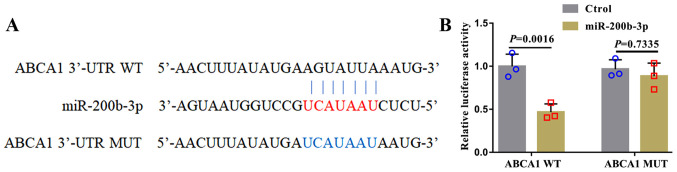Abstract
Atherosclerosis (As) is a chronic cardiovascular disease characterized by abnormal of lipid accumulation and cholesterol efflux. The present study aimed to investigate whether the micro-RNA (miR)-200b-3p could exacerbate As by promoting lipid accumulation and inhibiting cholesterol efflux via ATP-binding cassette transporter A1 (ABCA1) in macrophage-derived foam cells. Blood samples from 30 patients with As and 30 healthy people were collected at Quanzhou First Hospital. RAW264.7 cells were used to establish foam cells using oxidized low-density lipoprotein. The expression of miR-200b-3p and ABCA1 was evaluated by reverse transcription quantitative PCR and western blotting. Lipid accumulation was analyzed by Oil Red O staining and cholesterol content was assessed by ELISA. A targeting relationship between miR-200b-3p and ABCA1 was demonstrated by luciferase reporter assays. Compared with healthy volunteers and RAW264.7 cells, the expression level of miR-200b-3p was significantly increased whereas the expression level of ABCA1 was significantly decreased in patients with As and foam cells. Furthermore, miR-200b-3p expression was negatively correlated with ABCA1 expression in the blood of the patients with As. Lipid content was significantly decreased and cholesterol efflux was significantly increased in foam cells transfected with the miR-200b-3p inhibitor compared with inhibitor control cells. In addition, ABCA1 was shown to be targeted by miR-200b-3p. Furthermore, the lipid content in foam cells transfected with the miR-200b-3p inhibitor and small interfering-ABCA1 was significantly increased, while the cholesterol efflux was significantly decreased compared with foam cells transfected with the miR-200b-3p inhibitor. In conclusion, the findings from the present study indicated that inhibition of miR-200b-3p may alleviate lipid accumulation and promote cholesterol efflux by targeting ABCA1 in macrophage-derived foam cells.
Keywords: ATP-binding cassette transporter A1, micro-RNA-200b-3p, atherosclerosis, cholesterol efflux, lipid accumulation
Introduction
Atherosclerosis (As) is a chronic cardiovascular disease that seriously endangers human health. Numerous studies have confirmed that abnormal levels of blood lipids and cholesterol are closely related to the occurrence of As (1). Elevated plasma lipids can promote cholesterol to enter and deposit in the arterial wall, leading to thickening of the vessel intima, narrowing of the vessels and atherosclerotic lesions (2). The main clinical manifestations of As are mononuclear infiltration, foam cell formation, lipid accumulation in the vascular wall, plaque formation and inflammation (3,4). Foam cell formation is characterized by accumulation of lipids and cholesterol efflux obstruction in macrophages (4). Reverse cholesterol transport (RCT), an important physiological mechanism for maintaining intracellular cholesterol balance (5), is also a crucial process in the occurrence and development of As.
ATP-binding cassette transporter A1 (ABCA1), an integrated membrane protein, promotes the efflux of free cholesterol and phospholipids from cells (6). A previous study demonstrated that ABCA1 mutations can prevent intracellular cholesterol and phospholipids from being transported out of the cell, causing accumulation of macrophage-derived foam cells in various tissues, or even resulting in atherosclerotic lesions (7). It was reported that metastasis associated lung adenocarcinoma transcript 1 inhibition reduces the outflow of cholesterol and increases cholesterol accumulation in macrophages, accelerating therefore the process of As by downregulating the expression of ABCA1(8). A previous study demonstrated that urolithin A can increase ABCA1 expression and RCT in macrophages, thus alleviating the progression of As (9). Hesperidin can attenuate varenicline-induced oxidized low-density lipoprotein (ox-LDL) uptake in RAW264.7 cells by upregulating the expression of ABCA1(10). The accumulation of lipids and cholesterol is the first step in As formation (1,2). Although ABCA1 plays a crucial role in regulating cholesterol efflux (11), the underlying mechanism remains poorly understood.
Micro-RNAs (miRNAs) are non-coding RNAs of 20-25 nucleotides in length that are involved in gene transcription and mRNA stability. miRNAs are expressed throughout the body, and abnormal expression of miRNAs is correlated with numerous diseases, such as cardiovascular disease (12,13). For example, the expression of miR-200b-3p is lower in human cytomegalovirus (HCMV)-infected gastrointestinal tract and bronchi/lung tissue than in normal tissue, which indicates that low levels of miR-200b-3p are associated with inflammation due to HCMV infection in humans (14). miR-200b-3p inhibits prostate cancer cell proliferation and promotes cell apoptosis by targeting PPKAR2B (15). miR-200b-3p improves oxaliplatin resistance and induces migration, growth inhibition and apoptosis in oxaliplatin-resistant colorectal cancer cells by suppressing expression of TUBB3(16). Liu et al (17) demonstrated that miR-200b-3p is highly expressed in lung adenocarcinoma tissue and promotes the proliferation and metastasis of lung adenocarcinoma cells by targeting ABCA1. Furthermore, ABCA1 expression is associated with lipid accumulation and RCT in macrophage-derived foam cells and regulates the development of As (7). It is however unclear whether miR-200b-3p could exacerbate As by promoting lipid accumulation and inhibiting cholesterol efflux via ABCA1 in macrophage-derived foam cells.
In the present study, the aim was to investigate whether miR-200b-3p exacerbates As and the specific mechanism of miR-200b-3b-induced lipid accumulation and cholesterol efflux inhibition in macrophage-derived foam cells. The present study aimed provide a novel basis for targeting miR-200b-3p as a potential therapeutic strategy for patients with As.
Materials and methods
Clinical samples
Blood samples from 30 patients with As and 30 healthy volunteers were collected at Quanzhou First Hospital between December 2020 and January 2021. A total of 2 ml blood from each individual was collected in an EDTA-containing anticoagulant tube, then centrifuged at 1,000 x g at 4˚C for 10 min. The supernatant (plasma) was collected to detect the expression of miR-200b-3p and ABCA1 or stored at -20˚C for further experiments. The expression levels of miR-200b-3p and ABCA1 in these samples were evaluated by revers transcription quantitative (RT-q)PCR. The procedures were approved by the Institutional Ethics Committee of Quanzhou First Hospital (approval no. 2020-05). Patients and volunteers signed informed consent and the experiments were conducted according to the principles of the Declaration of Helsinki.
Cell culture
RAW264.7 cells were purchased from the American Type Culture Collection. Cells were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin-streptomycin (Sigma-Aldrich; Merck KGaA) placed at 37˚C in a humidified incubator containing 5% CO2.
Foam cells formation
RAW264.7 cells were seeded in 6-well plates at a density of 5x105 cells/well. When 60% confluence was reached, cells were exposed to ox-LDL (50 mg/ml; cat. no. 20605ES05; Shanghai Yeasen Biotechnology Co., Ltd.) for 48 h to induce the formation of foam cells (18,19).
RT-qPCR
Total RNA was isolated from cells and human plasma samples using TransZol Up Plus RNA kit (cat. no. ER501-01; TransGen Biotech Co., Ltd.) according to the manufacturers' protocol. The concentration of total RNA was determined by a NanoDrop ND-2000 spectrophotometer (Thermo Fisher Scientific, Inc.). RNA was reverse transcribed using an EasyScript First-Strand cDNA Synthesis SuperMix (AE301-02; TransGen Biotech Co., Ltd.) according to the manufacturers' instructions. Finally, the expression level of miR-200b-3p and ABCA1 was evaluated using PerfectStart™ Green qPCR SuperMix (AQ601-01; TransGen Biotech Co., Ltd.) on the Bio-Rad C1000 Touch Thermal Cycler CFX96 Real-Time System (Bio-Rad Laboratories, Inc.). RT-qPCR was performed as follows: Initial denaturation at 95˚C for 10 min; denaturation at 95˚C for 15 sec and annealing/extension at 60˚C for 60 sec (40 cycles). The relative expression levels of miR-200b-3p and ABCA1 were normalized to the endogenous controls 18S and U6 and were expressed as 2-∆∆Cq (20). The sequences of the primers for miR-200b-3p and ABCA1 (Sangon Biotech Co., Ltd.) are presented in Table I.
Table I.
Sequence of the primers used for reverse transcription quantitative PCR.
| Primer | Sequence (5'-3') |
|---|---|
| miR-200b-3p | |
| Forward | GCTGCTGAATTCCATCTAATTTCCAAAAG |
| Reverse | TATTATGGATCCGCCCCCAGGGCAATGGG |
| ABCA1 | |
| Forward | GGCATCGTGTATGAGAAGG |
| Reverse | CTGTAGGGCAGCAGGTTT |
| U6 | |
| Forward | GCTTCGGCAGCACATATACT |
| Reverse | GCAGGGTCCGAGGTATTC |
| 18S | |
| Forward | AGAAACGGCTACCACATCCA |
| Reverse | CACCAGACTTGCCCTCCA |
miR, micro-RNA; ABAC1, ATP-binding cassette transporter A1.
Cell transfection
miR-200b-3p inhibitor control (5'-CAGUACUUUUGUGUAGUACUAA-3'), miR-200b-3p inhibitor (5'-UCAUCAUUACCAGGCAGUAUUA-3'), small interfering (si)RNA-control (si-control, 5'-TAGT TAGACGCGUCACGTAGG-3') and si-ABCA1 (5'-GCAG TGCCTTTGTAGCCTATG-3') were synthesized by Shanghai GenePharma Co., Ltd. The ox-LDL-induced foam cells were seeded in 6-well plates at 5x105 cells/well. When 70-80% confluence was reached, cells were transfected with the miR-200b-3p inhibitor (2 µg) or co-transfected with miR-200b-3p inhibitor (1 µg) and si-ABCA1/si-control (1 µg) using Lipofectamine™ 3000 Transfection Reagent (cat. no. L3000015; Thermo Fisher Scientific, Inc.) according to the manufacturers' protocol. The transfected cells were incubated for 48 h in an incubator with 5% CO2 at 37˚C. After 48 h, the cells were collected for subsequent experiments.
Western blotting
Cells transfected with si-ABCA1 or miR-200b-3p inhibitor were collected, washed in cold PBS and lysed on ice for 30 min using RIPA buffer (cat. no. P0013B; Beyotime Institute of Biotechnology) supplemented with a protease inhibitor cocktail (cat. no. 04693132001; Roche Diagnostics). The cell lysate was centrifuged at 12,000 x g at 4˚C for 30 min. The supernatant was collected and the protein concentration was quantified by a NanoDrop ND-2000 spectrophotometer (Thermo Fisher Scientific, Inc.). Proteins (50 µg) were separated by 8% SDS-PAGE and transferred onto a polyvinylidene fluoride membrane. The membranes were blocked with 5% skim milk at room temperature for 1 h and then incubated with a mouse anti-ABCA1 antibody (1:1,000; cat. no. ab18180; Abcam) and mouse anti-β-actin antibody (1:2,000; cat. no. HC201-01; TransGen Biotech Co., Ltd.) at room temperature for 2 h. Subsequently, membranes were incubated with a secondary anti-mouse antibody (1:5,000; cat. no. 7076P2; Cell Signaling Technology, Inc.) at room temperature for 1 h. Enhanced chemiluminescence reagent (cat. no. 170-5061; Bio-Rad Laboratories, Inc.) was used to detect the signal on the membrane. The data were analyzed via densitometry using ImageJ software (1.52v, National Institutes of Health) and normalized to expression of the internal control β-actin.
Oil Red O stain
Oil Red O (0.5%; cat. no. O1391-250ML; Sigma-Aldrich; Merck KGaA) was diluted with distilled water at a ratio of 3:2 and filtered with filter paper to generate the working solution. The cells were fixed with 4% paraformaldehyde at room temperature for 30 min and washed three times with PBS. The cells were stained with Oil Red O working solution at 37˚C for 30 min and then rinsed with 65% isopropanol. Finally, the cells were observed under a 10x inverted microscope (magnification, x10).
Enzyme-linked immunosorbent assay (ELISA)
Total cholesterol (TC) and free cholesterol (FC) levels were evaluated using ELISA kits (cat. nos. E1015-105 and E1016-105, respectively; Applygen Technologies, Inc.) according to the manufacturers' protocol. Briefly, cells were washed in cold PBS and harvested. The cells were mixed with the appropriate lysis buffer and allowed to stand for 10 min for lysis. The protein concentration was measured in some lysates with an Easy II Protein Quantitative kit (BCA; TransGen Biotech Co., Ltd.). The rest of the lysates was centrifuged at 2,000 x g for 5 min at room temperature and supernatants were collected. TC and FC levels were measured with a microplate reader (Molecular Devices, LLC). The cholesterol ester (CE) level was calculated using the following equation: CE=TC-FC. The data were normalized to the total cellular protein concentration.
Luciferase reporter assay
miR-200b-3p mimic, wild-type (WT) and mutant (MUT) 3'-UTRs of ABCA1 were synthesized by Sangon Biotech Co, Ltd. The WT and MUT ABCA1 3'-UTR were inserted into the luciferase reporter vector (pGL3; Promega Corporation) to construct pGL3-ABCA1-WT or pGL3-ABCA1-MUT plasmids, respectively. The foam cells were co-transfected with plasmid DNA and miR-200b-3p mimic with Lipofectamine 3000 Transfection Reagent (cat. no. L300015, Thermo Fisher Scientific, Inc.) following the manufacturers' protocol. After transfection for 48 h, the cells were collected for analysis of luciferase activity. Luciferase activity was measured by a microplate reader (Tecan Infinite 200, Tecan Group, Ltd.) with Dual Luciferase Reporter Gene Assay kit (cat. no. RG027; Beyotime Institute of Biotechnology) in according to the manufacturers' protocol. The luciferase activity of cells was analyzed using Renilla luciferase activity as an internal reference.
Statistical analysis
All data from three independent experiments were presented as the means ± standard deviation and analyzed using GraphPad Prism 7.0 software (GraphPad Software, Inc.). Comparison between two groups was performed using Student's t-test. Spearman correlation analysis was used to analyze the correlation between the expression of miR-200b-3p and ABCA1 in patients with As. P<0.05 was considered to indicate a statistically significant difference.
Results
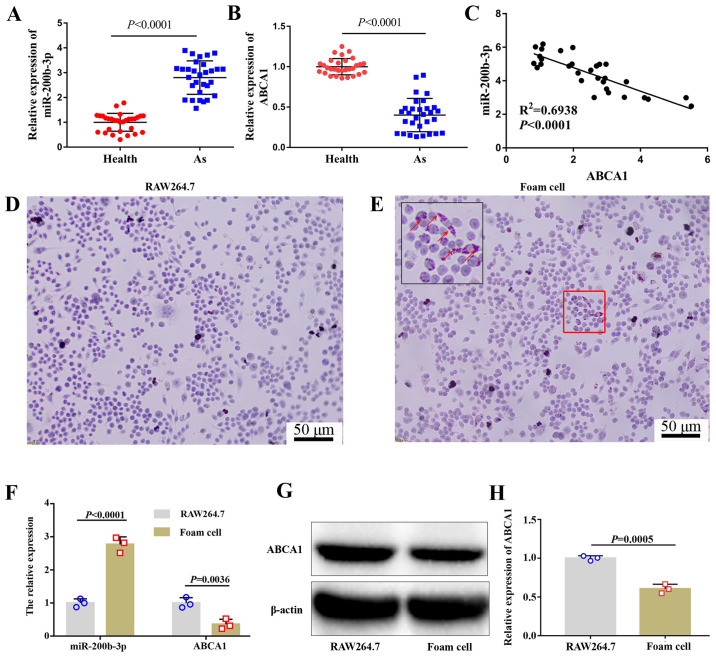
miR-200b-3p expression is upregulated and ABCA1 expression is downregulated in patients with As and foam cells
The expression of miR-200b-3p and ABCA1 in patients with or without As was evaluated by RT-qPCR. The results demonstrated that miR-200b-3p expression in patients with As was significantly higher compared with healthy volunteers (Fig. 1A), whereas the expression of ABCA1 in patients with As was significantly lower compared with healthy people (Fig. 1B). Furthermore, miR-200b-3p expression was negatively correlated with ABCA1 expression (Fig. 1C).
Figure 1.
Expression of ABCA1 and miR-200b-3p is evaluated in patients with As and in RAW264.7-derived foam cells. Expression of (A) miR-200b-3p and (B) ABCA1 was measured by RT-qPCR in patients with or without As (n=30). (C) miR-200b-3p expression was negatively correlated with ABCA1 expression, as analyzed by Spearman correlation analysis. RAW264.7 cells were treated with or without 50 mg/l ox-LDL for 48 h. Lipid accumulation was evaluated by Oil Red O staining in (D) RAW264.7 and (E) RAW264.7-derived foam cells. Red arrows indicate lipid particles. Data show that lipid accumulation was increased in foam cells. (F) Expression of miR-200b-3p and ABCA1 was measured by RT-qPCR in cells. (G and H) Protein expression of ABCA1 was measured by western blotting in cells. As, atherosclerosis; miR, micro-RNA; ABAC1, ATP-binding cassette transporter A1; RT-qPCR, reverse transcription quantitative PCR.
Based on previous research (8,11,18), RAW264.7 cells were selected to explore the specific molecular mechanism of miR-200b-3p in As. RAW246.7 cells were treated with ox-LDL for 48 h. To investigate whether foam cells were formed following this treatment, lipid content was evaluated by Oil Red O staining. The results demonstrated that the number of lipid particles was markedly increased in cells treated with ox-LDL (Fig. 1D and E), which confirmed the formation of RAW264.7-derived foam cells induced by ox-LDL treatment. The expression levels of miR-200b-3p and ABCA1 were measured by RT-qPCR. As shown in Fig. 1F, miR-200b-3p was upregulated whereas ABCA1 was downregulated in RAW264.7-derived foam cells compared with RAW264.7 cells. In addition, the protein expression of ABCA1 was significantly decreased in foam cells compared with RAW264.7 cells (Fig. 1G and H).
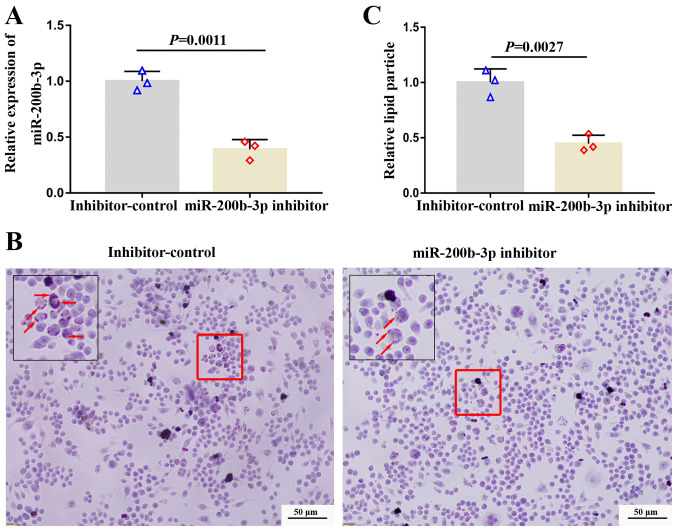
Inhibition of miR-200b-3p decreases lipid accumulation and enhances cholesterol efflux
To investigate the effect of miR-200b-3p on lipid accumulation and cholesterol efflux, foam cells were transfected with a miR-200b-3p inhibitor and the expression of miR-200b-3p was measured by RT-qPCR. The expression of miR-200b-3p was distinctly decreased in foam cells transfected with miR-200b-3p inhibitor compared with cells transfected with inhibitor-control (Fig. 2A), suggesting the successful transfection. Subsequently, lipid content was evaluated by Oil Red O staining. The results demonstrated that the number of orange particles in the cytoplasm was decreased in cells transfected with the miR-200b-3p inhibitor compared with cells transfected with inhibitor control (Fig. 2B and C), suggesting that miR-200b-3p inhibition could decrease lipid accumulation.
Figure 2.
Inhibition of miR-200b-3p decreases lipid accumulation in RAW264.7-derived foam cells. (A) Relative expression of miR-200b-3p was measured by reverse transcription quantitative PCR in the cells transfected with the miR-200b-3p inhibitor control and the miR-200b-3p inhibitor. Lipid accumulation was measured by Oil Red O staining in RAW264.7-derived foam cells. (B) Representative images showing lipid accumulation in the cells. Red arrows indicate lipid particles. (C) Inhibition of miR-200b-3p decreased relative lipid accumulation in foam cells. miR, micro-RNA
Furthermore, the levels of cholesterol in the foam cells and in foam cell culture media were analyzed by ELISA. The TC, FC and EC levels in the foam cells transfected with the miR-200b-3p inhibitor were significantly decreased compared with those in foam cells transfected with the inhibitor-control (P<0.05; Table II); however, the EC/TC ratio was not significantly changed in foam cells transfected with the miR-200b-3p inhibitor (Table II). Furthermore, FC level in the culture media of foam cells transfected with the miR-200b-3p inhibitor was significantly increased compared with that in the culture media of foam cells transfected with inhibitor control (P<0.05; Table III). To analyze the effect of miR-200b-3p on cholesterol efflux in foam cells derived from macrophages, the cholesterol efflux rate of the foam cells was calculated according to the following equation: Cholesterol efflux rate % = culture media FC/(FC in culture media + FC in foam cells) x100%. As seen in Table IV, the cholesterol outflow of the foam cells transfected with the miR-200b-3p inhibitor was significantly higher than that of the inhibitor-control cells (P<0,05), which indicated that miR-200b-3p inhibition may enhance cholesterol efflux in RAW264.7-derived foam cells.
Table II.
Effect of miR-200b-3p on intracellular cholesterol content in foam cells.
| Inhibitor control | miR-200b-3p inhibitor | P-value | |
|---|---|---|---|
| TC (nmol/mg total protein) | 362.74±11.04 | 285.28±8.11 | 0.0006 |
| FC (nmol/mg total protein) | 151.74±5.85 | 109.28±8.42 | 0.0020 |
| CE (nmol/mg total protein) | 211.00±5.37 | 176.04±11.30 | 0.0084 |
| CE/TC (%) | 58.17±0.41 | 61.69±3.04 | 0.1181 |
TC, total cholesterol; FC, free cholesterol; CE, cholesterol ester; miR, micro-RNA.
Table III.
Free cholesterol level in the culture media of foam cells.
| Inhibitor control | miR-200b-3p inhibitor | P-value | |
|---|---|---|---|
| FC (nmol/mg total protein) | 53.73±4.05 | 82.42±3.29 | 0.0007 |
FC, free cholesterol; miR, micro-RNA.
Table IV.
Effect of miR-200b-3p on cholesterol efflux in foam cells.
| Inhibitor control | miR-200b-3p inhibitor | P-value | |
|---|---|---|---|
| Cholesterol efflux (%) | 26.14±1.16 | 43.04±0.91 | <0.0001 |
miR, micro-RNA.
Taken together, these findings suggested that inhibition of miR-200b-3p may decrease lipid accumulation and enhance cholesterol efflux.
miR-200b-3p downregulates the expression of ABCA1 in foam cells
In human lung adenocarcinoma cell lines, miR-200b-3p targets ABCA1 and negatively regulates the expression of ABCA1(17). To investigate whether miR-200b-3p could target ABCA1 in RAW264.7-derived foam cells, WT and MUT ABCA1 3'-UTRs were designed (Fig. 3A) and co-transfected into foam cells. Then, luciferase activity was evaluated. As presented in Fig. 3B, luciferase activity in cells transfected with miR-200b-3p mimic and the WT ABCA1 3'-UTR was decreased compared with that in cells transfected with miR-200b-3p mimic and MUT ABCA1 3'-UTR.
Figure 3.
miR-200b-3p targets ABCA1 in RAW264.7 macrophages. (A) Sequences of the ABCA1 3'-UTR and miR-200b-3p. Red letters represent the base sequence where the 3'-UTR of ABCA1 binds to miR-200b-3p. Blue letters represent the base sequence of the MUT ABCA1 3'-UTR. RAW264.7 cells were co-transfected with miR-200b-3p mimic and the WT or MUT 3'-UTR of ABCA1. (B) Relative luciferase activity was measured to verify the direct target relationship between miR-200b-3p and ABCA1. miR, micro-RNA; ABAC1, ATP-binding cassette transporter A1; WT, wild-type; MUT, mutant.
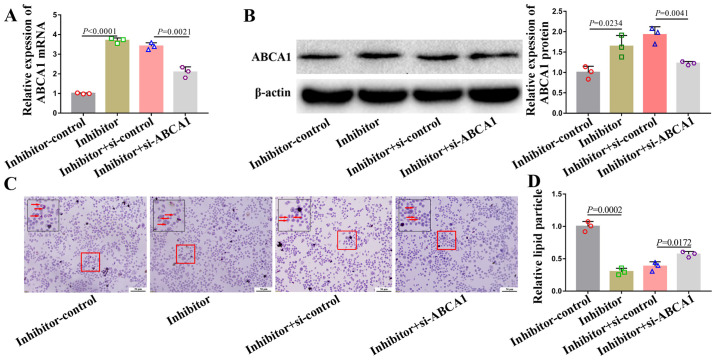
miR-200b-3p inhibitor alleviates lipid accumulation and accelerates cholesterol efflux by upregulating ABCA1 expression
A rescue experiment was performed to determine the relationship between miR-200b-3p and ABCA1. To do so, foam cells were co-transfected with a miR-200b-3p inhibitor and si-ABCA1. The mRNA and protein expression of ABCA1 in cells transfected with si-ABCA1 and miR-200b-3p inhibitor was partly downregulated (Fig. 4A and B). Furthermore, si-ABCA1 partly abrogated the alleviation of lipid accumulation (Fig. 4C and D) and acceleration of cholesterol efflux (Table V) induced by miR-200b-3p inhibitor.
Figure 4.
si-ABCA1 partly abolishes the miR-200b-3p inhibitor induced decrease in lipid accumulation in RAW264.7-derived foam cells. miR-200b-3p inhibitor or si-ABCA1 was transfected into foam cells. (A) mRNA and (B) protein expression of ABCA1was analyzed by reverse transcription quantitative PCR and western blotting. (C) Lipid accumulation in foam cells was analyzed by Oil Red O staining. Red arrows indicate lipid particles. (D) Lipid particles were quantified by ImageJ software in cells transfected with the miR-200b-3p inhibitor (inhibitor) control, inhibitor, inhibitor + si-control and inhibitor + si-ABCA1. miR, micro-RNA; ABAC1, ATP-binding cassette transporter A1; si, small interfering.
Table V.
si-ABCA1 partly abrogates miR-200b-3p inhibitor-induced accelerated cholesterol efflux in foam cells.
| Groups | Cholesterol efflux rate % | P-value |
|---|---|---|
| miR-200b-3p inhibitor control | 27.13±2.67 | 0.0019 |
| miR-200b-3p inhibitor | 47.71±4.12 | |
| miR-200b-3p inhibitor + si-control | 45.63±3.58 | 0.0310 |
| miR-200b-3p inhibitor + si-ABCA1 | 35.31±4.15 |
miR, micro-RNA; si, small interfering; ABAC1, ATP-binding cassette transporter A1.
Taken together, these data suggested that miR-200b-3p inhibitor may alleviate lipid accumulation and accelerate cholesterol efflux by positively regulating ABCA1 in RAW264.7-derived foam cells.
Discussion
As is the main cause of cardiovascular and cerebrovascular diseases (21,22). The pathogenesis of As involves multiple genetic and environmental factors. For example, abnormal of SOS3 methylation and histone results in As by disrupting the function of vascular endothelium (23). Smoking accelerates As formation by impairing endothelial function and increasing inflammation/oxidative stress (24). Lipid metabolism disorders are the main pathological bases of As and are mainly characterized by macrophage infiltration, foam cell formation, lipid accumulation in the blood vessel wall and plaque formation (3,4,25). Cholesterol outflow disorder is one of the main causes of macrophage lipid accumulation and foam cell formation and it leads to the development of As (25,26). Studying cholesterol efflux disorders is therefore beneficial when investigating the underlying mechanism of As and determining strategies for the prevention and treatment of As.
In the early development of As, macrophages aggregate under local inflammatory stimulation, excessive lipids accumulate intracellularly and cholesterol outflow decreases in macrophages, resulting in the formation of macrophage-derived foam cells (27). Macrophage-derived foam cells are a sign of plaque formation and early development of As. ABCA1, an integrin on the cytomembrane, plays an important role in high density lipoprotein (HDL) and very low-density lipoprotein production, inflammation, insulin-glucose imbalance and obesity (28,29). Furthermore, ABCA1 combined with the apolipoprotein apoA-I promotes the outflow of FC from macrophage cells (30). A previous study on the structure of ABCA1 protein demonstrated that the C-terminus of ABCA1 regulates cholesterol flippase activity and cholesterol efflux (31). Previous studies have reported that ABCA1 mutations can cause Tangier disease (32) and familial HDL deficiency (7). Although abnormalities in ABCA1 can cause a variety of diseases, the role of ABCA1 in the regulation of lipid accumulation and cholesterol efflux is a current focus of research. Previous studies have demonstrated that abnormal expression of ABCA1 disturbs lipid accumulation and cholesterol efflux in foam cells, resulting in As (33,34). ABCA1 is therefore considered as a biomarker for predicting As and a potential target gene for the treatment of As (35,36).
Previous studies have suggested that the targeting of ABCA1 by many miRNAs can exacerbate/attenuate the development of As, such as miR-30e/92a (37), miR-143/145(38) and miR-148a (39). Therefore, miRNAs can regulate the progression of As by affecting the expression of ABCA1.
In the present study, RAW264.7 macrophages were treated with ox-LDL to induce the formation of RAW264.7-derived foam cells. The expression of ABCA1 was significantly decreased in the foam cells, whereas the expression of miR-200b-3p was significantly increased. Results from Oil red O staining demonstrated that lipid accumulation in foam cells was significantly increased compared with that in RAW264.7 cells. Furthermore, in RAW264.7-derived foam cells transfected with miR-200b-3p inhibitor, lipid accumulation was alleviated, cholesterol efflux was enhanced, which suggested that miR-200b-3p may promote lipid accumulation and inhibit cholesterol efflux. Subsequently, miR-200b-3p may be considered as a biomarker for the prediction of As; however, the underlying mechanisms of miR-200b-3p remain to be further explored. Bioinformatics analysis and luciferase reporter experiments verified that miR-200b-3p could target ABCA1 in RAW264.7 macrophages, which was consistent with Liu et al (17). Rescue experiments revealed that miR-200b-3p exacerbated lipid accumulation, suppressed cholesterol efflux in macrophages and accelerated the formation of foam cells, which ultimately led to the formation of As.
This study presented some limitations. Ox-LDL was used for foam cell formation, which was recognized by the majority of scientific researchers. However, ox-LDL can elevate the oxidative stress in macrophages and potentially affect the expression of numerous genes not related to cholesterol efflux. Due to this limitation, other triggers, such as acetylated and aggregated LDL and intermediate-density lipoprotein, will be further assessed in cholesterol and lipid load into macrophages in the future. The ABCA1 mediated cholesterol efflux is most relevant for As, not only in macrophages, but also in liver, intestine and other organs. The ApoE-/- mice model with As will be used to further verify that miR-200b-3p could target ABCA1 to promote the formation and progression of As in macrophages, liver, intestine and other organs. In addition, regulation of cholesterol efflux by miR-200b-3p and ABCA1 in hepatocytes and intestinal cells will be investigated in the future.
In summary, the present study demonstrated that inhibition of miR-200b-3p alleviated lipid accumulation and promoted cholesterol efflux by targeting ABCA1 in macrophage-derived foam cells. miR-200b-3p could therefore be considered as a potential target gene for treating As.
Acknowledgements
Not applicable.
Funding Statement
Funding: No funding was received.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
ZJG and YBY designed the study and helped to revise the manuscript. YTW, JBL, HQL, GXZ, CMH and ML performed the experiments and analyzed the data. YTW wrote the manuscript. YTW, ZJG and YBY confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study adhered to the principles in the Declaration of Helsinki. All participants provided written informed consent. This study was approved by the Institutional Ethics Committee of Quanzhou First Hospital (approval no. 2020-05).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ference BA, Kastelein JJP, Catapano AL. Lipids and lipoproteins in 2020. JAMA. 2020;324:595–596. doi: 10.1001/jama.2020.5685. [DOI] [PubMed] [Google Scholar]
- 2.Aguilar-Ballester M, Herrero-Cervera A, Vinué Á, Martínez-Hervás S, González-Navarro H. Impact of cholesterol metabolism in immune cell function and atherosclerosis. Nutrients. 2020;12(12) doi: 10.3390/nu12072021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li D, Liu Y, Zhang X, Lv H, Pang W, Sun X, Gan LM, Hammock BD, Ai D, Zhu Y. Inhibition of soluble epoxide hydrolase alleviated atherosclerosis by reducing monocyte infiltration in Ldlr(-/-) mice. J Mol Cell Cardiol. 2016;98:128–137. doi: 10.1016/j.yjmcc.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Webb NR, Moore KJ. Macrophage-derived foam cells in atherosclerosis: Lessons from murine models and implications for therapy. Curr Drug Targets. 2007;8:1249–1263. doi: 10.2174/138945007783220597. [DOI] [PubMed] [Google Scholar]
- 5.Jin P, Bian Y, Wang K, Cong G, Yan R, Sha Y, Ma X, Zhou J, Yuan Z, Jia S. Homocysteine accelerates atherosclerosis via inhibiting LXRα-mediated ABCA1/ABCG1-dependent cholesterol efflux from macrophages. Life Sci. 2018;214:41–50. doi: 10.1016/j.lfs.2018.10.060. [DOI] [PubMed] [Google Scholar]
- 6.Demina EP, Miroshnikova VV, Schwarzman AL. Role of the ABC transporters A1 and G1, key reverse cholesterol transport proteins, in atherosclerosis. Mol Biol (Mosk) 2016;50:223–230. doi: 10.7868/S002689841602004X. (In Russian) [DOI] [PubMed] [Google Scholar]
- 7.Maranghi M, Truglio G, Gallo A, Grieco E, Verrienti A, Montali A, Gallo P, Alesini F, Arca M, Lucarelli M. A novel splicing mutation in the ABCA1 gene, causing Tangier disease and familial HDL deficiency in a large family. Biochem Biophys Res Commun. 2019;508:487–493. doi: 10.1016/j.bbrc.2018.11.064. [DOI] [PubMed] [Google Scholar]
- 8.Liu L, Tan L, Yao J, Yang L. Long non-coding RNA MALAT1 regulates cholesterol accumulation in ox-LDL-induced macrophages via the microRNA-17-5p/ABCA1 axis. Mol Med Rep. 2020;21:1761–1770. doi: 10.3892/mmr.2020.10987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han QA, Su D, Shi C, Liu P, Wang Y, Zhu B, Xia X. Urolithin A attenuated ox-LDL-induced cholesterol accumulation in macrophages partly through regulating miR-33a and ERK/AMPK/SREBP1 signaling pathways. Food Funct. 2020;11:3432–3440. doi: 10.1039/c9fo02471a. [DOI] [PubMed] [Google Scholar]
- 10.Koga M, Kanaoka Y, Inada K, Omine S, Kataoka Y, Yamauchi A. Hesperidin blocks varenicline-aggravated atherosclerotic plaque formation in apolipoprotein E knockout mice by downregulating net uptake of oxidized low-density lipoprotein in macrophages. J Pharmacol Sci. 2020;143:106–111. doi: 10.1016/j.jphs.2020.01.012. [DOI] [PubMed] [Google Scholar]
- 11.Ren K, Li H, Zhou HF, Liang Y, Tong M, Chen L, Zheng XL, Zhao GJ. Mangiferin promotes macrophage cholesterol efflux and protects against atherosclerosis by augmenting the expression of ABCA1 and ABCG1. Aging (Albany NY) 2019;11:10992–11009. doi: 10.18632/aging.102498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang F, Zhang R, Zhang X, Wu Y, Li X, Zhang S, Hou W, Ding Y, Tian J, Sun L, et al. Comprehensive analysis of circRNA expression pattern and circRNA-miRNA-mRNA network in the pathogenesis of atherosclerosis in rabbits. Aging (Albany NY) 2018;10:2266–2283. doi: 10.18632/aging.101541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laffont B, Rayner KJ. MicroRNAs in the pathobiology and therapy of atherosclerosis. Can J Cardiol. 2017;33:313–324. doi: 10.1016/j.cjca.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee KH, Lim BJ, Ferreira VH, Min SY, Hong YM, Jo JH, Han SH. Expression of human miR-200b-3p and -200c-3p in cytomegalovirus-infected tissues. Biosci Rep. 2018;38(BSR20180961) doi: 10.1042/BSR20180961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xia L, Han Q, Chi C, Zhu Y, Pan J, Dong B, Huang Y, Xia W, Xue W, Sha J. Transcriptional regulation of PRKAR2B by miR-200b-3p/200c-3p and XBP1 in human prostate cancer. Biomed Pharmacother. 2020;124(109863) doi: 10.1016/j.biopha.2020.109863. [DOI] [PubMed] [Google Scholar]
- 16.Wu YZ, Lin HY, Zhang Y, Chen WF. miR-200b-3p mitigates oxaliplatin resistance via targeting TUBB3 in colorectal cancer. J Gene Med. 2020;22(e3178) doi: 10.1002/jgm.3178. [DOI] [PubMed] [Google Scholar]
- 17.Liu K, Zhang W, Tan J, Ma J, Zhao J. MiR-200b-3p functions as an oncogene by targeting ABCA1 in lung adenocarcinoma. Technol Cancer Res Treat. 2019;18(1533033819892590) doi: 10.1177/1533033819892590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao H, Jia Q, Yan L, Chen C, Xing S, Shen D. Quercetin suppresses the progression of atherosclerosis by regulating MST1-mediated autophagy in ox-LDL-induced RAW264.7 macrophage foam cells. Int J Mol Sci. 2019;20(6093) doi: 10.3390/ijms20236093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin M, Lu J, Guo Z, Zhang Y, Liu J, Wu T, Guo K, Luo T, Guo Z. Reduced SULT2B1b expression alleviates ox-LDL-induced inflammation by upregulating miR-148-3P via inhibiting the IKKβ/NF-κB pathway in macrophages. Aging (Albany NY) 2021;13:3428–3442. doi: 10.18632/aging.202273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Rahman MS, Woollard K. Atherosclerosis. Adv Exp Med Biol. 2017;1003:121–144. doi: 10.1007/978-3-319-57613-8_7. [DOI] [PubMed] [Google Scholar]
- 22.Kalaria RN. The pathology and pathophysiology of vascular dementia. Neuropharmacology. 2018;134:226–239. doi: 10.1016/j.neuropharm.2017.12.030. [DOI] [PubMed] [Google Scholar]
- 23.Grimaldi V, Vietri MT, Schiano C, Picascia A, De Pascale MR, Fiorito C, Casamassimi A, Napoli C. Epigenetic reprogramming in atherosclerosis. Curr Atheroscler Rep. 2015;17(476) doi: 10.1007/s11883-014-0476-3. [DOI] [PubMed] [Google Scholar]
- 24.Lechner K, von Schacky C, McKenzie AL, Worm N, Nixdorff U, Lechner B, Kränkel N, Halle M, Krauss RM, Scherr J. Lifestyle factors and high-risk atherosclerosis: Pathways and mechanisms beyond traditional risk factors. Eur J Prev Cardiol. 2020;27:394–406. doi: 10.1177/2047487319869400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chistiakov DA, Bobryshev YV, Orekhov AN. Macrophage-mediated cholesterol handling in atherosclerosis. J Cell Mol Med. 2016;20:17–28. doi: 10.1111/jcmm.12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boshuizen MC, Hoeksema MA, Neele AE, van der Velden S, Hamers AA, Van den Bossche J, Lutgens E, de Winther MP. Interferon-β promotes macrophage foam cell formation by altering both cholesterol influx and efflux mechanisms. Cytokine. 2016;77:220–226. doi: 10.1016/j.cyto.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 27.Yuan Y, Li P, Ye J. Lipid homeostasis and the formation of macrophage-derived foam cells in atherosclerosis. Protein Cell. 2012;3:173–181. doi: 10.1007/s13238-012-2025-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Babashamsi MM, Koukhaloo SZ, Halalkhor S, Salimi A, Babashamsi M. ABCA1 and metabolic syndrome; a review of the ABCA1 role in HDL-VLDL production, insulin-glucose homeostasis, inflammation and obesity. Diabetes Metab Syndr. 2019;13:1529–1534. doi: 10.1016/j.dsx.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 29.Jung D, Cao S, Liu M, Park S. A Meta-analysis of the associations between the ATP-binding cassette transporter ABCA1 R219K (rs2230806) polymorphism and the risk of type 2 diabetes in Asians. Horm Metab Res. 2018;50:308–316. doi: 10.1055/a-0583-0201. [DOI] [PubMed] [Google Scholar]
- 30.Liu M, Mei X, Herscovitz H, Atkinson D. N-terminal mutation of apoA-I and interaction with ABCA1 reveal mechanisms of nascent HDL biogenesis. J Lipid Res. 2019;60:44–57. doi: 10.1194/jlr.M084376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okamoto Y, Tomioka M, Ogasawara F, Nagaiwa K, Kimura Y, Kioka N, Ueda K. C-terminal of ABCA1 separately regulates cholesterol floppase activity and cholesterol efflux activity. Biosci Biotechnol Biochem. 2020;84:764–773. doi: 10.1080/09168451.2019.1700775. [DOI] [PubMed] [Google Scholar]
- 32.Puntoni M, Sbrana F, Bigazzi F, Sampietro T. Tangier disease: Epidemiology, pathophysiology, and management. Am J Cardiovasc Drugs. 2012;12:303–311. doi: 10.2165/11634140-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 33.Ouimet M, Barrett TJ, Fisher EA. HDL and reverse cholesterol transport. Circ Res. 2019;124:1505–1518. doi: 10.1161/CIRCRESAHA.119.312617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Price NL, Rotllan N, Zhang X, Canfrán-Duque A, Nottoli T, Suarez Y, Fernández-Hernando C. Specific disruption of Abca1 targeting largely mimics the effects of miR-33 knockout on macrophage cholesterol efflux and atherosclerotic plaque development. Circ Res. 2019;124:874–880. doi: 10.1161/CIRCRESAHA.118.314415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu YR, Shi XY, Ma CY, Zhang Y, Xu RX, Li JJ. Liraglutide improves lipid metabolism by enhancing cholesterol efflux associated with ABCA1 and ERK1/2 pathway. Cardiovasc Diabetol. 2019;18(146) doi: 10.1186/s12933-019-0954-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan YL, Ou HX, Zhang M, Gong D, Zhao ZW, Chen LY, Xia XD, Mo ZC, Tang CK. Tanshinone IIA promotes macrophage cholesterol efflux and attenuates atherosclerosis of apoE-/- mice by Omentin-1/ABCA1 pathway. Curr Pharm Biotechnol. 2019;20:422–432. doi: 10.2174/1389201020666190404125213. [DOI] [PubMed] [Google Scholar]
- 37.Wang Z, Zhang J, Zhang S, Yan S, Wang Z, Wang C, Zhang X. MiR-30e and miR-92a are related to atherosclerosis by targeting ABCA1. Mol Med Rep. 2019;19:3298–3304. doi: 10.3892/mmr.2019.9983. [DOI] [PubMed] [Google Scholar]
- 38.Yue Y, Zhang Z, Zhang L, Chen S, Guo Y, Hong Y. miR-143 and miR-145 promote hypoxia-induced proliferation and migration of pulmonary arterial smooth muscle cells through regulating ABCA1 expression. Cardiovasc Pathol. 2018;37:15–25. doi: 10.1016/j.carpath.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 39.Goedeke L, Rotllan N, Canfrán-Duque A, Aranda JF, Ramírez CM, Araldi E, Lin CS, Anderson NN, Wagschal A, de Cabo R, et al. MicroRNA-148a regulates LDL receptor and ABCA1 expression to control circulating lipoprotein levels. Nat Med. 2015;21:1280–1289. doi: 10.1038/nm.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.