Abstract
Histamine is a pleiotropic mediator involved in a broad spectrum of (patho)-physiological processes, one of which is the regulation of inflammation. Compounds acting on three out of the four known histamine receptors are approved for clinical use. These approved compounds comprise histamine H1-receptor (H1R) antagonists, which are used to control allergic inflammation, antagonists at H2R, which therapeutically decrease gastric acid release, and an antagonist at H3R, which is indicated to treat narcolepsy. Ligands at H4R are still being tested pre-clinically and in clinical trials of inflammatory diseases, including rheumatoid arthritis, asthma, dermatitis, and psoriasis. These trials, however, documented only moderate beneficial effects of H4R ligands so far. Nevertheless, pre-clinically, H4R still is subject of ongoing research, analyzing various inflammatory, allergic, and autoimmune diseases. During inflammatory reactions in gut tissues, histamine concentrations rise in affected areas, indicating its possible biological effect. Indeed, in histamine-deficient mice experimentally induced inflammation of the gut is reduced in comparison to that in histamine-competent mice. However, antagonists at H1R, H2R, and H3R do not provide an effect on inflammation, supporting the idea that H4R is responsible for the histamine effects. In the present review, we discuss the involvement of histamine and H4R in inflammatory and inflammation-associated diseases of the gut.
Keywords: inflammation, cancer, colitis, mast cell, epithelial cell, gut
1. Introduction
The biogenic amine histamine (2-(4-imidazolyl)-ethylamine) is a short-acting local mediator present in virtually all mammalian tissues. It is generated by the enzyme L-histidine decarboxylase (HDC) [1], and degraded either intracellularly by methylation (histamine N-methyltransferase; HNMT) [2] or extracellularly by oxidation (diamine oxidase; DAO) [3]. Cells commonly known to produce histamine are mast cells and basophils [4,5], intestinal enterochromaffine-like cells [6], and histaminergic neuronal cells [7]. These cells constitutively express HDC and produce histamine, which is bound to acidic polysaccharides such as heparin and stored in granules [8,9]. Release of histamine from these cells can take place by different mechanisms: piecemeal degranulation, kiss-and-run exocytosis, and conventional full fusion exocytosis [10,11]. Conventional exocytosis is induced by a specific stimulus, e.g., Fcε receptor-mediated activation of mast cells, and results in the rapid but transient occurrence of high local histamine concentrations. In contrast, piecemeal degranulation in mast cells is observed in diseased tissues and results in sustained presence of histamine, however, at lower concentration. Other cells, such as monocytes, dendritic cells, and T cells [12], produce significant amounts of histamine only during inflammatory reactions [13,14], which induce expression of HDC. Histamine produced in these cells is immediately released instead of being stored. In comparison to the release from storage granules, this leads to slower but sustained release kinetics and resulting tissue concentrations remain lower [15,16].
The biological functions of histamine are pleiotropic and comprise, but are not restricted to, the regulation of gastric acid release, neurotransmission, and pain perception/hypersensitization as well as the contribution to inflammation. Due to the widely used anti-histamines, the most commonly known role of histamine in inflammation is to promote allergic reactions. But beyond that, histamine is involved in manifold key aspects of inflammation, such as shaping the Th1/Th17/Th2-balance and regulating access of other mediators and inflammatory cells to inflamed tissue by acting on resident immune cells and vascular endothelial cells. Moreover, pain and itch sensation from inflamed tissue are modulated by histamine acting on neuronal cells. The pleiotropic effects of histamine on inflammatory processes have been reviewed in detail elsewhere [17]. These different biological functions of histamine rely on the existence of four histamine receptor subtypes and their tissue and cellular distribution [18]. All the histamine receptor subtypes belong to the class A rhodopsin-like G protein-coupled receptors (GPCRs) and have been named histamine H1-receptor (H1R), H2R, H3R, and H4R in order of their discovery. While H1R and H2R are ubiquitously expressed and bind histamine with relatively low potency (pEC50 ~6), the expression of high potency (pEC50 ~8) histamine receptor subtypes H3R and H4R is rather restricted [19]. While H3R is found on mammalian neuronal cells [20], H4R originally has been identified in immune cells, such as mucosa-associated immune cells of the intestine. A more detailed summary of immune cells expressing H4R is presented in Table 1 (Table 1); note that caution has to be paid, since not all of the data provided in the cited literature have been verified by two independent methods (see discussion below). Meanwhile also non-immune cells, such as epithelial cells of the gut, have been shown to express H4R [21]. Thus, due to its expression pattern, H4R is an attractive pharmacological target to treat e.g., inflammatory reactions of the gut [22,23,24,25].
Table 1.
Expression of H4R in immune cells (mRNA and/or functional testing).
| Cell Type | Mouse | Human | Reference |
|---|---|---|---|
| CD4+ T cell | x | [26] | |
| Th2 | x | [27] | |
| Th9 | x | [28] | |
| Th17 | x | x | [29,30,31] |
| Treg | x | x | [32,33] |
| γδ T cell | x | [34] | |
| CD8+ T cell | x | [35] | |
| DC | x | x | [36,37,38,39,40,41,42,43,44,45,46,47] |
| NKT | x | [48] | |
| Mast cell | x | x | [26,49,50,51,52,53,54,55,56,57,58,59] |
| Neutrophil | x | x | [23,26,60,61] |
| Monocyte | x | [26,36,60,62,63] | |
| MΦ | x | [64] | |
| M1 | x | [65,66] | |
| M2 | x | [67] | |
| Basophil | x | [49,68,69] | |
| Eosinophil | x | x | [26,38,49,60,70,71,72,73,74,75,76,77] |
| NK cell | x | [43,78,79] |
2. H4R Basics
The existence of a distinct histamine receptor mediating calcium mobilization in eosinophils has been claimed already in 1994 [80]. Discovery and cloning of the corresponding H4R were published in the year 2000 quasi-contemporaneously by several independent groups. In essence, H4R was identified based on its relative high genetic homology (37%) with H3R, while showing less than 30% homology with H1R, H2R, and other biogenic amine receptors [26,60,70,81,82,83]. Engagement of agonists entails recruitment of Gαi proteins to H4R that subsequently leads to activation of phospholipase C as well as to inhibition of membrane-bound adenylyl cyclase activity. As a consequence, calcium ([Ca2+]i) is mobilized from intracellular stores and the concentration of cytosolic cAMP is diminished. Independent of G protein-signaling, β-arrestin 2 is recruited to agonist-bound H4R, initiating mitogen-activated protein kinase (MAPK) cascade activation as well as receptor desensitization and internalization [84,85,86,87]. Activation of the H4R signaling pathways lead to induction of pro-inflammatory AP-1 in Th2 cells and monocyte-derived dendritic cells (MoDC) [27]. Furthermore, production of the Th1-associated cytokines IL-12 and IP10 in MoDC is reduced by engaging the H4R, indicating that histamine might shape a Th2-biased immune response via these mechanisms [36,37,88]. Attraction and activation of eosinophils, mast cells, and dendritic cells could also be shown to be mediated by H4R activation, thus further amplifying the Th2 inflammatory phenotype [37,38,49]. If activation of H4R-signalling can be exploited to treat Th1/Th17-driven pathologies such as Crohn’s disease by inducing a Th2-bias remains controversial [89,90].
Pharmacological ligands, either agonists or antagonists/inverse agonists, or in vivo models using genetically modified animals, are the main tools to identify H4R functions on specific cells or tissues in health and disease. Compound JNJ7777120 (1-[(5-Chloro-1H-indol-2-yl)carbonyl]-4-methylpiperazine) was the first selective H4R antagonist developed and it has been used in a large number of laboratories in in vivo and in vitro analyses [91]. JNJ7777120 has turned out a biased ligand at the human H4R, inversely agonizing G protein-dependent signaling, while agonizing G protein-independent signaling. Thus, the results gained so far must be re-evaluated accordingly [92,93]. Moreover, it was claimed that JNJ7777120, while acting as inverse agonist at the human H4R, is an agonist at the mouse and rat H4R. These results, however, were obtained using membrane preparations of insect cells expressing recombinant human H4R together with selected G proteins and have never been reproduced in a native setting [94]. Unfortunately, a comprehensive evaluation of the activities of JNJ7777120 in different species remains to be performed. Alternative to JNJ7777120 other ligands at the H4R are available, providing agonistic and inverse agonistic/antagonistic activities. However, their use may be afflicted with problems concerning specificity, effectivity, and potency, too. Thus, in a complex biological system, studies on receptor function should not be based on a single selected ligand. At least chemical different ligands have to be employed or complementary experimental approaches must be applied. For example, genetic manipulation of the receptor to be studied can be used in addition to ligand-driven experimental systems. Of course, also vice versa, data generated by a genetic-based approach alone should be very carefully validated unless proved by an independent experimental system.
Mice are widely used to model human diseases since their handling is relatively easy and inexpensive. Furthermore, a huge array of different strains with defined genotypes is available. Among these are mice with a targeted mutation in the Hhr4 locus, which encodes the mouse H4R. These mice do not express a functional H4R and without any manipulation they do not demonstrate any biological abnormalities as compared to their wild type (WT) counterparts [49]. However, upon experimental induction of allergic diseases such as ovalbumin-induced asthma or experimental dermatitis, the lack of functional H4R leads to reduced symptoms [39,40,95]. These results have been reproduced by applying pharmacologic blockade of the H4R, and thus, the involvement of H4R in experimental asthma and dermatitis in mice is widely accepted. Unfortunately, in humans H4R antagonists did not gain the anticipated results regarding effectiveness and safety [96] (clinical trials.gov: NCT01823016 (asthma), NCT01493882 (asthma), NCT03517566 (dermatitis)). This discrepancy may be based on the H4R expression pattern, which is similar, but not necessarily identical in mice and man, as exemplarily demonstrated in Table 1 for immune cell types (Table 1). Whether this holds true for other tissues and cell types such as the gut as well remains to be analyzed in detail. Thus, if mice are able to serve as model for human H4R pathophysiology, has to be evaluated in a context (i.e., tissue/cell type)-dependent way. Using different approaches to manipulate H4R function, it became evident that of all different immune cells, H4R mainly regulates human and mouse mast cells and eosinophils, e.g., their migration and degranulation [38,49,50,71,72,97,98]. Because mast cells are the main producers of histamine and at the same time respond to histamine via the H4R, it is intriguing to speculate about the biological meaning of this auto-regulatory cycle. In eosinophils, the H4R seems to be only a minor player in comparison to classical activators and may be responsible for the cellular activities’ fine tuning [73]. For the evaluation of histamine effects, it also has to be taken into account that most cells do not exclusively express H4R. The observed effects elicited by histamine in cells, tissues, or animals are always the sum of effects elicited by all histamine receptor subtypes present, which are H1R, H2R, and H4R in the case of eosinophils and mast cells. This scenario becomes even more complicated, since, as described above, the histamine receptor subtypes possess different affinities for histamine and effective interstitial concentrations of histamine cannot be reliably measured so far.
3. Histamine in the Intestine
Inflammatory bowel diseases (IBD) are idiopathic, chronic-recurring diseases of the gut. Their two main manifestations, ulcerative colitis (UC) and Crohn’s disease (CD), differ in their clinical, endoscopic, and histologic appearance. In CD, the inflammation appears in diffuse lesions that can be found all over the digestive tract and deeply penetrates the intestinal wall, possibly affecting all layers. In contrast, inflammatory lesions in UC start in the rectum, proceed upwards but do not exceed the colon, and remain superficial at the mucosa [99,100,101,102]. CD and UC also differ regarding the immune response: while CD is largely associated with a Th1/Th17-dominated response, UC is mostly defined by a Th2-response [103,104,105]. Nevertheless, both ailments evoke a series of similar symptoms (e.g., mucosal lesions, ulcera, edema, diarrhea, bloody stool, abdominal pain) severely affecting the quality of patients’ lives and eventually limiting their life expectancy through extra- and intra-intestinal complications such as colorectal cancer (CRC) [100,101,106]. The risk to develop such complications is further augmented by the currently applied treatment schemes, which are based on immunosuppressive drugs such as 5-aminosalicylic acid or glucocorticoid receptor agonists. These drug demonstrate remission rates of only 50%, but long-term treatment can induce and/or support immunosuppression-related disorders [100,101,107].
It has been known for quite a long time that in affected colorectal mucosal samples obtained from CD and UC patients histamine concentrations are elevated in comparison to samples from unaffected sections or healthy control persons [108,109]. A first experimental hint that histamine may be involved in inflammatory diseases of the gut was identified in a model of acute colon inflammation, i.e., dextran sulfate sodium (DSS)-induced colitis. Comparing wild type mice to mice deficient in HDC expression, lack of HDC-mediated histamine synthesis resulted in reduced clinical symptoms, while inflammation of the colon was not affected. The authors attributed these findings to the reduced number of IL-10-producing lymphocytes in the colonic mucosa, probably reducing, but not abolishing, a Th2 response, and the altered composition of fecal bacteria [110]. Unfortunately, the explanation why reduced IL-10 production, commonly regarded as an anti-inflammatory cytokine, leads to reduced clinical symptoms but does not affect colonic inflammation was somewhat sparse. In fact, the possibility that IL-10 evokes differential functions in seemingly identical settings is quite well described. While mice devoid of IL-10 production commonly serve as models that develop severe enterocolitis, the susceptibility to develop colitis largely depends on the complex gut microbiota [111,112]. Thus, for the study performed by Bene et al. [110], while they excluded the contamination of their system by alimentary-provided histamine due to feeding a histamine-free diet, it would have been advantageous to analyze the microbiota of the mice, or, at least, the presence of histamine-producing species such as E. coli, M. morganii, or L. vaginalis [113].
4. Histamine Receptors in the Intestine
The detection of functional expression of H4R as well as of other GPCR is a still ongoing discussion that currently lacks impetus. Since in 2012 the specificity of H4R-selective antibodies was questioned [114,115], a comprehensive processing of this issue has not taken place. Thus, antibodies recognizing H4R that have been rigorously evaluated according to some general rules [116] are still missing (or at least the data demonstrating the rigorous evaluation have not been provided). These kinds of problems are not specific for the detection of H4R proteins by selective antibodies. Other methods such as mRNA detection by RT-(q)PCR or functional assessment using agonistic or antagonistic ligands also bear uncertainties. RT-PCR is a very sensitive detection method, but, besides the risk that trace amounts of contaminating genomic DNA may pose the presence of specific mRNA [117], it is not known which degree of mRNA expression would enable effective receptor expression [118]. The use of receptor ligands is afflicted with problems similar to those of the antibodies; selectivity, which is strictly concentration-dependent, is the major concern [18,119]. Thus, eventually at least two independent techniques should be provided in order to reliable demonstrate functional H4R expression. Alternatively, genetic approaches (knock out/knock down cells, tissues, or animals) are to be used to support expression data gained by other methods. In particular, targeted knock-out approaches would be able to foster our knowledge on the cell type-specific expression of H4R, being applicable not only to the intestine but to all other tissues as well.
Since histamine seemed to be functionally involved in gut (patho)physiology, studies aimed at comprehensively identifying the histamine receptor subtypes expressed in the intestinal tract using tissues derived from different species [24,120,121,122,123]. Summarizing these and other studies, all histamine receptors except H3R are expressed throughout the intestinal tract from mice, rats, monkeys, and humans (Table 2), while the data gained in dogs are questionable due to the use of not fully reliable methods [120]. From a quantitative point of view, it also appeared that H4R expression is significantly less abundant in comparison to H1R and H2R, at least on the mRNA level [24,121]. Subsequently, a possible function of H4R was investigated using pharmacological and genetic approaches. In rats suffering from trinitrobenzene sulfonic acid (TNBS)-induced colitis, the binding of H4R by JNJ7777120 resulted in reduced inflammatory symptoms [124], indicating indeed a pathological involvement of H4R in the colon. Following studies using mice either or not deficient in H4R expression and using JNJ7777120 in models of chemically-induced colitis confirmed this indication [89,90]. The data discussed so far strongly indicate that the local mediator histamine is present in high concentrations at inflamed sites of gut inflammatory diseases and that histamine affects the inflammatory pathology. However, they do not provide any evidence on the cell types involved.
Table 2.
Studies on H4R in the gastrointestinal tract.
| Main Findings | Species | Ref. |
|---|---|---|
| H2R and H4R are pro-proliferative and pro-angiogenic in HT29, Caco-2, and HCT116 colon cancer cell lines | human | [125] |
| H4R possesses pro-inflammatory role in TNBS-induced colitis in rats | rat | [124] |
| H1R, H2R, H4R are expressed in the human gastrointestinal tract; expression is altered in patients with gastrointestinal diseases | human | [24] |
| H1R, H2R, H3R, H4R activation excite human enteric neurons | human | [25] |
| H1R, H2R, and H4R are expressed in colon carcinoma and in adjacent normal mucosa; H1R and H4R expression is reduced in carcinoma compared to normal colon | human | [126] |
| H1R, H2R, and H4R are expressed in simian colon smooth muscle cells | mouse, monkey | [121] |
| H4R activity contributes to radiation-induced cytotoxic and genotoxic damages in small intestine | rat | [127] |
| H4R stimulates the release of DAO, contributing to histamine deamination during fat absorption | rat | [128] |
| H4R and H1R contribute to post-inflammatory visceral hypersensitivity | rat | [129] |
| H4R possesses a pro-inflammatory role in DSS-induced colitis in mice | mouse | [89] |
| H1R and H4R regulate the DC-CD4+ T-cell axis in peanut-induced intestinal allergic responses | mouse | [130] |
| H4R possesses an anti-inflammatory role in TNBS-induced colitis in mice | mouse | [90] |
| H4R possesses a pro-inflammatory role in experimental colitis in mice | mouse | [23] |
| H4R is functionally expressed on colon epithelial cells, affecting epithelial barrier integrity | mouse | [22] |
| H4R is involved in carcinogenesis of chemically-induced colitis-associated colorectal carcinoma | mouse | [131] |
5. Histamine-Responsive Cells
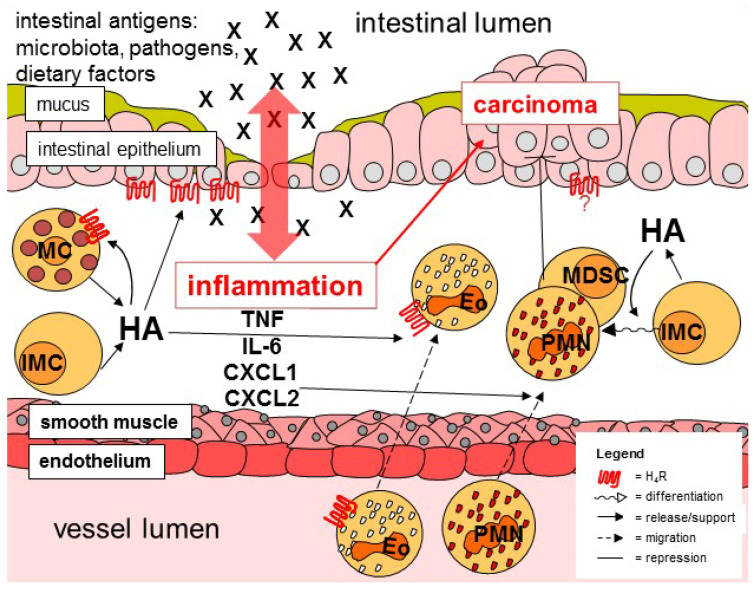
As discussed above, mast cells, allocated to the innate immune system, do not only produce histamine, but also respond to stimulation of the H4R. Besides the contribution of mast cells to allergic inflammation [132], they also seem to be involved in non-allergic inflammatory diseases of the gut, since in the intestine of IBD patients, parallel to mucosal histamine concentrations, number and activity of mast cells are increased [133,134]. Moreover, few data indicate that the application of cromolyn, a mast cell-stabilizing drug, affects experimentally-induced pathogenesis of gastric cancer and ischemia/reperfusion injury of the ileum [135,136]. In the pathogenesis of experimental colitis in mice, however, mast cells seem to be dispensable [137,138]. Thus, histamine apparently is generated also by sources other than mast cells (Figure 1). Indeed, using mast cell-deficient rats, it was demonstrated that the absence of intestinal mast cells reduced mucosal histamine concentrations, but did not nullify them [139]. Expression of the histamine-generating enzyme HDC can be induced in myeloid cells and probably also in virtually every other cell type [140,141]. As a possible alternative source of histamine in mouse intestinal pathology, CD11b+Ly6G+ immature myeloid cells have been described, since they are involved in inflammation-associated carcinogenesis in the gut [142]. Whether or not a human counterpart does exist, at least a functional one, has not been elaborated so far. Moreover, although originally identified in another model, dendritic cells are thought to be able to generate histamine [39,40,98,143]. Thus, dendritic cell-generated histamine may contribute to intestinal inflammation, too. This, although initial data argue against it [23], remains to be elucidated.
Figure 1.
Schematic representation of inflammatory and carcinogenic mechanisms in the colon. MC, mast cell; IMC, immature myeloid cell; HA, histamine; Eo, eosinophil; PMN, neutrophil; MDSC, myeloid-derived suppressor cells.
Besides mast cells, enhanced numbers of eosinophils have been identified in the inflamed gut mucosa of IBD patients, too [144,145]. The role of these innate immune cells in the pathogenesis of colitis, however, is still controversially discussed, either promoting pathogenesis or limiting inflammation [145,146]. Eosinophils are generated in the bone marrow, released into blood circulation, and eventually migrate to target tissues with the gut being a main destination. Eosinophil migration is mainly regulated by the chemokine eotaxin-1. As already mentioned above, it has been shown in vitro that histamine affects this process, albeit to a lesser extent, by its interaction with H4R on eosinophils [38,71,72,73,80]. Thus, one may speculate that this process is active in the inflamed colon, facilitating histamine to add to the attraction of eosinophils (Figure 1). Consequently, this hypothesis favors a pro-inflammatory function of eosinophils, since blockade of H4R activity, which diminishes experimental colitis, would also reduce number and, thus, activity of eosinophils in the affected intestinal areas. [21,147,148].
Neutrophils belong to those cells first recruited to the site of inflammation, where they combat the potentially harmful intruders applying rather unspecific mechanisms, condoning collateral damage at healthy cells and tissues. Consequently, neutrophils can be detected in high numbers in the colitic mucosa, probably dependent on the enhanced expression of IL-6 and downstream CXCL-chemokines (Figure 1), mediators that activate/attract these cells [149,150]. In H4R knockout mice as compared to WT mice, reduced numbers of colon-infiltrating neutrophils as well as lower colon tissue concentrations of IL-6, CXCL1, and CXCL2 have been detected upon induction of experimental colitis [23,89]. Thus, it is most probable that histamine via H4R increases neutrophil infiltration during colitis, presumably in an indirect manner [151,152]. However, the precise cellular sources of histamine/H4R-regulated production of IL-6, CXCL1, and CXCL2 remain ill defined. The related chemokines CXCL1 and CXCL2, which both bind to the receptor CXCR2, are synthesized by variety of immune cells including macrophages and neutrophils themselves, and by epithelial cells. Macrophages have been shown to respond to histamine via H4R [64,65], however, not in the context of intestinal diseases so far. Whether or not neutrophils functionally express H4R is still matter of debate. Eventually, epithelial cells are the most probable source of CXCL1 and CXCL2 in the context of colonic inflammation.
Colonic epithelial cells comprise a group of cells forming a tightly linked monolayer epithelium lining the inner surface of the intestine and providing, by several means, a barrier between the gut lumen and the host tissue. Impairment of this barrier would result in the luminal content gaining access to intra- or sub-epithelial areas, followed by an inflammatory reaction [153,154]. New intestinal epithelial cells are generated throughout the life of the host by proliferation of intestinal epithelial stem cells residing in the base of each intestinal crypt. Due to the continuous proliferation of these stem cells, the resulting daughter cells move forward towards the tip of the crypt or villus, where they eventually are scaled off and die. During this movement, the cells differentiate into specialized lineages, providing the different functions of the intestinal epithelium. The majority of developing cells become absorptive enterocytes, responsible for selective uptake, metabolism, and transport of nutrients. Other cells differentiate into mucus-secreting goblet cells that produce two layers of mucus with different densities at the luminal surface, adding to the colonic barrier function. The integrity of the cellular barrier is provided by close intercellular junctions created by tight junction proteins, adherens junction proteins, and desmosomes, omitting epithelial transfer by diffusion while enabling regulated uptake by either trans- or para-cellular pathways (reviewed in [155,156]). There are some indications that colon epithelial cells of mouse and men express H4R [89], although others conclude controversially [23]. Nevertheless, using organoid-derived mouse colonic epithelial cells, a direct in vitro effect of histamine on the trans-epithelial electric resistance is detected. Since the employed cell culture setting is devoid of any other cell type than epithelial cells, a direct effect of histamine on the epithelial cells could be concluded (Figure 1). Finally, this effect was inhibited by either the application of JNJ7777120 or by the use of cells collected from organoids formed of H4R-deficient colon epithelial cells, both pointing to a direct effect of histamine at H4R expressed on mouse colon epithelial cells [89]. Moreover, human colon epithelial cell-derived organoids that were micro-injected with FITC-dextran and either or not stimulated with histamine indicate that histamine promotes leakage of the dyed macromolecule out of the organoid’s lumen (unpublished).
In summary, it seems probable that histamine via H4R contributes to colonic inflammation, mostly to the innate arm by regulating production and release of the pro-inflammatory mediators TNF, IL-6, CXCL1, and CXCL2 promoting neutrophil recruitment. In addition, in vitro the epithelial cell barrier is battered by histamine/H4R as well, possibly contributing to inflammation. Whether or not colon epithelial cells also are responsible for the histamine/H4R-triggered enhanced release of the mediators mentioned above is not definitively settled so far.
6. Histamine and Carcinogenesis
Cancer, in general, is a major public health problem and does not only account for a high physical strain of the patients but also for intensive emotional pressure in both patients and their relatives and, finally, for a large number of deaths. Although anti-cancer therapies, including new strategies and compounds, have advanced strongly in the past decades, they are still loaded with detrimental effects, such as toxicity or poor response [157]. These effects are, at least in part, due to the high pathophysiological and genetic heterogeneity of the diverse forms of cancer, rendering it necessary to develop new therapeutic options that meet the different requirements of different cancers. A common feature of all forms of cancer is the inadequate cellular proliferation, resulting in the tumor. Thus, anti-cancer drugs aim at inhibiting proliferation and/or inducing death of cancer cells, and a major task for modern therapeutics is to provide these functions in a cell type- and probably differentiation status-specific manner [158]. A tumor cell can be defined by the presence of a specific pattern of molecules, which may represent targets for a specific tumor therapy. A single cellular receptor hardly defines a specific cell type/differentiation status, but may create a group of cells that reasonably can be targeted, avoiding severe adverse effects occurring when using broad acting anti-neoplastic drugs/therapies such as 5-fluoruracil, hydroxycarbamide, or ionizing radiation. Moreover, cellular receptors may be used as targets for adjunctive drugs, able to support the main drug and/or to reduce its effective dose, and thereby in summary reducing the risk of adverse effects, as exemplified by the H2R antagonist cimetidine in colorectal cancer patients [159,160]. In this respect, the endogenous agonist histamine in combination with IL-2 is approved to treat patients in the post-chemotherapy phase of acute myeloid leukemia [161], in which it reduces the risk of relapse. Although neither the molecular mechanism nor the histamine receptor subtype(s) involved have been determined so far, these clinical data place histamine in the list of possible anti-cancer drugs [162]. Thus, histaminergic drugs may be used in other types of cancers and substances selectively addressing specific histamine receptor subtypes as agonists or antagonists may provide therapeutic advantages [163,164].
CRC is the third most common type of cancer in humans and accounts for high mortality. UC patients are at a 2- to 3-fold higher risk to develop CRC in comparison to individuals not suffering from IBD and severity, extent, and persistence of inflammation positively correlates to carcinogenesis. In addition to colitis-associated CRC, CRC appears also sporadic and hereditary [165,166]. Although in these cases inflammation rarely precedes CRC, anti-inflammatory drugs are effective in preventing or delaying the disease [167,168]. Thus, inflammatory reactions seem to be involved in tumorigenesis of non-colitis-associated CRC, too. Consequently, it is tempting to speculate that histamine, in parallel to colon inflammation, promotes CRC, and, thus, that interventions blocking histamine activity reduce carcinogenesis. Consistently, in the colonic mucosa of CRC patients, HDC activity as well as histamine content are increased in comparison to normal samples [125,169]. However, in mice with experimentally induced CRC, deletion of HDC resulted in enhanced tumorigenesis as compared to wild type mice, pointing towards an anti-carcinogenic effect of histamine, which is supported by the finding that gut microbiota-derived histamine suppresses colorectal tumorigenesis [170]. The multiple effects histamine may exert in CRC, at least in animal models, can explain this contradiction. On the one hand, histamine promotes the underlying inflammatory process, leading to tumor initiation. On the other hand, histamine in the tumor’s tissue may affect differentiation of immature myeloid cells towards neutrophils and myeloid-derived suppressor cells, both resulting in tumor regression (Figure 1) [142,171]. While the effect of histamine on the differentiation towards neutrophils is a direct one, the differentiation of myeloid-derived suppressor cells is affected by IL-17, which is produced by tumor-associated mast cells upon histamine stimulation. This anti-cancer effect of histamine is supported by similar findings obtained in models of esophageal squamous carcinoma [172]. Interestingly, mast cells have been found to be abundant in colon carcinoma and to promote carcinogenesis in chemically-induced CRC in mice [173], and are associated with a poor prognosis in human CRC patients [174]. Others, however, reported that blocking histamine receptor function in a mouse model of CRC ameliorates disease symptoms [175]. These differences may be due to the different models used, concerning the experimental schedule and the species. Regarding the modes of action of histamine discussed above, the question arises if specific receptor subtypes are responsible for these different effects of histamine on inflammatory cells and tumor-regulating cells. In analogy to the pro-inflammatory effect of histamine via the H4R that has already been discussed above, the absence of H4R expression also leads to a reduction of chemically-induced carcinogenesis in mice [131]. Whether this is due to the reduced inflammation promoting carcinogenesis or to an effect of histamine on the tumor cell itself is unknown at present. However, some indications arise from the observation that the expression of H4R decreases in gastric carcinoma during progression, accompanied by the attenuated histamine-induced suppression of proliferation [172,176]. This idea is supported by our own unpublished data indicating that human colon cancer cell lines, which mostly arise from late stage carcinomas, do not functionally express H4R. It is worth noting that others have found H4R expression in such cell lines [125], which may reflect differences in details of the detection systems or the development of laboratory-specific cellular sub-lines. In summary, these data strongly indicate that the function of histamine in general and of the H4R in detail is far from being finally settled.
7. Conclusions
Histamine is a mediator that is mainly recognized due to its function in allergy, but takes part also in non-allergic inflammatory reactions. In the gut, the involvement of histamine and H4R in IBD, especially in the colon, has been demonstrated in different model systems by several laboratories, who mainly conclude a pro-inflammatory function (Figure 1). Cellular and molecular details of the H4R function in colon inflammation, however, remain elusive. In addition, tissues with minor indications on histamine/H4R responsiveness (smooth muscles, neurons) have not been discussed in this review. Even less differentiated is the picture on the effect of H4R in colon carcinogenesis. It is likely that H4R due to its pro-inflammatory function promotes the pathogenicity of colitis-associated CRC, but an additional effect of H4R on tumor or accessory cells currently can neither be excluded nor confirmed. Therefore, still a lot of work on histamine function on colon carcinogenesis has to be envisaged.
Author Contributions
Conceptualization and literature analysis, B.S. and D.N.; writing—original draft preparation, B.S. and D.N.; writing—review and editing, B.S. and D.N.; preparation of figures and tables, B.S. and D.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Schayer R.W. The metabolism of histamine in various species. Br. J. Pharmacol. Chemother. 1956;11:472–473. doi: 10.1111/j.1476-5381.1956.tb00020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schayer R.W., Karjala S.A. Ring N methylation; a major route of histamine metabolism. J. Biol. Chem. 1956;221:307–313. doi: 10.1016/S0021-9258(18)65250-0. [DOI] [PubMed] [Google Scholar]
- 3.Best C.H., McHenry E.W. The inactivation of histamine. J. Physiol. 1930;7:349–372. doi: 10.1113/jphysiol.1930.sp002700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riley J.F., West G.B. The presence of histamine in tissue mast cells. J. Physiol. 1953;120:528–537. doi: 10.1113/jphysiol.1953.sp004915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graham H.T., Lowry O.H., Wahl N., Priebat M.K. Mast cells as sources of tissue histamine. J. Exp. Med. 1955;102:307–318. doi: 10.1084/jem.102.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hakanson R., Owman C. Concomitant histochemical demonstration of histamine and catecholamines in enterochromaffin-like cells of gastric mucosa. Life Sci. 1967;6:759–766. doi: 10.1016/0024-3205(67)90133-6. [DOI] [PubMed] [Google Scholar]
- 7.Kwiatkowski H. Histamine in nervous tissue. J. Physiol. 1943;102:32–41. doi: 10.1113/jphysiol.1943.sp004011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fawcott D.W. Cytological and pharmacological observations on the release of histamine by mast cells. J. Exp. Med. 1954;100:217–224. doi: 10.1084/jem.100.2.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aborg C.H., Novotny J., Uvnäs B. Ionic binding of histamine in mast cell granules. Acta Physiol. Scand. 1967;69:276–283. doi: 10.1111/j.1748-1716.1967.tb03523.x. [DOI] [PubMed] [Google Scholar]
- 10.Dvorak A.M., McLeod R.S., Ondcrdonk A., Monahan-Earley R.A., Cullen J.B., Antonioli D.A., Morgari E., Blair J.E., Estrella P., Cisneros R.L., et al. Ultrastructural evidence for piecemeal and anaphylactic degranulation of human gut mucosal mast cells in vivo. Int. Arch. Allergy Immunol. 1992;99:74–83. doi: 10.1159/000236338. [DOI] [PubMed] [Google Scholar]
- 11.Sahid M.N.A., Kiyoi T. Mast cell activation markers for in vitro study. J. Immunoass. Immunochem. 2020;41:7798–7816. doi: 10.1080/15321819.2020.1769129. [DOI] [PubMed] [Google Scholar]
- 12.Jutel M., Akdis M., Akdis C.A. Histamine, histamine receptors and their role in immune pathology. Clin. Exp. Allergy. 2009;39:1786–18000. doi: 10.1111/j.1365-2222.2009.03374.x. [DOI] [PubMed] [Google Scholar]
- 13.Deng X., Wu X., Yu Z., Arai I., Sasano T., Sugawara S., Endo Y. Inductions of histidine decarboxylase in mouse tissues following systemic antigen challenge: Contributions made by mast cells, non-mast cells and IL-1. Int. Arch. Allergy Immunol. 2007;144:69–78. doi: 10.1159/000102617. [DOI] [PubMed] [Google Scholar]
- 14.Wu X., Yoshida A., Sasano T., Iwakura Y., Endo Y. Histamine production via mast cell-independent induction of histidine decarboxylase in response to lipopolysaccharide and interleukin-1. Int. Immunopharmacol. 2004;4:516–520. doi: 10.1016/j.intimp.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 15.Hirasawa N. Expression of histidine decarboxylase and its roles in inflammation. Int. J. Mol. Sci. 2019;20:376. doi: 10.3390/ijms20020376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moriguchi T., Takai J. Histamine and histidine decarboxylase: Immunomodulatory functions and regulatory mechanisms. Genes to Cells. 2020;25:443–449. doi: 10.1111/gtc.12774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calvielli Castelo Branco A.C., Seiti F., Yoshikawa Y., Pietrobon A.J., Sato M.N. Role of histamine in modulating the immune response and inflammation. Mediat. Inflamm. 2018;2018:e9524075. doi: 10.1155/2018/9524075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seifert R., Strasser A., Schneider E.H., Neumann D., Dove S., Buschauer A. Molecular and cellular analysis of human histamine receptor subtypes. Trends Pharmacol. Sci. 2013;34:33–58. doi: 10.1016/j.tips.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tiligada E., Ennis M. Histamine pharmacology: From Sir Henry Dale to the 21st century. Br. J. Pharmacol. 2020;177:469–489. doi: 10.1111/bph.14524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panula P. Histamine, histamine H3 receptor, and alcohol use disorder. Br. J. Pharmacol. 2020;177:634–641. doi: 10.1111/bph.14634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schirmer B., Lindemann L., Bittkau K.S., Isaev R., Bösche D., Juchem M., Seifert R., Neumann D. Mouse colonic epithelial cells functionally express the histamine H4 receptor. J. Pharmacol. Exp. Ther. 2020;373:167–174. doi: 10.1124/jpet.119.264408. [DOI] [PubMed] [Google Scholar]
- 22.Mehta P., Miszta P., Rzodkiewicz P., Michalak O., Krzeczyński P., Filipek S. Enigmatic histamine receptor H4 for potential treatment of multiple inflammatory, autoimmune, and related diseases. Life. 2020;10:50. doi: 10.3390/life10040050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wechsler J.B., Szabo A., Hsu C.L., Krier-Burris R.A., Schroeder H.A., Wang M.Y., Carter R.G., Velez T.E., Aguiniga L.M., Brown J.B., et al. Histamine drives severity of innate inflammation via histamine 4 receptor in murine experimental colitis. Mucosal Immunol. 2018;11:861–870. doi: 10.1038/mi.2017.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sander L.E., Lorentz A., Sellge G., Coëffier M., Neipp M., Veres T., Frieling T., Meier P.N., Manns M.P., Bischoff S.C. Selective expression of histamine receptors H1R, H2R, and H4R, but not H3R, in the human intestinal tract. Gut. 2006;55:498–504. doi: 10.1136/gut.2004.061762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Breunig E., Michel K., Zeller F., Seidl S., Weyhern C.W., Schemann M. Histamine excites neurones in the human submucous plexus through activation of H1, H2, H3 and H4 receptors. J. Physiol. 2007;583:731–742. doi: 10.1113/jphysiol.2007.139352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu Y., Michalovich D., Wu H., Tan K.B., Dytko G.M., Mannan I.J., Boyce R., Alston J., Tierney L.A., Li X., et al. Cloning, expression, and pharmacological characterization of a novel human histamine receptor. Mol. Pharmacol. 2001;59:434–441. doi: 10.1124/mol.59.3.434. [DOI] [PubMed] [Google Scholar]
- 27.Gutzmer R., Mommert S., Gschwandtner M., Zwingmann K., Stark H., Werfel T. The histamine H4 receptor is functionally expressed on TH2 cells. J. Allergy Clin. Immunol. 2009;123:619–625. doi: 10.1016/j.jaci.2008.12.1110. [DOI] [PubMed] [Google Scholar]
- 28.Schaper-Gerhardt K., Wohlert M., Mommert S., Kietzmann M., Werfel T., Gutzmer R. Stimulation of histamine H4 receptors increases the production of IL-9 in Th9 polarized cells. Br. J. Pharmacol. 2020;177:614–622. doi: 10.1111/bph.14566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mommert S., Gschwandtner M., Koether B., Gutzmer R., Werfel T. Human memory Th17 cells express a functional histamine H4 receptor. Am. J. Pathol. 2012;180:177–185. doi: 10.1016/j.ajpath.2011.09.028. [DOI] [PubMed] [Google Scholar]
- 30.Cowden J.M., Yu F., Banie H., Farahani M., Ling P., Nguyen S., Riley J.P., Zhang M., Zhu J., Dunford P.J., et al. The histamine H4 receptor mediates inflammation and Th17 responses in preclinical models of arthritis. Ann. Rheum. Dis. 2014;73:600–608. doi: 10.1136/annrheumdis-2013-203832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han S.H., Hur M.S., Kim M.J., Kim B.M., Kim K.W., Kim H.R., Choe Y.B., Ahn K.J., Lee Y.W. Preliminary study of histamine H4 receptor expressed on human CD4+ T cells and its immunomodulatory potency in the IL-17 pathway of psoriasis. J. Dermatol. Sci. 2017;88:29–35. doi: 10.1016/j.jdermsci.2017.05.011. [DOI] [PubMed] [Google Scholar]
- 32.Morgan R.K., McAllister B., Cross L., Green D.S., Kornfeld H., Center D.M., Cruikshank W.W. Histamine 4 receptor activation induces recruitment of FoxP3+ T cells and inhibits allergic asthma in a murine model. J. Immunol. 2007;178:8081–8089. doi: 10.4049/jimmunol.178.12.8081. [DOI] [PubMed] [Google Scholar]
- 33.del Rio R., Noubade R., Saligrama N., Wall E.H., Krementsov D.N., Poynter M.E., Zachary J.F., Thurmond R.L., Teuscher C. Histamine H4 receptor optimizes T regulatory cell frequency and facilitates anti-inflammatory responses within the central nervous system. J. Immunol. 2012;188:541–547. doi: 10.4049/jimmunol.1101498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Truta-Feles K., Lagadari M., Lehmann K., Berod L., Cubillos S., Piehler S., Herouy Y., Barz D., Kamradt T., Maghazachi A., et al. Histamine modulates γδ-T lymphocyte migration and cytotoxicity, via Gi and Gs protein-coupled signalling pathways. Br. J. Pharmacol. 2010;161:1291–1300. doi: 10.1111/j.1476-5381.2010.00639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gantner F., Sakai K., Tusche M.W., Cruikshank W.W., Center D.M., Bacon K.B. Histamine H4 and H2 receptors control histamine-induced interleukin-16 release from human CD8+ T cells. J. Pharmacol. Exp. Ther. 2002;303:300–307. doi: 10.1124/jpet.102.036939. [DOI] [PubMed] [Google Scholar]
- 36.Glatzer F., Mommert S., Köther B., Gschwandtner M., Stark H., Werfel T., Gutzmer R. Histamine downregulates the Th1-associated chemokine IP-10 in monocytes and myeloid dendritic cells. Int. Arch. Allergy Immunol. 2014;163:11–19. doi: 10.1159/000355960. [DOI] [PubMed] [Google Scholar]
- 37.Gutzmer R., Diestel C., Mommert S., Köther B., Stark H., Wittmann M., Werfel T. Histamine H4 receptor stimulation suppresses IL-12p70 production and mediates chemotaxis in human monocyte-derived dendritic cells. J. Immunol. 2005;174:5224–5232. doi: 10.4049/jimmunol.174.9.5224. [DOI] [PubMed] [Google Scholar]
- 38.Ling P., Ngo K., Nguyen S., Thurmond R.L., Edwards J.P., Karlsson L., Fung-Leung W.P. Histamine H4 receptor mediates eosinophil chemotaxis with cell shape change and adhesion molecule upregulation. Br. J. Pharmacol. 2004;142:161–171. doi: 10.1038/sj.bjp.0705729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dunford P.J., O’Donnell N., Riley J.P., Williams K.N., Karlsson L., Thurmond R.L. The histamine H4 receptor mediates allergic airway inflammation by regulating the activation of CD4+ T cells. J. Immunol. 2006;176:7062–7070. doi: 10.4049/jimmunol.176.11.7062. [DOI] [PubMed] [Google Scholar]
- 40.Hartwig C., Munder A., Glage S., Wedekind D., Schenk H., Seifert R., Neumann D. The histamine H4 receptor (H4R) regulates eosinophilic inflammation in ovalbumin-induced experimental allergic asthma in mice. Eur. J. Immunol. 2015;45:1129–1140. doi: 10.1002/eji.201445179. [DOI] [PubMed] [Google Scholar]
- 41.Lundberg K., Broos S., Greiff L., Borrebaeck C.A.K., Lindstedt M. Histamine H4 receptor antagonism inhibits allergen-specific T-cell responses mediated by human dendritic cells. Eur. J. Pharmacol. 2011;651:197–204. doi: 10.1016/j.ejphar.2010.10.065. [DOI] [PubMed] [Google Scholar]
- 42.Gschwandtner M., Mommert S., Köther B., Werfel T., Gutzmer R. The histamine H4 receptor is highly expressed on plasmacytoid dendritic cells in psoriasis and histamine regulates their cytokine production and migration. J. Investig. Dermatol. 2011;131:1668–1676. doi: 10.1038/jid.2011.72. [DOI] [PubMed] [Google Scholar]
- 43.Damaj B.B., Becerra C.B., Esber H.J., Wen Y., Maghazachi A.A. Functional expression of H4 histamine receptor in human natural killer cells, monocytes, and dendritic cells. J. Immunol. 2007;179:7907–7915. doi: 10.4049/jimmunol.179.11.7907. [DOI] [PubMed] [Google Scholar]
- 44.Dijkstra D., Stark H., Chazot P.L., Shenton F.C., Leurs R., Werfel T., Gutzmer R. Human inflammatory dendritic epidermal cells express a functional histamine H4 receptor. J. Investig. Dermatol. 2008;128:1696–1703. doi: 10.1038/sj.jid.5701250. [DOI] [PubMed] [Google Scholar]
- 45.Bäumer W., Wendorff S., Gutzmer R., Werfel T., Dijkstra D., Chazot P., Stark H., Kietzmann M. Histamine H4 receptors modulate dendritic cell migration through skin—Immunomodulatory role of histamine. Allergy. 2008;63:1387–1394. doi: 10.1111/j.1398-9995.2008.01720.x. [DOI] [PubMed] [Google Scholar]
- 46.Simon T., Jelinek I., Apponyi G., László V., Rajnavölgyi É., Falus A. Expression and function of histamine H4 receptor in mouse splenic dendritic cells. Inflamm. Res. 2010;59(Suppl. S2):S201–S203. doi: 10.1007/s00011-009-0130-7. [DOI] [PubMed] [Google Scholar]
- 47.Gschwandtner M., Schäkel K., Werfel T., Gutzmer R. Histamine H4 receptor activation on human slan-dendritic cells down-regulates their pro-inflammatory capacity. Immunology. 2011;132:49–56. doi: 10.1111/j.1365-2567.2010.03336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leite-de-Moraes M.C., Diem S., Michel M.-L., Ohtsu H., Thurmond R.L., Schneider E., Dy M. Cutting Edge: Histamine receptor H4 activation positively regulates in vivo IL-4 and IFN-γ production by invariant NKT cells. J. Immunol. 2009;182:1233–1236. doi: 10.4049/jimmunol.182.3.1233. [DOI] [PubMed] [Google Scholar]
- 49.Hofstra C.L., Desai P.J., Thurmond R.L., Fung-Leung W.P. Histamine H4 receptor mediates chemotaxis and calcium mobilization of mast cells. J. Pharmacol. Exp. Ther. 2003;305:1212–1221. doi: 10.1124/jpet.102.046581. [DOI] [PubMed] [Google Scholar]
- 50.Jemima E.A., Prema A., Thangam E.B. Functional characterization of histamine H4 receptor on human mast cells. Mol. Immunol. 2014;62:19–28. doi: 10.1016/j.molimm.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 51.Kay L.J., Suvarna S.K., Peachell P.T. Histamine H4 receptor mediates chemotaxis of human lung mast cells. Eur. J. Pharmacol. 2018;837:38–44. doi: 10.1016/j.ejphar.2018.08.028. [DOI] [PubMed] [Google Scholar]
- 52.Honda T., Nishio Y., Sakai H., Asagiri M., Yoshimura K., Inui M., Kuramasu A. Calcium/calmodulin-dependent regulation of Rac GTPases and Akt in histamine-induced chemotaxis of mast cells. Cell. Signal. 2021;83:1099733. doi: 10.1016/j.cellsig.2021.109973. [DOI] [PubMed] [Google Scholar]
- 53.Lippert U., Artuc M., Grützkau A., Babina M., Guhl S., Haase I., Blaschke V., Zachmann K., Knosalla M., Middel P., et al. Human skin mast cells express H2 and H4, but not H3 receptors. J. Investig. Dermatol. 2004;123:116–123. doi: 10.1111/j.0022-202X.2004.22721.x. [DOI] [PubMed] [Google Scholar]
- 54.Godot V., Arock M., Garcia G., Capel F., Flys C., Dy M., Emilie D., Humbert M. H4 histamine receptor mediates optimal migration of mast cell precursors to CXCL12. J. Allergy Clin. Immunol. 2007;120:827–834. doi: 10.1016/j.jaci.2007.05.046. [DOI] [PubMed] [Google Scholar]
- 55.Desai P., Thurmond R.L. Histamine H4 receptor activation enhances LPS-induced IL-6 production in mast cells via ERK and PI3K activation. Eur. J. Immunol. 2011;41:1764–1773. doi: 10.1002/eji.201040932. [DOI] [PubMed] [Google Scholar]
- 56.Mirzahosseini A., Dalmadi B., Csutora P. Histamine receptor H4 regulates mast cell degranulation and IgE induced FcεRI upregulation in murine bone marrow-derived mast cells. Cell. Immunol. 2013;283:38–44. doi: 10.1016/j.cellimm.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 57.Aldi S., Takano K.I., Tomita K., Koda K., Chan N.Y.K., Marino A., Salazar-Rodriguez M., Thurmond R.L., Levi R. Histamine H4-receptors inhibit mast cell renin release in ischemia/reperfusion via protein kinase Cε-dependent aldehyde dehydrogenase type-2 activation. J. Pharmacol. Exp. Ther. 2014;349:508–517. doi: 10.1124/jpet.114.214122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ebenezer A.J., Arunachalam P., Elden B.T. H4R activation utilizes distinct signaling pathways for the production of RANTES and IL-13 in human mast cells. J. Recept. Signal Transduct. 2017;37:133–140. doi: 10.1080/10799893.2016.1203938. [DOI] [PubMed] [Google Scholar]
- 59.Kuramasu A., Wakabayashi M., Inui M., Yanai K. Distinct roles of small GTPases RAC1 and RAC2 in histamine H4 receptor–mediated chemotaxis of mast cells. J. Pharmacol. Exp. Ther. 2018;367:9–19. doi: 10.1124/jpet.118.249706. [DOI] [PubMed] [Google Scholar]
- 60.Oda T., Morikawa N., Saito Y., Masuho Y., Matsumoto S. Molecular cloning and characterization of a novel type of histamine receptor preferentially expressed in leukocytes. J. Biol. Chem. 2000;275:36781–36786. doi: 10.1074/jbc.M006480200. [DOI] [PubMed] [Google Scholar]
- 61.Dib K., Perecko T., Jenei V., McFarlane C., Comer D., Brown V., Katebe M., Scheithauer T., Thurmond R.L., Chazot P.L., et al. The histamine H4 receptor is a potent inhibitor of adhesion-dependent degranulation in human neutrophils. J. Leukoc. Biol. 2014;96:411–418. doi: 10.1189/jlb.2AB0813-432RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dijkstra D., Leurs R., Chazot P., Shenton F.C., Stark H., Werfel T., Gutzmer R. Histamine downregulates monocyte CCL2 production through the histamine H4 receptor. J. Allergy Clin. Immunol. 2007;120:300–307. doi: 10.1016/j.jaci.2007.03.024. [DOI] [PubMed] [Google Scholar]
- 63.Peng H., Wang J., Ye X.Y., Cheng J., Huang C.Z., Li L.Y., Li T.Y., Li C.W. Histamine H4 receptor regulates IL-6 and INF-γ secretion in native monocytes from healthy subjects and patients with allergic rhinitis. Clin. Transl. Allergy. 2019;9:49. doi: 10.1186/s13601-019-0288-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Czerner C.P., Klos A., Seifert R., Neumann D. Histamine induces chemotaxis and phagocytosis in murine bone marrow-derived macrophages and RAW 264.7 macrophage-like cells via histamine H4-receptor. Inflamm. Res. 2014;63:239–247. doi: 10.1007/s00011-013-0694-0. [DOI] [PubMed] [Google Scholar]
- 65.Mommert S., Ratz L., Stark H., Gutzmer R., Werfel T. The histamine H4 receptor modulates the differentiation process of human monocyte-derived M1 macrophages and the release of CCL4/MIP-1β from fully differentiated M1 macrophages. Inflamm. Res. 2018;67:503–513. doi: 10.1007/s00011-018-1140-0. [DOI] [PubMed] [Google Scholar]
- 66.Mommert S., Hüer M., Schaper-Gerhardt K., Gutzmer R., Werfel T. Histamine up-regulates oncostatin M expression in human M1 macrophages. Br. J. Pharmacol. 2020;177:600–613. doi: 10.1111/bph.14796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mommert S., Aslan D., Ratz L., Stark H., Gutzmer R., Werfel T. The anaphylatoxin C3a receptor expression on human M2 macrophages is down-regulated by stimulating the histamine H4 receptor and the IL-4 receptor. J. Innate Immun. 2018;10:349–362. doi: 10.1159/000490426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schneider E., Rolli-Derkinderen M., Arock M., Dy M. Trends in histamine research: New functions during immune responses and hematopoiesis. Trends Immunol. 2002;23:255–263. doi: 10.1016/S1471-4906(02)02215-9. [DOI] [PubMed] [Google Scholar]
- 69.Mommert S., Kleiner S., Gehring M., Eiz-Vesper B., Stark H., Gutzmer R., Werfel T., Raap U. Human basophil chemotaxis and activation are regulated via the histamine H4 receptor. Allergy. 2016;71:1264–1273. doi: 10.1111/all.12875. [DOI] [PubMed] [Google Scholar]
- 70.Liu C., Ma X., Jiang X., Wilson S.J., Hofstra C.L., Blevitt J., Pyati J., Li X., Chai W., Carruthers N., et al. Cloning and pharmacological characterization of a fourth histamine receptor (H4) expressed in bone marrow. Mol. Pharmacol. 2001;59:420–426. doi: 10.1124/mol.59.3.420. [DOI] [PubMed] [Google Scholar]
- 71.O’Reilly M., Alpert R., Jenkinson S., Gladue R.P., Foo S., Trim S., Peter B., Trevethick M., Fidock M. Identification of a histamine H4 receptor on human eosinophils-role in eosinophil chemotaxis. J. Recept. Signal Transduct. Res. 2002;22:431–448. doi: 10.1081/RRS-120014612. [DOI] [PubMed] [Google Scholar]
- 72.Buckland K.F., Williams T.J., Conroy D.M. Histamine induces cytoskeletal changes in human eosinophils via the H4 receptor. Br. J. Pharmacol. 2003;140:1117–1127. doi: 10.1038/sj.bjp.0705530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reher T.M., Neumann D., Buschauer A., Seifert R. Incomplete activation of human eosinophils via the histamine H4-receptor: Evidence for ligand-specific receptor conformations. Biochem. Pharmacol. 2012;84:192–203. doi: 10.1016/j.bcp.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 74.Schaper-Gerhardt K., Köther B., Wolff L., Kabatas A., Gehring M., Nikolouli E., Mommert S., Werfel T., Gutzmer R. The H4R is highly expressed on eosinophils from AD patients and IL-4 upregulates expression and function via the JAK/STAT pathway. Allergy. 2021;76:1261–1264. doi: 10.1111/all.14599. [DOI] [PubMed] [Google Scholar]
- 75.Grosicki M., Adami M., Micheloni C., Głuch-Lutwin M., Siwek A., Latacz G., Łażewska D., Więcek M., Reiner-Link D., Stark H., et al. Eosinophils adhesion assay as a tool for phenotypic drug screening—The pharmacology of 1,3,5–triazine and 1H-indole like derivatives against the human histamine H4 receptor. Eur. J. Pharmacol. 2021;890:173611. doi: 10.1016/j.ejphar.2020.173611. [DOI] [PubMed] [Google Scholar]
- 76.Nakayama T., Kato Y., Hieshima K., Nagakubo D., Kunori Y., Fujisawa T., Yoshie O. Liver-expressed chemokine/CC chemokine ligand 16 attracts eosinophils by interacting with histamine H4 receptor. J. Immunol. 2004;173:2078–2083. doi: 10.4049/jimmunol.173.3.2078. [DOI] [PubMed] [Google Scholar]
- 77.Barnard R., Barnard A., Salmon G., Liu W., Sreckovic S. Histamine-induced actin polymerization in human eosinophils: An imaging approach for histamine H4 receptor. Cytometry A. 2008;73:299–304. doi: 10.1002/cyto.a.20514. [DOI] [PubMed] [Google Scholar]
- 78.Mommert S., Dittrich-Breiholz O., Stark H., Gutzmer R., Werfel T. The histamine H4 receptor regulates chemokine production in human natural killer cells. Int. Arch. Allergy Immunol. 2015;166:225–230. doi: 10.1159/000381340. [DOI] [PubMed] [Google Scholar]
- 79.Ehling S., Roßbach K., Dunston S.M., Stark H., Bäumer W. Allergic inflammation is augmented via histamine H4 receptor activation: The role of natural killer cells in vitro and in vivo. J. Dermatol. Sci. 2016;83:106–115. doi: 10.1016/j.jdermsci.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 80.Raible D.G., Lenahan T., Fayvilevich Y., Kosinski R., Schulman E.S. Pharmacologic characterization of a novel histamine receptor on human eosinophils. Am. J. Respir. Crit. Care Med. 1994;149:1506–1511. doi: 10.1164/ajrccm.149.6.8004306. [DOI] [PubMed] [Google Scholar]
- 81.Morse K.L., Behan J., Laz T.M., West R.E., Greenfeder S.A., Anthes J.C., Umland S., Wan Y., Hipkin R.W., Gonsiorek W., et al. Cloning and characterization of a novel human histamine receptor. J. Pharmacol. Exp. Ther. 2001;296:1058–1066. [PubMed] [Google Scholar]
- 82.Nakamura T., Itadani H., Hidaka Y., Ohta M., Tanaka K. Molecular cloning and characterization of a new human histamine receptor, HH4R. Biochem. Biophys. Res. Commun. 2000;279:615–620. doi: 10.1006/bbrc.2000.4008. [DOI] [PubMed] [Google Scholar]
- 83.Nguyen T., Shapiro D.A., George S.R., Setola V., Lee D.K., Cheng R., Rauser L., Lee S.P., Lynch K.R., Roth B.L., et al. Discovery of a novel member of the histamine receptor family. Mol. Pharmacol. 2001;59:427–433. doi: 10.1124/mol.59.3.427. [DOI] [PubMed] [Google Scholar]
- 84.Verweij E.W.E., Al Araaj B., Prabhata W.R., Prihandoko R., Nijmeijer S., Tobin A.B., Leurs R., Vischer H.F. Differential role of serines and threonines in intracellular loop 3 and C-terminal tail of the histamine H4 receptor in β-arrestin and G protein-coupled receptor kinase interaction, internalization, and signaling. ACS Pharmacol. Transl. Sci. 2020;3:321–333. doi: 10.1021/acsptsci.0c00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Beermann S., Bernhardt G., Seifert R., Buschauer A., Neumann D. Histamine H1- and H4-receptor signaling cooperatively regulate MAPK activation. Biochem. Pharmacol. 2015;98:432–439. doi: 10.1016/j.bcp.2015.09.011. [DOI] [PubMed] [Google Scholar]
- 86.Beermann S., Vauth M., Hein R., Seifert R., Neumann D. Distinct Signalling Pathways of Murine Histamine H1- and H4-receptors expressed at comparable levels in HEK293 cells. PLoS ONE. 2014;9:e107481. doi: 10.1371/journal.pone.0107481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schneider E.H., Seifert R. Pharmacological characterization of human histamine receptors and histamine receptor mutants in the Sf9 cell expression system. In: Hattori Y., Seifert R., editors. Histamine and Histamine Receptors in Health and Disease. Volume 241. Springer; Cham, Switzerland: 2017. pp. 63–118. Handbook of Experimental Pharmacology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rovedatti L., Kudo T., Biancheri P., Sarra M., Knowles C., Rampton D., Corazza G., Monteleone G., Di Sabatino A., Macdonald T. Differential regulation of interleukin 17 and interferon gamma production in inflammatory bowel disease. Gut. 2009;58:1629–1636. doi: 10.1136/gut.2009.182170. [DOI] [PubMed] [Google Scholar]
- 89.Schirmer B., Rezniczek T., Seifert R., Neumann D. Proinflammatory role of the histamine H4 receptor in dextrane sodium sulfate-induced acute colitis. Biochem. Pharmacol. 2015;98:102–109. doi: 10.1016/j.bcp.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 90.Wunschel E.J., Schirmer B., Seifert R., Neumann D. Lack of histamine H4-receptor expression aggravates TNBS-induced acute colitis symptoms in mice. Front. Pharmacol. 2017;8:642. doi: 10.3389/fphar.2017.00642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Thurmond R.L., Desai P.J., Dunford P.J., Fung-Leung W.-P.P., Hofstra C.L., Jiang W., Nguyen S., Riley J.P., Sun S., Williams K.N., et al. A potent and selective histamine H4 receptor antagonist with anti-inflammatory properties. J. Pharmacol. Exp. Ther. 2004;309:404–413. doi: 10.1124/jpet.103.061754. [DOI] [PubMed] [Google Scholar]
- 92.Rosethorne E.M., Charlton S.J. Agonist-biased signaling at the histamine H4 receptor: JNJ7777120 recruits β-arrestin without activating G proteins. Mol. Pharmacol. 2011;79:749–757. doi: 10.1124/mol.110.068395. [DOI] [PubMed] [Google Scholar]
- 93.Seifert R., Schneider E.H., Dove S., Brunskole I., Neumann D., Strasser A., Buschauer A. Paradoxical stimulatory effects of the “standard” histamine H4-receptor antagonist JNJ7777120: The H4 receptor joins the club of 7 transmembrane domain receptors exhibiting functional selectivity. Mol. Pharmacol. 2011;79:631–638. doi: 10.1124/mol.111.071266. [DOI] [PubMed] [Google Scholar]
- 94.Wifling D., Löffel K., Nordemann U., Strasser A., Bernhardt G., Dove S., Seifert R., Buschauer A. Molecular determinants for the high constitutive activity of the human histamine H4 receptor: Functional studies on orthologues and mutants. Br. J. Pharmacol. 2015;172:785–798. doi: 10.1111/bph.12801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cowden J.M., Riley J.P., Ma J.Y., Thurmond R.L., Dunford P.J. Histamine H4 receptor antagonism diminishes existing airway inflammation and dysfunction via modulation of Th2 cytokines. Respir. Res. 2010;11:86. doi: 10.1186/1465-9921-11-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kollmeier A., Francke K., Chen B., Dunford P.J., Greenspan A.J., Xia Y., Xu X.L., Zhou B., Thurmond R. The H4 receptor antagonist, JNJ 39758979, is effective in reducing histamine-induced pruritus in a randomized clinical study in healthy subjects. J. Pharmacol. Exp. Ther. 2014;350:181–187. doi: 10.1124/jpet.114.215749. [DOI] [PubMed] [Google Scholar]
- 97.Reher T.M., Brunskole I., Neumann D., Seifert R. Evidence for ligand-specific conformations of the histamine H2 receptor in human eosinophils and neutrophils. Biochem. Pharmacol. 2012;84:1174–1185. doi: 10.1016/j.bcp.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 98.Xie G., Wang F., Peng X., Liang Y., Yang H., Li L. Modulation of mast cell toll-like receptor 3 expression and cytokines release by histamine. Cell. Physiol. Biochem. 2018;46:2401–2411. doi: 10.1159/000489646. [DOI] [PubMed] [Google Scholar]
- 99.Podolsky D.K. Inflammatory bowel disease. N. Engl. J. Med. 2002;247:417–429. doi: 10.1056/NEJMra020831. [DOI] [PubMed] [Google Scholar]
- 100.Baumgart D.C., Sandborn W.J. Inflammatory bowel disease: Clinical aspects and established and evolving therapies. Lancet. 2007;369:1641–1657. doi: 10.1016/S0140-6736(07)60751-X. [DOI] [PubMed] [Google Scholar]
- 101.Baumgart D.C., Carding S.R. Inflammatory bowel disease: Cause and immunobiology. Lancet. 2007;369:1627–1640. doi: 10.1016/S0140-6736(07)60750-8. [DOI] [PubMed] [Google Scholar]
- 102.Xavier R.J., Podolsky D.K. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 103.Cobrin G.M., Abreu M.T. Defects in mucosal immunity leading to Crohn’s disease. Immunol. Rev. 2005;206:277–295. doi: 10.1111/j.0105-2896.2005.00293.x. [DOI] [PubMed] [Google Scholar]
- 104.Elson C.O., Weaver C.T. Inflammatory Bowel Disease: From Bench to Bedside. Springer; Berlin/Heidelberg, Germany: 2006. Experimental mouse models of inflammatory bowel disease: New insights into pathogenic mechanisms. [Google Scholar]
- 105.Pizarro T.T., Cominelli F. Cytokine therapy for Crohn’s disease: Advances in translational research. Annu. Rev. Med. 2007;58:433–444. doi: 10.1146/annurev.med.58.121205.100607. [DOI] [PubMed] [Google Scholar]
- 106.Eaden J.A., Abrams K.R., Mayberry J.F. The risk of colorectal cancer in ulcerative colitis: A meta-analysis. Gut. 2001;48:526–535. doi: 10.1136/gut.48.4.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rahier J.F. Prevention and management of infectious complications in IBD. Dig. Dis. 2012;30:408–414. doi: 10.1159/000338143. [DOI] [PubMed] [Google Scholar]
- 108.Baenkler H.W., Lux G., Günthner R., Kohlhäufl M., Matek W. Biopsy histamine in ulcerative colitis and Crohn’s disease. Hepatogastroenterology. 1987;34:289–290. [PubMed] [Google Scholar]
- 109.Raithel M., Matek M., Baenkler H.W., Jorde W., Hahn E.G. Mucosal histamine content and histamine secretion in Crohn’s disease, ulcerative colitis and allergic enteropathy. Int. Arch. Allergy Immunol. 1995;108:127–133. doi: 10.1159/000237129. [DOI] [PubMed] [Google Scholar]
- 110.Bene L., Sápi Z., Bajtai A., Buzás E., Szentmihályi A., Arató A., Tulassay Z., Falus A. Partial protection against dextran sodium sulphate induced colitis in histamine-deficient, histidine decarboxylase knockout mice. J. Pediatr. Gastroenterol. Nutr. 2004;39:171–176. doi: 10.1097/00005176-200408000-00009. [DOI] [PubMed] [Google Scholar]
- 111.Keubler L.M., Buettner M., Häger C., Bleich A. A multihit model: Colitis lessons from the interleukin-10-deficient mouse. Inflamm. Bowel Dis. 2015;21:1967–1975. doi: 10.1097/MIB.0000000000000468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Buechler G., Wos-Oxley M.L., Smoczek A., Zschemisch N.-H., Neumann D., Pieper D.H., Hedrich H.J., Bleich A. Strain-specific colitis susceptibility in IL10-deficient mice depends on complex gut microbiota-host interactions. Inflamm. Bowel Dis. 2012;18:943–954. doi: 10.1002/ibd.21895. [DOI] [PubMed] [Google Scholar]
- 113.Barcik W., Pugin B., Westermann P., Perez N.R., Ferstl R., Wawrzyniak M., Smolinska S., Jutel M., Hessel E.M., Michalovich D., et al. Histamine-secreting microbes are increased in the gut of adult asthma patients. J. Allergy Clin. Immunol. 2016;138:1491–1494. doi: 10.1016/j.jaci.2016.05.049. [DOI] [PubMed] [Google Scholar]
- 114.Beermann S., Seifert R., Neumann D. Commercially available antibodies against human and murine histamine H4 receptor lack specificity. Naunyn Schmiedebergs Arch. Pharmacol. 2012;385:125–135. doi: 10.1007/s00210-011-0700-4. [DOI] [PubMed] [Google Scholar]
- 115.Gutzmer R., Werfel T., Bäumer W., Kietzmann M., Chazot P.L., Leurs R. Well characterized antihistamine 4 receptor antibodies contribute to current knowledge of the expression and biology of the human and murine histamine 4 receptor. Naunyn Schmiedebergs Arch. Pharmacol. 2012;385:853–860. doi: 10.1007/s00210-012-0744-0. [DOI] [PubMed] [Google Scholar]
- 116.Michel M.C., Wieland T., Tsujimoto G. How reliable are G-protein-coupled receptor antibodies? Naunyn Schmiedebergs Arch. Pharmacol. 2009;379:385–388. doi: 10.1007/s00210-009-0395-y. [DOI] [PubMed] [Google Scholar]
- 117.Hashemipetroudi S.H., Nematzadeh G., Ahmadian G., Yamchi A., Kuhlmann M. Assessment of DNA contamination in RNA samples based on ribosomal DNA. J. Vis. Exp. 2018;22:55451. doi: 10.3791/55451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Maier T., Güell M., Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583:3966–3973. doi: 10.1016/j.febslet.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 119.Strasser A., Wittmann H.J., Buschauer A., Schneider E.H., Seifert R. Species-dependent activities of G-protein-coupled receptor ligands: Lessons from histamine receptor orthologs. Trends Pharmacol. Sci. 2013;34:13–32. doi: 10.1016/j.tips.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 120.Sullivant A., Mackin A., Pharr T., Cooley J., Wills R., Archer T. Identification of histamine receptors in the canine gastrointestinal tract. Vet. Immunol. Immunopathol. 2016;182:29–36. doi: 10.1016/j.vetimm.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 121.Kim H., Dwyer L., Song J.H., Martin-Cano F.E., Bahney J., Peri L., Britton F.C., Sanders K.M., Koh S.D. Identification of histamine receptors and effects of histamine on murine and simian colonic excitability. Neurogastroenterol. Motil. 2011;23:949–e409. doi: 10.1111/j.1365-2982.2011.01760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Smolinska S., Groeger D., Perez N.R., Schiavi E., Ferstl R., Frei R., Konieczna P., Akdis C.A., Jutel M., OʼMahony L. Histamine receptor 2 is required to suppress innate immune responses to bacterial ligands in patients with inflammatory bowel disease. Inflamm. Bowel Dis. 2016;22:1575–1586. doi: 10.1097/MIB.0000000000000825. [DOI] [PubMed] [Google Scholar]
- 123.Deiteren A., De Man J.G., Pelckmans P.A., De Winter B.Y. Histamine H₄ receptors in the gastrointestinal tract. Br. J. Pharmacol. 2015;172:1165–1178. doi: 10.1111/bph.12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Varga C., Horvath K., Berko A., Thurmond R.L., Dunford P.J., Whittle B.J. Inhibitory effects of histamine H4 receptor antagonists on experimental colitis in the rat. Eur. J. Pharmacol. 2005;522:130–138. doi: 10.1016/j.ejphar.2005.08.045. [DOI] [PubMed] [Google Scholar]
- 125.Cianchi F., Cortesini C., Schiavone N., Perna F., Magnelli L., Fanti E., Bani D., Messerini L., Fabbroni V., Perigli G., et al. The role of cyclooxygenase-2 in mediating the effects of histamine on cell proliferation and vascular endothelial growth factor production in colorectal cancer. Clin. Cancer Res. 2005;11:6807–6815. doi: 10.1158/1078-0432.CCR-05-0675. [DOI] [PubMed] [Google Scholar]
- 126.Boer K., Helinger E., Helinger A., Pocza P., Pos Z., Demeter P., Baranyai Z., Dede K., Darvas Z., Falus A. Decreased expression of histamine H1 and H4 receptors suggests disturbance of local regulation in human colorectal tumours by histamine. Eur. J. Cell Biol. 2008;87:227–236. doi: 10.1016/j.ejcb.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 127.Lamas D.J.M., Carabajal E., Prestifillipo J.P., Rossi L., Elverdin J.C., Merani S., Bergoc R.M., Rivera E.S., Medina V.A. Protection of radiation-unduced damage to the hematopoietic system, small intestine and salivary glands in rats by JNJ7777120 compound, histamine H4 ligand. PLoS ONE. 2013;8:e69106. doi: 10.1371/journal.pone.0069106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ji Y., Sakata Y., Li X., Zhang C., Yang Q., Xu M., Wollin A., Langhans W., Tso P. Lymphatic diamine oxidase secretion stimulated by fat absorption is linked with histamine release. Am. J. Physiol. Gastrointest. Liver Physiol. 2013;304:G732–G740. doi: 10.1152/ajpgi.00399.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Deiteren A., De Man J.G., Ruyssers N.E., Moreels T.G., Pelckmans P.A., De Winter B.Y. Histamine H4 and H1 receptors contribute to postinflammatory visceral hypersensitivity. Gut. 2014;63:1873–1882. doi: 10.1136/gutjnl-2013-305870. [DOI] [PubMed] [Google Scholar]
- 130.Wang M., Han J., Domenico J., Shin Y.S., Jia Y., Gelfand E.W. Combined blockade of the histamine H1 and H4 receptor suppresses peanut-induced intestinal anaphylaxis by regulating dendritic cell function. Allergy. 2016;71:1561–1574. doi: 10.1111/all.12904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schirmer B., Rother T., Bruesch I., Bleich A., Werlein C., Jonigk D., Seifert R., Neumann D. Genetic deficiency of the histamine H4-receptor reduces experimental colorectal carcinogenesis in mice. Cancers. 2020;12:912. doi: 10.3390/cancers12040912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Thurmond R.L., Gelfand E.W., Dunford P.J. The role of histamine H1 and H4 receptors in allergic inflammation: The search for new antihistamines. Nat. Rev. Drug Discov. 2008;7:41–53. doi: 10.1038/nrd2465. [DOI] [PubMed] [Google Scholar]
- 133.Rijnierse A., Nijkamp F.P., Kraneveld A.D. Mast cells and nerves tickle in the tummy: Implications for inflammatory bowel disease and irritable bowel syndrome. Pharmacol. Ther. 2007;116:207–235. doi: 10.1016/j.pharmthera.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 134.van Hoboken E.A., Thijssen A.Y., Verhaaren R., van der Veek P.P., Prins F.A., Verspaget H.W., Masclee A.A. Symptoms in patients with ulcerative colitis in remission are associated with visceral hypersensitivity and mast cell activity. Scand. J. Gastroenterol. 2011;46:981–987. doi: 10.3109/00365521.2011.579156. [DOI] [PubMed] [Google Scholar]
- 135.Karhausen J., Qing M., Gibson A., Moeser A.J., Griefingholt H., Hale L.P., Abraham S.N., MacKensen G.B. Intestinal mast cells mediate gut injury and systemic inflammation in a rat model of deep hypothermic circulatory arrest. Crit. Care Med. 2013;41:e200–e210. doi: 10.1097/CCM.0b013e31827cac7a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Eissmann M.F., Dijkstra C., Jarnicki A., Phesse T., Brunnberg J., Poh A.R., Etemadi N., Tsantikos E., Thiem S., Huntington N.D., et al. IL-33-mediated mast cell activation promotes gastric cancer through macrophage mobilization. Nat. Commun. 2019;10:2735. doi: 10.1038/s41467-019-10676-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chin K.W., Barrett K.E. Mast cells are not essential to inflammation in murine model of colitis. Dig. Dis. Sci. 1994;39:513–525. doi: 10.1007/BF02088336. [DOI] [PubMed] [Google Scholar]
- 138.Minocha A., Thomas C., Omar R. Lack of crucial role of mast cells in pathogenesis of experimental colitis in mice. Dig. Dis. Sci. 1995;40:1757–1762. doi: 10.1007/BF02212698. [DOI] [PubMed] [Google Scholar]
- 139.Araki Y., Andoh A., Fujiyama Y., Bamba T. Development of dextran sulphate sodium-induced experimental colitis is suppressed in genetically mast cell-deficient Ws/Ws rats. Clin. Exp. Immunol. 2000;119:264–269. doi: 10.1046/j.1365-2249.2000.01094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Higuchi S., Tanimoto A., Arima N., Xu H., Murata Y., Hamada T., Makishima K., Sasaguri Y. Effects of histamine and interleukin-4 synthesized in arterial intima on phagocytosis by monocytes/macrophages in relation to atherosclerosis. FEBS Lett. 2001;505:217–222. doi: 10.1016/S0014-5793(01)02823-X. [DOI] [PubMed] [Google Scholar]
- 141.Zwadlo-Klarwasser G., Vogts M., Hamann W., Belke K., Baron J., Schmutzler W. Generation and subcellular distribution of histamine in human blood monocytes and monocyte subsets. Inflamm. Res. 1998;74:434–439. doi: 10.1007/s000110050357. [DOI] [PubMed] [Google Scholar]
- 142.Yang X.D., Ai W., Asfaha S., Bhagat G., Friedman R.A., Jin G., Park H., Shykind B., Diacovo T.G., Falus A., et al. Histamine deficiency promotes inflammation-associated carcinogenesis through reduced myeloid maturation and accumulation of CD11b+Ly6G+ immature myeloid cells. Nat. Med. 2011;17:87–95. doi: 10.1038/nm.2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Amaral M.M., Davio C., Ceballos A., Salamone G., Cañones C., Geffner J., Vermeulen M. Histamine improves antigen uptake and cross-presentation by dendritic cells. J. Immunol. 2007;179:3425–3433. doi: 10.4049/jimmunol.179.6.3425. [DOI] [PubMed] [Google Scholar]
- 144.Bischoff S.C., Gebhardt T. Role of mast cells and eosinophils in neuroimmune interactions regulating mucosal inflammation in inflammatory bowel disease. Adv. Exp. Med. Biol. 2006;579:177–208. doi: 10.1007/0-387-33778-4_12. [DOI] [PubMed] [Google Scholar]
- 145.Masterson J.C., McNamee E.N., Fillon S.A., Hosford L., Harris R., Fernando S.D., Jedlicka P., Iwamoto R., Jacobsen E., Protheroe C., et al. Eosinophil-mediated signalling attenuates inflammatory responses in experimental colitis. Gut. 2015;64:1236–1247. doi: 10.1136/gutjnl-2014-306998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Loktionov A. Eosinophils in the gastrointestinal tract and their role in the pathogenesis of major colorectal disorders. World J. Gastroenterol. 2019;24:3503–3526. doi: 10.3748/wjg.v25.i27.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Grosicki M., Wójcik T., Chlopicki S., Kieć-Kononowicz K. In vitro study of histamine and histamine receptor ligands influence on the adhesion of purified human eosinophils to endothelium. Eur. J. Pharmacol. 2016;777:49–59. doi: 10.1016/j.ejphar.2016.02.061. [DOI] [PubMed] [Google Scholar]
- 148.Schirmer B., Bringmann L., Seifert R., Neumann D. In vivo evidence for partial activation of eosinophils via the histamine H4 receptor: Adoptive transfer experiments using eosinophils from H4R−/− and H4R+/+ mice. Front. Immunol. 2018;9:2119. doi: 10.3389/fimmu.2018.02119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gay J., Kokkotou E., O’Brien M., Pothoulakis C., Karalis K.P. Interleukin-6 Genetic ablation protects from trinitrobenzene sulfonic acid-induced colitis in mice. Neuroimmunomodulation. 2006;13:114–121. doi: 10.1159/000096656. [DOI] [PubMed] [Google Scholar]
- 150.Huang E., Liu R., Lu Z., Liu J., Liu X., Zhang D., Chu Y. NKT cells mediate the recruitment of neutrophils by stimulating epithelial chemokine secretion during colitis. Biochem. Biophys. Res. Commun. 2016;474:252–258. doi: 10.1016/j.bbrc.2016.04.024. [DOI] [PubMed] [Google Scholar]
- 151.Werner K., Neumann D., Buschauer A., Seifert R. No evidence for histamine H4 receptor in human monocytes. J. Pharmacol. Exp. Ther. 2014;351:519–526. doi: 10.1124/jpet.114.218107. [DOI] [PubMed] [Google Scholar]
- 152.Takeshita K., Sakai K., Bacon K.B., Gantner F. Critical role of histamine H4 receptor in leukotriene B4 production and mast cell-dependent neutrophil recruitment induced by zymosan in vivo. J. Pharmacol. Exp. Ther. 2003;307:1072–1078. doi: 10.1124/jpet.103.057489. [DOI] [PubMed] [Google Scholar]
- 153.Peterson L.W., Artis D. Intestinal epithelial cells: Regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 2014;14:141–153. doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- 154.Odenwald M.A., Turner J.R. The intestinal epithelial barrier: A therapeutic target? Nat. Rev. Gastroenterol. Hepatol. 2017;14:9–21. doi: 10.1038/nrgastro.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Landy J., Ronde E., English N., Clark S.K., Hart A.L., Knight S.C., Al-Hassi H.O. Tight junctions in inflammatory bowel diseases and inflammatory bowel disease associated colorectal cancer. World J. Gastroenterol. 2016;22:3117–3126. doi: 10.3748/wjg.v22.i11.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Buckley A., Turner J.R. Cell biology of tight junction barrier regulation and mucosal disease. Cold Spring Harb. Perspect. Biol. 2018;10:a029314. doi: 10.1101/cshperspect.a029314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Otegbeye E.E., Mitchem J.B., Park H., Chaudhuri A.A., Kim H., Mutch M.G., Ciorba M.A. Immunity, immunotherapy, and rectal cancer: A clinical and translational science review. Transl. Res. 2021;231:124–138. doi: 10.1016/j.trsl.2020.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kimiz-Gebologlu I., Gulce-Iz S., Biray-Avci C. Monoclonal antibodies in cancer immunotherapy. Mol. Biol. Rep. 2018;45:2935–2940. doi: 10.1007/s11033-018-4427-x. [DOI] [PubMed] [Google Scholar]
- 159.Matsumoto S., Imaeda Y., Umemoto S., Kobayashi K., Suzuki H., Okannoto T. Cimetidine increases survival of colorectal cancer patients with high levels of sialyl Lewis-X and sialyl Lewis-A epitope expression on tumour cells. Br. J. Cancer. 2002;86:161–167. doi: 10.1038/sj.bjc.6600048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ali A.H., Hale L., Yalamanchili B., Ahmed M., Ahmed M., Zhou R., Wright S.E. The effect of perioperative cimetidine administration on time to colorectal cancer recurrence. Am. J. Ther. 2018;25:e405–e411. doi: 10.1097/MJT.0000000000000547. [DOI] [PubMed] [Google Scholar]
- 161.Yang L.P.H., Perry C.M. Spotlight on histamine dihydrochloride in acute myeloid leukaemia. Drugs Aging. 2011;28:325–329. doi: 10.2165/11206810-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 162.Medina V.A., Rivera E.S. Histamine as a potential adjuvant to immuno and radiotherapy for cancer treatment: Discovering new functions for the oldest biogenic amine. Curr. Immunol. Rev. 2010;6:357–370. doi: 10.2174/1573395511006040357. [DOI] [Google Scholar]
- 163.Massari N.A., Nicoud M.B., Medina V.A. Histamine receptors and cancer pharmacology: An update. Br. J. Pharmacol. 2020;177:516–538. doi: 10.1111/bph.14535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Medina V.A., Coruzzi G., Lamas D.J.M., Massari N., Adami M., Levi-Schaffer F., Ben-Zimra M., Schwelberger H., Rivera E.S. Histamine H4 Receptor: A Novel Drug Target in Immunoregulation and Inflammation. De Gruyter Open Poland; Warsaw, Poland: 2013. Histamine in cancer. [Google Scholar]
- 165.Vacante M., Ciuni R., Basile F., Biondi A. Gut microbiota and colorectal cancer development: A closer look to the adenoma-carcinoma sequence. Biomedicines. 2020;8:489. doi: 10.3390/biomedicines8110489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Janney A., Powrie F., Mann E.H. Host–microbiota maladaptation in colorectal cancer. Nature. 2020;585:509–517. doi: 10.1038/s41586-020-2729-3. [DOI] [PubMed] [Google Scholar]
- 167.Mathers J.C., Movahedi M., Macrae F., Mecklin J.-P., Moeslein G., Olschwang S., Eccles D., Evans G., Maher E.R., Bertario L., et al. Long-term effect of resistant starch on cancer risk in carriers of hereditary colorectal cancer: An analysis from the CAPP2 randomised controlled trial. Lancet Oncol. 2012;13:1242–1249. doi: 10.1016/S1470-2045(12)70475-8. [DOI] [PubMed] [Google Scholar]
- 168.Chan A.T., Lippman S.M. Aspirin and colorectal cancer prevention in Lynch syndrome. Lancet. 2011;378:2051–2052. doi: 10.1016/S0140-6736(11)61216-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Masini E., Fabbroni V., Giannini L., Vannacci A., Messerini L., Perna F., Cortesini C., Cianchi F. Histamine and histidine decarboxylase up-regulation in colorectal cancer: Correlation with tumor stage. Inflamm. Res. 2005;54:S80–S81. doi: 10.1007/s00011-004-0437-3. [DOI] [PubMed] [Google Scholar]
- 170.Gao C., Ganesh B.P., Shi Z., Shah R.R., Fultz R., Major A., Venable S., Lugo M., Hoch K., Chen X., et al. Gut microbe–mediated suppression of inflammation-associated colon carcinogenesis by luminal histamine production. Am. J. Pathol. 2017;187:232–236. doi: 10.1016/j.ajpath.2017.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Chen X., Churchill M.J., Nagar K.K., Tailor Y.H., Chu T., Rush B.S., Jiang Z., Wang E.B.C., Renz B.W., Wang H., et al. IL-17 producing mast cells promote the expansion of myeloid-derived suppressor cells in a mouse allergy model of colorectal cancer. Oncotarget. 2015;6:32966–32979. doi: 10.18632/oncotarget.5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.He G.H., Ding J.Q., Zhang X., Xu W.M., Lin X.Q., Huang M.J., Feng J., Wang P., Cai W.K. Activation of histamine H4 receptor suppresses the proliferation and invasion of esophageal squamous cell carcinoma via both metabolism and non-metabolism signaling pathways. J. Mol. Med. 2018;96:951–964. doi: 10.1007/s00109-018-1676-z. [DOI] [PubMed] [Google Scholar]
- 173.Tanaka T., Ishikawa H. Mast cells and inflammation-associated colorectal carcinogenesis. Semin. Immunopathol. 2013;35:245–254. doi: 10.1007/s00281-012-0343-7. [DOI] [PubMed] [Google Scholar]
- 174.Malfettone A., Silvestris N., Saponaro C., Ranieri G., Russo A., Caruso S., Popescu O., Simone G., Paradiso A., Mangia A. High density of tryptase-positive mast cells in human colorectal cancer: A poor prognostic factor related to protease-activated receptor 2 expression. J. Cell. Mol. Med. 2013;17:1025–1037. doi: 10.1111/jcmm.12073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Tanaka T., Kochi T., Shirakami Y., Mori T., Kurata A., Watanabe N., Moriwaki H., Shimizu M. Cimetidine and clobenpropit attenuate inflammation-associated colorectal carcinogenesis in male ICR Mice. Cancers. 2016;8:25. doi: 10.3390/cancers8020025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zhang C., Xiong Y., Li J., Yang Y., Liu L., Wang W., Wang L., Li M., Fang Z. Deletion and down-regulation of HRH4 gene in gastric carcinomas: A potential correlation with tumor progression. PLoS ONE. 2012;7:e31207. doi: 10.1371/journal.pone.0031207. [DOI] [PMC free article] [PubMed] [Google Scholar]