Abstract
In this study, we investigated the diversity of diatrypaceous fungi from six regions in China based on morpho-molecular analyses of combined ITS and tub2 gene regions. We accept 23 genera in Diatrypaceae with 18 genera involved in the phylogram, and the other five genera are lacking living materials with sequences data. Eleven species included in four genera (viz. Allocryptovalsa, Diatrype, Diatrypella, and Eutypella) have been isolated from seven host species, of which nine novel species (viz. Allocryptovalsa castaneae, A. castaneicola, Diatrype betulae, D. castaneicola, D. quercicola, Diatrypella betulae, Da. betulicola, Da. hubeiensis, and Da. shennongensis), a known species of Diatrypella favacea, and a new record of Eutypella citricola from the host genus Morus are included. Current results show the high diversity of Diatrypaceae which are wood-inhabiting fungi in China.
Keywords: Allocryptovalsa, Diatrype, Diatrypella, Eutypella, fungal diversity, phylogeny, taxonomy
Introduction
Diatrypaceae is an important family in Xylariales (Sordariomycetes, Ascomycota), containing many taxa with a worldwide distribution (Glawe and Jacobs, 1987; Mayorquin et al., 2016; Senwanna et al., 2017; Moyo et al., 2018a, b; Konta et al., 2020; Wijayawardene et al., 2020). Species of the Diatrypaceae are frequently saprobic on the decaying wood of angiosperms (Tendulkar, 1970; Acero et al., 2004; de Almeida et al., 2016). However, an endophyte of Picea abies, a gymnosperm host, was identified as an asexual morph of Diatrypaceae (Libertella sp.) by Caroll et al. (1977). Few endophytes such as Diatrypella frostii and Peroneutypa scoparia were reported later (de Errasti et al., 2010; Vieira et al., 2011). Strikingly, several plant pathogens of Diatrypaceae were reported causing canker, dieback, and grapevine trunk diseases, e.g., Cryptosphaeria populina was linked to canker in Populus species (Glawe and Rogers, 1984); Cryptosphaeria pullmanensis caused canker disease in Populus alba and Salix alba (Ma et al., 2016); Cryptovalsa ampelina caused grapevine trunk disease on Vitis species (Luque et al., 2006); Eutypa lata was isolated from Prunus armeniaca and Vitis species with canker and dieback symptoms (Lardner et al., 2005); Eutypa leptoplaca contributed to the dieback of grapevines (Trouillas and Gubler, 2004; Catal et al., 2007); and Eutypella parasitica caused canker in Acer species (Rappaz, 1987).
Nitschke (1867) proposed the first important study of Diatrypaceae (as Diatrypeae) including five genera (Supplementary Table 1). Kirk et al. (2001) accepted nine genera. Later, the family was considered to accommodate 13 genera by Kirk et al. (2008) (Supplementary Table 1). Diatrypasimilis, Monosporascus, and Pedumispora have been added to the family in subsequent studies (Abdel-Wahab et al., 2014; Klaysuban et al., 2014; Maharachchikumbura et al., 2015). Wijayawardene et al. (2018) reported 17 genera in the family (Supplementary Table 1). Subsequently, Dayarathne et al. (2016), Senwanna et al. (2017), Phookamsak et al. (2019) respectively introduced Allocryptovalsa, Halodiatrype, and Neoeutypella, and accepted them as members of Diatrypaceae. Wijayawardene et al. (2020) listed 20 genera in the family. Later, Dayarathne et al. (2020a, b), Konta et al. (2020) respectively added Allodiatrype, Halocryptosphaeria, and Halocryptovalsa to this family.
The sexual morph members of Diatrypaceae are characterized by perithecial ascomata usually with ostiolar necks, 8-spored or polysporous asci with a very long pedicel and J-/J+ apical apparatus, and allantoid ascospores (Senanayake et al., 2015; Dayarathne et al., 2016; de Almeida et al., 2016). Several asexual genera included in coelomycetes or hyphomycetes (viz. Cytosporina, Libertella, and Phaeoisaria) have been linked to the family Diatrypaceae (Dayarathne et al., 2016; de Almeida et al., 2016; Mehrabi et al., 2016; Shang et al., 2017). However, the asexual morphs of many species were still indistinguishable (Acero et al., 2004). Senanayake et al. (2015) summarized that the asexual morph of this family had acervular and astromatic conidiomata, branched conidiophores and filiform, allantoid or rarely straight conidia with flattened base and blunt apex. However, in most cases, it is difficult to differentiate diatrypaceous species based on asexual morphs (Glawe and Rogers, 1986; de Almeida et al., 2016).
Due to overlapping phenotypic characters in Diatrypaceae, polyphasic approaches to solve the taxonomy of fungi were very common in recent studies (Wijayawardene et al., 2016; Norphanphoun et al., 2017; Fan et al., 2018, 2020; Lawrence et al., 2018; Zhu et al., 2020). The first molecular phylogenetic analysis of Diatrypaceae based on ITS showed that Cryptosphaeria, Diatrype, Diatrypella, Eutypa, and Eutypella were polyphyletic (Acero et al., 2004). Recently, the identification and classification of diatrypaceous taxa were performed by the multiple sequence data (mostly ITS and tub2) and morphological characters (Phookamsak et al., 2019; Dayarathne et al., 2020a; Hyde et al., 2020b; Konta et al., 2020). Moreover, Konta et al. (2020) introduced one new genus Allodiatrype and five new species belonging to Allocryptovalsa, Allodiatrype, and Diatrypella from palms (Arecaceae) based on this criterion.
During the investigation of forest pathogens in China, 86 diatrypaceous specimens associated with various disease symptoms were collected from Beijing City, Xinjiang Uygur Autonomous Region, and four other provinces in China viz. Hubei, Hebei, Jiangsu, and Yunnan. The objectives were to supplement a multi-gene DNA dataset of Diatrypaceae including ITS and tub2, improve the phylogenetic systematics of this family, and provide a theoretical basis for the identification of diseases and pathogens.
Methods
Isolates
Symptomatic branches or twigs were collected from seven tree hosts (Betula albosinensis, B. davurica, B. platyphylla, Castanea mollissima, Juglans regia, Morus alba, and Quercus mongolica) from Beijing City, Xinjiang Uygur Autonomous Region, and four other provinces in China viz. Hubei, Hebei, Jiangsu, and Yunnan. Eighty-six fresh specimens of Diatrypaceae were put into envelopes with records of their altitude, collector, collecting time, host, longitude, and latitude. A total of 21 representative isolates were obtained by removing the ascospores or conidial mass from fresh specimens on the surface of 1.8% potato dextrose agar (PDA) and incubating at 25°C for 24 h. Single germinating spore was transferred onto a fresh PDA plate. Specimens and isolates were deposited in the Beijing Forestry University (BJFU) and the Beijing Museum of Natural History (BJM). Strains of the new species are maintained in the China Forestry Culture Collection Centre (CFCC).
Morphological Analysis
Species identification was based on morphological features of fruiting bodies and micromorphology supplemented by cultural characteristics. Macro-morphological observations including structure and size of stromata, ectostromatic disc, and ostioles were determined using a Leica stereomicroscope (M205 FA) (Leica Microsystems, Wetzlar, Germany). Micro-morphological photographs were captured using a Nikon Eclipse 80i microscope (Nikon Corporation, Tokyo, Japan), including conidiophores, asci, and conidia/ascospores. Adobe Bridge CS v. 6 and Adobe Photoshop CS v. 5 were used for manual editing. At least 10 conidiomata/ascomata, 10 asci, and 30 conidia/ascospores were randomly selected for measurement to calculate the mean width/length and respective standard deviations (SD). Cultural characteristics of strains incubated in the dark at 25°C were recorded. Colony morphology was described using the color charts of Rayner (1970). Nomenclatural novelties were deposited in the MycoBank (1 Crous et al., 2004).
DNA Extraction, PCR Amplification, and Sequencing
Fungal mycelium grown on the cellophane on PDA was scraped for the extraction of genomic DNA following the modified CTAB method (Doyle and Doyle, 1990). Two loci were amplified, including the internal transcribed spacer (ITS) region and partial beta-tubulin (tub2) using the primer pairs ITS1/ITS4 (White et al., 1990) and T1/Bt2b (Glass and Donaldson, 1995; O’Donnell and Cigelnik, 1997), respectively. The additional combination of Bt2a and Bt2b (Glass and Donaldson, 1995) was used in case of amplification failure of the primer T1 and Bt2b. The polymerase chain reaction (PCR) assay was conducted as described in Fan et al. (2020). PCR amplification products were estimated via electrophoresis in 2% agarose gels. DNA sequencing was performed using an ABI PRISM® 3730XL DNA Analyzer with a BigDye Terminater Kit v. 3.1 (Invitrogen, United States) at the Shanghai Invitrogen Biological Technology Company Limited (Beijing, China).
DNA Sequence Analysis
The initial identities of our strains sequenced were obtained by morphological observations and nucleotide BLAST search. To clarify the phylogenetic position, the alignment based on a combined matrix using ITS and tub2 sequences was performed to compare with other available species in Diatrypaceae. Reference sequences were selected based on ex-type or ex-epitype sequences available from relevant recently published literature (de Almeida et al., 2016; Senwanna et al., 2017; Shang et al., 2017, 2018; Hyde et al., 2019, 2020a; Phookamsak et al., 2019; Dayarathne et al., 2020a, b; Konta et al., 2020; Supplementary Table 2). Xylaria hypoxylon (CBS 122620) was selected as the outgroup. For each gene, sequences were aligned using MAFFT v. 7 (Katoh and Standley, 2013) and manually improved where necessary using MEGA v. 6 (Tamura et al., 2013). Ambiguously aligned sequences were excluded from the analysis. Alignments were used to infer a preliminary phylogenetic relationship for our sequences based on Maximum Parsimony (MP) with PAUP v. 4.0b10 (Swofford, 2003), Maximum Likelihood (ML) with PhyML v. 3.0 (Guindon et al., 2010), and Bayesian Inference (BI) analyses with MrBayes v. 3.1.2 (Ronquist and Huelsenbeck, 2003).
Maximum parsimony analysis was performed using a heuristic search option of 1,000 random-addition sequences. The tree bisection and reconnection (TBR) was selected as option to the branch swapping algorithm (Swofford, 2003). The branches of zero length were collapsed, and all equally parsimonious trees were saved. Clade stability was assessed with a bootstrap analysis of 1,000 replicates (Hillis and Bull, 1993). Tree length (TL), consistency index (CI), retention index (RI), and rescaled consistency (RC) were calculated (Swofford, 2003). ML analysis including 1,000 bootstrap replicates (Hillis and Bull, 1993) was conducted with a general time reversible (GTR) model of site substitution, including gamma-distributed rate heterogeneity and a proportion of invariant sites (Guindon et al., 2010). The nucleotide model of evolution for each of the data partitions were estimated by MrModeltest v. 2.3 (Posada and Crandall, 1998) before the Bayesian analysis. BI analysis was performed using a Markov Chain Monte Carlo (MCMC) algorithm with Bayesian posterior probabilities (Rannala and Yang, 1996). Two MCMC chains were run for 1,000,000 generations with a sampling frequency at every 100th generation. The first 25% of trees were discarded as the burn-in phase of each analysis, and the posterior probabilities (BPP) were calculated to assess the remaining trees (Rannala and Yang, 1996). The branch support from MP and ML analyses were evaluated with a bootstrapping (BS) method of 1,000 replicates (Hillis and Bull, 1993). The resulting trees were plotted in Figtree v. 1.4.4 and edited in Adobe Illustrator CS6 v. 16.0.0. All sequences from this study were deposited in GenBank (Supplementary Table 2). The multi-gene sequence alignment files were submitted to TreeBASE (2 accession number: S27126).
Results
Phylogenetic Analyses
The phylogenetic analysis combined ITS and tub2 contained 146 ingroup strains with 1,175 characters including gaps (713 for ITS and 462 for tub2), of which 471 were constant, 191 variable characters were parsimony-uninformative, and 513 characters were variable and parsimony-informative. The MP analysis resulted 500 parsimonious trees, and the first tree (TL = 3,637, CI = 0.362, RI = 0.771, RC = 0.279) was presented in Figure 1. For BI analyses, the best-fit model of nucleotide evolution was deduced on the AIC (ITS: GTR + I + G; tub2: HKY + I + G). Tree topologies of ML and BI analyses did not significantly differ from the MP. Topology of the phylogenetic analyses were similar to the relevant recently published literature (Senwanna et al., 2017; Shang et al., 2017, 2018; Hyde et al., 2019, 2020a; Phookamsak et al., 2019; Dayarathne et al., 2020a; Konta et al., 2020).
FIGURE 1.

Phylogram of Diatrypaceae based on combined ITS and tub2 sequence data. The MP and ML bootstrap support values above 70% are shown at the first and second positions, respectively. Thickened branches represent posterior probabilities above 0.95 from the BI. Ex-type strains are in bold, type species are denoted with the superscript “TS” and the disputable type species are denoted with the superscript “TSQ.” Strains from the current study are in blue.
Based on phylogenetic analyses, the phylogram included 27 lineages, representing 21 known (Allocryptovalsa/Eutypella sensu lato, Allodiatrype, Anthostoma, Cryptosphaeria 1, Cryptosphaeria 2, Cryptovalsa, Diatrypasimilis, Diatrype sensu stricto, Diatrypella 1, Diatrypella 2, Eutypa sensu lato, Eutypella sensu stricto, Eutypa sensu stricto, Halocryptosphaeria, Halocryptovalsa, Halodiatrype, Monosporascus, Neoeutypella, Pedumispora, Peroneutypa, Quaternaria) and six incertae sedis clades. Eleven lineages are, herein, described as nine new species and two known species belonging to four genera in Diatrypaceae (Figure 1).
Some confused taxa were excluded in the current phylogram after the primary analyses. Diatrype decorticata (ANM 1498), Diatrype enteroxantha (HUEFS 155116), Diatrype macowaniana (CBS 214.8), Diatrype oregonensis (DCA600), Diatrype polycocca (CBS 213.87), Diatrype prominens (ATCC MYA-4410), Diatrype whitmanensis (CDB011), Eutypella parasitica (CBS 210.39), and Eutypella prunastri (CBS 277.87) are not the type strains, which have single clade in phylogenetic tree or mixed with in clade of other genera. And the sequence data of Halocryptovalsa avicenniae (MAW 2017a) is inconsistent with the position of genus.
Clade 06 (Diatrypella 2): This clade comprises nine newly generated strains and other six Diatrypella strains with strong statistical supports (MP/ML/BI = 96/99/1) in Figure 1. Diatrypella hubeiensis (CFCC 52413) was the basal subclade close to Da. yunnanensis. Diatrypella shennongensis (CFCC 52414 and CFCC 52415) formed a single clade with high support (MP/ML/BI = 100/100/1). Diatrypella betulae (CFCC 52404, CFCC 52405 and CFCC 52406) clustered close to Da. shennongensis. Diatrypella betulicola (CFCC 52411 and CFCC 52412) also formed a distinct strongly supported clade (MP/ML/BI = 95/96/1). The isolate CFCC 52409 clustered with Diatrypella favacea (CBS 198.49, R191, and DL26C), which was recognized as known species.
Clade 09 (Diatrype sensu stricto): The type species of Diatrype, D. disciformis (CBS 205.87 and GB 5815), and other Diatrype and Diatrypella species grouped with strong statistical supports (MP/ML/BI = 100/97/1) in Figure 1. Our six new strains clustered in this clade as three different subclades viz. Diatrype quercicola (CFCC 52418, CFCC 52419, and CFCC 52420), D. castaneicola (CFCC 52425 and CFCC 52426) and D. betulae (CFCC 52416). Diatrype betulae was the basal subclade close to D. undulata. Diatrype castaneicola was the internal clade with high support (MP/ML/BI = 99/100/1) close to D. stigma. Diatrype quercicola formed a single clade with high support (MP/ML/BI = 89/98/1) and grouped with D. virescens with no support value.
Clade 12 (Allocryptovalsa/Eutypella sensu lato): This clade comprises Allocryptovalsa and part Eutypella species clustered with strong support values (MP/ML/BI = 87/94/1) in Figure 1. Two isolates (CFCC 52433 and CFCC 52434) grouped together with Eutypella citricola (HVVIT07 and HVGRF01) with strong support (MP/ML/BI = 100/100/1). The isolates CFCC 52432 formed a separate branch separated from Eutypella citricola and Eutypella vitis (MP/ML/BI = 95/96/1), which represented a new species Allocryptovalsa castaneicola. Allocryptovalsa castaneae (CFCC 52427, CFCC 52428, and CFCC 52429) also regarded as a new species with the distinct strongly supported clade (MP/ML/BI = 96/99/1), which was clustered with Allocryptovalsa cryptovalsoidea, A. elaeidis, A. polyspora, and A. truncata (MP/ML/BI = 99/100/1).
Taxonomy
Allocryptovalsa Senwanna, Phookamsak & K.D. Hyde, Mycosphere 8(10): 1839 (2017).
Type: Allocryptovalsa polyspora Senwanna, Phookamsak & K.D. Hyde, Mycosphere 8(10): 1840 (2017).
Known distribution: Australia, China, Germany, India, Thailand, and United States (Saccardo, 1882; Trouillas et al., 2011; Senwanna et al., 2017; Hyde et al., 2020b; Konta et al., 2020; This study).
Notes: Allocryptovalsa typified with A. polyspora was originally introduced to accommodate another two new combination species (A. cryptovalsoidea and A. rabenhorstii), which was with the character of immersed perithecia, polysporous asci, and allantoid ascospores (Senwanna et al., 2017). Later, Hyde et al. (2020a); Konta et al. (2020) reported A. elaeidis and A. truncata isolated from Elaeis guineensis and decaying twig, respectively. In this study, we introduce two additional species, Allocryptovalsa castaneae and A. castaneicola, based on morphological coupled with molecular data (Figure 1; clade 12).
Allocryptovalsa castaneae N. Jiang & X.L. Fan sp. nov. Figure 2.
FIGURE 2.

Holomorph of Allocryptovalsa castaneae (BJFU CF2020518, holotype). (A) Ascomata on the host. (B) Ascoma on the host. (C) Transverse section of ascoma. (D) Longitudinal section through ascoma. (E) Conidiomata on the host. (F) Conidioma on the host. (G) Asci and ascospores. (H) Ascospores. (I,J) Conidia attatch to conidiogenous cells. (K) Conidia. Scale bars: (A,F) = 1 mm; (B−E) = 500 μm; (G−K) = 10 μm.
MycoBank MB 837777.
Typification: CHINA. Hebei Province, Qinhuangdao City, Qinglong County, 119°11′52.25″ E 40°22′52.13″ N, 246 m msl., from branches of Castanea mollissima, 16 Oct. 2017, C.M. Tian & N. Jiang, holotype BJFU CF2020518, ex-type culture CFCC 52428. Hebei Province, Qinhuangdao City, Qinglong County, 119°11′52.25″ E 40°22′52.13″ N, 246 m msl., from branches of Castanea mollissima, 16 Oct. 2017, C.M. Tian & N. Jiang, isotype BJM 240506, ex-isotype culture CFCC 52429.
Etymology: Named after the host genus from which it was collected, Castanea.
Diagnosis: Phylogenetically sister to Allocryptovalsa rabenhorstii, differs by the smaller size of ascospores (8–11 × 2.5–3.5 vs. 13.5–15 × 4–5 μm).
Descriptions: Necrotrophic on branches of Castanea mollissima. Sexual morph: Stromata solitary to gregarious, immersed in the bark, erumpent through the surface of bark, with 3–5 perithecia arranged irregularly (0.3–)0.5–0.8 mm (av. = 0.6 ± 0.2 mm, n = 10) in diam. Ectostromatic disc orange, unconspicuous, circular to oblong, with 3–5 ostioles arranged irregularly per disc. Ostioles numerous, brown to black, at the same level as the disc, scattered (85–)120–130 μm (av. = 122.4 ± 14.0 μm, n = 10) in diam. Perithecia outer surface lacking powdery entostroma, black, flask-shaped to spherical, with discrete perithecial necks (320–)360–400(−420) μm (av. = 379.7 ± 19.7 μm, n = 10) in diam. Asci clavate to elongate obovoid, polysporous, thin-walled, short pedicellate, apically rounded (52–)60–83(–92) × (11–)12–17(–25) μm (av. = 71.5 ± 11.4 × 14.4 ± 2 μm, n = 30). Ascospores elongate-allantoid, thin-walled, pale yellowish to pale brown at maturity, slightly curved, aseptate, 8–11(–13) × 2.5–3.5 (–4) μm (av. = 10.1 ± 0.8 × 3.1 ± 0.4 μm, n = 30). Asexual morph: Coelomycetous. Conidiomata pycnidial, immersed in the bark, scattered, erumpent through the surface of bark. Ectostromatic disc flat or concave, orange, surrounded by bark flaps, circular to ovoid, with 8–10 ostioles arranged circularly on per disc (200–)260–320(–370) μm (av. = 280.8 ± 43.3 μm, n = 10) in diam. Ostioles black, at the same level as the disc surface (45–)60–70 μm (av. = 64.1 ± 9.8 μm, n = 10) in diam. Conidiogenous cells holoblastic conidiogenesis, approximately cylindrical, hyaline, integrated, arising from pseudoparenchymatous cells, unicellular, with wide base producing conidia at the apex (15–)19–30(–31) × (1–)1.5–2(–2.5) μm (av. = 24.5 ± 4.9 × 1.7 ± 0.2 μm, n = 30). Conidia hyaline, elongate-allantoid, not curved, smooth, aseptate (4–)5–7(–8) × (1–) 1.5–2(–2.5) μm (av. = 5.9 ± 0.9 × 1.8 ± 0.2 μm, n = 30).
Culture characteristics: Cultures are initially white with irregular margin, becoming dark green at the margin and stopping growing with 7 cm in diam. after 2 weeks, comprising dense, irregular, flat mycelium.
Known host and distribution: Known on Castanea mollissima and Juglans regia in China.
Additional collection examined: CHINA. Yunnan Province, Chuxiong Yi Autonomous Prefecture, Dayao County, 101°20′15.7″ E 25°44′47.19″ N, 2,002 m msl., from branches of Juglans regia, August 07, 2015, N. Zhao, paratype BJFU CF2020516, ex-paratype culture CFCC 52427.
Notes: Three new strains isolated from branches of Castanea mollissima and Juglans regia, show high support value (MP/ML/BI = 99/100/1) with the closely clustered isolates in Allocryptovalsa (Figure 1; Clade 12: Allocryptovalsa/Eutypella sensu lato). Moreover, this species has different morphological characters. Morphological comparison of members of Allocryptovalsa is provided in Supplementary Table 3. Other species of this genus were not reported with asexual morph. Therefore, Allocryptovalsa castaneae showed coelomycetous asexual morph from host in China for the first time. In addition, Allocryptovalsa castaneae differs from A. castaneicola (from Castanea mollissima), A. cryptovalsoidea (from Ficus carica), A. elaeidis (from Elaeis guineensis), A. polyspora (from Hevea brasiliensis), and A. rabenhorstii (from Vitis vinifera and Sambuscus nigra) in host association (Saccardo, 1882; Trouillas et al., 2011; Senwanna et al., 2017; Konta et al., 2020).
Allocryptovalsa castaneicola N. Jiang & X.L. Fan sp. nov. Figure 3.
FIGURE 3.
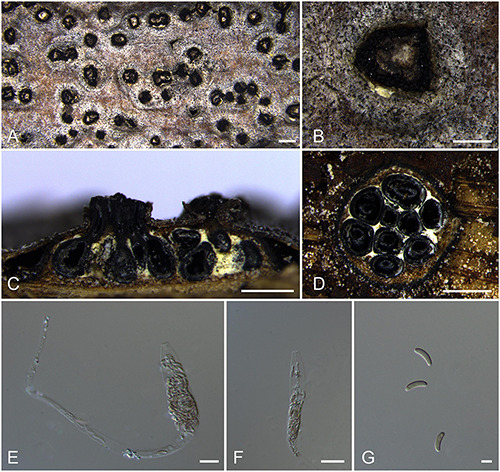
Sexual morph of Allocryptovalsa castaneicola (BJFU CF2020519, holotype). (A) Ascomata on the host. (B) Ascoma on the host. (C) Longitudinal section through ascoma. (D) Transverse section of ascoma. (E,F) Ascus and ascospores. (G) Ascospores. Scale bars: (A) = 1 mm; (B–D) = 500 μm; (E–G) = 10 μm.
MycoBank MB 837787.
Typification: CHINA. Hebei Province, Chengde City, Kuancheng Manchu Nationality Autonomous County, 118°27′54″ E 40°38′37″ N, 450 m msl., from branches of Castanea mollissima, 14 Oct. 2017, C.M. Tian & N. Jiang, holotype BJFU CF2020519, ex-type culture CFCC 52432. Hebei Province, Chengde City, Kuancheng Manchu Nationality Autonomous County, 118°27′54″ E 40°38′37″ N, 450 m msl., from branches of Castanea mollissima, 14 Oct. 2017, C.M. Tian & N. Jiang, isotype BJM 240515.
Etymology: Named after the host genus from which it was collected, Castanea.
Diagnosis: Allocryptovalsa castaneicola differs from other Allocryptovalsa species by its polysporous asci and larger size of ascospores (22–25 × 5–6 μm).
Descriptions: Necrotrophic on branches of Castanea mollissima. Sexual morph: Stromata scattered to gregarious, immersed in the bark, erumpent through the surface of bark, with 8–10 perithecia arranged irregularly (1.5–)1.7–2.0 mm (av. = 1.8 ± 0.2 mm, n = 10) in diam. Ectostromatic disc brown, circular to oblong, with more than 10 ostioles arranged circularly per disc, 0.7–0.9(–1.0) mm (av. = 0.8 ± 0.1 mm, n = 10) in diam. Ostioles numerous, gregarious, umbilicate, 4-sulcate dark brown to black, at the same level as the disc, 104–120(–140) μm (av. = 114.9 ± 11.3 μm, n = 10) in diam. Perithecia outer surface coated with yellow, powdery entostromablack, flask-shaped, perithecial necks erumpent in groups (200–)250–320(–380) μm (av. = 282.7 ± 49.8 μm, n = 10) in diam. Asci clavate to elongate obovoid, polysporous, thin-walled, long pedicellate, apically flat, 194–202 × 15–21 μm (av. = 198.4 ± 3.3 × 18.9 ± 0.5 μm, n = 10). Ascospores elongate-allantoid, thin-walled, pale yellowish to pale brown at maturity, slightly curved, aseptate, smooth, 22–25 × 5–6 μm (av. = 23.8 ± 1.1 × 5.4 ± 0.3 μm, n = 30). Asexual morph: not observed.
Culture characteristics: Colonies are initially white, uniform, becoming dark after 2 weeks.
Known host and distribution: Known only on Castanea mollissima in Hebei Province, China.
Notes: The new species displays some features of morphology typical of the recent genus Allocryptovalsa (well-developed ascostromata producing polysporous asci and allantoid ascospores) (Senwanna et al., 2017), although it appears closer placed in Eutypella sensu lato (Figure 1; Clade 12). Morphologically, Allocryptovalsa castaneicola differs from the closest species Eutypella australiensis by larger size of asci (194–202 × 15–21 vs. 40–50 × 7–8.5 μm) and ascospores (22–25 × 5–6 vs. 8–10 × 3 μm) (Trouillas et al., 2010a). Allocryptovalsa castaneicola was also distinguished from other Eutypella species resembles by having polysporous asci rather than the 8-spored asci (Trouillas et al., 2010b). Thus, we introduce it here as a new species in genus Allocryptovalsa.
Moreover, Allocryptovalsa castaneicola can differ from Cryptovalsa species by having a yellow rather than white powdery entostroma appeared on the ascomatal outer surface (Dayarathne et al., 2020b). Also, phylogenetic analyses show affinities of this fungus with strains from Eutypella spp. Therefore, the assignment of the strains to the genus Eutypella sensu lato (Figure 1; Clade 12) may require future reconsideration.
Diatrype Fr., Summa veg. Scand., Sectio Post. (Stockholm): 384, 1849.
Type: Diatrype disciformis (Hoffm.) Fr., Summa veg. Scand., Sectio Post. (Stockholm): 385 (1849).
Known distribution: Asia, Europe, North America, Oceania, and South Africa (Doidge, 1950; Munk, 1957; Conners, 1967; Rappaz, 1987; Mulenko et al., 2008; Trouillas et al., 2010a, b).
Notes: The genus Diatrype was established by Fries (1849) with Diatrype disciformis as the generic type, which have often been regarded as saprobes on decaying wood and have a strong ability to resist harsh conditions (Senanayake et al., 2015). Due to the taxonomic confusion, Diatrype may require a thorough revision together with the entire family in the future.
Diatrype betulae H.Y. Zhu & X.L. Fan sp. nov. Figure 4.
FIGURE 4.

Asexual morph of Diatrype betulae (BJFU CF2020510, holotype). (A) Conidiomata on the host. (B) Conidioma on the host. (C,D) Transverse section of conidioma. (E) Longitudinal section through conidioma. (F) Conidiogenous cells. (G,H) Conidia. Scale bars: (A) = 1 mm; (B−E) = 500 μm; (F−H) = 10 μm.
MycoBank MB 837784.
Typification: CHINA. Beijing City, Mentougou District, Mount Dongling, Xiaolongmen Forestry Centre, 115°26′51.27″ E 39°58′19.62″ N, 1,302 m msl., from branches of Betula davurica, 21 Aug. 2017, H.Y. Zhu & X.L. Fan, holotype BJFU CF2020510, ex-type culture CFCC 52416. Beijing City, Mentougou District, Mount Dongling, Xiaolongmen Forestry Centre, 115°26′51.27″ E 39°58′19.62″ N, 1,302 m msl., from branches of Betula davurica, 21 Aug. 2017, H.Y. Zhu & X.L. Fan, isotype BJM 240512.
Etymology: Named after the host genus from which it was collected, Betula.
Diagnosis: Phylogenetically, Diatrype betulae formed a separate clade.
Descriptions: Necrotrophic on branches of Betula davurica. Sexual morph: not observed. Asexual morph: Coelomycetous. Conidiomata pycnidial, immersed in the bark, scattered, erumpent slightly through the surface of bark, with multiple locules and orange colloid conidial drops exuding from the ostioles. Locules numerous, buff, circular to ovoid, 1.0–1.4 mm (av. = 1.2 ± 0.2 mm, n = 10) in diam. Conidiogenous cells approximately cylindrical, mostly straight, discrete or integrated, arising from pseudoparenchymatous cells, hyaline, unicellular, with wide base producing conidia at the apex, holoblastic conidiogenesis (12–)14–20 × 1–2 μm (av. = 16.6 ± 3.5 × 1.2 ± 0.2 μm, n = 30). Conidia hyaline, filiform, smooth or rough, aseptate, 10–13 × 1–2 μm (av. = 11.7 ± 1.2 × 1.5 ± 0.1 μm, n = 30).
Culture characteristics: Cultures are white, uniform, dense, slow growing, reaching 4 cm after 2 weeks, not produced pigmentation on PDA media.
Known host and distribution: Known only on Betula davurica in Beijing City, China.
Notes: Diatrype betulae was isolated from branches of Betula davurica in Beijing, China. One strain of Diatrype betulae (CFCC 52416) clusters as a single lineage (Figure 1). Diatrype albopruinosa, D. undulata, and D. stigma were also reported from Betula sp. (Tiffany and Gilman, 1965; Chlebicki, 2005; Mulenko et al., 2008; Goos, 2010). However, Diatrype betulae can be easily distinguished from the other three. Diatrype albopruinosa and D. undulata lack the asexual morph and sequences of D. albopruinosa are unavailable. Diatrype stigma was known on various hosts with worldwide distribution, but it can differ from D. betulae by smaller conidia (4.5–7.5 × 1–2 vs. 10–13 × 1–2 μm) (Rappaz, 1987). Diatrype betulae is also phylogenetically closely related to D. bullata, D. castaneicola, D. disciformis, D. iranensis, D. macrospora, D. quercicola, D. quercina, D. spilomea, and D. virescens. Diatrype betulae can differ from D. bullata that is a common species isolated from willows in the northern hemisphere in host association (Vasilyeva and Ma, 2014). Diatrype iranensis, D. macrospora, D. quercicola, and D. quercina were only reported from Quercus sp. (Croxall, 1950; Mehrabi et al., 2015, 2016). Diatrype betulae can easily be distinguished from them by its plant host (Betula sp.) and smaller conidia (18–38 × 0.6–0.8 μm in D. iranensis and 20–40 × 0.6–0.8 μm in D. macrospora) (Mehrabi et al., 2015, 2016). In morphology, D. betulae can be differentiated from D. disciformis and D. virescens by having asexual morph. Moreover, D. betulae differs from other closest species D. castaneicola and D. spilomea by the size of conidia (10–13 × 1–2 vs. 4–6 × 1–1.5, 13–18.5 × 1–1.2 μm) (Rappaz, 1987).
Diatrype castaneicola N. Jiang & X.L. Fan sp. nov. Figure 5.
FIGURE 5.
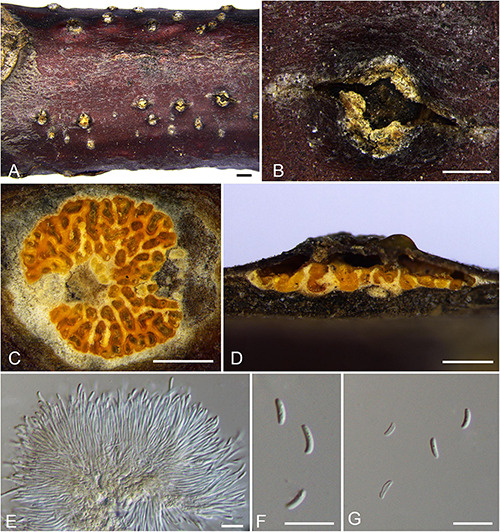
Asexual morph of Diatrype castaneicola (BJFU CF2020515, holotype). (A) Conidiomata on the host. (B) Conidioma on the host. (C) Transverse section of conidioma. (D) Longitudinal section through conidioma. (E) Conidiogenous cells. (F,G) Conidia. Scale bars: (A) = 1 mm; (B–D) = 500 μm; (E–G) = 10 μm.
MycoBank MB 837785.
Typification: CHINA. Hebei Province, Qinhuangdao City, Qinglong County, 119°11′52.25″ E 40°22′52.13″ N, 246 m msl., from branches of Castanea mollissima, 16 Oct. 2017, C.M. Tian & N. Jiang, holotype BJFU CF2020515, ex-type culture CFCC 52425. Hebei Province, Qinhuangdao City, Qinglong County, 119°11′52.25″ E 40°22′52.13″ N, 246 m msl., from branches of Castanea mollissima, 16 Oct. 2017, C.M. Tian & N. Jiang, isotype BJM 240513, ex-isotype culture CFCC 52426.
Etymology: Named after the host genus from which it was collected, Castanea.
Diagnosis: Phylogenetically, Diatrype castaneicola formed a separate clade.
Descriptions: Necrotrophic on branches of Castanea mollissima. Sexual morph: not observed. Asexual morph: Coelomycetous. Conidiomata pycnidial, immersed in the bark, scattered, erumpent slightly through the surface of bark, with multiple locules (0.7–)0.8–1.2(–1.6) mm (av. = 1.0 ± 0.2 mm, n = 10). Ectostromatic disc brown, unconspicuous, circular to ovoid. Ostiole unconspicuous, gray to black, at the same the level as the disc surface, covered by ectostroma tissue. Locules numerous, circular to ovoid, 1.0–1.4 mm (av. = 1.2 ± 0.2 mm, n = 10) in diam. Conidiogenous cells approximately cylindrical, mostly straight, discrete or integrated, arising from pseudoparenchymatous cells, hyaline, unicellular, with wide base producing conidia at the apex, holoblastic conidiogenesis (15–)18–26(–33) × 1–1.5 μm (av. = 22.5 ± 3.5 × 1.2 ± 0.2 μm, n = 30). Conidia hyaline, elongate-allantoid, slightly curved, smooth, aseptate, multiguttulate, often containing guttules per cell, 4–6 × 1–1.5 μm (av. = 5.3 ± 0.6 × 1.3 ± 0.2 μm, n = 30).
Culture characteristics: Colonies are white, dense, not produced pigmentation on PDA media. Pycnidia distributed irregularly on colony surface with yellow cream conidial drops exuding from the ostioles.
Known host and distribution: Known only on Castanea mollissima in Hebei Province, China.
Notes: Diatrype castaneicola was isolated from branches of Castanea mollissima in Hebei Province, China. Our new isolates (CFCC 52425 and CFCC 52426) grouped in Diatrype sensu stricto as a separate clade with high statistical support (MP/ML/BI = 99/100/1) (Figure 1). Diatrype castaneicola differs from the closely related one, D. stigma, by its smaller conidia (4–6 × 1–1.5 vs. 4.5–7.5 × 1–2 mm) (Rappaz, 1987). Currently, Diatrype castaneicola is reported with only the asexual morph, thus more studies are essential to report the sexual morph.
Diatrype quercicola H.Y. Zhu & X.L. Fan sp. nov. Figure 6.
FIGURE 6.

Sexual morph of Diatrype quercicola (BJFU CF2020512, holotype). (A) Ascomata on the host. (B) Ascoma on the host. (C) Transverse section of ascoma. (D) Longitudinal section through ascoma. (E,F) Ascus and ascospores. (G) Ascospores. Scale bars: (A) = 1 mm; (B–D) = 500 μm; (E–G) = 10 μm.
MycoBank MB 837786.
Typification: CHINA. Beijing City, Mentougou District, Mount Dongling, Xiaolongmen Forestry, 115°26′51.27″ E 39°58′19.62″ N, 1,267 m msl., from branches of Quercus mongolica, 21 Aug. 2017, H.Y. Zhu & X.L. Fan, holotype BJFU CF2020512, ex-type culture CFCC 52418. Beijing City, Mentougou District, Mount Dongling, Xiaolongmen Forestry, 115°26′51.27″ E 39°58′19.62″N, 1,267 m msl., from branches of Quercus mongolica, 21 Aug. 2017, H.Y. Zhu & X.L. Fan, isotype BJM 240514, ex-isotype culture CFCC 52419.
Etymology: Named after the host genus from which it was collected, Quercus.
Diagnosis: Phylogenetically, Diatrype quercicola formed a separate clade. Howeverits asci are polysporous and differ from the common 8-ascospores asci in Diatrype.
Descriptions: Necrotrophic on branches of Quercus mongolica. Sexual morph: Stromata solitary, immersed in the bark, erumpent through the surface of bark, with more than 10 perithecia arranged irregularly (2.3–)2.5–2.9 mm (av. = 2.7 ± 0.2 mm, n = 10) in diam. Ectostromatic disc brown, circular to oblong, with more than 10 ostioles arranged regularly per disc (1.5–)1.7–2.4 mm (av. = 2.0 ± 0.3 mm, n = 10) in diam. Ostioles dark brown to black, at the same level as the disc, scattered, 210–275(–335) μm (av. = 253.3 ± 22.1 μm, n = 10) in diam. Perithecia outer surface coated with yellow, powdery entostromablack, flask-shaped, with discrete perithecial necks (475–)515–620(–665) μm (av. = 566.6 ± 53.2 μm, n = 10) in diam. Asci clavate to elongate obovoid, polysporous, thin-walled, long pedicellate, apically rounded, 172–183 × (16–)20–43 μm (av. = 178 ± 3.3 × 31.7 ± 10.9 μm, n = 10). Ascospores elongate-allantoid, thin-walled, pale yellowish to pale brown at maturity, slightly curved, aseptate, multiguttulate, often containing 1–3 symmetrical guttules per cell, 17–27 × 4–6 μm (av. = 22.6 ± 2.6 × 5.4 ± 0.6 μm, n = 30). Asexual morph: not observed.
Culture characteristics: Colonies are white, irregular, reaching 9 cm after 7 days, not produced pigmentation on PDA media.
Known host and distribution: Known only on Quercus mongolica in Beijing City, China.
Additional collection examined: CHINA. Beijing City, Mentougou District, Mount Dongling, Xiaolongmen Forestry, 115°26′51.27″ E 39°58′19.62″N, 1,267 m msl., from branches of Quercus mongolica, 21 Aug. 2017, H.Y. Zhu & X.L. Fan, BJFU CF2020513, living culture CFCC 52420.
Notes: Diatrype quercicola is a unique Diatrype species isolated from Quercus mongolica in China. Asci of this species are polysporous and differ from the common 8-ascospores asci of Diatrype, including the closely related taxa, D. virescens. Moreover, D. quercicola can differ from D. virescens by larger asci and ascospres (172–183 × 20–43 vs. 35–40 × 4–6 μm; 17–27 × 4–6 vs. 12–14 × 2.5–3 μm) (Vasilyeva and Stephenson, 2005). Nevertheless, based on phylogeny analyses, this taxon appears best placed in Diatrype. Therefore, the assignment of this species to Diatrype may require reconsideration due to the taxonomic confusion around Diatrypaceae.
Diatrype albopruinosa, D. standleyi, and D. stigmaoides were also isolated from Quercus sp. (Tiffany and Gilman, 1965; Rappaz, 1987; Mendez-Mayboca et al., 2008; Vasilyeva and Stephenson, 2009). Diatrype albopruinosa and D. standleyi differ from D. quercicola by its 8-ascospores asci (Vasilyeva and Ma, 2014). Diatrype stigmaoides differs from D. quercicola by its hyaline ascospores (Vasilyeva and Stephenson, 2009).
Diatrypella (Ces. & De Not.) Nitschke, Pyr. Germ: 69, 1867.
Type: Diatrypella verruciformis (Ehrh.) Nitschke, Pyrenomyc. Germ. 1: 76, 1867 [Current name: Diatrypella favacea (Fr.)].
Known distribution: Asia, Europe, North America, Oceania, South Africa, and South America (Unamuno, 1941; Doidge, 1950; Rao, 1966; Conners, 1967; Hanlin, 1992; Crous et al., 2016).
Notes: Diatrypella was introduced by Nitschke (1867) to accommodate Diatrype sect. Diatrypella Ces. & De Not. (1863). The type species is disputable that Croxall (1950) relegated Da. verruciformis (as D. verrucaeformis) to synonymy with Da. favacea, whereas Munk (1957) recognized both species as different fungi appear to have been frequently included under Da. verruciformis. Glawe and Rogers (1984) believed Diatrypella was well distinguished genus as its well-delveloped stromata and the single host affiliation (Da. verruciformis on Alnus and Da. favacea on Betula). Further studies are needed to clarify them. Vasilyeva and Stephenson (2005) pointed out that Diatrypella morphologically resembled Cryptovalsa. Diatrypella and Cryptovalsa were mentioned as the polysporous complement of Diatrype and Eutypa (Vasilyeva and Stephenson, 2005). Nevertheless, it is still difficult to determine the differences between Diatrypella and Cryptovalsa based on morphological characters (Acero et al., 2004; Vasilyeva and Stephenson, 2005). Therefore, multilocus phylogeny including more representative taxa are needed to clarify the relationship among species in Diatrypella (Mehrabi et al., 2015).
Newly generated nine isolates show affinities to Diatrypella favacea clade based on phylogenetic analyses. Therefore, we prefer the assignment of the strains to the genus Diatrypella (Figure 1; Clade 06) preliminarily and tentatively, as it may require future reconsideration after the typification work on type species of Diatrypella.
Diatrypella betulae H.Y. Zhu & X.L. Fan sp. nov. Figure 7.
FIGURE 7.
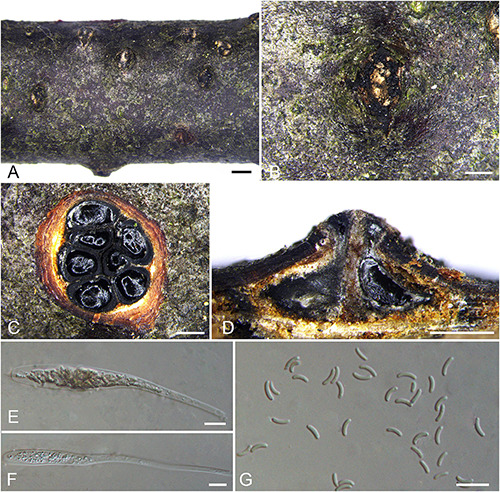
Sexual morph of Diatrypella betulae (BJFU CF2020501, holotype). (A) Ascomata on the host. (B) Ascoma on the host. (C) Transverse section of ascoma. (D) Longitudinal section through ascoma. (E) Ascus and ascospores. (F) ascus. (G) Ascospores. Scale bars: (A) = 1 mm; (B–D) = 500 μm; (E–G) = 10 μm.
MycoBank MB 837778.
Typification: CHINA. Hubei Province, Shennongjia Forest District, Shennong Stream, 110°17′51.54″ E 31°28′15.79″ N, 2273 m msl., from branches of Betula albosinensis, 17 Aug. 2017, Z. Du & Q. Yang, holotype BJFU CF2020501, ex-type culture CFCC 52406. Hubei Province, Shennongjia Forest District, Shennong Stream, 110°17′51.54″ E 31°28′15.79″ N, 2273 m msl., from branches of Betula albosinensis, 17 Aug. 2017, Z. Du & Q. Yang, isotype BJM 240507, ex-isotype culture CFCC 52404.
Etymology: Named after the host genus from which it was collected, Betula.
Diagnosis: Phylogenetically sister to Diatrypella shennongensis, differ by the number of perithecia.
Descriptions: Necrotrophic on branches of Betula albosinensis. Sexual morph: Stromata solitary, immersed in the bark, erumpent through the surface of bark, with 7–10 perithecia arranged irregularly (1.0–)1.6–2.1 mm (av. = 1.8 ± 0.2 mm, n = 10) in diam. Ectostromatic disc orange, circular to oblong, with 7–10 ostioles arranged regularly per disc, 0.8–1.3(–1.5) mm (av. = 1.0 ± 0.3 mm, n = 10) in diam. Ostioles numerous, scattered, umbilicate, sulcate, dark brown to black, at the same level as the disc (125–)175–240 μm (av. = 190.4 ± 38.0 μm, n = 10) in diam. Perithecia outer surface lacking powdery entostroma, black, flask-shaped, with long discrete perithecial necks (430–)530–830(–870) μm (av. = 680.2 ± 152.7 μm, n = 10) in diam. Asci clavate to elongate obovoid, polysporous, thin-walled, long pedicellate, apically rounded to flat, 132–140 × 7.5–10.5(–11.5) μm (av. = 136.2 ± 3 × 9.5 ± 1.4 μm, n = 10). Ascospores elongate-allantoid, thin-walled, pale yellowish to pale brown at maturity, slightly curved, smooth, aseptate (4.5–)5–7 × 1–2 μm (av. = 5.7 ± 0.5 × 1.5 ± 0.2 μm, n = 30). Asexual morph: not observed.
Culture characteristics: Cultures are flat, reaching 9 cm diam. after 7–10 days. Colonies white, rough on surface, not produced pigmentation on PDA media.
Known host and distribution: Known only on Betula albosinensis in Hubei Province, China.
Additional collection examined: CHINA. Hubei Province, Shennongjia Forest District, Shennong Stream, 110°17′51.54″ E 31°28′15.79″ N, 2,273 m msl., from branches of Betula albosinensis, 17 Aug. 2017, Z. Du & Q. Yang, BJFU CF2020502, living culture CFCC 52405.
Notes: Three strains representing Diatrypella betulae appear most closely related to Da. shennongensis reported from the same host plant Betula sp., which also clustered in a well-supported clade (MP/ML/BI = 92/100/1). In morphology of asci and ascospores, Diatrypella betulae resembles Da. shennongensis by the size of asci and ascospores (132–140 × 7.5–10.5 vs. 129–140 × 8–12; 5–7 × 1–2 vs. 5–6.5 × 1–1.5 μm), but they can be distinguished by the number of perithecia (less than 10 vs. more than 10). Moreover, Diatrypella betulae differs from Da. shennongensis based on ITS and tub2 loci (67/665 in ITS and 13/416 in tub2).
Diatrypella betulicola H.Y. Zhu & X.L. Fan sp. nov. Figure 8.
FIGURE 8.

Sexual morph of Diatrypella betulicola (BJFU CF2020505, holotype). (A) Ascomata on the host. (B) Ascoma on the host. (C) Transverse section of ascoma. (D) Longitudinal section through ascoma. (E,F) Ascus and ascospores. (G) Ascospores. Scale bars: (A) = 1 mm; (B–D) = 500 μm; (E–G) = 10 μm.
MycoBank MB 837779.
Typification: CHINA. Beijing City, Mentougou District, Mount Dongling, Xiaolongmen Forestry Centre, 115°26′51.27″ E 39°58′19.62″ N, 1,209 m msl., from branches of Betula davurica, 21 Aug. 2017, H.Y. Zhu & X.L. Fan, holotype BJFU CF2020505, ex-type culture CFCC 52411. Beijing City, Mentougou District, Mount Dongling, Xiaolongmen Forestry Centre, 115°26′51.27″ E 39°58′19.62″ N, 1,209 m msl., from branches of Betula davurica, 21 Aug. 2017, H.Y. Zhu & X.L. Fan, isotype BJM 240508.
Etymology: Named after the host genus from which it was collected, Betula.
Diagnosis: Diatrypella betulicola is different from other species of Diatrypella on host association and the size of asci and ascospores.
Descriptions: Necrotrophic on branches of Betula davurica and B. platyphylla. Sexual morph: Stromata solitary, immersed in the bark, erumpent through the surface of bark, with 7–10 perithecia arranged regularly (1.3–)1.5–1.9(–2.1) mm (av. = 1.7 ± 0.2 mm, n = 10) in diam. Ectostromatic disc brown to black, circular to oblong, with more than 10 ostioles arranged regularly per disc (0.5–)0.6–1.2(–1.4) mm (av. = 0.9 ± 0.3 mm, n = 10) in diam. Ostioles numerous, scattered, umbilicate, sulcate, dark brown to black, at the same level as the disc (145–)185–240(–270) μm (av. = 211.0 ± 31.1 μm, n = 10) in diam. Perithecia outer surface lacking powdery entostroma, black, flask-shaped, with short discrete perithecial necks (520–)600–730(–790) μm (av. = 667.8 ± 61.1 μm, n = 10) in diam. Asci clavate to elongate obovoid, polysporous, thin-walled, long pedicellate, apically flat,117–133 × 10–12 μm (av. = 124.9 ± 7.7 × 10.6 ± 0.5 μm, n = 10). Ascospores elongate-allantoid, thin-walled, pale yellowish to pale brown at maturity, slightly curved, aseptate, smooth or multiguttulate, occasionally containing one guttule per cell, 5–8 × 1–2 μm (av. = 6.6 ± 0.6 × 1.6 ± 0.2 μm, n = 30). Asexual morph: not observed.
Culture characteristics: Cultures are fluffy, reaching 9 cm after 7 days, becoming pale yellow at the margin after 2 weeks. Colonies dense with aerial mycelium at the center, sparse at the margin.
Known host and distribution: Known on Betula davurica and B. platyphylla in Beijing City, China.
Additional collection examined: CHINA. Beijing City, Mentougou District, Mount Dongling, Xiaolongmen Forestry, 115°26′51.27″ E 39°58′19.62″ N, 1,209 m msl., from branches of Betula platyphylla, 21 Aug. 2017, H.Y. Zhu & X.L. Fan, paratype BJFU CF2020506, ex-paratype culture CFCC 52412.
Notes: Diatrypella betulicola was isolated from branches of Betula davurica and Betula platyphylla in China. Phylogenetically, two strains representing Diatrypella betulicola cluster in a well-supported clade (MP/ML/BI = 95/96/1) in Diatrypella 2 clade (Figure 1). Diatrypella betulae, Da. favacea, and Da. shennongensis are the most closely related species. Diatrypella betulicola can be differentiated from Da. betulicola and Da. favacea by the number of ostioles (more than 10 ostioles in Diatrypella betulae and less than 10 ostioles in Da. betulicola and Da. favacea). Moreover, Da. betulicola can be easily distinguished from Da. shennongensis by unique size of asci (117–133 × 10–12 vs. 129–140 × 8–12 μm) and ascospore (5–8 × 1–2 vs. 5–6.5 × 1–1.5 μm). Moreover, it is different from other species of Diatrypella on host association and the size of asci and ascospores (Supplementary Table 4).
Diatrypella favacea (Fr.) Ces. & De Not., Sfer. Ital.: 29 (1863). Figure 9.
FIGURE 9.
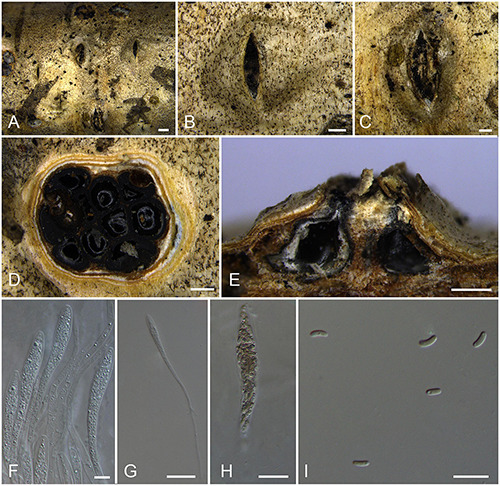
Sexual morph of Diatrypella favacea (BJFU CF2020504). (A) Ascomata on the host. (B,C) Ascoma on the host. (D) Transverse section of ascoma. (E) Longitudinal section through ascoma. (F) Asci. (G) Ascus. (H) Ascus and ascospores. (I) Ascospores. Scale bars: (A) = 1 mm; (B–E) = 500 μm; (F–I) = 10 μm.
Descriptions: Necrotrophic on branches of Betula platyphylla. Sexual morph: Stromata solitary, consisting of an inconspicuous pale yellow ectostromatic disc, immersed in the bark, erumpent through the surface of bark, with 7–10 perithecia arranged irregularly (1.9–)2.9–3.3 mm (av. = 3.1 ± 0.2 mm, n = 10) in diam. Ectostromatic disc brown to black, circular to oblong, with 5–7 ostioles arranged irregularly per disc (0.5–)1.8–2.3(–2.4) mm (av. = 2.1 ± 0.2 mm, n = 10) in diam. Ostioles numerous, gregarious, umbilicate, sulcate, dark brown to black, at the same level as the disc (230–)240–260(–280) μm (av. = 252.4 ± 16.6 μm, n = 10) in diam. Paraphyses elongate cylindrical, 118–125 × 2–3.5 μm (av. = 121.6 ± 2.7 × 2.7 ± 0.7 μm, n = 10). Perithecia outer surface lacking powdery entostroma, black, flask-shaped to spherical, with long discrete perithecial necks (690–)780–850(940) μm (av. = 821.9 ± 34.1 μm, n = 10) in diam. Asci clavate to elongate obovoid, polysporous, thin-walled, long pedicellate, apically rounded, 64–124 × (9–)9.5–12(–12.5) μm (av. = 92.7 ± 14.5 × 10.3 ± 1.7 μm, n = 10). Ascospores short-allantoid, thin-walled, pale yellowish to pale brown at maturity, slightly curved, aseptate, multiguttulate, often containing one guttulae per cell (3.5–)4–5.5(–6) × 1.5–2 μm (av. = 4.8 ± 0.5 × 1.8 ± 0.2 μm, n = 30). Asexual morph: not observed.
Culture characteristics: Cultures are white, uniform, attaining 9 cm in 7 days. Colonies sparse at the center, medium dense at the margin, rough on the surface, not produced pigmentation on PDA media.
Known host and distribution: Known on various hosts with worldwide distribution3.
Collection examined: CHINA. Xinjiang Uygur Autonomous Region, Bortala Mongol Autonomous Prefecture, Wenquan County, 81°46′22.96″ E 45°13′08.47″ N, 1,439 m msl., from branches of Betula platyphylla, July 15, 2017, C.M. Tian & R. Ma, BJFU CF2020504, living culture CFCC 52409.
Notes: Diatrypella favacea was reported to be restricted to Betula spp. in previous study (Saccardo, 1882; Glawe and Rogers, 1984), but then it got involved in the problematic species concept and delimitation with Da. verruciformis (Farr et al., 1989). Diatrypella favacea and Da. pulvinata clustered in a single clade until Hyde et al. (2020b) reported Da. yunnanensis. The strain CFCC 52409 clusters with Diatrypella favacea (CBS 198.49, DL26C, and R191) in a separate lineage. Morphologically, our strain is similar to those previously reported in terms of the size of asci (64–124 × 9.5–12 vs. 70–90 × 8–12 μm) and ascospores (4–5.5 vs. 6–8 μm) (Vasilyeva and Stephenson, 2005). The current definition of Diatrypella favacea seems to be difficult due to the lack of type material with available living culture or DNA sequence data. Thus, the current identification is preliminary and awaits further studies of typification.
Diatrypella hubeiensis H.Y. Zhu & X.L. Fan sp. nov. Figure 10.
FIGURE 10.

Sexual morph of Diatrypella hubeiensis (BJFU CF2020507, holotype). (A) Ascomata on the host. (B) Ascoma on the host. (C) Transverse section of ascoma. (D) Longitudinal section through ascoma. (E,F) ascus. (G, H) Ascospores. Scale bars: (A) = 1 mm; (B–D) = 500 μm; (E–H) = 10 μm.
MycoBank MB 837781.
Typification: CHINA. Hubei Province, Shennongjia Forest District, Tianyan Scenic Area, 110°27′36.71″ E 31°42′59.10″ N, 2,140 m msl., from branches of Betula davurica, 16 Aug. 2017, Z. Du & Q. Yang, holotype BJFU CF2020507, ex-type culture CFCC 52413. Hubei Province, Shennongjia Forest District, Tianyan Scenic Area, 110°27′36.71″ E 31°42′59.10″ N, 2,140 m msl., from branches of Betula davurica, 16 Aug. 2017, Z. Du & Q. Yang, isotype BJM 240510.
Etymology: Named after the location where it was collected, Hubei Province.
Diagnosis: Phylogenetically sister to Diatrypella yunnanensis, differ by smaller size of ascospores (6–8.5 × 1–2 vs. 18–22 × 3–4 μm).
Descriptions: Necrotrophic on branches of Betula davurica. Sexual morph: Stromata solitary, immersed in the bark, erumpent through the surface of bark, with 5–7 perithecia arranged regularly (1.4–)1.7–2.5(–3.0) mm (av. = 2.1 ± 0.4 mm, n = 10) in diam. Ectostromatic disc brown to black, circular to oblong, with 5–7 ostioles arranged irregularly per disc (0.9–)1.0–2.0(–2.6) (av. = 1.5 ± 0.4 mm, n = 10) in diam. Ostioles numerous, gregarious, umbilicate, sulcate, dark brown to black, at the same level as the disc (120–)130–170(–200) μm (av. = 143.1 ± 30.4 μm, n = 10) in diam. Perithecia outer surface lacking powdery entostroma, black, flask-shaped to spherical, with long discrete perithecial necks, 500–680(–720) μm (av. = 619.3 ± 71.3 μm, n = 10) in diam. Asci clavate to elongate obovoid, occasionally similar to an inverted volumetric flask, polysporous, thin-walled, long pedicellate, apically rounded to flat, 189–240 × 18–21 μm (av. = 213.4 ± 13.3 × 19.8 ± 0.5 μm, n = 10). Ascospores elongate-allantoid, thin-walled, slightly curved, aseptate, multiguttulate, often containing two symmetrical guttules per cell, 6–8.5(–9) × 1–2 μm (av. = 7.4 ± 0.7 × 1.6 ± 0.2 μm, n = 30). Asexual morph: not observed.
Culture characteristics: Cultures are white, fluffy, fast growing, attaining 9 cm in 7 days. Colonies dense, slightly raised with aerial mycelium, not produced pigmentation on PDA media.
Known host and distribution: Known only on Betula davurica in Hubei Province, China.
Notes: The only strain CFCC 52413 representing Diatrypella hubeiensis clusters with Da. pulvinata and Da. yunnanensis. However, Diatrypella pulvinata was introduced as an asexual fungus in Quercus garryana. Diatrypella hubeiensis differs from its closest relative Da. yunnanensis by larger asci (189–240 × 18–21 vs. 105–210 × 15–30 μm) and smaller size of ascospores (6–8.5 × 1–2 vs. 18–22 × 3–4 μm) (Hyde et al., 2020b).
Diatrypella shennongensis H.Y. Zhu & X.L. Fan sp. nov. Figure 11.
FIGURE 11.

Sexual morph of Diatrypella shennongensis (BJFU CF2020508, holotype). (A) Ascomata on the host. (B,C) Ascoma on the host. (D) Transverse section of ascoma. (E) Longitudinal section through ascoma. (F,G) Ascus and ascospores. (H) Ascospores. Scale bars: (A) = 1 mm; (B–E) = 500 μm; (F–H) = 10 μm.
MycoBank MB 837780.
Typification: CHINA. Hubei Province, Shennongjia Forest District, Shennong Ding, Shennong Camp, 110°17′28.39″ E 31°26′34.59″ N, 2,647 m msl., from branches of Betula albosinensis, 17 Aug. 2017, Z. Du & Q. Yang, holotype BJFU CF2020508, ex-type culture CFCC 52415. Hubei Province, Shennongjia Forest District, Shennong Ding, Shennong Camp, 110°17′28.39″ E 31°26′34.59″ N, 2,647 m msl., from branches of Betula albosinensis, 17 Aug. 2017, Z. Du & Q. Yang, isotype BJM 240509, ex-isotype culture CFCC 52414.
Etymology: Named after the location where it was collected, Shennong Ding.
Diagnosis: Phylogenetically, sister to Diatrypella betulae, differ by ITS and tub2 loci (67/665 in ITS and 13/416 in tub2).
Descriptions: Necrotrophic on branches of Betula albosinensis. Sexual morph: Stromata solitary, immersed in the bark, erumpent through the surface of bark, causing a pustulate bark surface, with more than 10 perithecia arranged irregularly 1.9–2.8(–3.7) mm (av. = 2.5 ± 0.6 mm, n = 10) in diam. Ectostromatic disc orange, circular to oblong, with more than 10 ostioles arranged regularly to irregularly per disc (1–)1.1–2.2(–2.6) mm (av. = 1.7 ± 0.6 mm, n = 10) in diam. Ostioles numerous, scattered, umbilicate, sulcate, dark brown to black, at the same level as the disc (160–)170–200(–215) μm (av. = 184.6 ± 5.4 μm, n = 10) in diam. Perithecia outer surface lacking powdery entostroma, black, flask-shaped to spherical, with long discrete perithecial necks (525–)630–850(–970) μm (av. = 748.4 ± 119.4 μm, n = 10) in diam. Asci clavate to elongate obovoid, polysporous, thin-walled, long pedicellate, apically rounded to flat, 129–140 × (5–)8–12 μm (av. = 135 ± 4.5 × 9.8 ± 1.7 μm, n = 10). Ascospores elongate-allantoid, thin-walled, pale yellowish to pale brown at maturity, slightly curved, smooth, aseptate (4.5–)5–6.5(–7) × 1–1.5 μm (av. = 5.8 ± 0.6 × 1.3 ± 0.2 μm, n = 30). Asexual morph: not observed.
Culture characteristics: Cultures are white, dense, uniform, fluffy, growing up to 4 cm in diam. After 3 days, and reaching 9 cm within 10 days. Colonies do not produced pigmentation on PDA media.
Known host and distribution: Known only on Betula albosinensis in Hubei Province, China.
Notes: Diatrypella shennongensis can be distinguished from its closest relative, Da. betulae, by its number of perithecia (less than 10 vs. more than 10) in one stroma and base number difference (67/665 in ITS and 13/416 in tub2). In addition, the multigene phylogenetic analyses support this species as a new species with high statistical support (MP/ML/BI = 100/100/1).
Eutypella (Nitschke) Sacc., Atti Soc. Veneto-Trent. Sci. Nat., Padua, Sér. 44: 80, 1875.
Type: Eutypella cerviculata (Fr.) Sacc., Syll. Fung. (Abellini) 1: 146, 1882.
Known distribution: Asia, Europe, North America, Oceania, South Africa, and South America (Doidge, 1950; Rappaz, 1987; Trouillas et al., 2011; Jayawardena et al., 2018).
Notes: The genus Eutypella was established by Saccardo (1875) with E. cerviculata as the type species (Saccardo, 1882). Eutypella species was mainly associated with canker diseases in Vitis vinifera (Vasilyeva and Stephenson, 2006; Trouillas et al., 2011; Luque et al., 2012). Although 251 species epithets of Eutypella have been listed in Index Fungorum (2021), the most of species are lacking DNA sequence. In phylogenetic analyses of Diatrypaceae, it showed that Eutypella was polyphyletic (Acero et al., 2004; Chacón et al., 2013; de Almeida et al., 2016; Shang et al., 2017). Thus, further studies of the taxa in Eutypella is needed.
Eutypella citricola Speg., Anales del Museo Nacional de Buenos Aires 6: 245, 1898.
Descriptions: see Trouillas et al. (2011).
Known host and distribution: Known from Citrus limon, C. sinensis, C. paradisi, Schinus molle var. areira, Ulmus procera in Australia; Vitis vinifera in United States, Morus alba in China.
Collections examined: CHINA. Jiangsu Province, Yangzhou City, 119°28′11.81″ E, 32°47′25.10″ N, 1 m msl., from branches of Morus alba, 12 Nov. 2017, C.M. Tian & N. Jiang, BJFU CF2020520, living culture CFCC 52433; ibid., BJFU CF2020521, living culture CFCC 52434.
Notes: The current strains CFCC 52433 and CFCC 52434 cluster in a well-supported clade (MP/ML/BI = 100/100/1) with Eutypella citricola (HVVIT07 and HVGRF01), residing in Allocryptovalsa/Eutypella sensu lato (Figure 1; Clade 12). In morphology, this fungus can be identified by its 3–4 sulcate ostioles, 8-spored, 55–80 × 7.5–9 μm asci and (9–)10.5–12(–13) × 2–3 μm ascospores (Trouillas et al., 2011). This study represents the first record of this species from Morus alba. Species of Allocryptovalsa represents similarity to Cryptovalsa in morphology by having polysporous asci and is different from the eight spored asci of Eutypella (Senwanna et al., 2017). Unfortunately, we did not observe any microscopic feature from premature ascostromata. Thus, the current identification is preliminary and tentative, as it requires typification before a stable species concept achieved.
Other Genera Included in Diatrypaceae
In here, we follow Hyde et al. (2020a); Wijayawardene et al. (2020) to list the genera in Diatrypaceae. A taxonomic key to distinguish 23 genera of Diatrypaceae is provided.
Allodiatrype Konta & K.D. Hyde, Mycosphere 11(1): 247, 2020.
MycoBank MB 1814.
Type: Allodiatrype arengae Konta & K.D. Hyde, Mycosphere 11(1): 249, 2020.
Known distribution: Thailand (Li et al., 2016; Konta et al., 2020).
Notes: Allodiatrype was established by Konta et al. (2020) and typified by A. arengae. In the meanwhile, A. elaeidicola, A. elaeidis, and A. thailandica were also accommodated to this genus (Konta et al., 2020). The genus shares most similarities with Diatrype, whereas they can be distinguished by the shape and size of stromata (Konta et al., 2020).
Anthostoma Nitschke, Pyrenomyc. Germ. 1: 110, 1867.
MycoBank MB 224.
Type: Anthostoma decipiens (DC.) Nitschke, Pyrenomyc. Germ. 1: 111, 1867.
Known distribution: Asia, Europe, North America, and South America (Cash, 1952; Ahmad, 1969; Farr, 1973; Rappaz, 1995).
Notes: Both sexual and asexual morphs of the type species Anthostoma decipiens were studied by Rappaz (1992, 1993). Phylogenetic analyses of sequence data also supported these results (MP/ML/BI = 100/100/1) (Rocchi et al., 2010; Jaklitsch et al., 2014). Species of Anthostoma own dark brown to dark, globose to subglobose ascomata, cylindrical, prominent ostioles, cylindrical to clavate asci with apically rounded to truncate apices and a short pedicel and brown to black-brown ascospores (Nitschke, 1867).
Cryptosphaeria Ces. & De Not., Comm. Soc. Crittog. Ital. 1(4): 231, 1863.
MycoBank MB 26092.
Type: Cryptosphaeria millepunctata Grev., Fl. Edin.: 360, 1824.
Known distribution: Asia, Europe, North America, Oceania, and South Africa (Teodoro, 1937; Trouillas et al., 2015; Jayawardena et al., 2018; Moyo et al., 2019).
Notes: Cryptosphaeria is widely accepted in Diatrypaceae (Nitschke, 1867, 1870; Rappaz, 1989; Trouillas et al., 2010b, 2015). The genus comprises corticolous species and is characterized by widely effuse and poorly developed stromata, often covered by a periderm, with separately emerging ostioles, spindle-shaped, long pedicellate asci, and sub-olivaceous to brown ascospores (Glawe, 1984; Rappaz, 1987).
Cryptovalsa Ces. & De Not. ex Fuckel, Jb. Nassau. Ver. Naturk. 23–24: 212, 1870.
MycoBank MB 1340.
Type: Cryptovalsa protracta (Pers.) De Not., Hedwigia 2: 178, 1863.
Known distribution: Asia, Europe, North America, Oceania, South Africa, and South America (Teodoro, 1937; Trouillas et al., 2010b, 2011; Jayawardena et al., 2018; Moyo et al., 2019).
Notes: Cryptovalsa was established by Cesati and De Notaris (1863) to accommodate C. protracta, C. ampelina, C. nitschkei, and C. effusa. The genus is characterized by eutypoid stromata that are rather variable, when erumpent separately diatrypelloid, often immersed in wood, but sometimes invading bark tissues. Asci are cylindrical or clavate, polysporous, with short or long pedicels. Ascospores are crowded, allantoid, and yellowish (Spooner, 1981; Vasilyeva and Stephenson, 2005). Fifty-five species epithets of Cryptovalsa are listed in Index Fungorum (2021), but only Cryptovalsa ampelina has sequence data.
Diatrypasimilis J.J. Zhou & Kohlm., Mycologia 102(2): 432, 2010.
MycoBank MB 515026.
Type: Diatrypasimilis australiensis J.J. Zhou & Kohlm., Mycologia 102(2): 432, 2010.
Known distribution: Australia (Chalkley et al., 2010).
Notes: Diatrypasimilis was established to accommodate D. australiensis as type species from mangroves based on conventional taxonomic criteria and molecular phylogeny by Chalkley et al. (2010). The genus is characterized by carbonaceous, black stromata, 8-spored, cylindrical asci, and ellipsoidal, dark brown ascospores with a germ slit.
Dothideovalsa Speg., Anal. Mus. Nac. B. Aires, Ser. 3(12): 414, 1909.
MycoBank MB 1697.
Type: Dothideovalsa tucumanensis Speg., Anal. Mus. Nac. B. Aires, Ser. 3(12): 414, 1909.
Known distribution: Argentina, Brazil, and United States (Rappaz, 1987; Hanlin, 1992).
Notes: Dothideovalsa was introduced to accommodate D. diantherae, D. eutypoides, D. tucumanensis, and D. turnerae. At present, only four species epithets of Dothideovalsa were listed in Index Fungorum (2021). However, there was no available sequence data in GenBank. Therefore, this genus is still doubtful and needs further studies.
Endoxylina Romell, Bot. Notiser 1892: 173, 1892.
MycoBank MB 1814.
Type: Endoxylina stellulata Romell, Bot. Notiser: 173 (1892).
Known distribution: Asia, Europe, and North America (Tai, 1979; Ju et al., 1996).
Notes: Endoxylina was introduced and assigned to Diatrypales (Current name: Xylariales fide Kirk et al., 2008) without assigning the familial position by Romell (1892). Based on previous morphological literature and herbarium studies, Hyde et al. (2017) transferred Endoxylina to Diatrypaceae. The concept of genus Endoxylina is rather broad, and it is characterized as having stromata of valsoid or eutypoid configurations and 8-spored, long pedicellate, asci with J-, apical ring, as well as uni- to triseptate, ascospores (Romell, 1892; Ju et al., 1996; Vasilyeva, 2010; Hyde et al., 2017). At present, 21 species epithets of Endoxylina have been described in Index Fungorum (2021), however, some of these Endoxylina species have now been transferred and synonymized with other genera (Ellis and Everhart, 1892; Wehmeyer, 1975; Rappaz, 1987; Barr, 1993). Therefore, the number of species recognized for this genus is not well-established.
Eutypa Tul. & C. Tul., Select. Fung. Carpol. (Paris) 2: 52, 1863.
MycoBank MB 1950.
Type: Eutypa lata (Pers.) Tul. & C. Tul., Select. Fung. Carpol. (Paris) 2: 56, 1863.
Known distribution: Asia, Europe, North America, Oceania, South Africa, and South America (Ahmad, 1978; Rappaz, 1987; Schmid-Heckel, 1988; Eriksson and Yue, 1998; Mulenko et al., 2008; Moyo et al., 2019).
Notes: Species of Eutypa are the causal agents of dieback of grapevine, apricots, and cherries (Trouillas and Gubler, 2004, 2010; Baumgartner et al., 2010; Camps et al., 2010; Blanco-Ulate et al., 2013; Camps et al., 2014). The genus is characterized by stromata which are irregular in shape, as confluent bumps, with conspicuous, scattered, roundish to prominent ostioles on the host surface. Asci are 8-spored, clavate, apically rounded to truncate, with indistinct apical rings and long pedicels. Ascospores are allantoid to ellipsoidal, aseptate, and pale yellowish.
Halocryptosphaeria Dayar., Devadatha, V.V. Sarma & K.D. Hyde, Mycosphere 11(1): 136, 2020.
MycoBank MB 556800.
Type: Halocryptosphaeria bathurstensis (K.D. Hyde & Rappaz) Dayar. & K.D. Hyde, Mycosphere 11(1): 136, 2020.
Known distribution: India (Dayarathne et al., 2020a).
Notes: Halocryptosphaeria was established by Dayarathne et al. (2020a) to accommodate only one species, H. bathurstensisspecies. Although Halocryptosphaeria has some morphological similarities with Cryptosphaeria, they can be easily distinguished by phylogenetical analyses.
Halocryptovalsa Dayar. & K.D. Hyde, Cryptog. Mycol. 41(3): 49, 2020.
MycoBank MB 824308.
Type: Halocryptovalsa avicenniae (Abdel-Wahab, Bahkali & E.B.G. Jones) Dayar. & K.D. Hyde, Cryptog. Mycol. 41(3): 50, 2020.
Known distribution: India, Saudi Arabia, and Thailand (Abdel-Wahab et al., 2017; Dayarathne et al., 2020a, b).
Notes: Halocryptovalsa was established by Dayarathne et al. (2020b) to accommodate species resembling Cryptovalsa from marine environments namely Cryptovalsa avicenniae and a new species Halocryptovalsa salicorniae. The genus is characterized by poorly developed stromata and poly-spored asci, with a J-, cylindrical, conspicuous apical or subapical ring. Ascospores are hyaline or yellow-brown to brown, allantoid, with small, fat globules at the end (Dayarathne et al., 2020b).
Halodiatrype Dayar. & K.D. Hyde, Phytotaxa 7(5): 617, 2016.
MycoBank MB 552254.
Type: Halodiatrype salinicola Dayar. & K.D. Hyde, Mycosphere 7(5): 617, 2016.
Known distribution: Thailand (Dayarathne et al., 2016, 2020b).
Notes: Halodiatrype was established by Dayarathne et al. (2016) to accommodate H. avicenniae and H. salinicola isolated from mangroves. The characteristic of this genus is having ascomata lacking stromatal tissues, 8-spored, cylindrical to clavate, pedicellate asci, oblong to allantoid or sub-inequilateral, larger ascospores with septa, and libertella-like asexual morphs, which can easily identify Halodiatrype from other genera in Diatrypaceae (Dayarathne et al., 2016; 2020b).
Leptoperidia Rappaz, Mycol. Helv. 2(3): 544, 1987.
MycoBank MB 25186.
Type: Leptoperidia macropunctata (Rehm) Rappaz, Mycol. Helv. 2(3): 545, 1987.
Known distribution: Congo, Mexico, Philippines (Ellis and Everhart, 1896; Rehm, 1913).
Notes: Leptoperidia was introduced to accommodate L. applanata, L. asperrima, L. macropunctata, and L. trifida (Rappaz, 1987). The genus is characterized by relatively small stroma, asci and ascospores, perithecia with very thin and slightly melanized walls (Rappaz, 1987). At present, only four species epithets of Leptoperidia are listed in Index Fungorum (2021). Sequence data are unavailable in GenBank.
Libertella Desm., Annls Sci. Nat., Bot. 19: 275, 1830.
MycoBank MB 8769.
Type: Libertella betulina Desm., Annls Sci. Nat., Bot. 19: 276, 1830.
Known distribution: Asia, Europe, North America, Oceania, South Africa, and South America (Doidge, 1950; Conners, 1967; Ahmad, 1969; Farr, 1973; Sosnowski et al., 2007; Mulenko et al., 2008).
Notes: Libertella was introduced by Desmazières (1830) to accommodate L. betulina, L. faginea, and L. rosae. This genus was mostly reported as the asexual morph of Diatrypella, however, some species were reported as the asexual morph of Eutypa, Eutypella, Diaporthe, and Polystigma (Kirk et al., 2001). The genus is characterized by subcortical, erumpent and yellow to red acervula conidiomata, and branched conidiophores that produce hyaline, 1-celled, filiform conidia (Barnett and Hunter, 1972; Sutton, 1980; von Arx, 1981).
Monosporascus Pollack & Uecker, Mycologia 66(2): 348, 1974.
MycoBank MB 3260.
Type: Monosporascus cannonballus Pollack & Uecker, Mycologia 66(2): 348, 1974.
Known distribution: Africa, Asia, Europe, North America, and South America (Pande, 2008; Sales et al., 2010; Chew-Madinaveitia et al., 2012; Salem et al., 2013; Aleandri et al., 2017).
Notes: Monosporascus was introduced by Pollack and Uecker (1974) with M. cannonballus as the type species. The genus is characterized by pyriform asci and the formation of one (rarely two) single large, sphaerical ascospores (Pollack and Uecker, 1974).
Neoeutypella M. Raza, Q.J. Shang, Phook. & L. Cai, Fungal Diversity 95: 167, 2019.
MycoBank MB 555373.
Type: Neoeutypella baoshanensis M. Raza, Q.J. Shang, Phook. & L. Cai, Fungal Diversity 95: 168, 2019.
Known distribution: China and France (Phookamsak et al., 2019).
Notes: Neoeutypella was introduced by Phookamsak et al. (2019) to accommodate two fungal strains under the name Eutypella caricae and a new strain isolated from Pinus armandii (Pinaceae). Neoeutypella is characterized by carbonaceous stromata, erumpent through host epidermis, producing yellow pigments surrounding the stroma, 8-spored, spindle-shaped asci with long pedicellate, and overlapping 1–3-seriate, allantoid, aseptate, slightly or moderately curved ascospores, with a libertella-like asexual morph (Phookamsak et al., 2019).
Pedumispora K.D. Hyde & E.B.G. Jones, Mycol. Res. 96(1): 78, 1992.
MycoBank MB 25433.
Type: Pedumispora rhizophorae K.D. Hyde & E.B.G. Jones, Mycol. Res. 96: 78, 1992.
Known distribution: Australia, India, and Thailand (Hyde and Jones, 1992; Pande, 2008; Dayarathne et al., 2020b).
Notes: Pedumispora was established by Hyde and Jones (1992) to accommodate a taxon from mangrove habitats. A phylogenetic study based on nuclear ITS and LSU regions showed that the taxonomic position of Pedumispora was in Diatrypaceae (Klaysuban et al., 2014). The genus is characterized by 8-spored, fusiform, pedicellate, unitunicate, apically truncate asci. Ascospores are filiform, mutli-septate, curved, longitudinally striate, with tapering poles, with one or both ends crook-like (Hyde and Jones, 1992).
Peroneutypa Berl., Icon. Fung. 3: 80, 1902.
MycoBank MB 3834.
Type: Peroneutypa bellula (Desm.) Berl., Icon. Fung. 3: 81, 1902.
Known distribution: Asia, Europe, South Africa, and South America (Teodoro, 1937; Moyo et al., 2018a, Castilla-Cayuman et al., 2019; Moyo et al., 2019).
Notes: Peroneutypa was introduced by Berlese (1902) to accommodate P. bellula, P. corniculata, and P. heteracantha without designating the type species. Rappaz (1987) proposed P. bellula as the type species and synonymized Peroneutypa under Eutypella. However, Carmarán et al. (2006) resurrected Peroneutypa based on morphological characters and phylogenetic. The genus is described as valsoid stromata, with packed, long prominent necks, sessile to long pedicels, small asci with truncate apices, and allantoid ascospores (Carmarán et al., 2006, 2014; Senwanna et al., 2017; Shang et al., 2017).
Quaternaria Tul. & C. Tul., Select. Fung. Carpol. 2: 104, 1863.
MycoBank MB 4632.
Type: Quaternaria persoonii Tul. & C. Tul., Select. Fung. Carpol. 2: 105, 1863.
Known distribution: Asia, Europe, and South America (Mujica and Vergara, 1945; Munk, 1957; Srinivasulu and Sathe, 1970).
Notes: Quaternaria was introduced by Tulasne and Tulasne (1863) and was typified by Q. persoonii. Clements and Shear (1931) lectotypified the illegitimate name Q. quaternata to Q. persoonii and considered Quaternaria as a synonym of Eutypella (Tulasne and Tulasne, 1863). Based on molecular phylogeny and the discussion of Gams (1994), Quaternaria was considered to be an independent genus by Acero et al. (2004). The genus is characterized by stromata was cryptosphaeroid in appearance and developed within the bark parenchyma.
Rostronitschkia Fitzp., Mycologia 11(4): 165, 1919.
MycoBank MB 4793.
Type: Rostronitschkia nervincola Fitzp., Mycologia 11(4): 166, 1919.
Known distribution: Puerto Rico and Virgin Islands (Stevenson, 1975).
Notes: Rostronitschkia was introduced to accommodate the type species R. nervincola. At present, only the type species was listed in Index Fungorum (2021). However, there were no available sequence data and strains. Therefore, this genus is still doubtful and needs to further study.
Key to genera of Diatrypaceae
-
1
Sexual morph absent………………………………………………………. 2
-
1
Sexual morph present…………………………………………………….. 3
-
2
Conidiomata acervuli, yellow to red………………….. Libertella
-
2
Conidiomata pycnidial, brownish yellow, watery, bubble-like…………………………………………………………………….. Diatrype
-
3
Ascospores globose, fusiform, or oblong to ellipsoidal…… 4
-
3
Ascospores allantoid……………………………………………………… 7
-
4
Ascospores fusiform, septate………………………. Pedumispora
-
4
Ascospores aseptate……………………………………………………….. 5
-
5
Ascospores globose, with 1–2 spores in each ascus………… ………………………………………………………………… Monosporascus
-
5
Ascospores oblong to ellipsoidal, with 8 spores in each ascus……………………………………………………………………………… 6
-
6
Ascospores with a germ slit……………………… Diatrypasimilis
-
6
Ascospores lacking a germ slit………………………. Anthostoma
-
7
Asci with more than 8 spores………………………………………… 8
-
7
Asci with 8 spores………………………………………………………….. 9
-
8
Stromata erumpent through host surface, discoid……. ……………………………………………………………………… Diatrypella
-
8
Stromata immersed in wood but sometimes invading bark tissues, eutypoid…………………………………………………………… 10
-
9
Perithecia outer surface lacking powdery entostroma………. ………………………………………………………………… Allocryptovalsa
-
9
Perithecia outer surface coated with white, powdery entostroma……………………………………………………. Cryptovalsa
-
10
Habitats marine…………………………………….. Halocryptovalsa
-
10
Habitats terrestrial……………………………………………………….. 11
-
11
Ascospores 0-1 septate…………………………………………………. 12
-
11
Ascospores uni- to triseptate………………………….. Endoxylina
-
12
Stromata semi-immersed to erumpent through the host periderm (ectostromatic)…………………………………………….. 13
-
12
Stromata deeply immersed in the host periderm (entostromatic)…………………………………………………………….. 15
-
13
Asci lacking an apical ring…………………………… Halodiatrype
-
13
Asci with J-, cylindrical, conspicuous apical ring…………. 14
-
14
Ascospores hyaline becoming yellowish at maturity…….. ……………………………………………………………………… Allodiatrype
-
14
Ascospores hyaline…………………………………………….. Diatrype
-
15
Ascomata clustered, forming valsoid configuration, breaking through entostroma by short to long necks….. 16
-
15
Ascomata scattered, arranged in linear entostroma, with short to long necks………………………………………………………. 18
-
16
Entostromata immersed in the host, with individually protruding necks at the center…………………….. Quaternaria
-
16
Entostromata slightly raised on the host, with long cylindrical, packed necks……………………………………………. 17
-
17
Hosts range wide……………………………………………. Eutypella
-
17
Host Pinus armandii…………………………………… Neoeutypella
-
18
Ascomata forming very long necks, through the host surface………………………………………………………… Peroneutypa
-
18
Ascomata forming short papilla protruding the host surface……………………………………………………………………….. 19
-
19
Peridium thin-walled, composed of a single layer of melanized cells, difficult to separate from the entostroma………………………………………………… Leptoperidia
-
19
Peridium thick-walled, composed of not one distinct layers, separating from entostroma………………………………………… 20
-
20
Asci cylindric-clavate, with pale yellow ascospores… Eutypa
-
20
Asci generally spindle-shaped, with sub-olivaceous to brown ascospores………………………………………………………… 21
-
21
Peridium compose of two distinct layers… Cryptosphaeria
-
21
Peridium compose of three distinct layers…………………… ………………………………………………………….. Halocryptosphaeria
Key to Diatrypaceae species on Betula spp.
-
1
Sexual morph absent……………………………… Diatrype betulae
-
1
Sexual morph present………………………………………………….. 2
-
2
Asci containing 8 biseriate ascospores…………………………… 3
-
2
Asci polysporous…………………………………………………………… 5
-
3
Apical ring amyloid…………………………….. Diatrype undulata
-
3
Apical ring indistinguishable………………………………………….. 4
-
4
Size of asci more than 25 μm………. Cryptosphaeria venusta
-
4
Size of asci less than 25 μm………………. Diatrype platystoma
-
5
Stromata immersed in wood but sometimes invading bark tissues, eutypoid……………………………….. Eutypella halseyana
-
5
Stromata erumpent through host surface, discoid………….. 6
-
6
Asci cylindrical………………………………… Diatrypella favacea
-
6
Asci clavate to spindle shaped………………………………………… 7
-
7
More than 10 perithecia arranged irregularly………. ……………………………………………….. Diatrypella shennongensis
-
7
Less than 10 perithecia arranged regularly……………………. 8
-
8
More than one host………………………… Diatrypella betulicola
-
8
Only one host…………………………………………………………………. 9
-
9
Length of asci less than 140 μm……………………… Diatrypella betulae
-
9
Length of asci more than 140 μm………………………………. ……………………………………………………… Diatrypella hubeiensis
Discussion
In this study, 21 isolates were identified as diatrypaceous fungi from Betula albosinensis, B. davurica, and B. platyphylla (Betulaceae), Castanea mollissima and Quercus mongolica (Fagaceae), Juglans regia (Juglandaceae), and Morus alba (Moraceae). Morpho-molecular analyses confirmed that these strains belong in four genera (viz. Allocryptovalsa, Diatrype, Diatrypella, and Eutypella) including nine novel species (viz. Allocryptovalsa castaneae, A. castaneicola, Diatrype betulae, D. castaneicola, D. quercicola, Diatrypella betulae, Da. betulicola, Da. hubeiensis, and Da. shennongensis) and two known species (viz. Diatrypella favacea and Eutypella citricola). Eutypella citricola was reported from Morus host for the first time.
The generic concepts of Diatrypaceae have been unstable, thus many species were transferred from one genus to another (Phookamsak et al., 2019; Konta et al., 2020). The current study revised the Diatrypaceae and accepted 23 genera in this family (Supplementary Table 1). However, there only exist 18 genera in current phylogenetic analyses due to availability of molecular data (Figure 1). In China, 11 genera and 62 species belong in Diatrypaceae have been recorded (Supplementary Table 5). However, 40 species (66.67%) do not have molecular data until now.
Birch is of high economic, medicinal, and ornamental value. Six species of Diatrypaceae were recorded from Betula spp. with DNA sequences in the current study, including Diatrype betulae, Diatrypella betulae, Da. betulicola, Da. favacea, Da. hubeiensis, and Da. shennongensis. All species of Diatrypella 2 clade were isolated from Betula spp., except for Da. pulvinata and Da. yunnanensis isolated from an unidentified plant. It showed that many Diatrypella species may have obvious host specificity. The other four species (viz. Cryptosphaeria venusta, Diatrype platystoma, D. undulata, and Eutypella halseyana) of Diatrypaceae were recorded from Betula spp. in China but no materials with DNA sequences, including Cryptosphaeria venusta, Diatrype platystoma, D. undulata, and Eutypella halseyana (Teng, 1996; Vasilyeva and Ma, 2014). A morphological key was provided to separate them in the current study.
The Allocryptovalsa species clustered within the same clade as Eutypella species in our phylogenetic analyses (Figure 1; Clade 12: Allocryptovalsa/Eutypella sensu lato). Allocryptovalsa was introduced by Senwanna et al. (2017) which resembles Cryptovalsa in morphology by having polysporous asci different from the 8-spored asci of Eutypella. The number of ascospores per ascus (eight spores vs. multiple spores) has been used traditionally to differentiate the genera of Diatrypaceae (Diatrype vs. Diatrypella and Cryptovalsa vs. Eutypella). However, the recent studies indicated that the polysporous ascus feature maybe not significant in Diatrypaceae (Acero et al., 2004; Trouillas et al., 2011; Chacón et al., 2013; Liu et al., 2015). A thorough revision is needed to resolve the problematic situation of Diatrypaceae, which includes a mass of misidentified genus/species, as a result of the unstable phylogenetic frame with type materials. Therefore, it seems to better if the future work could treate the strains from clade 12 into one genus Allocryptovalsa.
The strains of Diatrype formed a clade with high support values (Figure 1; Clade 09: Diatrype sensu stricto). However, in this clade, some strains of Diatrypella (Da. quercina, Da. iranensis, and Da. macrospora) were placed between Diatrype species. Acero et al. (2004) reported that Cryptosphaeria, Diatrype, Diatrypella, Eutypa, and Eutypella were polyphyletic and confused probably due to lack of tub2 gene sequences or misidentified species. However, these five genera are still polyphyletic within the family from previous studies (de Almeida et al., 2016; Shang et al., 2017; Mehrabi et al., 2019; Dayarathne et al., 2020a, b; Konta et al., 2020) based on the ITS and tub2 sequences data. Allodiatrype and Halodiatrype reported their morphological resemblance to Diatrype (Dayarathne et al., 2016; Konta et al., 2020). However, asci of Halodiatrype lack an apical ring, the asci of Allodiatrype and Diatrype have J-, cylindrical, conspicuous apical ring (Dayarathne et al., 2016; Maharachchikumbura et al., 2016; Konta et al., 2020). Additionally, Diatrype can be differentiated from Allodiatrype by the color of ascospores. The ascospores of Diatrype are hyaline becoming yellowish ascospores at maturity, whereas the ascospores of Allodiatrype are hyaline (Maharachchikumbura et al., 2016; Konta et al., 2020). Though Diatrype and Diatrypella are not polyphyletic any more, some strains of Diatrype and Diatrypella species still need further study.
In some cases, some strains of Diatrype species (D. brunneospora and D. palmicola), Diatrypella species (Da. banksiae), Eutypa species (E. flavovirens and E. guttulata) and Eutypalla species (E. cearensis) formed distinct lineages within Diatrypaceae (Figure 1; Incertae sedis). Diatrypella banksiae is closely related to the genus Neoeutypella with high support values from the phylogenetic analyses (MP/ML/BI = 98/100/1) (Figure 1), which produced an asexual morph (Crous et al., 2016; Phookamsak et al., 2019). Nevertheless, Diatrypella banksiae having spindle-shaped conidia can be easily differentiated from Neoeutypella baoshanensis having filiform conidia (Crous et al., 2016; Phookamsak et al., 2019). In the future study, sexual morph of Diatrypella banksiae also remains to be studied, because the asexual morphs of Diatrypaceae are not generally useful in separating species (Glawe and Rogers, 1982, 1984; Rappaz, 1987; Konta et al., 2020). Therefore, Diatrypella banksiae probably belongs to the genus Neoeutypella. Further study of the relationship between Neoeutypella and Diatrypella banksiae is needed.
Eutypa guttulata is closely related to the genus Halodiatrype and Pedumispora from the phylogenetic analyses (Figure 1) as a basal branch. Eutypa guttulata can be distinguished by fusiform ascospores, whereas Pedumispora has allantoid ascospores (Hyde and Jones, 1992). It is also different from Halodiatrype because of lacking an apical ring (Dayarathne et al., 2016). Diatrype brunneospora is closed to Eutypa guttulata (Figure 1), which is morphologically similar to members of Eutypa spp. (Trouillas et al., 2010b). Therefore, the assignment of this isolate to the genus Diatrype may require reconsideration in the future. Eutypa flavovirens is closely related to the genus Cryptosphaeria with no support (Figure 1). Diatrypella can be distinguished by hyaline to subhyaline, rarely pale olivaceous ascospores, whereas Diatrype palmicola has pale yellowish to pale brown ascospores at maturity (Liu et al., 2015). For those species, the assignment of these isolates still remain unclear, which may require reconsideration in the future.
Eutypa microasca appeared in a strongly supported clade along with two Peroneutypa species with fusiform asci in Figure 1 (Clade 19: Peroneutypa). Carmarán et al. (2006) suggested that the morphology of the ascus could explain the phylogenetic relationships within Diatrypaceae better than stromata, although our study indicates that it is not entirely supported in Peroneutypa. The phylogenetic signal of the ascus shape and the phylogenetic placement of Eutypa microasca should be further tested in future study.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
XF and CT conceived and designed the experiments. HZ, MP, and RM performed the experiment. HZ and MP analyzed the data. NW and DD provided some materials and polished the language. HZ wrote the manuscript. XF revised and approved the final version of the manuscript. All authors contributed extensively to the work presented in the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors want to thank the Experimental Teaching Centre (College of Forestry, Beijing Forestry University) for providing installed scientific equipment during the whole process. NW and DD thank Province Universities of the Diversity and Ecological Adaptive Evolution for Animals and Plants on the Yun-Gui Plateau, the National Natural Science Foundation of China, and the Thousand Talents Plan, Youth Project of Yunnan Provinces. NW gratefully acknowledges the State Key Laboratory of Functions and Applications of Medicinal Plants, Guizhou Medical University; Science and Technology Department of Guizhou Province.
Funding. This study was financed by the National Natural Science Foundation of China (No. 31670647) and the Fundamental Research Funds for the Central Universities (No. 2019ZY23).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.646262/full#supplementary-material
Placement of genera in Diatrypaceae by different authors.
Isolates and GenBank accession numbers used in the phylogenetic analyses of Diatrypaceae.
Synopsis of species of Allocryptovalsa.
Synopsis of species of Diatrypella from Betula spp.
Distribution of diatrypaceous fungi on plants in China.
References
- Abdel-Wahab M. A., Dayarathne M. C., Suetrong S., Guo S. Y., Alias S. A., Bahkali A. H., et al. (2017). New saprobic marine fungi and a new combination. Bot. Mar. 60 469–488. 10.1515/bot-2016-0118 [DOI] [Google Scholar]
- Abdel-Wahab M. A., Hodhod M. S., Bahkali A. H. A., Jones E. B. G. (2014). Marine fungi of Saudi Arabia. Bot. Mar. 57 323–335. 10.1515/bot-2014-0010 [DOI] [Google Scholar]
- Acero F. J., González V., Sánchez-Ballesteros J., Rubio V., Checa J., Bills G. F., et al. (2004). Molecular phylogenetic studies on the Diatrypaceae based on rDNA-ITS sequences. Mycologia 96 249–259. 10.2307/3762061 [DOI] [PubMed] [Google Scholar]
- Ahmad S. (1969). Fungi of West Pakistan. Biological society of Pakistan monograph. supplement I. Biol. Lahore 5 1–110. [Google Scholar]
- Ahmad S. (1978). Ascomycetes of Pakistan, Part I. Biol. Soc. Pakistan Monogr. 7 1–236. [Google Scholar]
- Aleandri M. P., Martignoni D., Reda R., Alfaro-fernandez A., Font M. I., Armengol J., et al. (2017). Involvement of Olpidium bornovanus and O. virulentus in the occurence of melon root rot and vine decline caused by Monosporascus cannonballus in Central Italy. J. Plant Pathol. 99 169–176. 10.4454/jpp.v99i1.3787 32896216 [DOI] [Google Scholar]
- Barnett H. L., Hunter B. B. (1972). llustrated Genera of Imperfect Fungi, 3rd Edn. Minneapolis, MN: Burgess Publishing Co. 241. [Google Scholar]
- Barr M. E. (1993). Redisposition of some taxa described by JB Ellis. Mycotaxon 46 45–76. [Google Scholar]
- Baumgartner K., Bergemann S. E., Fujiyoshi P., Rolshausen P. E., Gubler W. D. (2010). Microsatellite markers for the grapevine pathogen, Eutypa lata. Mol. Ecol. Resour. 9 222–224. 10.1111/j.1755-0998.2008.02405.x [DOI] [PubMed] [Google Scholar]
- Blanco-Ulate B., Rolshausen P. E., Cantu D. (2013). Draft genome sequence of the grapevine dieback fungus Eutypa lata UCR-EL1. Genome Announc. 1:e00228-13. 10.1128/genomeA.00228-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps C., Kappel C., Lecomte P., Léon C., Coutos-Thevenot P., Delrot S., et al. (2014). Identification of grapevine marker genes for early, non-destructive Eutypa lata infection diagnosis. Plant Pathol. 63 323–333. 10.1111/ppa.12101 [DOI] [Google Scholar]
- Camps C., Kappel C., Lecomte P., Léon C., Gomés E., Coutos-Thévenot P., et al. (2010). A transcriptomic study of grapevine (Vitis vinifera cv. Cabernet-Sauvignon) interaction with the vascular ascomycete fungus Eutypa lata. J. Exp. Bot. 61 1719–1737. 10.1093/jxberq040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmarán C. C., Robles C. A., D’Jonsiles M. F., Ceriani E., Español E., López S., et al. (2014). Clave para la identificación de especies de la familia Diatrypaceae (Xylariales, Ascomycota). Lilloa Fundación Miguel Lillo Tucumán Argentina 51 20–32. [Google Scholar]
- Carmarán C. C., Romero A. I., Giussani L. M. (2006). An approach towards a new phylogenetic classification in Diatrypaceae. Fungal Divers. 23 67–87. [Google Scholar]
- Caroll F. E., Müller E., Sutton B. C. (1977). Prelirninary studies on the incidence of needle endophytes in sorne European conifers. Sydowia 29 87–103. [Google Scholar]
- Cash E. K. (1952). A Record of the Fungi Named by J.B. Ellis. Part I, Acanthostigma-Gymnosporangium. USA. Beltsville, MD: United States Department Of Agriculture. [Google Scholar]
- Castilla-Cayuman A., Lolas M., Diaz G. A. (2019). First report of Peroneutypa scoparia causing cane dieback in kiwifruit in Chile. Plant Dis. 103:373. 10.1094/PDIS-06-18-1092-PDN [DOI] [Google Scholar]
- Catal M., Jordan S. A., Butterworth S. C., Schilder A. M. C. (2007). Detection of Eutypa lata and Eutypella vitis in grapevine by nested multiplex polymerase chain reaction. Phytopathology 97 737–747. 10.1094/PHYTO-97-6-0737 [DOI] [PubMed] [Google Scholar]
- Cesati V., De Notaris G. (1863). Schema di classificazione degli sferiacei italici aschigeri: più omeno appartenenti al genere Sphaerianell’antico significato attribuitogli da Persoon. Commentario Soc. Crittogamol. Ital. 1 177–240. [Google Scholar]
- Chacón S., Dorge D., Weisenborn J., Piepenbring M. (2013). A new species and a new record of Diatrypaceae from Panama. Mycologia 105 681–688. 10.3852/12-131 [DOI] [PubMed] [Google Scholar]
- Chalkley D. B., Suh S. O., Volkmann-Kohlmeyer B., Zhou J. J. (2010). Diatrypasimilis australiensis, a novel xylarialean fungus from mangrove. Mycologia 102 430–437. 10.3852/09-142 [DOI] [PubMed] [Google Scholar]
- Chew-Madinaveitia Y. I., Gaytan-Mascorro A., Herrera-Perez T. (2012). First report of Monosporascus cannonballus on melon in Mexico. Plant Dis. 96:1068. 10.1094/PDIS-02-12-0180-PDN [DOI] [PubMed] [Google Scholar]
- Chlebicki A. (2005). Some species of the genus Diatrype from the Czech Republic preserved in PRM, BRNM and KRAM. Czech Mycol. 57 117–138. 10.33585/cmy.57107 [DOI] [Google Scholar]
- Clements F. E., Shear C. L. (1931). Genera of Fungi. New York, NY: Hafner Publishing Company. [Google Scholar]
- Conners I. L. (1967). An Annotated Index of Plant Diseases in Canada and Fungi Tecorded on Plants in Alaska, Canada and Greenland, Canada. Ottawa, Ont: Canadian Department of Agriculture. [Google Scholar]
- Crous P. W., Gams W., Stalpers J. A., Robert V., Stegehuis G. (2004). MycoBank: an online initiative to launch mycology into the 21st century. Stud. Mycol. 50 19–22. [Google Scholar]
- Crous P. W., Wingfield M. J., Burgess T. I., St. Hardy J., Crane C., Barrett S., et al. (2016). Fungal planet description sheets: 469–557. Persoonia 37 218–403. 10.3767/003158516X694499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croxall H. E. (1950). Studies on British pyrenomycetes III. The British species of the genus Diatrypella Cesati & de Notaris. Trans. Br. Mycol. Soc. 33 45–72. [Google Scholar]
- Dayarathne M. C., Jones E. B. G., Maharachchikumbura S. S. N., Devadatha B., Sarma V. V., Khongphinitbunjong K., et al. (2020a). Morpho-molecular characterization of microfungi associated with marine based habitats. Mycosphere 11 1–188. 10.5943/mycosphere/11/1/1 [DOI] [Google Scholar]
- Dayarathne M. C., Phookamsak R., Hyde K. D., Manawasinghe I. S., Toanun C., Jones E. B. G. (2016). Halodiatrype, a novel diatrypaceous genus from mangroves with H. salinicola and H. avicenniae spp. nov. Mycosphere 7 612–627. 10.5943/mycosphere/7/5/7 [DOI] [Google Scholar]
- Dayarathne M. C., Wanasinghe D. N., Devadatha B., Abeywickrama P., Gareth J. E. B., Chomnunti P., et al. (2020b). Modern taxonomic approaches to identifying diatrypaceous fungi from marine habitats, with a novel genus Halocryptovalsa Dayarathne & K.D. Hyde, gen. nov. Cryptogam. Mycol. 41 21–67. 10.5252/cryptogamie-mycologie2020v41a3 33311142 [DOI] [Google Scholar]
- de Almeida D. A. C., Gusmão L. F. P., Miller A. N. (2016). Taxonomy and molecular phylogeny of Diatrypaceae (Ascomycota, Xylariales) species from the Brazilian semi-arid region, including four new species. Mycol. Prog. 15 1–27. 10.1007/s11557-016-1194-8 [DOI] [Google Scholar]
- de Errasti A., Carmarán C., Novas M. V. (2010). Diversity and significance of fungal endophytes from living stems of naturalized trees from Argentina. Fungal Divers. 41 29–40. 10.1007/s13225-009-0012-x [DOI] [Google Scholar]
- Desmazières J. B. H. J. (1830). Monographie du genre N aemaspora des auteurs modernes, et du genre Libertella. Ann. Sci. Nat. 19 269–279. [Google Scholar]
- Doidge E. M. (1950). The South African fungi and lichens to the end of 1945. Bothalia 5 1–1094. [Google Scholar]
- Doyle J. J., Doyle J. L. (1990). Isolation of plant DNA from fresh tissue. Focus 12 13–15. [Google Scholar]
- Ellis J. B., Everhart B. M. (1892). The North American Pyrenomycetes. A Contribution to Mycologic Botany. Newfield, NJ: Ellis & Everhart. [Google Scholar]
- Ellis J. B., Everhart B. M. (1896). New species of tropical fungi. Bull. Lab. Nat. Hist. State Univ. Iowa 4 67–72. [Google Scholar]
- Eriksson O. E., Yue J. Z. (1998). Bambusicolous pyrenomycetes, an annotated check-list. Myconet 1 25–78. [Google Scholar]
- Fan X. L., Bezerra J. D. P., Tian C. M., Crous P. W. (2018). Families and genera of diaporthalean fungi associated with canker and dieback of tree hosts. Persoonia 40 119–134. 10.3767/persoonia.2018.40.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X. L., Bezerra J. D. P., Tian C. M., Crous P. W. (2020). Cytospora (Diaporthales) in China. Persoonia 45 1–45. 10.3767/persoonia.2020.45.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr D. F., Bills G. F., Chamuris G. P., Rossman A. Y. (1989). Fungi on Plants and Plant Products in the United States. St Paul, MN: American Phytopathological Society. [Google Scholar]
- Farr M. L. (1973). An Annotated List of Spegazzini’s Fungus Taxa 1. Michigan, IN: J. Cramer. [Google Scholar]
- Fries E. M. (1849). Summa vegetabilium Scandinaviae. Sect. Posterior 2 259–572. [Google Scholar]
- Gams W. (1994). Report of the committee for fungi: 4. Taxon 43 265–267. [Google Scholar]
- Glass N. L., Donaldson G. C. (1995). Development of primer sets designed for use with the PCR to amplify conserved genes from flamentous ascomycetes. Appl. Environ. Microbiol. 61 1323–1330. 10.1002/bit.260460112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glawe A., Jacobs K. A. (1987). Taxonomic notes on Eutypella vitis, Cryptosphaeria populina, and Diatrype stigma. Mycologia 79 135–139. [Google Scholar]
- Glawe D. A. (1984). Cryptosphaeria pullmanensis, a new species from Washington state. Mycologia 76 166–169. [Google Scholar]
- Glawe D. A., Rogers J. D. (1982). Observations on the anamorphs of six species of Diatrype and Diatrypella. Can. J. Bot. 60 245–251. [Google Scholar]
- Glawe D. A., Rogers J. D. (1984). Diatrypaceae in the Pacific Northwest. Mycotaxon 20 401–460. [Google Scholar]
- Glawe D. A., Rogers J. D. (1986). Conidial states of some species of Diatrypaceae and Xylariaceae. Can. J. Bot. 64 1493–1498. [Google Scholar]
- Goos R. D. (2010). The mycota of Rhode Island: a checklist of the fungi recorded in Rhode Island (including lichens and myxomycetes). R. I. Nat. Hist. Surv. 4:222. [Google Scholar]
- Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W., Gascuel O., et al. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59 307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Hanlin R. T. (1992). Index to genera and species of ascomycetes described by A. P. Viegas. Mycotaxon 43 207–230. [Google Scholar]
- Hillis D. M., Bull J. J. (1993). An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 42 182–192. [Google Scholar]
- Hyde K. D., Dong Y., Phookamsak R., Jeewon R., Jeewon R., Bhat D. J., et al. (2020a). Fungal diversity notes 1151–1276: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 100 5–277. 10.1007/s13225-020-00439-5 [DOI] [Google Scholar]
- Hyde K. D., Jones E. B. G. (1992). Intertidal mangrove fungi: Pedumispora gen. nov. (Diaporthales). Mycol. Res. 96 78–80. [Google Scholar]
- Hyde K. D., Norphanphoun C., Abreu V. P., Bazzicalupo A., Chethana K. W. T., Clericuzio M., et al. (2017). Fungal diversity notes 603–708: taxonomic and phylogenetic notes on genera and species. Fungal Divers. 87 1–235. 10.1007/s13225-017-0391-3 [DOI] [Google Scholar]
- Hyde K. D., Norphanphoun C., Maharachchikumbura S. S. N., Bhat D. J., Jones E. B. G., Bundhun D., et al. (2020b). Refined families of Sordariomycetes. Mycosphere 11 305–1059. 10.5943/mycosphere/11/1/7 [DOI] [Google Scholar]
- Hyde K. D., Tennakoon D. S., Jeewon R., Bhat D. J., Maharachchikumbura S. S. N., Rossi W., et al. (2019). Fungal diversity notes 1036–1150: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 96 1–242. 10.1007/s13225-019-00429-2 [DOI] [Google Scholar]
- Jaklitsch W. M., Fournier J., Rogers J. D., Voglmayr H. (2014). Phylogenetic and taxonomic revision of Lopadostoma. Persoonia Mol. Phylogeny Evol. Fungi 32 52–82. 10.3767/003158514X679272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayawardena R. S., Purahong W., Zhang W., Wubet T., Li X. H., Liu M., et al. (2018). Biodiversity of fungi on Vitis vinifera L. revealed by traditional and high-resolution culture-independent approaches. Fungal Divers. 90 1–84. 10.1007/s13225-018-0398-4 [DOI] [Google Scholar]
- Ju Y. M., Rogers J. D., Huhndorf S. M. (1996). Valsaria and notes on Endoxylina, Pseudothyridaria, Pseudovalsaria, and Roussoella. Mycotaxon 58 419–481. [Google Scholar]
- Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk P. M., Cannon P. F., David J. C., Stalpers J. A. (2001). Ainsworth and Bisby’s Dictionary of the Fungi, 9th Edn. London: CABI Publishing. [Google Scholar]
- Kirk P. M., Cannon P. F., Minter D. W., Stalper J. A. (2008). Ainsworth and Bisby’s Dictionary of the Fungi, 10th Edn. London: CAB International. [Google Scholar]
- Klaysuban A., Sakayaroj J., Jones E. G. (2014). An additional marine fungal lineage in the Diatrypaceae, Xylariales: Pedumispora rhizophorae. Bot. Mar. 57 413–420. 10.1515/bot-2014-0017 [DOI] [Google Scholar]
- Konta S., Maharachchikumbura S. S. N., Senanayake I. C., Mckenzie E. H. C., Stadler M., Boonmee S., et al. (2020). A new genus Allodiatrype, five new species and a new host record of diatrypaceous fungi from palms (Arecaceae). Mycosphere 11 239–268. 10.5943/mycosphere/11/1/4 27409875 [DOI] [Google Scholar]
- Lardner R., Stummer B. E., Sosnowski M. R., Scott E. S. (2005). Molecular identification and detection of Eutypa lata in grapevine. Mycol. Res. 109 799–808. 10.1017/S0953756205002893 [DOI] [PubMed] [Google Scholar]
- Lawrence D. P., Holland L. A., Nouri M. T., Travadon R., Trouillas F. P. (2018). Molecular phylogeny of Cytospora species associated with canker diseases of fruit and nut crops in California, with the descriptions of ten new species and one new combination. IMA Fungus 9 333–369. 10.5598/imafungus.2018.09.02.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G. J., Hyde K. D., Zhao R. L., Hongsanan S., Abdel-Aziz F. A., Abdel-Wahab M. A., et al. (2016). Fungal diversity notes 253–366: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 78 1–237. 10.1007/s13225-016-0366-9 [DOI] [Google Scholar]
- Liu J. K., Hyde K. D., Jones E. B. G., Ariyawansa H. A., Bhat D. J., Boonmee S., et al. (2015). Fungal diversity notes 1–110: taxonomic and phylogenetic contributions to fungal species. Fungal Divers. 72 1–197. 10.1007/s13225-015-0324-y [DOI] [Google Scholar]
- Luque J., Garcia-Figueres F., Legorburu F. J., Muruamendiaraz A., Armengol J., Trouillas F. P. (2012). Species of Diatrypaceae associated with grapevine trunk diseases in Eastern Spain. Phytopathol. Mediterr. 51 528–540. [Google Scholar]
- Luque J., Sierra D., Torres E., Garcia F. (2006). Cryptovalsa ampelina on grapevines in NE Spain: identification and pathogenicity. Phytopathol. Mediterr. 45 101–109. 10.1520/STP37480S 33159212 [DOI] [Google Scholar]
- Ma R., Zhu Y. F., Fan X. L., Tian C. M. (2016). Canker disease of willow and poplar caused by Cryptosphaeria pullmanensis recorded in China. For. Pathol. 46 327–335. 10.1111/efp.12261 [DOI] [Google Scholar]
- Maharachchikumbura S. S. N., Hyde K. D., Jones E. B. G., McKenzie E. H. C. (2015). Towards a natural classification and backbone tree for Sordariomycetes. Fungal Divers. 72 199–301. 10.1007/s13225-015-0331-z [DOI] [Google Scholar]
- Maharachchikumbura S. S. N., Hyde K. D., Jones E. B. G., McKenzie E. H. C., Bhat D. J., Dayarathne M. C., et al. (2016). Families of Sordariomycetes. Fungal Divers. 79 1–317. 10.1007/s13225-016-0369-6 [DOI] [Google Scholar]
- Mayorquin J. S., Wang D. H., Twizeyimana M., Eskalen A. (2016). Identification, distribution, and pathogenicity of Diatrypaceae and Botryosphaeriaceae associated with Citrus branch canker in the Southern California desert. Plant Dis. 100 2402–2413. 10.1094/PDIS-03-16-0362-RE [DOI] [PubMed] [Google Scholar]
- Mehrabi M., Bita A., Roghayeh H. (2019). Two new species of Eutypella and a new combination in the genus Peroneutypa (Diatrypaceae). Mycol. Prog. 18 1057–1069. 10.1007/s11557-019-01503-4 [DOI] [Google Scholar]
- Mehrabi M., Hemmati R., Vasilyeva L. N., Trouillas F. P. (2015). A new species and a new record of Diatrypaceae from Iran. Mycosphere 6 60–68. 10.5943/mycosphere/6/1 [DOI] [Google Scholar]
- Mehrabi M., Hemmati R., Vasilyeva L. N., Trouillas F. P. (2016). Diatrypella macrospora sp. nov. and new records of diatrypaceous fungi from Iran. Phytotaxa 252 43–55. 10.11646/phytotaxa.252.1.4 [DOI] [Google Scholar]
- Mendez-Mayboca F. R., Chacon S., Esqueda M., Coronado M. L. (2008). Ascomycetes of Sonora, Mexico. 1: the Ajos-Bavispe National forest reserve and wildlife refuge. Mycotaxon 103 87–95. [Google Scholar]
- Moyo P., Damm U., Mostert L., Halleen F. (2018a). Eutypa, Eutypella, and Cryptovalsa species (Diatrypaceae) associated with Prunus species in South Africa. Plant Dis. 102 1402–1409. 10.1094/PDIS-11-17-1696-RE [DOI] [PubMed] [Google Scholar]
- Moyo P., Mostert L., Halleen F. (2019). Diatrypaceae species overlap between vineyards and natural ecosystems in South Africa. Fungal Ecol. 39 142–151. 10.1016/j.funeco.2018.11.015 [DOI] [Google Scholar]
- Moyo P., Mostert L., Spies C. F. J., Damm U., Halleen F. (2018b). Diversity of Diatrypaceae species associated with dieback of grapevines in South Africa, with the description of Eutypa cremea sp. nov. Plant Dis. 102 220–230. 10.1094/PDIS-05-17-0738-RE [DOI] [PubMed] [Google Scholar]
- Mujica F., Vergara C. (1945). Flora Fungosa Chilena. Indice Preliminar de los Huéspedes de los Hongos Chilenos y sus Referencias Bibliográficas. Santiago: Ministerio de Agricultura. [Google Scholar]
- Mulenko W., Majewski T., Ruszkiewicz-Michalska M. (2008). A Preliminary Checklist of Micromycetes in Poland. Kraków: W. Szafer Institute of Botany, Polish Academy of Sciences. [Google Scholar]
- Munk A. (1957). Danish pyrenomycetes. A preliminary flora. Dan. Bot. Arkiv 17 1–491. [Google Scholar]
- Nitschke T. (1867). Pyrenomycetes Germanici 1. Breslau: E. Trewendt. [Google Scholar]
- Nitschke T. (1870). Pyrenomycetes Germanici 2. Breslau: E. Trewendt. [Google Scholar]
- Norphanphoun C., Doilom M., Daranagama D. A., Phookamsak R., Wen T. C., Bulgakov T. S., et al. (2017). Revisiting the genus Cytospora and allied species. Mycosphere 8 51–97. 10.5943/mycosphere/8/1/7 [DOI] [Google Scholar]
- O’Donnell K., Cigelnik E. (1997). Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol. Phylogenet. Evol. 7 103–116. [DOI] [PubMed] [Google Scholar]
- Pande A. (2008). Ascomycetes of Peninsular India. Jodhpur: Scientific Publishers. [Google Scholar]
- Phookamsak R., Hyde K. D., Jeewon R., Bhat D. J., Jones E. B. G., Maharachchikumbura S. S. N., et al. (2019). Fungal diversity notes 929–1035: taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Divers. 95 1–273. 10.1007/s13225-019-00421-w [DOI] [Google Scholar]
- Pollack F. G., Uecker F. A. (1974). Monosporascus cannonballus an unusual ascomycete in cantaloupe roots. Mycologia 66 346–349. [Google Scholar]
- Posada D., Crandall K. A. (1998). Modeltest: testing the model of DNA substitution. Bioinformatics 14 817–818. [DOI] [PubMed] [Google Scholar]
- Rannala B., Yang Z. (1996). Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. J. Mol. Evol. 43 304–311. 10.1007/pl00006090 [DOI] [PubMed] [Google Scholar]
- Rao R. (1966). Some additions to fungi of India. I. Mycopathol. Mycol. Appl. 28 45–48. [DOI] [PubMed] [Google Scholar]
- Rappaz F. (1987). Taxonomie et nomenclatijre des Diatrypaceae à asques octospores. Mycol. Helv. 2 285–648. [Google Scholar]
- Rappaz F. (1989). Proposal to conserve Cryptosphaeria Ces. & De Not. (1863) against Cryptosphaeria Grev. (1822) (Ascomycotina). Taxon 38:664. 10.2307/1222663 [DOI] [Google Scholar]
- Rappaz F. (1992). Anthostoma decipiens et sa position systématique. Mycol. Helv. 5 21–32. [Google Scholar]
- Rappaz F. (1993). Germination conidienne des Diatrypacées: rôle du substrat et de l’inoculum misen évidence chez Anthostoma decipiens. Sydowia 44 294–306. [Google Scholar]
- Rappaz F. (1995). Anthostomella and related xylariaceous fungi on hard wood from Europe and North America. Mycol. Helv. 7 99–168. [Google Scholar]
- Rayner R. W. (1970). A Mycological Colour Chart. London: Commonwealth Mycological Institute & British Mycological Society. [Google Scholar]
- Rehm H. (1913). Ascomycetes philippinenses IV. Leafl. Philipp. Bot. 6 1935–1947. [Google Scholar]
- Rocchi F., Quaroni S., Sardi P., Saracchi M. (2010). Studies on Anthostoma decipiens involved in Carpinus betulus decline. J. Plant Pathol. 92 637–644. [Google Scholar]
- Romell L. (1892). Nagra ord om Sphaeria astroidea, eutypa, leioplaca, lata, polycocca, aspera och Bertia collapsa. Bot. Notiser 1 170–178. [Google Scholar]
- Ronquist F., Huelsenbeck J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Saccardo P. A. (1875). Conspectus generum pyrenomycetum italicorum additis speciebus fungorum venetorum novis vel criticis, systemate carpologico dispositorum. Atti Soc. Veneziana Trentina Istriana Sci. Nat. 4 77–100. [Google Scholar]
- Saccardo P. A. (1882). Sylloge Fungorum 1. Padua: Sumptibus auctoris. [Google Scholar]
- Salem B., Correia K. C., Boughalleb N., Michereff S. J., Leon M., Abad-Campos P., et al. (2013). Monosporascus eutypoides, a cause of root rot and vine decline in Tunisia, and evidence that Monosporascus cannonballus and Monosporascus eutypoides are distinct species. Plant Dis. 97 737–743. 10.1094/PDIS-05-12-0464-RE [DOI] [PubMed] [Google Scholar]
- Sales R., Santana C. V. S., Nogueira D. R. S., Silva K. J. P., Guimaraes I. M., Michereff S. J., et al. (2010). First report of Monosporascus cannonballus on watermelon in Brazil. Plant Dis. 94:278. 10.1094/PDIS-94-2-0278B [DOI] [PubMed] [Google Scholar]
- Schmid-Heckel H. (1988). Pilze in den Berchtesgadener Alpen. Forschungsberichte Nationalpark Berchtesgaden 15 1–136. [Google Scholar]
- Senanayake I. C., Maharachchikumbura S. S. N., Hyde K. D., Bhat D. J., Jones E. B. G., McKenzie E. H. C., et al. (2015). Towards unraveling relationships in Xylariomycetidae (Sordariomycetes). Fungal Divers. 73 73–144. 10.1007/s13225-015-0340-y [DOI] [Google Scholar]
- Senwanna C., Phookamsak R., Doilom M., Hyde K. D., Cheewangkoon R. (2017). Novel taxa of Diatrypaceae from Para rubber (Hevea brasiliensis) in northern Thailand; introducing a novel genus Allocryptovalsa. Mycosphere 8 1835–1855. 10.5943/mycosphere/8/10/9 [DOI] [Google Scholar]
- Shang Q. J., Hyde K. D., Jeewon R., Khan S., Promputtha I., Phookamsak R., et al. (2018). Morpho-molecular characterization of Peroneutypa (Diatrypaceae, Xylariales) with two novel species from Thailand. Phytotaxa 356 1–18. 10.11646/phytotaxa.356.1.1 [DOI] [Google Scholar]
- Shang Q. J., Hyde K. D., Phookamsak R., Doilom M., Bhat D. J., Maharachchikumbura S. S. N., et al. (2017). Diatrypella tectonae and Peroneutypa mackenziei spp. nov. (Diatrypaceae) from northern Thailand. Mycol. Prog. 16 463–476. 10.1007/s11557-017-1294-0 [DOI] [Google Scholar]
- Sosnowski M. R., Lardner R., Wicks T. J., Scott E. S. (2007). The influence of grapevine cultivar and isolate of Eutypa lata on wood and foliar symptoms. Plant Dis. 91 924–931. 10.1094/PDIS-91-8-0924 [DOI] [PubMed] [Google Scholar]
- Spooner B. M. (1981). New records and species of British microfungi. Trans. Br. Mycol. Soc. 76 265–301. [Google Scholar]
- Srinivasulu B. V., Sathe P. G. (1970). Genus Quaternaria from India. Sydowia 24 302–304. [Google Scholar]
- Stevenson J. A. (1975). Fungi of Puerto Rico and the American Virgin Islands. Contrib. Reed Herbarium 24:522. 10.2307/1219515 [DOI] [Google Scholar]
- Sutton B. C. (1980). The Coelomycetes. Fungi Imperfecti with Pycnidia, Acervuli and Stromata. Kew: Commonwealth Mycological Institute. [Google Scholar]
- Swofford D. L. (2003). PAUP∗: Phylogenetic Analysis using Parsimony. Version 4.0 Beta. Sunderland, MA: Sinauer Associates. [Google Scholar]
- Tai F. L. (1979). Sylloge Fungorum Sinicorum. Beijing: Science Press. [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tendulkar J. S. (1970). Four new species of Diatrype from India. Sydowia 24 282–285. [Google Scholar]
- Teng S. C. (1996). Fungi of China. Ithaca, NY: Mycotaxon Ltd. [Google Scholar]
- Teodoro N. G. (1937). An Enumeration of Philippine Fungi. Manila: Bureau of printing. [Google Scholar]
- Tiffany L. H., Gilman J. C. (1965). Iowa ascomycetes IV. Diatrypaceae. Iowa State Coll. J. Sci. 40 121–161. [Google Scholar]
- Trouillas F. P., Gubler W. D. (2004). Identification and characterization of Eutypa leptoplaca, a new pathogen of grapevine in Northern California. Mycol. Res. 108 1195–1204. 10.1017/S0953756204000863 [DOI] [PubMed] [Google Scholar]
- Trouillas F. P., Gubler W. D. (2010). Host range, biological variation, and phylogenetic diversity of Eutypa lata in California. Phytopathology 100 1048–1056. 10.1094/PHYTO-02-10-0040 [DOI] [PubMed] [Google Scholar]
- Trouillas F. P., Hand F. P., Inderbitzin P., Gubler W. D. (2015). The genus Cryptosphaeria in the western United States: taxonomy, multilocus phylogeny and a new species, C. multicontinentalis. Mycologia 107 1304–1313. 10.3852/15-115 [DOI] [PubMed] [Google Scholar]
- Trouillas F. P., Pitt W. M., Sosnowski M. R., Huang R., Peduto F., Loschiavo A., et al. (2011). Taxonomy and DNA phylogeny of Diatrypaceae associated with Vitis vinifera and other woody plants in Australia. Fungal Divers. 49 203–223. 10.1007/s13225-011-0094-0 [DOI] [Google Scholar]
- Trouillas F. P., Sosnowski M. R., Gubler W. D. (2010a). Two new species of Diatrypaceae from coastal wattle in Coorong National Park, South Australia. Mycosphere 1 183–188. [Google Scholar]
- Trouillas F. P., Urbez-Torres J. R., Gubler W. D. (2010b). Diversity of diatrypaceous fungi associated with grapevine canker diseases in California. Mycologia 102 319–336. 10.3852/08-185 [DOI] [PubMed] [Google Scholar]
- Tulasne L. R., Tulasne C. (1863). Selecta Fungorum Carpologia 2. Paris: Imperial Press. [Google Scholar]
- Unamuno P. L. M. (1941). Enumeracion y disbucion geografica de los ascomicetos de la Peninsula lberica y de las lslas Baleares. Mem. Real Acad. Ci Exact Madrid 8 1–403. [Google Scholar]
- Vasilyeva L. N. (2010). New species of pyrenomycetes from eastern Russia: Endoxylina rufula (Diatrypaceae). Mycosphere 1 163–166. [Google Scholar]
- Vasilyeva L. N., Ma H. (2014). Diatrypaceous fungi in north-eastern China. 1. Cryptosphaeria and Diatrype. Phytotaxa 186 261–270. 10.11646/phytotaxa.186.5.3 [DOI] [Google Scholar]
- Vasilyeva L. N., Stephenson S. L. (2005). Pyrenomycetes of the Great Smoky Mountains National Park. II. Cryptovalsa Ces. et De Not. and Diatrypaceae (Ces. et De Not.) Nitschke (Diatrypaceae). Fungal Divers. 19 189–200. [Google Scholar]
- Vasilyeva L. N., Stephenson S. L. (2006). Pyrenomycetes of the Great Smoky Mountains National Park. III. Cryptosphaeria, Eutypa and Eutypella (Diatrypaceae). Fungal Divers. 22 243–254. 10.1111/j.1439-0507.2006.01249.x [DOI] [PubMed] [Google Scholar]
- Vasilyeva L. N., Stephenson S. L. (2009). The genus Diatrype (Ascomycota, Diatrypaceae) in Arkansas and Texas (USA). Mycotaxon 107 307–313. 10.1063/1.119340 [DOI] [Google Scholar]
- Vieira M. L. A., Hughes A. F. S., Gil V. B., Vaz A. B. M., Alves T. M. A., Zani C. L., et al. (2011). Diversity and antimicrobial activities of the fungal endophyte community associated with the traditional Brazilian medicinal plant Solanum cernuum Vell. (Solanaceae). Can. J. Microbiol. 58 54–56. 10.1139/w11-105 [DOI] [PubMed] [Google Scholar]
- von Arx J. A. (1981). The Genera of Fungi Sporulating in Pure Culture, 3rd Edn. Vaduz: J Cramer. [Google Scholar]
- Wehmeyer L. E. (1975). The pyrenomycetous fungi. Mycol. Mem. 6 1–250. [Google Scholar]
- White T. J., Bruns T., Lee S., Taylor J. (1990). Amplifcation and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. Guide Methods Appl. 18 315–322. [Google Scholar]
- Wijayawardene N. N., Hyde K. D., Al-Ani L. K. T., Tedersoo L., Haelewaters D., Rajeshkumar K. C., et al. (2020). Outline of fungi and fungus-like taxa. Mycosphere 11 1060–1456. 10.5943/mycosphere/11/1/8 10549654 [DOI] [Google Scholar]
- Wijayawardene N. N., Hyde K. D., Lumbsch H. T., Liu J. K., Maharachchikumbura S. S. N., Ekanayaka A. H., et al. (2018). Outline of ascomycota: 2017. Fungal Divers. 88 167–263. 10.1007/s13225-018-0394-8 [DOI] [Google Scholar]
- Wijayawardene N. N., Hyde K. D., Wanasinghe D. N., Papizadeh M., Goonasekara I. D., Camporesi E., et al. (2016). Taxonomy and phylogeny of dematiaceous coelomycetes. Fungal Divers. 77 1–316. 10.1007/s13225-016-0360-2 [DOI] [Google Scholar]
- Zhu H. Y., Pan M., Bezerra J. D. P., Tian C. M., Fan X. L. (2020). Discovery of Cytospora species associated with canker disease of tree hosts from Mount Dongling of China. MycoKeys 62 97–121. 10.3897/mycokeys.62.47854 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Placement of genera in Diatrypaceae by different authors.
Isolates and GenBank accession numbers used in the phylogenetic analyses of Diatrypaceae.
Synopsis of species of Allocryptovalsa.
Synopsis of species of Diatrypella from Betula spp.
Distribution of diatrypaceous fungi on plants in China.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.


