Abstract
MicroRNAs (miRNAs) are involved in the development of non-small cell lung cancer (NSCLC). However, the biological roles of several aberrantly expressed miRNAs have not been explored yet. In the present study, miR-4491 was identified as a novel upregulated miRNA in NSCLC tissues and cell lines. Downregulation of miR-4491 by a miR-4491 inhibitor inhibited the proliferation and triggered the apoptosis of NSCLC cells. Tripartite motif containing 7 (TRIM7), a tumor suppressor gene expressed in NSCLC, was demonstrated in the present study to be directly targeted by miR-4491. This finding was verified by bioinformatics analysis, reverse transcription-quantitative PCR, western blotting and dual luciferase reporter assays. Furthermore, downregulation of miR-4491 inactivated nuclear factor-κB signaling via induction of TRIM7. In addition, TRIM7 silencing attenuated the effect of miR-4491 inhibitor in NSCLC cells. The decreased TRIM7 level in NSCLC tissues was negatively correlated with miR-4491 expression in NSCLC tissues. In conclusion, the findings from this study demonstrated that miR-4491 expression was upregulated in NSCLC tissues and cells and that miR-4491 may promote NSCLC progression via targeting TRIM7.
Keywords: non-small cell lung cancer, cell proliferation, cell apoptosis, miR-4491, tripartite motif containing 7
Introduction
Lung cancer (LC) is a life-threatening disease with 1,735,350 new cancer cases diagnosed and 609,640 mortality cases in 2018 in the United States and ranks second among all types of cancer worldwide (1,2). The major risk factor for LC is smoking, which accounts for 75–80% of LC-associated mortality (3). Non-small cell lung cancer (NSCLC) accounts for 85% of LC cases and can be further divided into two main subtypes, including lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD) (4). The occurrence and development of NSCLC involve a multistep carcinogenic process, which includes numerous variations of gene expression, for instance, V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog, and signal transduction, including nuclear factor-κB (NF-κB), ERK1/2 and AKT (5). The combination of surgery and chemotherapy remains the primary treatment method for patients with NSCLC (6). Despite significant improvements in the detection, diagnosis and targeted therapy, the 5-year survival rate of patients with NSCLC is only ~15% (7). The underlying mechanisms promoting the development of NSCLC are very specific and require further investigation in order to develop novel therapeutic strategies.
MicroRNAs (miRNAs) are non-coding RNA molecules of ~20 nucleotides in length that bind to the 3′ untranslated region (UTR) of the mRNA. This miRNA-mRNA interaction results in mRNA degradation or translation repression, which regulates target gene expression (8). Furthermore, it has been verified that miRNAs serve as regulators in multiple physiological and pathological conditions (9). The expression of specific miRNAs is commonly dysregulated in tumor tissues, indicating that miRNAs may serve as oncogenes or tumor suppressors according to their target genes (10), leading to the regulation of cell proliferation, cell death and metastasis (11). Based on their functions, miRNAs are considered as therapeutic agents (8,12).
Numerous miRNAs have been reported to be involved in the development of NSCLC. For example, miR-486 was found to be associated with the overall survival of patients with advanced-stage NSCLC (13), whereas miRNA-148a can inhibit NSCLC cell invasion and migration in vitro as well as cancer metastasis in vivo (14). Furthermore, based on the analysis of the miRNA expression profile from the peripheral blood of healthy controls and patients with LC, miR-4491 was shown to be increased (15). However, its role in the development of NSCLC remains unknown.
Materials and methods
Collection of tissue samples
Tumor and matched normal tissues were collected from 78 patients (62 men and 16 women) diagnosed with NSCLC at the Shanxi Provincial Cancer Hospital between March 2016 and October 2018. The age range of patients was 45–76 years, 33 patients were ≤65 years and 45 patients were >65 years. The clinicopathological characteristics of patients are listed in Table I. A total of 52 patients were at I–II stages, 26 patients were at III–IV stages, 44 patients were diagnosed with LUAD, 34 patients were diagnosed with LUSC, 37 patients had no lymph node metastasis and 41 patients exhibited lymph node metastasis. All patients provided written informed consent prior to study enrollment. The present study was approved by the Ethics Committee of Shanxi Provincial Cancer Hospital (IRB no. SXPCP20160302).
Table I.
Clinicopathological characteristics of patients with non-small cell lung cancer.
| Characteristics | Patient number |
|---|---|
| Age | |
| ≤65 | 33 |
| >65 | 45 |
| Sex | 62 |
| Male | 16 |
| Female | |
| TNM stage | |
| I–II | 52 |
| III–IV | 26 |
| Pathological type | |
| Adenocarcinoma | 44 |
| Squamous cell carcinoma | 34 |
| Lymph node metastasis | |
| No | 37 |
| Yes | 41 |
Cell lines
The bronchial epithelium transformed cell line BEAS-2B and the LUAD cell lines A549 and NCI-H1650 were purchased from the American Type Culture Collection. The cells were maintained in RPMI-1640 medium (HyClone; Cytiva), which was supplemented with 10% fetal bovine serum (HyClone) and placed at 37°C in a humidified incubator containing 5% CO2.
Cell transfections
miR-negative control mimic (NC, 50 nM, 5′-UAGUCUCGGGAGACUCACUACC-3′), miR-NC inhibitor (50 nM, 5′-UAACCGAAUUCACAUGGUCCUA-3′), miR-4491 inhibitor (50 nM, 5′-UUUGGUCACACCAGUCCACAUU-3′), miR-4491 mimic (50 nM, 5′-AAUGUGGACUGGUGUGACCAAA-3′), si-control (100 nM, 5′-TTCTCCGAACGTGTCACGTTT-3′) and si-Tripartite motif containing 7 (TRIM7, 100 nM, 5′-CGGAAAAGAAGGAGAGCAA-3′) were synthesized by GenePharma. Cell transfections were performed by Lipofectamine 2000® solution (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Transfections were performed at 37°C for 48 h and cells were collected for subsequent experiments.
miR-4491 expression profile
The Starbase V2.0 project (http://starbase.sysu.edu.cn/) was used for the analysis of miR-4491 expression levels in NSCLC and normal tissues from The Cancer Genome Atlas (TCGA) datasets (TCGA-LUAD and TCGA-LUSC) (16).
mRNA and miRNA quantification
Total RNA was isolated from tumor and normal tissues derived from patients with NSCLC and from BEAS-2B, A549 and NCI-H1650 cells using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.). cDNA synthesis was performed using TaqMan microRNA reverse transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) and TransScript First-Strand cDNA Synthesis SuperMix (Beijing TransGen Biotech Co., Ltd.). Reverse transcription-quantitative PCR (RT-qPCR) experiments were performed on an ABI7500 system (Applied Biosystems; Thermo Fisher Scientific, Inc.) under the following conditions: 95°C for 5 min, followed by 40 cycles of 95°C for 10 sec, 60°C for 30 sec and 72°C for 1 sec. miR-4491 levels were normalized to those of U6. TRIM7 levels were normalized to those of GAPDH. The primers were synthetized by GenScript. The sequences of primers were as follows: GAPDH, forward 5′-GAAGGTGAAGGTCGGAGTC-3′, reverse 5′-GAAGATGGTGATGGGATTTC-3′; U6, forward 5′-GCTTCGAGGCAGGTTACATG-3′, reverse 5′-GCAACACACAACATCTCCCA-3′; TRIM7, forward 5′-ATTATATAGGGTGTCCACATA-3′, reverse 5′-TATGTGGACACCCTATATAAT-3′; miR-4491, forward 5′-AATGTGGACTGGTGTGACCAAA-3′ and reverse 5′-TTTGGTCACACCAGTCCACATT-3′. The relative expression levels were normalized to endogenous control and were expressed as 2−ΔΔCq (17).
Western blotting
A549 and NCI-H1650 cells were lysed using RIPA on ice buffer (Sigma-Aldrich; Merck KGaA). The protein concentration was determined using the BCA method (Sigma-Aldrich; Merck KGaA). Proteins (20 µg) were separated by 8% SDS-PAGE and were transferred onto PVDF membranes. Membranes were blocked by 5% skimmed milk for 2 h at room temperature and incubated with primary antibodies against TRIM7 (cat. no. ab105330; 1:1,000; Abcam), p65 (cat. no. ab32536; 1:1,000; Abcam) and β-actin (cat. no. 4970; 1:1,000; Cell Signaling Technology, Inc.) overnight at 4°C. Membranes were then incubated with goat-anti-rabbit secondary antibody (cat. no. 7074; 1:2,000, Cell Signaling Technology, Inc.) for 2 h at room temperature. The protein blots were developed by ECL Western Blot Kit (Pierce; Thermo Fisher Scientific, Inc.). Band intensities were analyzed by Image Lab™ (version 4.0; Bio-Rad Laboratories, Inc.). Relative expression levels were normalized to endogenous control β-actin using ImageLab™ (version 3.0; Bio-Rad Laboratories, Inc.).
Cell proliferation assay
The proliferation of A549 and NCI-H1650 cells was evaluated using the Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.). Briefly, A549 and NCI-H1650 cells (3×104) were seeded in 96-well plates and incubated at 37°C for 48 h. Subsequently, cells were incubated with CCK-8 reagent (10 µl) for 3 h at 37°C. The absorbance was measured at 450 nm by spectrophotometry.
Cell apoptosis assay
A549 and NCI-H1650 cell apoptosis was assessed with the Annexin V-FITC/propidium (PI) apoptosis detection kit (Sigma-Aldrich; Merck KGaA). A549 and NCI-H1650 cells (1×106) were seeded in 6-well plates and incubated at 37°C for 48 h. Subsequently, cells were stained with Annexin V (5 µl) and PI (1 µl) for 10 min at room temperature in the dark. The number of apoptotic cells was estimated by a FACSCalibur flow cytometer (BD Biosciences). The data were analyzed by the CellQuest software (version 3.3; BD Biosciences).
Prediction of target genes
miRWalk (http://mirwalk.umm.uni-heidelberg.de/) and TargetScan (http://www.targetscan.org/vert_72/) softwares were used to predict miR-4491 targets. The targets predicted by miRWalk were further selected for score analysis. A score of 1.0 was considered as optimal. The 50 top ranked targets of miR-4491 predicted by TargetScan were also selected. The two gene sets were overlapped by online Vein Map (http://bioinformatics.psb.ugent.be/webtools/Venn/). The functions of the 7 overlapped genes were subsequently investigated by literature review.
Dual luciferase reporter assays
Wild-type or mutant TRIM7 3′UTR were ligated into pGL3-luciferase reporter plasmids (Promega Corporation), co-transfected with miR-4491 mimic or miR-NC into A549 and NCI-H1650 cells by Lipofectamine® 2000 solution (Invitrogen; Thermo Fisher Scientific, Inc.) and incubated for 48 h at 37°C. Subsequently, A549 and NCI-H1650 cells were trypsinized and transferred to 24-well plates. Luciferase reporter activities were examined by the Dual Luciferase Assay System (Promega Corporation). The luciferase reporter activities were normalized to the Renilla luciferase activity (Promega Corporation).
Statistical analysis
The data were analyzed by GraphPad Prism 6 (GraphPad Software, Inc.) and were expressed as the means ± standard deviation. The differences between two groups from tissues and cell lines were analyzed using paired or unpaired Student's t-test, respectively. The differences between three groups were analyzed by one-way ANOVA and Newman Keuls analysis. The correlation between miR-4491 and TRIM7 expression was analyzed by Pearson Correlation analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-4491 levels are increased in patients with NSCLC
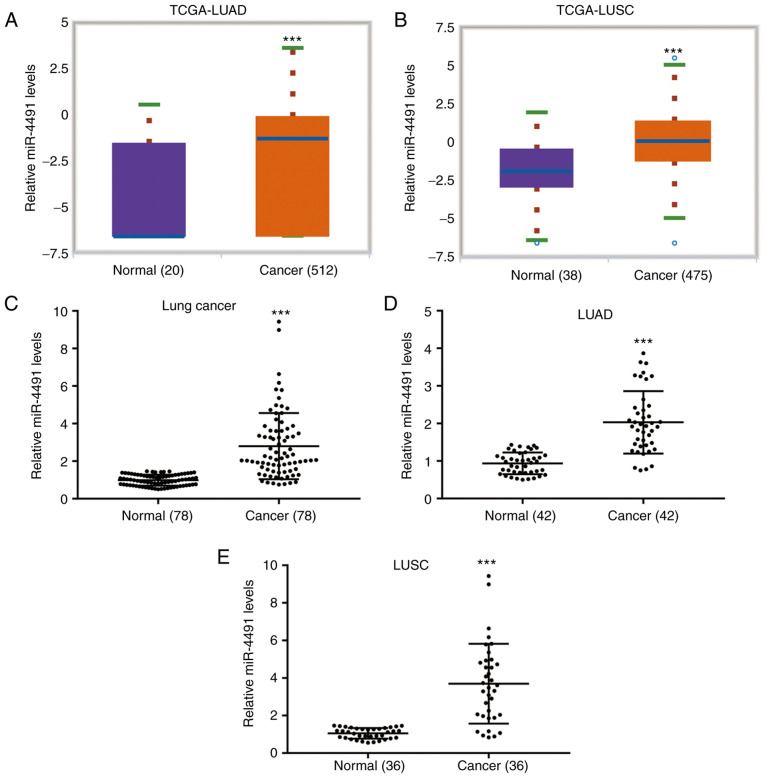
The data derived from TCGA indicated that miR-4491 levels were significantly increased in cancer tissues from patients with LUAD (n=512) compared with adjacent normal tissues from cancer patients (n=20; Fig. 1A). In addition, miR-4491 levels were significantly increased in cancer tissues from patients with LUSC (n=475) compared with those in adjacent normal tissues from cancer patients (n=38; Fig. 1B). The data derived from patient tissues collected in the present study indicated that the miR-4491 levels in NSCLC tissues (n=78) were significantly higher compared with those in the adjacent normal tissues (Fig. 1C). A similar expression profile of miR-4491 was also observed in patients with LUAD (n=42; Fig. 1D) and LUSC (n=36; Fig. 1E).
Figure 1.
miR-4491 expression was increased in patients with non-small cell lung cancer. Data from The Cancer Genome Atlas database indicated that miR-4491 expression was significantly increased in patients with (A) LUAD and (B) LUSC. (C) Data derived from NSCLC patient tissues demonstrated that miR-4491 expression was significantly higher in tumor tissues compared with those in normal tissues. Analysis included patients with (D) LUAD and (E) LUSC. ***P<0.001 vs. normal. miR, microRNA; NSCLC, non-small cell lung cancer; LUSC, lung squamous cell carcinoma; LUAD, lung adenocarcinoma.
Downregulation of miR-4491 negatively affects the proliferation and induces the apoptosis of NSCLC cells
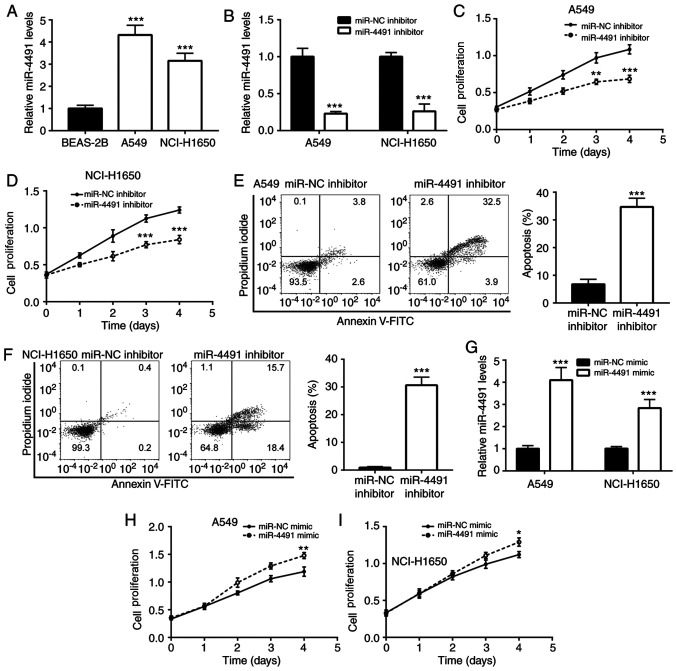
To detect the expression profile of miR-4491 in NSCLC cell lines, A549 and NCI-H1650 cells were selected. The results indicated that miR-4491 expression was significantly increased in A549 and NCI-H1650 cells compared with BEAS-2B cells (Fig. 2A).
Figure 2.
Downregulation of miR-4491 decreased the proliferation and induced the apoptosis of NSCLC cell lines. (A) miR-4491 expression was significantly increased in A549 and NCI-H1650 cells compared with that of BEAS-2B cells. A549 and NCI-H1650 cell transfection miR-4491 inhibitor decreased (B) miR-4491 expression and (C and D) cell proliferation and (E and F) increased cell apoptosis. A549 and NCI-H1650 cell transfection with miR-4491 mimic increased (G) miR-4491 expression and (H and I) cell proliferation. *P<0.05, **P<0.01 and ***P<0.001 vs. miR-NC mimic, miR-NC inhibitor or BEAS-2B cells. miR, microRNA; NSCLC, non-small cell lung cancer; NC, negative control.
Furthermore, the effects of the downregulation of miR-4491 in NSCLC cell lines were determined. The downregulation of miR-4491 expression by the miR-4491 inhibitor was confirmed in A549 and NCI-H1650 cells (Fig. 2B). Subsequently, transfection of A549 and NCI-H160 cell with miR-4491 inhibitor resulted in a decrease in cell proliferation (Fig. 2C and D) in a time-dependent manner compared with that of the miR-NC group. In addition, miR-4491 inhibitor induced apoptosis of A549 and NCI-H1650 cells (Fig. 2E and F).
In addition, the effects of miR-4491 overexpression were determined in NSCLC cell lines. The overexpression of miR-4491 in A549 and NCI-H1650 cells using miR-4491 mimic was verified by RT-qPCR analysis (Fig. 2G). The effects caused by the miR-4491 mimic were opposite to those caused by the miR-4491 inhibitor. miR-4491 mimic increased A549 and NCI-H1650 cell proliferation (Fig. 2H and I) in a time-dependent manner.
TRIM7 is targeted by miR-4491 in NSCLC cell lines
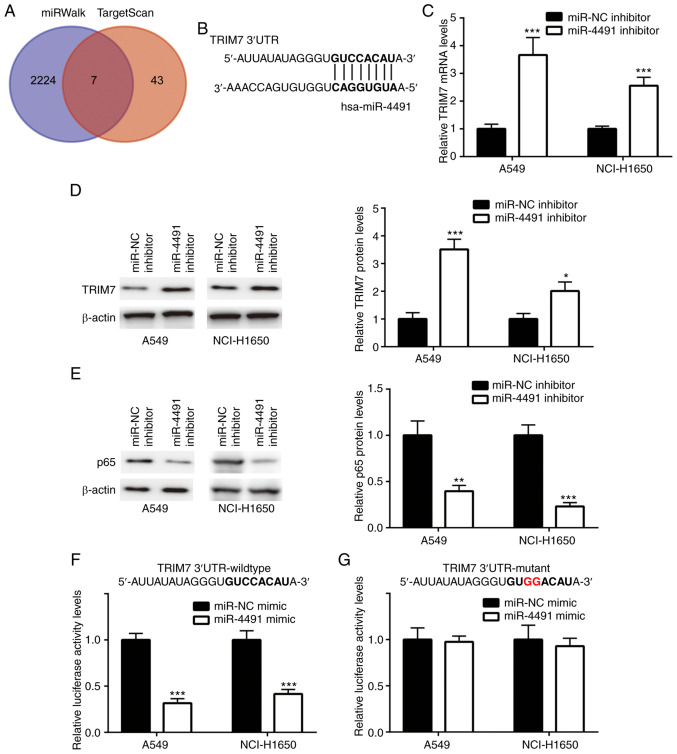
The gene sets from miRWalk (n=2,231) and TargetScan (n=51) were retrieved and 7 overlapping genes (Fig. 3A) were identified. The complementary sites between TRIM7 and miR-4491 are presented in Fig. 3B.
Figure 3.
TRIM7 was targeted by miR-4491 in non-small cell lung cancer cell lines. (A) Target genes of miR-4491. (B) Complementary sites between TRIM7 and miR-4491. (C and D) miR-4491 inhibitor significantly increased TRIM7 expression level and (E) decreased p65 protein expression in A549 and NCI-H1650 cells. miR-4491 mimic significantly decreased the relative luciferase activity of (F) TRIM7 wild-type but not (G) TRIM7 mutant variants in A549 and NCI-H1650 cells. *P<0.05, **P<0.01 and ***P<0.001 vs. miR-NC mimic or miR-NC inhibitor. TRIM7, tripartite motif containing 7; miR, microRNA; NC, negative control.
Cell transfection with miR-4491 inhibitor induced a significant upregulation of TRIM7 mRNA and protein levels (Fig. 3C and D) and a concomitant significant downregulation of the protein levels of p65 (Fig. 3E) in A549 and NCI-H1650 cells. Furthermore, miR-4491 mimic significantly decreased the relative luciferase activity of TRIM7 wild-type (Fig. 3F) but not TRIM7 mutant (Fig. 3G) in A549 and NCI-H1650 cells.
Downregulation of miR-4491 decreases p65 levels by targeting TRIM7 in NSCLC cells
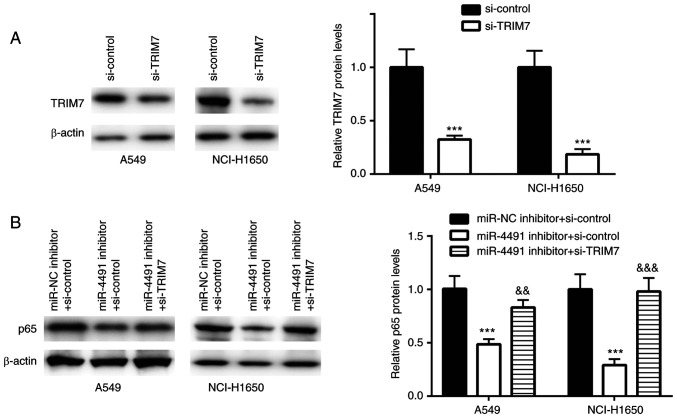
TRIM7 was successfully downregulated using siRNA transfection in A549 and NCI-H1650 cells (Fig. 4A).
Figure 4.
Downregulation of miR-4491 inhibited p65 protein expression by targeting TRIM7 in non-small cell lung cancer cell lines. (A) si-TRIM7 significantly decreased TRIM7 protein expression in A549 and NCI-H1650 cells. (B) Effect of si-TRIM7 was rescued by the miR-4491 inhibitor and resulted in decreased p65 protein expression in A549 and NCI-H1650 cells. ***P<0.001 vs. si-control or miR-NC inhibitor + si-control. &&P<0.01 and &&&P<0.001 vs. miR-4491 inhibitor + si-control. miR, microRNA; TRIM7, tripartite motif containing 7; NC, negative control; si, small interfering.
In A549 and NCI-H1650 cells transfected with miR-4491 inhibitor, p65 protein expression was significantly decreased, an effect that was rescued following co-transfection with si-TRIM7 (Fig. 4B).
Downregulation of miR-4491 negatively affects the proliferation and triggers the apoptosis of NSCLC cells by targeting TRIM7
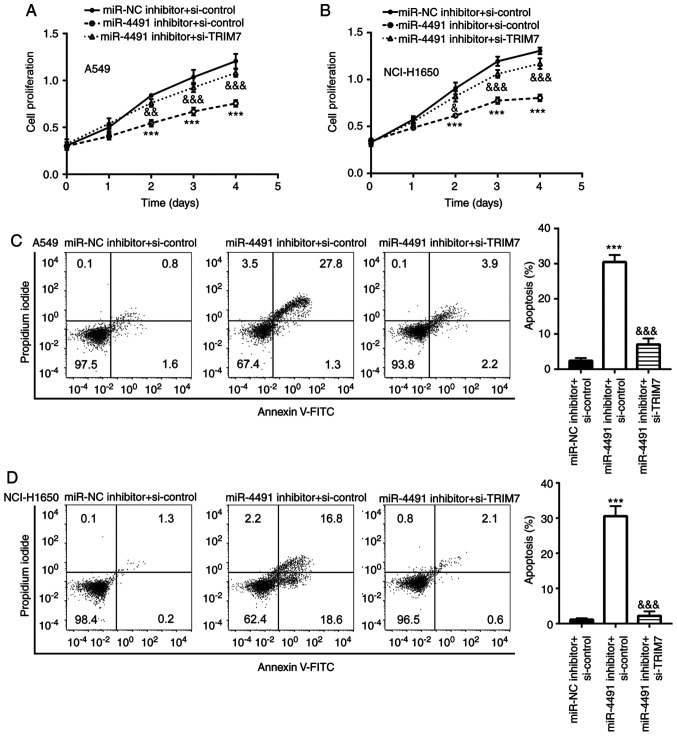
The miR-4491 inhibitor inhibited the proliferation of A549 and NCI-H1650 cells (Fig. 5A and B) in a time-dependent manner compared with that of the miR-NC + si-control group, which was rescued following the co-transfection with si-TRIM7. In addition, the miR-4491 inhibitor induced A549 and NCI-H1650 cell apoptosis (Fig. 5C and D), which was reversed following the co-transfection with si-TRIM7.
Figure 5.
Downregulation of miR-4491 inhibited cell proliferation and induced cell apoptosis by targeting TRIM7 in non-small cell lung cancer cell lines. Effect of si-TRIM7 was rescued by miR-4491 inhibitor and resulted in (A and B) decreased proliferation and (C and D) increased apoptosis of A549 and NCI-H1650 cells. ***P<0.001 vs. miR-NC + si-control. &P<0.05, &&P<0.01 and &&&P<0.001 vs. miR-4491 inhibitor + si-control. miR, microRNA; TRIM7, tripartite motif containing 7; NC, negative control; si, small interfering.
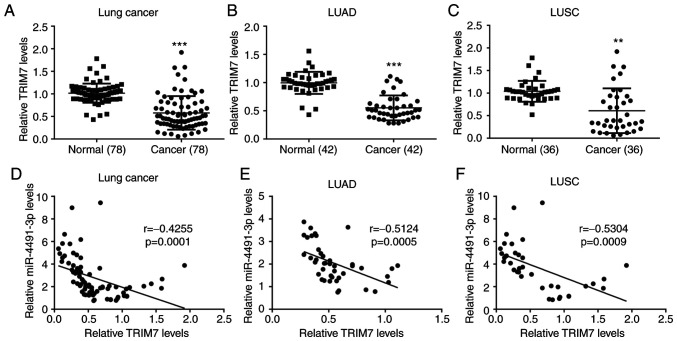
miR-4491 expression is negatively correlated with TRIM7 expression in NSCLC tissues
The data derived from patient tissues collected in the present study indicated that TRIM7 expression in NSCLC tissues (n=78) was significantly lower compared with that in the adjacent normal tissues (Fig. 6A). A similar expression profile of TRIM7 was also observed in patients with LUAD (n=42; Fig. 6B) and LUSC (n=36; Fig. 6C). In addition, miR-4491 expression was negatively correlated with TRIM7 expression in NSCLC tissues (Fig. 6D), LUAD tissues (Fig. 6E) and LUSC tissues (Fig. 6F).
Figure 6.
miR-4491 expression is negatively correlated with TRIM7 expression in patients with NSCLC. TRIM7 expression level in tumor tissues from patients with (A) NSCLC, (B) LUAD and (C) LUSC was significantly lower than those in adjacent normal tissues. There was a negative correlation between miR-4491 and TRIM7 expression in (D) NSCLC tissues, (E) LUAD tissues and (F) LUSC tissues. **P<0.01 and ***P<0.001, vs. normal. miR, microRNA; TRIM7, tripartite motif containing 7; NSCLC, non-small cell lung cancer; LUSC, lung squamous cell carcinoma; LUAD, lung adenocarcinoma.
Discussion
miRNAs are emerging biomarkers used in the diagnosis and treatment of patients with NSCLC (18,19). In particular, miR-21 (20), miR-9 (21) and miR-143 (22) have been used as biomarkers of NSCLC. miR-4491 has shown potential diagnostic value for several diseases, as it was reported to be downregulated in ischemic stroke (23), while it is upregulated in gastric cancer (24) and chronic heart failure (25). A recent study reported a diagnostic value of miR-4491 in LC (15). However, its therapeutic value in NSCLC has yet to be reported.
In the present study, the data from TCGA and from the collected tumor tissues indicated that miR-4491 expression was increased in tumor tissues of patients with NSCLC compared with normal matched tissues. Furthermore, miR-4491 downregulation negatively affected the proliferation and triggered the apoptosis of NSCLC cells. These findings suggested that miR-4491 may have an oncogenic role in NSCLC.
The miRNA-mRNA interaction regulates target gene expression by inducing mRNA degradation or translational repression (8). This interaction might therefore regulate cell proliferation, cell death and metastasis (11). The involvement of miR-4491 target genes in NSCLC requires thus further investigation in order to determine the function of miR-4491 in NSCLC.
In the present study, miRWalk and TargetScan were used to identify 7 overlapping genes, including INTS3 and NABP interacting protein (INIP), protein O-mannose kinase (POMK), muscle RAS oncogene homolog (MRAS), alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase B (MGAT4B), angiotensin II receptor type 1 (AGTR1), chromosome 1 open reading frame 116 (C1orf116) and TRIM7, which could all be targeted by miR-4491. Among these genes, there has not been any reports about the function of INIP or POMK in the development of cancer. MRAS and MGAT4B however were reported as oncogenic genes, and MRAS can initiate tumor formation in lungs (26). MGAT4B transcripts are also upregulated in diethylnitrosamine-induced mouse model for hepatocellular carcinoma (27). There are only few reports about the expression profile of AGTR1 and C1orf116 in patients with lung cancer. AGTR1 expression is shown to be decreased in LUAD tissues compared with adjacent normal tissues (28), and downregulation of C1orf116 is associated with a poor prognosis of patients with lung cancer (29). TRIM7 belongs to the TRIM protein family, which is involved in cell proliferation (30), cell apoptosis (31) and immunity (32), and is a well-known tumor suppressor involved in the development of various types of cancer. For example, TRIM7 inhibits the progression of hepatocellular carcinoma by negatively regulating Src (33), and TRIM7 expression is decreased in tumor tissues from patients with LC (34). Subsequently, TRIM7 was selected for the subsequent experimentations in the present study.
In the present study, TRIM7 was targeted and negatively regulated by miR-4491. However, the downstream targets of TRIM7 were not detected.
NF-κB consists of 5 subunits, including NF-κB1 (p50 and its precursor p105), NF-κB2 (p52 and its precursor p100), RelA (p65), RelB and c-Rel. In addition, the p50/65 heterodimer is enriched in nearly all types of cells (35,36). NF-κB can target genes that stimulate cell proliferation, inflammation, angiogenesis, metastasis and cancer cell resistance to chemotherapy and radiotherapy (36). NF-κB p65 is commonly activated in LC (37). TRIM7 has been initially identified to degrade p65 in LC (34). The present study demonstrated that miR-4491 inhibitor decreased p65 protein expression in NSCLC cells. This effect was reversed following transfection with si-TRIM7. These observations were consistent with previous findings.
However, whether TRIM7 could affect the role of miR-4491 in the proliferation and apoptosis of NSCLC cells remains unknown. Cell transfection with miR-4491 inhibitor negatively affected cell proliferation and triggered apoptosis, whereas these effects were reversed by si-TRIM7 in NSCLC cells.
The present study demonstrated also that TRIM7 expression in cancer tissues from patients with NSCLC, including LUAD and LUSC, was downregulated compared with the adjacent normal tissues. In addition, miR-4491 expression was negatively correlated with TRIM7 expression in NSCLC tissues, including LUAD and LUSC.
Taken together, the results from the present study suggested that miR-4491 may enhance cell proliferation and resistance to apoptosis as well as the activation of p65 in NSCLC by targeting TRIM7.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The data and materials are available from the corresponding author on reasonable request.
Authors' contributions
FH, GC, YG, BL, YS, XQ, HT and XZ conducted the experimentations, data interpretation and data analysis. HZ designed the experimentations, performed the data interpretation and data analysis, and wrote the article. FH, GC, YG, BL, YS, XQ, HT, XZ and HZ confirmed the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Shanxi Provincial Cancer Hospital (IRB no. SXPCP20160302). All patients provided written informed consent prior to study enrollment.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Cersosimo RJ. Lung cancer: A review. Am J Health Syst Pharm. 2002;59:611–642. doi: 10.1093/ajhp/59.7.611. [DOI] [PubMed] [Google Scholar]
- 4.Inamura K. Lung cancer: Understanding its molecular pathology and the 2015 WHO classification. Front Oncol. 2017;7:193. doi: 10.3389/fonc.2017.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalari KR, Rossell D, Necela BM, Asmann YW, Nair A, Baheti S, Kachergus JM, Younkin CS, Baker T, Carr JM, et al. Deep sequence analysis of non-small cell lung cancer: Integrated analysis of gene expression, alternative splicing, and single nucleotide variations in lung adenocarcinomas with and without oncogenic KRAS mutations. Front Oncol. 2012;2:12. doi: 10.3389/fonc.2012.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ, Jr, Wu YL, Paz-Ares L. Lung cancer: Current therapies and new targeted treatments. Lancet. 2017;389:299–311. doi: 10.1016/S0140-6736(16)30958-8. [DOI] [PubMed] [Google Scholar]
- 7.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Sosman JA, Atkins MB, Leming PD, et al. Five-year survival and correlates among patients with advanced melanoma, renal cell carcinoma, or non-small cell lung cancer treated with nivolumab. JAMA Oncol. 2019;5:1411–1420. doi: 10.1001/jamaoncol.2019.2187. (Online ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shah MY, Ferrajoli A, Sood AK, Lopez-Berestein G, Calin GA. Microrna therapeutics in cancer-an emerging concept. EBioMedicine. 2016;12:34–42. doi: 10.1016/j.ebiom.2016.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchan JR, Parker R. Molecular biology. The two faces of miRNA. Science. 2007;318:1877–1878. doi: 10.1126/science.1152623. [DOI] [PubMed] [Google Scholar]
- 10.Goh JN, Loo SY, Datta A, Siveen KS, Yap WN, Cai W, Shin EM, Wang C, Kim JE, Chan M, et al. microRNAs in breast cancer: Regulatory roles governing the hallmarks of cancer. Biol Rev Camb Philos Soc. 2016;91:409–428. doi: 10.1111/brv.12176. [DOI] [PubMed] [Google Scholar]
- 11.Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011;223:102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henry JC, Azevedo-Pouly ACP, Schmittgen TD. MicroRNA replacement therapy for cancer. Pharm Res. 2011;28:3030–3042. doi: 10.1007/s11095-011-0548-9. [DOI] [PubMed] [Google Scholar]
- 13.Hu Z, Chen X, Zhao Y, Tian T, Jin G, Shu Y, Chen Y, Xu L, Zen K, Zhang C, Shen H. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol. 2010;28:1721–1726. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- 14.Li J, Yu T, Cao J, Liu L, Liu Y, Kong HW, Zhu MX, Lin HC, Chu DD, Yao M, Yan MX. MicroRNA-148a suppresses invasion and metastasis of human non-small-cell lung cancer. Cell Physiol Biochem. 2015;37:1847–1856. doi: 10.1159/000438546. [DOI] [PubMed] [Google Scholar]
- 15.He Q, Fang Y, Lu F, Pan J, Wang L, Gong W, Fei F, Cui J, Zhong J, Hu R, et al. Analysis of differential expression profile of miRNA in peripheral blood of patients with lung cancer. J Clin Lab Anal. 2019;33:e23003. doi: 10.1002/jcla.23003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cancer Genome Atlas Research Network. Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Q, Huang SX, Zhang F, Li SJ, Liu C, Xi YY, Wang L, Wang X, He QQ, Sun CC, Li DJ. MicroRNAs: A novel potential biomarker for diagnosis and therapy in patients with non-small cell lung cancer. Cell Prolif. 2017;50:e12394. doi: 10.1111/cpr.12394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petrek H, Yu AM. MicroRNAs in non-small cell lung cancer: Gene regulation, impact on cancer cellular processes, and therapeutic potential. Pharmacol Res Perspect. 2019;7:e00528. doi: 10.1002/prp2.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bica-Pop C, Cojocneanu-Petric R, Magdo L, Raduly L, Gulei D, Berindan-Neagoe I. Overview upon miR-21 in lung cancer: Focus on NSCLC. Cell Mol Life Sci. 2018;75:3539–3551. doi: 10.1007/s00018-018-2877-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei W, Dong Z, Gao H, Zhang YY, Shao LH, Jin LL, Lv YH, Zhao G, Shen YN, Jin SZ. MicroRNA-9 enhanced radiosensitivity and its mechanism of DNA methylation in non-small cell lung cancer. Gene. 2019;710:178–185. doi: 10.1016/j.gene.2019.05.050. [DOI] [PubMed] [Google Scholar]
- 22.Jiang Q, Yuan Y, Gong Y, Luo X, Su X, Hu X, Zhu W. Therapeutic delivery of microRNA-143 by cationic lipoplexes for non-small cell lung cancer treatment in vivo. J Cancer Res Clin Oncol. 2019;145:2951–2967. doi: 10.1007/s00432-019-03051-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu X, Liu X, Liu Y, Chang W, Song Y, Zhu S. Uncovering the potential differentially expressed miRNAs and mRNAs in ischemic stroke based on integrated analysis in the gene expression omnibus database. Eur Neurol. 2020;9:404–414. doi: 10.1159/000507364. [DOI] [PubMed] [Google Scholar]
- 24.Yang B, Jing C, Wang J, Guo X, Chen Y, Xu R, Peng L, Liu J, Li L. Identification of microRNAs associated with lymphangiogenesis in human gastric cancer. Clin Transl Oncol. 2014;16:374–379. doi: 10.1007/s12094-013-1081-6. [DOI] [PubMed] [Google Scholar]
- 25.Li H, Fan J, Yin Z, Wang F, Chen C, Wang DW. Identification of cardiac-related circulating microRNA profile in human chronic heart failure. Oncotarget. 2016;7:33–45. doi: 10.18632/oncotarget.6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mishra J, Kumar A, Sinha A, Das S, Srivastava A. Ingenuity in pattern recognition: A novel bioinformatics approach towards lung cancer identification. Int J Bioinform Res Appl. 2010;6:531–541. doi: 10.1504/IJBRA.2010.038735. [DOI] [PubMed] [Google Scholar]
- 27.Blomme B, Heindryckx F, Stassen JM, Geerts A, Colle I, Van Vlierberghe H. Serum protein N-glycan alterations of diethylnitrosamine-induced hepatocellular carcinoma mice and their evolution after inhibition of the placental growth factor. Mol Cell Biochem. 2013;372:199–210. doi: 10.1007/s11010-012-1461-1. [DOI] [PubMed] [Google Scholar]
- 28.Goldstein B, Trivedi M, Speth RC. Alterations in gene expression of components of the renin-angiotensin system and its related enzymes in lung cancer. Lung Cancer Int. 2017;2017:6914976. doi: 10.1155/2017/6914976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parsana P, Amend SR, Hernandez J, Pienta KJ, Battle A. Identifying global expression patterns and key regulators in epithelial to mesenchymal transition through multi-study integration. BMC Cancer. 2017;17:447. doi: 10.1186/s12885-017-3413-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen L, Chen DT, Kurtyka C, Rawal B, Fulp WJ, Haura EB, Cress WD. Tripartite motif containing 28 (Trim28) can regulate cell proliferation by bridging HDAC1/E2F interactions. J Biol Chem. 2012;287:40106–40118. doi: 10.1074/jbc.M112.380865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zaman MM, Nomura T, Takagi T, Okamura T, Jin W, Shinagawa T, Tanaka Y, Ishii S. Ubiquitination-deubiquitination by the TRIM27-USP7 complex regulates tumor necrosis factor alpha-induced apoptosis. Mol Cell Biol. 2013;33:4971–4984. doi: 10.1128/MCB.00465-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Versteeg GA, Benke S, Garcia-Sastre A, Rajsbaum R. InTRIMsic immunity: Positive and negative regulation of immune signaling by tripartite motif proteins. Cytokine Growth Factor Rev. 2014;25:563–576. doi: 10.1016/j.cytogfr.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu L, Qin C, Li T, Ma X, Qiu Y, Lin Y, Ma D, Qin Z, Sun C, Shen X, et al. The E3 ubiquitin ligase TRIM7 suppressed hepatocellular carcinoma progression by directly targeting Src protein. Cell Death Differ. 2020;27:1819–1831. doi: 10.1038/s41418-019-0464-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin J, Lu Z, Wang X, Liu Y, Han T, Wang Y, Wang T, Gan M, Xie C, Wang J, Yu B. E3 ubiquitin ligase TRIM7 negatively regulates NF-kappa B signaling pathway by degrading p65 in lung cancer. Cell Signal. 2020;69:109543. doi: 10.1016/j.cellsig.2020.109543. [DOI] [PubMed] [Google Scholar]
- 35.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 36.Park MH, Hong JT. Roles of NF-κB in cancer and inflammatory diseases and their therapeutic approaches. Cells. 2016;5:15. doi: 10.3390/cells5020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bassères DS, Ebbs A, Levantini E, Baldwin AS. Requirement of the NF-kappaB subunit p65/RelA for K-Ras-induced lung tumorigenesis. Cancer Res. 2010;70:3537–3546. doi: 10.1158/0008-5472.CAN-09-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data and materials are available from the corresponding author on reasonable request.