm monomer was confirmed by the TLC analysis Supplementary Information A (Fig. S1) that showed that HIPAAm was successfully prepared with two main secondary products. Column chromatography was used for HIPAAm purification as shown also in Fig. S1, and then finally, confirmation of HIPAAm synthesis was done using 1H NMR results (Fig. S2).
3.2. Confirmation and characterization of P(NIPAAm-co-HIPAAm) & P (NIPAAm-co-HIPAAm-co-SAKIPAAm)
1H NMR analysis showed and confirmed the successful polymerization of HIPAAm with NIPAAm with the ratio 3.6: 96.4, where PNIPAAm LCST that is well known to be around 32 °C was increased after introducing HIPAAm hydroxy group to be 37.4 °C and the successful conjugation of the strained alkyne, SAK group to form our temperature-responsive polymer P(NIPAAm-co-HIPAAm-co-SAKIPAAm) with ratio 96.4:1.2:2.4. Moreover, regarding the molecular weight, GPC analysis showed that the molecular weight of the synthesized polymer was shifted from 1.904 × 104 to 2.014 × 104 (g/mol) due to the insertion of the strained alkyne, SAK group and the LCST was shifted from 37.4 to 30.1° C as shown in Figs. S3–S5, Fig. 2 & Table 1.
Fig. 2.
Lower critical solution temperature (LCST) (Thermal-response) of the synthesized P(NIPAAm-co-HIPAAm) (A), P(NIPAAm-co-HIPAAm-co-SAKIPAAm) (B) and Polymer antibody conjugate (c). (Solvent: PBS (pH = 7.4), Polymer conc.: 2.0 mg/mL, Heating rate: 0.2 °C/min, Wavelength: 450 nm).
Table 1.
Characterization of the synthesized polymers.
| Polymer | Structure according to 1H NMR (mol/mol/mol) |
LCST °C | Molecular weight (g/mol) using GPC | ||
|---|---|---|---|---|---|
| NIPAAm | HIPAAm | SAKIPAAm | |||
| P(NIPAAm-HIPAAm) | 96.4 | 3.6 | 0 | 37.4 | 1.904 × 104 |
| P(NIPAAm-co-HIPAAm-coSAKIPAAm) | 96.4 | 1.2 | 2.4 | 30.1 | 2.014 × 104 |
H NMR (Solvent: D2O, DMSO‑d6), GPC (Solvent:DMF, Standard: PSt), LCST was measured by Spectrophotometer (Solvent: PBS (pH = 7.5), Polymer conc.: 2.0 mg/mL, Heating rate: 0.2 °C/min, Wavelength: 450 nm).
3.3. Synthesis of azido-Anti- SARS-CoV-2 antibody
Introduction of Azido-groups to Anti- SARS-CoV-2 antibody was performed by binding of azido-(EG)4-NHS to the antibody lysine residues. Azido-(EG)4-NHS confirmation and quantification were done using the fluorescamine reduction method where more conjugation results in fewer available lysine residues and amine groups and vice versa. Increasing the azido-(EG)4-NHS feeding resulted in the gradual increase of azido group conjugation as follows (0, 6.7, 10.7, 20.9) and gradual decreasing of amine group conjugation as follows (21.1, 14.7, 10.7, 0.2) p-value < 0.001 as shown in (Fig. 3).
Fig. 3.
Introduction of azido-(EG)4-NHS per SARS-CoV-2 antibody measured by fluorescamine. (mean ± SD, n = 3).
3.4. Confirmation of the synthesized Anti- SARS-CoV-2 polymer conjugate prepared via click reaction
Sodium dodecyl sulfate gel electrophoresis (SDS-PAGE) gel images of different concentrations of polymer with a constant concentration of anti-SARS-CoV-2 antibody. Different anti-SARS-CoV-2 antibody: polymer ratios – 1:1, 1:2, 1:4, 1:8, 1:15, and 1:30 (lanes 2 to 7) were investigated as shown in Fig. 4, increasing the amount of polymer in the reaction solution caused more conjugation to the antibody and finally, more band broadening and increasing in the conjugate molecular weight. Compared to lane 8 that showed no band where it contained only free polymer and (lane 9:10) that showed the denatured light and heavy chains of both free SARS-CoV-2 antibody and azido SARS-CoV-2 antibody. These results indicate successful anti-SARS-CoV-2 conjugated polymer synthesis.
Fig. 4.
SDS-PAGE for evaluation of Click reaction between the antibody and different concentrations of the synthesized polymer (1) Protein ladder (2) Antibody: polymer = 1:1 (mol/mol) (3) 1:2 (4) 1:4 (5) 1:8 (6) 1:15 (7) 1:30 (8) free polymer (9) Azido- Antibody (10) Antibody (11) protein ladder.
3.5. SARS-CoV-2 antibody-temperature-responsive polymer enrichment efficacy evaluation
SARS-CoV-2 antibody-temperature-responsive polymer supported with free polymer showed a highly significant enrichment capacity after applying thermal precipitation compared to the control one without applying thermal precipitation, p-value < 0.001 as shown in Fig. 5 where no. of enrichment folds was increased to reach around 71-fold after thermal induction.
Fig. 5.
Conjugate enrichment folds before and after thermal induction. (mean ± SD, n = 3).
3.6. Antigen enrichment using SARS-CoV-2 antibody-temperature-responsive polymer
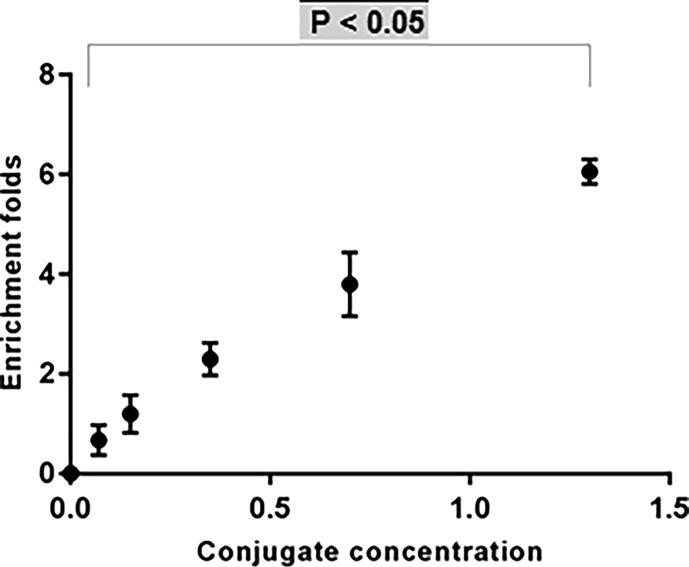
As shown in Fig. 6, by increasing the concentration of the purified SARS-CoV-2 antibody polymer conjugate, the number of recombinant SARS-CoV-2 nucleocapsid protein folds were shifted from 0 to reach to about 6 folds of the original antigenic concentration with the highest conjugate concentration (1.3 mg/mL) p-value < 0.05 that indicated the capability of our developed system to enrich low viral load samples.
Fig.6.
Recombinant SARS-CoV-2 nucleocapsid protein enrichment folds with different concentrations of antibody conjugates. (mean ± SD, n = 3).
3.7. Evaluation of antigen enrichment using lateral flow immunoassay strip
As shown in Fig. 7A, the minimum detection limit of recombinant SARS-CoV-2 nucleocapsid protein (48 kDa) using LFIA was around 1.08 × 10−15 mol/mL (a5) and 2.08 × 10−15 mol/mL (a4) showed a weak faint band at the test zone while concentrations 4.17 × 10−15 to 20.83 × 10−15 mol/mL showed strong positive bands as shown in Fig. 7a1, a2, a3 respectively. However, both false-negative concentrations showed strong positive bands after enrichment using SARS-CoV-2 antibody polymer conjugate as shown in Fig. 7B
Fig. 7.
Lateral flow immune assay (LFIA) evaluation of SARS-CoV-2 antibody-temperature-responsive polymer enrichment efficacy, Fig. A Different concentrations of recombinant SARS-CoV-2 nucleocapsid protein was tested using lateral flow immunoassay, a1: 20.83 × 10−15, a2: 10.42 × 10−15, a3 :4.17 × 10−15, a4: 2.08 × 10−15and a5: 1.04 × 10−15 mol/mL. Fig. B Comparing samples before and after enrichment using SARS-CoV-2 antibody-temperature-responsive polymer, b1: 1.04 × 10−15 mol/mL without enrichment, b2: 1.04 × 10−15 mol/mL after enrichment, b3: 2.08 × 10−15 mol/mL without enrichment, b4: 2.08 × 10−15 mol/mL after enrichment.
3.8. The designed material strategy can be proceeded regardless of body fluid heterogenicity
As shown in Fig. 8, Sodium dodecyl sulfate gel electrophoresis (SDS-PAGE) gel image for the designed polymer conjugation in different body fluids including nasopharyngeal, oropharyngeal, and urine samples compared to PBS (lanes 5–12) and compared with antibody alone, polymer conjugates light and heavy chains were shifted to higher molecular weights, with a marked band broadening and smeared migration patterns in the loading well. Compared to lane 4 that showed no band where it contained only free polymer and (lane 2:3) that showed the denatured light and heavy chains of both free antibody and azido antibody. These results indicate successful polymeric conjugation in different body fluids including the upper respiratory tract samples that are commonly used for SARS-CoV-2 detection regardless of the heterogenicity and complexity of these samples.
Fig. 8.
SDS-PAGE for evaluation of designed material strategy and polymeric conjugation in real biological samples (1) Protein ladder (2) Antibody (3) Azido- Antibody (4) only polymer (5) PBS (6) polymer conjugate in PBS (7) Nasopharyngeal sample (8) polymer conjugate in Nasopharyngeal sample (9) Oropharyngeal sample (10) Polymer conjugate in Oropharyngeal sample (11) Urine sample (12) Polymer conjugate in urine sample (13) protein ladder.
4. Discussion
Most of SARS-CoV-2 commercially available diagnostic methods including PCR, ELISA, and LFIA based techniques have many limitations, especially high false-negative rates and poor sensitivity [2], [3], [4]. Low viral load in the tested specimens is the main cause of this misdiagnosis [8], [9]. Especially, nasopharyngeal and oropharyngeal (NP/OP) specimens are the common samples used for SARS-CoV-2 detection despite their problems [10]. Our findings were in agreement with these previous reports Supplementary Information(B)
On the other hand, the special nature of the current pandemic affects this situation where SARS-CoV-2 viral load peaked approximately at day 10 after the onset of infection [14], [15], [16]. SARS-CoV-2 antibodies and antigenic proteins in blood plasma were reported to be<40% among individuals after the first week of infection onset [17]. Viral antibody immune response may not be enhanced in some patients until the 4th week after the onset of infection, especially in asymptomatic patients [18], [19], [20], [21]. Thus, our great challenge during this third wave and the upcoming waves is how to reduce the misdiagnosis rates of COVID-19 patients to decrease viral transmission.
Poly NIPAAm (PNIPAAm), is a temperature-responsive polymer that was selected in our study for its water solubility below its LCST of 32 °C where the PNIPAAm hydrophilic amide conjugates with water molecules by hydrogen-bonding and water molecules surrounding the hydrophobic isopropyl group performs hydrophobic hydration by hydrogen bonds causing the complete water solubility [33]. On the other hand, it aggregates above the LCST 32 °C where the hydrophobic hydration turns to an unstable state by increasing the temperature above 32 °C, causing PNIPAAm dehydration and aggregation due to hydrophobic interaction [34].
Therefore, depending on the previous, we can easily control the solubility state of PNIPAAm in water by thermal induction. Moreover, decoration of PNIPAAm with specific antibodies can enable immuno-affinity trapping and finally enrichment of different antigenic targets by applying thermal induction.
In our study, we used click chemistry for enabling polymer and SARS-CoV-2 antibody conjugation. Therefore, we labelled anti-SARS-CoV-2 nucleocapsid antibody with azido groups and conjugated the thermo-responsive polymer with a cyclic strained alkyne group (SAK group) for enabling the Huisgen cycloaddition click reaction [35] between the antibody azido group and SAK group. Click reaction forms a triazole ring by a 1,3-dipolar cycloaddition of the azido group and the alkyne group [36]. This reaction is highly selective and specific and can proceed regardless of the surrounding reaction environment including impurities, temperature, and reaction solution [37]. These properties explain why we preferred the click chemistry for the polymeric antibody conjugation.
In this study, P(NIPAAm-co-HIPAAm) was prepared using RAFT polymerization to enable the introduction of the strained alkyne group into NIPAAm. The synthesis was confirmed using 1H NMR, also, LCST measurements confirmed the polymeric synthesis where the PNIPAAm LCST that is well known to be around 32 °C was increased after introducing HIPAAm hydroxy group to be 37.4 °C and decreased by introducing the hydrophobic SAKIPAAm strained alkyne group to be 30.1 °C, in addition to GPC measurements that confirmed the molecular weight change. To the best of our knowledge, this may be the first report that introduces the strained alkyne group into NIPAAm copolymer, but these findings were agreed with our previous report where we introduced carboxy isopropyl acrylamide into the NIPAAm copolymer and mentioned the relative LCST elevation [38] also, our findings were parallel with Maeda et al., who introduced HIPAAm into the NIPAAm copolymer [39]. SARS-CoV-2 antibody conjugation with azido groups was achieved by the addition of azido-(EG)4-NHS to the antibody lysine residues that were confirmed by the fluorescamine reduction method where increasing azido-(EG)4-NHS feeding during the reaction showed a higher azido conjugation to the antibody and fewer amine residues, these results were in agreement with Byeong results who labelled the anti-Her2 IgG to azido groups previously [40]. Our SDS-PAGE gel images showed different polymeric concentrations after conjugation with a constant concentration of SARS-CoV-2 antibody. Increasing the amount of polymer in the reaction solution caused more conjugation to the antibody and finally, more band broadening and increasing in the conjugate molecular weight, which confirmed the successful conjugate synthesis. Finally, we decided to use the antibody to polymer ratio 1:1 to avoid any kind of antibody denaturation due to over polymeric conjugation and to keep the antibody affinity as much as we can in combination with the free polymer to enhance conjugate sedimentation, these data were inconsistent with Huisgen click reaction findings [33].
SARS-CoV-2 antibody-temperature-responsive polymer supported with free polymers showed a high significant recovery after applying thermal precipitation that enabled recombinant SARS-CoV-2 nucleocapsid protein enrichment to about 6 folds of its original concentration that indicated the capability of our developed system to enrich low viral load samples, these data were parallel with Hoffman reports who succeeded in polymer antibody conjugate enrichment for biomarkers enrichment [34], [41] and, with Roy et al. who conjugated a stimuli-responsive polymer with anti-p24 IgG antibody with about 100% efficiency using RAFT and Tetrafluorophenyl active ester chemistry [42]. The strength of this work is that to the best of our knowledge, this report may be the first to try to functionalize SARS-CoV-2 antibody to a thermo-responsive polymer for increasing the screening sensitivity during the current pandemic. According to Fig. 7A & B, misdiagnosis of some SARS-CoV-2 samples, especially low viral load specimens happened, and these results were inconsistent with previous reports [43], [44], [45], [46] that discussed the same obstacle especially, during using LFIA diagnostic methods with different respiratory tract samples and this problem would have a great setback in SARS-CoV-2 transmission.
Our system succeeded in antigenic enrichment and avoiding misdiagnosis of low concentration specimens. The two concentrations 1.04 and 2.08 × 10−15 mol/mL were detected as negative samples, then detected as positive after our polymeric system treatment. Body fluids are complexed and heterogeneous solutions that contain a massive content of different biomolecules and impurities that may affect polymeric conjugation negatively [47]. To overcome this challenge without any adverse effects on the polymeric conjugation and affinity, we conducted our experiments using click chemistry that is considered highly selective and specific and can proceed regardless of the reaction environment [37]. This concept was clear in our findings where the results indicated successful polymeric conjugation in different body fluids including the upper respiratory tract samples that are commonly used for SARS-CoV-2 detection. Polymeric conjugation was succeeded in urine, nasopharyngeal, and oropharyngeal specimens regardless of the heterogenicity and complexity of these samples. This report may be the first to conduct polymeric conjugation for diagnosis purposes in real body fluids, especially upper respiratory tract samples that commonly used for SARS-CoV-2 detection.
The limitation of this work may be that we need to functionalize our polymeric strategy using not only anti-SARS-CoV-2 nucleocapsid antibody but also different antibodies to target different viral epitopes, especially some antigenic targets are hidden or highly glycosylated in native form. Moreover, we need more validation for these different SARS-CoV-2 antigenic targets to be used in many diagnostic approaches, especially PCR.
5. Conclusion
In our study, we developed a portable, fast, easy, and sensitive strategy based on the concepts of immune-affinity and responsive smart polymeric materials. This strategy was developed to be used as a preanalytical step for SARS-CoV-2 specimen enrichment to avoid the misdiagnosis that usually happens during SARS-CoV-2 diagnosis due to the low viral load in the tested samples. Our developed system succeeded in antigenic enrichment where it enriched recombinant SARS-CoV-2 nucleocapsid protein samples to about 6 folds in addition to avoiding misdiagnosis of low viral load specimens that were detected as negative samples, then detected positive after our polymeric system treatment. The strength of this work is that to the best of our knowledge, this report may be the first to try to functionalize SARS-CoV-2 antibodies to a thermo-responsive polymer for utilization of increasing the screening sensitivity during the current pandemic.
CRediT authorship contribution statement
A. Nabil: Conceptualization, Investigation, Methodology, Data curation, Formal analysis, Validation, Visualization, Writing - original draft, Writing - review & editing. E. Yoshihara: Investigation, Methodology, Formal analysis, Writing - review & editing. K. Hironaka: Investigation, Methodology, Formal analysis, Writing - review & editing. A.A. Hassan: Formal analysis, Writing - review & editing. G. Shiha: Formal analysis, Writing - review & editing. M. Ebara: Supervision, Resources, Funding acquisition, Project administration, Conceptualization, Data curation, Formal analysis, Validation, Visualization, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The author(s) received no financial support for the research, authorship, and/or publication of this article.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.06.016.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Iacobucci G. Covid-19: UK must prepare now for winter peak or risk many more deaths, scientists warn. BMJ. 2020;370 doi: 10.1136/bmj.m2825. [DOI] [PubMed] [Google Scholar]
- 2.Nabil A., Uto K., Elshemy M.M., Soliman R., Hassan A.A., Ebara M. Current coronavirus (SARS-CoV-2) epidemiological, diagnostic and therapeutic approaches: an updated review until June 2020. EXCLI J. 2020;19:992–1016. doi: 10.17179/excli2020-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fang Y., Zhang U., Xie J., Lin M., Ying L., Pang P. Sensitivity of Chest CT for COVID-19: Comparison to RT-PCR. Radiology. 2020 doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W. (COVID-19) in China: a report of 1014 cases. Radiology. 2019;2020 doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y., Yao L., Li J., Chen L., Song Y., Cai Z. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically 319 diagnosed with. COVID-19. 2020;92(7):903–908. doi: 10.1002/jmv.25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li D., Wang D., Dong J., Wang N., Huang H., Xu H. False-negative results of real-time reverse-transcriptase polymerase chain 321reaction for severe acute respiratory syndrome coronavirus 2: role of deep-learning-based 322CT diagnosis and insights from two cases. Korean J Radiol. 2020;21(4):505–508. doi: 10.3348/kjr.2020.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Z., Li Y., Wu B., Hou Y., Bao J., Deng X. A patient with COVID-19 presenting a false-negative reverse transcriptase polymerase Chain reaction result. Korean J Radiol. 2020;21(5):623–624. doi: 10.3348/kjr.2020.0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winichakoon P., Chaiwarith R., Liwsrisakun C., Salee P., Goonna A., Limsukon A. Negative nasopharyngeal and oropharyngeal swabs do not rule out COVID-19. J Clin Microbiol. 2020;58(5):e00297–e320. doi: 10.1128/JCM.00297-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zou L., Ruan E., Huang M., Liang L., Huang H., Hong Z. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382(12):1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X., Tan L., Wang X., Liu W., Lu Y., Cheng L. Comparison of nasopharyngeal and oropharyngeal swabs for SARS-CoV-2 detection in 353 patients received tests with both specimens simultaneously. Int J Infect Dis. 2020;94:107–109. doi: 10.1016/j.ijid.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, ZhaoX, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med, 2020; 382(8): p. 727-733. [DOI] [PMC free article] [PubMed]
- 12.Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis. 2020;20(4):411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao W.J., Li L.M. Advances on presymptomatic or asymptomatic carrier transmission of COVID-19. Zhonghua Liu Xing Bing Xue Za Zhi. 2020;41:485–488. doi: 10.3760/cma.j.cn112338-20200228-00207. [DOI] [PubMed] [Google Scholar]
- 14.Peeri N.C., Shrestha N., Rahman M.S., Zaki R., Tan Z., Bibi S. The SARS, MERS and novel coronavirus (COVID-19) epidemics, the newest 305and biggest global health threats: what lessons have we learned? Int J Epidemiol. 2020;49(3):717–726. doi: 10.1093/ije/dyaa033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prompetchara E., Ketloy C., Palaga T. Immune responses in COVID-19 and potential 307 vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol. 2020;38(1):1–9. doi: 10.12932/AP-200220-0772. [DOI] [PubMed] [Google Scholar]
- 16.Peiris J.S., Chu C.M., Cheng V.C.C., Chan K.S., Hung I.F.N., Poon L.L.M. Clinical progression and viral load in a community outbreak of 300coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361(9371):1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacofsky D., Jacofsky E.M., Jacofsky M. Understanding Antibody Testing for COVID-19. J Arthroplasty. 2020;35(7S):S74–S81. doi: 10.1016/j.arth.2020.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long Q.X., Tang X.J., Shi Q.L., Li Q., Deng H.J., Yuan J. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med. 2020;26(8):1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 19.Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S. Development and clinical Applicationof A rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J Med Virol. 2020 doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu F., Wang A., Liu M., Wang Q., Chen J., Xia S. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. MedRxiv. 2020 doi: 10.1101/2020.03.30.20047365. [DOI] [Google Scholar]
- 21.Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis. 2020:ciaa344. doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heng S., Kong F. Separation of antigens and antibodies by immunoaffinity chromatography. Pharm Biol. 2012;1038–1044 doi: 10.3109/13880209.2011.653493. [DOI] [PubMed] [Google Scholar]
- 23.Shu T., Hu L., Shen Q., Jiang L., Zhang Q., Serpe M.J. Stimuli-responsive polymer-based systems for diagnostic applications. J Mater Chem B. 2020;8(32):7042–7061. doi: 10.1039/d0tb00570c. [DOI] [PubMed] [Google Scholar]
- 24.Jauregui R., Srinivasan S., Vojtech L.N., Gammill H.S., Chiu D.T., Hladik F. Temperature-responsive magnetic nanoparticles for enabling affinity separation of extracellular vesicles. ACS Appl Mater Interfaces. 2018;10(40):33847–33856. doi: 10.1021/acsami.8b09751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Las Heras Alarcon C, Pennadam S, Alexander C. Stimuli responsive polymers for biomedical applications. Chem Soc Rev., 2005; 34(3):276-285. [DOI] [PubMed]
- 26.Yoneoka S., Park K.C., Nakagawa Y., Ebara M., Tsukahara T. Synthesis and evaluation of thermoresponsive boron-containing Poly(N-isopropylacrylamide) diblock copolymers for self-assembling nanomicellar boron carriers. Polymers (Basel) 2018;11(1):42. doi: 10.3390/polym11010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uto K., Ebara M. Magnetic responsive microparticles that switch shape at 37°C. Appl Sci. 2017;7(11):1203. [Google Scholar]
- 28.Maeda T., Kim Y.J., Aoyagi T., Ebara M. The design of temperature-responsive nano fiber meshes for cell Storage applications. Fibers. 2017;5(1):13. [Google Scholar]
- 29.Kotsuchibashi Y., Ebara M., Aoyagi T., Narain R. Recent advances in dual temperature responsive block copolymers and their potential as biomedical applications. Polymers (Basel) 2016;8(11):380. doi: 10.3390/polym8110380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ebara M., Uto K., Idota N., Hoffman J.M., Aoyagi T. Shape-memory surface with dynamically tunable nanogeometry activated by body heat. Adv Mater. 2012;24:273–278. doi: 10.1002/adma.201102181. [DOI] [PubMed] [Google Scholar]
- 31.Lai J.J., Hoffman J.M., Ebara M., Hoffman A.S., Estournes C., Wattiaux A. Dual magnetic-/temperature-responsive nanoparticles for microfluidic separations and assays. Langmuir. 2007;23:7385–7391. doi: 10.1021/la062527g. [DOI] [PubMed] [Google Scholar]
- 32.Maeda T., Kanda T., Yonekura Y., Yamamoto K., Aoyagi T. Hydroxylated poly (N-isopropylacrylamide) as functional thermoresponsive materials. Biomacromolecules. 2006;7:545–549. doi: 10.1021/bm050829b. [DOI] [PubMed] [Google Scholar]
- 33.Heskins M., Guillet J.E. Solution properties of poly(N-Isopropylacrylamide) J Macromol Sci Chem. 1968;2:1441–1455. [Google Scholar]
- 34.Hoffman A.S. Applications of thermally reversible polymers and hydrogels in therapeutics and diagnostics. J Control Release. 1987;6:297–305. [PubMed] [Google Scholar]
- 35.Huisgen R. 1,3-dipolar cycloadditions. Past and future. Angew Chem Int Ed. 1963;2(10):565–598. [Google Scholar]
- 36.Agard N.J., Prescher J.A., Bertozzi C.R. A strain-promoted [3 + 2] Azide−alkyne cycloaddition for covalent modification of biomolecules in living systems. J Am Chem Soc. 2004;126(46):15046–15047. doi: 10.1021/ja044996f. [DOI] [PubMed] [Google Scholar]
- 37.Kolb H.C., Sharpless K.B. The growing impact of click chemistry on drug discovery”. Angew Chem Int Ed. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 38.Ebara M., Aoyagi T., Sakai K., Okano T. Introducing reactive carboxyl side chains retains phase transition temperature sensitivity in N-isopropylacrylamide copolymer gels. Macromolecules. 2000;33(22):8312–8316. [Google Scholar]
- 39.Maeda T., Kanda T., Yonekura Y., Yamamoto K., Aoyagi T. Hydroxylated poly(N-isopropylacrylamide) as functional thermoresponsive materials. Biomacromolecules. 2006;7:545–549. doi: 10.1021/bm050829b. [DOI] [PubMed] [Google Scholar]
- 40.Lee B.S., Lee Y., Park J., Jeong B.S., Jo M., Jung S.T. Construction of an immunotoxin via site-specific conjugation of anti-Her2 IgG and engineered Pseudomonas exotoxin. J Biol Eng. 2019;13:56. doi: 10.1186/s13036-019-0188-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoffman J.M., Ebara M., Lai J.J., Hoffman A.S., Folch A., Stayton P.S. A helical flow, circular microreactor for separating and enriching ‘‘smart’’ polymer–antibody capture reagents. Lab Chip. 2010;10:3130–3138. doi: 10.1039/c004978f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roy D., Nehilla B.J., Lai J.J., Stayton P.S. Stimuli-responsive polymer-antibody conjugates via RAFT and tetrafluorophenyl active ester chemistry. ACS Macro Lett. 2013;2:132–136. doi: 10.1021/mz300620v. [DOI] [PubMed] [Google Scholar]
- 43.Adams E.R., Ainsworth M., Anand R. Antibody testing for COVID-19: a report from the National COVID scientific advisory panel [version 1; peer review: 1 approved] Wellcome Open Res. 2020;5:139. doi: 10.12688/wellcomeopenres.15927.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu W., Liu L., Kou G., Zheng Y., Ding Y., Ni W. Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J Clin Microbiol. 2020;58(6):e00461–e520. doi: 10.1128/JCM.00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo L., Ren L., Yang S., Xiao M., Chang D., Yang F. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin Infect Dis. 2020;71(15):778–785. doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nabil A., Elshemy M.M., Uto K., Soliman R., Hassan A.A., Shiha G. Coronavirus (SARS-CoV-2) in gastroenterology and its current epidemiological situation: an updated review until January 2021. EXCLI J. 2021;20:366–385. doi: 10.17179/excli2021-3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu S., Loo J.A., Wong D.T. Human body fluid proteome analysis. Proteomics. 2006;6(23):6326–6353. doi: 10.1002/pmic.200600284. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.