ABSTRACT
We report a coding-complete genome sequence of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) strain SARS-CoV-2/BGD/GC001, isolated from a Bangladeshi patient with respiratory symptoms. Phylogenetic analysis assigned this strain to lineage B.1.1.7, which presented a total of 36 mutations in the spike and other genomic regions compared to strain Wuhan Hu-1 (GenBank accession number NC_045512.2).
ANNOUNCEMENT
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) belongs to the family Coronaviridae and genus Betacoronavirus (1). Lineage B.1.1.7, which emerged in the United Kingdom, has attracted particular attention due to its high transmissibility and immune escape potential (2). Herein, we announce a coding-complete genome sequence of the strain SARS-CoV-2/BGD/GC001 (GC001) belonging to lineage B.1.1.7 and detected in a patient with respiratory symptoms who presented on 31 January 2021 in Dhaka, Bangladesh. Ethical approval was obtained from the icddr,b Research and Ethical Review Committee (protocol number PR-21040).
The specimen (naso-oropharyngeal swabs) from the symptomatic patient was processed for nucleic acid isolation utilizing a QIAamp viral RNA minikit (Qiagen, Germany). SARS-CoV-2 was confirmed using a TaqMan real-time PCR (RT-PCR) assay (3). A cDNA library was prepared utilizing the Illumina TruSeq stranded total RNA Gold low-throughput (LT) library preparation kit following the manufacturer’s instructions; rRNA reduction was performed using the Ribo-Zero Gold protocol. The libraries were normalized to 10 nM following Illumina’s standard normalization method and pooled using 10 μl of each library, which was then sequenced using the NextSeq v2.5 midoutput kit (2 × 150 cycles) on the NextSeq 500 instrument at the Genomics Center of the icddr,b in Dhaka, Bangladesh. There were 112,690,904 paired-end raw reads, whose quality was assessed using FastQC v0.11.11 (4). The adapters were trimmed using Trimmomatic v0.39 based on Q30 values with the following parameters: window size, 4; Phred quality, 15; and minimum read length, 40 (5). After trimming, 205,697,670 reads were used for reference-based alignment. The SARS-CoV-2-specific reads were mapped and filtered using SMALT v0.7.6 (http://www.sanger.ac.uk/science/tools/smalt-0) and SAMtools v1.9 (6). De novo assembly was performed with 17,324 reads using SPAdes v3.11.1 (7), and the quality was assessed using QUAST v5.0.2 (8). RATT was employed to annotate the genome with Wuhan-Hu-1 as the reference strain (GenBank accession number NC_045512.2) (9). Mutations were assessed utilizing the Genome Detective Coronavirus Typing Tool (10). The Nextstrain and PANGOLIN Web tools were used for comparative genomics (11, 12). MUSCLE v3.8.31 (13) and MEGA7 (14) software were used to generate a phylogenetic tree using the neighbor-joining method with 1,000 bootstraps (Fig. 1). Default parameters were applied for all tools unless otherwise mentioned.
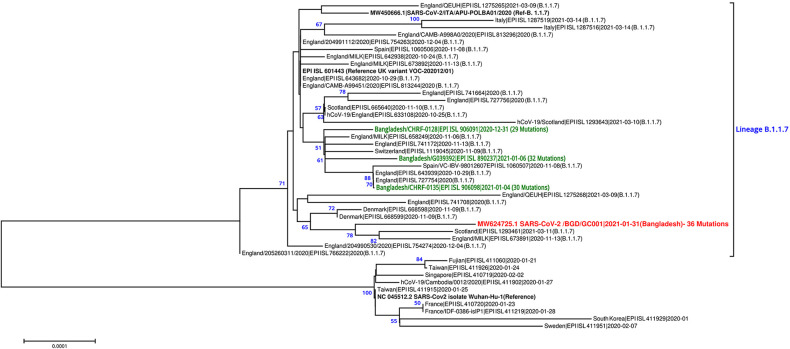
FIG 1.
Phylogenetic tree showing the genome sequence of SARS-CoV-2/BGD/GC001/2021 (GenBank accession number MW624725.1) together with 44 sequences, including 34 lineage B.1.1.7 sequences and 10 non-B.1.1.7 lineage sequences retrieved from GISAID and NCBI GenBank. The sequence reported in this announcement is highlighted in red; other B.1.1.7 lineage sequences from Bangladesh are shown in green. The percent bootstrap support values are indicated at each node (values of <50 are omitted).
The genome sequence of strain GC001 was 29,842 bp long with a G+C content of 37.98%. Phylogenetic analysis assigned strain GC001 to lineage B.1.1.7, which is a predominant lineage worldwide (Fig. 1). We identified 33 mutations and 3 deletions in GC001 in comparison to strain Wuhan-Hu-1 (GenBank accession number NC_045512.2) (Table 1). Strain GC001 showed a high number of mutations compared to other SARS-CoV-2 lineage B.1.1.7 strains identified in Bangladesh at the time of the analysis (Fig. 1). Moreover, strain GC001 did not cluster closely with other B.1.1.7 genomes identified from Bangladesh to date, which hints toward its independent introduction into the country. These observations suggest the importance of genome sequencing of SARS-CoV-2 samples from travelers, particularly those returning from high-risk countries.
TABLE 1.
Mutations of SARS-CoV-2 strain GC001 (GenBank accession number MW624725.1) in comparison to the reference strain (NC_045512.2)
| Gene or region | Mutation no. | CDS codon positiona | Amino acid change | Nucleotide position | Nucleotide change |
|---|---|---|---|---|---|
| 5′ untranslated region | 1 | 241 | C > T | ||
| ORF1ab | 2 | 216 | 913 | C > T | |
| 3 | 615 | 2110 | C > T | ||
| 4 | 924 | 3037 | C > T | ||
| 5 | 1001 | T > I | 3267 | C >T | |
| 6 | 1708 | A > D | 5388 | C > A | |
| 7 | 1907 | 5986 | C > T | ||
| 8 | 2230 | I > T | 6954 | T > C | |
| 9 | 2573 | 7984 | T > C | ||
| 10 | 3198 | 9857 | C > T | ||
| 11 | 3675–3677 | SGF deletion | 11288–11296 | Deletion TCTGGTTTT | |
| 12 | 4619 | P > L | 14120 | C > T | |
| 13 | 4715 | P > L | 14408 | C > T | |
| 14 | 4804 | 14676 | C > T | ||
| 15 | 505 | 15279 | C > T | ||
| 16 | 5304 | 16176 | T > C | ||
| 17 | 6376 | P > S | 19390 | C > T | |
| S | 18 | 69–70 | HV deletion | 21766–21771 | Deletion ACATGT |
| 19 | 75 | G > V | 21786 | G > T | |
| 20 | 144 | Y deletion | 21992–21994 | Deletion TAT | |
| 21 | 501 | N > Y | 23063 | A > T | |
| 22 | 570 | A > D | 23271 | C > A | |
| 23 | 614 | D > G | 23403 | A > G | |
| 24 | 681 | P > H | 23604 | C > A | |
| 25 | 716 | T > I | 23709 | C > T | |
| 26 | 982 | S > A | 24506 | T > G | |
| 27 | 1118 | D > H | 24914 | G > C | |
| ORF8 | 28 | 27 | Q > stop | 27972 | C > T |
| 29 | 52 | R > I | 28048 | G > T | |
| 30 | 68 | K > stop | 28095 | A > T | |
| 31 | 73 | Y > C | 28111 | A > G | |
| N | 32 | 3 | D > L | 28280–28282 | GAT > CTA |
| 33 | 203 | R > K | 28881–28882 | GG > AA | |
| 34 | 204 | G > R | 28883 | G > C | |
| 35 | 235 | S > F | 28977 | C > T | |
| 36 | 269 | 29080 | T > C |
CDS, coding DNA sequence.
Data availability.
The genome sequence of SARS-CoV-2/BGD/GC001 was deposited in the NCBI database under the BioProject accession number PRJNA702998, BioSample accession number SAMN17993365, and GenBank accession number MW624725.1. The Illumina raw reads have been deposited in the NCBI Sequence Read Archive under accession number SRR13744683.
ACKNOWLEDGMENTS
This research study was funded by core donors who provide unrestricted support to icddr,b for its operations and research. Current donors providing unrestricted support include the governments of Bangladesh, Canada, Sweden, and the United Kingdom. We gratefully acknowledge our core donors for their support and commitment to icddr,b’s research efforts.
Contributor Information
Dinesh Mondal, Email: din63d@icddrb.org.
John J. Dennehy, Queens College CUNY
REFERENCES
- 1.Wu F, Zhao S, Yu B, Chen Y-M, Wang W, Song Z-G, Hu Y, Tao Z-W, Tian J-H, Pei Y-Y, Yuan M-L, Zhang Y-L, Dai F-H, Liu Y, Wang Q-M, Zheng J-J, Xu L, Holmes EC, Zhang Y-Z. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rondinone V, Pace L, Fasanella A, Manzulli V, Parisi A, Capobianchi MR, Ostuni A, Chironna M, Caprioli E, Labonia M, Cipolletta D, Della Rovere I, Serrecchia L, Petruzzi F, Pennuzzi G, Galante D. 2021. VOC 202012/01 variant is effectively neutralized by antibodies produced by patients infected before its diffusion in Italy. Viruses 13:276. doi: 10.3390/v13020276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu X, Wang L, Sakthivel SK, Whitaker B, Murray J, Kamili S, Lynch B, Malapati L, Burke SA, Harcourt J, Tamin A, Thornburg NJ, Villanueva JM, Lindstrom S. 2020. US CDC real-time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis 26:1654–1665. doi: 10.3201/eid2608.201246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 5.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mikheenko A, Prjibelski A, Saveliev V, Antipov D, Gurevich A. 2018. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics 34:i142–i150. doi: 10.1093/bioinformatics/bty266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Otto TD, Dillon GP, Degrave WS, Berriman M. 2011. RATT: Rapid Annotation Transfer Tool. Nucleic Acids Res 39:e57. doi: 10.1093/nar/gkq1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cleemput S, Dumon W, Fonseca V, Abdool Karim W, Giovanetti M, Alcantara LC, Deforche K, De Oliveira T. 2020. Genome Detective Coronavirus Typing Tool for rapid identification and characterization of novel coronavirus genomes. Bioinformatics 36:3552–3555. doi: 10.1093/bioinformatics/btaa145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, Callender C, Sagulenko P, Bedford T, Neher RA. 2018. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rambaut A, Holmes EC, O’Toole Á, Hill V, McCrone JT, Ruis C, Du Plessis L, Pybus OG. 2021. Addendum: a dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol 6:415. doi: 10.1038/s41564-021-00872-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequence of SARS-CoV-2/BGD/GC001 was deposited in the NCBI database under the BioProject accession number PRJNA702998, BioSample accession number SAMN17993365, and GenBank accession number MW624725.1. The Illumina raw reads have been deposited in the NCBI Sequence Read Archive under accession number SRR13744683.