Abstract
The high incidence and mortality of lung cancer make early detection of lung cancer particularly important. At present, the diagnosis of lung cancer mainly depends on diagnostic imaging and tissue biopsy. However, current diagnostics are not satisfactory owing to the low specificity and inability of multiple sampling. Accumulating evidence indicates that circular RNAs (circRNAs) play a critical role in cancer progression and are promising cancer biomarkers. In particular, circRNAs are considered novel specific diagnostic markers for non-small cell lung cancer (NSCLC). Liquid biopsy is an important method in the early diagnosis of cancer due to its high sensitivity and specificity, as well as the possibility of performing multiple sampling. circRNAs are stably present in exosomes and sometimes become part of circulating nucleic acids, making them ideal for liquid biopsy. In this review, we summarize the advances in the research on circRNAs in NSCLC, and also highlight their potential applications for NSCLC detection.
Keywords: circular RNAs, exosome, liquid biopsy, non-small cell lung cancer, biomarker
Introduction
Cancer is a major public health problem worldwide. In China, lung cancer is one of the most common cancers and has the highest incidence and mortality rates among all cancers (Chen, 2015). The 5-year survival rate of patients with lung cancer is 54% when diagnosed at an early stage (Bach et al., 2012). However, this rate decreases to only 18% in patients with advanced cancer (Siegel et al., 2018). Non-small cell lung cancer (NSCLC), which is historically divided into adenocarcinoma, squamous cell carcinoma, and large cell carcinoma, accounts for approximately 80% of lung cancers. Using current diagnostics, NSCLC patients are usually diagnosed at the middle or late stages. To increase the survival rate of NSCLC patients, it is necessary to discover new biomarkers for its early diagnosis.
Precision oncology aims to improve the diagnosis and treatment of cancer (Ashley, 2016; Kumar-Sinha and Chinnaiyan, 2018). Studies on the comprehensive molecular signatures of cancers at the DNA, RNA, protein, and epigenetic levels deepen our understanding of the molecular events in different cancer types. Accurate identification of tumor subtypes and drug targets based on molecular signature profiling is a prerequisite for precision oncology (Heitzer et al., 2019). In the past few decades, molecular signatures have usually been identified by tissue biopsies. Histological examination is the gold standard for tumor diagnosis and grading (Chen, 2015). However, it is difficult to sample tumor tissues multiple times, and it is also difficult to conduct real-time tumor monitoring. In addition, cancer heterogeneity impedes the complete revelation of tumor information. Therefore, researchers have become increasingly interested in liquid biopsy, because liquid biopsy not only provides information equivalent to that provided by tissue biopsy, but also performs better than tissue biopsy in the minimally invasive, time-sensitive, and dynamic monitoring of cancers. With the advancement of research, liquid biopsy has played a significant role in the clinical testing of cancers.
In oncology, liquid biopsy refers to the sampling and measurement of analytes in various biological fluids (e.g., blood, urine, ascites, pleural effusions) (Siravegna et al., 2017). Peripheral blood analytes include circulating tumor cells (CTCs), circulating nucleic acids, circulating extracellular vesicles (EVs), tumor-educated platelets (TEPs), proteins, and metabolites. They can reveal several types of tumor information that are usually obtained by traditional tissue biopsy. Recent studies show that liquid biopsy could be a powerful tool for the early diagnosis of cancer (Cohen et al., 2018; Pantel and Alix-Panabićres, 2019).
Circular RNAs (circRNAs) are a type of endogenous non-coding RNAs (Shabaninejad et al., 2019; Borran et al., 2020; Naeli et al., 2020). Other types of non-coding RNAs include microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) (Vafadar et al., 2019; Sadri Nahand et al., 2020; Razavi et al., 2021). circRNAs are stably present in exosomes or circulating RNAs, making them an important target for liquid biopsy. Significant circRNA expression has been found in patients with esophageal, gastric, liver, and lung cancer (Zhang et al., 2013; Jeck and Sharpless, 2014). In addition, circRNAs that carry tumor tissue information can be transported from tumor tissues to the blood by exosomes (Tang et al., 2018). Therefore, tumor tissue-derived exosomal circRNAs (exo-circRNAs)s could serve as novel circulating tumor biomarkers. This review summarizes the role of circRNAs in the progression of NSCLC, and discusses the feasibility of using circRNAs as NSCLC biomarkers.
Characteristics and Functions of Circular RNAs
Biogenesis and Characteristics of circRNAs
Circular RNAs were discovered by Nigro et al. (1991) in their analysis of human DCC gene sequencing. The 3′ and 5′ ends of circRNAs are not free, but covalently linked to each other. This linking occurs at a site flanked by canonical splice signals. To yield a circRNA, a splice donor must be linked to an upstream splice acceptor (Jeck et al., 2013; Memczak et al., 2013). This splicing product can be divided into two categories according to the differences in their structure and function: (1) circRNAs consisting exclusively of coding sequences and (2) circRNAs in which an intron is retained (exon-intron ciRNAs, EIciRNAs) (Xia et al., 2017).
circRNAs are widespread in eukaryotic cells. Evidence suggests that circRNAs have the following characteristics: (1) high stability. circRNAs are more stable than lncRNAs because of their circular structure. They are not easily degraded by exonucleases and thus have a longer half-life (Bachmayr-Heyda et al., 2015; Wu et al., 2019). (2) High abundance. Memczak et al. (2015) observed a higher expression of circRNAs relative to corresponding linear RNAs, as evidenced by the high circular-to-linear RNA ratio in the blood. In some cases, the ratio was more than 10-fold. (3) Evolutionary conservation. The sequences of circRNAs are evolutionarily conserved among different species (Jeck et al., 2013). (4) Specific expression. circRNAs are often expressed in a tissue- or developmental-stage-specific manner (Memczak et al., 2013).
Functions of circRNAs
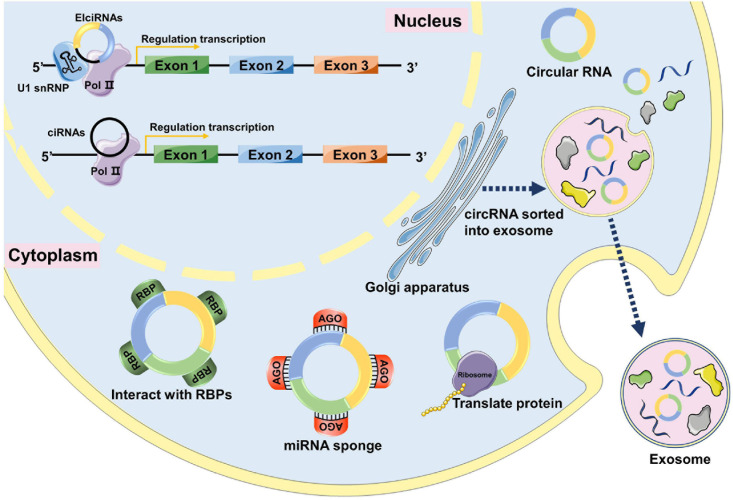
Previous studies have indicated that circRNAs have four major functions: act as miRNA sponges, interact with RNA-binding proteins, regulate gene transcription, and encode polypeptides (Figure 1).
FIGURE 1.
The functions of circRNAs. In the nucleus, circRNAs can interact with transcription complexes in the promoter region of their host gene to induce gene transcription by interacting with U1 snRNP. circRNAs can directly interact with transcription complexes on host genes to induce their transcription. In the cytoplasm, circRNAs can interact with RBPs and affect their functions and translocations. circRNAs can act as miRNA sponges to inhibit miRNA activity by interacting with miRNA-Ago2 complexes. Some circRNAs have protein-coding capacity and can encode proteins. circRNAs can be loaded into exosomes, released by donor cells, and enter recipient cells through endocytosis, thus modulating gene expression in recipient cells.
Acting as miRNA Sponges
MicroRNA sponges are RNA transcripts containing multiple high-affinity binding sites that associate with and sequester specific miRNAs to prevent them from interacting with their target messenger RNAs (mRNAs). Some circRNAs contain a large number of miRNA binding sites and function as miRNA sponges to indirectly regulate gene expression. One of the representative circRNAs of this type is the ciRS-7 (Hansen et al., 2013). CirS7 has been shown to contain 470 conserved binding sites of miR7, thus it is considered a “sponge” of miR7 (Hansen et al., 2013). However, only a small fraction of circRNAs function as potential miRNA sponges in mammals (Chen, 2016).
Interacting With RNA-Binding Proteins
Several circRNAs can bind to RNA-binding proteins (RBPs) and function as RBP sponges. For example, circFoxo3 has been reported to interact with cyclin-dependent kinase 2 (Cdk2) and p21 to inhibit the G1-S transition in the cell cycle, resulting in cell cycle arrest (Du et al., 2016).
Regulating Gene Transcription
Circular RNAs can also regulate gene transcription. For instance, EIciRNAs (Li Z. et al., 2015) interact with U1 small nuclear RNA (snRNA) to enhance the expression of their parental genes in cis (Li Z. et al., 2015). circRNAs that regulate gene transcription are mainly located in the nucleus.
Encoding Polypeptides
Although circRNAs are non-coding RNAs, a few circRNAs encode polypeptides that exert regulatory functions. This feature of circRNAs increases the complexity of the transcriptome and proteome. circ-ZNF609 can be translated into proteins in a splicing-dependent and cap-independent manner (Legnini et al., 2017). Further analysis revealed that consensus m6A motifs were enriched in circRNAs, and a single m6A site was sufficient to drive translation initiation. The study also stated that the m6A-driven translation of circRNAs is widespread, and hundreds of endogenous circRNAs have translational potential (Yang et al., 2017).
Identification and Validation of Circular RNAs
Identification of circRNAs
RNA sequencing (RNA-seq) is an important method for genome-wide circRNA research. High-throughput sequencing allows for deeper sequencing and longer read lengths, making circRNA detection possible (Jeck and Sharpless, 2014). RNA-seq library preparation and bioinformatics analysis of RNA-seq data are the main challenges of high-throughput circRNA sequencing (Szabo and Salzman, 2016). RNA purification is crucial for library preparation and subsequent circRNA detection. Currently, circRNAs are typically purified by either poly(A)-selected or depleted ribosomal RNA (Szabo and Salzman, 2016; López-Jiménez et al., 2018). Various software packages, such as CIRI and circRNA_finder, can be used to analyze circRNAs from RNA-seq data, including 11 algorithms (Zeng et al., 2017). However, complicated data analysis methods and high cost severely limit the application of RNA-seq in circRNA detection. Compared with RNA-seq, a circRNA microarray is a more sensitive and efficient platform for circRNA identification. Combined with enzymatic linear RNA removal, a circRNA microarray uses unique circular junction-specific probes to enrich, capture, and quantify circRNAs with high sensitivity and specificity (Xu et al., 2011).
Validation of circRNAs
Real-time fluorescent quantitative polymerase chain reaction (RT-qPCR) is essential for the verification of differentially expressed circRNAs after genome-wide sequencing or microarray analysis (Starke et al., 2015; Panda et al., 2017). In addition, RT-qPCR can conveniently quantify the expression of circRNAs used as biomarkers in clinical research (Guria et al., 2020). In a typical RT-qPCR analysis, total RNAs including linear RNAs (such as mRNAs and miRNAs) are extracted from cells, and linear RNAs are removed by ribosomal RNA (rRNA) consumption or ribonuclease R (RNase R) (an exoribonuclease) digestion. Next, complementary DNAs (cDNAs) of circRNAs are obtained by reverse transcription (RT). Finally, the fluorescent signal is accumulated and acquired by PCR amplification (Jeck and Sharpless, 2014). However, current methods for rRNA consumption and RNase R digestion cannot completely remove linear RNAs, especially linear RNAs with extensive secondary structures. Amplification of residual linear RNAs hinders quantitative and qualitative analysis of circRNAs (Panda et al., 2017). Furthermore, during the RT process, rolling circle amplification (RCA) using circRNAs as templates produces technical artifacts, leading to irreversible false-positive results (Szabo and Salzman, 2016).
Northern blotting enables the specific detection and characterization of circRNAs (Hansen et al., 2013; Zhang et al., 2014). Detection of specific circRNAs by northern blotting can be accomplished by short probes spanning the circular splice junction, or by longer probes covering as much as an entire circularized exon (Yang et al., 2018). Compared with RT-qPCR, northern blotting is laborious and time-consuming, but it is still an important tool for circRNA characterization (Schneider et al., 2018). circRNA in situ hybridization (RNA-ISH) can locate and quantify circRNAs by using specific probes that bind to back-spliced junction sites (Yang et al., 2020). However, it is also time-consuming and labor-intensive. In addition, RNA-ISH requires complex experimental steps and yields low detection signals. Therefore, advanced microscopic technology is required to enhance the signals to detectable levels.
Considering the growing interest in the comprehensive characterization of circRNAs, it is essential to develop better circRNA detection methods. The detection and imaging method based on catalyzed hairpin assembly (CHA) is a promising strategy. In CHA, two complementary nucleic acid hairpins are designed to kinetically hinder their spontaneous hybridization by embedding complementary regions in hairpin stems. In the presence of a target input chain, one hairpin is opened as a result of the toehold-mediated strand displacement, which further induces the assembly of the two hair clips. Finally, similar to a catalyst, the target chain is spontaneously replaced and recycled to trigger more hairpin assembly events (Jung and Ellington, 2014). The CHA-based method has high sensitivity and programmability. It has been proven to be a versatile tool for the detection of various molecules and events, including nucleic acids, small molecules, proteins, enzyme activity, metal ions, and cancer cells (Liu J. et al., 2019). Using this method, Jin et al. constructed a linear DNA nanostructure (LDN) to directly detect circRNAs in complex samples and even in cells (Jiao et al., 2020). They can accurately identify circRNAs without linear RNA-derived interference and image intracellular circRNAs in situ. Hence, CHA provides a simple, effective, and stable method for the detection and quantification of circRNAs. This may facilitate the clinical application of circRNAs as biomarkers.
Circular RNAs in NSCLC
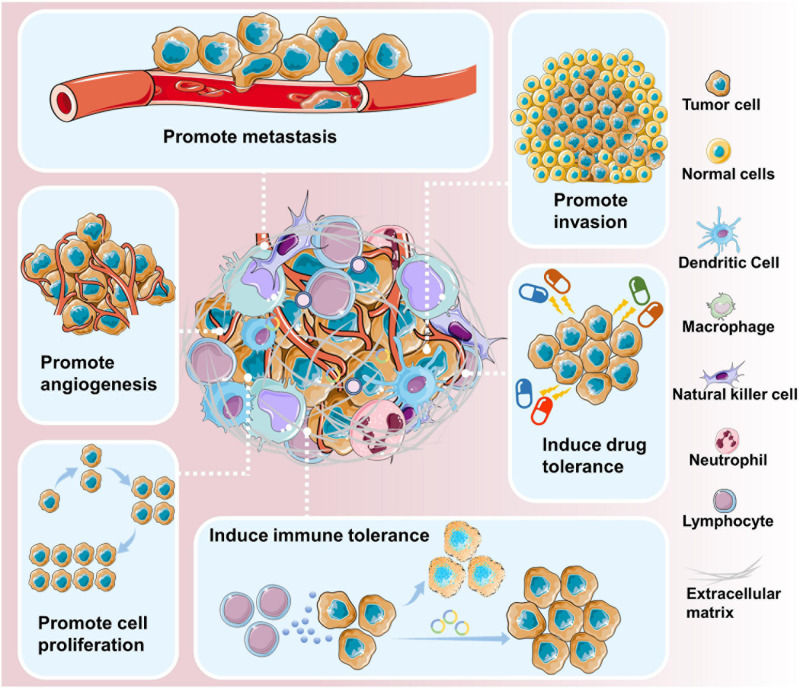
Circular RNAs are closely related to the initiation and development of cancer (Figure 2). Genome-wide analysis has revealed that circRNAs are differentially expressed in various cancer tissues and cell lines (Dou et al., 2016; Shang et al., 2016; Wan et al., 2016; Chen et al., 2017; Zhao et al., 2017). Cancer cell lines express a more diverse pattern of circRNAs than non-cancer cell lines (Hsiao et al., 2017; Liu X-X. et al., 2019).
FIGURE 2.
circRNAs are involved in cancer development and progression. circRNAs have been shown to contribute to various aspects of cancer progression, including promoting metastasis, promoting invasion, inducing drug tolerance, inducing immune tolerance, promoting cell proliferation, and promoting angiogenesis.
Circular RNAs may be involved in the onset of lung cancer. By comparing the expression of circRNAs between tumor samples and paired adjacent normal tissues, researchers found that 357 circRNAs were dysregulated in patients with early-stage lung adenocarcinoma (Zhao et al., 2017). Indeed, high expression of certain circRNAs is positively correlated with the progression of lung cancer. For example, circ_0043278 promotes NSCLC cell proliferation, invasion, and migration by directly inhibiting miR-520f to increase the expression of ROCK1, CDKN1B, and AKT3 (Cui et al., 2019). Furthermore, there is a close correlation between the upregulated circRNA_100876, lymph node metastasis and tumor staging in NSCLC. The overall survival time of NSCLC patients with high circRNA_100876 expression was significantly shorter than that of patients with low circRNA_100876 expression (Yao et al., 2017).
Circular RNAs can also play a role in cancer inhibition (Wan et al., 2016). For instance, ectopic expression of cir-ITCH markedly elevated the expression of its parental cancer-suppressive gene ITCH and inhibited the proliferation of lung cancer cells. The molecular mechanism underlying this effect is that cir-ITCH acts as a sponge of oncogenic miR-7 and miR-214 to enhance ITCH expression, thus suppressing the activation of Wnt/β-catenin signaling.
In conclusion, some circRNAs are closely related to the pathogenesis of NSCLC. Therefore, they may serve as targets for NSCLC therapy.
Circular RNAs as Promising Biomarkers for NSCLC
Circular RNAs can be easily and repeatedly detected in blood samples, making them promising biomarker candidates for human diseases (Memczak et al., 2015). Furthermore, the discovery of circRNAs in saliva indicates the presence of circRNAs in other body fluids (Bahn et al., 2015). Compared to traditional biomarkers found in tumor tissues, circRNAs in body fluids could be used as novel biomarkers in the more convenient and non-invasive “liquid biopsy” for the diagnosis of tumors at different stages. Until recently, efforts have been made to explore the significance of circRNAs as biomarkers for various types of cancers (Wang et al., 2016). At present, there are many studies on the application of circRNA expression in peripheral blood in the diagnosis of NSCLC, which proves the effectiveness of circRNA in the diagnosis of NSCLC (Table 1).
TABLE 1.
circRNAs as biomarkers in liquid biopsy for the diagnosis of NSCLC.
| circRNA | Expression level | Intersection molecules and/or pathway | Effect | Sample | AUC | References |
| circ_0008928 | Up | miR-488/HK2 axis | Regulates cisplatin sensitivity, tumor progression, and glycolysis metabolism. | Serum exosomes | Shi et al., 2021 | |
| circ_PIP5K1A | Up | miR-101/ABCC1 axis | Regulates the progression of NSCLC and cisplatin sensitivity. | Serum and serum exosomes | Shao et al., 2021 | |
| circCDYL | Down | miR-185-5p/TNRC6A axis | Inhibited cell viability, proliferation, and induced apoptosis. | Plasma | Bian et al., 2021 | |
| hsa_circ_0014235 | Up | miR-520a-5p/CDK4 axis | Promotes cisplatin chemoresistance and deteriorates the development of NSCLC. | Serum exosomes | Xu et al., 2020 | |
| has_circ_0060937 | Up | – | Closely associated with bone metastasis in NSCLC. | Serum | Zhang J. et al., 2020 | |
| hsa_circ_0046264 | Up | – | Notably associated with the patient’s age, tumor size, TNM stage, and lymph node metastasis. | Serum | 0.915 | Liu et al., 2020 |
| circ-MEMO1 | Up | miR-101-3p/KRAS axis | Promotes the progression and aerobic glycolysis of NSCLC. | Serum exosomes | 0.760 | Ding et al., 2020 |
| circARHGAP10 | Up | miR-638/FAM83F axis | Promotes proliferation, migration, invasion, and glycolysis. | Serum exosomes | – | Fang et al., 2020 |
| circPVT1 | Up | – | Associated with chemotherapy resistance. | Serum | – | Lu et al., 2020 |
| hsa_circ_0002130 | Up | miR-498/GLUT1/HK2/LDHA axis | Facilitates osimertinib resistance. | Serum exosomes | – | Ma et al., 2020 |
| the panel of circ_0047921, circ_0056285, and circ_0007761 | Up, Up, Down | – | Distinguishing early-stage NSCLC cases from healthy controls, chronic obstructive pulmonary disease controls, or tuberculosis controls. | Serum exosomes | 0.919 | Xian et al., 2020 |
| circSATB2 | Up | miR-326/FSCN1 axis | Promotes the proliferation, migration, and invasion of NSCLC cells. | Serum exosomes | – | Zhang N. et al., 2020 |
| hsa_circRNA_012515 | Up | – | May be a mechanism leading to gefitinib resistance in NSCLC patients. | Peripheral blood | – | Fu et al., 2020 |
| hsa_circ_0134501 combined with hsa_circ_0109320 | Down | – | Biomarker candidates for predicting the therapeutic effect of gefitinib in NSCLC. | Plasma | 0.79 | Liu Y.-T. et al., 2019 |
| circ FECR1 | Up | miR584-3p/ROCK1 axis | Predicts survival outcomes and predicts the response to chemotherapies. | Serum exosomes | – | Li et al., 2019 |
| F-circEA | Exist | – | Could be a novel “liquid biopsy” biomarker to monitor the EML4-ALK fusion gene in NSCLC. | Plasma | – | Tan et al., 2018 |
| hsa_circ_0102533 | Up | – | Possesses an oncogenic property in the carcinogenesis. | Whole blood | 0.744 | Zhou et al., 2018 |
| circFARSA | Up | miR-330-5p/FASN or miR-326/FASN axis | Promotes cell migration and invasion. | Plasma | 0.71 | Hang et al., 2018 |
| hsa_circ_0013958 | Up | miR-134/CCND1 axis | Promotes cell proliferation and invasion and inhibits cell apoptosis. | Plasma | 0.794 | Zhu et al., 2017 |
TNM, tumor-node-metastasis; AUC, area under the ROC curve (AUC).
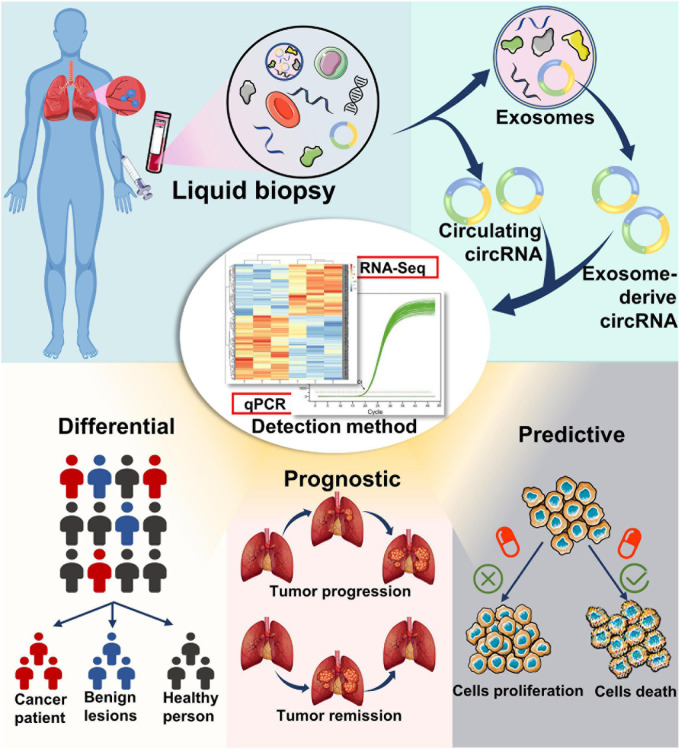
In the past decades, numerous studies have investigated the expression profiles of miRNAs and lncRNAs in patients with NSCLC to assess if these RNA types can serve as biomarkers for the early detection of NSCLC (Wei and Zhou, 2016; Han and Li, 2018). However, the results remain controversial. Compared with other non-coding RNAs, circRNAs have superior features, such as higher stability, abundance, and evolutionary conservation, making them more suitable diagnostic biomarkers (Figure 3).
FIGURE 3.
Potential application of circular RNAs as liquid biopsy biomarkers. circRNA biomarkers can be isolated from blood, as well as from the exosomes in blood. Detection and analysis of circRNAs can be completed using RNA-seq, qPCR, and other methods. circRNAs may serve as potential biomarkers for diagnosis, prognosis, and therapy selection of cancer.
Circular RNAs as Differential Diagnosis Biomarkers for NSCLC
At present, the most commonly used serum tumor markers for the diagnosis of NSCLC are embryonic antigen (CEA), squamous cell carcinoma antigen (SCCA), and cytokeratin 19 fragment (CYFRA21-1) (Ye et al., 2020). However, the sensitivity and specificity of detecting a single circulating biomarker in the diagnosis of lung cancer are not satisfactory, especially in the early stages (Kamel et al., 2019). circRNAs can stably exist in peripheral blood, and the detection of circRNAs expression in the blood has gradually become the focus of research in the diagnosis of NSCLC. Circulating miRNAs have shown potential advantages for early screening of NSCLC. A study found that circ_0047921, circ_0056285, and circ_0007761 in serum exosomes display significant diagnostic validity in distinguishing early-stage NSCLC cases from healthy controls, chronic obstructive pulmonary disease controls, and tuberculosis controls. In distinguishing NSCLC cases from healthy controls, the panel of the aforementioned circRNAs exhibited area under the curve (AUC) values of 0.926 [95% confidence interval (CI), 0.895–0.956] in the training set and 0.919 (95% CI, 0.877–0.962) in the validation set (Xian et al., 2020). A study on hsa_circ_0046264 showed that the area under the ROC curve (AUC) of hsa_circ_0046264 in serum was 0.915, the specificity was 0.957, and the sensitivity was 0.927 (Liu et al., 2020). The recent discovery of circRNAs in liquid biopsy samples suggests a novel and potentially useful tool for non-invasive NSCLC diagnosis.
Circular RNAs as Prognostic Biomarkers for NSCLC
Prognostic biomarkers should provide information about the patient’s overall disease outcome, such as disease recurrence, metastasis, or progression, and may help identify high-risk cases that require active measures. circ_0067934 has been reported to function as an oncogenic circRNA in NSCLC by promoting the proliferation, migration, and invasion of NSCLC cells. High circ_0067934 expression is significantly associated with TNM stage, lymph node status, and distant metastasis, thus serving as a potential prognostic biomarker and therapeutic target for NSCLC (Wang and Li, 2018). Another study showed that by acting as a sponge for miR-1287 and regulating GAGE1 expression, circ_0016760 promoted the growth and metastasis of NSCLC cells, while inhibiting apoptosis. Moreover, the upregulation of circ_0016760 is associated with advanced TNM stages, lymph node metastasis, and poor prognosis in patients with NSCLC (Li Y. et al., 2018). Similarly, the upregulation of circRNA_100876 was tightly correlated with lymph node metastasis and advanced tumor stages in NSCLC. In addition, NSCLC patients with high circRNA_100876 expression had significantly shorter survival times than those with low circRNA_100876 expression. These observations strongly suggest the role of circRNAs as prognostic biomarkers.
Circular RNAs as Predictive Biomarkers for Cancer Therapy
In addition, circRNAs may be useful for predicting tumor responsiveness to chemotherapy or different therapeutic approaches. Resistance to targeted therapy or immunotherapy is an ongoing problem in the successful treatment of NSCLC. It is increasingly necessary to develop biomarkers to monitor and predict the efficacy of targeted therapy or immunotherapy in patients with NSCLC. Recent studies suggested that circRNAs are likely to serve this purpose.
A study demonstrated that hsa_circ_0004015 increased the resistance of the NSCLC cell line HCC827 to gefitinib (an EGFR tyrosine kinase inhibitor) (Zhou et al., 2019). In addition, the EGFR-resistant LADC cell line H1975 expresses high levels of circRNA CCDC66 (Joseph et al., 2018). A high-throughput circRNA microarray showed that in comparison with parental cells, the paclitaxel-resistant lung adenocarcinoma cell line A549 had higher levels of 2,909 circRNAs and lower levels of 8,372 circRNAs (Xu et al., 2018). A recently identified circRNA, termed F-circEA, results from the fusion of the ALK gene with the EML4 gene in NSCLC patients. For EML4-ALK-positive NSCLC patients, F-circEA could be both a novel diagnostic biomarker and a predictive biomarker of the response to targeted therapy (Tan et al., 2018). Another study showed that overexpression of circFGFR1 resulted in resistance of NSCLC to treatment with anti-programmed cell death protein-1 (PD-1) (Zhang et al., 2019). In general, these studies suggest that dysregulated circRNAs lead to resistance to targeted therapy and immunotherapy in NSCLC patients.
Circular RNAs also influence chemotherapy resistance in patients with NSCLC. It has been reported that circRNA cESRP1 enhances SCLC8 sensitivity to chemotherapy agents by inhibiting miR-93-5p (Huang et al., 2020). Downregulation of hsa_circ_0001946 increases NSCLC cell resistance to the chemotherapeutic drug cisplatin (Huang et al., 2019), whereas overexpression of circ-ABCB10 increases lung cancer resistance to cisplatin (Wu et al., 2020). Therefore, circRNAs may be used as biomarkers to predict the sensitivity or resistance to platinum-based chemotherapy.
Exosomal circRNAs as NSCLC Biomarkers
Exosomes are tiny vesicles secreted by cells. They carry a variety of biomarkers such as DNA, RNA, proteins, and metabolites (Théry et al., 2002; Kahlert and Kalluri, 2013; Kumar et al., 2018). Exosomal contents are functional in recipient cells and highly variable depending on the origin of the cells. Cells can produce different exosomes under distinct physiological or pathological conditions (Fujita et al., 2016). Cancer cells produce more exosomes than their non-malignant counterparts (Rodríguez et al., 2014). Cancer cell-released exosomes induce phenotypic or functional alterations in recipient cells, thereby playing a role in cancer growth, angiogenesis, and metastasis (Kosaka et al., 2016).
Exosomes can mediate cell-to-cell communication via the direct exchange of genetic material between cells (Villarroya-Beltri et al., 2014). Exosomes can be isolated from diverse biofluids, such as blood (Caby et al., 2005), urine (Bryzgunova et al., 2016), and saliva (Ogawa et al., 2008). In recent years, circulating cancer-derived exosomes have emerged as promising biomarkers for monitoring cancer progression in non-invasive cancer diagnosis (Peinado et al., 2012; Xiao et al., 2012; Camacho et al., 2013; Zhou et al., 2014; Sundararajan et al., 2018). Studies on exosomal miRNAs and lncRNAs have contributed to the great progress in cancer screening, diagnosis, and prognostic evaluation (Liu et al., 2015; Li B. et al., 2018; Lee et al., 2019). However, our understanding of exo-circRNAs remains unclear.
Jae et al. first reported and validated circRNAs in human body fluids (Bahn et al., 2015). Later, another study confirmed abundant circRNAs in exosomes. RNA-seq analyses revealed the enrichment of circRNAs in the exosomes. It has been shown that more than 1,000 circRNAs are present in human serum exosomes (Li Y. et al., 2015), highlighting circRNAs as a novel class of stable RNA molecules in exosomes. Upon the uptake of EVs by neighboring or distant cells, exo-circRNAss can contribute to cell-to-cell communication. The existence of exo-circRNAs supports the idea that the expulsion of circRNAs from cells into the extracellular space, such as through EV release, is a probable mechanism by which circRNAs are cleared (Li Y. et al., 2015; Lasda and Parker, 2016).
Interestingly, serum exo-circRNAs might distinguish cancer patients from healthy individuals, illustrating the significant possibility of translating exo-circRNAs into cancer diagnostic biomarkers (Li Y. et al., 2015). A study found that circ-MEMO1 was higher in exosomes generated from the serum of NSCLC patients than in the healthy volunteers. The AUC reached a value of approximately 0.76, with a diagnostic sensitivity and specificity of 56.67 and 96%, respectively (95% CI 0.6259–0.8941) (Ding et al., 2020). Another study showed that circ_0008928 expression was increased in serum exosomes of cisplatin-resistant NSCLC patients compared with that in the cisplatin-sensitive NSCLC patients (Shi et al., 2021). In addition, a study showed that serum exosomal circFECR1 was associated with poor survival (P = 0.038) and clinical response to chemotherapy (Li et al., 2019). In summary, exo-circRNAs can be used as a promising biomarker for the diagnosis of NSCLC.
Concluding Remarks and Perspectives
Liquid biopsy is a rapidly growing field, and advances in detection technology provide the possibility for the discovery and clinical translation of circulating cancer biomarkers. Although there has been extensive research on the discovery of circulating markers for cancer diagnosis, most biomarkers are still in the experimental stage. Before circulating biomarkers can be used in clinical practice, many biological, technical, and clinical challenges need to be addressed.
circRNAs have a clear functional role in the occurrence and development of NSCLC, and tumor-related differential expression has been detected in the peripheral blood of NSCLC patients. Due to the complexity of blood samples, it is necessary to develop a new methodology that can accurately extract and reproducibly measure NSCLC-related circulating circRNAs and conduct extensive verification in a large number of clinical samples to prove its clinical effectiveness. Furthermore, a study reported that it is obvious that the “multiple biomarker profile” has higher sensitivity and specificity than a single biomarker (Gulati et al., 2014). In the future, it is expected to improve the accuracy of circRNA in the diagnosis, prognosis, and prediction of NSCLC through the beneficial combination of circRNAs and different levels of other molecules (combining genome, transcriptome, and proteome). The transition from a single-biomarker view to a multi-biomarker view will help promote the development of liquid biopsy.
Circular RNAs have potential applications in both laboratory and clinical cancer research. However, there are still many questions regarding circRNAs. One major question is whether the products of endogenous circRNAs have specific functions in eukaryotic cells. Another interesting question is how circRNAs are degraded. To answer these questions, circRNAs should be studied in depth.
Moreover, circRNAs may be considered as novel therapeutic targets. Although there are no such reports to date, the effects of circRNAs on lung cancer proliferation, metastasis, and drug resistance indicate potential for targeting of circRNAs in anti-cancer therapy (Chi et al., 2019; Dong et al., 2019; Tian et al., 2019). For example, it is possible to deliver small interfering RNAs (siRNAs) or vectors into the body to target certain circRNAs in cancer patients. In addition, research on circRNA biogenesis might identify an approach to regulate the upstream pathways of circRNA expression (Verduci et al., 2019).
Taken together, the ultimate goal of circRNA research is to develop new effective diagnostic or therapeutic strategies for cancer treatment. With the advancement of elaborate research, our understanding of circRNAs will significantly contribute to cancer prevention, diagnosis, and treatment in the coming decades.
Author Contributions
LH wrote the manuscript. YR conceived the review and performed bibliographic research. XT and KY prepared the figures and table. JW and FW contributed to the critical interpretation of the manuscript. All authors read and approved the final version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
- circRNAs
circular RNAs
- NSCLC
non-small cell lung cancer
- CTCs
circulating tumor cells
- EVs
extracellular vesicles
- TEPs
tumor-educated platelets
- miRNAs
microRNAs
- lncRNAs
long non-coding RNAs
- mRNAs
messenger RNAs
- RBPs
RNA-binding proteins
- Cdk2
cyclin-dependent kinase 2
- snRNA
small nuclear RNA
- RNA-seq
RNA sequencing
- RT-Qpcr
real-time fluorescent quantitative polymerase chain reaction
- rRNA
ribosomal RNA
- RNase R
ribonuclease R
- cDNAs
complementary DNAs
- RT
reverse transcription
- RCA
rolling circle amplification
- RNA-ISH
RNA in situ hybridization
- CHA
catalyzed hairpin assembly
- LDN
linear DNA nanostructure
- CEA
embryonic antigen
- SCCA
squamous cell carcinoma antigen
- CYFRA21-1
cytokeratin 19 fragment
- ROC
receiver operating characteristic
- AUC
area under the ROC curve
- CI
confidence interval
- PD-1
programmed cell death protein-1
- exo-circRNAs
exosomal circRNAs
- siRNA
small interfering RNA.
Footnotes
Funding. This work was supported by the Improvement Project for Theranostic Ability on Difficulty Miscellaneous Disease (Tumor) and the Science and Technology Innovation Fostering Foundation of Zhongnan Hospital of Wuhan University (No. ZLYNXM202008, znpy2019050, znpy2019004, and znpy2018027). This work was also funded by the Medical Top-Talented Youth Development Project of Hubei Province (No. WJ2019Q049).
References
- Ashley E. A. (2016). Towards precision medicine. Nat. Rev. Genet. 17 507–522. 10.1038/nrg.2016.86 [DOI] [PubMed] [Google Scholar]
- Bach P. B., Mirkin J. N., Oliver T. K., Azzoli C. G., Berry D. A., Brawley O. W., et al. (2012). Benefits and harms of CT screening for lung cancer: a systematic review. JAMA 307 2418–2429. 10.1001/jama.2012.5521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmayr-Heyda A., Reiner A. T., Auer K., Sukhbaatar N., Aust S., Bachleitner-Hofmann T., et al. (2015). Correlation of circular RNA abundance with proliferation–exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci. Rep. 5:8057. 10.1038/srep08057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn J. H., Zhang Q., Li F., Chan T.-M., Lin X., Kim Y., et al. (2015). The landscape of MicroRNA, Piwi-interacting RNA, and circular RNA in Human Saliva. Clin. Chem. 61 221–230. 10.1373/clinchem.2014.230433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian W.-X., Xue F., Wang L.-Y., Xing X.-F. (2021). Circular RNA CircCDYL regulates proliferation and apoptosis in non-small cell lung cancer cells by sponging miR-185-5p and upregulating TNRC6A. Cancer Manag. Res. 13 633–642. 10.2147/CMAR.S280315 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Borran S., Ahmadi G., Rezaei S., Anari M. M., Modabberi M., Azarash Z., et al. (2020). Circular RNAs: new players in thyroid cancer. Pathol. Res. Pract. 216 153217. 10.1016/j.prp.2020.153217 [DOI] [PubMed] [Google Scholar]
- Bryzgunova O. E., Zaripov M. M., Skvortsova T. E., Lekchnov E. A., Grigor’eva A. E., Zaporozhchenko I. A., et al. (2016). Comparative study of extracellular vesicles from the urine of healthy individuals and prostate cancer patients. PLoS One 11:e0157566. 10.1371/journal.pone.0157566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caby M. P., Lankar D., Vincendeau-Scherrer C., Raposo G., Bonnerot C. (2005). Exosomal-like vesicles are present in human blood plasma. Int. Immunol. 17 879–887. 10.1093/intimm/dxh267 [DOI] [PubMed] [Google Scholar]
- Camacho L., Guerrero P., Marchetti D. (2013). MicroRNA and protein profiling of brain metastasis competent cell-derived exosomes. PLoS One 8:e73790. 10.1371/journal.pone.0073790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.-L. (2016). The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 17:205. 10.1038/nrm.2015.32 [DOI] [PubMed] [Google Scholar]
- Chen S., Li T., Zhao Q., Xiao B., Guo J. (2017). Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin. Chim. Acta 466 167–171. 10.1016/j.cca.2017.01.025 [DOI] [PubMed] [Google Scholar]
- Chen W. (2015). Cancer statistics: updated cancer burden in China. Chin J Cancer Res. 27 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Y., Luo Q., Song Y., Yang F., Wang Y., Jin M., et al. (2019). Circular RNA circPIP5K1A promotes non-small cell lung cancer proliferation and metastasis through miR-600/HIF-1α regulation. J. Cell. Biochem. 120 19019–19030. 10.1002/jcb.29225 [DOI] [PubMed] [Google Scholar]
- Cohen J. D., Li L., Wang Y., Thoburn C., Afsari B., Danilova L., et al. (2018). Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 359 926. 10.1126/science.aar3247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li W., Liu G., Chen X., Gao X., Lu H., et al. (2019). A novel circular RNA, hsa_circ_0043278, acts as a potential biomarker and promotes non-small cell lung cancer cell proliferation and migration by regulating miR-520f. Artif. Cells Nanomed. Biotechnol. 47 810–821. 10.1080/21691401.2019.1575847 [DOI] [PubMed] [Google Scholar]
- Ding C., Xi G., Wang G., Cui D., Zhang B., Wang H., et al. (2020). Exosomal Circ-MEMO1 promotes the progression and aerobic glycolysis of non-small cell lung cancer through targeting MiR-101-3p/KRAS Axis. Front. Genet. 11:962. 10.3389/fgene.2020.00962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Xu T., Zhong S., Wang B., Zhang H., Wang X., et al. (2019). Circ_0076305 regulates cisplatin resistance of non-small cell lung cancer via positively modulating STAT3 by sponging miR-296–5p. Life Sci. 239:116984. 10.1016/j.lfs.2019.116984 [DOI] [PubMed] [Google Scholar]
- Dou Y., Cha D. J., Franklin J. L., Higginbotham J. N., Jeppesen D. K., Weaver A. M., et al. (2016). Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci. Rep. 6:37982. 10.1038/srep37982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W. W., Yang W., Liu E., Yang Z., Dhaliwal P., Yang B. B. (2016). Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 44 2846–2858. 10.1093/nar/gkw027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang K., Chen X., Qiu F., Xu J., Xiong H., Zhang Z. (2020). Serum-derived exosomes-mediated circular RNA ARHGAP10 modulates the progression of non-small-cell lung cancer through the miR-638/FAM83F Axis. Cancer Biother. Radiopharm. 10.1089/cbr.2019.3534 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Fu Y., Huang L., Tang H., Huang R. (2020). hsa_circRNA_012515 is highly expressed in NSCLC patients and affects its prognosis. Cancer Manag. Res. 12 1877–1886. 10.2147/CMAR.S245525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Yoshioka Y., Ochiya T. (2016). Extracellular vesicle transfer of cancer pathogenic components. Cancer Sci. 107 385–390. 10.1111/cas.12896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati S., Martinez P., Joshi T., Birkbak N. J., Santos C. R., Rowan A. J., et al. (2014). Systematic evaluation of the prognostic impact and intratumour heterogeneity of clear cell renal cell carcinoma biomarkers. Eur. Urol. 66 936–948. 10.1016/j.eururo.2014.06.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guria A., Sharma P., Natesan S., Pandi G. (2020). Circular RNAs-the road less traveled. Front. Mol. Biosci. 6:146. 10.3389/fmolb.2019.00146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Li H. (2018). miRNAs as biomarkers and for the early detection of non-small cell lung cancer (NSCLC). J. Thorac. Dis. 10 3119–3131. 10.21037/jtd.2018.05.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang D., Zhou J., Qin N., Zhou W., Ma H., Jin G., et al. (2018). A novel plasma circular RNA circFARSA is a potential biomarker for non-small cell lung cancer. Cancer Med. 7 2783–2791. 10.1002/cam4.1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T. B., Jensen T. I., Clausen B. H., Bramsen J. B., Finsen B., Damgaard C. K., et al. (2013). Natural RNA circles function as efficient microRNA sponges. Nature 495 384–388. 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- Heitzer E., Haque I. S., Roberts C. E. S., Speicher M. R. (2019). Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 20 71–88. 10.1038/s41576-018-0071-5 [DOI] [PubMed] [Google Scholar]
- Hsiao K.-Y., Lin Y.-C., Gupta S. K., Chang N., Yen L., Sun H. S., et al. (2017). Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 77 2339–2350. 10.1158/0008-5472.CAN-16-1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M.-S., Liu J.-Y., Xia X.-B., Liu Y.-Z., Li X., Yin J.-Y., et al. (2019). Hsa_circ_0001946 inhibits lung cancer progression and mediates cisplatin sensitivity in non-small cell lung cancer via the nucleotide excision repair signaling pathway. Front. Oncol. 9:508. 10.3389/fonc.2019.00508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Yang Y., Wu J., Niu Y., Yao Y., Zhang J., et al. (2020). Circular RNA cESRP1 sensitises small cell lung cancer cells to chemotherapy by sponging miR-93-5p to inhibit TGF-β signalling. Cell Death Differ. 27 1709–1727. 10.1038/s41418-019-0455-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck W. R., Sharpless N. E. (2014). Detecting and characterizing circular RNAs. Nat. Biotechnol. 32 453–461. 10.1038/nbt.2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck W. R., Sorrentino J. A., Wang K., Slevin M. K., Burd C. E., Liu J., et al. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19 141–157. 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao J., Xiang Y., Duan C., Liu Y., Li C., Li G. (2020). Lighting Up CircRNA using a linear DNA nanostructure. Anal. Chem. 92 12394–12399. 10.1021/acs.analchem.0c02146 [DOI] [PubMed] [Google Scholar]
- Joseph N. A., Chiou S.-H., Lung Z., Yang C.-L., Lin T.-Y., Chang H.-W., et al. (2018). The role of HGF-MET pathway and CCDC66 cirRNA expression in EGFR resistance and epithelial-to-mesenchymal transition of lung adenocarcinoma cells. J. Hematol. Oncol. 11:74. 10.1186/s13045-018-0557-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C., Ellington A. D. (2014). Diagnostic applications of nucleic acid circuits. Acc. Chem. Res. 47 1825–1835. 10.1021/ar500059c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlert C., Kalluri R. (2013). Exosomes in tumor microenvironment influence cancer progression and metastasis. J. Mol. Med. (Berl.) 91 431–437. 10.1007/s00109-013-1020-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamel L. M., Atef D. M., Mackawy A. M. H., Shalaby S. M., Abdelraheim N. (2019). Circulating long non-coding RNA GAS5 and SOX2OT as potential biomarkers for diagnosis and prognosis of non-small cell lung cancer. Biotechnol. Appl. Biochem. 66 634–642. 10.1002/bab.1764 [DOI] [PubMed] [Google Scholar]
- Kosaka N., Yoshioka Y., Fujita Y., Ochiya T. (2016). Versatile roles of extracellular vesicles in cancer. J. Clin. Invest. 126 1163–1172. 10.1172/JCI81130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Michael I. J., Park J., Granick S., Cho Y. K. (2018). Cloaked exosomes: biocompatible, durable, and degradable encapsulation. Small 14:e1802052. 10.1002/smll.201802052 [DOI] [PubMed] [Google Scholar]
- Kumar-Sinha C., Chinnaiyan A. M. (2018). Precision oncology in the age of integrative genomics. Nat. Biotechnol. 36 46–60. 10.1038/nbt.4017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasda E., Parker R. (2016). Circular RNAs co-precipitate with extracellular vesicles: a possible mechanism for circRNA clearance. PLoS One 11:e0148407. 10.1371/journal.pone.0148407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. R., Kim G., Tak W. Y., Jang S. Y., Kweon Y. O., Park J. G., et al. (2019). Circulating exosomal noncoding RNAs as prognostic biomarkers in human hepatocellular carcinoma. Int. J. Cancer 144 1444–1452. 10.1002/ijc.31931 [DOI] [PubMed] [Google Scholar]
- Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., et al. (2017). Circ-ZNF609 Is a circular RNA that can be translated and functions in myogenesis. Mol. Cell 66 22–37.e9. 10.1016/j.molcel.2017.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Mao R., Liu C., Zhang W., Tang Y., Guo Z. (2018). LncRNA FAL1 promotes cell proliferation and migration by acting as a CeRNA of miR-1236 in hepatocellular carcinoma cells. Life Sci. 197 122–129. 10.1016/j.lfs.2018.02.006 [DOI] [PubMed] [Google Scholar]
- Li L., Li W., Chen N., Zhao H., Xu G., Zhao Y., et al. (2019). Exonic circular RNAs as a novel oncogenic driver to promote tumor metastasis in small cell lung cancer. Clin. Cancer Res. 25 1302–1317. 10.1158/1078-0432.CCR-18-1447 [DOI] [PubMed] [Google Scholar]
- Li Y., Hu J., Li L., Cai S., Zhang H., Zhu X., et al. (2018). Upregulated circular RNA circ_0016760 indicates unfavorable prognosis in NSCLC and promotes cell progression through miR-1287/GAGE1 axis. Biochem. Biophys. Res. Commun. 503 2089–2094. 10.1016/j.bbrc.2018.07.164 [DOI] [PubMed] [Google Scholar]
- Li Y., Zheng Q., Bao C., Li S., Guo W., Zhao J., et al. (2015). Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 25 981–984. 10.1038/cr.2015.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., et al. (2015). Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Amp. Mol. Biol. 22:256. 10.1038/nsmb.2959 [DOI] [PubMed] [Google Scholar]
- Liu J., Zhang Y., Xie H., Zhao L., Zheng L., Ye H. (2019). Applications of catalytic hairpin assembly reaction in biosensing. Small 15:1902989. 10.1002/smll.201902989 [DOI] [PubMed] [Google Scholar]
- Liu W.-H., Ren L.-N., Wang X., Wang T., Zhang N., Gao Y., et al. (2015). Combination of exosomes and circulating microRNAs may serve as a promising tumor marker complementary to alpha-fetoprotein for early-stage hepatocellular carcinoma diagnosis in rats. J. Cancer Res. Clin. Oncol. 141 1767–1778. 10.1007/s00432-015-1943-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.-X., Yang Y.-E., Liu X., Zhang M.-Y., Li R., Yin Y.-H., et al. (2019). A two-circular RNA signature as a noninvasive diagnostic biomarker for lung adenocarcinoma. J. Transl. Med. 17:50. 10.1186/s12967-019-1800-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.-T., Han X.-H., Xing P.-Y., Hu X.-S., Hao X.-Z., Wang Y., et al. (2019). Circular RNA profiling identified as a biomarker for predicting the efficacy of Gefitinib therapy for non-small cell lung cancer. J. Thorac. Dis. 11 1779– 1787. 10.21037/jtd.2019.05.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. H., Yang S. Z., Chen X. T., Shao M. R., Dong S. Y., Zhou S. Y., et al. (2020). Correlations of hsa_circ_0046264 expression with onset, pathological stage and chemotherapy resistance of lung cancer. Eur. Rev. Med. Pharmacol. Sci. 24 9511–9521. [DOI] [PubMed] [Google Scholar]
- López-Jiménez E., Rojas A. M., Andrés-León E. (2018). “RNA sequencing and prediction tools for circular RNAs analysis,” in Circular RNAs: Biogenesis and Functions, ed. Xiao J. (Singapore: Springer Singapore; ), 17–33. 10.1007/978-981-13-1426-1_2 [DOI] [PubMed] [Google Scholar]
- Lu H., Xie X., Chen Q., Cai S., Liu S., Bao C., et al. (2020). Clinical significance of circPVT1 in patients with non-small cell lung cancer who received cisplatin combined with gemcitabine chemotherapy. Tumori 3008916209 41940. 10.1177/0300891620941940 [DOI] [PubMed] [Google Scholar]
- Ma J., Qi G., Li L. A. (2020). Novel serum exosomes-based biomarker hsa_circ_0002130 facilitates osimertinib-resistance in non-small cell lung cancer by sponging miR-498. Onco Targets Ther. 13 5293–5307. 10.2147/OTT.S243214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495:333. 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- Memczak S., Papavasileiou P., Peters O., Rajewsky N. (2015). Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS One 10:e0141214. 10.1371/journal.pone.0141214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeli P., Pourhanifeh M. H., Karimzadeh M. R., Shabaninejad Z., Movahedpour A., Tarrahimofrad H., et al. (2020). Circular RNAs and gastrointestinal cancers: epigenetic regulators with a prognostic and therapeutic role. Crit. Rev. Oncol. Hematol. 145:102854. 10.1016/j.critrevonc.2019.102854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigro J. M., Cho K. R., Fearon E. R., Kern S. E., Ruppert J. M., Oliner J. D., et al. (1991). Scrambled exons. Cell 64 607–613. 10.1016/0092-8674(91)90244-S [DOI] [PubMed] [Google Scholar]
- Ogawa Y., Kanai-Azuma M., Akimoto Y., Kawakami H., Yanoshita R. (2008). Exosome-Like vesicles with dipeptidyl peptidase IV in human saliva. Biol. Pharm. Bull. 31 1059–1062. 10.1248/bpb.31.1059 [DOI] [PubMed] [Google Scholar]
- Panda A. C., De S., Grammatikakis I., Munk R., Yang X., Piao Y., et al. (2017). High-purity circular RNA isolation method (RPAD) reveals vast collection of intronic circRNAs. Nucleic Acids Res 45:e116. 10.1093/nar/gkx297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantel K., Alix-Panabićres C. (2019). Liquid biopsy and minimal residual disease — latest advances and implications for cure. Nat. Rev. Clin. Oncol. 16 409–424. 10.1038/s41571-019-0187-3 [DOI] [PubMed] [Google Scholar]
- Peinado H., Alećkovič M., Lavotshkin S., Matei I., Costa-Silva B., Moreno-Bueno G., et al. (2012). Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 18 883–891. 10.1038/nm.2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razavi Z. S., Tajiknia V., Majidi S., Ghandali M., Mirzaei H. R., Rahimian N., et al. (2021). Gynecologic cancers and non-coding RNAs: epigenetic regulators with emerging roles. Crit. Rev. Oncol. Hematol. 157:103192. 10.1016/j.critrevonc.2020.103192 [DOI] [PubMed] [Google Scholar]
- Rodríguez M., Silva J., López-Alfonso A., López-Muřiz M. B., Peña C., Domínguez G., et al. (2014). Different exosome cargo from plasma/bronchoalveolar lavage in non-small-cell lung cancer. Genes Chromosomes Cancer 53 713–724. 10.1002/gcc.22181 [DOI] [PubMed] [Google Scholar]
- Sadri Nahand J., Moghoofei M., Salmaninejad A., Bahmanpour Z., Karimzadeh M., Nasiri M., et al. (2020). Pathogenic role of exosomes and microRNAs in HPV-mediated inflammation and cervical cancer: a review. Int. J. Cancer 146 305–320. 10.1002/ijc.32688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider T., Schreiner S., Preußer C., Bindereif A., Rossbach O. (2018). “Northern blot analysis of circular RNAs,” in Circular RNAs: Methods and Protocols, eds Dieterich C., Papantonis A. (New York, NY: Springer New York; ), 119–133. 10.1007/978-1-4939-7562-4_10 [DOI] [PubMed] [Google Scholar]
- Shabaninejad Z., Vafadar A., Movahedpour A., Ghasemi Y., Namdar A., Fathizadeh H., et al. (2019). Circular RNAs in cancer: new insights into functions and implications in ovarian cancer. J. Ovarian Res. 12:84. 10.1186/s13048-019-0558-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang X., Li G., Liu H., Li T., Liu J., Zhao Q., et al. (2016). Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular crcinoma development. Medicine (Baltimore) 95:e3811. 10.1097/MD.0000000000003811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao N., Song L., Sun X. (2021). Exosomal circ_PIP5K1A regulates the progression of non-small cell lung cancer and cisplatin sensitivity by miR-101/ABCC1 axis. Mol. Cell Biochem. 476 2253–2267. 10.1007/s11010-021-04083-8 [DOI] [PubMed] [Google Scholar]
- Shi Q., Ji T., Ma Z., Tan Q., Liang J. (2021). Serum exosomes-based biomarker circ_0008928 regulates cisplatin sensitivity, tumor progression, and glycolysis metabolism by miR-488/HK2 axis in cisplatin-resistant nonsmall cell lung carcinoma. Cancer Biother. Radiopharm. 10.1089/cbr.2020.4490 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Siegel R. L., Miller K. D., Jemal A. (2018). Cancer statistics, 2018. CA Cancer J. Clin. 68 7–30. 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- Siravegna G., Marsoni S., Siena S., Bardelli A. (2017). Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 14 531–548. 10.1038/nrclinonc.2017.14 [DOI] [PubMed] [Google Scholar]
- Starke S., Jost I., Rossbach O., Schneider T., Schreiner S., Hung L.-H., et al. (2015). Exon circularization requires canonical splice signals. Cell Rep. 10 103–111. 10.1016/j.celrep.2014.12.002 [DOI] [PubMed] [Google Scholar]
- Sundararajan V., Sarkar F. H., Ramasamy T. S. (2018). The multifaceted role of exosomes in cancer progression: diagnostic and therapeutic implications [corrected]. Cell. Oncol. (Dordr.) 41 223–252. 10.1007/s13402-018-0378-4 [DOI] [PubMed] [Google Scholar]
- Szabo L., Salzman J. (2016). Detecting circular RNAs: bioinformatic and experimental challenges. Nat. Rev. Genet. 17 679–692. 10.1038/nrg.2016.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S., Gou Q., Pu W., Guo C., Yang Y., Wu K., et al. (2018). Circular RNA F-circEA produced from EML4-ALK fusion gene as a novel liquid biopsy biomarker for non-small cell lung cancer. Cell Res. 28 693–695. 10.1038/s41422-018-0033-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Fu K., Sun H., Rong D., Wang H., Cao H. (2018). CircRNA microarray profiling identifies a novel circulating biomarker for detection of gastric cancer. Mol. Cancer 17:137. 10.1186/s12943-018-0888-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C., Zitvogel L., Amigorena S. (2002). Exosomes: composition, biogenesis and function. Nat. Rev. Immunol. 2 569–579. 10.1038/nri855 [DOI] [PubMed] [Google Scholar]
- Tian X., Zhang L., Jiao Y., Chen J., Shan Y., Yang W. (2019). CircABCB10 promotes nonsmall cell lung cancer cell proliferation and migration by regulating the miR-1252/FOXR2 axis. J. Cell. Biochem. 120 3765–3772. 10.1002/jcb.27657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafadar A., Shabaninejad Z., Movahedpour A., Mohammadi S., Fathullahzadeh S., Mirzaei H. R., et al. (2019). Long non-coding RNAs as epigenetic regulators in cancer. Curr. Pharm. Des. 25 3563–3577. 10.2174/1381612825666190830161528 [DOI] [PubMed] [Google Scholar]
- Verduci L., Strano S., Yarden Y., Blandino G. (2019). The circRNA-microRNA code: emerging implications for cancer diagnosis and treatment. Mol. Oncol. 13 669–680. 10.1002/1878-0261.12468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarroya-Beltri C., Baixauli F., Gutiérrez-Vázquez C., Sánchez-Madrid F., Mittelbrunn M. (2014). Sorting it out: regulation of exosome loading. Semin. Cancer Biol. 28 3–13. 10.1016/j.semcancer.2014.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L., Zhang L., Fan K., Cheng Z.-X., Sun Q.-C., Wang J.-J. (2016). Circular RNA-ITCH suppresses lung cancer proliferation via inhibiting the Wnt/β-catenin pathway. Biomed. Res. Int. 2016:1579490. 10.1155/2016/1579490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Nazarali A. J., Ji S. (2016). Circular RNAs as potential biomarkers for cancer diagnosis and therapy. Am. J. Cancer Res. 6 1167–1176. [PMC free article] [PubMed] [Google Scholar]
- Wang J., Li H. (2018). CircRNA circ_0067934 silencing inhibits the proliferation, migration and invasion of NSCLC cells and correlates with unfavorable prognosis in NSCLC. Eur. Rev. Med. Pharmacol. Sci. 22 3053–3060. [DOI] [PubMed] [Google Scholar]
- Wei M.-M., Zhou G.-B. (2016). Long non-coding rnas and their roles in non-small-cell lung cancer. Genomics Proteomics Bioinformatics 14 280–288. 10.1016/j.gpb.2016.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Qi X., Liu L., Hu X., Liu J., Yang J., et al. (2019). Emerging Epigenetic Regulation of Circular RNAs in Human Cancer. Mol Ther Nucleic Acids. 16 589–596. 10.1016/j.omtn.2019.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Gong Q., Yu Y., Zhu J., Li W. (2020). Knockdown of circ-ABCB10 promotes sensitivity of lung cancer cells to cisplatin via miR-556-3p/AK4 axis. BMC Pulm. Med. 20:10. 10.1186/s12890-019-1035-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S. Y., Feng J., Lei L. J., Hu J., Xia L. J., Wang J., et al. (2017). Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Brief Bioinform. 18 984–992. [DOI] [PubMed] [Google Scholar]
- Xian J., Su W., Liu L., Rao B., Lin M., Feng Y., et al. (2020). Identification of three circular RNA cargoes in serum exosomes as diagnostic biomarkers of Non–small-cell lung cancer in the chinese population. J. Mol. Diagnos. 22 1096–1108. 10.1016/j.jmoldx.2020.05.011 [DOI] [PubMed] [Google Scholar]
- Xiao D., Ohlendorf J., Chen Y., Taylor D. D., Rai S. N., Waigel S., et al. (2012). Identifying mRNA, microRNA and protein profiles of melanoma exosomes. PLoS One 7:e46874. 10.1371/journal.pone.0046874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N., Chen S., Liu Y., Li W., Liu Z., Bian X., et al. (2018). Profiles and bioinformatics analysis of differentially expressed circrnas in taxol-resistant non-small cell lung cancer cells. Cell. Physiol. Biochem. 48 2046– 2060. 10.1159/000492543 [DOI] [PubMed] [Google Scholar]
- Xu W., Seok J., Mindrinos M. N., Schweitzer A. C., Jiang H., Wilhelmy J., et al. (2011). Human transcriptome array for high-throughput clinical studies. Proc. Natl. Acad. Sci. U.S.A. 108 3707–3712. 10.1073/pnas.1019753108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Tao R., Sun L., Ji X. (2020). Exosome-transferred hsa_circ_0014235 promotes DDP chemoresistance and deteriorates the development of non-small cell lung cancer by mediating the miR-520a-5p/CDK4 pathway. Cancer Cell Int. 20:552. 10.1186/s12935-020-01642-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Li X., Meng Q., Sun H., Wu S., Hu W., et al. (2020). CircPTK2 (hsa_circ_0005273) as a novel therapeutic target for metastatic colorectal cancer. Mol. Cancer 19:13. 10.1186/s12943-020-1139-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Fan X., Mao M., Song X., Wu P., Zhang Y., et al. (2017). Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 27:626. 10.1038/cr.2017.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Gao X., Zhang M., Yan S., Sun C., Xiao F., et al. (2018). Novel Role of FBXW7 Circular RNA in repressing glioma tumorigenesis. J. Natl. Cancer Inst. 110 304–315. 10.1093/jnci/djx166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J.-T., Zhao S.-H., Liu Q.-P., Lv M.-Q., Zhou D.-X., Liao Z.-J., et al. (2017). Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol. Res. Pract. 213 453–456. 10.1016/j.prp.2017.02.011 [DOI] [PubMed] [Google Scholar]
- Ye R., Tang R., Gan S., Li R., Cheng Y., Guo L., et al. (2020). New insights into long non-coding RNAs in non-small cell lung cancer. Biomed. Pharmacother. 131:110775. 10.1016/j.biopha.2020.110775 [DOI] [PubMed] [Google Scholar]
- Zeng X., Lin W., Guo M., Zou Q. (2017). A comprehensive overview and evaluation of circular RNA detection tools. PLoS Comput. Biol. 13:e1005420. 10.1371/journal.pcbi.1005420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Mao W., Chen Z., Gu H., Lian C. (2020). Clinical significance of Has_circ_0060937 in bone metastasis of NSCLC. Int. J. Gen. Med. 13 1115–1121. 10.2147/IJGM.S279023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Nan A., Chen L., Li X., Jia Y., Qiu M., et al. (2020). Circular RNA circSATB2 promotes progression of non-small cell lung cancer cells. Mol. Cancer 19:101. 10.1186/s12943-020-01221-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P.-F., Pei X., Li K.-S., Jin L.-N., Wang F., Wu J., et al. (2019). Circular RNA circFGFR1 promotes progression and anti-PD-1 resistance by sponging miR-381-3p in non-small cell lung cancer cells. Mol. Cancer 18:179. 10.1186/s12943-019-1111-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.-O., Wang H.-B., Zhang Y., Lu X., Chen L.-L., Yang L. (2014). Complementary sequence-mediated exon circularization. Cell 159 134–147. 10.1016/j.cell.2014.09.001 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhang X. O., Chen T., Xiang J. F., Yin Q. F., Xing Y. H., et al. (2013). Circular intronic long noncoding RNAs. Mol. Cell 51 792–806. 10.1016/j.molcel.2013.08.017 [DOI] [PubMed] [Google Scholar]
- Zhao J., Li L., Wang Q., Han H., Zhan Q., Xu M. (2017). CircRNA expression profile in early-stage lung adenocarcinoma patients. Cell. Physiol. Biochem. 44 2138–2146. 10.1159/000485953 [DOI] [PubMed] [Google Scholar]
- Zhou W., Fong M. Y., Min Y., Somlo G., Liu L., Palomares M. R., et al. (2014). Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 25 501–515. 10.1016/j.ccr.2014.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Liu H.-Y., Wang W.-Y., Zhao H., Wang T. (2018). Hsa_circ_0102533 serves as a blood-based biomarker for non-small-cell lung cancer diagnosis and regulates apoptosis in vitro. Int. J. Clin. Exp. Pathol. 11 4395–4404. [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Zheng X., Xu B., Chen L., Wang Q., Deng H., et al. (2019). Circular RNA hsa_circ_0004015 regulates the proliferation, invasion, and TKI drug resistance of non-small cell lung cancer by miR-1183/PDPK1 signaling pathway. Biochem. Biophys. Res. Commun. 508 527–535. 10.1016/j.bbrc.2018.11.157 [DOI] [PubMed] [Google Scholar]
- Zhu X., Wang X., Wei S., Chen Y., Chen Y., Fan X., et al. (2017). hsa_circ_0013958: a circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J. 284 2170–2182. 10.1111/febs.14132 [DOI] [PubMed] [Google Scholar]