Abstract
Background
Lung cancer is a common tumor and a leading cause of death worldwide. DEAD/H box RNA helicases (DDX) include several family members which regulate mRNA translation in cancer cells. In this study, we demonstrated that DEAD/H box RNA helicase 10 (DDX10) was significantly upregulated in lung cancer tissues compared with adjacent nontumor tissues.
Methods
DDX10 expression was knocked down with shRNA in order to investigate the impact on A549 lung cancer cell growth and related molecular mechanisms in vitro and in vivo. DDX10 expression in lung cancer was assessed using online databases and patient samples.
Results
DDX10 knockdown significantly inhibited the proliferation of lung cancer cells in vitro and in vivo. Furthermore, the bioinformatic tool indicated the putative downstream protein U3 small nucleolar ribonucleoprotein 4 (IMP4). Our data showed a positive correlation between IMP4 and DDX10. We found that IMP4 overexpression could reverse the effect of DDX10 knockdown on the proliferation and apoptosis of A549 cells.
Conclusions
The findings of this study suggest that DDX10/IMP4 might be a novel target for lung cancer diagnosis and treatment.
Keywords: DDX10, DEAD/H box RNA helicases, IMP4, lung cancer, U3 small nucleolar ribonucleoprotein
DEAD/H box RNA helicase 10 (DDX10) is significantly upregulated in lung cancer. DDX10 knockdown significantly inhibited the proliferation of lung cancer cells in vitro and in vivo. IMP4 overexpression could reverse the effect of DDX10 knockdown on the proliferation and apoptosis of lung cancer cells.
INTRODUCTION
About 1.6 million lung cancer patients die every year. Lung cancer is the most commonly diagnosed cancer and most common cause of cancer death, and is a global problem and public health issue. 1 In recent years, a series of neoadjuvant chemotherapies and immunotherapies have become available. 2 With the development of personalized therapy (precision medicine), lung cancer screening, immune checkpoint, and tumorigenic mechanism studies have been reported with particular emphasis on new targeted therapies. 3 Previous studies have identified several biomarkers for early diagnosis, predicting prognosis, and monitoring the treatment of lung cancer. The development of new treatment is based on explorations of the molecular mechanisms of cancer cell proliferation, metastasis, and regulation.
DEAD/H box RNA helicases (DDX) are a group of proteins with several family members regulating mRNA translation in cancer. 4 Wang et al. previously reported that DDX5 is involved in lung cancer progression through direct interaction with β‐catenin. 5 Furthermore, gene mutation and change in DDX10 expression have also been previously investigated in osteosarcoma, hepatocellular carcinoma, leukemia, and ovarian cancer. 6 , 7 , 8 , 9 Accumulating evidence suggests that DDX family proteins play a crucial role in tumorigenesis of cancer cells and stem cell differentiation. 10 However, little is known about the detailed molecular mechanisms of DDX protein regulation.
Therefore, in this study, we knocked down DDX10 expression with shRNA to investigate the impact on A549 lung cancer cell growth and related molecular mechanism in vitro and in vivo. We assessed the DDX10 expression in lung cancer with online databases and patient samples, and found that IMP4 overexpression could reverse the effect of DDX10 knockdown on the proliferation and apoptosis of A549 cells.
METHODS
Patients and tumor samples
The formalin‐fixed and paraffin‐embedded tumor and adjacent nontumor tissues from 31 patients with lung cancer who underwent primary surgical resection between 2015–2020 at the Department of Thoracic Surgery, Beijing Friendship Hospital, Capital Medical University, China, were analyzed.
Cell line and transfection
Human epithelial adenocarcinoma A549 cells and H1299 were purchased from American Type Culture Collection (ATCC). A549 cells and H1299 were cultured in Dulbecco's modified Eagle's medium (DMEM, Gibco) with 10% fetal bovine serum (Gibco) at 37°C. Plasmid encoding IMP4, DDX10, empty vector, or shRNA were purchased from Genechem. Transfection with shRNAs or plasmid DNAs was performed using Lipofectamine 2000 (Thermo) according to the manufacturer's instructions.
Animal model
Female Balb/c nude mice (6–8 weeks, Beijing Hfk Bioscience) were used to establish an animal model by subcutaneous injection of A549 cells (107 cells/mouse, 4–5 mice/group) into the back of mice. The tumor growth in the mice were closely monitored every four days.
Gene expression profiling interactive analysis (GEPIA)
GEPIA is a large‐scale expression profiling and interactive analysis web server designed by Tang et al. 11 and updated to GEPIA 2 in 2019 12 (http://gepia2.cancer-pku.cn/). In this study, we employed the gene expression boxplot and survival analysis in lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD).
Immunohistochemistry
Tumor tissues and adjacent nontumor tissues were embedded in paraffin and cut into 4 μm‐thick sections. Slides were deparaffinized and antigen retrieval was performed using LPH Buffer (BioCare Medical) for 60 min at 93°C followed by 20 min cooling. The DDX10 polyclonal antibody (Thermo Fisher, 1:100) was incubated at 4°C overnight. The slices were reacted with horseradish peroxidase‐labeled anti‐rabbit secondary antibody (Beyotime Biotechnology) for 1 h at room temperature. The slices were stained with hematoxylin and sealed with neutral gel.
Real‐time PCR
Total RNA was extracted from cultured cells with a total RNA kit (Qiagen), and cDNA was synthesized with a cDNA reverse transcription kit (Takara). The primers (forward and reverse) for ß‐actin were: 5′‐CTGGAACGGTGAAGGTGACA‐3′ and 5′‐AAGGGACTTCCTGTAACAATGCA‐3′, for DDX10 were: 5′‐GTGCGGAGCTTCAATCGCT‐3′ and 5′‐CCTGCCATTCGGGTTTCTTCA‐3′ and for IMP4 were: 5′‐GAAAACCGCCTGATTCCCACT‐3′ and 5′‐GTGGCTGGTCACACCTTCA‐3′. Quantitative real‐time PCR was performed using 7500 fast real‐time system (Thermo). The relative mRNA expressions were analyzed by the comparative Ct method and normalized by the abundance of ß‐actin.
Cell proliferation assay
The proliferation of A549 cells was measured using the MTT (3‐[4,5‐dimethylthiazol‐2‐yl]‐2,5‐diphenyltetrazolium bromide) assay. To perform the MTT assay, A549 cells were incubated in the medium containing MTT (5 mg/ml, Sigma) for 4 h at 37°C. The supernatants were then removed and the cells were solubilized in DMSO (Sigma) for 10 min. Finally, the absorbance was determined using a microplate reader at 490 nm.
Flow cytometer
To assess the apoptosis of A549 cells, the apoptosis detection kit (BD) was used according to the manufacturer's protocol. The cell cycle was analyzed using propidium iodide (PI) staining. A549 cells were harvested and fixed in 70% ice‐cold ethanol for 12 h at 4°C. After being washed with PBS, A549 cells were stained for DNA content by PI. The cells were incubated at room temperature for 30 min. Stained cells were immediately analyzed on a BD FACScanto II flow cytometer and further analyzed with Flowjo software (Becton Dickinson).
GeneMANIA analysis
Pathway and network analysis of the DDX10 gene was performed using GeneMania (http://www.genemania.org) 13 with the following network interactions: coexpression, pathway, physical interaction, and shared protein domains. DDX10 gene was submitted to the website to predict the related gene. DDX10 was indicated with stripes; related genes added by GeneMANIA are represented in black, and colored links represent the interactions between genes. The thickness of links indicates the strength/weight of the interaction.
Statistical analysis
Data were compared using a one‐way ANOVA or a Student's t test. p < 0.05 was considered statistically significant.
RESULTS
Lung tumor tissues express high levels of DDX10
There was a tendency toward higher expression of DDX10 in LUAD and LUSC tissues when compared with normal tissues (Figure 1(a)). We further analyzed the potential associations between the expression levels of DDX10 and the survival of patients with lung cancer. As shown in Figure 1(b), high expression levels of DDX10 may contribute to a worse prognosis in patients with LUAD (p < 0.01). Immunohistochemistry results showed that DDX10 expression in tumor tissues was obviously higher than that in adjacent nontumor tissues (Figure 1(c)). Similarly, high levels of DDX10 mRNA expression were observed in tumor tissues by real‐time PCR (p < 0.01, Figure 1(d)).
FIGURE 1.
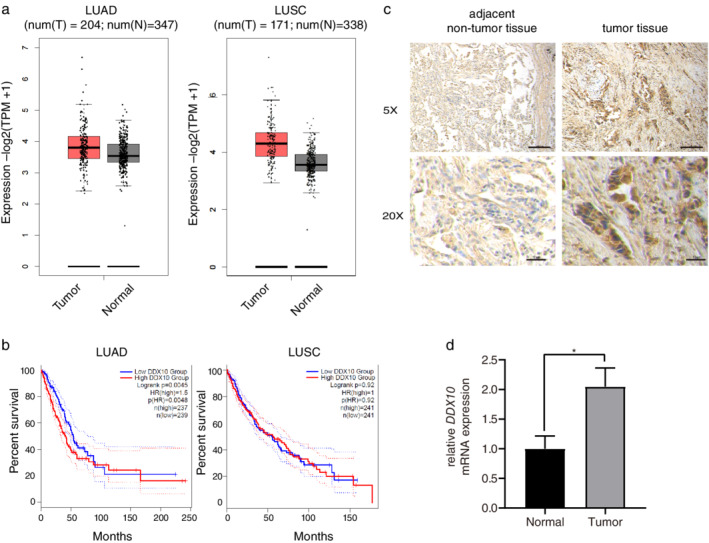
Lung cancer tissue expresses high levels of DDX10. (a) Expression of DDX10 was compared between normal and tumor samples and is given as log2 (TPM + 1) using the interactive web server gene expression profiling interactive analysis (GEPIA). (b) Overall survival associated with the DDX10 gene based on The Cancer Genome Atlas (TCGA) database via GEPIA. (c) Immunohistochemistry of DDX10 in tumor and adjacent nontumor tissue. (d) Relative mRNA expression of DDX10 was assessed by real‐time PCR. Results are represented as mean ± SEM of three independent experiments. LUSC, lung squamous cell carcinoma; LUAD, lung adenocarcinoma. *p < 0.05
DDX10 impacts the proliferation, apoptosis, and cell cycle of lung cancer cells
To investigate the impact of DDX10 on lung cancer cells, we transfected the A549 and H1299 cell lines with shDDX10 or DDX10 overexpressed plasmid. The production of DDX10 mRNA was significantly reduced or increased after shDDX10 or DDX10 overexpressed plasmid transfection (Figure 2(a)). A549 proliferation was determined using the MTT assay. As shown in Figure 2(b), DDX10 significantly promotes the proliferation of A549 and H1299 cells (p < 0.05). Moreover, DDX10 significantly inhibited the apoptosis of A549 and H1299 cells (p < 0.01, Figure 2(c) and (d)). To determine if cell cycle progression could possibly be regulated by DDX10, we performed cell cycle analysis using propidium iodide staining. A greater proportion of A549 and H1299 cells transfected shDDX10 were engaged in the G0/G1‐phase (Figure 2(e)). Similarly, compared with control, DDX10 overexpression promoted more DNA synthesis and cell division in these two cell lines (Figure 2(f)).
FIGURE 2.
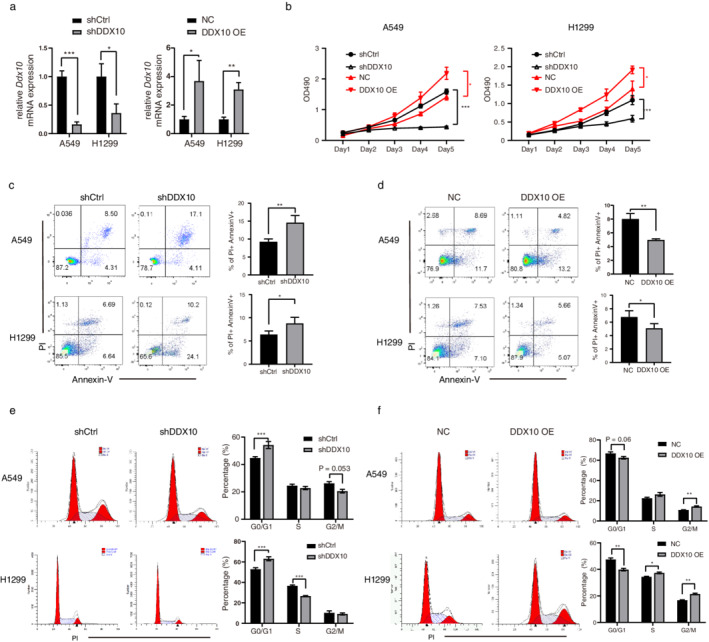
DDX10 affected the proliferation, cell cycle, and apoptosis of A549 cell in vitro. (a) The knockdown and overexpression of DDX10 significantly modulated the mRNA expression of DDX10 in A549 and H1299 cells. (b) Cell viability of A549 was measured by MTT assay. (c–d) Apoptosis and (e–f) cell cycle of A549 and H1299 cells were measured after DDX10 knockdown or overexpression at day 5 by flow cytometry. *p < 0.05, **p < 0.01, ***p < 0.001
DDX10 knockdown reduces the growth of A549 cells in vivo
The efficacy of DDX10 in vivo was evaluated using subcutaneously xenografted A549 cells. All nude mice injected with shRNA transfected A549 cells developed subcutaneous xenografted tumors (Figure 3(a)). The tumor volume and weight of the shDDX10 group were lower than those of the shCtrl group (Figure 3(b) and (c)), although we did not observe a significant difference in bodyweight between the two groups (Figure 3(d)).
FIGURE 3.
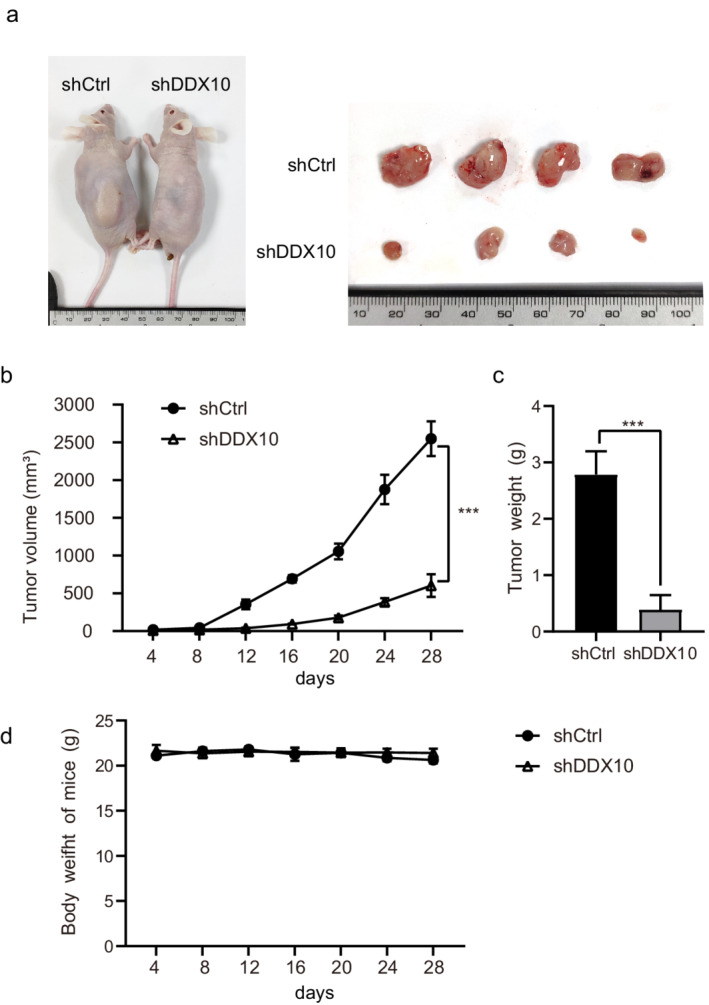
Knockdown of DDX10 reduces A549 tumor growth in vivo. (a) Representative photographs of tumors derived from shCtrl and shDDX10 transfected A549 cells on day 28 after implantation. (b) Tumor growth kinetics of subcutaneously implanted nude mice. (c) Weight of tumor at time of sacrifice. (d) The bodyweight of mice represented at different points. Error bars represent mean ± SEM. ***p < 0.001
DDX10 promotes the proliferation of A549 cell via IMP4
To explore the molecular mechanism of DDX10 in lung cancer cells, we generated the gene interaction network of DDX10 using geneMANIA. The predicted network showed that IMP4 and DDX10 may be highly coexpressed in human cells (Figure 4(a)). The Cancer Genome Atlas (TCGA) database verified the positive correlation between DDX10 and IMP4 in LUAD and LUSC tissues (Figure 4(b)). Compared with normal lung tissue, the mRNA expression of IMP4 was also higher in tumor tissue (p < 0.05, Figure 4(c)). We also identified the significant downregulation of IMP4 in shDDX10 transfected A549 cells compared to the shCtrl group (p < 0.001, Figure 4(d)). Moreover, DDX10 overexpression increased the expression of IMP4 (p < 0.05, Figure 4(e)). To investigate the influence of IMP4 in the DDX10 induced tumor cell proliferation, we overexpressed IMP4 in shDDX10 transfected A549 cells. Compared to the shDDX10 group, the shDDX10 + IMP4 group significantly recovered the proliferation of A549 cells (p < 0.001, Figure 4(f)). Furthermore, IMP4 overexpression also reduced apoptosis and promoted cell division of shDDX10 transfected A549 cells (p < 0.05, Figure 4(g) and (h)).
FIGURE 4.
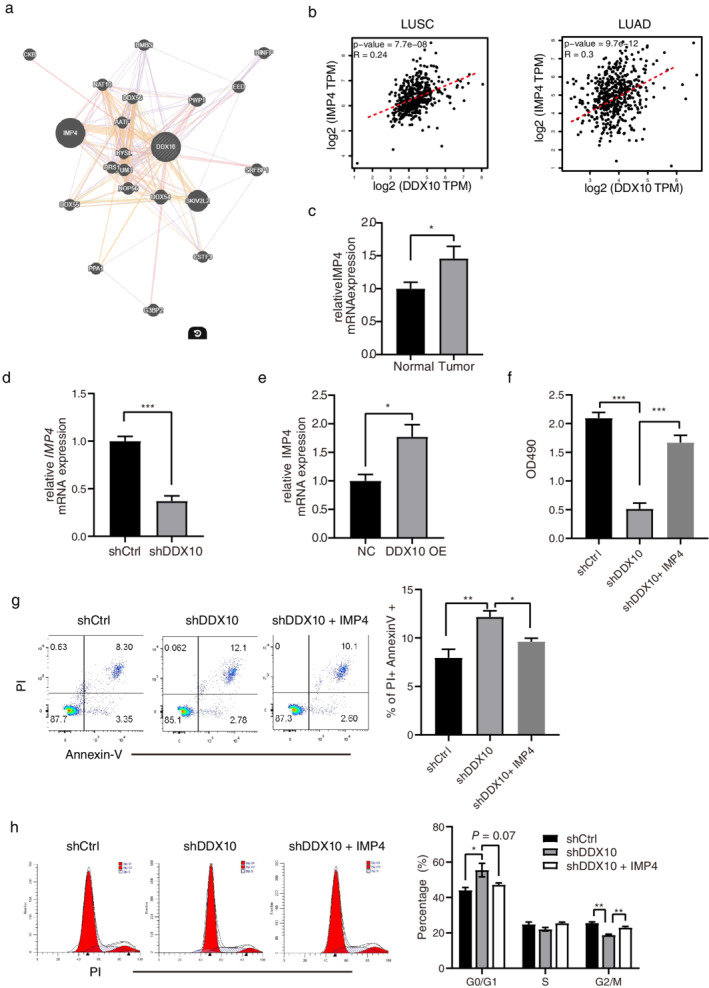
DDX10 promotes the proliferation of A549 cells via IMP4. (a) Gene interaction network of DDX10 was generated using the geneMANIA tool (http://www.genemania.org). The thickness of the line indicates the extent of the relationship between two genes. (b) Correlation between DDX10 and IMP4 in LUSC and LUAD. Expression levels are derived from the TCGA mRNA database, and test statistics from Pearson product–moment correlation. (c) Relative DDX10 mRNA expression was detected by real‐time PCR in normal or tumor tissue. (d–e) Relative IMP4 mRNA expression was detected by real‐time PCR in DDX10 knockdown or overexpressed A549 cells. A549 cells were transfected by shCtrl, shDDX10 or shDDX10 + IMP4. (f) Cell viability, (g) apoptosis, and (h) cell cycle of A549 were measured by MTT assay and flow cytometry, respectively. Error bars represent mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001
DISCUSSION
Lung cancer is the most common tumor and a leading cause of death worldwide. 14 Lung cancer therapy has achieved unprecedented progress in recent years. Recently, DEAD box protein DDX5 has been reported to play a tumor‐promoting role in lung cancer progression. 5 Furthermore, several studies have demonstrated the crucial roles of DDX family protein in tumorigenesis. In the current study, we demonstrated that a high expression of DDX10 in tumor tissue was related to the proliferation, apoptosis and cell cycle of tumor cells. To investigate the underlying mechanisms of DDX10, we predicted the protein IMP4 interacted with DDX10 via online bioinformatic tools. In vitro experiments verified the positive correlation between IMP4 and DDX10, which showed the pivotal role of IMP4 in DDX10 promoted proliferation of lung cancer cells.
The U3 small nucleolar RNA (snoRNA) is required for the early cleavage of pre‐rRNA processing, which is essential for the formation of the small ribosomal subunit RNA (18S rRNA) and the survival of eukaryocytes. 15 A number of proteins have been identified that are specifically associated with the U3 snoRNA, such as IMP4, IMP3, Dhr1, Lcp5, Rcl1, and Sof1. 16 IMP4 family is characterized by the σ70‐like motif that confers binding to RNA, and all the proteins in this family interact with pre‐rRNA processing intermediates. 17 Compared with other members of this family, IMP4 protein is associated with the U3 snoRNA and is required for the early cleavage of pre‐rRNA processing. 17 , 18
In the interaction of RNA helicase and small nucleolar RNA, Soltanieh and his colleagues showed that DEAD‐box RNA helicase Dbp4 is required for small‐subunit processome formation and production of 18S ribosomal RNA, which are crucial in the translation of proteins. 19 , 20 Moreover, DEAD‐box RNA helicase DHX37 and Dhr1 promoted the maturation of the small ribosomal subunit, which is linked to cancer and genetic diseases. 21 , 22 In this study, our data also indicated a positive correlation between DDX10 and IMP4 in lung cancer patients. IMP4 overexpression reversed the suppressive effect of DDX10 knockdown on the proliferation of lung cancer cells. Related studies have also unraveled the coexpression and protein–protein interaction between DDX10 and IMP4. 23 , 24 Thus, it is possible that DDX10 participates in the RNA unwinding of IMP4 and induces IMP4 protein production. Further studies investigating the role of IMP4 and the interaction between DDX protein and IMP4 on lung cancer are needed.
In this study, we discovered the different mortality DDX10 caused between LUAD and LUSC. As summarized in the study by Herbst et al., the most common genetic alterations are disparate in LUAD and LUSC. 23 The most commonly mutated genes in LUAD include KRAS and EGFR, while TP53 and CDKN2A have been identified as the most common mutated genes in LUSC. As DDX family proteins participate in various biological processes, we suspect the gene mutation status between the two kinds of NSCLC may play a role in the function of DDX10. As an RNA helicase, DDX family proteins participate in various biological processes, which are important to normal cell metabolism and maintenance. Wang et al. also indicated that DDX10 promotes the activity of classical Wnt/β‐catenin signaling in HCC cell lines, which plays a central role in cell proliferation and survival. 7 There are concerns about normal cell toxicity of translation inhibitors. To date, it has been previously reported that DDX3 inhibitor RK‐33 does not cause toxicity in mice at effective dose levels. 25 Thus, DDX family protein inhibitors have the potential to be used in cancer therapy, without unacceptable normal cell toxicity. However, more clinical trials in humans and more studies in the mechanisms involved need to be undertaken in the future.
In conclusion, the present study demonstrates that DDX10 promotes the proliferation of lung cancer cells in vitro and in vivo. Online databases and patient samples verified the high expression of DDX10 and IMP4 in tumor tissue. IMP4 overexpression could inhibit apoptosis of A549 cells induced by DDX10 silencing. These data show the role of the DDX10/IMP4 pathway in lung cancer cells which may provide a new target for lung cancer diagnosis and therapy.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
ACKNOWLEDGMENTS
We thank all the patients who have participated in the current study. The study was supported by the Research Foundation of Beijing Friendship Hospital, Capital Medical University (no. yyqdkt2018‐27).
Liu C, Tang J, Duan X, Du Y, Wang X, Cui Y. DDX10 promotes human lung carcinoma proliferation by U3 small nucleolar ribonucleoprotein IMP4. Thorac Cancer. 2021;12:1873–1880. 10.1111/1759-7714.13976
Funding information Research Foundation of Beijing Friendship Hospital, Capital Medical University, Grant/Award Number: yyqdkt2018‐27
REFERENCES
- 1. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–86. 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2. Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ Jr, Wu YL, et al. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389(10066):299–311. 10.1016/S0140-6736(16)30958-8. [DOI] [PubMed] [Google Scholar]
- 3. Provencio M, Nadal E, Insa A, Garcia‐Campelo MR, Casal‐Rubio J, Domine M, et al. Neoadjuvant chemotherapy and nivolumab in resectable non‐small‐cell lung cancer (NADIM): an open‐label, multicentre, single‐arm, phase 2 trial. Lancet Oncol. 2020;21:1413–22. 10.1016/S1470-2045(20)30453-8. [DOI] [PubMed] [Google Scholar]
- 4. Heerma Van Voss MR, van Diest PJ, Raman V. Targeting RNA helicases in cancer: the translation trap. Biochim Biophys Acta Rev Cancer. 2017;1868(2):510–20. 10.1016/j.bbcan.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang Z, Luo Z, Zhou L, Li X, Jiang T, Fu E. DDX5 promotes proliferation and tumorigenesis of non‐small‐cell lung cancer cells by activating beta‐catenin signaling pathway. Cancer Sci. 2015;106(10):1303–12. 10.1111/cas.12755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shi JH, Hao YJ. DDX10 overexpression predicts worse prognosis in osteosarcoma and its deletion prohibits cell activities modulated by MAPK pathway. Biochem Biophys Res Commun. 2019;510(4):525–9. 10.1016/j.bbrc.2019.01.114. [DOI] [PubMed] [Google Scholar]
- 7. Wang Y, Xiao W, Wei S, Chen X, Wei L, Tian R, et al. Highly expressed DDX10 promotes hepatocellular carcinoma cell proliferation through Wnt/β‐catenin signaling. Int J Clin Exp Pathol. 2017;10(5):6047–53. [Google Scholar]
- 8. Gorello P, Nofrini V, Brandimarte L, Pierini V, Crescenzi B, Nozza F, et al. Inv(11)(p15q22)/NUP98‐DDX10 fusion and isoforms in a new case of de novo acute myeloid leukemia. Cancer Genet. 2013;206(3):92–6. 10.1016/j.cancergen.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 9. Gai M, Bo Q, Qi L. Epigenetic down‐regulated DDX10 promotes cell proliferation through Akt/NF‐kappaB pathway in ovarian cancer. Biochem Biophys Res Commun. 2016;469(4):1000–5. 10.1016/j.bbrc.2015.12.069. [DOI] [PubMed] [Google Scholar]
- 10. Yassin ER, Abdul‐Nabi AM, Takeda A, Yaseen NR. Effects of the NUP98‐DDX10 oncogene on primary human CD34+ cells: role of a conserved helicase motif. Leukemia. 2010;24(5):1001–11. 10.1038/leu.2010.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large‐scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–W60. 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Warde‐Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Romaszko AM, Doboszynska A. Multiple primary lung cancer: A literature review. Adv Clin Exp Med. 2018;27(5):725–30. 10.17219/acem/68631. [DOI] [PubMed] [Google Scholar]
- 15. Hughes JM, Ares M Jr. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre‐ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991;10(13):4231–9. 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Terns MP, Terns RM. Small nucleolar RNAs: versatile trans‐acting molecules of ancient evolutionary origin. Gene Expr. 2002;10(1–2):17–39. [PMC free article] [PubMed] [Google Scholar]
- 17. Wehner KA, Baserga SJ. The sigma(70)‐like motif: a eukaryotic RNA binding domain unique to a superfamily of proteins required for ribosome biogenesis. Mol Cell. 2002;9(2):329–39. 10.1016/S1097-2765(02)00438-0. [DOI] [PubMed] [Google Scholar]
- 18. Lee SJ, Baserga SJ. Imp3p and Imp4p, two specific components of the U3 small nucleolar ribonucleoprotein that are essential for pre‐18S rRNA processing. Mol Cell Biol. 1999;19(8):5441–52. 10.1128/MCB.19.8.5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Soltanieh S, Osheim YN, Spasov K, Trahan C, Beyer AL, Dragon F. DEAD‐box RNA helicase Dbp4 is required for small‐subunit processome formation and function. Mol Cell Biol. 2015;35(5):816–30. 10.1128/MCB.01348-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Soltanieh S, Lapensee M, Dragon F. Nucleolar proteins Bfr2 and Enp2 interact with DEAD‐box RNA helicase Dbp4 in two different complexes. Nucleic Acids Res. 2014;42(5):3194–206. 10.1093/nar/gkt1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Choudhury P, Hackert P, Memet I, Sloan KE, Bohnsack MT. The human RNA helicase DHX37 is required for release of the U3 snoRNP from pre‐ribosomal particles. RNA Biol. 2019;16(1):54–68. 10.1080/15476286.2018.1556149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sardana R, Liu X, Granneman S, Zhu J, Gill M, Papoulas O, et al. The DEAH‐box helicase Dhr1 dissociates U3 from the pre‐rRNA to promote formation of the central pseudoknot. PLoS Biol. 2015;13(2):e1002083. 10.1371/journal.pbio.1002083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Herbst RS, Morgensztern D, Boshoff C. The biology and management of non‐small cell lung cancer. Nature. 2018;553(7689):446–54. 10.1038/nature25183. [DOI] [PubMed] [Google Scholar]
- 24. Wang Q, Diskin S, Rappaport E, Attiyeh E, Mosse Y, Shue D, et al. Integrative genomics identifies distinct molecular classes of neuroblastoma and shows that multiple genes are targeted by regional alterations in DNA copy number. Cancer Res. 2006;66(12):6050–62. [DOI] [PubMed] [Google Scholar]
- 25. Bol GM, Vesuna F, Xie M, Zeng J, Aziz K, Gandhi N, et al. Targeting DDX3 with a small molecule inhibitor for lung cancer therapy. EMBO Mol Med. 2015;7(5):648–69. 10.15252/emmm.201404368. [DOI] [PMC free article] [PubMed] [Google Scholar]


